Abstract
The T cell-specific DNA binding protein TCF-1 is a central regulator of T cell development and function along multiple stages and lineages. Because it interacts with β-catenin, TCF-1 has been classically viewed as a downstream effector of the canonical Wnt signaling, although there is strong evidence for β-catenin-independent TCF-1 functions. TCF-1 co-binds accessible regulatory regions containing or lacking its conserved motif and cooperates with other nuclear factors to establish, context-dependent epigenetic and transcription programs, that are essential for T cell development and for regulating immune responses to infection, autoimmunity, and cancer. While it has mostly been associated with positive regulation of chromatin accessibility and gene expression, TCF-1 has the potential to reduce chromatin accessibility and thereby suppress gene expression. In addition the binding of TCF-1 bends the DNA and affects the chromatin conformation genome-wide. This review discusses the current understanding of the multiple roles of TCF-1 in T cell development and function and their mechanistic underpinnings.
Introduction
T lymphocyte development is a highly ordered stepwise process that depends on the cooperative and highly orchestrated action of multiple transcription and epigenetic regulators1. These regulators organize cooperating complexes to establish the specific chromatin landscape and transcription profile required in each T cell lineage and developmental stage. The DNA binding protein TCF-1, encoded by the Tcf7 gene, has emerged as a central player in these processes2–4. A recent review by Xue and colleagues elegantly discusses findings on TCF-1 across the fields of T cell immunity reflecting on the potential application of this knowledge to therapeutic intervention in viral infections and antitumor immunity5. The present review focuses on our current understanding of the molecular mechanisms through which TCF-1 leverages its diverse functions to shape T cell immunity.
TCF-1 is a member of the TCF/LEF family of high-mobility group (HMG) domain-containing proteins that have been conventionally viewed as effectors of the canonical Wnt signaling pathway (for reviews see6–8). The Wnt cascade is activated in response to signals induced by the binding of extracellular Wnt ligands to Frizzled and LRP5/6 receptors on the cell surface. This results in the stabilization of β-catenin, which is then transported to the nucleus where it binds to TCF/LEF factors and promotes chromatin accessibility and gene expression. In the absence of Wnt signals, DNA-bound TCF/LEF factors interact with repressors of the Grg/TLE family that reduce chromatin accessibility and suppress transcription. This simplified view does not consider that in addition to the full-length TCF-1 protein, which can interact with β-catenin, TCF-1 also expresses isoforms that lack the β-catenin-interacting domain9. These short isoforms originally presumed to have dominant-negative regulatory functions10, are sufficient to support thymocyte maturation11,12 and the generation of memory CD8+ T cells in response to acute infection13. The long isoform, on the other hand, supports thymocyte survival and is needed for the optimal maturation of central memory CD8+ T cells13. However, it is unclear whether the specific functions of the long isoform involve the canonical Wnt signaling, since ablating β-catenin, γ-catenin, or manipulating the expression and function of Wnt pathway components does not impair normal T cell development and function14–18. Altogether these findings suggest that the functions of TCF-1 in T cell development are largely independent of the classical Wnt signaling.
By contrast, in leukemia, autoimmunity, and cancer, uncontrolled pathological activation of Wnt signaling in T cells, engages TCF-1 to promote aberrant developmental progression and transformation of thymocytes as well as immune imbalance19–25. Stabilizing mutations of β-catenin and activation of the Wnt signaling pathway have been reported in human T cell malignancies, including precursor (T-ALL), peripheral (PTCL), cutaneous (CTCL), and adult T cell leukemia (ATL)26–30. Similarly, conditional stabilization of β-catenin in mouse double-positive (DP) thymocytes induced leukemias with recurrent chromosomal translocations like the ones seen in human T-ALL, providing mechanistic validation of the findings in human leukemias20,25,31. Importantly, we found that TCF-1 is responsible for the transformation of DP thymocytes with stabilized β-catenin since conditional ablation of TCF-1 in these cells abrogates leukemogenesis (submitted for publication). Furthermore, in chronic inflammatory conditions including Inflammatory Bowel Disease (IBD) and Multiple Sclerosis (MS) as well as in Colon Cancer, the regulatory T (TREG) cells were found to express high levels of β-catenin and to acquire proinflammatory properties22–24,32. Conditional stabilization of β-catenin in mouse TREG cells induced an IPEX-like syndrome and experimental autoimmune encephalomyelitis (EAE) which models MS. Mechanistically, the pathologies were linked to β-catenin/TCF-1-mediated changes in chromatin accessibility and gene expression23,24.
Altogether, these findings urge the need to better understand how TCF-1 leverages its Wnt-dependent versus Wnt-independent functions in T cells under physiological conditions and in the context of autoimmunity and cancer.
TCF-1 in T cell lineage specification and thymocyte differentiation
Notch activation enforces T cell specification on early thymic progenitors (ETPs) upon their entry into the thymus33,34 (Fig 1). Tcf7 is among the first genes upregulated in ETPs in direct response to Notch signals35–38. We and others have established that the specification of ETPs to the T cell lineage requires TCF-1 and its loss results in an early block of T cell development35,36. TCF-1 positively regulates T lineage genes including Gata3, Il2ra, and Bcl11b leading to T cell commitment. In an elegant recent study, Rothenberg and colleagues validated these original findings by single-cell CRISPR perturbation of the genes encoding early acting transcription factors combined with single-cell RNA-seq39. These analyses established that TCF-1 was profoundly needed for progression through the earliest phase of the ETP stage, while GATA-3 becomes especially important after the initiating role of TCF-1 in T cell specification. TCF-1 was further found to play a critical role in the γδ versus αβ T cell lineage separation. While the gradual upregulation of TCF-1 facilitates the αβ T cell fate after assembly of the pre-T cell antigen receptor (pre-TCR)40,41, reduced TCF-1 expression favors the γδ T lineage fate42,43. In the γδ T cell lineage TCF-1 also controls the γδ T cell effector fate, and essentially all TCF-1-deficient γδ cells convert into interleukin 17 (IL-17)-producing γδ effectors43. More recent investigations using conditional CD4-Cre mediated ablation of Tcf7, which targets CD4+CD8+ DP thymocytes, revealed functions of TCF-1 in these later stages of thymic development. Our own studies established that TCF-1 co-binds DNA regulatory sites together with other nuclear factors including Ikaros, Runx1, HEB, and in particular it cooperates with HEB to establish the molecular profile of DP thymocytes44. Following positive selection, the DP thymocytes face a lineage choice decision to the CD4+ or the CD8+ T cell lineage. This binary decision is guided by the transcription factors TH-POK and RUNX3, respectively45–47. Loss of TCF-1 impairs the CD4 lineage choice through insufficient TH-POK induction and elevated RUNX3 expression48. Thus, TCF-1 is a T cell specification and fate-determining factor at multiple stages of thymocyte development.
