Abstract
The poly(A) tail enhances translation and transcript stability, and tail length is under dynamic control during cell state transitions. Tail regulation plays essential roles in translational timing and fertilization in early development, but poly(A) tail dynamics have not been fully explored in post-embryonic systems. Here, we examined the landscape and impact of tail length control during macrophage activation. Upon activation, more than 1500 mRNAs, including proinflammatory genes, underwent distinctive changes in tail lengths. Increases in tail length correlated with mRNA levels regardless of transcriptional activity, and many mRNAs that underwent tail extension encode proteins necessary for immune function and post-transcriptional regulation. Strikingly, we found that ZFP36, whose protein product destabilizes target transcripts, undergoes tail extension. Our analyses indicate that many mRNAs undergoing tail lengthening are, in turn, degraded by elevated levels of ZFP36, constituting a post-transcriptional feedback loop that ensures transient regulation of transcripts integral to macrophage activation. Taken together, this study establishes the complexity, relevance, and widespread nature of poly(A) tail dynamics, and the resulting post-transcriptional regulation during macrophage activation.
Keywords: poly(A) tail, TED-seq, macrophage activation, RNA regulation, cytoplasmic polyadenylation, ZFP36
INTRODUCTION
Polyadenylation refers to the 3′ extension of mRNAs with adenosines and occurs after nascent transcript cleavage (Millevoi and Vagner 2010). The newly synthesized poly(A) tail is up to 250 nt long and serves as a binding site for poly(A)-binding proteins (PABPs), which control multiple events throughout the RNA lifecycle, including nuclear export, translation and mRNA stability (Gallie 1991; Jalkanen et al. 2014). The length of the tail changes throughout the mRNA lifecycle, and many of these changes are mediated by interactions between 3′-UTR regulatory sequences and RNA binding proteins or microRNAs (miRNAs). 3′-UTR-trans factors often recruit deadenylases to shorten the tail but can also recruit cytoplasmic polyadenylases (Braun et al. 2011; Weill et al. 2012). Deadenylation is associated with mRNA decay, translational repression and altered localization, and deadenylation impacts most mRNAs (Zheng et al. 2008; Eichhorn et al. 2016; Park et al. 2016). However, exceptions exist: shortened poly(A) tails can be reelongated in the cytoplasm to stabilize mRNAs and promote translation. Maternal mRNAs in early embryogenesis and viral RNAs in host cells undergo poly(A) tail extension, mediated by 3′-UTR sequence or secondary structures that recruit noncanonical poly(A) polymerases (Lim et al. 2016; Kim et al. 2020). While these examples show the significance of poly(A) tail regulation (Wu et al. 1998; Wells et al. 2001; Weill et al. 2012; Lim et al. 2016), the extent and the importance of cytoplasmic polyadenylation in somatic cells has been unclear.
Multiple genome-wide poly(A) tail profiling methods exist, including PAL-seq (Subtelny et al. 2014), TAIL-seq (Chang et al. 2014), TED-seq (Woo et al. 2018), and FLAM-seq (Legnini et al. 2019). Using these techniques, many studies found a large variation of tail lengths in steady-state post-embryonic transcriptomes. However, in contrast to early embryos, there were only weak associations of tail size to translation efficiency, mRNA stability, abundance, and PABP binding (Subtelny et al. 2014; Lima et al. 2017; Rissland et al. 2017). For example, while miRNAs increase deadenylation rates of target mRNAs (Giraldez et al. 2006; Wu et al. 2006; Eisen et al. 2020a), tail length changes were only captured by presteady-state measurements (Eisen et al. 2020a). In steady-state post-embryonic systems, tail changes were masked as a consequence of the rapid decay of deadenylated intermediates (Eisen et al. 2020a,b). These studies highlight the complexity of steady-state poly(A) tail lengths in post-embryonic systems, and suggest that understanding poly(A) tail length regulation requires discrete presteady-state measurements across a gene regulatory response. However, most systematic, genome-wide investigations of transient-state poly(A) tail length control had been limited to specific biological contexts, such as oocyte fertilization (Lim et al. 2016). Prefertilization maternal mRNA is stored as an un-adenylated or partially adenylated form, and thus repressed in translation. Upon fertilization, they are polyadenylated in the cytoplasm, and their translation initiates globally (Lim et al. 2016). The absence of zygotic transcription makes oocytes a tractable system to study post-transcriptional regulation, as it becomes possible to examine changes in poly(A) tail lengths without the confounding influence of new transcripts. The extent and significance of poly(A) tail length control upon a developmental cue in transcriptionally active somatic cells has been difficult to examine.
