Abstract
In-situ hybridization has been a robust method for detection of mRNA expression within whole-mount samples or tissue sections for more than 50 years. Recent technical advances for in-situ hybridization methods have incorporated oligo-based probes that attain greater tissue penetration and signal amplification steps with restricted localization for visualization of specific mRNAs within single cells. One such method is Third Generation in-situ Hybridization Chain Reaction (V3HCR) developed by Choi et al. that was first described in 2018. Here, we report an optimized protocol for V3HCR detection of gene expression using sectioned frozen tissues from mouse and human on microscope slides. Our methods and modifications for cryo-sectioning, tissue fixation, and processing steps over a three-day V3HCR protocol are detailed along with recommendations for aliquoting and storing V3HCR single-stranded DNA probes and hairpin amplifiers. In addition, we describe a method for blocking background signal from lipofuscin, a highly auto-fluorescent material that is widespread in human neurons and often complicates imaging efforts. After testing multiple strategies for reduction of lipofuscin we determined that application of lipofuscin quencher dye is compatible with V3HCR in contrast to other methods like cupric sulfate quenching or Sudan Black B blocking that cause V3HCR signal loss. This adaptation enables application of V3HCR for in-situ detection of gene expression among human neuronal populations that are otherwise problematic due to lipofuscin background auto-fluorescence.
Basic Protocol 1:
Mouse and Human Fresh-Frozen Tissue In-situ Hybridization Chain Reaction on Microscope Slides
Support Protocol 1:
Aliquoting HCR Probes and Hairpins from Molecular Instruments
Keywords: Third generation hybridization chain reaction (V3HCR), RNA in situ, tissue sections, single cell, Pax3, Elavl4
Introduction
Single cell RNA-Sequencing (scRNA-Seq) has substantially expanded knowledge of gene expression within individual cells. However, generation of scRNA-Seq data requires dissociation of cells from tissues, resulting in loss of positional context. The ability to validate scRNA-Seq data in situ within tissues hinges on implementation of methods for RNA localization that offer single cell resolution. Fortunately, recent technical advances among in-situ hybridization methods have acheived RNA localization with much higher resolution than previously possible (H. M. T. Choi et al., 2018; H. M. T. Choi, Schwarzkopf, & Pierce, 2020; Zhang, Svensson Akusjarvi, Sonnerborg, & Neogi, 2018). Third Generation Hybridization Chain Reaction (V3HCR) is one approach with multiple benefits including small probe size for greater penetration into tissues, adjacent oligonucleotide hybridization that suppresses background signal, multiplex imaging of distinct mRNA transcripts within a sample, and non-enzymatic signal amplification that produces tightly localized fluorescence within individual cells (H. M. T. Choi et al., 2018; H. M. T. Choi et al., 2020).We have adapted this protocol for validation of scRNA-Seq data on tissue sections for both human and mouse (May-Zhang et al., 2021). As a result of using single-strand oligonucleotide probe pairs designed to anneal to a targeted section of mRNA sequence, probes can be designed to avoid regions of homology between gene family members or repetitive sequences in untranslated mRNA regions. Probe pairs are separated by a two-nucleotide gap in between hybridization sequences and extended by an overhang “amplifier” sequence that folds back over the 2-nucleotide gap to form an initiator complex. Addition of fluorescently tagged DNA hairpins that hybridize to the extended overhangs of the initiator complex accomplish detection and signal amplification. This strategy allows visualization of intense fluorescent signal at the location of the RNA molecule to which the probes and amplifiers are bound. Moreover, multiple probe pairs can be designed to anneal along the length of an RNA molecule in a “shotgun” approach, to increase the intracellular fluorescence signal intensity. This facilitates the detection of lowly expressed messages.
The following protocol details utilization of V3HCR for frozen tissue sections adapted from the Molecular Instruments online protocol. We describe the processing of tissue including fixation and dehydration, slide handling for the method, and detailed processing through the V3HCR reaction protocol (Figure 1). The three-day V3HCR protocol includes hybridization, washing to remove unbound probe, amplification steps that bind amplifiers to the hybridized probe, with subsequent washing to remove excess amplifiers and several possible mounting options to allow flexibility in imaging. We also include steps for blocking background fluorescence due to lipofuscin, a lysosomal product that is widespread in human neurons and produces non-specific fluorescent puncta that can confound analysis of in situ hybridization results (Katz & Robison, 2002). These procedures can be used in parallel on human and mouse tissues to identify species differences as we previously published for analysis of the enteric nervous system in May-Zhang et al. (2021).
Figure 1:

Overview of sample processing for tissue preparation and procedures to complete V3HCR.
Strategic Planning
When performing multiplex detection of distinct mRNAs, attention must be paid to the selection of initiator sequences present in each probe. Up to four distinct amplifiers (B1, B2, B3, B4) can be applied to detect hybridized probes as long as those probes contain the corresponding initiator sequences. Using probes that the include the same initiator sequences, for instance with two probes both including B1, will cause those probe signals to be indistinguishable.
All solutions should be certified free from RNase or made up in baked bottles with RNase-free water. Some solutions need to be made up fresh and some reagents such as hydrogen peroxide are limited in the time they can be used after opening.
The steps involved in V3HCR sample processing require close attention to sample handling. It is recommended that the first time the protocol is attempted that other concurrent experiments be avoided.
Before proceeding with sample processing, treat equipment or surfaces that will be used with a RNase decontamination solution to avoid any RNase contamination that could reduce the fluorescent signal obtained from your sample.
Prepare in advance a humidification chamber (either constructed or purchased) to hold slides with tissue sections oriented upwards and kept level so that the reagents do not run off the sides during overnight incubation periods. This chamber should be sealed to prevent evaporation through use of parafilm or other sealing material such as Glad Cling Wrap.
Prepare “cover slips” of ParaFilm for use in covering tissue sections on slides during pre-hybridization, hybridization, and amplification steps. These should be cut from a roll of ParaFilm using scissors pre-treated with RNase decontamination solution. Handle these by their edges and avoid touching the surface that will be over the tissue sections on the slides.
Check that the incubator being used for slide incubation stably achieves 37°C in the location where the humidified chamber will be positioned as there can be temperature differences within incubators. Adjust the incubator set temperature accordingly to attain 37°C at the desired position by monitoring with thermometers positioned adjacent to where the humidification chamber will sit.
The type of mounting media used can influence how long fluorescent signal is preserved in samples. If studies necessitate use of an aqueous mounting media for later unmounting, it is best to perform confocal imaging within a week of sample mounting to avoid signal loss. It has been our experience that V3HCR signal can fade with some aqueous mounting medias.
Basic Protocol 1
Mouse and Human Fresh-Frozen Tissue In-situ Hybridization Chain Reaction on Microscope Slides
The following protocol utilizes V3HCR reaction for fluorescent visualization of RNA transcripts in mouse and human tissue sections. This protocol has been optimized for adult intestinal tissues although it can be readily applied to other tissues, stages, and sample thicknesses. Samples used in the protocol are first flash frozen, sectioned, fixed, dehydrated, and stored at −80°C. After this initial sample preparation, the first day of the HCR protocol can be performed, in which probe sets are hybridized to one or more RNA transcripts within tissue sections inside a humidification chamber. After an overnight hybridization step at 37°C, the samples are washed to remove unbound probe, and then amplifier DNA hairpins are added to generate fluorescent signal in an overnight incubation at room temperature. On the third day, samples are further washed to remove excess amplifiers, stained with DAPI (if desired), and coverslips are added for imaging.