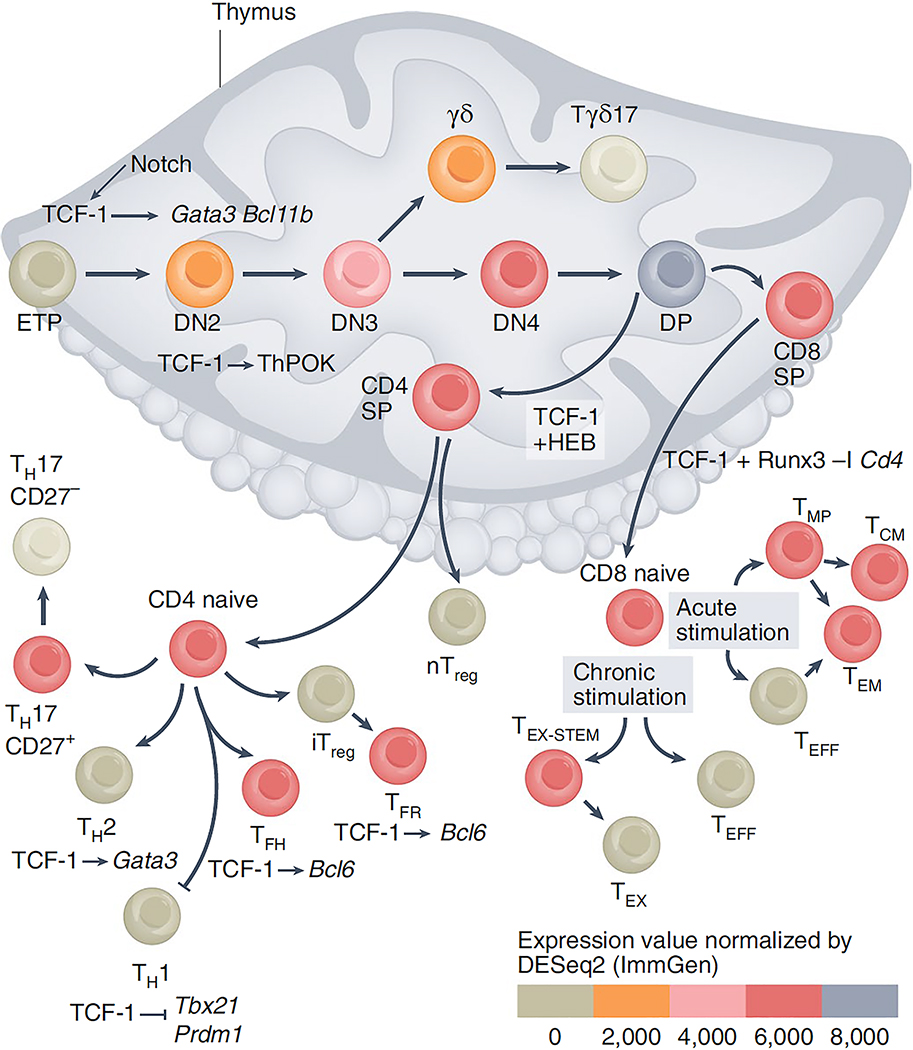
Figure 1: T cell development and the stages of TCF-1 implication.
Notch signaling upregulates Tcf7 expression in early ETPs 35–39. TCF-1 induces the expression of genes encoding transcription factors critical for T cell specification, including Gata3 and Bcl11b. The levels of TCF-1 increases progressively up to the CD4+CD8+ (DP) stage. In CD4+CD8+ (DP) thymocytes TCF-1 cooperates with HEB to define their epigenetic landscape and trasncription profile 44. Following thymic selection the DP cells become either CD4+ or CD8+ SP cells. TCF-1 fosters the CD4+ T cell fate by promoting Zbtb7b (TH-POK) expression, and although TCF-1 is not required for commitment to the CD8+ T cell lineage it ensures CD8+ T cell stability by cooperating with RUNX3 to suppress Cd4 gene expression 48. TCF-1 also suppresses Rorc (RORγT) and Il17 expression in DP thymocytes preventing their conversion to CD4+ TH17 and CD8+ TC17 cells 50,113. After thymic egress CD4+ T cells differentiate into TH subsets. TCF-1 promotes TFH differentiation by inducing Bcl6 and supressing Prdm1 (BLIMP-1) expression and limits TH1 differentiation by suppressing Tbx21 (T-bet) expression 63–65. TCF-1 promotes TH2 differentiation by directly upregulating Gata3 59. TCF-1 is expressed by stem cell-like CD27+ TH17 cells, which persist in models of MS 54. In TREG cell subsets, TCF-1 cooperates with Foxp3 to limit expression of T cell activation and TH17 differentiation genes. It also enhances Bcl6 expression to promote TFR differentiation. CD8+ T cells are critical for defense against acute and chronic viral infections and cancers. In acute viral infection TCF-1 is important in driving the development of TMP and TMEM cells. Both in acute and chronic LCMV infection downregulation of TCF-1 is needed for the differentiation of TEFF and TEX cells. In chronic viral infections and cancer, TCF-1 and BCL-6 support the differentiation and maintenance of TEX-STEM cells that express PD-1 and can respond to anti-PD-1 therapy.
TCF-1 in differentiation of peripheral CD4+ T helper cell lineages
After thymic egress, the circulating naïve CD4+ T cells have the potential to differentiate into several T helper (TH) cell lineages in response to antigen stimulation. These include the TH1, TH2, TH17, and T follicular helper (TFH) cell lineages that act to control the source of the foreign antigen49. TCF-1 orchestrates the development, equilibrium, and function of all these CD4+ T cell lineages (for reviews see5,50,51). TH17 cells have critical roles in host defense against bacteria and specific pathogens and also drive pathogenic inflammation in autoimmunity and cancer52. Early studies suggested that germline deletion of Tcf7 resulted in increased IL-17 gene expression both in thymus and peripheral T cells, which led to enhanced Th17 differentiation 53. However, these mice also have abnormall T-cell development due to their consitutive lack of TCF-1. Moreover, in the context of experimental autoimmune encephalomyelitis (EAE), TCF-1 expression was shown to mark a CD27+ TH17 progenitor-like cell subset, which upon activation can give rise to a disease-promoting TCF-1lo CD27− TH17 cell subset54. TH2 cells are critical for the immune response against extracellular parasites, and are activated by allergens and toxins55. They can induce allergy and cooperate with TH17 cells to mount tumor-promoting inflammation involving IL-33 and IL-1056–58. TCF-1 promotes the TH2 cell polarization by transactivating GATA-3, the signature regulator of the TH2 cell lineage59. TH1 cells, differentiate in response to infection by intracellular pathogens, and have a shared transitional stage during their early differentiation steps with TFH cells, which help B cells produce antibodies60–62. TCF-1 controls the bifurcation between the TH1 and TFH cell lineages in favor of TFH, by upregulating the BCL-6, which drives TFH cell differentiation and downregulating BLIMP-1, which normally suppresses BCL-6 (Fig 1). BCL-6 limits TH1 cell differentiation by directly suppressing the expression of critical TH1 cell differentiation genes, including Tbx21 (encoding T-bet), which is the signature TH1 lineage transcription factor63–65. Thus, starting from a common naïve CD4+ T cell TCF-1 guides the maturation and the equilibrium of T helper cell lineages by selectively modulating the expression of key regulatory genes.
The specific roles of TCF-1 in T regulatory cells
Among CD4+ T cells, TREG cells, have indispensable roles in resolving tissue inflammation and preserving tolerance to self-antigens66,67. The majority of TREG cells are generated in the thymus while a smaller group is converted in the periphery from naïve CD4+ T cells. The function of TCF-1 in TREG cells is complex. Earlier studies using a germline Tcf7 knockout model showed that heterozygous deletion of Tcf7 increases the number of thymically generated TREG cells68, suggesting that TCF-1 negatively regulates TREG cell development and T cell receptor affinity. Consistent with this finding, the TREG cell lineage determining factor FOXP3 was reported to suppress Tcf7 gene expression55. More recent studies demonstrated that TCF-1 interacts with Foxp3 and chronic activation of Wnt signaling impairs Foxp3 functions through TCF-169. Subsequently, several studies verified that in TREG cells, TCF-1 shares a large number of DNA binding sites with Foxp3 and regulates the expression of FOXP3 target genes to fine tune TREG properties23,70,71. In line with these findings, we found that the targeted ablation of TCF-1 in TREG cells enhanced their suppression of CD8+ T cell cytotoxicity but compromised their anti-inflammatory functions72. Interestingly, scRNA-Seq analysis identified numerous TREG cell subsets and established that loss of TCF-1 has greater impact in the peripherally induced as compared to the thymic TREG cells72. TREG cell-specific ablation of both TCF-1 and LEF1 induced autoimmune pathologies70, whereas loss of TCF-1 alone did not by itself render the mice unhealthy. However, in mice predisposed to polyposis the TREG cell-specific loss of TCF-1 exacerbated pathogenic inflammation driving aggressive tumor growth72. Consistently, TREG cells infiltrating human colorectal cancer tumors expressed lower levels of TCF-1 compared to those in the healthy margins or peripheral blood72. These findings suggest that TCF-1 alters TREG cell functions in response to environmental cues to produce appropriate or, in the case of autoimmunity and cancer, inappropriate and pathogenic immune responses72. A distinct sub-category of TREG cells are the T follicular regulatory (TFR) cells that act in the germinal centers (GCs) to maintain immune homeostasis by suppressing excessive TFH and B cell responses73. TREG cell-specific loss of TCF-1 diminished the number of TFR cells and impaired immune regulation within GCs70. This could contribute to the enhanced inflammation observed in mice with TCF-1-deficient TREG cells70,72. Thus, TCF-1 expression in TREG and TFH cells is essential to selectively regulate TREG functions that control inflammation.