Immune responses often require rapid and adaptable gene regulation, features suited to post-transcriptional control (Carpenter et al. 2014; Corbett 2018). Exposure of macrophages to lipopolysaccharides (LPS) induces rapid expression of inflammatory cytokines, such as tumor necrosis factor-α (TNF; Kontoyiannis et al. 1999; Parameswaran and Patial 2010). Upon LPS stimulation, TNF transcripts are stabilized and their translation is substantially enhanced; this post-transcriptional switch is associated with TNF tail lengthening (Crawford et al. 1997). Importantly, cytoplasmic adenylation has been implicated as the mechanism of TNF tail length control (Crawford et al. 1997), suggesting that a transcript-specific cytoplasmic poly(A) polymerase may engage in the regulation of macrophage activation. Intriguingly, the LPS-induced acute immune response is marked by rapid, short-term expression of inflammatory cytokines, followed by their rapid inhibition. This rapid shutdown of proinflammatory genes is critical for the prevention of chronic inflammation, and post-transcriptional inhibition by the RNA-binding protein ZFP36 is one component of this phenomenon (Mukherjee et al. 2014; Tiedje et al. 2016). ZFP36 guides proinflammatory mRNAs for degradation and translational repression, by recruiting deadenylation complexes to the poly(A) tail of target mRNAs (Sandler et al. 2011; Brooks and Blackshear 2013). These studies suggest that macrophage activation can be a model system to examine poly(A) tail length dynamics in a post-embryonic context. However, TNF has been the only example of poly(A) tail length control during the macrophage immune response, and several questions remain unsolved: are there other transcripts regulated by poly(A) tail control, and what are the implications and consequences of poly(A) tail length control during the macrophage immune response? Finally, and perhaps most interestingly, what is the extent and the importance of readenylation during macrophage activation?
In this study, we characterize poly(A) tail dynamics across a time-course using a cell-line model of human macrophage activation. We apply a combination of transcriptome-wide methods to profile nascent RNA synthesis (Kwak et al. 2013), poly(A) tail lengths (Woo et al. 2018), mRNA levels, and 3′-UTR isoform preferences (Fu et al. 2011). These methods generate accurate profiles of poly(A) tail length dynamics with 3′ isoform resolution, allowing a comprehensive understanding of post-transcriptional regulation in macrophages during a time-resolved immune response. We discover widespread and complex patterns of regulation mediated, in part, by changes in poly(A) tail length. We find evidence of extensive poly(A) tail lengthening, which is most pronounced in immune-related genes and factors involved in post-transcriptional regulation itself. Notably, our data suggests that readenylation of ZFP36, along with other mRNAs bound by ZFP36 protein, is an important early event during macrophage activation. Interestingly, these ZFP36-interacting transcripts show rapid tail shortening later in the response, likely as a consequence of elevated ZFP36. Thus, readenylation first stabilizes a set of genes implicated in macrophage function, and then results in their inhibition. Taken together, we show that macrophage activation entails extensive post-transcriptional regulation involving poly(A) tail length control.