Materials
| Reagent, Equipment | Vendor | Catalog Number |
|---|---|---|
| TFM/OCT | General Data Healthcare | TFM-(COLOR) |
| NEG-50 (tissue freezing media used for mouse) | Thermo Scientific | 6502 |
| Parafilm | VWR | 52858-032 |
| Glad Cling Wrap Press’NSeal | local grocery store | https://www.glad.com/food-protection/food-wraps/ |
| Superfrost™ Plus Microscope Slides | Fisher Scientific | 12-550-15 |
| Marker II/Superfrost (Histo-pen) | Secureline | 14905-30 |
| RNaseZAP | Sigma-Aldrich | R2020 |
| Cryostat | Leica Biosystems | |
| Kimwipes | Kimberly-Clark | 34155-EA |
| DEPC | Sigma-Aldrich | D5758 |
| PFA | Electron Microscopy Sciences | RT 19200 Prill |
| Sucrose | Fisher Scientific | S5-3 |
| DMSO | Sigma-Aldrich | D2650 |
| Hydrogen Peroxide | Sigma | H1009 |
| Ethanol | Pharmco by Greenfield Global | 111000200 |
| Molecular Seives | Acros Organics | AC197255000 |
| Methanol | Thermo Fisher Scientific | 176845000 |
| 20xSSC | Fisher Bioreagents | BP1325-20 |
| Tween® 20 Detergent, molecular biology grade | EMD Milipore Corp. | 655204-100ML |
| 10xPBS, pH 7.4 | Gibco | 70011-044 |
| ColorpHast Test Strips, pH range 6.5–10 | Fisher Scientific | M1095430001 |
| Pall-Gelman Membrane Forcep (51147) | VWR | 30033-042 |
| Proteinase K | Gorjira Fine Chemicals, LLC | PK1001 |
| HCR Probe Hybridization Buffer (30% Formamide) | Molecular Instruments | HCR Buffer Tissue Sections |
| HCR Probe Wash Buffer | Molecular Instruments | HCR Buffer Tissue Sections |
| Fluorescent HCR amplifier hairpins | Molecular Instruments, Alexa Fluor® | HCR Amplifiers |
| HCR Amplification Buffer | Molecular Instruments | HCR Buffer Tissue Sections |
| DAPI | ThermoFisher Scientific | 62247 |
| 5-place slide mailer | Electron Microscope Sciences | 71557-02 |
| TrueBlack® Lipofuscin Autofluorescence Quencher | Biotium | 23007 |
| Mounting media - Aqua-Poly/Mount (aqueous mount) | Polysciences | 18606-20 |
| Mounting media - Prolong Gold (hard mount) | ThermoFisher Scientific | P10144 |
Tissue Preparation: Sectioning
-
1
Embed tissue of interest in TFM/OCT freezing medium as previously illustrated (May-Zhang, Deal, & Southard-Smith, 2018) and store at −80°C wrapped in foil with an overlay of Cling Wrap to prevent dehydration until ready to use. If thoroughly coated with TFM/OCT and stored properly, samples have been successfully used even after being stored for multiple years at −80°C.
-
2
Pre-cool the Cryostat that will be used for sectioning the tissue to the optimal cutting temperature (−18 to −22°C).
-
3
Fill an insulated container with dry ice (CO2) for preparing tissue.
-
4
Thaw 4% PFA/3% Sucrose that was previously prepared (see Reagents and Solutions). Usually, 15 minutes in a 37°C water bath works well. Avoid heating the PFA beyond the point when it is melted and once thawed keep it on ice until use.
-
5
Fill another insulated container with wet ice to keep paraformaldehyde (PFA) chilled.
-
6
Prepare (or if storing long-term, thaw) and fill 5-place slide mailers with 15 ml of 4% PFA each and chill in the insulated container on wet ice (See Figure 2).
-
7
Label microscope slides with a Histo-Pen to correspond to each tissue type and section number as planned in advance. A Histo-Pen is used so that alcohols and other reagents used in sample processing do not remove the labels.
-
8
Prepare the cryostat for use by wiping down surfaces with 100% ethanol and treating the blade and brush tray with RNase decontamination solution.
-
9
Chill a microscope slide on top of a Kimwipe inside the Cryostat for approximately 10 seconds (or until chilled completely). This is to make sure that samples melt very slowly when adhered, which ensures minimal degradation of RNA.
-
10
Section the tissue of choice at 10 μm thickness and adhere to a pre-chilled microscope slide. If adhering more than one section per slide, keep all tissue sections in the cryostat on the metal base plate where tissue sections rest immediately after cutting. This will keep the sections frozen. Then position the sections adjacent to one another and non-overlapping so they can all be adhered to the slide simultaneously.
Figure 2: Use of slide mailers for processing slides through V3HCR using minimal solution volumes.
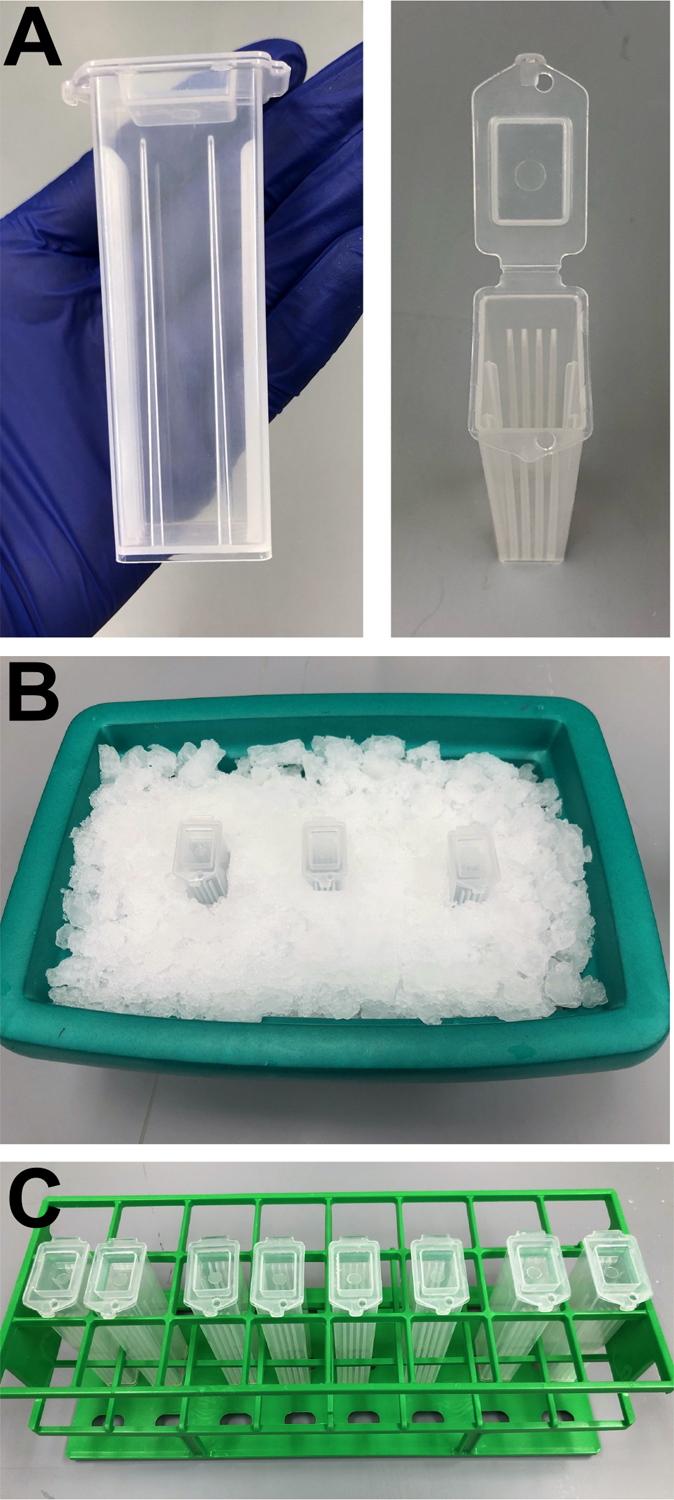
(A) Side and top views of slide mailer shown with slots from holding maximally five slides at a time. (B) Positions of slide mailers on wet ice in insulated container to keep samples chilled while fixing in PFA. (C) Arrangement of slide mailers in a plastic rack to prevent tipping so that slides can be quickly moved from one wash solution to another.