TCF-1 in CD8+ T cell survival and progenitor states
Expression of TCF-1 in CD8+ T cells distinguishes antigen-stimulated cells that have progenitor potential from their terminally differentiated couterparts. This distinction applies to T cells that are subjected to acute or chronic TCR stimulation.
TCF-1 is downregulated in mouse CD8+ T cells cells that respond to acute lymphocytic choriomeningitis virus (LCMV) infection, as they progress from naïve to terminally differentiated effector cells (TEFF). However, expression of TCF-1 is maintained in a subset of T cell precursors with stem cell-like features74, that differentiate to long-lived central memory cells (TCM)75–77. Several subsets of antigen experienced CD8+ T cells are defined based on their longevity, progenitor properties, and effector functions as well as expression of nuclear factors and cell surface markers (for review see78). Hybrid phenotypes have also been described based on the expression of markers for migration and tissue residence (for review see79). TCF-1 deficiency alone diminishes but does not ablate CD8+ TCM cells, while loss of both TCF-1 and LEF1 almost completely eliminates the CD8 TCM precursors (TMP), suggesting that TCF1 together with LEF1 contribute to the TMP cell phenotype80.
In mice with chronic LCMV infection, persistent TCR signaling drives the downstream activation of transcription factors that promote the expression of inhibitory receptors, such as PD-1, LAG-3, and TIGIT, and reduce expression of KLRG181, resulting in exhausted CD8+ T cells (TEX). These transcription factors include the calcineurin-dependent nuclear factor of activated T cells (NFAT)5, interferon regulatory factor-4 (IRF4), basic leucine zipper transcription factor, ATF-like (BATF), nuclear receptor subfamily 4 group A (NR4A), and thymocyte selection-associated HMG BOX (TOX). TOX is the master regulator of the exhausted epigenetic state. TEX cells were initially thought to be dysfunctional, however the loss of TOX also limits the percistence of functionally active antigen-specific CD8+ T cells, indicating that a subset of TEX cells have progenitor properties82–88. This subset has elevated expression of TCF-1 and controls viral spread in chronically infected mice. These observations are consistent with TCF-1 modulating the epigenetic outcomes of TOX activity.
The tumor-induced T cell dysfunction occurs in early stages of carcinogenesis and is driven by the presence and persistence of tumor antigen89. Tumor infiltrating CD8+ T cells become increasingly dysfunctional with time, expressing high levels of genes associated with reduced immune function such as those encoding the transcriptional repressors Egr1, Batf, Blimp-1, and the inhibitory receptors PD1, LAG3, 2B4, and TIM3. Dysfunctional CD8+ T cells fail to produce effector cytokines in response to cognate antigen. By contrast, non-tumor-specific T cells that infiltrate the same tumor do not upregulate inhibitory receptors, and produce interferon-γ (IFN-γ) and tumor necrosis factor (TNF)89. This finding suggests that chronic TCR stimulation by tumor antigens drives the dysfunctional CD8+ T cell phenotype. There is growing consensus that elevated expression of TCF-1 identifies the tumor antigen-specific CD8+ T cell subset that maintains long-term functional responses (TEX-STEM) from the terminally differentiated TEX subset90–92 (for review see5). This concept may be oversimplified as discussed below.
TOX and TCF-1 are both members of the family of transcription factors that contain the conserved high mobility group box (HMG-box) region. TCF-1 partners with TOX to maintain an epigenetic state in CD8+ TEX-STEM cells that supports long term survival and progenitor potential but not necessarily their functionality83,85,93,94. Expression of TCF-1 is under the control of TOX. In the absence of TOX chronically stimulated CD8+ T cells maintain elevated levels of TCF-1 and fail to upregulate exhaustion markers. However, loss of TOX is not enough for expression of IFN-γ and TNF by tumor-infiltrating CD8+ T cells. This finding has led to the suggestion that the expression of exhaustion markers is uncoupled from the loss of effector functions, but rather serves to prevent the overstimulation of T cells and activation-induced cell death85. This finding, brings into question the immediate benefits of targeting TOX or TCF-1 for immunotherapy of cancer95. Further research is needed to establish the mechanisms responsible for functionall inactivity of tumor-infiltrating cells, as well as the similarities and differences between dysfunctional CD8+ T cells in cancer and exhausted T cells in chronic viral infection96.
Functions of TCF-1 in the Wnt cascade and the option of Wnt independence
It is intriguing that TCF-1 is critically required in all stages of T cell development and maturation including various context-dependent T cell functions. The developmental stage specific and T cell lineage-specific transcriptional profiles and biological outcomes attributed to the action of TCF-1 raise intriguing questions about its’ Wnt dependent versus independent functions, and the roles of molecular partners of TCF-1 that help mediate diverse outcomes.
In the logic of the WNT cascade, DNA-bound TCF-1 enables directly interacting transcription and epigenetic factors to access specific genomic regions in order to regulate the chromatin landscape and transcription of the associated genes. In this scenario β-catenin interacts with the full-length TCF-1 protein in complex with epigenetic and transcription regulators that enhance chromatin accessibility and gene transcription (Fig 2a). In the absence of β-catenin, repressors of the Grg/TLE family, that bind TCF-1 at a region proximal to the HMG DNA binding domain6,97, reduce chromatin accessibility, and suppress gene transcription (Fig 2a)98. The extent to which β-catenin or repressors of the Grg/TLE family are involved in T cell development and function is uncertain and needs further study (for review see5). It is possible that other regulators, which can bind TCF-1 outside the context of Wnt signaling, also follow this scenario to modulate the chromatin landscape and transcription profile of the cells, but the identities of such regulators remain to be elucidated.
Figure 2. Understanding the fundamental molecular functions of TCF-1.