RESULTS
Determination of isoform-specific poly(A) tail lengths in THP-1 cells
To study poly(A) tail length (PAL) control upon macrophage activation, we stimulated differentiated human THP-1 cells with LPS, and followed the resulting acute inflammatory stage of the immune response over a 4-h time course (Fig. 1A). Induction of TNF and IL1B confirmed proper activation (Fig. 1B). We used tail end displacement sequencing (TED-seq) to measure PAL transcriptome-wide prior to stimulation (0 h), and at three subsequent time-points (1, 2, and 4 h) with biological replicates. TED-seq estimates PAL by an accurate size selection of the sequencing libraries, which include the poly(A) tail region (Fig. 1C, top; Supplemental Fig. S1A; Woo et al. 2018). We sized libraries at 300 nt, thus, PAL is derived by subtracting the distance between the 5′ end of TED-seq reads and the 3′ cleavage and polyadenylation site (PAS) from 300 nt. The 3′-UTR isoform-specific poly(A) tail lengths are intuitively visualized: for transcripts with longer poly(A) tails, TED-seq reads map closer to the PAS, whereas those with shorter tails map further from the PAS and into the 3′ UTR. The collection of reads derived from each 3′-UTR isoform generates a clustered distribution of poly(A) tail lengths for that isoform (Fig. 1C, bottom). For example, SPSB1, an interferon-stimulated gene expressed preferentially in macrophages, has two annotated 3′-UTR isoforms. In unstimulated THP-1 cells, TED-seq reads mapped to SPSB1 indicate distinct distributions of poly(A) tails for both 3′-UTR isoforms, with mean poly(A) tail lengths of 91 and 94 nt, respectively (Fig. 1D). TED-seq accuracy and precision were validated by four spike-in standards with different poly(A) tail lengths (40, 80, 120, and 160 nt), displaying sharp PAL distributions with expected median sizes (Fig. 1E; Supplemental Fig. S1B).
FIGURE 1.
Determination of PAL with isoform specificity. (A) Schematic of activation time-course and sequencing strategy. (B) Validation of macrophage activation using qRT-PCR. In each sample, expression values were normalized to GAPDH expression. For each gene, plotted is fold change of gene expression (post-stimulation/unstimulated). Data is representative of two biological replicates, each performed with four technical replicates. (C) Structure of TED-seq libraries (top) and an illustration of PAL visualization by TED-seq on genome browser track (bottom). (D) Genome browser tracks of TED-seq (5′ terminus of reads) for SPSB1, indicating PAS previously annotated and determined by 3′-seq. Mean PAL (nt) and CPM values displayed for each 3′-UTR isoform in TED-seq and 3′-seq tracks, respectively. Arrow marks on top of the genome browser track indicate PAL from reference point for each 3′-UTR isoform. (E) Cumulative distribution (CDF) of spike-in PALs (x-axis). Representative data from a single library. The results from all time points are shown in Supplemental Figure S1B. (F) Histogram of PAS counts per gene. (G) PAS counts grouped by distance to reference PAS (nearest annotated PAS), for 3′-UTR sites and those within 1 kb downstream (n = 30,141). (H) Genome browser tracks of TED-seq and 3′-seq on ANTXR1 3′ UTR. Mean PAL (nt) and read count (CPM) displayed, with TED-seq read distributions magnified (inset box), and relative position of 3′-seq peaks to reference PAS shown under the PAS track (minus indicates upstream). (I) Genome browser tracks of TED-seq and 3′-seq on CD83 3′ UTR. De novo PAS isoforms track (PAS) shows the positions of PAS and their distances from annotated PAS, as in H. Mean PAL (nt) and read count (CPM) displayed as in C. Two biological replicates of TED-seq and 3′-seq libraries were prepared at each time point. One of two 0 h biological replicates is shown for TED-seq and 3′-seq data as a representative on the genome browser for D, H, and I. See also Supplemental Figure S1.