Tissue Preparation: Fixation of Sections
-
11
Immediately after tissue sections are adhered to the slide, each slide should be quickly immersed in in the 4% PFA fixative inside the plastic slide mailer(s) that are chilled on ice (Figure 2). Leave the tissue sections submerged for approximately 30 minutes.
-
12
Move PFA-filled slide mailers containing slides with tissue sections to room temperature (RT) and allow it to equilibrate (reaching room temperature could take 30–50 minutes). This is to ensure that the RNA is maximally preserved during the rest of the protocol, which is done at room temperature.
-
13
When the solution reaches RT, pour out the PFA in the slide mailers into a proper chemical waste disposal container by placing a pair of forceps across the opening of the slide mailer to keep the slides inside.
-
14
Rinse slides with DEPC water (see Reagents and Solutions) briefly two times. This is done by pouring DEPC water into the slide mailer to a level that covers all the tissue sections and then pouring it off again. A pair of forceps can be placed across the opening of the slide mailer to keep the slides in place. Or alternatively, slide mailers can be placed adjacent to one another filled with the appropriate solutions and the slides can be moved from one mailer to another using forceps to lift each slide (See Figure 2).
-
15
Rinse slides in 1x PBS (diluted in DEPC water) for 10 minutes at room temperature.
Tissue Preparation: Dehydration of Sections
Notes: The following ethanol dehydration steps should be performed in the same plastic slide mailer containers. Ethanol used for this process should be completely anhydrous (200 proof), which is best sourced from a newly opened bottle of 100% ethanol. Alternatively, water can be removed from open bottles of ethanol by addition of molecular sieves. By adding molecular sieves (8–12 mesh) to the bottle of 100% ethanol prior to the start of the procedure any water absorbed from the atmosphere is absorbed (Clement-Ziza, Munnich, Lyonnet, Jaubert, & Besmond, 2008).
-
16
After the final rinse of 1x PBS (DEPC water-diluted), pour off 1x PBS and pour to fill to the brim with 50% ethanol in the slide mailer(s) with slides. Incubate for 5 minutes at RT.
-
17
Pour off the 50% ethanol and fill the slide mailer to the brim with 75% ethanol. Incubate for 5 minutes at RT.
-
18
Pour off the 75% ethanol and fill the slide mailer to the brim with 100% ethanol. Incubate for 5 minutes at RT.
-
19
Replace the 100% ethanol in the slide mailer with fresh 100% ethanol. Incubate for 1 hour or more at room temperature.
-
20
Store samples for long-term storage in the 100% ethanol solution at −80°C. Samples should be used within 4 months following storage.
Note: Storage for a minimum of 24 hours in ethanol is required for success of the protocol. Do not attempt to decrease this incubation time.
Day 1 HCR: Slide Preparation and Hybridization
Notes: The following steps should be performed first in the plastic slide mailers, and then in the humidification chamber. The slide mailers used in this protocol have 5 slide spaces, so batches of 5 were used in this process. With batches of 5 slides, you will need 25 ml of wash buffer, 4 ml of hybridization buffer, and 4 ml of amplification buffer. Make sure to perform steps in the humidification chamber quickly to prevent sample drying.
-
21
Arrange the humidification chamber with Whatman paper at the bottom with Milli-Q filtered deionized H2O just soaking the paper (excess water must not be pooling in the chamber; see picture, Figure 3A).
-
22
Set an incubator to 37°C. Be sure to check that the location where your humid chamber will be positioned in the incubator is actually at 37°C as there can be positional differences in temperature within an incubator.
-
23
Retrieve samples that were previously stored in 100% ethanol at −80°C and allow them to equilibrate to room temperature for processing. These may also be thawed at 4°C for more sensitive treatment.
-
24
If a full slide mailer is not being used and just a few samples are being processed, place the selected slides into a −80°C-prechilled slide mailer filled with 100% EtOH and then allow that slide mailer to equilibrate to room temperature.
-
25
(Optional Dent’s Bleach Treatment can be performed to quench autofluorescence background due to presence of red blood cells in the tissue.) Remove slides from the 100% EtOH with forceps and place them in a fresh slide mailer of 100% MeOH. After 5 minutes, remove the slides and place them face up in the humidification chamber. Place 400 μl of Dent’s bleach (see Reagents and Solutions) on the surface of each slide and cover with parafilm cut into “coverslips” on top for 45 minutes (See Figure 3A). Parafilm coverslips are made by cutting parafilm into slide sized strips with scissors treated with RNase decontamination solution. Remove the paper backing from the strip and orient the strip so that the surface that was covered is placed down against the tissue section on the slide. Handle strips by the edges and avoid touching the central area that will be over the tissue sections. Follow the Dent’s bleach treatment with 1xPBST (see recipe) washes three times for five minutes each.
-
26
Thaw a frozen aliquot of 4% PFA that was previously prepared (see Reagents and Solutions) until it is just thawed. Usually, 15 minutes in a 37°C water bath works well. Avoid heating the PFA and once thawed keep it on ice until use.
-
27
After equilibration to room temperature, wash slides in RT 1x PBS two times for 5 minutes, then once for 10 minutes.
-
28
During this or one of the following incubation periods, set up a slide drying station with lint free absorbent wipes, like Kimwipes (see Figure 3C). This area will be used for removing excess liquid from the slides when transitioning into the humidification chamber.
-
29
After washes, thaw 1 μg/ml Proteinase K as previously prepared (see Reagents in Reagents and Solutions), then immediately pour off 1x PBS wash from the sample(s) inside slide mailer(s) and pour in the Proteinase K. Incubate for 10 minutes at room temperature.
-
30
Pour off proteinase K solution and inactivate the residual enzyme by pouring the thawed 4% PFA into the sample slide mailer. Incubate at room temperature for 10 minutes.
-
31
Pour off 4% PFA into the appropriate chemical waste container.
-
32
Immediately wash slides at RT in 1x PBS three times for 5 minutes each.
-
33
Remove the slides from the slide mailer by using forceps to lift them from the labeled end.
-
34
Place the slides on the drying station with the tissue sections and label facing up.
-
35
Take a 11×21 cm Kimwipe and fold it several times until there is a folded angle (see Figure 3C).
-
36
Pick up the slide with forceps and run the folded angle of the Kimwipe along the edges of the slide to wick away solution without touching the sample.
-
37
Return the slide(s) to the humidification chamber sample side up.
-
38
Perform a pre-hybridization step by adding 300–400 μl of HCR Probe Hybridization Buffer (see Materials list) to the samples on the slides in the humidification chamber. The volume of HCR Probe Hybridization Buffer used will depend on the size of the tissue section and should cover the entire sample. Close the chamber and incubate at 37°C for 30 minutes. Covering the pre-hybridization solution on the slides is not necessary since this incubation is of a short duration.
-
39
During the pre-hybridization period, prepare probe dilutions into HCR Probe Hybridization Buffer. Remove a tube of aliquoted probe from −20°C storage (see Support Protocol 1). Dilute each probe being used so that 0.8 pmols of each is added into the HCR Probe Hybridization Buffer. For example: If applying three probes to a slide, 0.8 μl of each probe totaling 2.4 μl will be added to a final volume of 200 μL per slide.
-
40
Following the 30-minute pre-hybridization process, use the drying station again, including a new folded Kimwipe, to remove the initial HCR Probe-Hybridization Buffer used in the pre-hybridization process from the slide(s). Make sure to avoid touching the sample/tissue during this process.
-
41
Return the slide(s) to the humidification chamber with the tissue section sample side up.
-
42
Apply probe solutions to each slide at 200 μl per slide.
-
43
Cover the tissue sections with the newly diluted probe in HCR Probe Hybridization Buffer using a parafilm “coverslip” cut to fit the slide’s sample to the slides over the probe solutions as described above in step 25. Covering the sections in this manner will spread the solution and help keep it from evaporating.
-
44
Make sure the Whatman paper is well soaked with Milli-Q H2O in the bottom of the chamber, then shut the chamber.
-
45
Seal the chamber to prevent moisture loss during hybridization (Figure 3B). This can be done by applying Parafilm or Glad Cling Wrap to the edges of the chamber if the chamber being used does not have a built-in mechanism.
-
46
Incubate the humidification chamber overnight (12 to 16 hours) at 37°C. This time interval is a minimum period for hybridization. If circumstances require, the hybridization can be left for a longer period up to 72 hours as long as the slides remain moist and evaporation of solutions does not occur.
Figure 3: Humidification chamber and slide drying setup for V3HCR processing.

(A) Humidification chamber is configured from a square tissue culture plate (24.2 × 24.2 × 2.2 cm) in which 5 ml plastic pipettes are glued to the bottom of the plate. Strips of Whatman paper are placed in the bottom and saturated with water to provide a humid environment. Slides are positioned on top of the pipette supports and can be temporarily coverslipped with parafilm strips at critical steps of the procedure to reduce evaporation from the slide surface. (B) The entire chamber is sealed with ParaFilm (Glad ClingWrap can also be used) wrapped around the edges to prevent evaporation. (C) Drying station made of lint-free disposable wipes, like KimWipes. The folding of a small wipe into a triangular shape as shown facilitates removal of solution from the edges of the slides.
Day 2 HCR: Washing to remove probe and Amplification
-
47
Turn on a heat block, or slide warmer, and set it to 37°C.
-
48
Set up a fresh slide drying station.
-
49
Place HCR Probe Wash Buffer into a water bath set to 37°C. Wait to proceed until the temperature of the wash buffer has equilibrated to 37°C.
-
50
Once the heat block is at 37°C, take out the humidification chamber from the incubator, open the seal, and place the chamber on top of the heat block or slide warmer.
-
51
Pick up slides carefully from the chamber with forceps and pour off the hybridization-probe solution from the overnight incubation into an appropriate chemical waste container. Removal of hybridization solution does not have to be complete and it is better to leave the slides moist to avoid drying.
-
52
Immediately place the slides back into the chamber sample side up.
-
53
Immediately add HCR Probe Wash Buffer to the samples at 400 μl per slide.
-
54
Shut the humidification chamber and incubate the chamber at 37°C for 30–60 minutes.
-
55While the slides are in the initial wash buffer (step 54 above) incubation, prepare fresh mixtures of HCR Probe Wash Buffer with 5XSSC-T and warm them to 37°C in a water bath:
- ≥15 ml (for a 5-space plastic slide mailer) 75% HCR Probe Wash Buffer/25% 5XSSC-T
- ≥15 ml 50% HCR Probe Wash Buffer/50% 5XSSC-T
- ≥15 ml 25% HCR Probe Wash Buffer/75% 5XSSC-T
- ≥15 ml 100% 5XSSC-T
-
56
As the slides are incubating in step 54 above, prepare ≥15 ml 100% 5XSSC-T at RT.
-
57
After the initial wash (step 54), pour off the HCR Probe Wash Buffer and use the drying station and folded Kimwipe to remove excess solution. Make sure to not contact the sample during this process and to pick up the slide with forceps.
-
58
Place slide(s) into a new plastic slide mailer(s) and fill with pre-warmed 75% HCR Probe Wash Buffer/25% 5XSSC-T and incubate for 15 minutes in a 37°C water bath. If reagents are limited, this step and the next three wash buffer/5XSSC-T incubations can be done on the slide surfaces with approximately 400 μL of the solution per slide.
-
59
For the next wash, pour out the 75% wash buffer/25% 5XSSC-T mixture and replace with pre-warmed 50% HCR Probe Wash Buffer/50% 5XSSC-T. Incubate for 15 minutes at 37°C in the water bath.
-
60
For the next wash, pour out the 50% wash buffer/50% 5x SSC-T mixture and replace with pre-warmed 25% HCR Probe Wash Buffer/75% 5XSSC-T. Incubate for 15 minutes at 37°C in the water bath. The next step (61) must be performed during this incubation time.
-
61Denaturation of amplifiers:
- Remove the HCR amplification buffer (Materials list) from 4°C storage and aliquot approximately 4 ml into a tube.
- Thaw hairpins as ordered from Molecular Instruments and aliquoted using the support protocol in this manuscript.
- Turn on a thermocycler and enter a program to run at 95°C for 90 seconds.
- Calculate the required volume of hairpins that will be needed for amplification. For example, 1.5–2 μl per slide of hairpins are used in a volume of 75 μl of HCR Amplification Buffer (see Step 67) therefore, 1.5 μl or 2 μl hairpins x n slides is the appropriate volume of hairpins to retrieve from the stocks.
- Pipet the calculated volume of each hairpin into separate PCR tubes allowing for use of 1.5–2 μl of each hairpin per slide. For instance, denature 2 μl of hairpin H1 in one PCR tube, while 2 μl of hairpin H2 is denatured in a separate tube in parallel.
- Denature the hairpins by incubating the PCR tubes through the thermocycler program.
- Transfer tubes to a dark location (a drawer works well for this) and let cool to room temperature for 30 minutes. This 30-minute cooling time overlaps with the next steps.
-
62
After the 15-minute incubation at 37°C in 25% HCR Probe Wash Buffer/75% 5XSSC-T in step 60, replace the solution with the 100% 5XSSC-T pre-equilibrated to 37°C and incubate the slide mailer and samples at 37°C for ≥15 minutes.
-
63
After the step 62 15-minute incubation in 100% 5XSSC-T at 37°C, replace the solution with the 100% 5XSSC-T at room temperature and incubate the slide mailer and samples at room temperature for ≥5 minutes (incubation time can be extended if needed).
-
64
Use forceps to remove slides from the slide mailer and use a folded Kimwipe to blot dry the slides very briefly and transfer them into the humidification chamber.
-
65
Immediately add 400 μl of room temperature HCR amplification buffer as each slide is being dried with step 64.
-
66
Shut the chamber and incubate at room temperature for 30 minutes once each slide has 400 μl of HCR amplification buffer.
-
67
After letting the hairpins cool to room temperature, dilute each set of hairpins 1:50 in HCR amplification buffer. For example, if using 1.5 μl of hairpins per slide (3.0 μl total hairpins), dilute each set of hairpins in 75μl HCR amplification buffer. Similarly, if making this dilution for 5 slides, 7.5 μl of each hairpin (15 μl total for the hairpin set) would be diluted into 375 μl of HCR amplification buffer. As a result of this process: the hairpins are in one volume that can be distributed evenly amongst the slides. Refer to step 61d for instructions on calculating the amount of hairpins to denature.
-
68
Handling one slide at a time, pour off amplification buffer briefly and immediately apply the solution containing the denatured, diluted hairpins to the slide. If tissue sections are not immediately covered, the slides begin to dry, which can cause a background autoflourescent glow over the entire slide.
-
69
Cover slides with small pieces of parafilm pre-cut to the size of a coverslip (see Figure 3A inset) to ensure even coverage of the hairpin solution.
-
70
Shut the humidification chamber and seal it as described in step 45, making sure the Whatman paper is sufficiently soaked with MilliQ-H2O to allow for overnight incubation.
-
71
Cover the humidification chamber with aluminum foil or place it in a dark drawer to protect from light. Incubate overnight at room temperature (12–16 hours). Note that this 16-hour period is a minimum time, and the amplification incubation can be extended if circumstances require it up to 72 hours as long as the slides are kept moist and solutions remain on the slides covering the tissue sections.
Day 3 HCR: Washing, DAPI Staining, and Coverslipping
-
72
Set up a fresh slide drying station as previously described in step 28.
-
73
Open the humidification chamber containing the slides in HCR Amplification buffer, dry slides as previously described in step 64, and transfer to fresh plastic slide mailers that are pre-filled with room temperature 5XSSC-T.
-
74
Wash slides in the mailer 2 times for 30 minutes each with 5x SSC-T at room temperature in a dark drawer.
-
75
Apply 200 μl of DAPI DNA stain (diluted 1:50000 in 1x PBS) to slides for 5 minutes at room temperature.
-
76
Pour off DAPI solution from the slides and rinse in 1x PBS for 5 minutes in a fresh slide mailer.
-
77(Optional treatment with TrueBlack® lipofuscin quencher dye is recommended if HCR processing is being performed on human tissue where lipofuscin in neurons is a frequent problem :
- Briefly (30 seconds) apply minimally 150 μl TrueBlack® dye solution (see Reagents and Solutions) to a slide. While the stain is on the slide, perform gentle continuous agitation by hand, holding the slide level with a swirling motion, to keep the stain moving over the surface of the tissue section.
- De-stain the slide by dipping three times into 30 ml of 45% ethanol in a conical tube for approximately 10 seconds with gentle agitation.
- Wash the slides three times for 5 minutes in 50 ml of 1x PBS.
-
78Mount and coverslip:
- If using TrueBlack® and processing human tissue, it is necessary to use compatible mounting medias that do not remove the TrueBlack® blocking. Some mounting medias are incompatible with TrueBlack®, causing it to quickly leach out of the sample. We have found that short term mounting (7–10 days) in 1X PBS is compatible and sustains the TrueBlack® blocking on the tissue. Alternatively other mounting medias recommended by Biotium for mounting after TrueBlack® use may be applied (see https://biotium.com/faqs/what-mounting-media-can-be-used-with-trueblack/)
- If blocking of lipofuscin is not necessary, standard mounting medias may be applied. Hard mount medias such as ProLong Gold are compatible with V3HCR and maintain the signal for months. Alternatively, a brief post-fix in 4% PFA after washing to remove excess amplifiers (step 74) performed at RT for 20 minutes, followed by two more washes in 5XSSC-T at room temperature locks the signal in place so that aqueous mounting media, like AquaPolyMount can be used.
-
79
Use nail polish to seal the around the edges of the coverslips onto the slides if mounting with PBS or an aqueous media.
-
80
If mounted in 1X PBS store slides at 4°C in a humidified chamber and perform imaging as quickly as possible.
Support Protocol 1
Aliquoting HCR Probes and Hairpins from Molecular Instruments
The purpose of this protocol is to serve as a guide for aliquoting HCR probes and HCR amplifiers that are distinct essential reagents for V3HCR sample processing. Both HCR probes and HCR amplifiers may be purchased from Molecular Instruments. Aliquoting HCR probes is done to enable ease of use over multiple experiments, as aliquots can be rapidly retrieved from freezer storage during an experiment. Storage of HCR amplifiers in aliquots avoids repeated freezing and thawing that could affect the fluorescence moieties on these molecules. HCR probes may be purchased directly from Molecular Instruments or synthesized by individual laboratories following the principles described by Choi et al., 2018. Some groups have generated informatics pipelines that facilitate the HCR probe design process (Lovely et al., 2022 In Press). HCR amplifiers are available for purchase from Molecular Instruments. For the aliquoting process, tubes are first prelabeled with the appropriate information, and equipment and surfaces are treated with RNase decontamination solution. Calculations are performed to have optimal concentrations of probes, and probes are diluted, aliquoted, and put into larger tubes or boxes for organization purposes. For example, if HCR probes are obtained from Molecular Instruments at 1μ M, and batches of 5 slides are being processed at a time, then probes should be aliquoted to approximately 6 μl per PCR tube to ensure sufficient probe is present in a single aliquot for application to five slides. Amplifiers and probes are aliquoted and distributed into larger conical tubes or freezer storage boxes for organizational purposes.
Materials
| Reagent, Equipment | Vendor | Catalog Number |
|---|---|---|
| RNaseZAP | Sigma-Aldrich | R2020 |
| 1.5ml Rnase-free tubes (3 per probe set) | ||
| HCR ssDNA Probes | Molecular Instruments | |
| Fluorescent HCR amplifier hairpins | Molecular Instruments, Alexa Fluor® | HCR Amplifier |
| Nuclease-free H2O | Ambion | AM9932 |
| Negative 20°C freezer for storage | ||
| An excess of 1–10 μl pipette tips | ||
| 8-channel 1–10 μl pipette | ||
| Repeat piettor with 200 μl capacity | ||
| PCR tubes, 8-tube strip with attached caps | ||
| 50 ml conical tubes (2 per probe set) | ||
| 1000 μl pipette | ||
| 96-well plastic tube rack (treat with bleach, then RNase decontamination solution) |
HCR Probe Aliquoting:
-
1
Thaw probes on ice. Probes from Molecular Instruments are received in a 1 μM stock solution. For each slide 0.8 μl of probe is used thus a five-slide experiment would consume a total of 4 μl of probe.
-
2
Remove 3 to 4 PCR tube strips (8-tubes with attached caps) from their container and place in a 96-well plastic tube rack (previously RNase decontaminated) at room temperature.
-
3Label the tube caps with the following information:
- Target gene symbol
- Species from which the tissue was obtained
- Adapter for which the probes are designed (B1–4)
-
4
Use the repeat pipettor to make 6 μl aliquots in each of the tubes that were labeled.
-
5
Separate the tubes from one another so a single aliquot can be used at a time.
-
6
Transfer to a previously labeled 50 ml conical tube.
-
7
Store at −20°C. Avoid repeated freezing/thawing.
Diluting and aliquoting HCR Amplifiers from Molecular Instruments:
-
8Pre-label a 50 ml conical tube with the following information:
- Adapter designation (B1–4)
- Wavelength corresponding to the adapter
- Hairpin aliquot designation (h1 or h2)
-
9
Wrap the 50 ml conical tube with aluminum foil and add a label with the same information from the previous step.
-
10
Remove amplifiers from −20°C storage and place them in an area treated with RNase decontamination solution that is away from direct light.
-
11
Place 4 PCR 8-strip tubes into an RNase decontamination-treated 96-tube rack. Position the strip tubes so they are spaced evenly apart.
-
12
Use the repeat pipettor to remove the full 100 μl for each h1 or h2 amplifier hairpin stock.
-
13Alternate tubes when aliquoting 5.5 μl for each amplifier per tube (see Figure 4).
- Note: The 5.5 μl volume is used here because if a 100 μl solution is made for the main protocol and 5 slides are being used, this corresponds to 1 μl per slide plus 0.5 μl to account for pipetting error.
-
14After aliquoting, close the tubes and label them with the following information:
- Adapter sequence (B1, B2, B3, B4)
- Wavelength corresponding to the hairpin
- Hairpin designation (h1 or h2)
-
15
Make three cuts per 8-tube strips on the connections between tubes so that they are divided into h1 and h2 pairs.
-
16
Place tubes into a pre-labeled 50 ml conical tube corresponding to the hairpin amplifiers being aliquoted.
Figure 4: Diagram of Aliquoting Process for HCR Amplifiers.

Place strip tubes in alternating rows of 96-place rack. Aliquot H1 and H2 amplifiers into every other tube in the strip tube so they are alternating. Cut between each pair of H1/H2 tubes as indicated ( ). Store the separated pairs of tubes in −20°C for future use.
). Store the separated pairs of tubes in −20°C for future use.
Reagents and Solutions
Diethyl pyrocarbonate (DEPC)-treated H2O
DEPC (Sigma Aldrich, D5758) is a potent alkylating agent and hazardous chemical with a maximum solubility of 1% in water and a half-life of ~15 minutes in water. All handling should be performed in an appropriate chemical fume hood while wearing appropriate PPE. DEPC treatment of water and glassware is performed to remove RNAse activity. When the procedure is followed as described below no residual DEPC that might interfere with hybridization or sample processing remains. Often times protocols for DEPC treatment call for autoclaving DEPC-treated water to remove the DEPC. Autoclaving is insufficient to completely remove DEPC and can lead to problems with chemical reactions performed in those solutions. Use of DEPC treated water is often less costly than purchase of RNAse-free water from commercial vendors. Begin by adding DEPC to a final concentration of 0.05% to deionized H2O in a 4L glass Erlenmeyer flask. Addition of 1.75ml of DEPC per 3.5 liters of H2O is an appropriate dilution and allows for head space in the flask to accommodate stirring and heating. Include a few Teflon boiling chips in the flask to facilitate boiling. After addition of DEPC, stir for 10 minutes, then boil for 2 hours to inactivate any remaining DEPC. Cover with saran wrap (avoid covering with foil) while cooling then filter sterilize through 0.2 micron and store or use to generate other solutions. The final solution after cooling should NOT have any residual odor of DEPC, which smells like green apple candy. An alternative to this process is purchase of certified RNAse-free water from a variety of vendors.
Dent’s Fixative
Make 4 parts 100% Methanol (MeOH, Thermo-Fisher, 176845000) and 1 part DMSO (Sigma-Aldrich, D2650). For example, 4 ml MeOH combined with 1 ml DMSO for a final volume of 5ml. Make this fresh for each use.
Dent’s Bleach
Make 1 part hydrogen peroxide (H2O2, Sigma-Aldrich, H1009), 2 parts Dent’s Fixative. For example, 1 ml 30% H2O2 mixed with 2 ml of the above freshly made Dent’s fixative. Make this fresh for each use. *Note that 30% Hydrogen Peroxide Stock solutions should be opened within the last 30 days and discarded 30 days after opening.
1xPBST
Make 1xPBS (diluted from 10xPBS, Gibco, 70011–044) with 0.1% Tween 20 (EMD Milipore Corp., 655204–100ML). For example, if 500 ml are needed, add 0.5 ml of Tween 20 to 500ml of 1XPBS.
RNase-Free 1x PBS in DEPC-treated H2O
Make a 1:10 dilution of 10x PBS in DEPC-treated H2O.
Proteinase K in RNase-free 1x PBS
Prepare a dilution of Proteinase K (Gorjira Fine Chemicals, PK1001) in 1x PBS (made up in DEPC-treated H2O) to a concentration of 1 μg/ml from a source stock that is 10mg/ml. Source stocks of Proteinase K are made up in DEPC-treated water to 10mg/ml, aliquoted to smaller volumes, and stored at −20°C long term. Do not thaw source stocks of Proteinase K more than twice. For softer tissue or embryonic tissue, a final concentration of 0.1 μg/ml 1x PBS is recommended.
4% Paraformaldehyde (PFA), 3% Sucrose
PFA is a crystallized polymer of formaldehyde that is a hazardous chemical (Electron Microscopy Sciences, RT 19200 Prill). PFA should be weighed in a chemical fume hood while wearing appropriate PPE. To make 1 L of solution, heat 800 ml of 1x PBS in a flat bottom glass container on a heating plate with a loosely capped to ~55–60°C. More rapid heating can be accomplished in a microwave, however, avoid heating the solution over 60°C. Heating PFA to temperatures above 65°C results in breakdown of the molecule. After the 1X PBS is warmed, add 30 g of sucrose and stir to dissolve. Add the 40 g of EM-grade PFA to the solution and continue stirring to dissolve. Check the pH of the solution with pH Test Strips. Adjust the pH as needed to obtain pH 7.4 using either 5N NaOH or 1N HCl. Aliquot and store PFA solution at −20°C for up to 3 months. Thaw PFA for use by warming in a 37°C water bath. Do not thaw more than three times. Thawed PFA should be used within a few days. Chill thawed PFA on ice prior to use for tissue fixation.
5XSSC-T
250 ml 20X Saline Sodium Citrate (SSC) (Fisher Scientific, BP13254–4L) diluted with 750 ml DEPC H2O (see above). Add 1 ml of Tween® 20 Detergent, molecular biology grade (EMD Milipore Corp, 655204–100ML). This solution can be made and stored at RT long term for convenience.
TrueBlack® Lipofuscin Quencher Dye Dilution
Dilute 2.75 μl of TrueBlack® (Biotium, 23007) in 150 μl of 70% EtOH.
DAPI (4′,6-diamidino-2-phenylindole) DNA Stain
Dilute DAPI (Thermo Fisher Scientific, 62247) 1:50000 in 1x PBS.
Commentary
Background Information
In-situ hybridization chain reaction RNA-FISH (HCR), first developed by Choi et al. in 2010, was designed to solve the difficulties associated with imaging multiple mRNA target transcripts simultaneously. This protocol design took advantage of a previous concept that introduced by Dirks and Pierce in which DNA monomers assemble on exposure to some target fragment of DNA (Dirks & Pierce, 2004). The protocol has since been improved upon with the addition of DNA hairpins (H. M. Choi, Beck, & Pierce, 2014) and then single-stranded oligonucleotide probes (H. M. T. Choi et al., 2018) to replace the less stable and more expensive RNA reagents that were used in the first versions of the protocol. Next-generation HCR introduced higher signal-to-background ratios and the addition of DNA hairpins for lower cost (H. M. Choi et al., 2014). Next-generation HCR also introduced quantitative imaging techniques and digital quantitation of mRNA with digital HCR. Third generation HCR introduced single-stranded oligonucleotide probes and augmented amplifiers that have automatic background suppression, ensuring that non-specific binding of probes and amplifiers cause background signal. (H. M. T. Choi et al., 2018). RNAScope, another method developed in an effort to achieve single cell localization, uses a branched DNA method in which a label binds to an exposed substrate instead of self-amplifying DNA hairpins (Wang et al., 2012). There is potential for the RNAScope technique to produce non-specific binding of reagents as a result of left-over unstructured branches. This contrasts with V3HCR, which has a simplified two-stage scheme that only involves a detection and amplification step without a pre-amplifier bridge middle step (H. M. T. Choi et al., 2018). In our hands V3HCR has proven robust for detection of multiple mRNA transcripts across multiple tissue types and can be readily modified for whole-mount in situ detection as well (Ibarra-Garcia-Padilla, Howard, Singleton, & Uribe, 2021).
Critical Parameters
To achieve optimal application of V3HCR, the tissue type, strategy for achieving an RNase-free environment, management of a humidified chamber, the possibility of lipofusin in human tissues and selection of mounting media must be considered. Application of V3HCR to human tissues containing neurons requires that methods for blocking background fluorescence from lipofusin be applied. Lipofuscin is an autofluorescent pigment that accumulates in the lysosome of some cell types, particularly neurons, and has punctate fluorescent pattern (Brehmer, Blaser, Seitz, Schrodl, & Neuhuber, 2004; Schnell, Staines, & Wessendorf, 1999). Because lipofuscin is present at high levels in neurons and neuron-specific genes may be expressed at low levels, lipofuscin can overlap the signal from HCR, making it difficult to decipher true signal from noise (Figure 5). This problem is circumvented by use of a derivative of Sudan Black B (TrueBlack®) that serves as a lipofuscin autofluorescence quencher (Figure 5). We tested multiple alternatives including Cupric Sulfate quenching, bleaching in Dent’s fixative, detergent extraction, Murray’s clearing, treatment with 8% SDS, and standard Sudan Black blocking all of which failed to block lipofuscin (data not shown).
Figure 5: Lipofuscin in human neuronal cells can be blocked through use of TrueBlack®.

(A) Presence of lipofuscin in human enteric ganglia hybridized with V3HCR probe for SNAP25 shows prominent lipofuscin signal in the 488 channel, where no signal should be detected. This lipofuscin has strong overlap with neuronal specific probes and complicates interpretation of images. (B) Images of human myenteric ganglia prior to treatment with lipofuscin (pre-treatment at left) show prominent signal in the 488 channel. After blocking with TrueBlack® nonspecific signal of lipofuscin is not observed and SNAP25 HCR probe signal is confined to neuronal bodies in the 594 channel. Note that images at left and right are of independent ganglia as this effect was consistently observed and the large size of human tissue sections made locating the same ganglia before and after blocking challenging.
While TrueBlack® accomplishes lipofuscin blocking, it is not compatible with use of some aqueous mounting medias. We observed that some aqueous mounting medias, like AquaPolyMount, cause diffusion of TrueBlack® from the tissue and reappearance of the lipofuscin signal. We found that use of 1x PBS for mounting and short-term storage over a week to 10 days is sufficient to retain blocking of lipofuscin and allows time for subsequent imaging. However, this work around does not maintain the HCR signal for long and thus confocal imaging must be accomplished rapidly within a week. It has also been our observation that use of AquaPolyMount results in loss of HCR signal loss over the period of a few weeks, unless samples are subjected to a brief post-fixation step that fixes the HCR probes and amplifiers in place. Multiple “hard set” mounting medias are compatible for mounting HCR samples and yield durable signal retention over several weeks to months. These include: Fluoromount-G® (Southern Biotech), SlowFade Antifade Reagents of “several types” (ThermoFisher Scientific), and ProLong Gold/Diamond/Glass (ThermoFisher Scientific). However, we have not tested all of these for compatibility with TrueBlack® and find that a quick post-fix step is the most robust solution in our experience.
One essential aspect to maintain during the V3HCR procedure is rapid sample processing of slides to avoid drying of tissue sections or reagents onto the slide surface. We observed an autofluorescence glow over the slide surface when the sections on the slide have their incubation solutions removed too thoroughly or slowly before replacing them with reagents from the next step of the procedure. Rapid replacement of solutions at all stages avoids this background autofluorescence effect.
An RNase-free environment is critical for the detection of RNA transcripts within cells or tissues. This protocol uses RNase-free reagents throughout. All surfaces and equipment used in this protocol are treated multiple times throughout the procedure with RNase decontamination solution to ensure the environment stays RNase-free. In our experience we found RNase decontamination solution used liberally accomplished this and did not interfere with sample processing.
Use of a humidified chamber is essential to avoid drying of reagents on slides. If a “do-it-yourself” chamber is used there are several important metrics that must be considered. First, the chamber must be constructed in such a way to allow for a tight seal. This is to retain humidity inside the chamber so that tissues and reagents critical to the protocol do not dry out. One can maintain this seal by using ParaFilm. The second important consideration requires that there be a raised, level surface inside the chamber on which the slides holding the tissue can sit. A level support surface is necessary for making sure that the reagents do not wick off the slides during the incubation periods. We have found that having all slides oriented the same direction (label on the left and tissue on the right) and propping the chamber up to a very small incline on the “label” side keeps the solution reagents on the tissue.
Troubleshooting
The concentration of probes in HCR hybridization buffer, as well as incubation time for proteinase K and PFA inactivation, may be changed according to the thickness and density of the tissue of interest. The times and concentrations recommended in this protocol are optimized for 10 μm sections of human and mouse intestine although adjustments to these aspects may be needed for optimal detection in other tissue types.
Statistical Analyses
We have used third-generation HCR for verifying enteric ganglia neuron subtypes from single-nucleus RNA-sequencing data. Since this does not require quantitative imaging, only verification of the presence of a transcript or transcripts, we used binary data for each fluorescent probe. We recommend visiting the Molecular Instruments website for more information on quantitative imaging (https://www.molecularinstruments.com/qhcr-dhcr-comparison).
Understanding Results
Fluorescent signal for HCR appears as small dots in a granular pattern over the cell body or localized to a specific cellular region that depends on mRNA localization (Figure 6).
Figure 6: Detection of mRNA in single cells of adult and fetal tissues.

(A) V3HCR applied to detect marker genes for discrete types of enteric neurons localizes to individual neurons within sections of myenteric ganglia in mouse (left) and human (right) adult intestinal tissue. Signal for Snap25 (pan-neuronal marker), Dlx3, (enteric neuron subtype marker), and Otof (enteric neuron subtype marker) are localized over cell bodies of mouse enteric neurons. Signal for human probes SNAP25, KLHL1, and NMU are similarly restricted over neuronal cell bodies in human tissue section of myenteric ganglia. (B) V3HCR applied to sections of fetal mouse lower urinary tract localizes Pax3 mRNA within developing neurons that co-express Elavl4. Abbreviations: bl, bladder lumen; bw, bladder wall; pg, pelvic ganglion; u, urothelium; and uro, urothelium.
Time Considerations
The protocol takes minimally three days to perform, so planning for this period is recommended. Incubation periods are also utilized as time to perform additional steps (steps 58–63), so be sure to have equipment ready for these steps before that part of the protocol is performed.
Significance Statement.
Third generation in-situ Hybridization Chain Reaction (V3HCR) is a technique that enables visualization of specific RNA transcripts within tissues. V3HCR is advantageous because it has much more refined localization of signal within single cells compared to prior in situ hybridization methods that rely on precipitation of colorimetric substrate where the signal can diffuse across tissue. We demonstrate the usefulness of this method for single cell localization in mouse and human tissues. We also demonstrate elimination of background fluorescent puncta that naturally occur in the human neurons through use of an optimized lipofuscin-blocking process.
Acknowledgement
We are grateful to Dr. Karen Deal for assistance with imaging V3HCR samples. Optimization of this procedure was funded by National Institutes of Health awards OT2 OD023850 and U01 DK110804 to EMS2. AAM was supported by T32-DK007673. JTB was supported by T32 HD007502. The authors state that they have no conflicts of interest to declare.
Literature Cited
- Brehmer A, Blaser B, Seitz G, Schrodl F, & Neuhuber W (2004). Pattern of lipofuscin pigmentation in nitrergic and non-nitrergic, neurofilament immunoreactive myenteric neuron types of human small intestine. Histochem Cell Biol, 121(1), 13–20. doi: 10.1007/s00418-003-0603-7 [DOI] [PubMed] [Google Scholar]
- Choi HM, Beck VA, & Pierce NA (2014). Next-generation in situ hybridization chain reaction: higher gain, lower cost, greater durability. ACS Nano, 8(5), 4284–4294. doi: 10.1021/nn405717p [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi HMT, Chang JY, Trinh LA, Padilla JE, Fraser SE, and Pierce NA (2010). Programmable in situ amplification for multiplexed imaging of mRNA expression. Nature Biotechnology Nov;28(11):1208–12. doi: 10.1038/nbt.1692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi HMT, Schwarzkopf M, Fornace ME, Acharya A, Artavanis G, Stegmaier J, … Pierce NA (2018). Third-generation in situ hybridization chain reaction: multiplexed, quantitative, sensitive, versatile, robust. Development, 145(12). doi: 10.1242/dev.165753 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi HMT, Schwarzkopf M, & Pierce NA (2020). Multiplexed Quantitative In Situ Hybridization with Subcellular or Single-Molecule Resolution Within Whole-Mount Vertebrate Embryos: qHCR and dHCR Imaging (v3.0). Methods Mol Biol, 2148, 159–178. doi: 10.1007/978-1-0716-0623-0_10 [DOI] [PubMed] [Google Scholar]
- Clement-Ziza M, Munnich A, Lyonnet S, Jaubert F, & Besmond C (2008). Stabilization of RNA during laser capture microdissection by performing experiments under argon atmosphere or using ethanol as a solvent in staining solutions. RNA, 14(12), 2698–2704. doi: 10.1261/rna.1261708 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dirks RM, & Pierce NA (2004). Triggered amplification by hybridization chain reaction. Proc Natl Acad Sci U S A, 101(43), 15275–15278. doi: 10.1073/pnas.0407024101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ibarra-Garcia-Padilla R, Howard A. G. A. t., Singleton EW, & Uribe RA (2021). A protocol for whole-mount immuno-coupled hybridization chain reaction (WICHCR) in zebrafish embryos and larvae. STAR Protoc, 2(3), 100709. doi: 10.1016/j.xpro.2021.100709 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katz ML, & Robison WG Jr. (2002). What is lipofuscin? Defining characteristics and differentiation from other autofluorescent lysosomal storage bodies. Arch Gerontol Geriatr, 34(3), 169–184. doi: 10.1016/s0167-4943(02)00005-5 [DOI] [PubMed] [Google Scholar]
- Lovely AM, Duerr TJ, Qingchao Q, Galvan S, Voss SR, & Monaghan JR (2022. In Press). Wnt signaling coordinates gene expression of limb patterning genes during axolotl forelimb development and regeneration. Front Cell Dev Biol. DOI: 10.3389/fcell.2022.814250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- May-Zhang AA, Deal KK, & Southard-Smith EM (2018). Optimization of Laser-Capture Microdissection for the Isolation of Enteric Ganglia from Fresh-Frozen Human Tissue. J Vis Exp(136). doi: 10.3791/57762 [DOI] [PMC free article] [PubMed] [Google Scholar]
- May-Zhang AA, Tycksen E, Southard-Smith AN, Deal KK, Benthal JT, Buehler DP, … Southard-Smith EM (2021). Combinatorial Transcriptional Profiling of Mouse and Human Enteric Neurons Identifies Shared and Disparate Subtypes In Situ. Gastroenterology, 160(3), 755–770 e726. doi: 10.1053/j.gastro.2020.09.032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schnell SA, Staines WA, & Wessendorf MW (1999). Reduction of lipofuscin-like autofluorescence in fluorescently labeled tissue. J Histochem Cytochem, 47(6), 719–730. doi: 10.1177/002215549904700601 [DOI] [PubMed] [Google Scholar]
- Wang F, Flanagan J, Su N, Wang LC, Bui S, Nielson A, … Luo Y (2012). RNAscope: a novel in situ RNA analysis platform for formalin-fixed, paraffin-embedded tissues. J Mol Diagn, 14(1), 22–29. doi: 10.1016/j.jmoldx.2011.08.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang W, Svensson Akusjarvi S, Sonnerborg A, & Neogi U (2018). Characterization of Inducible Transcription and Translation-Competent HIV-1 Using the RNAscope ISH Technology at a Single-Cell Resolution. Front Microbiol, 9, 2358. doi: 10.3389/fmicb.2018.02358 [DOI] [PMC free article] [PubMed] [Google Scholar]
Key Reference
- Choi HMT, Schwarzkopf M, Fornace ME, Acharya A, Artavanis G, Stegmaier J, Cunha A, and Pierce NA 2018. Third-generation in situ hybridization chain reaction: multiplexed, quantitative, sensitive, versatile, robust. Development. 145(12):dev165753. 10.1242/dev.156869. [DOI] [PMC free article] [PubMed] [Google Scholar]