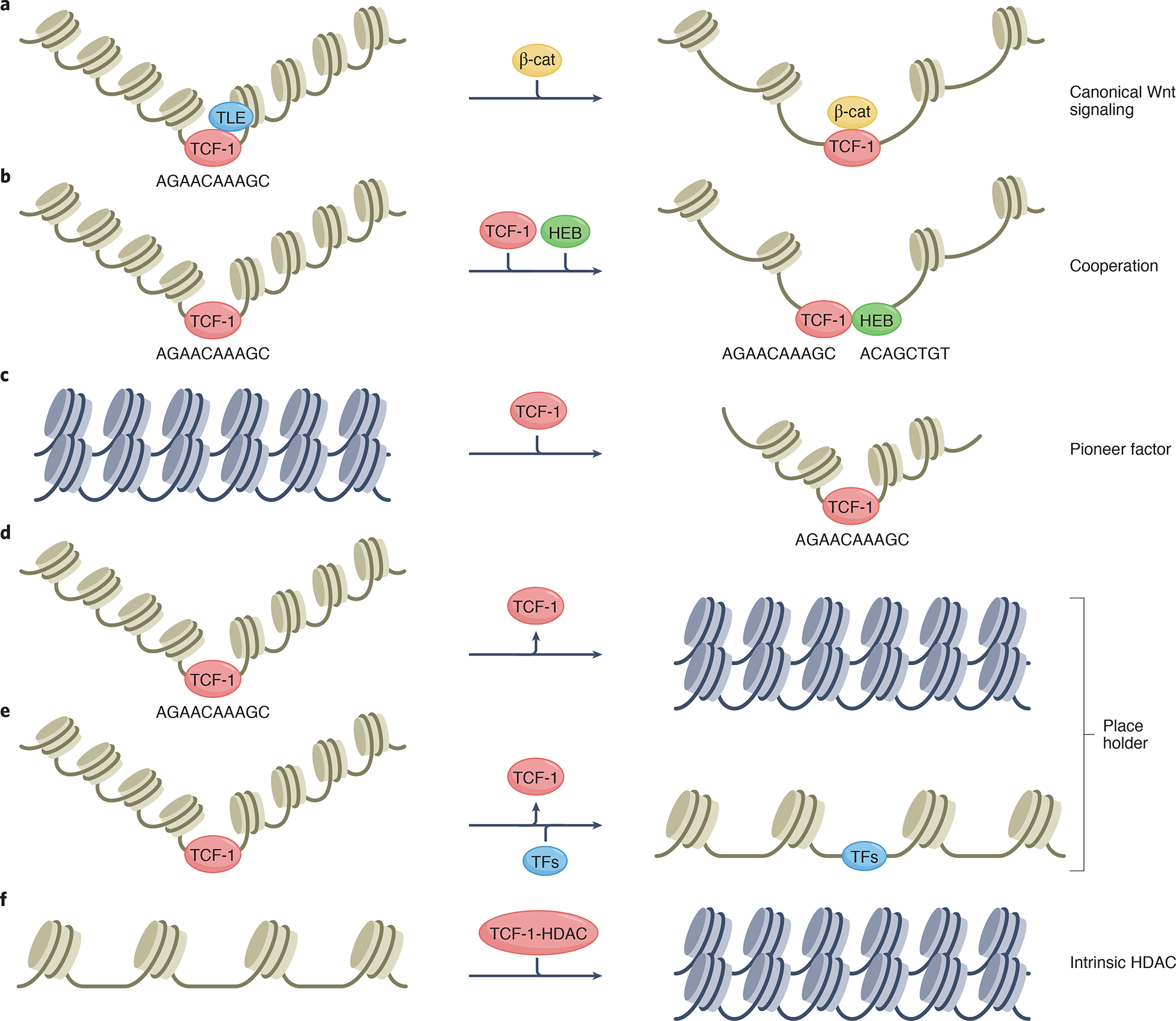
TCF-1 has multiple molecular functions that are superimposed on its’ ability to bend the DNA upon binding and regulate the 3D chromatin conformation.
(a) In the absence of β-catenin, TCF-1 can bind with the repressors of the Grg/TLE family at a region proximal to the HMG DNA binding domain 6,97 to reduce chromatin accessibility, and suppress gene transcription 98. Extracellular Wnts stabilize β-catenin by disrupting its’ degradatioin complex, allowing its’ nuclear translocation and interaction with the full-length TCF-1 protein. This results in the recruitment of epigenetic and transcription regulators that enhance chromatin accessibility and gene transcription. (b) TCF-1 cooperates with other transcription factors to establish context and developmental stage-specific epigenetic and transcription profiles through co-binding to common DNA sites. (c) TCF-1 could function as a pioneer-like factor and initiates changes in the chromatin landscape that establish the T cell lineage identity101. (d,e) TCF-1 may function as a “Place Holder”, to promote/maintain T cell-specific chromatin accessibility, independently of Wnt signals. (f) TCF-1 has intrinsic histone deacetylase activity (HDAC) and can directly reduce chromatin accessibility.
Cooperation of TCF-1 with other transcription and epigenetic regulators
In addition to its direct interaction with regulators, TCF-1 also cooperates with other transcription factors to establish context and developmental stage-specific epigenetic and transcription profiles through co-binding to common DNA sites. Regulation through co-binding is supported by findings that accessible chromatin sites along thymocyte development are not only enriched for the TCF/LEF motif, but also motifs for ETS, RUNX and E2A families of transcription factors 44,99. Comparing the genome-wide binding of TCF-1 to that of IKAROS, RUNX1 and HEB in DP thymocytes demonstrated an extensive overlap between TCF-1-occupied sites and sites bound by these transcription factors. Detailed studies focusing on the co-binding of TCF-1 and HEB, showed that TCF-1 cooperates with HEB to establish and maintain the epigenetic and transcription profile of DP thymocytes (Fig 2b)44. Furthermore, in TREG cells TCF-1 binding significantly overlaps with the binding of Foxp323,71. We found that this overlapping binding limited the expression of genes involved in TH17 inflammation, transforming growth factor-β (TGF-β) signaling, and T cell activation72. The genetic ablation of Tcf7 led to the upregulation of these genes without compromising the core TREG gene expression signature, and even upregulated Foxp3 expression 72. The interaction of TCF-1 with Foxp3 was further shown to involve Foxp3-mediated downregulation of Tcf771. Thus, these new findings establish that TCF-1 engages transcription factors from several different families as its cooperating partners to shape the molecular profiles of T cells during development and function.
TCF-1 preserves chromatin accessibility
It has been suggested that in T cell development TCF-1 promotes or preserves chromatin accessibility44,71,99,100. Vahedi and colleagues found that the upregulation of TCF-1 in early thymocytes undergoing T cell specification, coincides with increased accessibility of chromatin sites that are enriched in the conserved TCF-1 DNA binding motif99. Also during hematopoietic development, from hematopoietic stem cells to CD4+ and CD8+ single-positive (SP) thymocytes, the TCF-1 binding motif becomes progressively more enriched at accessible chromatin sites. The increase in chromatin accessibility parallels the progressive upregulation of Tcf7 gene expression in developing thymocytes99. These findings led to suggestions that TCF-1 may act as a pioneer-like factor101 that initiates the T cell-specific chromatin landscape and establishes the T cell lineage identity (Fig 2c). A detailed molecular analysis of TCF-1 sufficient and deficient lymphoid precursors cultured for a short time in T cell differentiation conditions (as described in39) could help clarify the precise role of TCF-1 in T cell specification and may provide direct support for the role of TCF-1 as a pioneer factor. Other studies showed that loss of TCF-1 in DP thymocytes44, or in CD8+ T cells100, resulted in overall reduced chromatin accessibility at sites previously bound by TCF-1. At least in DP thymocytes, these accessibility changes were more evident in TCF-1 bound enhancer sites that contained its conserved motif, and loss of TCF-1 correlated with an overall reduction of target gene expression 44. Along this line, Schietinger and colleagues showed that the transition of CD8+ TEX-STEM tumor-infiltrating lymphocytes (TILs), which express TCF-1, into terminal CD8+ TEX TILs, which do not, is associated with extensive changes in chromatin accessibility. Sites that progressively lose accessibility during this process are highly enriched for the TCF/LEF binding motifs 93, suggesting that TCF-1 sustains the open chromatin state in TEX-STEM TILs (Fig 2d), and can serve as a “place holder” for other transcription factors (Fig 2e). Similarly, following CD4+ and CD8+ T cell activation, TCF-1 is downregulated and its canonical binding sites lose accessibility. Exceptions are TCF-1 sites that are co-bound by Ets1, or by activation induced transcription factors, indicating that TCF-1 may act as a place-holder in these sites102.
Collectively, these findings suggest that TCF-1 predominantly maintains and potentially also promotes chromatin accessibility in T cell development and support the notion that this function underlies its ability to maintain some differentiation potential or “stemness” in T cells. These functions are likely independent of Wnt signals since in contrast to the critical need for TCF-1, β-catenin has minimal impact on normal T cell development.
TCF-1 directly modifies the chromatin through its intrinsic HDAC activity
Deciphering how TCF-1 functions in T lymphocytes has become more challenging by the finding of Xue and colleagues that TCF-1, in addition to shaping the chromatin landscape through its interacting partners it also has an intrinsic histone deacetylase activity (HDAC). This activity, which was mapped to a region between the N terminal β-catenin binding domain and the central HMG DNA-binding domain directly upstream of the Grg/TLE binding domain, can directly reduce chromatin accessibility (Fig 2f). It has been suggested that the TCF-1 HDAC activity is essential for establishing the CD8+ T cell identity98 and for sustaining the ability of TFH cells to provide B cell help103. Therefore, in order to determine the context-dependent functions of TCF-1 on chromatin accessibility, it is important to understand how its intrinsic HDAC activity orchestrates with the activities of regulators that directly interact with TCF-1 particularly the ones that promote chromatin accessibility.