It is recognized that reference PAS annotations in mammals are incomplete, and PAS usage is highly cell-type specific (MacDonald and McMahon 2010; Smibert et al. 2012; Zhang et al. 2020). Since the accuracy of TED-seq depends on correct PAS annotation, we experimentally determined PAS in the differentiated THP-1 cells. We performed 3′-seq (Fu et al. 2011) in two biological replicates using the same LPS stimulation time points examined by TED-seq (Fig. 1A; Supplemental Fig. S1C,D). 3′-seq identifies PAS by initiating reverse-transcription at the start of the poly(A) tail, and the resulting read counts correspond to the abundance of the 3′-UTR isoform ending at the given PAS. After filtering out nonspecific, internally priming-derived reads, we identified 47,986 PASs, 95% of which (n = 44,791) are located within annotated genes (n = 12,336), and only 5% mapped to intergenic sequences (Supplemental Fig. S1E). Most (64%) intragenic PASs mapped to annotated 3′ UTRs or within 1000 nt downstream (Supplemental Table S1). The remaining sites (Supplemental Fig. S1E) mapped to CDS (12%), intron (18%), or 5′-UTR (1%) regions, proportions equivalent to those found in other contexts (Jia et al. 2017). Sixty-three percent of genes with at least one site exhibited multiple PASs (Fig. 1F). Thirty three percent of de novo PASs mapped within 10 nt of annotated PASs (Fig. 1G; Supplemental Fig. S1F), and 67% PASs are discrepant with annotated sites (33% within 10 to 300 nt, 35% more than 300 nt away from any annotated site; Fig. 1G). These fractions indicate the prevalence of novel isoforms, which are similar to 3′-seq studies in other cell types (Katsanou et al. 2005; Zhang et al. 2020). For example, ANTXR1 uses de novo PAS at 160 nt upstream of the reference PAS (Fig. 1H), whereas CD83 exhibited two novel tandem 3′-UTR isoforms instead of the single annotated PAS (Fig. 1I). These results demonstrate the need to establish cell-type specific PAS usage for studies where comprehensive 3′-UTR isoform annotations are required, such as TED-seq. Therefore, we used our experimentally determined PAS sites in THP-1 cells instead of annotated PAS in TED-seq analysis. We generated a customized annotation of 30,141 3′-UTR isoforms in 10,589 genes (Supplemental Table S2), which were used to calculate isoform-specific PAL profiles. Biological replicates of TED-seq at each time point correlated well (Supplemental Fig. S1G; Pearson correlation coefficient, R = 0.99), and we used mean poly(A) tail lengths from two replicates for subsequent analyses.
We used our 3′-seq data (Supplemental Table S3) to assess whether 3′-UTR usage changed across the activation time-course (Supplemental Fig. S2A,B). Genes often contain multiple PAS in their 3′-UTR regions, generating alternative 3′-UTR isoforms with different 3′-UTR lengths through alternative polyadenylation (APA; Tian et al. 2007; Mayr and Bartel 2009; Mayr 2016). The degree of APA isoform usage between any two time points was assessed using the 3′-UTR switch index (USI; Harrow et al. 2012). Shifts toward distal APA (longer 3′ UTR) isoforms results in positive USI values (USI > 0.1) and proximal APA (shorter 3′ UTR) isoforms results in negative USI values (USI < −0.1, Supplemental Fig. S2B). Our data show that LPS stimulation induces extensive changes in isoform usage, for both distal (n = 566, FDR < 0.1) and proximal (n = 464, FDR < 0.1) switches, with a gradual increase in the proportion of proximal switching (Supplemental Figs. S1D, S2C, left) over the time course. Notably, the functions of genes that exhibit 3′-UTR isoform switching are enriched in immune responses, metabolic processes and protein transport/localization (Supplemental Fig. S2C, right). Thus, macrophage activation involves extensive changes in 3′-UTR isoform usage, which are potentially relevant to physiological changes during macrophage activation.
PAL dynamics during macrophage activation
Global poly(A) tail profiling studies revealed that many human mRNAs have mean PALs between 50–100 nt, shorter than thought previously (Chang et al. 2014; Jalkanen et al. 2014; Subtelny et al. 2014). In addition, modest tail length changes of 10–20 nt can impact RNA fates and thus be consequential (Jalkanen et al. 2014; Eisen et al. 2020a). Therefore, we strove to ensure that our PAL calculations from TED-seq data were accurate and high resolution. Shifts in APA isoform preferences occurring within the 300 nt library-sizing window (referred to as local PAS switch hereafter) complicate determinations of which PAS the TED-seq reads derive from. To exclude such potential errors we stringently removed transcript isoforms that showed LPS-induced local PAS switches within a tandem PAS cluster (multiple PAS isoforms within 300 nt). This leaves us with 6269 major isoforms in 5079 genes, which corresponded to ∼70% of TED-seq reads that can be unambiguously assigned to PASs (Supplemental Fig. S2D,E). We calculated PALs for these major isoforms and the changes in PALs between end points, linear time-points, as well as any two time point comparisons (0 h to 4 h, 0 h to 1 h, 1 h to 2 h, 2 h to 4 h, 0 h to 2 h, and 1 h to 4 h intervals) to comprehensively identify isoforms with tail length changes, and to resolve transient changes (Supplemental Table S4). This approach was necessary to capture transient PAL changes (e.g., between 0 h and 2 h). Thus, we generated an inclusive view of PAL dynamics during macrophage activation, identifying 1520 transcript isoforms with PAL changes (length differences ≥10 nt) in at least one interval comparison (Fig. 2A; Supplemental Fig. S2F, illustrating the 0 h to 1 h comparison and all other comparisons, respectively). Transcripts from 237 genes exhibited PAL increases during the time-course, although the majority (n = 1286) of transcripts with significant changes (K–S test, FDR < 0.1) exhibited PAL decreases. Notably, the tail length changes were independent of initial tail lengths (Supplemental Fig. S3A). To validate these results, we selected examples of transcripts with PAL increases (CCL4, CCRL2, and ZFP36) and no PAL changes (HNRNPF and ACTB), and used a PCR based poly(A) tail (PAT) assay to validate the results (Fig. 2B).
FIGURE 2.
PAL dynamics during macrophage activation. (A) Scatterplot of PALs between 0 (x-axis) and 1 h post-activation (y-axis), averaged from two biological replicates. Each point denotes an isoform identified by 3′-seq. Point density color-coded from blue to orange (low to high). Red points indicate isoforms with significant changes (|ΔPAL| ≥ 10 nt and K–S test FDR < 0.1 in both replicates). (B) Validation of TED-seq results by PAT assay for indicated genes. Final PAT-PCR products were analyzed on 6% nondenaturing polyacrylamide-TBE gel, followed by SYBR Gold staining. Deadenylated form (A0 mRNA; asterisk) was generated by treating total RNA with RNase H and Oligo dT. (C) Genome browser tracks of TNF: TED-seq profiles and mean PAL (top 4), 3′-seq profiles and percentage of PAS isoform (PPI; next 4), during time-course. Values in square brackets indicate read count (y-axis) range. De novo PAS isoforms track (PAS) is displayed on the ninth lane. For D and E, mean poly(A) tail lengths from two biological replicates were averaged for a given transcript isoform. The averaged PAL at each time point was mean-normalized by subtracting mean of the averaged PALs across all time points. The resulting mean-normalized PAL values were plotted. (D) Distinct poly(A) tail length changing patterns during macrophage activation. GO terms enriched (Fisher's exact test, FDR < 0.2) in genes of each PAL-changing pattern listed. (E) Heatmap of mean-normalized PALs in clusters 4, 5, and 6; labels identify genes and isoform index. (F) Genome browser tracks of genes from clusters 4, 5, and 6 (colored in blue in E, x-axis) and IL1B as a negative control with no PAL change. One of two biological replicates is shown for TED-seq and 3′-seq data as a representative on the genome browser for C and F. Mean PAL displayed per track. See also Supplemental Figures S2 and S3.
TNF had been proposed to undergo readenylation upon stimulation in mouse macrophage cells, exhibiting a PAL increase 1 h post activation (Crawford et al. 1997). