Abstract
The human lung plays vital roles in respiration, host defense and basic physiology. Recent technological advancements such as single-cell RNA sequencing and genetic lineage tracing have revealed novel cell types and enriched functional properties of existing cell types in lung. The time has come to take a new census. Initiated from members of the NHLBI-funded LungMAP Consortium and aided by experts in the lung biology community, we synthesized current data into a comprehensive and practical cellular census of the lung. Identities of cell types in the normal lung are captured in individual cell cards with delineation of function, markers, developmental lineages, heterogeneity, regenerative potential, disease links and key experimental tools. This publication will serve as the starting point of a live, up-to-date guide for lung research at https://lungmap.net/cell-cards-browser/. We hope that this Lung CellCards will promote the community-wide effort to establish, maintain and restore respiratory health.
eTOC:
Sun et al. takes a cellular census of the normal human lung by accounting for not only the recent explosion of single cell datasets, but also by annotating function and developmental origin. It is intended as a blueprint practical guide to initiate harmonization of cell nomenclature across lung biology studies.
INTRODUCTION
The human lung is vital for survival starting at first breath. Serving as the primary gas exchange organ, an average adult lung is composed of ∼480 million gas-exchange units called alveoli, comprising a surface area of ∼130m2 (Ochs, et al., 2004; Weibel, 2009). Inhaled air reaches alveoli through an elaborate branched network of conducting airways (Fig. 1). Once there, oxygen and CO2 exchange occurs across the alveolar epithelium-capillary interface, before air is exhaled through the same airways. Cells of the immune system are also critical components of the lung with innate immune cell types taking up residence early in development. Other immune cells traverse across the vasculature in response to infection or injury. The lung is also wired by nerves of both the afferent and efferent types, indicating that this large organ serves as both the signaling source and effector target of the nervous system.
Figure 1: Overall lung structure and regional niches.
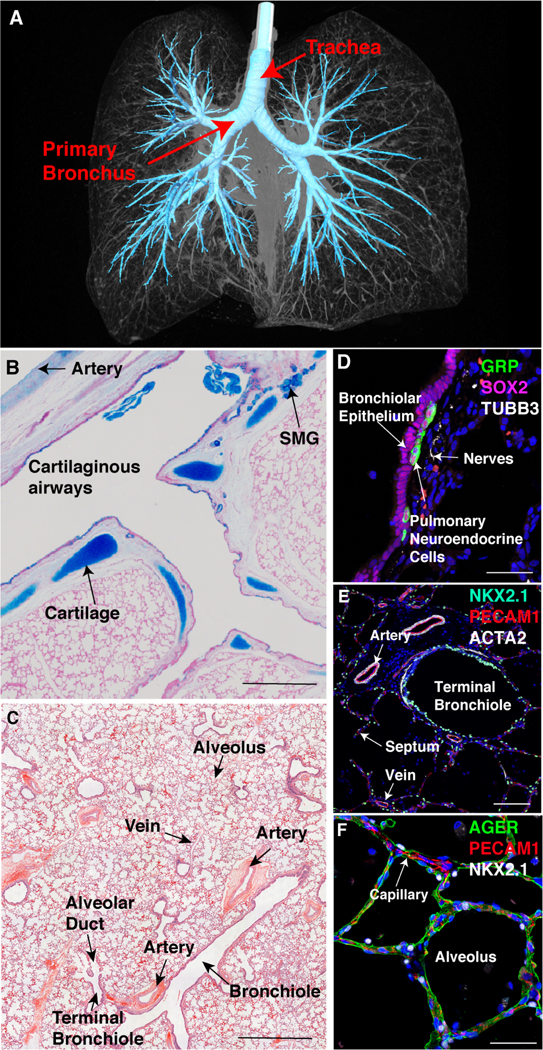
(A) Computer tomography of an intact human lung with the trachea and conducting airways highlighted. (B) A tile scan of a section of the normal human lung after alcian blue staining identifies proximal airway structures. SMG (submucosal gland). (C) A tile scan of a distal section of normal human lung after H& E staining identifies bronchial and alveolar structures. (D-F) Lung sections were stained with cell-selective markers and imaged by immunofluorescence confocal microscopy to identify diverse cells within airways and alveolar regions. Most conducting airway epithelial cells express SOX2 (D). Clusters of GRP+ pulmonary neuroendocrine cells were localized along the airways, innervated by TUBB3+ nerves (D). NKX2.1+ identifies epithelial cells in airways and alveoli (E, F). Smooth muscle and myofibroblasts express ACTA2 in bronchioles, pulmonary arteries, pulmonary veins, and alveolar septa (E,F). AGER+ alveolar type I cells (F) line the lumen of the alveoli. Scale bars are 1000 mm (B, C), 40 mm in (D, F) and 100 mm in (E).
Lung disease is a leading cause of morbidity and mortality in the world. Its impact on human health is newly demonstrated by the devastation of the COVID-19 pandemic. Only in the coming years will we learn how this pandemic has injured the lungs of so many, and what the long term consequences are. In 2008, a key paper was published following an NHLBI workshop on the cellular composition of the lung (Franks, et al., 2008). In the years since, multiple groundbreaking technological advancements have occurred such as single-cell RNA sequencing (scRNA-seq), in vivo genetic lineage tracing, and newer methods of cell fate tracking generated through CRISPR/Cas9-based genome editing. These technologies have led to discoveries of not only new cell types, but also a more precise definition of cellular properties in lung. Several of these cell types are unique to the lung (e.g. alveolar type 2 or AT2 cells). Others, while having counterparts in other tissues (e.g. capillary endothelial cells), are customized to accommodate unique demands in the respiratory system. In particular, the impressive accumulation of scRNA-seq datasets has led to the putative identification of new cell types or states in the mouse and human lung. Some of these efforts have been directed towards surveying the normal lung (Raredon, et al., 2019; Reyfman, et al., 2019; Deprez, et al., 2020; Travaglini, et al., 2020), while others have interrogated abnormal lungs from devastating diseases such as idiopathic pulmonary fibrosis and cystic fibrosis (Reyfman, et al., 2019; Adams, et al., 2020; Habermann, et al., 2020; Carraro, et al., 2021). While the datasets reported in these studies are critical to our understanding of normal lung development and the response to injury and disease, there remains a significant need for conformational studies to better support and understand these findings. One of the core directives of the LungMAP Consortium is to utilize single cell techniques to map and characterize human lung cell lineages across the lifespan. However, as a group, we understand the limitations of evolving single cell technologies and have chosen to focus on cell types which have been validated with further experimentation in this CellCards document (Fig. 2). These confirmatory approaches include lineage tracing, organoid assays, and extensive subsequent gene and protein expression studies. As further studies are performed and reported, we hope and expect that additional validated cell types will be added to the present collection. Until such rigorous interrogation is performed, however, we have defined some of the novel cell populations that have emerged from single cell transcriptome studies under the entry of “heterogeneity and cellular states”.
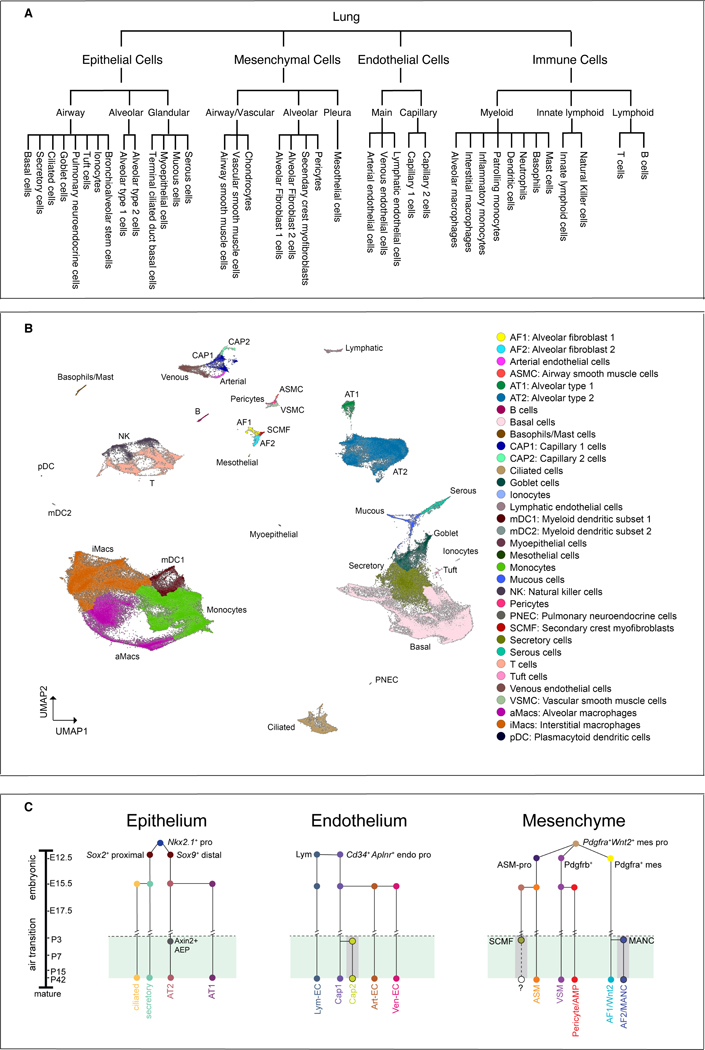
Figure 2: Lung cell types by region, by single cell clustering and by lineage.
(A) A schematic of cell types featured in CellCards, delineated by general regions of the lung where they reside. (B) UMAP visualization of human lung single cells (n=259,565) colored by their predicted cell identities. Data were from collected 5 published scRNA-seq cohorts (Reyfman, et al., 2019; Adams, et al., 2020; Deprez, et al., 2020; Goldfarbmuren, et al., 2020; Habermann, et al., 2020). Data integration and analysis were performed using Monocle 3 (Cao, et al., 2019). (C) Cell circuitry dendrogram showing developmental cell lineage relationships described in the various resident cell types of the mouse lung. This does not include all identified cell types or states in the various scRNA-seq analysis, rather those that have been confirmed across mouse development using complementary techniques including lineage fate mapping techniques and high resolution imaging across developmental time points. Adapted from (Zepp, et al., 2021), with additional input from (Rawlins, et al., 2009a; Zepp, et al., 2017; Zacharias, et al., 2018; Frank, et al., 2019; Gillich, et al., 2020).
While this document initiated from LungMAP investigators, the product is the result of a community-wide effort with extensive inputs from numerous leaders of lung biology. We have chosen to use a “cell cards” format, with the intent to provide comprehensive, streamlined, and easy-to-update information for practical assessment of lung cell types. Data are derived from both mouse and human lungs. We have utilized common names or terms to apply to well annotated cell types present in both the human and mouse lungs. In some instances, names or terms that have primarily been used for mouse cell types have been replaced with more generalizable names that can apply to multiple species, i.e. using the name secretory cell to encompass “club” cells, which is a term more specific to the mouse than human lung. We cover topics including cellular function, markers for identification, developmental origin, regenerative potential, and links to disease (Fig.3). In a section following the individual cards, we summarize emerging data from advanced proteomic and lipidomic approaches. While these streamlined cards are not intended to provide an in-depth assessment of lung cell types, in each section, we suggest key references for landmark primary studies and comprehensive reviews. For additional syntheses of existing cell type characteristics, we refer readers to several excellent reviews (Hogan, et al., 2014; Tata and Rajagopal, 2017; Basil, et al., 2020; Riccetti, et al., 2020; Ushakumary, et al., 2021).
Figure 3: Histology of normal and diseased human lung.
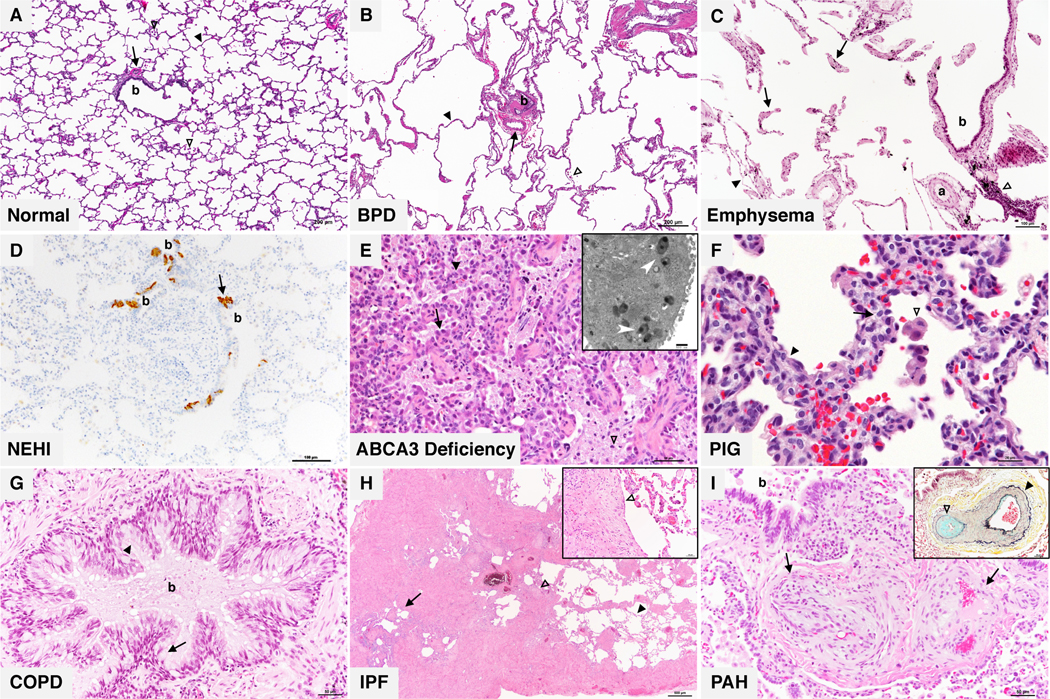
(A) Normal infant lung: Bronchiole (b) and accompanying artery (arrow) with surrounding alveoli (black arrowhead). Scattered alveolar macrophages (white arrowheads) are present within the alveolar spaces. (B) Bronchopulmonary dysplasia (BPD): large simplified alveoli separated by thin alveolar septa (black arrowhead) in the absence of significant airway injury. Enlargement of the alveoli can be appreciated when comparing to the size of the bronchiole (b) and accompanying artery (arrow). Accumulation of macrophages (white arrowhead) are frequently present. (C) Emphysema: isolated or “free floating” segments of viable alveolar septal tissue (arrows) are the characteristic histologic finding in emphysema. Lymphocytic inflammation with admixed macrophages containing anthracotic black pigment (white arrowhead) present adjacent to the bronchiole (b) and its accompanying artery (a) is a frequent finding in emphysema. (D) Neuroendocrine cell hyperplasia of infancy (NEHI): increased bombesin immunoreactive pulmonary neuroendocrine cells (PNEC) (arrow) within bronchioles (b) are the key diagnostic histologic feature of NEHI. NEHI is diagnosed in lung biopsies free of pathologic findings indicative of other disorders such as architectural disruption, diffuse or advanced airway injury, inflammation, and significant vascular changes. (E) Surfactant deficiency associated with ABCA3 mutation: diffuse AT2 cell hyperplasia (arrow) lining thickened alveolar septa (black arrowhead) and pulmonary alveolar proteinosis features including abundant granular eosinophilic material with admixed foamy macrophages (white arrowhead). Electron microscopic analysis (inset) reveals the characteristic electron dense inclusions within abnormal lamellar bodies described as resembling “fried eggs” (white arrows). (F) Pulmonary interstitial glycogenosis (PIG): alveolar septal expansion (black arrowhead) by glycogen laden mesenchymal cells with vacuolated cytoplasm and indistinct cell borders (arrow). Alveolar macrophages (white arrowhead) are also present. (G) Chronic obstructive pulmonary disease (COPD): large conducting bronchus (b) with increased goblet cells (arrow) and decreased ciliated cells (arrowhead) lining a mucus filled lumen. Chronic bronchitis and emphysema are the two cardinal features of smoking related COPD. (H) Idiopathic pulmonary fibrosis (IPF): nonuniform collagenous interstitial fibrosis associated with cystically dilated airways (arrow), fibroblastic foci (white arrowheads) alternating with areas of preserved alveoli with thin septa (black arrowhead). (I) Pulmonary arterial hypertension (PAH): muscular artery (arrows) with marked intimal cellular proliferations resulting in nearly total luminal occlusion and formation of multiple small vascular spaces. Another muscular artery in the same lung (inset) with prominent medial vascular smooth muscle hyperplasia (black arrowhead) and intimal cellular proliferation and fibrosis (white arrowhead) highlighted by Movat pentachrome stain.
This rigorously curated document is intended as the prototype of a frequently updated live version of Lung CellCards on the NHLBI-supported LungMAP.net website. We hope that this resource will promote lung research from both within the lung community and beyond.
As a vetted resource, the CellCards were created with the following considerations to limit repetition and enhance readability:
A Table was generated for transcript and protein markers, as well as mouse genetic tools for lung structural cell types (Table 1).
A figure was generated for immune cell type surface protein combinations for FACS and another for transcript markers (Fig. 6).
Markers for each cell type were selected based on existing markers in the literature as well as recent data from single-cell RNAseq experiments. Unless specified, entries (markers, cellular properties, etc.) apply to both mouse and human lungs. Human-specific markers are delineated with the superscript “H”, and mouse-specific markers are delineated with the superscript “M”. Under “Developmental origin” and “Regenerative potential”, unless otherwise stated, properties are based on findings from mouse studies.
To avoid repetition, under experimental validation: “standard approaches” refers to the following: immunofluorescence (IF) and immunohistochemical (IHC) staining; RNA in situ hybridization or RNAscope; bulk RNAseq of sorted cells, scRNA-seq.
Given space limitation, we restricted references to landmark primary studies and comprehensive reviews, in which additional important studies can be found.
Table 1.
Markers and cre lines:
| Cell lineage | Cell type | Marker Genes# | Surface Protein Genes# | Antibodies | Cre lines* |
|---|---|---|---|---|---|
| Epithelium | Basal cell | TP63, KRT5 | NGFRH ,TRPC6H | TRP63, KRT5 | Trp63creERT2(Lee, et al., 2014) Krt5creERT2(Van Keymeulen, et al., 2011) |
| Epithelium | Secretory cell |
SCGB1A1, SCGB3A2 |
SLC4A7H, SCUBE2H |
SCGB1A1 | Scgb1a1creERT2(Rawlins, et al., 2009b) |
| Epithelium | Ciliated cell | FOXJ1, RSPH1 | CDHR3, CDHR4 | FOXJ1, Acetylated- Tubulin | Foxj1creERT2(Rawlins and Hogan, 2008) |
| Epithelium | Goblet cell | MUC5AC, SPDEF | PCDH7H , SLC4A 11H | MUC5AC, AGR2 | |
| Epithelium | Pulmonary neuroendocrine cell | ASCL1, GRPH , Calca M |
NRXN1H, CDH18H |
GRPH/Bombesin, CGRPM |
Ascl1creERT2(Km, et al., 2011) Ca/cacreERT2(Song, et al., 2012) |
| Epithelium | Tuft cell | POU2F3, ASCL2, Dclk1 M | TRPM5 M | POU2F3, DCLKM | Dc/k1creERT2(Westphalen, et al., 2014) Pou2f3creERT2(McGinty, et al., 2020) |
| Epithelium | lonocyte | FOXI1, ASCL3, Cftr M | CFTRM | Asc/3EGFP-Cre(Bullard, et al., 2008) | |
| Epithelium | Bronchoalveolar stem cell | Co-express low level SFTPC and SCGB1A1 | Dual recombinases or split cre effector (Liu, et al., 2019; Salwig, et al., 2019) | ||
| Epithelium | Alveolar type 1 cell | AGERH, RTKN2, Hopx M | AGER, SEMA3B HTI-56H (MAB) | AGER, HOPX | HopxcreERT2(Jain, et al., 2015) AgercreERT2(Chung and Hogan, 2018) Aqp5cre(Flodby, et al., 2010) |
| Epithelium | Alveolar type 2 cell | SFTPC, LAMP3 | KCNJ15 HTII-280H (MAB) | SFTPC, ABCA3 | SftpccreERT2(Chapman, et al., 2011 ; Rock, et al., 2011) |
| Epithelium | Ductal basal cell | VIM, SOX9 | TRP63, KRT5 | Trp63creERT2(Lee, et al., 2014) Krt5creERT2(Van Keymeulen, et al., 2011) | |
| Epithelium | Myoepithelial cell | KRT14, MYH11 | ACTA2/SMA |
Acta2creERT2 (Anderson, et al., 2017; Lynch, et al., 2018; Tata, et al., 2018) Myh11-creERT2 (Anderson, et al., 2017). (Both are also active in smooth muscles and myofibroblasts.) |
|
| Epithelium | Mucous cell | MUC5B, SPDEF | MUC5B | ||
| Epithelium | Serous cell | LYZ, LTF | |||
| Mesenchyme | Airway smooth muscle cell | ACTA2, DES, LGR6 | LGR6 | ACTA2/SMA | Lgr6creERT2(Snippert, et al., 2010) Acta2creERT2(Moiseenko, et al., 2017; Zepp, et al., 2021) |
| Mesenchyme | Vascular smooth muscle cell | NTRK3, ITGA7, Cnn1 M | ITGA7, NTRK3 | ACTA2/SMA | Acta2creERT2(Moiseenko, et al., 2017; Zepp, et al., 2021) |
| Mesenchyme | Chondrocytes | COL2A1, HAPLN1 | SOX9 | Co/2a1creERT2(Zhu, et al., 2008) | |
| Mesenchyme | Alveolar fibroblast 1 | TCF21, WNT2 | PCDH15H | PLIN2/ADRP | Tcf21mercremer (Park, et al., 2019) Wt2creERT2(Zepp, et al., 2021) |
| Mesenchyme | Alveolar Fibroblast 2 | MFAP5, SCARA5 | CDON | ||
| Mesenchyme | Secondary crest myofibroblast cell | DACH2H , Fgf18 M | ITGBL1H | ACTA2/SMA | Fgf18creERT2(Hagan, et al., 2019a) PdgfrartTA(Li, et al., 2018) PdgfracreERT2 (Chung, et al., 2018) Acta2creERT2(Moiseenko, et al., 2017; Zepp, et al., 2021) |
| Mesenchyme | Pericyte | TRPC6. LAMC3 | TRPC6 | CSPG4/NG2 PDGFRb |
PdgfrbcreERT2 (Cuervo, et al., 2017) |
| Mesenchyme | Mesothelium | WT1, UPK3B, FREM2 | WT1 | Wt1creERT2(Zhou, et al., 2008) | |
| Endothelium | Arterial endothelial cell | DKK2, GJA5 | EFNB2 | VWF (also vein) | BmxcreERT2 (not active in small arterioles) (Ehling, et al., 2013) Sox17creERT2 (also labels capillaries) (Liao, et al., 2009) |
| Endothelium | Venous endothelial cell | ACKR1H, HDAC9H Slc6a2M | ACKR1 | VWF (also artery) Endomucin (also capillaries, but not artery) | |
| Endothelium | Lymphatic endothelial cell | PROX1, MMRN1 | LYVE1, NRP2 | LYVE1 | Prox1creERT2 (Bazigou et al., 2011) |
| Endothelium | Capillary cell 1 | IL7RH , Aplnr M , Gpihbp1 M | IL7RH , Aplnr M | AplnicreERT2 (Gillich, et al., 2020) | |
| Endothelium | Capillary cell 2 | EDNRB, HPGDH , Apln M , Car4 M | EDNRB | EDNRB, CA4M | AplncreERT2 (Gillich, et al., 2020) |
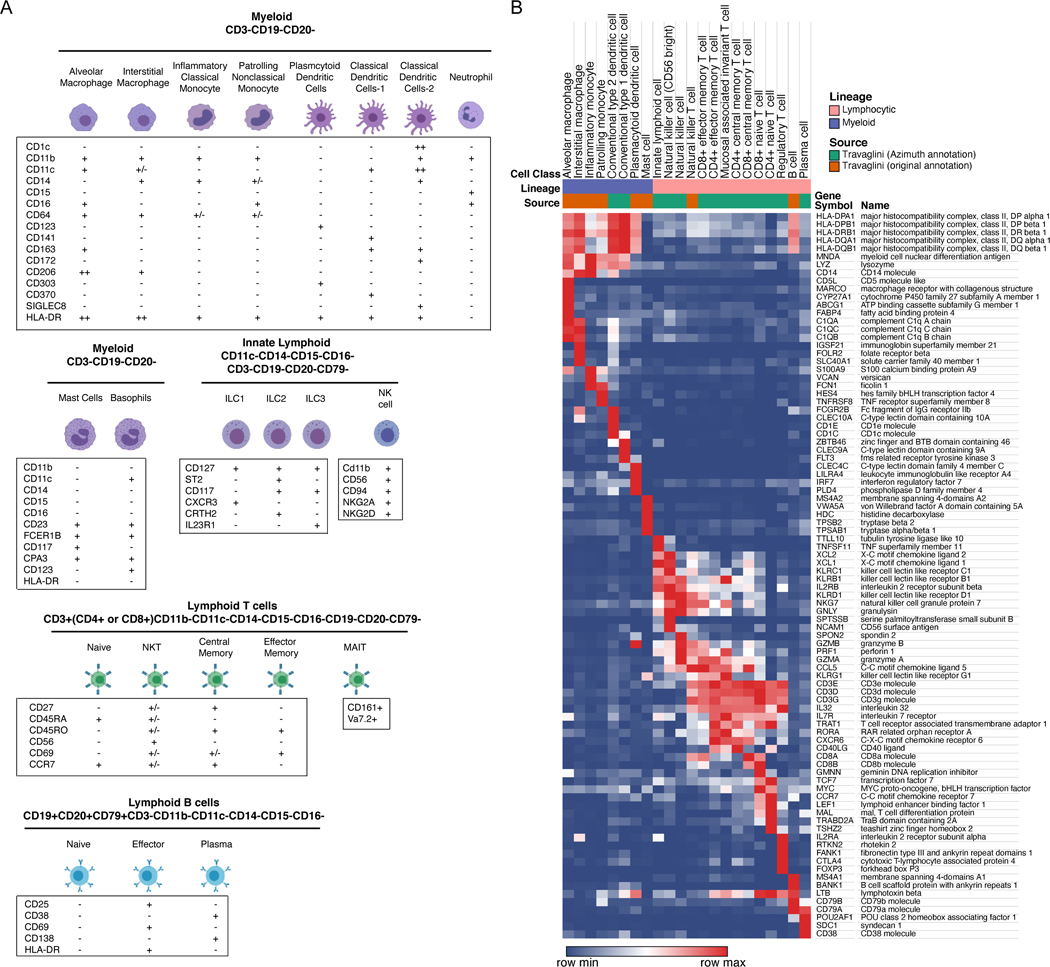
Figure 6. Immune cells of the human lung.
(A) Depiction of lung immune cell types featured in CellCards. They can be delineated based on surface protein expression, with all immune cells expressing CD45. Macrophages are CD11b+; dendritic cells are CD11c+; T cells are CD3+ and B cells are CD19+. Specialized T and B cell states can be identified using additional markers. (B) Heatmap shows expression levels of representative marker genes of lung immune cells using single cell RNAseq data (Travaglini, et al., 2020). Both original annotations and Azimuth-projected annotations were used for cell designations (Hao, et al., 2020). IGSF21+ dendritic cells and basophil/mast cells from the original publication were renamed as interstitial macrophages and mast cells based on the transcriptional profile. Cell ontology labels were used to better illustrate identities of cells. Average normalized values (log2(CPM+1)) for each cell type were used in the heatmap.
CELLCARDS BY LINEAGE
EPITHELIUM
The epithelium of the lungs and trachea are the externally facing cells of the respiratory system and as such have a wide variety of functions including gas exchange, clearance of foreign matters and pathogens, immunosurveillance, and transmission of external environmental cues to other cell types in the respiratory system (Fig. 4). The entirety of the respiratory epithelium is derived from the transcription factor NKX2–1-expressing endoderm specified in the early anterior ventral foregut (Kimura, et al., 1996; Minoo, et al., 1999). Early developmental events lead to the separation of the trachea and lung endoderm progenitors from the esophageal progenitors and rapid extension and branching of the primitive respiratory tree. Concurrent with these early events, the respiratory endoderm diverges along a proximal-distal axis to generate distinct progenitors essential for generating either airway or alveolar epithelium. After proximal-distal patterning, differentiation of the various mature epithelial lineages begins. These differentiation events have been characterized using histological analyses with multiple cell type-specific markers, cell lineage mapping techniques using indelible recombinase marking of both early and late endoderm/epithelial cell types, and most recently single-cell genomic analysis. While many of the distinct epithelial cell lineages in the mature lung have been identified, there remains much that is unknown about their cell fate relationships with each other during normal adult homeostasis and after injury. Questions regarding cellular plasticity versus stem/progenitor cell relationships have been reviewed previously (Hogan, et al., 2014; Tata and Rajagopal, 2017; Basil, et al., 2020). The cell lineages described below have been validated using multiple techniques including cell type-specific lineage tracing, loss- and gain-of-function genetic models, ex vivo models of cellular function including organoids, and single-cell genomic assessments (Fig. 2). We describe lung epithelial cells in the sequence of cell types in the airway, alveolar and glandular epithelium.
Figure 4: Epithelial cells in the human lung.
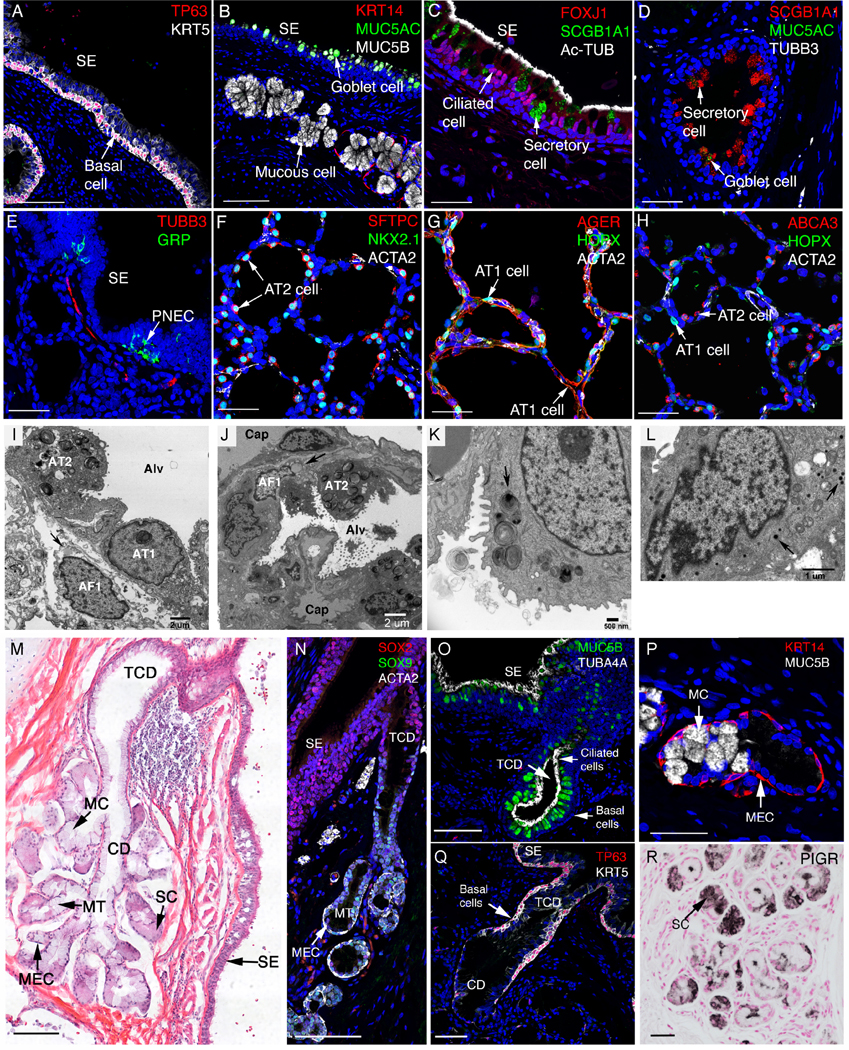
Lung sections were stained and imaged by immunofluorescence confocal microscopy (A) Trachea and larger conducting airways are lined by a pseudostratified epithelium comprised of TP63 and KRT5 stained basal cells. (B) Goblet cells expressing MUC5AC reside in surface epithelium (SE) while mucous cells expressing MUC5B reside in submucosal glands, adjacent to KRT14 stained myoepithelial cells. (C) SCGB1A1+ secretory and FOXJ1+, Ac-TUB+ ciliated cells intersperse along the airway. (D) A few SCGB1A1+ cells also express goblet marker MUC5AC. (E) Pulmonary neuroendocrine cells (PNEC) are found in clusters forming neuroendocrine bodies (NEB) that stain for GRP which are innervated by TUBB3+ nerves. (F-H) Alveolar regions are lined by AT 1 cells (AGER+, HOPX+) and AT2 cells (SFTPC+, NKX2.1+, and ABCA3+). AT 1 cells are closely opposed to capillary endothelial cells for efficient gas exchange. AT2 cells secrete pulmonary surfactant lipids and protein into the alveolus. ACTA2 stains alveolar myofibroblasts (F-H). (I) Alveolar septa with AT1 and AT2, capillaries and an alveolar fibroblast 1 cell (lipofibroblast) containing a lipid droplet (arrow). Alv (alveolar lumen), TEM X6900. (J) Alveolar septa with AT2, alveolar fibroblast 1 (lipofibroblast), capillaries (cap) lined with endothelial cells (en). Alv (alveolar lumen), TEM X2900. (K) Normal lamellar bodies in a AT2 cell, including ones with projection cores (arrow). TEM X12400. (L) A bronchiolar neuroendocrine cell containing multiple dense-core granules (arrows). TEM X19500. (M) H & E staining of a bronchial submucosal gland. Glands open to the airway lumen or surface epithelium (SE). TCD: terminal ciliated ducts; MC: mucous cell; CD: collecting ducts; MT: mucous tubules; SC: MEC: myoepithelial cells. (N) SMG cells in the ducts and most epithelial cells lining conducting airways express SOX2. SOX9 is selectively expressed in SMG epithelial cells. (O) Pseudostratified, terminal ciliated ducts (TCD) are lined by ciliated cells (TUB4A4+) and goblet cells (MUC5B+). (P) Collecting ducts are lined by mucous cells (expressing MUC5B but not MUC5AC). (Q) Pseudostratified, terminal ciliated ducts (TCD) are lined by basal cells (TP63+KRT5+). (R) Peripheral regions of the SMG are lined by serous cells (sc) expressing PIGR. Acini and ducts of the SMG are surrounded by myoepithelial cells (MEC) expressing ACTA2+ and KRT14+ (M, N, P). Scale bars: A, B (100 mm), C-H (40 mm), M, N and Q (100 mm), O and P (40 mm), R (50 mm).
AIRWAY EPITHELIUM
Airway epithelial cells line the conducting airways of the trachea, bronchi and bronchioles. Some of these cell types are common while others are rare. Each is functionally specialized to moisturize the air, clear inhaled particles, serve as progenitors in repair, or sense aerosolized signals. Collectively, the airways conduct air to and from the gas exchange alveoli with the epithelium serving as a mucosal barrier to pathogens.
Basal cells
Morphological features: epithelial cells that underline luminal cells.
Function: serve as progenitors for other airway epithelial cells in homeostasis and repair.
Other names: none.
Markers: genes and proteins: TP63, KRT5 (Table 1).
Location: reside in the basal layer of airway epithelium. In mouse, basal cells are common in the trachea and extrapulmonary main stem bronchi that are lined with cartilage. They are rare in intrapulmonary airways that are without cartilage (Yang, et al., 2018). In human, aside from trachea and extrapulmonary bronchi, basal cells are also found all along the intrapulmonary airways, with diminishing number towards respiratory bronchioles (Fig. 4A).
Experimental validation: standard approaches; air-liquid interface culture, organoids (Rock, et al., 2009; Mou, et al., 2016); mouse-lineage tracing using Trp63creERT2, Krt5creERT2; iPSC-derived human basal cells (Hawkins, et al., 2020).
Developmental origin: in mouse, differentiated basal cells were first detected in the basal layer of airway epithelium based on their expression of Krt5 at E15.5. During development, they arise from naïve airway epithelial progenitors (Yang, et al., 2018). During adult homeostasis and injury repair, they are replenished primarily by surviving basal cells (Montoro, et al., 2018). In mice, when all basal cells are genetically depleted, secretory (club) cells can de-differentiate into basal cells (Pardo-Saganta, et al., 2015). In regions with submucosal glands, lost surface epithelium basal cells can be replaced by myoepithelial cells (Anderson, et al., 2017; Lynch, et al., 2018; Tata, et al., 2018).
Heterogeneity and cellular states: Krt13-expressing hillock cells in the mouse trachea (Montoro, et al., 2018); suprabasal, squamous cells and additional subsets from scRNAseq of the human airway (Deprez, et al., 2020; Carraro, et al., 2021).
Regenerative potential: basal cells are the primary airway progenitor cells. They can generate secretory and ciliated cells during homeostasis and following airway epithelium injury such as naphthalene injury (Hong, et al., 2004; Montoro, et al., 2018). In mouse following severe H1N1 influenza infection, rare Trp63+ basal progenitor cells, or lineage negative epithelial progenitors (LNEPs), can give rise to alveolar Krt5+ pods which act as an emergency response to re-establish barrier in the lung (Xi, et al., 2017; Yang, et al., 2018). In mouse following bleomycin injury, Trp63+ basal cells contributed less efficiently to alveolar Krt5+ pods than Scgb1a1creERT2 lineaged cells (Cassandras, et al., 2020).
Link to disease: basal cell hyperplasia is a common feature of COPD. Basal cells can serve as precursors for NSCLC (Ferone, et al., 2020).
Key references: (Rock, et al., 2009; McCauley, et al., 2018; Yang, et al., 2018; Hawkins, et al., 2020)
Secretory cells
Morphological features: columnar or cuboidal luminal epithelial cells that contain secretory granules. In mouse airways, they exhibit dome-shaped apical surface.
Function: keep the airway moist through production of secretoglobins into airway lumen.
Other names: Clara cells, club cells.
Markers: genes: SCGB1A1, SCGB3A2, protein: SCGB1A1 (Table 1).
Location: luminal layer of airway epithelium (Fig. 4D).
Experimental validation: standard approaches; air-liquid interface culture; organoids; mouse lineage tracing using Scgb1a1creERT2 which is active in most secretory cells, and also labels a subset of AT2 cells (Rawlins, et al., 2009b; Ray, et al., 2016).
Developmental origin: in mouse, during development, secretory cell progenitors were first detected at E14.5 based on Scgb3a2 RNA expression (Guha, et al., 2012; Kiyokawa, et al., 2021). This is followed by the onset of Scgb1a1 expression at E16.5. During adult homeostasis and injury repair, they arise from basal cells or other secretory cells (Pardo-Saganta, et al., 2015; Montoro, et al., 2018).
Heterogeneity and cellular states: in mouse, a subset of variant secretory or club cells express reduced level of Cyp2f2, encoding Cytochrome P450, rendering them resistant to naphthalene-induced cell death (Hong, et al., 2001; Guha, et al., 2012). These surviving cells serve as a primary source of reparative cells. In mouse following bleomycin injury, H2-K1hi secretory-like cells have enhanced progenitor property and give rise to alveolar cells (Kathiriya, et al., 2020). Human and ferret lungs contain a SCGB3A2+ respiratory airway secretory cell (RASC) lineage unique to large mammal respiratory bronchioles, which are capable of regenerating AT2 cells (Basil et. al.-under revision).
Regenerative potential: secretory cells can self-renew and differentiate into ciliated cells during mouse airway development, homeostasis and airway epithelium injury repair (Rock, et al., 2009). Following severe lung injury such as bleomycin-induced damage, both Sox2creERT2-lineaged cells as well as Scgb1a1creERT2-lineaged cells can give rise to a small percentage of AT 1 and AT2 cells, as well as Krt5+ pods in the alveolar region (Yuan, et al., 2019; Cassandras, et al., 2020). As the common cell type that is lineage traced by both of these cre lines is secretory cells, these results suggest that secretory cells can give rise to a minority of alveolar epithelial cells following bleomycin-induced injury. The absolute number and precentage of alveolar epithelial cell generated from secretory cells after injury varies, depending on the severity and injury type including influenza and bleomycin based injuries (Ray, et al., 2016; Yuan, et al., 2019; Cassandras, et al., 2020; Kathiriya, et al., 2020). Use of the Scgb1a1creERT2 mouse line to lineage trace the contribution of secretory cells to the alveolar epithelium is confounded by the finding that this cre line marks a subset of AT2 cells at homeostasis in the uninjured lung (Rawlins, et al., 2009b; Ray, et al., 2016). The RASC lineage is capable of self-renewal and regenerating AT2 cells in the human lung (Basil et. al.-under revision).
Link to disease: can take on goblet cell characteristics and produce mucin in diseases such as asthma and chronic obstructive pulmonary disease (COPD).
Key references: (Giangreco, et al., 2009; Rock, et al., 2009; Pardo-Saganta, et al., 2015)
Ciliated cells
Morphological features: display multiple motile cilia on the apical surface.
Function: clear inhaled particles trapped by airway mucosal fluid.
Other names: multi-ciliated cells.
Markers: genes: FOXJ1, RSPH1; protein: FOXJ1, acetylated Tubulin (Table 1).
Location: luminal layer of airway epithelium (Fig. 4C).
Experimental validation: standard approaches; air-liquid interface culture; organoids; imaging beating of cilia, mouse lineage tracing using Foxj1creERT2 (Rawlins and Hogan, 2008).
Developmental origin: In mouse, ciliated cells were first detected in the airway epithelium based on their expression of FOXJ1 at embryonic day (E)15.5. During development, they arise from naïve airway epithelial progenitors (Rawlins and Hogan, 2008). During adult homeostasis and injury repair, they arise from either basal cells or secretory cells (Pardo-Saganta, et al., 2015; Montoro, et al., 2018).
Heterogeneity and cellular states: a subset of ciliated cells express Miwi2 (Wasserman, et al., 2017). Single cell RNAseq of the human airway revealed early, mature and immunoregulatory subsets (Carraro, et al., 2021), as well as a transitional cell cluster termed deuterosomal cells between secretory and multi-ciliated cells (Deprez, et al., 2020).
Regenerative potential: no regenerative potential reported. Ciliated cells are terminally differentiated.
Link to disease: signature cell type affected in primary ciliary dyskinesia. Disruption of ciliated cell function is also observed in many complex lung diseases, e.g., COPD, cystic fibrosis, and asthma.
Key references: (You, et al., 2004; Rawlins and Hogan, 2008; Carraro, et al., 2021)
Goblet cells
Morphological features: large cells that contain mucus granules.
Function: in a normal lung, goblet cells secrete mucus to the luminal surface to trap inhaled particles. In diseases such as asthma and COPD, over production/secretion of mucus obstructs the airway, leading to reduced air conductance and air trapping.
Other names: mucus cells.
Markers: genes: MUC5AC, SPDEF; proteins: MUC5AC, AGR2 (Table 1).
Location: luminal layer of airway epithelium. They are rare or absent in normal mouse airways but present in normal human airways (Fig. 4B). Are induced to form after allergen exposure or injury of the mouse airways.
Experimental validation: standard approaches; PAS staining, air-liquid interface culture, organoids.
Developmental origin: during development, they likely arise from naïve airway epithelial progenitors, although this has not been directly studied as they are rare in mouse airways. During pathogenesis such as in asthma models, they arise primarily from secretory cells that acquire goblet cell characteristics.
Heterogeneity and cellular states: by scRNAseq, mouse tracheal goblet cells are separated into three subsets: immature goblet, goblet 1 expressing mucosal genes Tff1, Tff2, Muc5ac and Muc5b, and goblet 2 expressing lectin-like secreted protein genes Dcpp1 and Dcpp2 (Montoro, et al., 2018). In human, MUC5AC is produced by goblet cells in surface epithelium while MUC5B is produced by goblet cells in both surface epithelium and submucosal glands (Okuda, et al.,2019).
Regenerative potential: unknown.
Link to disease: goblet cell metaplasia is a key feature of many airway diseases including COPD, asthma and CF.
Key references: (Chen, et al., 2009; Chen, et al., 2014a; Ostedgaard, et al., 2017)
Pulmonary neuroendocrine cells (PNECs)
Morphological features: rare airway epithelial cells with dense core vesicles that contain neuropeptides and neurotransmitters. Present either as solitary cells or in clusters in neuroepithelial bodies (NEBs). Solitary cells are spindle-like in morphology while clustered cells are wedge-like in morphology with wider apical than basal surface.
Function: act as airway sensor. PNECs are stimulated by signals such as allergen, nicotine and mechanical stretch, and respond by secreting neuropeptides and neurotransmitters.
Other names: none.
Markers: genes: ASCL1, GRPH(also termed Bombesin), CalcaM; proteins: GRPH, CGRPM (Table 1).
Location: reside in the tracheal and airway epithelium. In mice, NEBs are enriched at intrapulmonary airway branch points where inhaled particles congregate (Branchfield, et al., 2016b; Sui, et al., 2018). In human, clustered PNECs are less prevalent than in mice and their localization is less stereotypical (Fig. 4E,4L).
Experimental validation: standard approaches; air-liquid interface culture; organoids; mouse lineage tracing using Ascl1creERT2 (Kuo and Krasnow, 2015; Branchfield, et al., 2016b) and CalcacreERT2 (Song, et al., 2012).
Developmental origin: in mouse, specified PNECs were first detected by ASCL1 antibody staining at E12.5 (Kuo and Krasnow, 2015; Noguchi, et al., 2015). They likely arise from naïve airway epithelial progenitors. In mouse trachea during homeostasis or after hypoxia exposure, PNECs can originate from basal cells (Montoro, et al., 2018; Shivaraju, et al., 2021).
Heterogeneity and cellular states: heterogeneity in the expression of neuropeptides, Notch2 expression and proliferative potential (Ouadah, et al., 2019).
Regenerative potential: can give rise to club and ciliated cells following airway injury (Hong, et al., 2001; Song, et al., 2012).
Link to disease: PNECs are required for allergen induced asthmatic response (Kuo and Krasnow, 2015; Noguchi, et al., 2015; Branchfield, et al., 2016b; Sui, et al., 2018). Increase in PNEC number has been documented in a wide-spectrum of human lung diseases.
Key references: (Kuo and Krasnow, 2015; Noguchi, et al., 2015; Branchfield, et al., 2016b; Sui, et al., 2018)
Tuft cells
Morphological features: rare airway epithelial cells that are spindle in shape with microvilli on the apical surface.
Function: have chemosensory function. Tuft cells respond to signals by releasing cytokines such as IL25 as well as leukotrienes.
Other names: solitary chemosensory cells, brush cells.
Markers: genes: POU2F3, ASCL2, Dclk1M; proteins: POU2F3, DCLKM.
Location: in mouse during homeostasis, primarily found in the tracheal epithelium (Gerbe, et al., 2009; Saunders, et al., 2013; Bankova, et al., 2018). Following influenza infection, ectopic tuft cells arise in intrapulmonary airwarys and near alveolar Krt5+ pods (Saunders, et al., 2013; Montoro, et al., 2018; Rane, et al., 2019).
Experimental validation: standard approaches; mouse lineage tracing using Dclk1creERT2 (Westphalen, et al., 2014); lineage reporters Trpm5-GFP (Saunders, et al., 2013; Bankova, et al., 2018).
Developmental origin: the origin of tuft cells in prenatal development has not been determined. During postnatal development and homeostasis, lineage tracing and labeling of proliferative cells indicated that tuft cells can originate from basal cells (Saunders, et al., 2013; Bankova, et al., 2018; Montoro, et al., 2018).
Heterogeneity and cellular states: by scRNA-seq, mouse trachea tuft cells are separated into three subsets: immature tuft cells, tuft 1 (sensory) and tuft 2 (inflammatory) (Montoro, et al., 2018).
Regenerative potential: unknown. In intestine, tuft cells can promote proliferation of adjacent epithelial cells by producing paracrine signals (von Moltke, et al., 2016).
Link to disease: tracheal tuft cells play a role in allergen response (Bankova, et al., 2018).
Key references: (Saunders, et al., 2013; Bankova, et al., 2018; Montoro, et al., 2018; Rane, et al., 2019)
Ionocytes
Morphological features: rare columnar epithelial cells.
Function: maintains airway fluid balance. In mouse, ionocytes are the primary cells in the surface airway epithelium that express Cftr, the chloride channel gene mutated in cystic fibrosis (Montoro, et al., 2018; Plasschaert, et al., 2018).
Other names: none.
Markers: genes: FOXI1, ASCL3, CftM (Table 1).
Location: airway epithelium.
Experimental validation: standard approaches; air-liquid interface culture; mouse lineage tracing using Ascl3eGfp-cre (Bullard, et al., 2008); mouse reporter labeling using Foxi1-GFP (Montoro, et al., 2018).
Developmental origin: the origin of ionocytes in development has not been determined. During homeostasis, lineage tracing indicated that ionocytes can originate from basal cells (Montoro, et al., 2018).
Heterogeneity and cellular states: unknown.
Regenerative potential: unknown.
Link to disease: in the mouse trachea, ionocytes are the primary cell type that express the cystic fibrosis gene Cftr.
Key references: (Montoro, et al., 2018; Plasschaert, et al., 2018)
Bronchioalveolar stem cells (BASCs)
Morphological features: rare cuboidal lung epithelial cells that express both secretory and AT2 marker.
Function: progenitor cells that give rise to both airway and alveolar cells during repair (Kim, et al., 2005; Liu, et al., 2019; Salwig, et al., 2019).
Other names: none.
Markers: in mouse, co-express the AT2 marker Sftpc and secretory cell marker Scgb1a1 (Kim, et al., 2005; Liu, et al., 2019; Salwig, et al., 2019) (Table 1). Unclear whether this cell type, or a similar cell type, is present in human lungs.
Location: in mouse, reside at the bronchioalveolar junction (BADJ), a structure found in the mouse but not human lung.
Experimental validation: standard approaches; mouse dual recombinases or split cre effector tracing using Sftpc and Scgb1a1 drivers (Liu et al., 2019; Salwig et al., 2019).
Developmental origin: unknown.
Heterogeneity and cellular states: unknown.
Regenerative potential: can generate both alveolar and airway epithelial cells in lung repair in homeostasis and after injury (Kim, et al., 2005; Liu, et al., 2019; Salwig, et al., 2019).
Link to disease: BASCs were identified as a possible cell of origin for NSCLC (Kim, et al., 2005; Liu, et al., 2019; Salwig, et al., 2019).
Key references: (Kim, et al., 2005; Liu, et al., 2019; Salwig, et al., 2019)
ALVEOLAR EPITHELIUM
Alveolar epithelial cells line the alveoli. The large and squamous alveolar type 1 cells constitute ∼95% of the surface area. In comparison, alveolar type 2 cells secrete surfactant to reduce surface tension and promote alveoli expansion. Together, they perform the core function of gas exchange.
Alveolar type 1 (AT1) cells
Morphological features: large flattened epithelial cells.
Function: essential for gas exchange by forming large and thin gas-diffusible interface juxtaposing capillary endothelium. Also act as a potent signaling hub regulating postnatal alveologenesis (Zepp, et al., 2021).
Other names: alveolar epithelial cell 1 (AEC1).
Markers: genes: AGERH, HopxM, Rtkn2, proteins: AGER, HOPX (Table 1).
Location: alveoli (Fig. 4G,H,I; Fig. 5D).
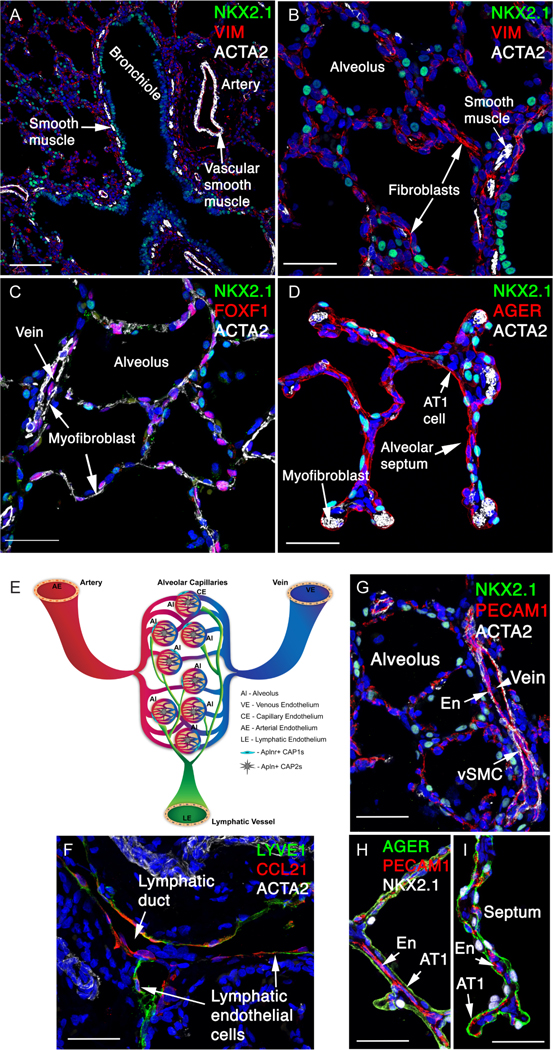
Figure 5: Mesenchymal and endothelial cells in the human lung.
(A,B) ACTA2+ airway smooth muscle cells line the NKX2–1+ bronchiled epithelium. ACTA2+ vascular smooth muscle cells line an artery. Vimentin (VIM+) stains a subset of fibroblasts. (C,D) ACTA2+ myofibroblasts in alveolar septae in close relationship to AT 1 cells (AGER+). FOXF1+ fibroblasts and endothelial cells are shown (C). (E) A schematic of the main types of pulmonary endothelial cells. Large vessels are lined by arterial endothelium (AE), venous endothelium (VE) and lymphatic endothelial (LE) cells. The alveolar microvasculature consists of two types of capillary endothelial cells: Aplnr+ CAP1 (gCAPs) and Apln+ CAP2 (aCAPs). (F) LYVE1+ and CCL21+ lymphatic endothelial cells line the lymphatic ducts. (G) PECAM1+ endothelial cells (En) are identified in vein and alveolar region, adjacent to vascular smooth muscle cells (vSMC). (H,I) PECAM1+ endothelial cells (En) in the alveolar region are shown adjacent to AGER+ AT1 epithelial cells in the alveoli, facilitating gas exchange. Scale bars: A (100 mm), B-D (40 mm), F-I (40 mm).
Experimental validation: standard approaches; organoids; mouse lineage tracing using HopxcreERT2 (Jain, et al., 2015), AgercreERT2 (Chung and Hogan, 2018) and Aqp5cre (Flodby, et al.,2010); fluorescence-activated cell sorting (FACS) using anti-HTI-56H antibody (Dobbs, et al., 1999).
Developmental origin: in mouse, by HopxcreERT2 lineage tracing and anti-HOPX antibody staining, specified AT 1 cells were first detected at E15.5, likely originating from multipotent Nkx2–1+ distal stalk cells (Frank, et al., 2019; Zepp, et al., 2021).
Heterogeneity and cellular states: in mouse, there are Igfbp2+ and Igfbp2-AT1 subtypes (Wang, et al., 2018).
Regenerative potential: while previously considered terminally differentiated, they exhibit extensive ability to reprogram into AT2 cells after hyperoxic injury (Penkala, et al., 2021), and to a lesser extent after pneumonectomy (Jain, et al., 2015). In postnatal homeostasis and following injury, lost AT1 cells are replaced by AT2 cells (Barkauskas, et al., 2013).
Link to disease: reduced in number and function in COPD and BPD.
Key references: (Jain, et al., 2015; Wang, et al., 2018; Frank, et al., 2019)
Alveolar type 2 (AT2) cells
Morphological features: cuboidal alveolar epithelial cells with lamellar bodies and specialized microvilli.
Function: produce surfactant, a protein and lipid mixture that reduces surface tension to allow lung expansion during inhalation, as well as promotes host defense by assisting the killing of pathogens.
Other names: alveolar epithelial cell 2 (AEC2).
Markers: genes: SFTPC, LAMP3; proteins: SFTPC, ABCA3 (Table 1).
Location: alveoli (Fig. 4F,I-K).
Experimental validation: standard approaches; organoids; mouse lineage tracing using SftpccreERT2 (Chapman, et al., 2011; Barkauskas, et al., 2013); FACS using anti-HTII-280H antibody (Dobbs, et al., 1999).
Developmental origin: in mouse, specified AT2s can be detected by Sftpc RNA in situ and lineage tracing at E14.5-E15.5, likely originating from Sox9/Id2+ distal tip cells (Frank, et al., 2019; Zepp, et al., 2021). In postnatal homeostasis and following injury, lost AT2 cells are primarily replaced by remaining AT2 cells.
Heterogeneity and cellular states: AT2s harbor a progenitor sub-lineage (WNT-responsive Axin2+ alveolar epithelial progenitors or AEPs) that are critical for alveolar regeneration after injury (Nabhan, et al., 2018; Zacharias, et al., 2018).
Regenerative potential: a key driver of alveolar epithelial regeneration. Can self-renew as well as differentiate into AT1, at a low frequency during homeostasis, and at extensive levels after alveolar injury.
Link to disease: surfactant deficiencies leads to respiratory distress in neonates. AT2 dysfunction and senescence is associated with pulmonary fibrosis (Nureki, et al., 2018; Katzen and Beers, 2020). AT2 cells are an important cell of origin for NSCLC adenocarcinoma (Desai, et al., 2014).
Key references: (Barkauskas, et al., 2013; Nabhan, et al., 2018; Zacharias, et al., 2018; Frank, et al., 2019)
GLANDULAR EPITHELIUM
Submucosal glands (SMG) are a unique microorgan found in the connective tissues lining the large proximal cartilaginous airways and are a major source of the protective mucus layer critical to normal innate defense (Widdicombe and Wine, 2015) (Fig. 4M-R). Morphologically, SMGs are a branched tubule-acinar structure similar to the salivary gland, consisting of a single duct that has undergone multiple dichotomous branching events and then terminates in bulbous acini. The resulting mature SMG can be divided into four structural domains along the proximal-to-distal axis: the terminal ciliated duct, collecting ducts, mucous tubules and the distal serous acini (Fig. 4M). The terminal ciliated duct links the SMG to the surface airway epithelium and is lined with cell types that parallel the surface airway counterparts including ciliated, goblet, secretory, basal cells and ionocytes (Fig. 4O). Ciliated cells are lost at the first bifurcation into the collecting ducts. Collecting ducts are lined by epithelial cells that transition from columnar to cuboidal to flattened morphology along the proximal-to-distal axis (Tos, 1966; Meyrick, et al., 1969; Meyrick and Reid, 1970; Matsuba, et al., 1972; Widdicombe, 2019). These cells are rich in mitochondria, a cytoplasm easily highlighted by eosin, and have centrally positioned nuclei. Mucous tubules are comprised of mucous cells while the most distal acini are comprised of serous cells. Surrounding the acini, mucous tubules, and collecting ducts are a thin layer of myoepithelial cells. The SMG is also known to be innervated and highly vascularized. In this section, we focus on specialized epithelial cells in the SMG that are distinct from those on the surface epithelium.
Terminal ciliated duct basal cells
Morphological features: single epithelial cell layer that may be pseudostratified.
Function: regenerate SMG and surface airway epithelial cells during normal homeostasis and following injury.
Other names: none.
Markers: aside from markers shared with surface airway basal cells, SMG terminal ciliated duct basal cells also express VIM, SOX9 (Hegab, et al., 2011; Hegab, et al., 2012b; Goldfarbmuren, et al., 2020) (Table 1).
Location: these basal cells are present along the basement membrane of the ciliated duct that opens into the airway (Fig. 4Q).
Experimental validation: standard approaches (Borthwick, et al., 2001; Hegab, et al., 2011; Hegab, et al., 2012b; Anderson, et al., 2017); mouse lineage tracing using Sox9preERT2/R26-LSL-tdTomato (Tata, et al., 2018); mouse reporter labeling using Krt5-eGFP strain (Schoch, et al., 2004); grafting experiments (Engelhardt, et al., 1995; Borthwick, et al., 2001; Hegab, et al., 2011).
Developmental origin: in humans, SMG ducts are first evident at ∼10 weeks of gestation. In mouse at postnatal day (P)0, placodes lined by naïve epithelial cells evaginate from the developing tracheal epithelium. These TRP63+/KRT5+/KRT14+/SOX9+/LEF1+ naïve epithelial cells give rise to basal cells and the other cell types within the SMG (Rawlins and Hogan, 2005; Lynch, et al., 2016). These buds elongate distally and become surrounded by myoepithelial like cells that are ACTA2+KRT5+KRT14+ (Anderson, et al., 2017). Following lung injury, SMG basal cells can arise from both remaining basal cells or myoepithelial cells of the SMG (Hegab, et al., 2011; Anderson, et al., 2017; Lynch, et al., 2018).
Heterogeneity and cell states: scRNA-seq analysis identified multiple subtypes of SMG basal cells (Goldfarbmuren, et al., 2020). Bulk sequencing of mouse surface airway basal versus SMG duct basal cells revealed distinct expression profiles (Hegab, et al., 2011).
Regenerative potential: serve as progenitors capable of repopulating cells within the SMG and the tracheal/bronchial epithelium (Engelhardt, et al., 1995; Borthwick, et al., 1999; Rawlins and Hogan, 2005; Hegab, et al., 2011; Anderson, et al., 2017; Lynch, et al., 2018; Tata, et al., 2018).
Link to Disease: This basal cell population is thought to contribute to the basal cell hyperplasia observed in smokers and gland hyperplasia observed in conditions like cystic fibrosis, asthma, and chronic bronchitis (Widdicombe and Wine, 2015). Depletion of this population may contribute to conditions including obliterative bronchitis (Swatek, et al., 2018).
Key references: (Hegab, et al., 2011; Hegab, et al., 2012a; Anderson, et al., 2017)
Myoepithelial cells (MECs)
Morphological features: flattened epithelial cells with elongated processes.
Function: provide structural support to acini to help maintain the hydrostatic pressure generated by fluid secretion. In response to stimulation by cholinergic agonists, myoepithelial cells contract to expel secreted contents of the glands. Following injury, these cells can regenerate multiple epithelial cell types within the SMG and on the airway surface.
Other names: none.
Markers: KRT14, MYH11 (Anderson, et al., 2017; Goldfarbmuren, et al., 2020) (Table 1).
Location: myoepithelial cells are found on the basal side of acini, mucous tubules and are scattered along the perimeter of ducts (Fig. 4N,P).
Experimental validation: standard approaches (Lynch, et al., 2018; Goldfarbmuren, et al., 2020); mouse lineage tracing using Acta2creERT2 (Anderson, et al., 2017; Lynch, et al., 2018; Tata, et al., 2018) and Myh11-creERT2 (Anderson, et al., 2017).
Developmental origin: myoepithelial cells are one of the earlier cell types that develop from naive SMG placode epithelium (Anderson, et al., 2017). Following lung injury, myoepithelial cells can arise from both basal and remaining myoepithelial cells of the SMG (Anderson, et al., 2017; Lynch, et al., 2018; Tata, et al., 2018).
Heterogeneity and cell states: unknown.
Regenerative potential: myoepithelial cells are capable of repopulating cells within the SMG and the tracheal/bronchial epithelium (Anderson, et al., 2017; Lynch, et al., 2018; Tata, et al., 2018).
Link to Disease: in patients with severe asthma, there is an increase in overall gland size and smooth muscle actin content of myoepithelial cells (Green, et al., 2010).
Key references: (Anderson, et al., 2017; Lynch, et al., 2018; Tata, et al., 2018; Goldfarbmuren, et al., 2020)
Mucous cells
Morphological features: cells with basally positioned nucleus and large densely packed electron opaque granules from the Golgi to the apical cell surface.
Function: in response to stimulation by basolateral neurotransmitters, these cells produce and secrete multiple mucins involved in host defense.
Other names: none.
Marker genes: MUC5B, SPDEF (Chen, et al., 2009) (Table 1). Mucous cells are identifiable by staining with the lectin Ulex Europaeus Agglutinin 1 (UEA-1)(Anderson, et al., 2017).
Location: line the mucous tubules that are located between the collecting ducts and distal serous acini (Fig. 4M,P,R).
Experimental validation: standard approaches (Audie, et al., 1993; Hegab, et al., 2012b; Widdicombe and Wine, 2015; Anderson, et al., 2017; Lynch, et al., 2018; Goldfarbmuren, et al., 2020), air-liquid interface trans-well cultures (Finkbeiner, et al., 2010).
Developmental origin: during development, mucous cells arise from either basal or myoepithelial precursors of the SMG (Engelhardt, et al., 1995; Hegab, et al., 2011; Anderson, et al., 2017). Similarly following lung injury, mucous cells can arise from either basal or myoepithelial cells of the SMG (Hegab, et al., 2012b; Lynch, et al., 2018; Tata, et al., 2018).
Heterogeneity and cell states: unknown.
Regenerative potential: unknown.
Link to Disease: mucous cells of the SMG are a primary source of excess mucus production in conditions like asthma and chronic bronchitis (Widdicombe and Wine, 2015).
Key references: (Chen, et al., 2009; Widdicombe and Wine, 2015)
Serous cells
Morphological features: contain small electron dense granules and intracellular canaliculi.
Function: produce a variety of mucins, antimicrobial peptides and other molecules involved in host defense. In addition, serous cells are a main source of fluid from the gland, driven by secretion of bicarbonate and chloride.
Other names: none.
Markers: LYZ, LTF (Lee and Foskett, 2010; Hegab, et al., 2012b) (Table 1). Serous cells are also identifiable following staining with the lectin Dolichos Biflorus Agglutinin (DBA) (Anderson, et al.,2017).
Location: distal acini of the SMG (Fig. 4R).
Experimental validation: standard approaches (Borthwick, et al., 1999; Lee and Foskett, 2010; Hegab, et al., 2012b; Anderson, et al., 2017; Lynch, et al., 2018; Goldfarbmuren, et al., 2020) (Engelhardt, et al., 1992; Audie, et al., 1993); air-liquid interface trans-well cultures (Finkbeiner, et al., 2010), fluid secretion assays (Lee and Foskett, 2010; Khansaheb, et al., 2011).
Developmental origin: during development, serous cells are derived from either basal or myoepithelial precursor cells of the SmG (Engelhardt, et al., 1995; Hegab, et al., 2011; Anderson, et al., 2017). Similarly following lung injury, serous cells are rederived from either basal or myoepithelial cells of the SMG (Hegab, et al., 2012b; Lynch, et al., 2018; Tata, et al., 2018).
Heterogeneity and cell states: unknown.
Regenerative potential: unknown.
Link to Disease: The lack of functional chloride channels encoded by mutations in CFTR are associated with reduced gland secretions from serous cells and mediate the pathogenesis observed in cystic fibrosis (OMIM:602421) (Widdicombe and Wine, 2015).
Key references: (Hegab, et al., 2012b; Widdicombe and Wine, 2015)
MESENCHYME
Starting from early development, the lung mesenchyme provides instructive cues to the epithelium to control proliferation, differentiation, and patterning along the proximal-distal axis. Classical tissue recombination experiments show that distal mesenchyme instructs distal epithelial specification, even when placed next to proximal epithelial cells (Shannon, 1994; Shannon, et al., 1998). The converse is true when proximal mesenchyme is placed next to distal epithelial cells. Subsequent studies have shown that reciprocal paracrine signaling between the mesenchyme and epithelium, so termed epithelium-mesenchyme crosstalk, coordinates the growth and differentiation of both cell populations (Shannon, 1994; Bellusci, et al., 1996; Bellusci, et al., 1997a; Bellusci, et al., 1997b; Naski, et al., 1998; Shannon, et al., 1998; Arman, et al., 1999; Weaver, et al., 1999). Throughout development, the pulmonary mesenchyme changes its paracrine and ECM-modulating functions to drive branching morphogenesis, sacculation and alveologenesis (Betsholtz, 1995; Olson and Soriano, 2011; Rock, et al., 2011; Zhang, et al., 2013; Endale, et al., 2017; Moiseenko, et al., 2017; Zepp, et al., 2021). In the adult lung, several mesenchymal cell types serve as niches and provide signals and ECM support for epithelial progenitors and their function in tissue regeneration (Lee, et al., 2017; Zepp, et al., 2017).
In mouse, the lung mesenchyme arises from a population of the lateral plate mesoderm called cardiopulmonary mesoderm progenitors (Peng, et al., 2013). Through multiple steps of development, specialized pulmonary mesenchymal cells emerge. Compared to the epithelial cell types, delineation of the different mesenchymal cell types remains less clear. As such, there is overlap between the different cell populations labeled using single gene reporter systems (Riccetti, et al., 2020). A recent study outlined the lineage progression of the multiple mesenchymal cell types (Zepp, et al., 2021). Aside from the mesenchymal cell cards outlined here, there are other cell groups identified from single cell studies or staining experiments that will require further validation to detail their unique characteristics (Dahlgren, et al., 2019; Travaglini, et al., 2020). Here, we focus on established mesenchymal cell types and describe them in the proximal-to-distal sequence of their localization.
Airway smooth muscle cells (ASMCs)
Morphological features: spindled mesenchymal cells in bundles subjacent to the airway epithelium in either a continuous circumferential pattern (in non-cartilaginous airway) or in stripes connecting the ends of cartilages (in cartilaginous airways) (Hines, et al., 2013; Cieri, 2019).
Function: contract to control airway tone and size.
Other names: none.
Markers: genes: ACTA2, DES, LGR6; protein ACTA2/SMA (Table 1).
Location: subjacent to airway epithelium in airway wall (Fig. 5A).
Experimental validation: standard approaches, mouse lineage tracing using Acta2creERT2 (Wendling, et al., 2009), Lgr6creERT2 (Lee, et al., 2017).
Developmental origin: in mouse lung, first detected at E10.5 by Acta2 expression. ASMCs are likely derived from cardiopulmonary mesoderm progenitor cells (Peng, et al., 2013; Ntokou, et al., 2015; El Agha, et al., 2017; Park, et al., 2019) and Acta2creERT2 E12.5 lineage traced cells (Zepp, et al., 2021). Wnt2 is necessary for the formation of ASMCs, but not vascular smooth muscle cells (Goss, et al., 2011).
Heterogeneity and cellular states: unknown.
Regenerative potential: direct regenerative potential unknown. Can serve as niche for regeneration of adjacent airway epithelium following injury (Rock, et al., 2011; Lee, et al., 2017; Volckaert, et al., 2017).
Link to disease: increase in cell number (hyperplasia) and cell size (hypertrophy) are found in asthma. Known to facilitate airway constriction in asthma.
Key references: (Lee, et al., 2017; Riccetti, et al., 2020).
Vascular smooth muscle cells (VSMCs)
Morphological features: spindled cells within the wall of pulmonary artery.
Function: contract to control pulmonary vessel tone and size.
Other names: none.
Markers: genes: NTRK3, ITGA7H, Cnn1M; protein: ACTA2/SMA (Table 1).
Location: prominent within the walls of pulmonary artery and a minor component of pulmonary vein (Fig. 5A,G).
Experimental validation: standard approaches, mouse lineage tracing using Acta2creERT2 (Sheikh, et al., 2015).
Developmental origin: in development, derived from the early (E8.5) cardiopulmonary mesoderm progenitors (Peng, et al., 2013) and the later (E12.5) Acta2creERT2 and PdgfrbcreERT2 expressing cells by lineage tracing (Zepp, et al., 2021). Unclear how cellular turnover during homeostasis is maintained.
Heterogeneity and cellular states: unknown.
Regenerative potential: unknown.
Link to disease: hyperplasia, hypertrophy and increase in constrictive property of VSMCs are linked to pulmonary hypertension. Source of signals for endothelium and vascular remodeling (de la Cuesta, et al., 2019).
Key references: (Greif, et al., 2012; Sheikh, et al., 2015; Steffes, et al., 2020)
Chondrocytes
Morphological features: round cells surrounded by thick matrix condensed together to form cartilage.
Function: give rise to cartilage that supports airway epithelium and prevent airway collapse at exhalation. Play a role in the development of tracheal epithelial cells (Hines, et al., 2013; Cieri, 2019).
Other names: cartilage cells.
Markers: gene: COL2A1, HAPLN1; protein: SOX9 (Table 1).
Location: in mouse, cartilage is only found in trachea and extrapulmonary bronchi. In human, airway cartilage is also present in intrapulmonary bronchi.
Experimental validation: standard approaches, mouse lineage tracing using Col2a1creERT2 (Hines, et al., 2019), alcian blue staining.
Developmental origin: in development, chondrocytes arise from SOX9+ cells in lung mesenchyme (Hines, et al., 2013).
Heterogeneity and cellular states: unknown.
Regenerative potential: unknown.
Link to disease: malformation of airway cartilage leads to tracheobronchomalacia. Altered airway cartilage pattern was identified as one of the earliest features of cystic fibrosis (Ogrinc, et al., 1998).
Key references: (Ogrinc, et al., 1998; Hines, et al., 2013; Chen, et al., 2014b)
Alveolar fibroblast 1 (AF1) cells
Morphological features: fibroblast-like morphology. Contain lipid granules in early postnatal mouse lung.
Function: a key source of signals such as FGF7 and FGF10 during lung development and postnatal homeostasis.
Other names: lipofibroblasts, matrix fibroblast 1.
Markers: genes: Tcf21, Wnt2M; proteins: PLIN2/ADRP.
Location: alveolar mesenchyme (Fig. 5B).
Experimental validation: standard approaches, organoids, lineage tracing using Tcf21mcrem (Park, et al., 2019), Wnt2creERT2 (Zepp, et al., 2021).
Developmental Origin: in mouse, these cells are derived from Wnt2+/Pdgfra+ precursors and obtain their mature phenotype by P3 (Zepp, et al., 2021).
Heterogeneity and cellular states: currently unknown but likely exists.
Regenerative potential: in mouse, proliferate in response to injury.
Link to disease: while some studies suggest that these cells can contribute to myofibroblasts in bleomycin model of lung fibrosis (El Agha, 2017), other studies using more specific cre drivers suggest they contribute to a minor fraction compared to Pdgfrb+ fibroblasts (pericytes-see below) (Torday and Rehan, 2016; Zepp, et al., 2017; Park, et al., 2019).
Key references: (Torday and Rehan, 2016; Zepp, et al., 2017; Park, et al., 2019; Ushakumary, et al., 2021)
Alveolar fibroblast 2 (AF2) cells
Morphological features: fibroblast-like morphology.
Function: a source of signals such as FGF7, FSTL1, WNT5A and IL6 that support AT2 cell proliferation and differentiation during postnatal lung maturation, homeostasis, and regeneration. Also a key source of ECM proteins.
Other names: matrix fibroblast 2, mesenchymal alveolar niche cell (MANC) (Torday and Rehan, 2016; Zepp, et al., 2017; Park, et al., 2019), type-2 associated stromal cell (TASC)(Chung, et al., 2018), adventitial fibroblast (Travaglini, et al., 2020).
Markers: genes: MFAP5, SCARA5 (Table 1).
Location: alveolar mesenchyme (Fig. 5B).
Experimental validation: standard approaches, organoids. In mouse, tracing and isolation using Axin2creERT2;PdgfraH2B-GFP approach (Torday and Rehan, 2016; Zepp, et al., 2017; Park, et al., 2019).
Developmental Origin: in development, these cells arise from cardiopulmonary progenitors cells together with several other lung mesenchymal cell types (Peng, et al., 2013; Zepp, et al., 2021).
Heterogeneity and cellular states: unknown but are a subset of Pdgfra+ mesenchymal cells.
Regenerative potential: proliferate after tissue injury. Axin2+Pdgfra+ AF2 cells supported AT2 cell organoid growth more efficiently than Wnt2+Pdgfra+ AF1 cells (Torday and Rehan, 2016; Zepp, et al., 2017; Park, et al., 2019).
Link to disease: unknown.
Key references: (Torday and Rehan, 2016; Zepp, et al., 2017; Park, et al., 2019; Ushakumary, et al., 2021; Zepp, et al., 2021).
Secondary Crest Myofibroblasts (SCMF)
Morphological features: elongated fibroblasts that contain long actin fibers and are contractile.
Function: a transient lineage that drives alveolar septa formation during alveologenesis, in part by receiving signals from AT 1 cells. Many disappear after alveologenesis through apoptosis, while those that remain in the adult lung no longer express markers such as ACTA2/SMA (Hagan, et al., 2019a; Zepp, et al., 2021).
Other names: myofibroblasts.
Markers: genes: DACH2H, Fgf18M; protein: ACTA2/SMA (Table 1).
Location: alveolar mesenchyme, underline nascent septal ridges and alveolar entrance rings (Fig. 5C,D) (Chen, et al., 2012; Li, et al., 2015; Branchfield, et al., 2016a; Hagan, et al., 2019b).
Experimental validation: standard approaches, mouse lineage tracing using Acta2creERT2 (Wendling, et al., 2009; Zepp, et al., 2021); Acta2 gene expression using the Acta2DsRedreporter (Zepp, et al., 2021), PdgfrartTA;tetO-cre (Li, et al., 2018); PdgfracreERT2 (Zepp, et al., 2021) and Fgf18creERT2 (Hagan, et al., 2019a) all at early postnatal stages when these cells are present as contractile cells.
Developmental origin: in mouse, first detected at E15.5 using Acta2DsRed reporter (Zepp, et al.,2021) or later by ACTA2+ staining in the alveolar region at the onset of alveologenesis at P3 (Chen, et al., 2012; Li, et al., 2015; Branchfield, et al., 2016a; Hagan, et al., 2019b). Lineage labeling in prenatal stages indicate that these cells originate from Pdgfra+ and Acta2+ lung mesenchyme as early as E15.5 (Moiseenko, et al., 2017; Li, et al., 2018; Zepp, et al., 2021).
Heterogeneity and cellular states: unknown.
Regenerative potential: as this is a transient lineage, their role in neonatal repair and regeneration remains unclear.
Link to disease: change in myofibroblast number and characteristics contributes to disruption of septa formation and alveolar simplification in BPD (Popova, et al., 2014; Branchfield, et al., 2016a).
Key references: (Chen, et al., 2012; Li, et al., 2015; Branchfield, et al., 2016a; Hagan, et al., 2019b; Ushakumary, et al., 2021; Zepp, et al., 2021)
Pericytes
Morphological features: fibroblast-like cells, often with long processes.
Function: adhere to the endothelium by gap, tight, and adherens junctions, enabling the endothelium to retain a tight barrier (Hung, et al., 2019).
Other names: mural cells, Axin2+ Myofibroblast Precursors (AMPs) (Torday and Rehan, 2016; Zepp, et al., 2017; Park, et al., 2019).
Markers: genes: TRPC6, LAMC3; protein:CSPG4 (NG2), PDGFRb (Table 1).
Location: adjacent to blood vessels and alveolar capillaries.
Experimental validation: standard approaches, organoids, mouse lineage tracing using PdgfrbcreERT2 (Cuervo, et al., 2017).
Developmental origin: arise from PdgfrbcreERT2 lineage tracing population during development (Zepp, et al., 2021).
Heterogeneity and cellular states: unknown.
Regenerative potential: unknown.
Link to disease: lineage-tracing using Foxd1-cre or Cspg4creERT2 labeled pericytes show that they can proliferate and express ACTA2 following bleomycin-induced injury (Rock, et al., 2011; Hung, et al., 2013; Wilson, et al., 2018).
Key references: (Chen, et al., 2012; Barkauskas, et al., 2013; Hung, et al., 2013; Lee, et al., 2017; Zepp, et al., 2017; Kato, et al., 2018; Biasin, et al., 2020; Zepp, et al., 2021)
Mesothelial cells
Morphological features: flat cells with epithelial morphology even though they are derived from mesenchymal cells.
Function: encase and protect the lung, and provide lubricant surface to allow smooth sliding between lung versus other organs and chest wall. Source of signaling molecules such as FGF9 during development and cytokines such as IL33 during homeostasis (Yin, et al., 2011; Mahlakoiv, et al., 2019).
Other names: lung pleura.
Markers: genes: WT1, UPK3B, FREM2; protein: WT1 (Table 1).
Location: external cell layer that wraps around the lung.
Experimental validation: standard approaches, mouse lineage tracing using Wt1creERT2(von Gise, et al., 2016).
Developmental origin: in mouse starting at E10.5, WT1+ mesothelial cells emerge from the mesodermal lineage (Que, et al., 2008).
Heterogeneity and cellular states: unknown.
Regenerative potential: during development, undergo epithelium-to mesenchyme transition and contribute to lung mesenchymal cells such as vascular smooth muscles and pericytes (Que, et al., 2008; von Gise, et al., 2016).
Link to disease: WT1 + mesothelial cells contribute to fibrotic myofibroblasts in models of fibrosis (Sontake, et al., 2015; Sontake, et al., 2018). Has1hi fibroblasts in regions of fibrosis express WT1, but it is unknown if these cells arose from WT1+ mesothelium or WT1-mesenchymal progenitors (Habermann, et al., 2019).
Key references: (Que, et al., 2008; Sontake, et al., 2015; von Gise, et al., 2016; Sontake, et al., 2018)
ENDOTHELIUM
Pulmonary endothelium facilitates gas exchange between air and blood, delivers nutrients and growth factors to the lung, plays key roles in inflammation, tissue fluid clearance, blood clotting, and serves as source of angiocrine signals for homeostasis and injury repair. The pulmonary vasculature in humans is composed of both pulmonary and bronchial circulations. The pulmonary circulation delivers deoxygenated blood from the right cardiac ventricle to the alveoli through the pulmonary artery and returns oxygenated blood to the left atrium through the pulmonary vein (Fig. 5E). The bronchial circulation originates from the aorta and provides oxygen and nutrient-rich blood to lung structural cells including conducting airways and surrounding tissues. The pulmonary lymphatic system consists of lymphatic vessels, nodes and blunt-ended capillaries, regulating host defense and tissue fluid clearance. Endothelial cells are heterogenous and are comprised of arterial, venous, lymphatic and microvascular/capillary endothelial cells (Fig. 5E), all of which share the expression of cell surface adhesion molecules CD31 (PECAM) (Fig. 1B,F; Fig. 5G,H) and CDH5 (VE-Cadherin), as well as the transcription factor ERG and signaling molecule PDGFB (Kalna, et al., 2019). In mouse, they can be traced using Cdh5creERT2 and PdgfbcreERT2, each label all endothelial cells (Bazigou, et al., 2011; Cai, et al., 2016). Based on scRNAseq studies in mice, pulmonary endothelial cells express a number of genes including Grtpl, Adrbl, Scn7a, Tmem100, Foxfl and lncRNA Fendrr that are unique to the lung and are not expressed in endothelial cells of other organs (Paik, et al., 2020). For established lung endothelial cell populations, we describe them in the sequence of cell types in the arterial and venous vessels, lymphatic vessels, and alveolar capillaries.
Arterial endothelial cells
Morphological features: thin flattened cells lining the interior surface of arteries and arterioles.
Function: conduct blood flow from the heart to the lung, regulate inflammatory responses, synthesize and secrete growth factors, regulate hemostasis and coagulation.
Other names: none.
Markers: genes: GJA5, DKK2; protein: VWF (also stains vein) (Table 1).
Location: pulmonary arteries are located near bronchi and bronchioles as part of the broncho-vascular bundles.
Experimental validation: standard approaches. In mouse, BmxcreERT2 labels arterial endothelium with the exception of smaller arterioles (Ehling, et al., 2013). Sox17creERT2 preferentially labels arteries than veins, and it also labels capillary endothelial cells (Liao, et al., 2009).
Developmental origin: at the initiation of lung development, they arise from cardiopulmonary progenitor cells (Peng, et al., 2013). Later on, they arise from endothelial progenitor cells (Bolte, et al., 2018). They can also arise from proliferation of existing arterial endothelial cells (Whitsett, et al., 2019).
Heterogeneity and cellular states: unknown.
Regenerative potential: can self-renew in mice (Ingram, et al., 2005; Zengin, et al., 2006).
Link to disease: linked to pulmonary arterial hypertension.
Key references: (De Val and Black, 2009; Peng, et al., 2013; Corada, et al., 2014; Ren, et al., 2019)
Venous endothelial cells
Morphological features: thin flattened cells lining the interior surface of veins and venules.
Function: conduct blood flow from the lung to the heart, regulate inflammatory responses, synthesize and secrete growth factors, regulate hemostasis and coagulation.
Other names: none.
Markers: genes: ACKR1H, HDAC9H, Slc6a2M; proteins: VWF (also artery), Endomucin (also capillaries, but not artery) (Table 1).
Location: in proximal lung, pulmonary veins are located next to the secondary bronchi and pulmonary arteries. In distal lung, pulmonary veins do not accompany smaller bronchioles and arteries but travel alone through the connective tissue septa.
Experimental validation: standard approaches; mouse reporter using Nr2f2lacZ (Coup-TFII-lacZ) knock-in allele labels venous endothelial cells (You, et al., 2005).
Developmental origin: at the initiation of lung development, they arise from cardiopulmonary progenitor cells (Peng, et al., 2013). Later on, they arise from endothelial progenitor cells (Bolte, et al., 2018). They can also arise from proliferation of specified venous endothelial cells (Ren, et al., 2019).
Heterogeneity and cellular states: unknown.
Regenerative potential: can self-renew in mice (Bolte, et al., 2018; Ren, et al., 2019).
Link to disease: linked to Pulmonary Veno-Occlusive Disease (PVOD), Alveolar Capillary Dysplasia with Misalignment of Pulmonary Veins (ACDMPV).
Key references: (You, et al., 2005; De Val and Black, 2009; Corada, et al., 2014; Neal, et al.,2019)
Lymphatic endothelial cells
Morphological features: thin flattened cells lining the interior surface of lymphatic vessels (Figure 1 C).
Function: maintain interstitial fluid homeostasis, conduct fluid away from lung interstitium, regulate immune responses.
Other names: lymphatics.
Markers: genes: PROX1, MMRN1; proteins: LYVE1 (Table 1).
Location: found along trachea, near airways, in the alveolar region and on the pleura.
Experimental validation: standard approaches, lineage tracing with Prox1creERT2 (Bazigou, et al., 2011).
Developmental origin: in mouse, specified lymphatic endothelial cells were first detected at E9.5 as Prox1-positive cells located within endothelium of the cardinal vein (Yang, et al., 2012). Their specific developmental origin is unknown.
Heterogeneity and cellular states: unknown.
Regenerative potential: can self-renew (Cui, et al., 2017).
Link to disease: linked to Lymphangiectasis, Lymphangioma, Lymphangioleiomyomatosis (LAM), Lymphatic malformation syndromes include Generalized Lymphatic Anomaly (GLA, also known as lymphangiomatosis), Gorham-Stout disease (GST) and Kaposiform lymphangiomatosis (Itkin and McCormack, 2016).
Key references: (Srinivasan, et al., 2014; Yao, et al., 2014; Stump, et al., 2017; Reed, et al.,2019)
Capillary 1 (CAP1) cells
Morphological features: thin flattened cells lining the interior surface of alveolar microvasculature.
Function: tissue perfusion and gas exchange with the external environment.
Other names: general capillary cells (gCAPs)
Markers: genes: IL7RH, AplnrM, Gpihbp1M (Table 1).
Location: form the microvascular network together with, and more proximal to Capillary 2 cells in the alveolar region based on makers PLVAP (CAP1) and CAR4 (CAP2) staining (Ren, et al., 2019; Ellis, et al., 2020; Gillich, et al., 2020; Kalucka, et al., 2020; Niethamer, et al., 2020).
Experimental validation: standard approaches, mouse lineage tracing with AplnrcreERT2 (Gillich, et al., 2020).
Developmental origin: these cells arise from Cdh5+ endothelial progenitors located in the lateral mesoderm at the beginning of lung development (Peng, et al., 2013). Later on, they arise from AplnrcreERT2 lineage-traced cells, which can also give rise to Capillary 2 cells (see below) (Gillich, et al., 2020).
Heterogeneity and cellular states: in the early postnatal mouse lung, in KIT+ cells which contain CAP1 cells, FOXF1-GFP+KIT+ subset exhibited higher colony forming potential than FOXF1-GFP- KIT+ subset (Wang, et al., 2021).
Regenerative potential: AplnrcreERT2-lineaged cells give rise to both CAP1 and CAP2 cells after elastase lung injury (Gillich, et al., 2020). Adoptive transfer of FOXF1-GFP+KIT+ cells which are enriched for CAP1 cells with endothelial progenitor cell potential, increased angiogenesis by differentiating into capillary, arterial and venous endothelium in a mouse model of alveolar capillary dysplasia with misalignment of pulmonary veins (Wang, et al., 2021).
Link to disease: linked to alveolar capillary dysplasia with misalignment of pulmonary veins, bronchopulmonary dysplasia, pulmonary capillary hemangiomatosis.
Key references: (Ren, et al., 2019; Ellis, et al., 2020; Gillich, et al., 2020; Kalucka, et al., 2020; Niethamer, et al., 2020) (Wang, et al., 2021).
Capillary 2 (CAP2) cells
Morphological features: thin flattened cells lining the interior surface of alveolar microvasculature.
Function: tissue perfusion and gas exchange with the external environment.
Other names: aerocytes, alveolar capillary cells (aCAPs), Car4+ capillary endothelial cells, CD34hi endothelial cells (mouse only).
Markers: genes: EDNRB, HPGDH, AplnM, Car4M; proteins: EDNRB, CA4.
Location: form the microvascular network together with and more distal to CAP1 cells in the alveolar region based on makers PLVAP (CAP1) and CAR4 (CAP2) staining (Ren, et al., 2019; Ellis, et al., 2020; Gillich, et al., 2020; Kalucka, et al., 2020; Niethamer, et al., 2020); located in close apposition to AT1 epithelial cells to facilitate gas exchange.
Experimental validation: standard approaches; CD34hi FACS (mouse) (Niethamer, et al., 2020), mouse lineage tracing with AplncreERT2 (Gillich, et al., 2020).
Developmental origin: Car4+/Ednrb+ cells appear just before birth and mark a subset of capillary endothelium enriched in expression of angiogenic factors (Ellis, et al., 2020). Aplnr-lineage labeled cells can give rise to CAP2 cells (Ren, et al., 2019; Ellis, et al., 2020; Gillich, et al., 2020; Kalucka, et al., 2020; Niethamer, et al., 2020).
Heterogeneity and cellular states: unknown.
Regenerative potential: in mouse, one study shows regenerative Car4/Ednrb/CD34hi cells in alveoli after influenza injury (Neithamer 2020), whereas a different study showed that AplncreERT2 lineage-traced cells do not proliferate after acute elastase lung injury in the mouse (Gillich, et al.,2020).
Link to disease: linked to Alveolar Capillary Dysplasia with Misalignment of Pulmonary Veins (ACDMPV), Bronchopulmonary Dysplasia (BPD), Pulmonary Capillary Hemangiomatosis (PCH).
Key references: (Ren, et al., 2019; Ellis, et al., 2020; Gillich, et al., 2020; Kalucka, et al., 2020; Niethamer, et al., 2020)
IMMUNE
The lung constitutes the largest surface of the body that is exposed to the outside environment. Pathogens can enter the lung either with inhaled air through the epithelial layer or via bloodstream through the endothelium. Cellular immunity is conducted by both immune cell types that reside in the lung interstitium, as well as immune cell types that patrol the extensive lung vasculature, ready to be recruited in response to infection or injury. Lung immune cells belong to either the innate or adaptive immune systems. In addition to defense against infection or injury, immune cells also play critical roles in tissue homeostasis.
Here, we will focus on immune cells in the healthy lung. Early in fetal development, the lung is seeded with immune cells that originate in the yolk sac, fetal liver or bone marrow (Holt and Jones, 2000; Tan and Krasnow, 2016; Ivanovs, et al., 2017; Dzierzak and Bigas, 2018; Ghosn, et al., 2019; Popescu, et al., 2019; Ardain, et al., 2020; Park, et al., 2020). Compared to lung structural cells in the epithelium, mesenchyme and endothelial lineages, immune cells are more dynamic in their gene expression and position. Thus, the entries of the cell cards in this section have been tailored to these cells. In addition to gene expression, cell surface molecules are widely used in FACS to study lung immune cells (Fig. 6). We describe lung immune cells in the myeloid lineage, followed by the lymphoid lineages (Fig. 2A).
Alveolar macrophages (AMs)
Morphological features: largest immune cells in lung, cytoplasm filled with lipid from surfactant.
Function: critical for surfactant turnover, phagocytosis of inhaled pathogens and particles. While they can present antigens to adaptive immune cells, their antigen presentation capacity is poor compared to other macrophages or dendritic cells.
Other names: aMac.
Markers: genes: SIGLEC1H, SiglecFM, ABCG1, FABP4, PPARG, MARCO (Fig. 6B); proteins: CD45, CD11b, CD11c, CD64, CD163, CD206, HLA-DR (Fig. 6A)
Location: luminal surface of alveoli in close proximity to epithelial cells.
Experimental validation: bulk RNAseq of sorted AMs in mice (Sajti, et al., 2020), flow cytometry with HLA-DR/CD64/CD68/CD206 in humans and SiglecF/CD11c/CD64 in mice (Fig. 6A) (Misharin, et al., 2013; Yu, et al., 2016; Hume, et al., 2020); parabiosis, clodronate depletion, adoptive transfer. Lineage tracing using Itgax-cre (Cd11c-cre, also target dendritic cell) (Caton, et al., 2007), Lyz2creERT2 also target interstitial macrophages and neutrophils (Canli, et al., 2017). Genetic ablation with Slco2b1flox/DTR crossed with Lyz2cre that labels all macrophage populations (Chakarov, et al., 2019).
Developmental origin: in mouse, they arise from fetal liver macrophage precursors (Guilliams, et al., 2013; Gomez Perdiguero, et al., 2015; Hoeffel, et al., 2015; Tan and Krasnow, 2016).
Heterogeneity and cellular states: unknown in normal healthy lung. In disease states, recruited macrophages join the resident AMs. In human fibrotic lungs, influenza, bacteria infected or LPS treated lungs, there is heterogeneity in AM profile (Mould, et al., 2019; Reyfman, et al., 2019).
Regenerative potential: regenerates itself during homeostasis and disease.
Link to disease: linked to infection, fibrosis, asthma, COPD and lung cancer (Misharin, et al., 2017). Key cell type in pulmonary alveolar proteinosis (Trapnell, et al., 2019).
Key references: (Ginhoux and Guilliams, 2016; Allard, et al., 2018)
Interstitial macrophages (IMs)
Morphological features: smaller in size than AMs, with smoother surface, irregularly shaped nucleus and numerous cytoplasmic vacuoles (Sabatel, et al., 2017).
Function: present antigens and induce adaptive immunity.
Other names: iMac.
Markers: genes: C1QA, C1QB, C1QC, HLA-DPA1, SLC40A1 (Fig. 6B); proteins: CD11b, CD64, CD206 (lower expression than aMAC), HLA-DR (Fig. 6A).
Location: by pan-macrophage marker staining, they are found in the lung interstitium around airways, vessels and nerves.
Experimental validation: bulk RNAseq of sorted IMs in mice (Sajti, et al., 2020), flow cytometry with CD206/CD64/CD68 in mice (Misharin, et al., 2013; Yu, et al., 2016; Hume, et al., 2020) (Fig. 6A). Mouse genetic lineage tracing using Lyz2creERT2 (also labels AMs and neutrophils) (Canli, et al., 2017).
Developmental origin: mixed origin, arise from embryonic yolk-sac or postnatal bone marrow (Tan and Krasnow, 2016). Parabiosis studies suggested that they can be replenished from blood monocytes in adults (Sabatel, et al., 2017).
Heterogeneity and cellular states: mouse lung: 3 different subpopulations based on surface markers: LYVE1loMHCIIhiCX3CR1hi (LYVE1loMHCIIhi) and LYVE1hiMHCIIloCX3CR1lo (LYVE1hiMHCIIlo) (Gibbings, et al., 2017; Chakarov, et al., 2019). Based on function: phagocytic and non-phagocytic subtypes and IL-10 producing subtype (Sabatel, et al., 2017).
Regenerative potential: unknown.
Link to disease: important in LPS induced lung inflammation (mouse model). LYVE1hiMHCIIlo IMs can ameliorate lung fibrosis (Chakarov, et al., 2019). Increased in numbers in the lung of smokers (Hume, et al., 2020). Decrease in number in asthma (Draijer, et al., 2017).
Key references: (Gibbings, et al., 2017; Schyns, et al., 2018; Chakarov, et al., 2019; Sajti, et al., 2020)
Inflammatory monocytes (iMONs)
Morphological features: smaller than macrophages, no other morphological distinctions.
Function: extravasate readily to sites of injury/inflammation, assemble antigens, and reenter circulation to present to other immune cells without differentiating into macrophages (Jakubzick, et al., 2013)..
Other names: classical monocytes.
Markers: genes: S100A8, S100A9, CD14, VCAN (Fig. 6B); proteins: CD11b, CD14, do not express CD16 (Fig. 6A).
Location: reside in blood vessels, retained even after extensive perfusion. Can extravasate to sites of injury/inflammation.
Experimental validation of cell type: flow cytometry markers: human CD14+CD16− (Fig. 6A), mouse LY6C+.
Developmental origin: bone marrow.
Heterogeneity and cellular states: circulating iMONs versus iMONs recruited to tissue.
Regenerative potential: cannot self-renew, live ∼2 days and are continuously replaced. Can give rise to macrophages or patrolling monocytes once they extravasate into the interstitium. Recruited inflammatory monocytes promote lung regeneration following pneumonectomy (Lechner, et al., 2017).
Link to disease: CX3CL1-CX3CR1 induced changes in iMONs have been shown in fibrotic lungs (Misharin, et al., 2017).
Key references: (Jakubzick, et al., 2013; Misharin, et al., 2017)
Patrolling monocytes (pMONs)
Morphological features: smaller than macrophages, no other morphological distinctions.
Function: patrol endothelium. In other organs, pMONs are known to remove damaged cells from the vasculature (Carlin, et al., 2013).
Other names: non-classical monocytes.
Markers: genes: CDKN1C, PTP4A3, HES4, TNFRSF8 (Fig. 6B); protein: CD11b, CD14 (dimmer than iMONs), CD16 (Fig. 6A)
Location: reside in the blood vessels and retained even after extensive perfusion. Engage in long-range migration along endothelium.
Experimental validation of cell type: flow cytometry markers: human CD14dimCD16+ (Fig. 6A), mouse Ly6c-.
Developmental origin: bone marrow, can arise from iMONs.
Heterogeneity and cellular states: unknown.
Regenerative potential: cannot self-renew, live ∼10 days and are continuously replaced from bone marrow.
Link to disease: in other organs, pMONs play a role in inflammation resolution (Carlin, et al., 2013).
Key references: (Carlin, et al., 2013; Sajti, et al., 2020)
Dendritic cells (DCs)
Morphological features: round to oval cell bodies with well-developed ER and multiple fine dendrites.
Function: at baseline the lung is populated by 3 types of DCs with distinct functions:
Plasmacytoid DC (pDC) are highly effective in sensing intracellular viral or self-DNA and RNA mainly via Toll-like receptors (TLRs). They in turn secrete a large amount of type I interferon to recruit lymphocytes and NK cells.
Classical DC subset 1 (cDC1) cross-present antigens to CD8+ T cells and produce IL-12 that support Th1 and cytotoxic responses.
Classical DC subset 2 (cDC2) can uptake, process and present antigens on MHC-I or -II molecules. They migrate to lymph nodes to cross-present to CD4+ T cells and can also support Th1, Th2, and Th17 polarization. cDC2 are more superior antigen presenter than pDC or cDC1.
Other names: cDC1 is also called myeloid dendritic cell 2 (mDC2), cDC2 is also called myeloid dendritic cell 1 (mDC1). Classical DCs are also called conventional DCs.
Markers: genes (Fig. 6B):
pDC: CLEC4C, LILRA4, IRF7, PLD4.
cDC1: FLT3, CLEC9A, ZBTB46.
cDC2: CD1C, CD1E, FCGR2B, CLEC10A.
protein (Fig. 6A):
pDC: CD123, CD303 (CLEC4C), HLA-DR
cDC1: CD141, CD163, CD370 (CLEC9A), HLA-DR
cDC2: CD1c, CD11b, CD14, CD172a (SIRPA), HLA-DR
Location: can be found below the epithelial cells of both the airway and alveoli with dendrites protruding into the air space to sample aerosol contents. After antigen uptake they migrate to the nearest lymph node (Patel and Metcalf, 2018).
Experimental validation: standard approaches; FACS: for human markers see above and Fig. 6A; mouse markers CD45R (B220), CD45RA, Ly-6C, Siglec-H, and BST2 (CD317). ZBTB46 is a transcription factor highly specific for cDCs. Batf3−/− mice lack mDC2s.
Developmental origin: pDCs develop from both common dendritic cell progenitors and LY6DhiCD2hi lymphoid progenitors. pDC development requires transcription factor IRF8, whereas pDC identity relies on TCF4 (Cisse, et al., 2008; Rodrigues, et al., 2018; Dress, et al., 2019). Both cDC1s and cDC2s develop from the common dendritic cell progenitor, with cDC1s dependent on IRF8 and cDC2s dependent on IRF4 (Schlitzer, et al., 2013).
Heterogeneity and cellular states: during steady state, CD11cdim pDCs are found in the conducting airways. During inflammation, CD11b+ monocyte-derived DCs, expressing LY6C and FceRI, are recruited. Heterogeneity of cDC-1 and cDC-2 are not known.
Regenerative potential: unknown. Short lived and replaced by blood-borne progenitors.
Link to disease: in animal model, pDCs triggered proinflammatory response can exacerbate asthma (Chairakaki, et al., 2018).
Key references: (GeurtsvanKessel and Lambrecht, 2008; Veres, et al., 2011; Patel and Metcalf, 2018; Rodrigues, et al., 2018; Balan, et al., 2019; Musumeci, et al., 2019)
Neutrophils
Morphological features: smallest cells among granulocytes, characteristic multilobed nucleus joined by thin strands, cytoplasm contains azurophilic granules with microbicidal agents.
Function: patrol for signs of microbial infections and respond to pathogens through three antimicrobial actions: phagocytosis, degranulation, and the release of neutrophil extracellular traps (NETs). In addition, neutrophils can modulate the activities of neighboring cells and contribute to the resolution of inflammation, regulate macrophages for long-term immune responses, actively participate in diseases including cancer, and play a role in innate immune memory (Rosales, 2018).
Other names: granulocytes, polymorphonuclear cells (PMNs).
Markers: genes: IL1B, CCR2, CSF1R; proteins: CD11b, CD15, in mice Ly6g (Fortunati, et al., 2009; Misharin, et al., 2013).
Location: marginalized pool in the capillary bed of the lung during steady state, recruited to the parenchyma of airways and alveoli during inflammation and infection.
Experimental validation: standard approaches. Neutrophils can be depleted by anti-Gr1 and anti-Ly6G antibody treatment. Genetic mouse models deficient in neutrophils include: hMrp8-Dtr mice, Mrp8-cre;Mcl1fl/fl mice, Lyz2cre;Mcl1f/fl mice, Csf3r(Gcsfr)−/− mice, Cxcr2−/− mice, Gfi1 hypomophic mutant Genista mice (Stackowicz, et al., 2020).
Developmental origin: bone marrow derived. Neutrophil development starts from granulocyte/monocyte progenitors which differentiate into myeloblasts, promyelocytes and myelocytes. Next, myelocytes give rise to non-proliferating metamyelocytes, band cells and finally mature neutrophils (Ng, et al., 2019).
Heterogeneity and cellular states: different maturational stages: metamyelocytes, band cells and mature neutrophils. During infection, the proportion of immature forms increases in blood.
Regenerative potential: none, die shortly after recruitment to lung.
Link to disease: linked to chronic inflammatory diseases (e.g. asthma, ARDS, COPD, etc) and cancer (Uribe-Querol and Rosales, 2015).
Key references: (Fortunati, et al., 2009; Misharin, et al., 2013; Uribe-Querol and Rosales, 2015; Rosales, 2018; Ng, et al., 2019; Stackowicz, et al., 2020)
Basophils
Morphological features: irregular blunt surface, condensed chromatin, and polylobed nuclei. Human basophils contain a large number of dense granules and are highly basophilic. In contrast, murine basophils lack obvious granules and are polymorphic and moderately basophilic (Lee and McGarry, 2007).
Function: basophils produce antimicrobial extracellular traps and regulate group 2 innate lymphoid cell (ILC2) responses (Lee and McGarry, 2007; Siracusa, 2016; Cohen, et al., 2018; Kabashima, et al., 2018; Inclan-Rico, et al., 2020; Marone, et al., 2020; Vivanco Gonzalez, et al., 2020). In addition, they produce effector molecules in response to stimuli such as inflammatory mediators (histamine, serotonin, platelet-activating factor, and leukotriene), cytokines (IL-4, IL-5, IL-6, IL-9, IL-13, and IL-15), and chemokines (CCL3, CCL4, CCL12, and CXCL12) (Siracusa, 2016; Inclan-Rico, et al., 2020).
Other names: none.
Markers: genes: MS4A2, TPSAB1, TPSB2, KIT, GATA2 (Fig. 6B); proteins: FCER1B, CD23, CD123, and lack of expression of CD117 (c-Kit) (Fig. 6A). In mice, basophils are identified by their expression of CD90, FcεRIa, CD200R, and CD49b and lack expression of CD117 (c-Kit) (Siracusa, 2016).
Location: resident basophils reside in the alveoli (Cohen, et al., 2018).
Experimental validation: standard approaches; FACS (Fig. 6A)
Developmental origin: bone marrow. Development starts as hematopoietic stem cells commit to the myeloid lineage. Later, they develop into granulocyte/monocyte precursor cells, and then to basophil/mast cell precursors, followed by basophil precursor cells that represent the direct precursors of terminally differentiated basophils.
Heterogeneity and cellular states: unknown in lung. Four subpopulations have been characterized in blood (Vivanco Gonzalez, et al., 2020)
Regenerative potential: no self-renewal potential (Yamanishi and Karasuyama, 2016).
Link to disease: unknown in lung.
Key references: (Lee and McGarry, 2007; Siracusa, 2016; Cohen, et al., 2018; Kabashima, et al., 2018; Inclan-Rico, et al., 2020; Marone, et al., 2020; Vivanco Gonzalez, et al., 2020)
Mast Cells
Morphological features: abundant granules in the cytoplasm, characteristic metachromatic staining with aniline dyes.
Function: stimulated to degranulate when they encounter antigen immunoglobulin E (IgE) antibody bound to high affinity IgE receptor, FcεRI. They then release chemical and biological mediators including histamine, ATP, prostaglandin, leukotriene, cytokines and angiogenic factors (de Souza Junior, et al., 2017; Ren, et al., 2020).
Aliases and acronyms: MCTC, mastocyte.
Markers: genes: MS4A2, TPSAB1, TPSB2, KIT, GATA2 (Fig. 6B); proteins FCER1B, CD117 (c-Kit) and lack of expression of CD123 (Fig. 6A).
Location: rare in human lung. When observed, they are found in bronchial epithelium, bronchial lamina propria or adjacent to blood vessels.
Experimental validation: standard approaches; FACS (Fig. 6A), mast cell-deficient KitW/Wv mice (Nakano, et al., 1985).
Developmental origin: bone marrow derived, SCF is the major growth factor essential for mast cell survival.
Heterogeneity and cellular states: each anatomical compartment of the lung harbors site-specific mast cell populations. Can be classified based on the content of their granules, tryptase only, chymase only or mixed. Connective tissue type mast cells (MCTC): IL-4+, IL-13+, TPSAB1+, CMA1+, CPA3+. Tissue resident mast cells (MCT): IL-5+, Il-6+, TPSAB1+, CMA1-, CPA3+.
Regenerative potential: unknown; long-lived cells.
Link to disease: found in higher numbers in asthma, BPD, COPD lungs. Also linked to IPF, ARDS, pulmonary hypertension, lung neoplasia.
Key references: (Kabashima, et al., 2018; Ravindran, et al., 2018; Komi, et al., 2020)
Innate lymphoid cells (ILCs)
Morphological features: lymphoid cell morphology.
Function: similar to Th cells in function but lack T cell receptor. There are three types of ILCs:
Innate lymphoid group 1 cells (ILC1): are involved in type I immune responses and host defense against viruses, intracellular microbes, and tumor cells. ILC1s respond to cytokines including IL-12, IL-18, and IL-15, and produce IFNγ and GM-CSF. Unlike NK cells, ILC1s are not cytotoxic.
Innate lymphoid group 2 cells (ILC2): are involved in type 2 immune responses and host defense against extracellular parasites, for example, helminths like N. brasiliensis and allergens. In response to cytokines such as IL-25, TSLP, and IL-33, ILC2s secrete IL-4, IL-5, IL-9, IL-13, amphiregulin (AREG) to repair damaged tissue.
Innate lymphoid group 3 cells (ILC3): possess similar function as Th17 cells and defend against extracellular microbes such as bacteria and fungi in lung. ILC3s can respond to IL-23, IL-1α, IL-1β, IL-7, TL1A, and prostaglandin E2 (PGE2). These cells produce GM-CSF, IL17, IL-22, TNF-α, and lymphotoxin α/β to promote epithelial stem cell proliferation.
Other names: Nuocytes (ILC2).
Markers: genes: FCGR3A, KLRB1, GZMB, PRF1 (Fig. 6B)
proteins (Fig. 6A)
ILC1s: in human CD127(IL-7Rα), CXCR3, and transcription factor (TF) T-bet and lack expression of TF eomesodermin (Eomes). In mouse, CD90.2, ILC1s express NKp46, NK1.1, CD49A, and T-bet, but lack Eomes.
ILC2s: in human CD127 (IL-7Rα), ST2, CD117, CRTH2, and TF GATA3. In mouse, CD90.2, CD127, CD117, CD90, and ST2.
ILC3s: in human CD127, IL23R1, CD56, and TFs, RORyT, and AHR. In mouse CD90.2, CD127, CD117, CD90, NKp46 and TFs T-bet, RORyT, and AHR (Yudanin, et al., 2019).
Location: resident in the airways, alveolar mucosa, and perivascular niches (ILC2 and ILC3). Can travel to lymph nodes for antigen presentation (ILC3) (Dahlgren, et al., 2019; Oherle, et al., 2020).
Experimental validation: standard approaches; FACS (Fig. 6A) (Eberl, et al., 2015b). In steady-state naïve mice, ILC2s can be labeled using Il5cre mice (von Moltke, et al., 2016); ILC3s can be labeled using Rorccre mice (Eberl, et al., 2004).
Developmental origin: ILCs develop from a common lymphoid progenitor (CLP), which further differentiates into a shared ILC precursor termed a-lymphoid precursor (αLP). Further differentiation of αLP generates different precursors with more restricted lineage repopulation capacity. Among them, the common helper ILC precursor (CHILP) give rise to ILC1, ILC2, and IlC3 (Constantinides, et al., 2014).
Heterogeneity and cell states: scRNAseq and ATACseq studies demonstrate considerable heterogeneity in transcriptome and epigenome in a tissue-dependent fashion (Yudanin, et al.,2019). Between ILC subtypes, the different ILCs can transdifferentiate from one to another (Vonarbourg, et al., 2010; Bal, et al., 2016). Within individual ILCs, pulmonary ILC2s exhibit two distinct subsets, lung resident natural ILC2s (nILC2s) and inflammatory ILC2s (iILC2s) recruited from gut to lung (Huang, et al., 2015; Huang, et al., 2018). ILC3s also show two subsets: natural cytotoxicity receptor-positive (NCR+) ILC3s or NCR negative (NCR-) ILC3s.
Regenerative potential: ILCs do not recirculate and are maintained predominantly via local self-renewal rather than through replenishment from blood-derived ILCs or their precursors from circulation and bone marrow (Gasteiger, et al., 2015).
Link to disease: ILC1s are implicated in host defense against influenza H1N1 and intracellular bacterial pathogens. ILC2s play a critical role in allergen response and respiratory viral, bacterial, and helminthic lung infections (Monticelli, et al., 2011; Barlow, et al., 2012). ILC3s are critical in respiratory viral (H1N1 influenza) and bacterial infections (K. pneumoniae, S. pneumoniae, and P. aeruginosa) (Chen, et al., 2011).
Key references: (Eberl, et al., 2004; Di Stefano, et al., 2009; Vonarbourg, et al., 2010; Chen, et al., 2011; Monticelli, et al., 2011; Barlow, et al., 2012; Klose, et al., 2013; Constantinides, et al., 2014; Eberl, et al., 2015b; Gasteiger, et al., 2015; Huang, et al., 2015; Zuo, et al., 2015; Bal, et al., 2016; Silver, et al., 2016; von Moltke, et al., 2016; Huang, et al., 2018; Dahlgren, et al., 2019; Schneider, et al., 2019; Yudanin, et al., 2019; Oherle, et al., 2020; Mazzurana, et al., 2021)
Natural Killer cells (NK cells)
Morphological features: large granular lymphocyte.
Function: NK cells are cytotoxic cells involved in type I immunity. They are the innate counterpart of CD8+ T cells. Similar to CD8+ T cells, NK cells employ the perforin/granzyme pathway, TRAIL, FAS/FASL interactions to kill cells. NK cells are also able to secrete IFNγ.
Other names: none.
Markers: genes: SPTSSB, NKG7, KLRD1, KLRC1 (Fig. 6B); proteins: CD11b, CD56, CD94, NKG2A, NKG2B (Fig. 6A)
Location: in circulating blood and in tissue.
Experimental validation: standard approaches; FACS (Fig. 6A). While NK cells have similarities with ILC1s, they differ by the expression of eomesodermin, which is present in NK cells (Eberl, et al., 2015a).
Developmental origin: common lymphoid progenitors differentiate into common innate lymphoid progenitors which give rise to NK progenitors.
Heterogeneity and cellular states: CD56 bright versus dim. Conventional NK cells that circulate versus tissue-resident NK cells.
Regenerative potential: none.
Link to disease: viral infection, lung cancer.
Key references: (Spits and Cupedo, 2012; Spits, et al., 2013; Artis and Spits, 2015; Eberl, et al., 2015a)
T cells
Morphological features: lymphoid morphology.
Function: there are multiple T cell types with distinct functions.
CD4+ (helper) T cells: help B cells mount antibody responses, co-stimulate DC, and support CD8+ T cells, perform direct cytotoxic functions, stimulate macrophages. Can form immunological memory.
CD8+ (cytotoxic) T cells: kill viral infected cells. A subset of the CD8 memory precursor cells in the lung will differentiate into tissue-resident memory T cells.
Th1: defend against viruses (influenza, RSV), M. tuberculosis, and fungi.
signature cytokines: IFNg, IL-2, TNFa
transcription factors: TBX21, STAT4
Th2: activated by allergen. Produce IL13 and induce airway hyperresponsiveness, goblet cell metaplasia, and mucus hypersecretion. Activate eosinophils through IL5 production. Defend against helminth parasites.
signature cytokines: IL-4, IL-5, IL-13
transcription factors: GATA3, STAT6
Th17: either protective or pro-inflammatory depending on infectious agent.
signature cytokines: IL-17
transcription factors: RORC, STAT3
Treg: control excess inflammation and maintain homeostasis.
signature cytokines: IL-10
transcription factors: FOXP3, STAT5
Mucosal-associated invariant T cells (MAIT): capable of mounting immediate response to bacteria by recognizing microbial ligands in conjunction with non-classical MHC-related protein MR1
NKT, iNKT, γδTcells: bridge innate and adaptive immunity.
Tissue resident effector memory T cells: provide local immune protection after reinfection. The majority of T cells in the human lung are memory phenotype (both CD4+ and CD8+T cells). These cells are CD45RA–CCR7–. They are different from blood TEM by expressing CD69. CD69+CD8+T cells also express CD103.
Other names: none.
Markers: genes: (Fig. 6B), proteins: (Fig. 6A)
Naïve: CD3E, MAL, CCR7
Central memory: CD3E, CD40LG
Effector memory: CD3E, CXCR6
T regulatory: FOXP3, CD3E, CTLA4, FANK1
MAIT: CD3E, KLRB1, IL7R
NKT: CD3E, CCL5
Location: found in the circulation and can be recruited to airway and alveoli. They do not recirculate.
Experimental Validation: genetic knockout models (T cell receptors, cytokines, chemokines, transcription factors), antibody-mediated depletion experiments, parabiosis experiments, cell transfer experiments, reporter gene animal models.
Developmental origin: precursors originate in the bone marrow and mature in the thymus.
Heterogeneity and cellular states: CD4+ T cells are composed of at least five distinct populations that display different surface markers, produce different effector cytokines and regulated by different transcription factors. T helper (Th)1, Th2 cells, and Th17 cells are well established cell states.
Regenerative potential: memory T cells can undergo clonal expansion. Other T cells have no self-renewal potential. Average half-life of a few weeks. Repopulated by blood borne cells.
Link to disease: linked to inflammatory lung diseases (e.g. Bronchopulmonary Dysplasia and fibrosis), infectious diseases, and cancer.
Key references: (Paul, et al., 2019; Snyder and Farber, 2019; Imanishi and Saito, 2020; Khan, 2020; Zemmour, et al., 2020; Bugya, et al., 2021; Wen, et al., 2021; Williams, et al., 2021).
B cells
Morphological features: no distinct morphological feature is used in characterization.
Function: produce antibodies and cytokines. Support and activate T helper cells.
Other names: plasma blasts, antibody secreting cells, antigen presenting B cells.
Markers: genes (Fig. 6B):
B cells: MS4A1, BANK1
Plasma cells: SdC1, CD38
Proteins (Fig. 6A) all expressing CD19, CD20, CD79;
Naïve B cells: CD25-, CD69-, HLA-DR-
Effector B cells: CD25+, CD69+, HLA-DR+
Plasma cells: CD38+, CD69-, CD138+, HLA-DR-
Location: found in circulation and can be recruited into parenchymal regions.
Experimental validation: standard approaches. In mouse, CD19 genetic ablation, transgenic B cell receptor animals, depletion studies.
Developmental origin: bone marrow.
Heterogeneity and cellular states: subsets of B effector (Be) cells can express transcription factors and cytokines akin to CD4 T cells (e.g. Be1, Be2, Be17 and Breg).
Regenerative potential: none.
Link to disease: increased in inflammation (COPD, allergies), produce autoantibodies in systemic sclerosis and IPF.
Key references: (Rickert, et al., 1997; Kato, et al., 2013; Tsuneto, et al., 2014; Chiu and Openshaw, 2015; Lam and Baumgarth, 2019; Sanz, et al., 2019; Allie and Randall, 2020; Grasseau, et al., 2020; Wang, et al., 2020b).
PROTEOMIC, LIPIDOMIC AND METABOLIC STUDIES OF THE LUNG
A variety of biological mechanisms directly impact the abundance of proteins relative to their transcripts, e.g., post-transcriptional regulation, different protein half-life, change in subcellular localization, etc. Lipids and metabolites are products of interactions between genes, transcripts, proteins, and the environment. In LungMAP, we have made initial efforts to delineate protein, lipid, and metabolomic pan-lineage markers at increasing resolutions using LC-MS/MS. Below, we summarize previous efforts and aggregated insights (Bandyopadhyay, et al., 2018; Kyle, et al., 2018; Du, et al., 2019) (Proteomics: https://lungmap.net/breath-omics-experiment-page/?experimentTypeId=LMXT0000000015&experimentId=LMEX0000000661; Lipidomics: https://lungmap.net/breath-omics-experiment-page/?experimentTypeId=LMXT0000000005&experimentId=LMEX0000001622)
Of the 3,320 proteins quantified, proteins unique or enriched in the pan-epithelial population include CD36/EPCAM and CDH1/E-Cadherin (Supplemental Table 1, link above). We also identified markers for ciliated cells RSPH1 and the Tektin family proteins (TEKT1, TEKT2, TEKT3, TEKT4); markers for goblet cells AGR2; the polymeric immunoglobulin receptor (PIGR) located at the apical surface of serous cells; the AT1 markers AGER/RAGE; the AT2 markers LAMP3, ABCA3, SFTPC. Proteins unique to or enriched in mesenchymal cells included pericyte marker PDGFR-β; or smooth muscle/myofibroblast factor TAGLN/SM22. Proteins unique or enriched in the pan-endothelial population include CD31/PECAM, CDH5/VE-Cadherin, PCDH1, CLDN5 and ICAM2 (Halai, et al., 2014). We also identified markers of capillary cells (e.g., SLC6A4, FCN3, CA4) and the venous cell marker EPHB4. Proteins unique or enriched in immune cells include the pan-immune marker CD45/PTPRC, and the pan-myeloid marker FCER1G. We also identified the panmacrophage marker CD68; alveolar macrophage transcription factor SPI1/PU.1; monocyte protein S100A8/A9, dendritic cell protein ribonuclease 6.
Less is known about lipid markers for the different pan-lineage populations in lung (Nakayasu, et al., 2016; Kyle, et al., 2018). Using a recently developed bioinformatic tool, Lipid Mini-On (Clair, et al., 2019), we uncovered structural traits enriched in the lipids over-represented in the different sorted cell populations (link above). Of the 286 lipid species quantified, 51 were more abundant in epithelial cells. They are enriched in long-chain fatty acids such as palmitic acid (16:0), and phosphatidylcholines with a total of 32 and 34 carbons in the fatty acid chains. As expected, epithelial lipids were enriched in phosphatidylcholines (PC) and phosphatidylglycerols (PG) that are the most abundant lipids found in pulmonary surfactants, e.g. PC(16:0/16:0), pC(14:0/16:1) PG(16:0/18:1). For the panendothelial population, 72 are over-represented. The endothelial lipids are enriched in ceramides, phosphatidylserines, phosphatidylethanolamines, and phosphatidylinositol lipids containing the fatty acid 20:4 (likely arachidonic acid). For the pan-mesenchymal population, 19 lipid species were unique or enriched in mesenchymal cells. The mesenchymal lipids were enriched in triglycerides with medium-chain fatty acids (C8-C12) and C16 or C18 saturated fatty acids (no double bonds). Finally, the 51 lipids over-represented in the pan-immune population are enriched in polyunsaturated fatty acids (most of which are 20:4 fatty acids), triglycerides that contain long-chain fatty acids, and ether-linked phosphatidylcholines. Interestingly, the lipid data also show that immune cells contain a lesser-known phospholipid and structural isomer of PGs, bis(monoacylglycerol)phosphates. Subcellularly, bis(monoacylglycerol)phosphates are localized to late endosomes and lysosomes (Kyle, et al., 2018).
CONCLUDING REMARKS
This collection of cell cards is intended as a foundational resource to unify and boost lung research, and will be posted as a live document on LungMAP.net and frequently updated. We anticipate that the number of cell cards will grow as new results become available. For example, cell types such as intrinsic neurons, glial cells and bronchial vascular endothelial cells will be defined as the number of cells captured by single-cell RNAseq increases. New cell types may also be elevated from currently listed cell states if evidence supports their persistence and distinguished function. Future studies will also iteratively refine these cell cards, generating additional markers from integrated analysis of multiple single-cell datasets. Currently, several cell types cannot be definitively traced using single gene lineage tracing tools (Table 1) (Riccetti, et al., 2020). The identification of more refined marker genes, along with the generation of intersectional dual recombinase tools (Liu et al., 2019; Salwig et al., 2019), should provide the wider lung community the ability to better characterize current and emerging cell lineages and states. Single-cell resolution spatial transcriptomics will reveal if heterogeneity within a cell type can be spatially distinguished. In addition, spatial data will generate new hypotheses for how ligand-receptor interactions may dictate cell fate specification and progenitor/niche collaboration. Epigenomic technologies such as single-nucleus ATACseq will reveal cell type resolved chromatin landscape and can be integrated with single cell transcriptomic data to better define the gene regulatory network that drives these cell fates (Wang, et al., 2020a; Zepp, et al., 2021).
Another dimension of knowledge growth will come from understanding of species differences. For example, it has been noted that basal cells in human are found all along the airway including intrapulmonary bronchi and bronchioles, while they are restricted to trachea and main stem bronchi of the mouse respiratory system (Rock, et al., 2010). Additional cellular differences in airway structure, bronchial circulation, immune investment will continue to be documented as the depths of study increases. Extensive gene expression differences are emerging from single cell RNAseq studies of the human, mouse, rat, ferret, pig and primate lungs, building the repertoire of data that could populate a new entry of “species differences” in future versions of cell cards (Raredon, et al., 2019). These cross-species cellular and molecular diversity will inform how lung function evolves and adapts to organismal size, energy demand and environment.
While focusing on healthy lungs here, these cell cards will serve as the foundation for comparisons with diseased lungs. Such comparison will yield changes in developmental lung diseases, a current focus of the NHLBI LungMAP consortium effort. These comparisons will also extend to chronic diseases of the adult lung including COPD and IPF. Overall, we hope that these cell cards will stimulate collaboration throughout the broad pulmonary research community, and help to build bridges to other tissue-focused mapping efforts.
METHODS
Histology and Immunofluorescence
7μm frozen embedded tissue sections of 4% paraformaldehyde fixed human lung tissue were equilibrated to room temperature and rehydrated in PBS. Slides were then re-fixed with 4% PFA for 5 minutes and washed in PBS. 5μm thick paraffin embedded sections of 10% formalin fixed human lung tissue were melted at 60°C for two hours, following rehydration through xylene and alcohol, and finally in PBS. Tissue sections were stained with H&E according to standard protocols. PIGR Immunohistochemistry was performed according to standard laboratory protocols. For immunofluorescence, antigen retrieval was performed in 0.1 M citrate buffer (pH 6.0) by microwaving. Slides were blocked for 2 hours at room temperature using either 4% normal goat serum (Jackson Immuno Research Laboratories) or 4% normal donkey serum (Jackson Immuno Research Laboratories) in PBS containing 0.2% Triton X-100, and then incubated with primary antibodies diluted in blocking buffer for approximately 16 hours at 4°C. Primary antibodies were listed in the key resources table. Appropriate secondary antibodies conjugated to Alexa Fluor 488, Alexa Fluor 555/568, or Alexa Fluor 633/647 (Thermo Fisher Scientific) were used at a dilution of 1:200 in blocking buffer for 1 hour at room temperature. Nuclei were counterstained with DAPI (1 μg/ml) (Thermo Fisher Scientific). Sections were mounted using ProLong Gold (Thermo Fisher Scientific) mounting medium and coverslipped.
KEY RESOURCES TABLE
| REAGENT | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| ABCA3 | Seven Hills Bioreagents | WMAB—17G524 |
| ACTA2 | Sigma-Aldrich | A5228 |
| AGER | R&D Systems | AF1145 |
| CCL21 | R&D Systems | AF366 |
| FOXF1 | R&D Systems | AF4798 |
| FOXJ1 | Thermo Fisher Scientific | 14-9965-02 |
| GRP | Thermo Fisher Scientific | 20073 |
| HOPX | Santa Cruz Biotechnology | SC-30216 |
| LYVE1 | ABCAM | AB36993 |
| KRT5 | Biolegend | PRB-160P-200 |
| KRT14 | ABCAM | AB7800 |
| MUC5AC | ABCAM | AB3649 |
| MUC5B | Santa Cruz Biotechnology | SC-20119 |
| NKX2.1 | Seven Hills Bioreagents | RB TTF-1 1231 |
| PECAM1 | R&D Systems | BBA7 |
| SCGB1A1 | Lifespan Bioscience | LS-B6822 |
| SOX2 | Santa Cruz Biotechnology | SC-365823 |
| SOX9 | Millipore | AB-5535 |
| TP63 | Biocare | CM163 |
| TUBA4A | Sigma Aldrich | T7451 |
| TUBB3 | Biolegend | 801201 |
| VIM | Santa Cruz Biotechnology | SC-7557 |
Confocal microscopy
Tissue sections stained by immunofluorescence were imaged on an inverted Nikon A1R confocal microscope (×20, 60X magnification) NA 1.27 objective using a 1.2 AU pinhole. Maximum intensity projections of multi-labeled Z stack images obtained sequentially using channel series across the sections were generated using Nikon NIS-Elements software. Brightfield images were captured using a Zeiss Axio ImagerA2 microscope utilizing Axiovision software.
Human lung single-cell data analysis
Single-cell data of Human Lung Cell Atlas (https://hlca.ds.czbiohub.org/) was downloaded and 10x sequencing data was further used for marker gene discoveries. Library size of each cell was normalized to 1 million and then log2 transformation was applied. To get better annotations of immune cells, especially lymphocytes, we applied Azimuth label projection strategy using human PBMC as the reference (https://azimuth.hubmapconsortium.org/) (Hao, et al., 2020). Average normalized expression values were calculated for each cell type and Morpheus was used for heatmap visualization (https://software.broadinstitute.org/morpheus/). Cell ontology terms were mapped to cell annotations manually (Meehan, et al., 2011). IGSF21+ dendritic cells from the original publication were reannotated as interstitial macrophages due to high levels of interstitial macrophages markers (such as C1QA, IGSF21 and SLC40A1), and low expression levels of dendritic cell markers (such as CD1C and CLEC9A). Basophil/Mast cells were reannotated as mast cells since they are mainly CD11c-(ITGAX), CD117+ (KIT), CD123- (IL3RA) cells.
A Census of the Lung: CellCards from LungMAP
DEVELOPMENTAL-CELL-D-21-00414R2
Highlights:
Rich single cell and lineage tracing data necessitate a new census of lung cell types
Lung CellCards serves as a curated, up-to-date, practical resource for lung research
CellCards annotates developmental origin, cellular function, regenerative potential etc.
Lung CellCards serves as a starting point for harmonization of lung nomenclature
ACKNOWLEGEM ENTS:
We thank all members of the LungMAP2 consortium for their discussion. We thank the LungMAP2 external advisory committee for their insights. This work is supported by NHLBI LungMAP2 grants 5U01HL148867 (X. Sun), 5U01HL148857 (E. Morrisey), 5U01HL148856 and U01HL134745 (J. Whitsett), 5U01HL148860 (J. Adkins and G. Clair), 5U01HL148861 and HL122700 (G. Pryhuber) and 5U24HL148865 (B. Aronow, N. Salomonis).
Footnotes
DECLARATION OF INTERESTS:
Dr. Xin Sun and Dr. Edward Morrisey are members of the advisory board for Developmental Cell.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES:
- Adams TS, Schupp JC, Poli S, Ayaub EA, Neumark N, Ahangari F, Chu SG, Raby BA, Deluliis G, Januszyk M, et al. (2020). Single-cell RNA-seq reveals ectopic and aberrant lung-resident cell populations in idiopathic pulmonary fibrosis. Sci Adv 6, eaba1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allard B, Panariti A, and Martin JG (2018). Alveolar Macrophages in the Resolution of Inflammation, Tissue Repair, and Tolerance to Infection. Front Immunol 9, 1777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allie SR, and Randall TD (2020). Resident Memory B Cells. Viral Immunol 33, 282–293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson PJ, Lynch TJ, and Engelhardt JF (2017). Multipotent Myoepithelial Progenitor Cells Are Born Early during Airway Submucosal Gland Development. Am J Respir Cell Mol Biol 56, 716–726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ardain A, Marakalala MJ, and Leslie A (2020). Tissue-resident innate immunity in the lung. Immunology 159, 245–256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arman E, Haffner-Krausz R, Gorivodsky M, and Lonai P (1999). Fgfr2 is required for limb outgrowth and lung-branching morphogenesis. Proc Natl Acad Sci U S A 96, 11895–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Artis D, and Spits H (2015). The biology of innate lymphoid cells. Nature 517, 293–301. [DOI] [PubMed] [Google Scholar]
- Audie JP, Janin A, Porchet N, Copin MC, Gosselin B, and Aubert JP (1993). Expression of human mucin genes in respiratory, digestive, and reproductive tracts ascertained by in situ hybridization. J Histochem Cytochem 41, 1479–85. [DOI] [PubMed] [Google Scholar]
- Bal SM, Bernink JH, Nagasawa M, Groot J, Shikhagaie MM, Golebski K, van Drunen CM, Lutter R, Jonkers RE, Hombrink P, et al. (2016). IL-1beta, IL-4 and IL-12 control the fate of group 2 innate lymphoid cells in human airway inflammation in the lungs. Nat Immunol 17, 636–45. [DOI] [PubMed] [Google Scholar]
- Balan S, Saxena M, and Bhardwaj N (2019). Dendritic cell subsets and locations. Int Rev Cell Mol Biol 348, 1–68. [DOI] [PubMed] [Google Scholar]
- Bandyopadhyay G, Huyck HL, Misra RS, Bhattacharya S, Wang Q, Mereness J, Lillis J, Myers JR, Ashton J, Bushnell T, et al. (2018). Dissociation, cellular isolation, and initial molecular characterization of neonatal and pediatric human lung tissues. Am J Physiol Lung Cell Mol Physiol 315, L576–L583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bankova LG, Dwyer DF, Yoshimoto E, Ualiyeva S, McGinty JW, Raff H, von Moltke J, Kanaoka Y, Austen KF, and Barrett NA (2018). The cysteinyl leukotriene 3 receptor regulates expansion of IL-25-producing airway brush cells leading to type 2 inflammation. Sci Immunol 3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barkauskas CE, Cronce MJ, Rackley CR, Bowie EJ, Keene DR, Stripp BR, Randell SH, Noble PW, and Hogan BLM (2013). Type 2 alveolar cells are stem cells in adult lung. J Clin Invest 123, 3025–3036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barlow JL, Bellosi A, Hardman CS, Drynan LF, Wong SH, Cruickshank JP, and McKenzie AN (2012). Innate IL-13-producing nuocytes arise during allergic lung inflammation and contribute to airways hyperreactivity. J Allergy Clin Immunol 129, 191–8 e1–4. [DOI] [PubMed] [Google Scholar]
- Basil MC, Katzen J, Engler AE, Guo M, Herriges MJ, Kathiriya JJ, Windmueller R, Ysasi AB, Zacharias WJ, Chapman HA, et al. (2020). The Cellular and Physiological Basis for Lung Repair and Regeneration: Past, Present, and Future. Cell Stem Cell 26, 482–502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bazigou E, Lyons OTA, Smith A, Venn GE, Cope C, Brown NA, and Makinen T (2011). Genes regulating lymphangiogenesis control venous valve formation and maintenance in mice. J Clin Invest 121, 2984–2992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bellusci S, Furuta Y, Rush MG, Henderson R, Winnier G, and Hogan BL (1997a). Involvement of Sonic hedgehog (Shh) in mouse embryonic lung growth and morphogenesis. Development 124, 53–63. [DOI] [PubMed] [Google Scholar]
- Bellusci S, Grindley J, Emoto H, Itoh N, and Hogan BLM (1997b). Fibroblast Growth Factor 10(FGF10) and branching morphogenesis in the embryonic mouse lung. Development 124, 4867–4878. [DOI] [PubMed] [Google Scholar]
- Bellusci S, Henderson R, Winnier G, Oikawa T, and Hogan BL (1996). Evidence from normal expression and targeted misexpression that bone morphogenetic protein (Bmp-4) plays a role in mouse embryonic lung morphogenesis. Development 122, 1693–702. [DOI] [PubMed] [Google Scholar]
- Betsholtz C (1995). Role of platelet-derived growth factors in mouse development. Int J Dev Biol 39, 817–25. [PubMed] [Google Scholar]
- Biasin V, Crnkovic S, Sahu-Osen A, Birnhuber A, El Agha E, Sinn K, Klepetko W, Olschewski A, Bellusci S, Marsh LM, et al. (2020). PdGfR alpha and alpha SMA mark two distinct mesenchymal cell populations involved in parenchymal and vascular remodeling in pulmonary fibrosis. Am J Physiol-Lung C 318, L684–L697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolte C, Whitsett JA, Kalin TV, and Kalinichenko VV (2018). Transcription Factors Regulating Embryonic Development of Pulmonary Vasculature. Adv Anat Embryol Cel 228, 1–20. [DOI] [PubMed] [Google Scholar]
- Borthwick DW, Shahbazian M, Krantz QT, Dorin JR, and Randell SH (2001). Evidence for stem-cell niches in the tracheal epithelium. Am J Respir Cell Mol Biol 24, 662–70. [DOI] [PubMed] [Google Scholar]
- Borthwick DW, West JD, Keighren MA, Flockhart JH, Innes BA, and Dorin JR (1999). Murine submucosal glands are clonally derived and show a cystic fibrosis gene-dependent distribution pattern. Am J Respir Cell Mol Biol 20, 1181–9. [DOI] [PubMed] [Google Scholar]
- Branchfield K, Li R, Lungova V, Verheyden JM, McCulley D, and Sun X (2016a). A three-dimensional study of alveologenesis in mouse lung. Dev Biol 409, 429–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Branchfield K, Nantie L, Verheyden JM, Sui PF, Wienhold MD, and Sun X (2016b). Pulmonary neuroendocrine cells function as airway sensors to control lung immune response. Science 351, 707–710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bugya Z, Prechl J, Szenasi T, Nemes E, Bacsi A, and Koncz G (2021). Multiple Levels of Immunological Memory and Their Association with Vaccination. Vaccines (Basel) 9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bullard T, Koek L, Roztocil E, Kingsley PD, Mirels L, and Ovitt CE (2008). Ascl3 expression marks a progenitor population of both acinar and ductal cells in mouse salivary glands. Developmental Biology 320, 72–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai Y, Bolte C, Le T, Goda C, Xu Y, Kalin TV, and Kalinichenko VV (2016). FOXF1 maintains endothelial barrier function and prevents edema after lung injury. Sci Signal 9, ra40. [DOI] [PubMed] [Google Scholar]
- Canli O, Nicolas AM, Gupta J, Finkelmeier F, Goncharova O, Pesic M, Neumann T, Horst D, Lower M, Sahin U, et al. (2017). Myeloid Cell-Derived Reactive Oxygen Species Induce Epithelial Mutagenesis. Cancer Cell 32, 869–883 e5. [DOI] [PubMed] [Google Scholar]
- Cao J, Spielmann M, Qiu X, Huang X, Ibrahim DM, Hill AJ, Zhang F, Mundlos S, Christiansen L, Steemers FJ, et al. (2019). The single-cell transcriptional landscape of mammalian organogenesis. Nature 566, 496–502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carlin LM, Stamatiades EG, Auffray C, Hanna RN, Glover L, Vizcay-Barrena G, Hedrick CC, Cook HT, Diebold S, and Geissmann F (2013). Nr4a1-dependent Ly6C(low) monocytes monitor endothelial cells and orchestrate their disposal. Cell 153, 362–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carraro G, Langerman J, Sabri S, Lorenzana Z, Purkayastha A, Zhang G, Konda B, Aros CJ, Calvert BA, Szymaniak A, et al. (2021). Transcriptional analysis of cystic fibrosis airways at single-cell resolution reveals altered epithelial cell states and composition. Nat Med 27, 806–814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cassandras M, Wang C, Kathiriya J, Tsukui T, Matatia P, Matthay M, Wolters P, Molofsky A, Sheppard D, Chapman H, et al. (2020). Gli1(+) mesenchymal stromal cells form a pathological niche to promote airway progenitor metaplasia in the fibrotic lung. Nat Cell Biol 22, 1295–1306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caton ML, Smith-Raska MR, and Reizis B (2007). Notch-RBP-J signaling controls the homeostasis of CD8-dendritic cells in the spleen. J Exp Med 204, 1653–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chairakaki AD, Saridaki MI, Pyrillou K, Mouratis MA, Koltsida O, Walton RP, Bartlett NW, Stavropoulos A, Boon L, Rovina N, et al. (2018). Plasmacytoid dendritic cells drive acute asthma exacerbations. J Allergy Clin Immunol 142, 542–556 e12. [DOI] [PubMed] [Google Scholar]
- Chakarov S, Lim HY, Tan L, Lim SY, See P, Lum J, Zhang XM, Foo S, Nakamizo S,Duan K, et al. (2019). Two distinct interstitial macrophage populations coexist across tissues in specific subtissular niches. Science 363. [DOI] [PubMed] [Google Scholar]
- Chapman HA, Li X, Alexander JP, Brumwell A, Lorizio W, Tan K, Sonnenberg A, Wei Y, and Vu TH (2011). Integrin alpha6beta4 identifies an adult distal lung epithelial population with regenerative potential in mice. J Clin Invest 121, 2855–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen G, Korfhagen TR, Karp CL, Impey S, Xu Y, Randell SH, Kitzmiller J, Maeda Y, Haitchi HM, Sridharan A, et al. (2014a). Foxa3 Induces Goblet Cell Metaplasia and Inhibits Innate Antiviral Immunity. Am J Resp Crit Care 189, 301–313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen G, Korfhagen TR, Xu Y, Kitzmiller J, Wert SE, Maeda Y, Gregorieff A, Clevers H, and Whitsett JA (2009). SPDEF is required for mouse pulmonary goblet cell differentiation and regulates a network of genes associated with mucus production. J Clin Invest 119, 2914–2924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen K, McAleer JP, Lin Y, Paterson DL, Zheng M, Alcorn JF, Weaver CT, and Kolls JK (2011). Th17 cells mediate clade-specific, serotype-independent mucosal immunity. Immunity 35, 997–1009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen L, Acciani T, Le Cras T, Lutzko C, and Perl AK (2012). Dynamic regulation of platelet-derived growth factor receptor alpha expression in alveolar fibroblasts during realveolarization. Am J Respir Cell Mol Biol 47, 517–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen M, Li S, Xie W, Wang B, and Chen D (2014b). Col2creer(T2), a Mouse Model for a Chondrocyte-Specific and Inducible Gene Deletion. Eur Cells Mater 28, 236–245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiu C, and Openshaw PJ (2015). Antiviral B cell and T cell immunity in the lungs. Nat Immunol 16, 18–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chung MI, Bujnis M, Barkauskas CE, Kobayashi Y, and Hogan BLM (2018). Niche-mediated BMP/SMAD signaling regulates lung alveolar stem cell proliferation and differentiation. Development 145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chung MI, and Hogan BLM (2018). Ager-CreER(T2): A New Genetic Tool for Studying Lung Alveolar Development, Homeostasis, and Repair. Am J Resp Cell Mol 59, 706–712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cieri RL (2019). Pulmonary Smooth Muscle in Vertebrates: A Comparative Review of Structure and Function. Integr Comp Biol 59, 10–28. [DOI] [PubMed] [Google Scholar]
- Cisse B, Caton ML, Lehner M, Maeda T, Scheu S, Locksley R, Holmberg D, Zweier, den Hollander NS, Kant SG, et al. (2008). Transcription factor E2–2 is an essential and specific regulator of plasmacytoid dendritic cell development. Cell 135, 37–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clair G, Reehl S, Stratton KG, Monroe ME, Tfaily MM, Ansong C, and Kyle JE (2019). Lipid Mini-On: mining and ontology tool for enrichment analysis of lipidomic data. Bioinformatics 35, 4507–4508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen M, Giladi A, Gorki AD, Solodkin DG, Zada M, Hladik A, Miklosi A, Salame TM, HαLPern KB, David E, et al. (2018). Lung Single-Cell Signaling Interaction Map Reveals Basophil Role in Macrophage Imprinting. Cell 175, 1031–1044.e18. [DOI] [PubMed] [Google Scholar]
- Constantinides MG, McDonald BD, Verhoef PA, and Bendelac A (2014). A committed precursor to innate lymphoid cells. Nature 508, 397–401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corada M, Morini MF, and Dejana E (2014). Signaling Pathways in the Specification of Arteries and Veins. Arterioscl Throm Vas 34, 2372–2377. [DOI] [PubMed] [Google Scholar]
- Cuervo H, Pereira B, Nadeem T, Lin M, Lee F, Kitajewski J, and Lin CS (2017). PDGFRbeta-P2A-CreER(T2) mice: a genetic tool to target pericytes in angiogenesis. Angiogenesis 20, 655–662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui Y, Liu KF, Lamattina AM, Visner G, and El-Chemaly S (2017). Lymphatic Vessels: The Next Frontier in Lung Transplant. Ann Am Thorac Soc 14, S226–S232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dahlgren MW, Jones SW, Cautivo KM, Dubinin A, Ortiz-Carpena JF, Farhat S, Yu KS, Lee K, Wang C, Molofsky AV, et al. (2019). Adventitial Stromal Cells Define Group 2 Innate Lymphoid Cell Tissue Niches. Immunity 50, 707–722 e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de la Cuesta F, Passalacqua I, Rodor J, Bhushan R, Denby L, and Baker AH (2019). Extracellular vesicle cross-talk between pulmonary artery smooth muscle cells and endothelium during excessive TGF-beta signalling: implications for PAH vascular remodelling. Cell Commun Signal 17, 143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Souza Junior DA, Mazucato VM, Santana AC, Oliver C, and Jamur MC (2017). Mast Cells Interact with Endothelial Cells to Accelerate In Vitro Angiogenesis. Int J Mol Sci 18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Val S, and Black BL (2009). Transcriptional Control of Endothelial Cell Development. Dev Cell 16, 180–195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deprez M, Zaragosi LE, Truchi M, Becavin C, Ruiz Garcia S, Arguel MJ, Plaisant M, Magnone V, Lebrigand K, Abelanet S, et al. (2020). A Single-Cell Atlas of the Human Healthy Airways. Am J Respir Crit Care Med 202, 1636–1645. [DOI] [PubMed] [Google Scholar]
- Desai TJ, Brownfield DG, and Krasnow MA (2014). Alveolar progenitor and stem cells in lung development, renewal and cancer. Nature 507, 190–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Stefano A, Caramori G, Gnemmi I, Contoli M, Vicari C, Capelli A, Magno F, D’Anna SE, Zanini A, Brun P, et al. (2009). T helper type 17-related cytokine expression is increased in the bronchial mucosa of stable chronic obstructive pulmonary disease patients. Clin Exp Immunol 157, 316–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dobbs LG, Gonzalez RF, Allen L, and Froh DK (1999). HTI56, an integral membrane protein specific to human alveolar type I cells. J Histochem Cytochem 47, 129–37. [DOI] [PubMed] [Google Scholar]
- Draijer C, Boorsma CE, Robbe P, Timens W, Hylkema MN, Ten Hacken NH, van den Berge M, Postma DS, and Melgert BN (2017). Human asthma is characterized by more IRF5+ M1 and CD206+ M2 macrophages and less IL-10+ M2-like macrophages around airways compared with healthy airways. J Allergy Clin Immunol 140, 280–283 e3. [DOI] [PubMed] [Google Scholar]
- Dress RJ, Dutertre CA, Giladi A, Schlitzer A, Low I, Shadan NB, Tay A, Lum J, Kairi M, Hwang YY, et al. (2019). Plasmacytoid dendritic cells develop from Ly6D(+) lymphoid progenitors distinct from the myeloid lineage. Nat Immunol 20, 852–864. [DOI] [PubMed] [Google Scholar]
- Du Y, Clair GC, Al Alam D, Danopoulos S, Schnell D, Kitzmiller JA, Misra RS, Bhattacharya S, Warburton D, Mariani TJ, et al. (2019). Integration of transcriptomic and proteomic data identifies biological functions in cell populations from human infant lung. Am J Physiol Lung Cell Mol Physiol 317, L347–L360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dzierzak E, and Bigas A (2018). Blood Development: Hematopoietic Stem Cell Dependence and Independence. Cell Stem Cell 22, 639–651. [DOI] [PubMed] [Google Scholar]
- Eberl G, Colonna M, Di Santo JP, and McKenzie AN (2015a). Innate lymphoid cells. Innate lymphoid cells: a new paradigm in immunology. Science 348, aaa6566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eberl G, Di Santo JP, and Vivier E (2015b). The brave new world of innate lymphoid cells. Nat Immunol 16, 1–5. [DOI] [PubMed] [Google Scholar]
- Eberl G, Marmon S, Sunshine MJ, Rennert PD, Choi Y, and Littman DR (2004). An essential function for the nuclear receptor RORgamma(t) in the generation of fetal lymphoid tissue inducer cells. Nat Immunol 5, 64–73. [DOI] [PubMed] [Google Scholar]
- Ehling M, Adams S, Benedito R, and Adams RH (2013). Notch controls retinal blood vessel maturation and quiescence. Development 140, 3051–61. [DOI] [PubMed] [Google Scholar]
- El Agha E, Moiseenko A, Kheirollahi V, De Langhe S, Crnkovic S, Kwapiszewska G, Szibor M, Kosanovic D, Schwind F, Schermuly RT, et al. (2017). Two-Way Conversion between Lipogenic and Myogenic Fibroblastic Phenotypes Marks the Progression and Resolution of Lung Fibrosis. Cell Stem Cell 20, 571. [DOI] [PubMed] [Google Scholar]
- Ellis LV, Cain MP, Hutchison V, Flodby P, Crandall ED, Borok Z, Zhou B, Ostrin EJ, Wythe JD, and Chen JC (2020). Epithelial Vegfa Specifies a Distinct Endothelial Population in the Mouse Lung. Dev Cell 52, 617-+. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Endale M, Ahlfeld S, Bao E, Chen X, Green J, Bess Z, Weirauch MT, Xu Y, and Perl AK (2017). Temporal, spatial, and phenotypical changes of PDGFR alpha expressing fibroblasts during late lung development. Dev Biol 425, 161–175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engelhardt JF, Schlossberg H, Yankaskas JR, and Dudus L (1995). Progenitor cells of the adult human airway involved in submucosal gland development. Development 121, 2031–46. [DOI] [PubMed] [Google Scholar]
- Engelhardt JF, Yankaskas JR, Ernst SA, Yang Y, Marino CR, Boucher RC, Cohn JA, and Wilson JM (1992). Submucosal glands are the predominant site of CFTR expression in the human bronchus. Nat Genet 2, 240–8. [DOI] [PubMed] [Google Scholar]
- Ferone G, Lee MC, Sage J, and Berns A (2020). Cells of origin of lung cancers: lessons from mouse studies. Genes Dev 34, 1017–1032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finkbeiner WE, Zlock LT, Mehdi I, and Widdicombe JH (2010). Cultures of human tracheal gland cells of mucous or serous phenotype. In Vitro Cell Dev Biol Anim 46, 450–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flodby P, Borok Z, Banfalvi A, Zhou BY, Gao DP, Minoo P, Ann DK, Morrisey EE, and Crandall ED (2010). Directed Expression of Cre in Alveolar Epithelial Type 1 Cells. Am J Resp Cell Mol 43, 173–178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fortunati E, Kazemier KM, Grutters JC, Koenderman L, and Van den Bosch VMM (2009). Human neutrophils switch to an activated phenotype after homing to the lung irrespective of inflammatory disease. Clin Exp Immunol 155, 559–566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frank DB, Penkala IJ, Zepp JA, Sivakumar A, Linares-Saldana R, Zacharias WJ, Stolz KG, Pankina J, Lu MQ, Wang QH, et al. (2019). Early lineage specification defines alveolar epithelial ontogeny in the murine lung. P Natl Acad Sci USA 116, 4362–4371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franks TJ, Colby TV, Travis WD, Tuder RM, Reynolds HY, Brody AR, Cardoso WV, Crystal RG, Drake CJ, Engelhardt J, et al. (2008). Resident cellular components of the human lung: current knowledge and goals for research on cell phenotyping and function. Proc Am Thorac Soc 5, 763–6. [DOI] [PubMed] [Google Scholar]
- Gasteiger G, Fan X, Dikiy S, Lee SY, and Rudensky AY (2015). Tissue residency of innate lymphoid cells in lymphoid and nonlymphoid organs. Science 350, 981–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerbe F, Brulin B, Makrini L, Legraverend C, and Jay P (2009). DCAMKL-1 expression identifies Tuft cells rather than stem cells in the adult mouse intestinal epithelium. Gastroenterology 137, 2179–80; author reply 2180–1. [DOI] [PubMed] [Google Scholar]
- GeurtsvanKessel CH, and Lambrecht BN (2008). Division of labor between dendritic cell subsets of the lung. Mucosal Immunol 1, 442–50. [DOI] [PubMed] [Google Scholar]
- Ghosn E, Yoshimoto M, Nakauchi H, Weissman IL, and Herzenberg LA (2019). Hematopoietic stem cell-independent hematopoiesis and the origins of innate-like B lymphocytes. Development 146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giangreco A, Arwert EN, Rosewell IR, Snyder J, Watt FM, and Stripp BR (2009). Stem cells are dispensable for lung homeostasis but restore airways after injury. Proc Natl Acad Sci U S A 106, 9286–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibbings SL, Thomas SM, Atif SM, McCubbrey AL, Desch AN, Danhorn T, Leach SM, Bratton DL, Henson PM, Janssen WJ, et al. (2017). Three Unique Interstitial Macrophages in the Murine Lung at Steady State. Am J Respir Cell Mol Biol 57, 66–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gillich A, Zhang F, Farmer CG, Travaglini KJ, Tan SY, Gu M, Zhou B, Feinstein JA, Krasnow MA, and Metzger RJ (2020). Capillary cell-type specialization in the alveolus. Nature 586, 785–789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ginhoux F, and Guilliams M (2016). Tissue-Resident Macrophage Ontogeny and Homeostasis. Immunity 44, 439–449. [DOI] [PubMed] [Google Scholar]
- Goldfarbmuren KC, Jackson ND, Sajuthi SP, Dyjack N, Li KS, Rios CL, Plender EG, Montgomery MT, Everman JL, Bratcher PE, et al. (2020). Dissecting the cellular specificity of smoking effects and reconstructing lineages in the human airway epithelium. Nat Commun 11, 2485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gomez Perdiguero E, Klapproth K, Schulz C, Busch K, Azzoni E, Crozet L, Garner H, Trouillet C, de Bruijn MF, Geissmann F, et al. (2015). Tissue-resident macrophages originate from yolk-sac-derived erythro-myeloid progenitors. Nature 518, 547–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goss AM, Tian Y, Cheng L, Yang J, Zhou D, Cohen ED, and Morrisey EE (2011). Wnt2 signaling is necessary and sufficient to activate the airway smooth muscle program in the lung by regulating myocardin/Mrtf-B and Fgf10 expression. Dev Biol 356, 541–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grasseau A, Boudigou M, Le Pottier L, Chriti N, Cornec D, Pers JO, Renaudineau Y, and Hillion S (2020). Innate B Cells: the Archetype of Protective Immune Cells. Clin Rev Allergy Immunol 58, 92–106. [DOI] [PubMed] [Google Scholar]
- Green FH, Williams DJ, James A, McPhee LJ, Mitchell I, and Mauad T (2010). Increased myoepithelial cells of bronchial submucosal glands in fatal asthma. Thorax 65, 32–8. [DOI] [PubMed] [Google Scholar]
- Greif DM, Kumar M, Lighthouse JK, Hum J, An A, Ding L, Red-Horse K, Espinoza FH, Olson L, Offermanns S, et al. (2012). Radial construction of an arterial wall. Dev Cell 23, 482–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guha A, Vasconcelos M, Cai Y, Yoneda M, Hinds A, Qian J, Li GH, Dickel L, Johnson JE, Kimura S, et al. (2012). Neuroepithelial body microenvironment is a niche for a distinct subset of Clara-like precursors in the developing airways. P Natl Acad Sci USA 109, 12592–12597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guilliams M, De Kleer I, Henri S, Post S, Vanhoutte L, De Prijck S, Deswarte K, Malissen B, Hammad H, and Lambrecht BN (2013). Alveolar macrophages develop from fetal monocytes that differentiate into long-lived cells in the first week of life via GM-CSF. J Exp Med 210, 1977–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Habermann AC, Gutierrez AJ, Bui LT, Yahn SL, Winters NI, Calvi CL, Peter L, Chung M-I, Taylor CJ, Jetter C, et al. (2019). Single-cell RNA-sequencing reveals profibrotic roles of distinct epithelial and mesenchymal lineages in pulmonary fibrosis. bioRxiv, 753806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Habermann AC, Gutierrez AJ, Bui LT, Yahn SL, Winters NI, Calvi CL, Peter L, Chung MI, Taylor CJ, Jetter C, et al. (2020). Single-cell RNA sequencing reveals profibrotic roles of distinct epithelial and mesenchymal lineages in pulmonary fibrosis. Sci Adv 6, eaba1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hagan AS, Boylan M, Smith C, Perez-Santamarina E, Kowalska K, Hung IH, Lewis RM, Hajihosseini MK, Lewandoski M, and Ornitz DM (2019a). Generation and validation of novel conditional flox and inducible Cre alleles targeting fibroblast growth factor 18 (Fgf18). Dev Dynam 248, 882–893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hagan AS, Zhang B, and Ornitz DM (2019b). Identification of an FGF18-expressing alveolar myofibroblast that is developmentally cleared during alveologenesis. Development. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halai K, Whiteford J, Ma B, Nourshargh S, and Woodfin A (2014). ICAM-2 facilitates luminal interactions between neutrophils and endothelial cells in vivo. J Cell Sci 127, 620–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hao Y, Hao S, Andersen-Nissen E, Mauck WM, Zheng S, Butler A, Lee MJ, Wilk AJ, Darby C, Zagar M, et al. (2020). Integrated analysis of multimodal single-cell data. bioRxiv, 2020.10.12.335331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hawkins FJ, Suzuki S, Beermann ML, Barilla C, Wang R, Villacorta-Martin C, Berical A, Jean JC, Le Suer J, Matte T, et al. (2020). Derivation of Airway Basal Stem Cells from Human Pluripotent Stem Cells. Cell Stem Cell. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hegab AE, Ha VL, Darmawan DO, Gilbert JL, Ooi AT, Attiga YS, Bisht B, Nickerson DW, and Gomperts BN (2012a). Isolation and in vitro characterization of basal and submucosal gland duct stem/progenitor cells from human proximal airways. Stem Cells Transl Med 1, 719–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hegab AE, Ha VL, Gilbert JL, Zhang KX, Malkoski SP, Chon AT, Darmawan DO, Bisht B, Ooi AT, Pellegrini M, et al. (2011). Novel stem/progenitor cell population from murine tracheal submucosal gland ducts with multipotent regenerative potential. Stem Cells 29, 1283–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hegab AE, Nickerson DW, Ha VL, Darmawan DO, and Gomperts BN (2012b). Repair and regeneration of tracheal surface epithelium and submucosal glands in a mouse model of hypoxic-ischemic injury. Respirology 17, 1101–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hines EA, Jones MKN, Verheyden JM, Harvey JF, and Sun X (2013). Establishment of smooth muscle and cartilage juxtaposition in the developing mouse upper airways. P Natl Acad Sci USA 110, 19444–19449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hines EA, Jones MN, Harvey JF, Perlyn C, Ornitz DM, Sun X, and Verheyden JM (2019). Crouzon syndrome mouse model exhibits cartilage hyperproliferation and defective segmentation in the developing trachea. Sci China Life Sci 62, 1375–1380. [DOI] [PubMed] [Google Scholar]
- Hoeffel G, Chen J, Lavin Y, Low D, Almeida FF, See P, Beaudin AE, Lum J, Low I, Forsberg EC, et al. (2015). C-Myb(+) erythro-myeloid progenitor-derived fetal monocytes give rise to adult tissue-resident macrophages. Immunity 42, 665–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hogan BL, Barkauskas CE, Chapman HA, Epstein JA, Jain R, Hsia CC, Niklason L, Calle E, Le A, Randell SH, et al. (2014). Repair and regeneration of the respiratory system: complexity, plasticity, and mechanisms of lung stem cell function. Cell Stem Cell 15, 123–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holt PG, and Jones CA (2000). The development of the immune system during pregnancy and early life. Allergy 55, 688–97. [DOI] [PubMed] [Google Scholar]
- Hong KU, Reynolds SD, Giangreco A, Hurley CM, and Stripp BR (2001). Clara cell secretory protein-expressing cells of the airway neuroepithelial body microenvironment include a label-retaining subset and are critical for epithelial renewal after progenitor cell depletion. Am J Resp Cell Mol 24, 671–681. [DOI] [PubMed] [Google Scholar]
- Hong KU, Reynolds SD, Watkins S, Fuchs E, and Stripp BR (2004). Basal cells are a multipotent progenitor capable of renewing the bronchial epithelium. Am J Pathol 164, 577–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang Y, Guo L, Qiu J, Chen X, Hu-Li J, Siebenlist U, Williamson PR, Urban JF Jr., and Paul WE (2015). IL-25-responsive, lineage-negative KLRG1(hi) cells are multipotential ‘inflammatory’ type 2 innate lymphoid cells. Nat Immunol 16, 161–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang Y, Mao K, Chen X, Sun MA, Kawabe T, Li W, Usher N, Zhu J, Urban JF Jr., Paul WE, et al. (2018). S1P-dependent interorgan trafficking of group 2 innate lymphoid cells supports host defense. Science 359, 114–119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hume PS, Gibbings SL, Jakubzick CV, Tuder RM, Curran-Everett D, Henson PM, Smith BJ, and Janssen WJ (2020). Localization of Macrophages in the Human Lung via Design-based Stereology. Am J Respir Crit Care Med 201, 1209–1217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hung C, Linn G, Chow YH, Kobayashi A, Mittelsteadt K, Altemeier WA, Gharib SA, Schnapp LM, and Duffield JS (2013). Role of Lung Pericytes and Resident Fibroblasts in the Pathogenesis of Pulmonary Fibrosis. Am J Resp Crit Care 188, 820–830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hung CF, Wilson CL, and Schnapp LM (2019). Pericytes in the Lung. Adv Exp Med Biol 1122, 41–58. [DOI] [PubMed] [Google Scholar]
- Imanishi T, and Saito T (2020). T Cell Co-stimulation and Functional Modulation by Innate Signals. Trends Immunol 41, 200–212. [DOI] [PubMed] [Google Scholar]
- Inclan-Rico JM, Ponessa JJ, Valero-Pacheco N, Hernandez CM, Sy CB, Lemenze AD, Beaulieu AM, and Siracusa MC (2020). Basophils prime group 2 innate lymphoid cells for neuropeptide-mediated inhibition. Nature Immunology 21, 1181–1193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ingram DA, Mead LE, Moore DB, Woodard W, Fenoglio A, and Yoder MC (2005). Vessel wall-derived endothelial cells rapidly proliferate because they contain a complete hierarchy of endothelial progenitor cells. Blood 105, 2783–2786. [DOI] [PubMed] [Google Scholar]
- Itkin M, and McCormack FX (2016). Nonmalignant Adult Thoracic Lymphatic Disorders. Clin Chest Med 37, 409-+. [DOI] [PubMed] [Google Scholar]
- Ivanovs A, Rybtsov S, Ng ES, Stanley EG, Elefanty AG, and Medvinsky A (2017). Human haematopoietic stem cell development: from the embryo to the dish. Development 144, 2323–2337. [DOI] [PubMed] [Google Scholar]
- Jain R, Barkauskas CE, Takeda N, Bowie EJ, Aghajanian H, Wang Q, Padmanabhan A, Manderfield LJ, Gupta M, Li D, et al. (2015). Plasticity of Hopx(+) type I alveolar cells to regenerate type II cells in the lung. Nat Commun 6, 6727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jakubzick C, Gautier EL, Gibbings SL, Sojka DK, Schlitzer A, Johnson TE, Ivanov S, Duan Q, Bala S, Condon T, et al. (2013). Minimal differentiation of classical monocytes as they survey steady-state tissues and transport antigen to lymph nodes. Immunity 39, 599–610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kabashima K, Nakashima C, Nonomura Y, Otsuka A, Cardamone C, Parente R, De Feo G, and Triggiani M (2018). Biomarkers for evaluation of mast cell and basophil activation. Immunol Rev 282, 114–120. [DOI] [PubMed] [Google Scholar]
- Kalna V, Yang YW, Peghaire CR, Frudd K, Hannah R, Shah AV, Almagro LO, Boyle JJ, Gottgens B, Ferrer J, et al. (2019). The Transcription Factor ERG Regulates Super-Enhancers Associated With an Endothelial-Specific Gene Expression Program. Circ Res 124, 1337–1349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalucka J, de Rooij LPMH, Goveia J, Rohlenova K, Dumas SJ, Meta E, Conchinha NV, Taverna F, Teuwen LA, Veys K, et al. (2020). Single-Cell Transcriptome Atlas of Murine Endothelial Cells. Cell 180, 764-+. [DOI] [PubMed] [Google Scholar]
- Kathiriya JJ, Brumwell AN, Jackson JR, Tang X, and Chapman HA (2020). Distinct Airway Epithelial Stem Cells Hide among Club Cells but Mobilize to Promote Alveolar Regeneration. Cell Stem Cell 26, 346–358 e4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kato A, Hulse KE, Tan BK, and Schleimer RP (2013). B-lymphocyte lineage cells and the respiratory system. J Allergy Clin Immunol 131, 933–57; quiz 958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kato K, Dieguez-Hurtado R, Park DY, Hong SP, Kato-Azuma S, Adams S, Stehling M, Trappmann B, Wrana JL, Koh GY, et al. (2018). Pulmonary pericytes regulate lung morphogenesis. Nature Communications 9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katzen J, and Beers MF (2020). Contributions of alveolar epithelial cell quality control to pulmonary fibrosis. J Clin Invest 130, 5088–5099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khan MA (2020). Regulatory T cells mediated immunomodulation during asthma: a therapeutic standpoint. J Transl Med 18, 456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khansaheb M, Choi JY, Joo NS, Yang YM, Krouse M, and Wine JJ (2011). Properties of substance P-stimulated mucus secretion from porcine tracheal submucosal glands. Am J Physiol Lung Cell Mol Physiol 300, L370–9. [DOI] [PubMed] [Google Scholar]
- Kim CFB, Jackson EL, Woolfenden AE, Lawrence S, Babar I, Vogel S, Crowley D, Bronson RT, and Jacks T (2005). Identification of bronchioalveolar stem cells in normal lung and lung cancer. Cell 121, 823–835. [DOI] [PubMed] [Google Scholar]
- Kim EJ, Ables JL, Dickel LK, Eisch AJ, and Johnson JE (2011). Ascl1 (Mash1) Defines Cells with Long-Term Neurogenic Potential in Subgranular and Subventricular Zones in Adult Mouse Brain. Plos One 6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimura S, Hara Y, Pineau T, FernandezSalguero P, Fox CH, Ward JM, and Gonzalez J (1996). The T/ebp null mouse thyroid-specific enhancer-binding protein is essential for the organogenesis of the thyroid, lung, ventral forebrain, and pituitary. Gene Dev 10, 60–69. [DOI] [PubMed] [Google Scholar]
- Kiyokawa H, Yamaoka A, Matsuoka C, Tokuhara T, Abe T, and Morimoto M (2021). Airway basal stem cells reutilize the embryonic proliferation regulator, Tgfbeta-Id2 axis, for tissue regeneration. Dev Cell 56, 1917–1929 e9. [DOI] [PubMed] [Google Scholar]
- Klose CS, Kiss EA, Schwierzeck V, Ebert K, Hoyler T, d’Hargues Y, Goppert N, Croxford AL, Waisman A, Tanriver Y, et al. (2013). A T-bet gradient controls the fate and function of CCR6-RORgammat+ innate lymphoid cells. Nature 494, 261–5. [DOI] [PubMed] [Google Scholar]
- Komi DEA, Mortaz E, Amani S, Tiotiu A, Folkerts G, and Adcock IM (2020). The Role of Mast Cells in IgE-Independent Lung Diseases. Clin Rev Allergy Immunol 58, 377–387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuo CS, and Krasnow MA (2015). Formation of a Neurosensory Organ by Epithelial Cell Slithering. Cell 163, 394–405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kyle JE, Clair G, Bandyopadhyay G, Misra RS, Zink EM, Bloodsworth KJ, Shukla AK, Du Y, Lillis J, Myers JR, et al. (2018). Cell type-resolved human lung lipidome reveals cellular cooperation in lung function. Sci Rep 8, 13455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lam JH, and Baumgarth N (2019). The Multifaceted B Cell Response to Influenza Virus. J Immunol 202, 351–359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lechner AJ, Driver IH, Lee J, Conroy CM, Nagle A, Locksley RM, and Rock JR (2017). Recruited Monocytes and Type 2 Immunity Promote Lung Regeneration following Pneumonectomy. Cell Stem Cell 21, 120–134 e7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee DK, Liu Y, Liao L, Wang F, and Xu J (2014). The prostate basal cell (BC) heterogeneity and the p63-positive BC differentiation spectrum in mice. Int J Biol Sci 10, 1007–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee JH, Tammela T, Hofree M, Choi J, Marjanovic ND, Han S, Canner D, Wu K, Paschini M, Bhang DH, et al. (2017). Anatomically and Functionally Distinct Lung Mesenchymal Populations Marked by Lgr5 and Lgr6. Cell 170, 1149–1163 e12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee JJ, and McGarry MP (2007). When is a mouse basophil not a basophil? Blood 109, 859–861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee RJ, and Foskett JK (2010). Mechanisms of Ca2+-stimulated fluid secretion by porcine bronchial submucosal gland serous acinar cells. Am J Physiol Lung Cell Mol Physiol 298, L210–31. [DOI] [PubMed] [Google Scholar]
- Li C, Li M, Li S, Xing Y, Yang CY, Li A, Borok Z, De Langhe S, and Minoo P (2015). Progenitors of secondary crest myofibroblasts are developmentally committed in early lung mesoderm. Stem Cells 33, 999–1012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li R, Bernau K, Sandbo N, Gu J, Preissl S, and Sun X (2018). Pdgfra marks a cellular lineage with distinct contributions to myofibroblasts in lung maturation and injury response. Elife 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao WP, Uetzmann L, Burtscher I, and Lickert H (2009). Generation of a Mouse Line Expressing Sox17-Driven Cre Recombinase With Specific Activity in Arteries. Genesis 47, 476–483. [DOI] [PubMed] [Google Scholar]
- Liu QZ, Liu K, Cui GZ, Huang XZ, Yao S, Guo WK, Qin Z, Li Y, Yang R, Pu WJ, et al. (2019). Lung regeneration by multipotent stem cells residing at the bronchioalveolarduct junction (vol 51, pg 728, 2019). Nat Genet 51, 766–766. [DOI] [PubMed] [Google Scholar]
- Lynch TJ, Anderson PJ, Rotti PG, Tyler SR, Crooke AK, Choi SH, Montoro DT, Silverman CL, Shahin W, Zhao R, et al. (2018). Submucosal Gland Myoepithelial Cells Are Reserve Stem Cells That Can Regenerate Mouse Tracheal Epithelium. Cell Stem Cell 22, 653–667 e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lynch TJ, Anderson PJ, Xie W, Crooke AK, Liu X, Tyler SR, Luo M, Kusner DM, Zhang Y, Neff T, et al. (2016). Wnt Signaling Regulates Airway Epithelial Stem Cells in Adult Murine Submucosal Glands. Stem Cells 34, 2758–2771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahlakoiv T, Flamar AL, Johnston LK, Moriyama S, Putzel GG, Bryce PJ, and Artis D (2019). Stromal cells maintain immune cell homeostasis in adipose tissue via production of interleukin-33. Sci Immunol 4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marone G, Schroeder JT, Mattei F, Loffredo S, Gambardella AR, Poto R, de Paulis A, Schiavoni G, and Varricchi G (2020). Is There a Role for Basophils in Cancer? Front Immunol 11, 2103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuba K, Takizawa T, and Thurlbeck WM (1972). Oncocytes in human bronchial mucous glands. Thorax 27, 181–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mazzurana L, Czarnewski P, Jonsson V, Wigge L, Ringner M, Williams TC, Ravindran A, Bjorklund AK, Safholm J, Nilsson G, et al. (2021). Tissue-specific transcriptional imprinting and heterogeneity in human innate lymphoid cells revealed by full-length singlecell RNA-sequencing. Cell Res. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCauley KB, Alysandratos K-DD, Jacob A, Hawkins F, Caballero IS, Vedaie M, Yang W, Slovik KJ, Morley M, Carraro G, et al. (2018). Single-Cell Transcriptomic Profiling of Pluripotent Stem Cell-Derived SCGB3A2+ Airway Epithelium. Stem cell reports 10, 1579–1595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGinty JW, Ting HA, Billipp TE, Nadjsombati MS, Khan DM, Barrett NA, Liang HE, Matsumoto I, and von Moltke J (2020). Tuft-Cell-Derived Leukotrienes Drive Rapid Anti-helminth Immunity in the Small Intestine but Are Dispensable for Anti-protist Immunity. Immunity 52, 528-+. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meehan TF, Masci AM, Abdulla A, Cowell LG, Blake JA, Mungall CJ, and Diehl AD (2011). Logical development of the cell ontology. BMC Bioinformatics 12, 6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyrick B, and Reid L (1970). Ultrastructure of cells in the human bronchial submucosal glands. J Anat 107, 281–99. [PMC free article] [PubMed] [Google Scholar]
- Meyrick B, Sturgess JM, and Reid L (1969). A reconstruction of the duct system and secretory tubules of the human bronchial submucosal gland. Thorax 24, 729–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minoo P, Su G, Drum H, Bringas P, and Kimura S (1999). Defects in tracheoesophageal and lung morphogenesis in Nkx2.1(−/−) mouse embryos. Dev Biol 209, 60–71. [DOI] [PubMed] [Google Scholar]
- Misharin AV, Morales-Nebreda L, Mutlu GM, Budinger GR, and Perlman H (2013). Flow cytometric analysis of macrophages and dendritic cell subsets in the mouse lung. Am J Respir Cell Mol Biol 49, 503–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Misharin AV, Morales-Nebreda L, Reyfman PA, Cuda CM, Walter JM, McQuattie-Pimentel AC, Chen CI, Anekalla KR, Joshi N, Williams KJN, et al. (2017). Monocyte-derived alveolar macrophages drive lung fibrosis and persist in the lung over the life span. J Exp Med 214, 2387–2404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moiseenko A, Kheirollahi V, Chao CM, Ahmadvand N, Quantius J, Wilhelm J, Herold S,Ahlbrecht K, Morty RE, Rizvanov AA, et al. (2017). Origin and characterization of alpha smooth muscle actin-positive cells during murine lung development. Stem Cells 35, 1566–1578. [DOI] [PubMed] [Google Scholar]
- Monticelli LA, Sonnenberg GF, Abt MC, Alenghat T, Ziegler CG, Doering TA, Angelosanto JM, Laidlaw BJ, Yang CY, Sathaliyawala T, et al. (2011). Innate lymphoid cells promote lung-tissue homeostasis after infection with influenza virus. Nat Immunol 12, 1045–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montoro DT, Haber AL, Biton M, Vinarsky V, Lin B, Birket SE, Yuan F, Chen S, Leung HM, Villoria J, et al. (2018). A revised airway epithelial hierarchy includes CFTR-expressing ionocytes. Nature 560, 319–324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mou H, Vinarsky V, Tata PR, Brazauskas K, Choi SH, Crooke AK, Zhang B,Solomon GM, Turner B, Bihler H, et al. (2016). Dual SmAD Signaling Inhibition Enables Long-Term Expansion of Diverse Epithelial Basal Cells. Cell Stem Cell 19, 217–231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mould KJ, Jackson ND, Henson PM, Seibold M, and Janssen WJ (2019). Single cell RNA sequencing identifies unique inflammatory airspace macrophage subsets. JCI Insight 4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Musumeci A, Lutz K, Winheim E, and Krug AB (2019). What Makes a pDC: Recent Advances in Understanding Plasmacytoid DC Development and Heterogeneity. Front Immunol 10, 1222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nabhan AN, Brownfield DG, Harbury PB, Krasnow MA, and Desai TJ (2018). Singlecell Wnt signaling niches maintain stemness of alveolar type 2 cells. Science 359, 1118–1123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakano T, Sonoda T, Hayashi C, Yamatodani A, Kanayama Y, Yamamura T, Asai H, Yonezawa T, Kitamura Y, and Galli SJ (1985). Fate of bone marrow-derived cultured mast cells after intracutaneous, intraperitoneal, and intravenous transfer into genetically mast cell-deficient W/Wv mice. Evidence that cultured mast cells can give rise to both connective tissue type and mucosal mast cells. J Exp Med 162, 1025–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakayasu ES, Nicora CD, Sims AC, Burnum-Johnson KE, Kim YM, Kyle JE, Matzke MM, Shukla AK, Chu RK, Schepmoes AA, et al. (2016). MPLEx: a Robust and Universal Protocol for Single-Sample Integrative Proteomic, Metabolomic, and Lipidomic Analyses. mSystems 1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naski MC, Colvin JS, Coffin JD, and Ornitz DM (1998). Repression of hedgehog signaling and BMP4 expression in growth plate cartilage by fibroblast growth factor receptor 3. Development 125, 4977–88. [DOI] [PubMed] [Google Scholar]
- Neal A, Nornes S, Payne S, Wallace MD, Fritzsche M, Louphrasitthiphol P, Wilkinson RN, Chouliaras KM, Liu K, Plant K, et al. (2019). Venous identity requires BMP signalling through ALK3. Nature Communications 10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ng LG, Ostuni R, and Hidalgo A (2019). Heterogeneity of neutrophils. Nature Reviews Immunology 19, 255–265. [DOI] [PubMed] [Google Scholar]
- Niethamer TK, Stabler CT, Leach JP, Zepp JA, Morley MP, Babu A, Zhou S, and Morrisey EE (2020). Defining the role of pulmonary endothelial cell heterogeneity in the response to acute lung injury. Elife 9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noguchi M, Sumiyama K, and Morimoto M (2015). Directed Migration of Pulmonary Neuroendocrine Cells toward Airway Branches Organizes the Stereotypic Location of Neuroepithelial Bodies. Cell Rep 13, 2679–86. [DOI] [PubMed] [Google Scholar]
- Ntokou A, Klein F, Dontireddy D, Becker S, Bellusci S, Richardson WD, Szibor M, Braun T, Morty RE, Seeger W, et al. (2015). Characterization of the platelet-derived growth factor receptor-alpha-positive cell lineage during murine late lung development. Am J Physiol Lung Cell Mol Physiol 309, L942–58. [DOI] [PubMed] [Google Scholar]
- Nureki SI, Tomer Y, Venosa A, Katzen J, Russo SJ, Jamil S, Barrett M, Nguyen V, Kopp M, Mulugeta S, et al. (2018). Expression of mutant Sftpc in murine alveolar epithelia drives spontaneous lung fibrosis. J Clin Invest 128, 4008–4024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ochs M, Nyengaard JR, Jung A, Knudsen L, Voigt M, Wahlers T, Richter J, and Gundersen HJ (2004). The number of alveoli in the human lung. Am J Respir Crit Care Med 169, 120–4. [DOI] [PubMed] [Google Scholar]
- Ogrinc G, Kampalath B, and Tomashefski JF (1998). Destruction and loss of bronchial cartilage in cystic fibrosis. Hum Pathol 29, 65–73. [DOI] [PubMed] [Google Scholar]
- Oherle K, Acker E, Bonfield M, Wang T, Gray J, Lang I, Bridges J, Lewkowich I, Xu Y, Ahlfeld S, et al. (2020). Insulin-like Growth Factor 1 Supports a Pulmonary Niche that Promotes Type 3 Innate Lymphoid Cell Development in Newborn Lungs. Immunity 52, 275294 e9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okuda K, Chen G, Subramani DB, Wolf M, Gilmore RC, Kato T, Radicioni G,Kesimer M, Chua M, Dang H, et al. (2019). Localization of Secretory Mucins MUC5AC and MUC5B in Normal/Healthy Human Airways. Am J Respir Crit Care Med 199, 715–727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olson LE, and Soriano P (2011). PDGFRbeta signaling regulates mural cell plasticity and inhibits fat development. Dev Cell 20, 815–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ostedgaard LS, Moninger TO, McMenimen JD, Sawin NM, Parker CP, Thornell IM, Powers LS, Gansemer ND, Bouzek DC, Cook DP, et al. (2017). Gel-forming mucins form distinct morphologic structures in airways. Proc Natl Acad Sci U S A 114, 6842–6847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ouadah Y, Rojas ER, Riordan DP, Capostagno S, Kuo CS, and Krasnow MA (2019). Rare Pulmonary Neuroendocrine Cells Are Stem Cells Regulated by Rb, p53, and Notch. Cell 179, 403–416 e23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paik DT, Tian L, Williams IM, Rhee S, Zhang H, Liu C, Mishra R, Wu SM, Red-Horse K, and Wu JC (2020). Single-Cell RNA Sequencing Unveils Unique Transcriptomic Signatures of Organ-Specific Endothelial Cells. Circulation 142, 1848–1862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pardo-Saganta A, Tata PR, Law BM, Saez B, Chow RDW, Prabhu M, Gridley T, and Rajagopal J (2015). Parent stem cells can serve as niches for their daughter cells. Nature 523, 597-+. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park J, Ivey MJ, Deana Y, Riggsbee KL, Sorensen E, Schwabl V, Sjoberg C, Hjertberg T, Park GY, Swonger JM, et al. (2019). The Tcf21 lineage constitutes the lung lipofibroblast population. Am J Physiol Lung Cell Mol Physiol 316, L872–L885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park JE, Jardine L, Gottgens B, Teichmann SA, and Haniffa M (2020). Prenatal development of human immunity. Science 368, 600–603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patel VI, and Metcalf JP (2018). Airway Macrophage and Dendritic Cell Subsets in the Resting Human Lung. Crit Rev Immunol 38, 303–331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paul AGA, Muehling LM, Eccles JD, and Woodfolk JA (2019). T cells in severe childhood asthma. Clin Exp Allergy 49, 564–581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng T, Tian Y, Boogerd CJ, Lu MM, Kadzik RS, Stewart KM, Evans SM, and Morrisey EE (2013). Coordination of heart and lung co-development by a multipotent cardiopulmonary progenitor. Nature 500, 589-+. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Penkala IJ, Liberti DC, Pankin J, Sivakumar A, Kremp MM, Jayachandran S, Katzen J, Leach JP, Windmueller R, Stolz K, et al. (2021). Age-dependent alveolar epithelial plasticity orchestrates lung homeostasis and regeneration. Cell Stem Cell. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plasschaert LW, Zilionis R, Choo-Wing R, Savova V, Knehr J, Roma G, Klein AM, and Jaffe AB (2018). A single-cell atlas of the airway epithelium reveals the CFTR-rich pulmonary ionocyte. Nature 560, 377-+. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Popescu DM, Botting RA, Stephenson E, Green K, Webb S, Jardine L, Calderbank EF, Polanski K, Goh I, Efremova M, et al. (2019). Decoding human fetal liver haematopoiesis. Nature 574, 365–371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Popova AP, Bentley JK, Cui TX, Richardson MN, Linn MJ, Lei J, Chen Q, Goldsmith AM, Pryhuber GS, and Hershenson MB (2014). Reduced platelet-derived growth factor receptor expression is a primary feature of human bronchopulmonary dysplasia. Am J Physiol-Lung C 307, L231–L239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Que J, Wilm B, Hasegawa H, Wang F, Bader D, and Hogan BL (2008). Mesothelium contributes to vascular smooth muscle and mesenchyme during lung development. Proc Natl Acad Sci U S A 105, 16626–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rane CK, Jackson SR, Pastore CF, Zhao G, Weiner AI, Patel NN, Herbert DR, Cohen NA, and Vaughan AE (2019). Development of solitary chemosensory cells in the distal lung after severe influenza injury. Am J Physiol-Lung C 316, L1141-L1149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raredon MSB, Adams TS, Suhail Y, Schupp JC, Poli S, Neumark N, Leiby KL, Greaney AM, Yuan Y, Horien C, et al. (2019). Single-cell connectomic analysis of adult mammalian lungs. Sci Adv 5, eaaw3851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ravindran A, Ronnberg E, Dahlin JS, Mazzurana L, Safholm J, Orre AC, Al-Ameri M, Peachell P, Adner M, Dahlen SE, et al. (2018). An Optimized Protocol for the Isolation and Functional Analysis of Human Lung Mast Cells. Front Immunol 9, 2193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rawlins EL, Clark CP, Xue Y, and Hogan BL (2009a). The Id2+ distal tip lung epithelium contains individual multipotent embryonic progenitor cells. Development 136, 3741–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rawlins EL, and Hogan BL (2005). Intercellular growth factor signaling and the development of mouse tracheal submucosal glands. Dev Dyn 233, 1378–85. [DOI] [PubMed] [Google Scholar]
- Rawlins EL, and Hogan BL (2008). Ciliated epithelial cell lifespan in the mouse trachea and lung. Am J Physiol Lung Cell Mol Physiol 295, L231–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rawlins EL, Okubo T, Xue Y, Brass DM, Auten RL, Hasegawa H, Wang F, and Hogan BL (2009b). The role of Scgb1a1+ Clara cells in the long-term maintenance and repair of lung airway, but not alveolar, epithelium. Cell Stem Cell 4, 525–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ray S, Chiba N, Yao C, Guan X, McConnell AM, Brockway B, Que L, McQualter JL, and Stripp BR (2016). Rare SOX2(+) Airway Progenitor Cells Generate KRT5(+) Cells that Repopulate Damaged Alveolar Parenchyma following Influenza Virus Infection. Stem Cell Reports 7, 817–825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reed HO, Wang LQ, Sonett J, Chen M, Yang JS, Li L, Aradi P, Jakus Z, D’Armiento J, Hancock WW, et al. (2019). Lymphatic impairment leads to pulmonary tertiary lymphoid organ formation and alveolar damage. J Clin Invest 129, 2514–2526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren XM, Ustiyan V, Guo MZ, Wang GL, Bolte C, Zhang YF, Xu Y, Whitsett JA, Kalin TV, and Kalinichenko VV (2019). Postnatal Alveologenesis Depends on FOXF1 Signaling in c-KIT+ Endothelial Progenitor Cells. Am J Resp Crit Care 200, 1164–1176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren Y, Lyu Y, Mereness JA, Wang S, Pang J, and Mariani TJ (2020). Rare Pulmonary Connective Tissue Type Mast Cells Regulate Lung Endothelial Cell Angiogenesis. Am J Pathol 190, 1763–1773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reyfman PA, Walter JM, Joshi N, Anekalla KR, McQuattie-Pimentel AC, Chiu S, Fernandez R, Akbarpour M, Chen CI, Ren Z, et al. (2019). Single-Cell Transcriptomic Analysis of Human Lung Provides Insights into the Pathobiology of Pulmonary Fibrosis. Am J Respir Crit Care Med 199, 1517–1536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riccetti M, Gokey JJ, Aronow B, and Perl AT (2020). The elephant in the lung: Integrating lineage-tracing, molecular markers, and single cell sequencing data to identify distinct fibroblast populations during lung development and regeneration. Matrix Biol 91–92, 51–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rickert RC, Roes J, and Rajewsky K (1997). B lymphocyte-specific, Cre-mediated mutagenesis in mice. Nucleic Acids Res 25, 1317–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rock JR, Barkauskas CE, Cronce MJ, Xue Y, Harris JR, Liang JR, Noble PW, and Hogan BLM (2011). Multiple stromal populations contribute to pulmonary fibrosis without evidence for epithelial to mesenchymal transition. P Natl Acad Sci USA 108, E1475-E1483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rock JR, Onaitis MW, Rawlins EL, Lu Y, Clark CP, Xue Y, Randell SH, and Hogan BLM (2009). Basal cells as stem cells of the mouse trachea and human airway epithelium. P Natl Acad Sci USA 106, 12771–12775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rock JR, Randell SH, and Hogan BL (2010). Airway basal stem cells: a perspective on their roles in epithelial homeostasis and remodeling. Dis Model Mech 3, 545–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodrigues PF, Alberti-Servera L, Eremin A, Grajales-Reyes GE, Ivanek R, and Tussiwand R (2018). Distinct progenitor lineages contribute to the heterogeneity of plasmacytoid dendritic cells. Nat Immunol 19, 711–722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosales C (2018). Neutrophil: A Cell with Many Roles in Inflammation or Several Cell Types? Front Physiol 9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sabatel C, Radermecker C, Fievez L, Paulissen G, Chakarov S, Fernandes C, Olivier S, Toussaint M, Pirottin D, Xiao X, et al. (2017). Exposure to Bacterial CpG DNA Protects from Airway Allergic Inflammation by Expanding Regulatory Lung Interstitial Macrophages. Immunity 46, 457–473. [DOI] [PubMed] [Google Scholar]
- Sajti E, Link VM, Ouyang Z, Spann NJ, Westin E, Romanoski CE, Fonseca GJ, Prince LS, and Glass CK (2020). Transcriptomic and epigenetic mechanisms underlying myeloid diversity in the lung. Nat Immunol 21, 221–231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salwig I, Spitznagel B, Vazquez-Armendariz AI, Khalooghi K, Guenther S, Herold S, Szibor M, and Braun T (2019). Bronchioalveolar stem cells are a main source for regeneration of distal lung epithelia in vivo. Embo J 38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanz I, Wei C, Jenks SA, Cashman KS, Tipton C, Woodruff MC, Hom J, and Lee FE (2019). Challenges and Opportunities for Consistent Classification of Human B Cell and Plasma Cell Populations. Front Immunol 10, 2458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saunders CJ, Reynolds SD, and Finger TE (2013). Chemosensory Brush Cells of the Trachea A Stable Population in a Dynamic Epithelium. Am J Resp Cell Mol 49, 190–196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schlitzer A, McGovern N, Teo P, Zelante T, Atarashi K, Low D, Ho AW, See P, Shin A, Wasan PS, et al. (2013). IRF4 transcription factor-dependent CD11b+ dendritic cells in human and mouse control mucosal IL-17 cytokine responses. Immunity 38, 970–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider C, Lee J, Koga S, Ricardo-Gonzalez RR, Nussbaum JC, Smith LK, Villeda SA, Liang HE, and Locksley RM (2019). Tissue-Resident Group 2 Innate Lymphoid Cells Differentiate by Layered Ontogeny and In Situ Perinatal Priming. Immunity 50, 14251438 e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schoch KG, Lori A, Burns KA, Eldred T, Olsen JC, and Randell SH (2004). A subset of mouse tracheal epithelial basal cells generates large colonies in vitro. Am J Physiol Lung Cell Mol Physiol 286, L631–42. [DOI] [PubMed] [Google Scholar]
- Schyns J, Bureau F, and Marichal T (2018). Lung Interstitial Macrophages: Past, Present, and Future. J Immunol Res 2018, 5160794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shannon JM (1994). Induction of alveolar type II cell differentiation in fetal tracheal epithelium by grafted distal lung mesenchyme. Dev Biol 166, 600–14. [DOI] [PubMed] [Google Scholar]
- Shannon JM, Nielsen LD, Gebb SA, and Randell SH (1998). Mesenchyme specifies epithelial differentiation in reciprocal recombinants of embryonic lung and trachea. Dev Dyn 212, 482–94. [DOI] [PubMed] [Google Scholar]
- Sheikh AQ, Misra A, Rosas IO, Adams RH, and Greif DM (2015). Smooth muscle cell progenitors are primed to muscularize in pulmonary hypertension. Sci Transl Med 7, 308ra159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shivaraju M, Chitta UK, Grange RMH, Jain IH, Capen D, Liao L, Xu J, Ichinose F, Zapol WM, Mootha VK, et al. (2021). Airway stem cells sense hypoxia and differentiate into protective solitary neuroendocrine cells. Science 371, 52–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silver JS, Kearley J, Copenhaver AM, Sanden C, Mori M, Yu L, Pritchard GH, Berlin AA, Hunter CA, Bowler R, et al. (2016). Inflammatory triggers associated with exacerbations of COPD orchestrate plasticity of group 2 innate lymphoid cells in the lungs. Nat Immunol 17, 626–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siracusa MC (2016). Basophils - Role in Immunity. In Encyclopedia of Immunobiology, Ratcliffe MJH, ed. (Oxford: Academic Press; ), pp. 326–333. [Google Scholar]
- Snippert HJ, Haegebarth A, Kasper M, Jaks V, van Es JH, Barker N, van de Wetering M, van den Born M, Begthel H, Vries RG, et al. (2010). Lgr6 Marks Stem Cells in the Hair Follicle That Generate All Cell Lineages of the Skin. Science 327, 1385–1389. [DOI] [PubMed] [Google Scholar]
- Snyder ME, and Farber DL (2019). Human lung tissue resident memory T cells in health and disease. Curr Opin Immunol 59, 101–108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song H, Yao E, Lin C, Gacayan R, Chen MH, and Chuang PT (2012). Functional characterization of pulmonary neuroendocrine cells in lung development, injury, and tumorigenesis. Proc Natl Acad Sci U S A 109, 17531–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sontake V, Kasam RK, Sinner D, Korfhagen TR, Reddy GB, White ES, Jegga AG, and Madala SK (2018). Wilms’ tumor 1 drives fibroproliferation and myofibroblast transformation in severe fibrotic lung disease. JCI Insight 3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sontake V, Shanmukhappa SK, DiPasquale BA, Reddy GB, Medvedovic M, Hardie WD, White ES, and Madala SK (2015). Fibrocytes Regulate Wilms Tumor 1-Positive Cell Accumulation in Severe Fibrotic Lung Disease. J Immunol 195, 3978–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spits H, Artis D, Colonna M, Diefenbach A, Di Santo JP, Eberl G, Koyasu S, Locksley RM, McKenzie AN, Mebius RE, et al. (2013). Innate lymphoid cells--a proposal for uniform nomenclature. Nat Rev Immunol 13, 145–9. [DOI] [PubMed] [Google Scholar]
- Spits H, and Cupedo T (2012). Innate lymphoid cells: emerging insights in development, lineage relationships, and function. Annu Rev Immunol 30, 647–75. [DOI] [PubMed] [Google Scholar]
- Srinivasan RS, Escobedo N, Yang Y, Interiano A, Dillard ME, Finkelstein D, Mukatira S,Gil HJ, Nurmi H, Alitalo K, et al. (2014). The Prox1-Vegfr3 feedback loop maintains the identity and the number of lymphatic endothelial cell progenitors. Gene Dev 28, 2175–2187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stackowicz J, Jönsson F, and Reber LL (2020). Mouse Models and Tools for the in vivo Study of Neutrophils. Frontiers in Immunology 10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steffes LC, Froistad AA, Andruska A, Boehm M, McGlynn M, Zhang F, Zhang WM, Hou D, Tian XF, Miquerol L, et al. (2020). A Notch3-Marked Subpopulation of Vascular Smooth Muscle Cells Is the Cell of Origin for Occlusive Pulmonary Vascular Lesions. Circulation 142, 1545–1561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stump B, Cui Y, Kidambi P, Lamattina AM, and El-Chemaly S (2017). Lymphatic Changes in Respiratory Diseases: More than Just Remodeling of the Lung? Am J Resp Cell Mol 57, 272–279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sui PF, Wiesner DL, Xu JH, Zhang Y, Lee J, Van Dyken S, Lashua A, Yu CY,Klein BS, Locksley RM, et al. (2018). Pulmonary neuroendocrine cells amplify allergic asthma responses. Science 360, 1086-+. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swatek AM, Lynch TJ, Crooke AK, Anderson PJ, Tyler SR, Brooks L, Ivanovic M, Klesney-Tait JA, Eberlein M, Pena T, et al. (2018). Depletion of Airway Submucosal Glands and TP63(+)KRT5(+) Basal Cells in Obliterative Bronchiolitis. Am J Respir Crit Care Med 197, 1045–1057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan SY, and Krasnow MA (2016). Developmental origin of lung macrophage diversity. Development 143, 1318–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tata A, Kobayashi Y, Chow RD, Tran J, Desai A, Massri AJ, McCord TJ, Gunn MD, and Tata PR (2018). Myoepithelial Cells of Submucosal Glands Can Function as Reserve Stem Cells to Regenerate Airways after Injury. Cell Stem Cell 22, 668–683 e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tata PR, and Rajagopal J (2017). Plasticity in the lung: making and breaking cell identity. Development 144, 755–766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torday JS, and Rehan VK (2016). On the evolution of the pulmonary alveolar lipofibroblast. Exp Cell Res 340, 215–219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tos M (1966). Development of the tracheal glands in man. Number, density, structure, shape, and distribution of mucous glands elucidated by quantitative studies of whole mounts. Acta Pathol Microbiol Scand 68, Suppl 185:3+. [PubMed] [Google Scholar]
- Trapnell BC, Nakata K, Bonella F, Campo I, Griese M, Hamilton J, Wang T, Morgan C, Cottin V, and McCarthy C (2019). Pulmonary alveolar proteinosis. Nat Rev Dis Primers 5, 16. [DOI] [PubMed] [Google Scholar]
- Travaglini KJ, Nabhan AN, Penland L, Sinha R, Gillich A, Sit RV, Chang S, Conley SD, Mori Y, Seita J, et al. (2020). A molecular cell atlas of the human lung from singlecell RNA sequencing. Nature 587, 619–625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsuneto M, Kajikhina E, Seiler K, Reimer A, Tornack J, Bouquet C, Simmons S, Knoll M, Wolf I, Tokoyoda K, et al. (2014). Environments of B cell development. Immunol Lett 157, 60–3. [DOI] [PubMed] [Google Scholar]
- Uribe-Querol E, and Rosales C (2015). Neutrophils in Cancer: Two Sides of the Same Coin. Journal of Immunology Research 2015, 983698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ushakumary MG, Riccetti M, and Perl AT (2021). Resident interstitial lung fibroblasts and their role in alveolar stem cell niche development, homeostasis, injury, and regeneration. Stem Cells Transl Med. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Keymeulen A, Rocha AS, Ousset M, Beck B, Bouvencourt G, Rock J, Sharma N, Dekoninck S, and Blanpain C (2011). Distinct stem cells contribute to mammary gland development and maintenance. Nature 479, 189–93. [DOI] [PubMed] [Google Scholar]
- Veres TZ, Voedisch S, Spies E, Tschernig T, and Braun A (2011). Spatiotemporal and functional behavior of airway dendritic cells visualized by two-photon microscopy. Am J Pathol 179, 603–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vivanco Gonzalez N, Oliveria JP, Tebaykin D, Ivison GT, Mukai K, Tsai MM, Borges L, Nadeau KC, Galli SJ, Tsai AG, et al. (2020). Mass Cytometry Phenotyping of Human Granulocytes Reveals Novel Basophil Functional Heterogeneity. iScience 23, 101724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Volckaert T, Yuan T, Chao CM, Bell H, Sitaula A, Szimmtenings L, El Agha E, Chanda D, Majka S, Bellusci S, et al. (2017). Fgf10-Hippo Epithelial-Mesenchymal Crosstalk Maintains and Recruits Lung Basal Stem Cells. Dev Cell 43, 48–59 e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Gise A, Stevens SM, Honor LB, Oh JH, Gao C, Zhou B, and Pu WT (2016). Contribution of Fetal, but Not Adult, Pulmonary Mesothelium to Mesenchymal Lineages in Lung Homeostasis and Fibrosis. Am J Respir Cell Mol Biol 54, 222–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Moltke J, Ji M, Liang HE, and Locksley RM (2016). Tuft-cell-derived IL-25 regulates an intestinal ILC2-epithelial response circuit. Nature 529, 221–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vonarbourg C, Mortha A, Bui VL, Hernandez PP, Kiss EA, Hoyler T, Flach M, Bengsch B, Thimme R, Holscher C, et al. (2010). Regulated expression of nuclear receptor RORgammat confers distinct functional fates to NK cell receptor-expressing RORgammat(+) innate lymphocytes. Immunity 33, 736–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang A, Chiou J, Poirion OB, Buchanan J, Valdez MJ, Verheyden JM, Hou X,Kudtarkar P, Narendra S, Newsome JM, et al. (2020a). Single-cell multiomic profiling of human lungs reveals cell-type-specific and age-dynamic control of SARS-CoV2 host genes. Elife 9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang G, Wen B, Ren X, Li E, Zhang Y, Guo M, Xu Y, Whitsett JA, Kalin TV, and Kalinichenko VV (2021). Generation of Pulmonary Endothelial Progenitor Cells for Cell-Based Therapy Using Interspecies Mouse-Rat Chimeras. Am J Respir Crit Care Med. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H, Morse HC 3rd, and Bolland S (2020b). Transcriptional Control of Mature B Cell Fates. Trends Immunol 41, 601–613. [DOI] [PubMed] [Google Scholar]
- Wang YJ, Tang Z, Huang HW, Li J, Wang Z, Yu YY, Zhang CW, Li J, Dai HP, Wang FC, et al. (2018). Pulmonary alveolar type I cell population consists of two distinct subtypes that differ in cell fate. P Natl Acad Sci USA 115, 2407–2412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wasserman GA, Szymaniak AD, Hinds AC, Yamamoto K, Kamata H, Smith NM, Hilliard KL, Carrieri C, Labadorf AT, Quinton LJ, et al. (2017). Expression of Piwi protein MIWI2 defines a distinct population of multiciliated cells. J Clin Invest 127, 3866–3876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weaver M, Yingling JM, Dunn NR, Bellusci S, and Hogan BL (1999). Bmp signaling regulates proximal-distal differentiation of endoderm in mouse lung development. Development 126, 4005–15. [DOI] [PubMed] [Google Scholar]
- Weibel ER (2009). What makes a good lung? Swiss Med Wkly 139, 375–86. [DOI] [PubMed] [Google Scholar]
- Wen X, Zhang X, Nian S, Wei G, Guo X, Yu H, Xie X, Ye Y, and Yuan Q (2021). Title of article: Mucosal-associated invariant T cells in lung diseases. Int Immunopharmacol 94, 107485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wendling O, Bornert JM, Chambon P, and Metzger D (2009). Efficient Temporally-Controlled Targeted Mutagenesis in Smooth Muscle Cells of the Adult Mouse. Genesis 47, 14–18. [DOI] [PubMed] [Google Scholar]
- Westphalen CB, Asfaha S, Hayakawa Y, Takemoto Y, Lukin DJ, Nuber AH, Brandtner A, Setlik W, Remotti H, Muley A, et al. (2014). Long-lived intestinal tuft cells serve as colon cancer-initiating cells. J Clin Invest 124, 1283–1295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitsett JA, Kalin TV, Xu Y, and Kalinichenko VV (2019). Building and Regenerating the Lung Cell by Cell. Physiological Reviews 99, 513–554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Widdicombe JH (2019). Early studies of airway submucosal glands. Am J Physiol Lung Cell Mol Physiol 316, L990–L998. [DOI] [PubMed] [Google Scholar]
- Widdicombe JH, and Wine JJ (2015). Airway Gland Structure and Function. Physiol Rev 95, 1241–319. [DOI] [PubMed] [Google Scholar]
- Williams M, Todd I, and Fairclough LC (2021). The role of CD8 + T lymphocytes in chronic obstructive pulmonary disease: a systematic review. Inflamm Res 70, 11–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson CL, Stephenson SE, Higuero JP, Feghali-Bostwick C, Hung CF, and Schnapp LM (2018). Characterization of human PDGFR-beta-positive pericytes from IPF and non-IPF lungs. Am J Physiol Lung Cell Mol Physiol 315, L991-L1002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xi Y, Kim T, Brumwell AN, Driver IH, Wei Y, Tan V, Jackson JR, Xu J, Lee DK, Gotts JE, et al. (2017). Local lung hypoxia determines epithelial fate decisions during alveolar regeneration. Nat Cell Biol 19, 904–914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamanishi Y, and Karasuyama H (2016). Basophils and mast cells in immunity and inflammation. Seminars in Immunopathology 38, 535–537. [DOI] [PubMed] [Google Scholar]
- Yang Y, Garcia-Verdugo JM, Soriano-Navarro M, Srinivasan RS, Scallan JP, Singh MK, Epstein JA, and Oliver G (2012). Lymphatic endothelial progenitors bud from the cardinal vein and intersomitic vessels in mammalian embryos. Blood 120, 2340–2348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Y, Riccio P, Schotsaert M, Mori M, Lu JN, Lee DK, Garcia-Sastre A, Xu JM, and Cardoso WV (2018). Spatial-Temporal Lineage Restrictions of Embryonic p63(+) Progenitors Establish Distinct Stem Cell Pools in Adult Airways. Dev Cell 44, 752-+. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao LC, Testini C, Tvorogov D, Anisimov A, Vargas SO, Baluk P, Pytowski B, Claesson-Welsh L, Alitalo K, and McDonald DM (2014). Pulmonary Lymphangiectasia Resulting From Vascular Endothelial Growth Factor-C Overexpression During a Critical Period. Circ Res 114, 806–822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin YJ, Wang F, and Ornitz DM (2011). Mesothelial- and epithelial-derived FGF9 have distinct functions in the regulation of lung development. Development 138, 3169–3177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- You LR, Lin FJ, Lee CT, DeMayo FJ, Tsai MJ, and Tsai SY (2005). Suppression of Notch signalling by the COUP-TFII transcription factor regulates vein identity. Nature 435, 98–104. [DOI] [PubMed] [Google Scholar]
- You Y, Huang T, Richer EJ, Schmidt JE, Zabner J, Borok Z, and Brody SL (2004). Role of f-box factor foxj1 in differentiation of ciliated airway epithelial cells. Am J Physiol Lung Cell Mol Physiol 286, L650–7. [DOI] [PubMed] [Google Scholar]
- Yu YR, Hotten DF, Malakhau Y, Volker E, Ghio AJ, Noble PW, Kraft M, Hollingsworth JW, Gunn MD, and Tighe RM (2016). Flow Cytometric Analysis of Myeloid Cells in Human Blood, Bronchoalveolar Lavage, and Lung Tissues. Am J Respir Cell Mol Biol 54, 13–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan T, Volckaert T, Redente EF, Hopkins S, Klinkhammer K, Wasnick R, Chao CM, Yuan J, Zhang JS, Yao C, et al. (2019). FGF10-FGFR2B Signaling Generates Basal Cells and Drives Alveolar Epithelial Regeneration by Bronchial Epithelial Stem Cells after Lung Injury. Stem Cell Reports 12, 1041–1055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yudanin NA, Schmitz F, Flamar AL, Thome JJC, Tait Wojno E, Moeller JB, Schirmer M, Latorre IJ, Xavier RJ, Farber DL, et al. (2019). Spatial and Temporal Mapping of Human Innate Lymphoid Cells Reveals Elements of Tissue Specificity. Immunity 50, 505519 e4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zacharias WJ, Frank DB, Zepp JA, Morley MP, Alkhaleel FA, Kong J, Zhou S, Cantu E, and Morrisey EE (2018). Regeneration of the lung alveolus by an evolutionarily conserved epithelial progenitor. Nature 555, 251-+. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zemmour D, Kiner E, and Benoist C (2020). CD4(+) teff cell heterogeneity: the perspective from single-cell transcriptomics. Curr Opin Immunol 63, 61–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zengin E, Chalajour F, Gehling UM, Ito WD, Treede H, Lauke H, Weil J,Reichenspurner H, Kilic N, and Ergun S (2006). Vascular wall resident progenitor cells: a source for postnatal vasculogenesis. Development 133, 1543–1551. [DOI] [PubMed] [Google Scholar]
- Zepp JA, Morley MP, Loebel C, Kremp MM, Chaudhry FN, Basil MC, Leach JP, Liberti DC, Niethamer TK, Ying Y, et al. (2021). Genomic, epigenomic, and biophysical cues controlling the emergence of the lung alveolus. Science 371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zepp JA, Zacharias WJ, Frank DB, Cavanaugh CA, Zhou S, Morley MP, andMorrisey EE (2017). Distinct Mesenchymal Lineages and Niches Promote Epithelial SelfRenewal and Myofibrogenesis in the Lung. Cell 170, 1134–1148 e10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang W, Menke DB, Jiang M, Chen H, Warburton D, Turcatel G, Lu CH, Xu W, Luo Y, and Shi W (2013). Spatial-temporal targeting of lung-specific mesenchyme by a Tbx4 enhancer. BMC Biol 11, 111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou B, Ma Q, Rajagopal S, Wu SM, Domian I, Rivera-Feliciano J, Jiang DW, von Gise A, Ikeda S, Chien KR, et al. (2008). Epicardial progenitors contribute to the cardiomyocyte lineage in the developing heart. Nature 454, 109–U5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu M, Chen M, Lichtler AC, O’Keefe RJ, and Chen D (2008). Tamoxifen-inducible Cre-recombination in articular chondrocytes of adult Col2a1-CreER(T2) transgenic mice. Osteoarthritis Cartilage 16, 129–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuo W, Zhang T, Wu DZ, Guan SP, Liew AA, Yamamoto Y, Wang X, Lim SJ, Vincent M, Lessard M, et al. (2015). p63(+)Krt5(+) distal airway stem cells are essential for lung regeneration. Nature 517, 616–20. [DOI] [PMC free article] [PubMed] [Google Scholar]