TCF-1 shapes the 3D chromatin conformation
The HMG domain family of proteins, including TCF-1, bind to the minor groove of the DNA helix and have the capacity to bend the DNA (reviewed in104). Early studies by Grosschedl and colleagues established that LEF-1, the relative of TCF-1 that is also expressed in T cells induces a significant DNA bend to its binding site105,106. In particular, LEF-1, likely through its ability to bend the DNA upon binding, facilitated interactions between proteins bound at nonadjacent sites and coordinated the assembly of a complex at the enhancer of the TCRα gene. Based on the similarity of their HMG domain, it is expected that TCF-1 also bends the DNA upon binding, and this could have significant implications on the ability of TCF-1 to regulate the 3D chromatin conformation (Fig 2). Newly developed technologies have made it possible to assess chromatin conformation changes genome-wide and associate them with lineage commitment stages. In this context, a comprehensive study combining high throughput chromatin conformation capture (HiC), with chromatin accessibility, and gene expression analyses, established that T cell commitment at the DN2 to DN3 thymocyte stage is associated with major chromatin conformation changes 107. Although this study did not implicate TCF-1, more recently Xue and colleagues integrated Hi-C with epigenetic and transcription data, to show that TCF-1 and LEF-1 promote the formation of extensively interconnected hubs by enforcing chromatin interaction and accessibility in CD8+ T cells 100. These findings open the way for further studies to independently assess the role of TCF-1 versus LEF-1 on chromatin conformation and to compare their differential impact in the various T cell lineages.
TCF-1 leverages its functions through abundance
The expression levels of TCF-1 in developing T cells are highly regulated. TCF-1 starts expressing at low levels in ETP thymocytes, and its progressively upregulated up to the DP stage where it reaches an expression level that is unusually high for a transcription factor. Past the DP stage TCF-1 expression levels are reduced but remain relatively high in all peripheral T cell lineages with the exception of the TREG cells, which express low levels of TCF-1. Polarization of CD4+ T cells to the TH lineages is invariably marked by downregulation of TCF-1. Similarly, progression of naïve CD8+ cells to the terminal effector or terminal exhaustion stages is associated with downregulation of TCF-1, while the progenitor exhausted CD8+ T-cells that are long lived and have progenitor propertors express high levels of TCF-1.
Recent studies have highlighted the molecular impact of TCF-1 downregulation. In particular, the reduced expression of TCF-1 after CD4 and CD8 T cell activation, was suggested to underlie the observed accessibility loss of sites uniquely bound by TCF-1102. The increased generation of thymic TREG cells after heterozygote TCF-1 deletion68, and the downregulation of TCF-1 by FOXP3, which reduces chromatin accessibility and associated gene expression71 in TREG cells, further support the suggestion that the levels of TCF-1 determine its functions. TCF-1 is also downregulated in colon tumor infiltrating TREG cells, indicating that TREG cells adapt to their environment by regulating the levels of TCF-1, which in this case contributed to their enhanced tumor promoting properties 72. Therefore, variations in TCF-1 levels can significantly impact the interplay between TCF-1 and other molecular partners affecting key biological processes including differentiation, cell fate decision, and function in health and disease.
Future perspectives
Deciphering how TCF-1 orchestrates its diverse functions during T cell development represents a challenging puzzle. This challenge is because TCF-1 has essential roles in multiple T cell lineages and developmental stages and its activity is context dependent. Therefore, any effort to dissect the molecular functions of TCF-1, must take into consideration in each T cell lineage and developmental stage: 1) the epigenetic and transcription profile of the cells, 2) the cooperating and interacting partners, and 3) the physiological levels of TCF-1. An additional parameter to consider in each context is the redundant functions of the transcription factor LEF1, the WNT responsive close relative of TCF-1 that is expressed in all T cells and shares significant structural homology with TCF-1. TCF-1 and LEF1 share the ability to bind β-catenin and Grg/TLE factors, and they both express shorter isoforms that do not bind β-catenin. The genome-wide DNA binding of LEF1 overlaps with that of TCF-1 in cells where it has been assessed, including DP thymocytes44 and TREG cells70. Simultaneous loss of both TCF-1 and LEF1 in many T cell lineages, and stages mainly strengthens the phenotypes observed by simply deleting TCF-148,63,70. It is even puzzling why the loss of TCF-1 has so much stronger impact on T cell development than the loss of LEF1 and it is unclear whether this is due to the lower levels of LEF1 expression, like in DP thymocytes, or to yet unknown functional differences. Although studies up to now have mainly identified functional similarities between TCF-1 and LEF-1 in T cell development it would be interesting to uncover their differences and the molecular basis of their functions.
Future investigations can take advantage of novel single cell and genome wide technologies. For example, single-cell technologies including scCrisprCAS9 perturbations39, scRNAseq and scATACseq have already provided a wealth of information on the roles of TCF-1 in T cell specification99, on TREG cell heterogeneity and on tumor-infiltrating T cells72,108–112. Similarly, integrating genome-wide transcription, epigenetic and chromosome conformation data is beginning to more accurately predict molecular functions of TCF-1. Addressing the molecular functions of TCF-1 will also require new model systems in which TCF-1 expression and/or protein levels can be temporally modulated in specific T cell developmental stages and lineages. Given that TCF-1 regulates multiple T cell properties, knowing how it functions in each situation will also provide the knowledge base to design context-specific therapeutic interventions in autoimmunity and cancer.
References
- 1.Hosokawa H & Rothenberg EV How transcription factors drive choice of the T cell fate. Nat Rev Immunol 21, 162–176, doi: 10.1038/s41577-020-00426-6 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Yang Q et al. T cell factor 1 is required for group 2 innate lymphoid cell generation. Immunity 38, 694–704, doi: 10.1016/j.immuni.2012.12.003 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ishizuka IE, Constantinides MG, Gudjonson H & Bendelac A The Innate Lymphoid Cell Precursor. Annu Rev Immunol 34, 299–316, doi: 10.1146/annurev-immunol-041015-055549 (2016). [DOI] [PubMed] [Google Scholar]
- 4.Kasal DN & Bendelac A Multi-transcription factor reporter mice delineate early precursors to the ILC and LTi lineages. J Exp Med 218, doi: 10.1084/jem.20200487 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zhao X, Shan Q & Xue HH TCF1 in T cell immunity: a broadened frontier. Nat Rev Immunol, doi: 10.1038/s41577-021-00563-6 (2021). [DOI] [PubMed] [Google Scholar]
- 6.Mosimann C, Hausmann G & Basler K Beta-catenin hits chromatin: regulation of Wnt target gene activation. Nat Rev. Mol. Cell BIol 10, 276–286, doi: 10.1038/nrm2654 (2009). [DOI] [PubMed] [Google Scholar]
- 7.MacDonald BT, Tamai K & He X Wnt/beta-catenin signaling: components, mechanisms, and diseases. Dev Cell 17, 9–26, doi: 10.1016/j.devcel.2009.06.016 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Clevers H Wnt/beta-catenin signaling in development and disease. Cell 127, 469–480, doi: 10.1016/j.cell.2006.10.018 (2006). [DOI] [PubMed] [Google Scholar]
- 9.van de Wetering M et al. The beta-catenin/TCF-4 complex imposes a crypt progenitor phenotype on colorectal cancer cells. Cell 111, 241–250 (2002). [DOI] [PubMed] [Google Scholar]
- 10.Cadigan KM & Waterman ML TCF/LEFs and Wnt signaling in the nucleus. Cold Spring Harb Perspect Biol 4, doi: 10.1101/cshperspect.a007906 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ioannidis V, Beermann F, Clevers H & Held W The beta-catenin--TCF-1 pathway ensures CD4(+)CD8(+) thymocyte survival. Nat Immunol 2, 691–697, doi: 10.1038/90623 (2001). [DOI] [PubMed] [Google Scholar]
- 12.Xu Z et al. Cutting Edge: beta-Catenin-Interacting Tcf1 Isoforms Are Essential for Thymocyte Survival but Dispensable for Thymic Maturation Transitions. J Immunol 198, 3404–3409, doi: 10.4049/jimmunol.1602139 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gullicksrud JA et al. Differential Requirements for Tcf1 Long Isoforms in CD8(+) and CD4(+) T Cell Responses to Acute Viral Infection. J Immunol 199, 911–919, doi: 10.4049/jimmunol.1700595 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Koch U et al. Simultaneous loss of beta- and gamma-catenin does not perturb hematopoiesis or lymphopoiesis. Blood 111, 160–164, doi: 10.1182/blood-2007-07-099754 (2008). [DOI] [PubMed] [Google Scholar]
- 15.Jeannet G et al. Long-term, multilineage hematopoiesis occurs in the combined absence of beta-catenin and gamma-catenin. Blood 111, 142–149, doi: 10.1182/blood-2007-07-102558 (2008). [DOI] [PubMed] [Google Scholar]
- 16.Zhao X et al. beta-catenin and gamma-catenin are dispensable for T lymphocytes and AML leukemic stem cells. Elife 9, doi: 10.7554/eLife.55360 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Liu J et al. Lrp5 and Lrp6 are required for maintaining self-renewal and differentiation of hematopoietic stem cells. FASEB J 33, 5615–5625, doi: 10.1096/fj.201802072R (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Prlic M & Bevan MJ Cutting edge: beta-catenin is dispensable for T cell effector differentiation, memory formation, and recall responses. J Immunol 187, 1542–1546, doi: 10.4049/jimmunol.1100907 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gekas C et al. beta-Catenin is required for T-cell leukemia initiation and MYC transcription downstream of Notch1. Leukemia 30, 2002–2010, doi: 10.1038/leu.2016.106 (2016). [DOI] [PubMed] [Google Scholar]
- 20.Guo Z et al. Beta-catenin stabilization stalls the transition from double-positive to single-positive stage and predisposes thymocytes to malignant transformation. Blood 109, 5463–5472, doi: 10.1182/blood-2006-11-059071 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Keerthivasan S et al. beta-Catenin promotes colitis and colon cancer through imprinting of proinflammatory properties in T cells. Sci Transl Med 6, 225ra228, doi: 10.1126/scitranslmed.3007607 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Blatner NR et al. Expression of RORgammat marks a pathogenic regulatory T cell subset in human colon cancer. Sci Transl Med 4, 164ra159, doi: 10.1126/scitranslmed.3004566 (2012). These studies demonstrate that pathogenic activation of β-catenin in inflamatory conditions renders TREG cells proinflammatory.
- 23. Quandt J et al. Wnt-beta-catenin activation epigenetically reprograms Treg cells in inflammatory bowel disease and dysplastic progression. Nat Immunol 22, 471–484, doi: 10.1038/s41590-021-00889-2 (2021). These studies demonstrate that pathogenic activation of β-catenin in inflamatory conditions renders TREG cells proinflammatory.
- 24.Sumida T et al. Activated beta-catenin in Foxp3(+) regulatory T cells links inflammatory environments to autoimmunity. Nat Immunol 19, 1391–1402, doi: 10.1038/s41590-018-0236-6 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gounari F et al. Somatic activation of beta-catenin bypasses pre-TCR signaling and TCR selection in thymocyte development. Nat Immunol 2, 863–869, doi: 10.1038/ni0901-863 (2001). [DOI] [PubMed] [Google Scholar]
- 26.Lento W, Congdon K, Voermans C, Kritzik M & Reya T Wnt signaling in normal and malignant hematopoiesis. Cold Spring Harb Perspect Biol 5, doi: 10.1101/cshperspect.a008011 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Groen RW et al. Illegitimate WNT pathway activation by beta-catenin mutation or autocrine stimulation in T-cell malignancies. Cancer Res 68, 6969–6977, doi: 10.1158/0008-5472.CAN-08-1322 (2008). [DOI] [PubMed] [Google Scholar]
- 28.Bellei B, Cota C, Amantea A, Muscardin L & Picardo M Association of p53 Arg72Pro polymorphism and beta-catenin accumulation in mycosis fungoides. Br. J. Dermatol 155, 1223–1229, doi: 10.1111/j.1365-2133.2006.07527.x (2006). [DOI] [PubMed] [Google Scholar]
- 29.Ram-Wolff C, Martin-Garcia N, Bensussan A, Bagot M & Ortonne N Histopathologic diagnosis of lymphomatous versus inflammatory erythroderma: a morphologic and phenotypic study on 47 skin biopsies. Am J Dermatopathol 32, 755–763, doi: 10.1097/DAD.0b013e3181cfbfbf (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ng OH et al. Deregulated WNT signaling in childhood T-cell acute lymphoblastic leukemia. Blood Cancer J 4, e192, doi: 10.1038/bcj.2014.12 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Dose M et al. beta-Catenin induces T-cell transformation by promoting genomic instability. Proc Natl Acad Sci U S A 111, 391–396, doi: 10.1073/pnas.1315752111 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gounaris E et al. T-regulatory cells shift from a protective anti-inflammatory to a cancer-promoting proinflammatory phenotype in polyposis. Cancer Res 69, 5490–5497, doi:69/13/5490 [pii] 10.1158/0008-5472.CAN-09-0304 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Rothenberg EV, Moore JE & Yui MA Launching the T-cell-lineage developmental programme. Nat Rev Immunol 8, 9–21, doi: 10.1038/nri2232 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Shah DK & Zuniga-Pflucker JC An overview of the intrathymic intricacies of T cell development. J Immunol 192, 4017–4023, doi: 10.4049/jimmunol.1302259 (2014). [DOI] [PubMed] [Google Scholar]
- 35. Germar K et al. T-cell factor 1 is a gatekeeper for T-cell specification in response to Notch signaling. Proc Natl Acad Sci U S A 108, 20060–20065, doi: 10.1073/pnas.1110230108 (2011). These studies establish that NOTCH1 induced TCF-1 expression in ETPs is essential for T cell specification.
- 36. Weber BN et al. A critical role for TCF-1 in T-lineage specification and differentiation. Nature 476, 63–68, doi: 10.1038/nature10279 (2011). These studies establish that NOTCH1 induced TCF-1 expression in ETPs is essential for T cell specification.
- 37.Harly C et al. A Shared Regulatory Element Controls the Initiation of Tcf7 Expression During Early T Cell and Innate Lymphoid Cell Developments. Front Immunol 11, 470, doi: 10.3389/fimmu.2020.00470 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kueh HY et al. Asynchronous combinatorial action of four regulatory factors activates Bcl11b for T cell commitment. Nat Immunol 17, 956–965, doi: 10.1038/ni.3514 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Zhou W, Gao F, Romero-Wolf M, Jo S & Rothenberg EV Single-cell perturbation dissects transcription factor control of progression speed and trajectory choice in early T-cell development. BioRxiv doi: 10.1101/2021.09.03.458944 (2021). These studies establish that NOTCH1 induced TCF-1 expression in ETPs is essential for T cell specification.
- 40.Okamura RM et al. Redundant regulation of T cell differentiation and TCRalpha gene expression by the transcription factors LEF-1 and TCF-1. Immunity 8, 11–20, doi: 10.1016/s1074-7613(00)80454-9 (1998). [DOI] [PubMed] [Google Scholar]
- 41.Yu S et al. The TCF-1 and LEF-1 transcription factors have cooperative and opposing roles in T cell development and malignancy. Immunity 37, 813–826, doi: 10.1016/j.immuni.2012.08.009 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Melichar HJ et al. Regulation of gammadelta versus alphabeta T lymphocyte differentiation by the transcription factor SOX13. Science 315, 230–233, doi: 10.1126/science.1135344 (2007). [DOI] [PubMed] [Google Scholar]
- 43.Fahl SP et al. The E protein-TCF1 axis controls gammadelta T cell development and effector fate. Cell Rep 34, 108716, doi: 10.1016/j.celrep.2021.108716 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Emmanuel AO et al. TCF-1 and HEB cooperate to establish the epigenetic and transcription profiles of CD4(+)CD8(+) thymocytes. Nat Immunol 19, 1366–1378, doi: 10.1038/s41590-018-0254-4 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wang L et al. Distinct functions for the transcription factors GATA-3 and ThPOK during intrathymic differentiation of CD4(+) T cells. Nat Immunol 9, 1122–1130, doi: 10.1038/ni.1647 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Egawa T & Littman DR ThPOK acts late in specification of the helper T cell lineage and suppresses Runx-mediated commitment to the cytotoxic T cell lineage. Nat Immunol 9, 1131–1139, doi: 10.1038/ni.1652 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Muroi S et al. Cascading suppression of transcriptional silencers by ThPOK seals helper T cell fate. Nat Immunol 9, 1113–1121, doi: 10.1038/ni.1650 (2008). [DOI] [PubMed] [Google Scholar]
- 48.Steinke FC et al. TCF-1 and LEF-1 act upstream of Th-POK to promote the CD4(+) T cell fate and interact with Runx3 to silence Cd4 in CD8(+) T cells. Nat Immunol 15, 646–656, doi: 10.1038/ni.2897 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Saravia J, Chapman NM & Chi H Helper T cell differentiation. Cell Mol Immunol 16, 634–643, doi: 10.1038/s41423-019-0220-6 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Mielke LA et al. TCF-1 limits the formation of Tc17 cells via repression of the MAF-RORgammat axis. J Exp Med 216, 1682–1699, doi: 10.1084/jem.20181778 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Escobar G, Mangani D & Anderson AC T cell factor 1: A master regulator of the T cell response in disease. Sci Immunol 5, doi: 10.1126/sciimmunol.abb9726 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Bettelli E, Korn T, Oukka M & Kuchroo VK Induction and effector functions of T(H)17 cells. Nature 453, 1051–1057, doi: 10.1038/nature07036 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Ma J, Wang R, Fang X, Ding Y & Sun Z Critical role of TCF-1 in repression of the IL-17 gene. PLoS One 6, e24768, doi: 10.1371/journal.pone.0024768 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Karmaus PWF et al. Metabolic heterogeneity underlies reciprocal fates of TH17 cell stemness and plasticity. Nature 565, 101–105, doi: 10.1038/s41586-018-0806-7 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Walker JA & McKenzie ANJ TH2 cell development and function. Nat Rev Immunol 18, 121–133, doi: 10.1038/nri.2017.118 (2018). [DOI] [PubMed] [Google Scholar]
- 56.Dennis KL et al. T-cell Expression of IL10 Is Essential for Tumor Immune Surveillance in the Small Intestine. Cancer Immuno Res 3, 806–814, doi: 10.1158/2326-6066.CIR-14-0169 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Saadalla A et al. Cell Intrinsic Deregulated β-Catenin Signaling Promotes Expansion of Bone Marrow Derived Connective Tissue Type Mast Cells, Systemic Inflammation, and Colon Cancer. Front Immunol 10, 2777, doi: 10.3389/fimmu.2019.02777 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Saadalla AM et al. Mast cells promote small bowel cancer in a tumor stage-specific and cytokine-dependent manner. Proc Natl Acad Sci U S A 115, 1588–1592, doi: 10.1073/pnas.1716804115 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Yu Q et al. T cell factor 1 initiates the T helper type 2 fate by inducing the transcription factor GATA-3 and repressing interferon-gamma. Nat Immunol 10, 992–999, doi: 10.1038/ni.1762 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Nakayamada S et al. Early Th1 cell differentiation is marked by a Tfh cell-like transition. Immunity 35, 919–931, doi: 10.1016/j.immuni.2011.11.012 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Crotty S T follicular helper cell differentiation, function, and roles in disease. Immunity 41, 529–542, doi: 10.1016/j.immuni.2014.10.004 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Oestreich KJ, Mohn SE & Weinmann AS Molecular mechanisms that control the expression and activity of Bcl-6 in TH1 cells to regulate flexibility with a TFH-like gene profile. Nat Immunol 13, 405–411, doi: 10.1038/ni.2242 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Choi YS et al. LEF-1 and TCF-1 orchestrate T(FH) differentiation by regulating differentiation circuits upstream of the transcriptional repressor Bcl6. Nat Immunol 16, 980–990, doi: 10.1038/ni.3226 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Wu T et al. TCF1 Is Required for the T Follicular Helper Cell Response to Viral Infection. Cell Rep 12, 2099–2110, doi: 10.1016/j.celrep.2015.08.049 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Xu L et al. The transcription factor TCF-1 initiates the differentiation of T(FH) cells during acute viral infection. Nat Immunol 16, 991–999, doi: 10.1038/ni.3229 (2015). [DOI] [PubMed] [Google Scholar]
- 66.Sakaguchi S, Yamaguchi T, Nomura T & Ono M Regulatory T cells and immune tolerance. Cell 133, 775–787, doi: 10.1016/j.cell.2008.05.009 (2008). [DOI] [PubMed] [Google Scholar]
- 67.Vignali DA, Collison LW & Workman CJ How regulatory T cells work. Nat Rev Immunol 8, 523–532, doi: 10.1038/nri2343 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Barra MM et al. Transcription Factor 7 Limits Regulatory T Cell Generation in the Thymus. J Immunol 195, 3058–3070, doi: 10.4049/jimmunol.1500821 (2015). [DOI] [PubMed] [Google Scholar]
- 69.van Loosdregt J et al. Canonical wnt signaling negatively modulates regulatory T cell function. Immunity 39, 298–310, doi: 10.1016/j.immuni.2013.07.019 (2013). [DOI] [PubMed] [Google Scholar]
- 70.Xing S et al. Tcf1 and Lef1 are required for the immunosuppressive function of regulatory T cells. J Exp Med 216, 847–866, doi: 10.1084/jem.20182010 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.van der Veeken J et al. The Transcription Factor Foxp3 Shapes Regulatory T Cell Identity by Tuning the Activity of trans-Acting Intermediaries. Immunity 53, 971–984 e975, doi: 10.1016/j.immuni.2020.10.010 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Osman A et al. TCF-1 controls Treg cell functions that regulate inflammation, CD8(+) T cell cytotoxicity and severity of colon cancer. Nat Immunol 22, 1152–1162, doi: 10.1038/s41590-021-00987-1 (2021). This study shows that TCF-1 differentially regulates Treg cell functions and cooperates with FOXP3 to supress TH17 and T cell activation programs.
- 73.Sage PT & Sharpe AH T follicular regulatory cells. Immunol Rev 271, 246–259, doi: 10.1111/imr.12411 (2016). [DOI] [PubMed] [Google Scholar]
- 74.Pais Ferreira D et al. Central memory CD8(+) T cells derive from stem-like Tcf7(hi) effector cells in the absence of cytotoxic differentiation. Immunity 53, 985–1000 e1011, doi: 10.1016/j.immuni.2020.09.005 (2020). [DOI] [PubMed] [Google Scholar]
- 75.Jeannet G et al. Essential role of the Wnt pathway effector Tcf-1 for the establishment of functional CD8 T cell memory. Proc Natl Acad Sci U S A 107, 9777–9782, doi: 10.1073/pnas.0914127107 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Zhou X et al. Differentiation and persistence of memory CD8(+) T cells depend on T cell factor 1. Immunity 33, 229–240, doi: 10.1016/j.immuni.2010.08.002 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Lin W.-Hsuan W. et al. CD8+ T Lymphocyte Self-Renewal during Effector Cell Determination. Cell Reports 17, 1773–1782, doi: 10.1016/j.celrep.2016.10.032 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Kaech SM & Cui W Transcriptional control of effector and memory CD8+ T cell differentiation. Nat Rev Immunol 12, 749–761, doi: 10.1038/nri3307 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Mueller SN, Gebhardt T, Carbone FR & Heath WR Memory T cell subsets, migration patterns, and tissue residence. Annu Rev Immunol 31, 137–161, doi: 10.1146/annurev-immunol-032712-095954 (2013). [DOI] [PubMed] [Google Scholar]
- 80.Zhou X & Xue HH Cutting edge: generation of memory precursors and functional memory CD8+ T cells depends on T cell factor-1 and lymphoid enhancer-binding factor-1. J Immunol 189, 2722–2726, doi: 10.4049/jimmunol.1201150 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Wherry EJ et al. Molecular signature of CD8+ T cell exhaustion during chronic viral infection. Immunity 27, 670–684, doi: 10.1016/j.immuni.2007.09.006 (2007). [DOI] [PubMed] [Google Scholar]
- 82.Yao C et al. Single-cell RNA-seq reveals TOX as a key regulator of CD8(+) T cell persistence in chronic infection. Nat Immunol 20, 890–901, doi: 10.1038/s41590-019-0403-4 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Khan O et al. TOX transcriptionally and epigenetically programs CD8(+) T cell exhaustion. Nature 571, 211–218, doi: 10.1038/s41586-019-1325-x (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Seo H et al. TOX and TOX2 transcription factors cooperate with NR4A transcription factors to impose CD8(+) T cell exhaustion. Proc Natl Acad Sci U S A 116, 12410–12415, doi: 10.1073/pnas.1905675116 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Scott AC et al. TOX is a critical regulator of tumour-specific T cell differentiation. Nature 571, 270–274, doi: 10.1038/s41586-019-1324-y (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Chen J et al. NR4A transcription factors limit CAR T cell function in solid tumours. Nature 567, 530–534, doi: 10.1038/s41586-019-0985-x (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Man K et al. Transcription Factor IRF4 Promotes CD8(+) T Cell Exhaustion and Limits the Development of Memory-like T Cells during Chronic Infection. Immunity 47, 1129–1141.e1125, doi: 10.1016/j.immuni.2017.11.021 (2017). [DOI] [PubMed] [Google Scholar]
- 88.Alfei F et al. TOX reinforces the phenotype and longevity of exhausted T cells in chronic viral infection. Nature 571, 265–269, doi: 10.1038/s41586-019-1326-9 (2019). [DOI] [PubMed] [Google Scholar]
- 89.Schietinger A et al. Tumor-Specific T Cell Dysfunction Is a Dynamic Antigen-Driven Differentiation Program Initiated Early during Tumorigenesis. Immunity 45, 389–401, doi: 10.1016/j.immuni.2016.07.011 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90. Utzschneider DT et al. T Cell Factor 1-Expressing Memory-like CD8(+) T Cells Sustain the Immune Response to Chronic Viral Infections. Immunity 45, 415–427, doi: 10.1016/j.immuni.2016.07.021 (2016). This study clearly defines characteristics of TEX- STEM cells and establishes their dependence on TCF-1 in the context of chronic viral infection.
- 91.Im SJ et al. Defining CD8+ T cells that provide the proliferative burst after PD-1 therapy. Nature 537, 417–421, doi: 10.1038/nature19330 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Wu T et al. The TCF1-Bcl6 axis counteracts type I interferon to repress exhaustion and maintain T cell stemness. Sci Immunol 1, doi: 10.1126/sciimmunol.aai8593 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93. Philip M et al. Chromatin states define tumour-specific T cell dysfunction and reprogramming. Nature 545, 452–456, doi: 10.1038/nature22367 (2017). This report associates tumour-specific T cell dysfunction to progressive loss of accessibility in chromatin sites enriched for the TCF motif.
- 94.Abdel-Hakeem MS et al. Epigenetic scarring of exhausted T cells hinders memory differentiation upon eliminating chronic antigenic stimulation. Nat Immunol 22, 1008–1019, doi: 10.1038/s41590-021-00975-5 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Liang C, Huang S, Zhao Y, Chen S & Li Y TOX as a potential target for immunotherapy in lymphocytic malignancies. Biomark Res 9, 20, doi: 10.1186/s40364-021-00275-y (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Blank CU et al. Defining ‘T cell exhaustion’. Nature Reviews Immunology 19, 665–674, doi: 10.1038/s41577-019-0221-9 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Arce L, Pate KT & Waterman ML Groucho binds two conserved regions of LEF-1 for HDAC-dependent repression. BMC Cancer 9, 159, doi: 10.1186/1471-2407-9-159 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98. Xing S et al. Tcf1 and Lef1 transcription factors establish CD8(+) T cell identity through intrinsic HDAC activity. Nat Immunol 17, 695–703, doi: 10.1038/ni.3456 (2016). This work identified and mapped self intrinsic HDAC activity in TCF-1 and LEF1, and determined its contribution in establishing the CD8+ T cell identity.
- 99.Johnson JL et al. Lineage-Determining Transcription Factor TCF-1 Initiates the Epigenetic Identity of T Cells. Immunity 48, 243–257 e210, doi: 10.1016/j.immuni.2018.01.012 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100. Shan Q et al. Tcf1 and Lef1 provide constant supervision to mature CD8(+) T cell identity and function by organizing genomic architecture. Nat Commun 12, 5863, doi: 10.1038/s41467-021-26159-1 (2021). This study identifies roles of TCF-1 and LEF-1 in shaping the chromatin conformation of CD8+ T cells
- 101.Zaret KS & Carroll JS Pioneer transcription factors: establishing competence for gene expression. Genes Dev 25, 2227–2241, doi: 10.1101/gad.176826.111 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102. Zhong Y et al. Hierarchical regulation of the resting and activated T cell epigenome by major transcription factor families. Nat Immunol 23, 122–134, doi: 10.1038/s41590-021-01086-x (2022). This paper leverages the genetic variability between B6 and CAST mice and integration of expression, transcription factor binding and epigenetic data to derive the central functions of trancription factor families in T cell activation.
- 103. Li F et al. TFH cells depend on Tcf1-intrinsic HDAC activity to suppress CTLA4 and guard B-cell help function. Proc Natl Acad Sci U S A 118, doi: 10.1073/pnas.2014562118 (2021). This study established that LEF 1 Bends the DNA and this facilitates the assembly of Functional Nucleoprotein complexes
- 104.Grosschedl R, Giese K & Pagel J HMG domain proteins: architectural elements in the assembly of nucleoprotein structures. Trends Genet 10, 94–100, doi: 10.1016/0168-9525(94)90232-1 (1994). [DOI] [PubMed] [Google Scholar]
- 105.Giese K, Cox J & Grosschedl R The HMG domain of lymphoid enhancer factor 1 bends DNA and facilitates assembly of functional nucleoprotein structures. Cell 69, 185–195, doi: 10.1016/0092-8674(92)90129-z (1992). [DOI] [PubMed] [Google Scholar]
- 106.Love JJ et al. Structural basis for DNA bending by the architectural transcription factor LEF-1. Nature 376, 791–795, doi: 10.1038/376791a0 (1995). [DOI] [PubMed] [Google Scholar]
- 107.Hu G et al. Transformation of Accessible Chromatin and 3D Nucleome Underlies Lineage Commitment of Early T Cells. Immunity 48, 227–242 e228, doi: 10.1016/j.immuni.2018.01.013 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Miller BC et al. Subsets of exhausted CD8(+) T cells differentially mediate tumor control and respond to checkpoint blockade. Nat Immunol 20, 326–336, doi: 10.1038/s41590-019-0312-6 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Brummelman J et al. High-dimensional single cell analysis identifies stem-like cytotoxic CD8(+) T cells infiltrating human tumors. J Exp Med 215, 2520–2535, doi: 10.1084/jem.20180684 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Kurtulus S et al. Checkpoint Blockade Immunotherapy Induces Dynamic Changes in PD-1(−)CD8(+) Tumor-Infiltrating T Cells. Immunity 50, 181–194 e186, doi: 10.1016/j.immuni.2018.11.014 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Siddiqui I et al. Intratumoral Tcf1(+)PD-1(+)CD8(+) T Cells with Stem-like Properties Promote Tumor Control in Response to Vaccination and Checkpoint Blockade Immunotherapy. Immunity 50, 195–211 e110, doi: 10.1016/j.immuni.2018.12.021 (2019). [DOI] [PubMed] [Google Scholar]
- 112.Vodnala SK et al. T cell stemness and dysfunction in tumors are triggered by a common mechanism. Science 363, doi: 10.1126/science.aau0135 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Zhang J et al. TCF-1 Inhibits IL-17 Gene Expression To Restrain Th17 Immunity in a Stage-Specific Manner. J Immunol 200, 3397–3406, doi: 10.4049/jimmunol.1800193 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]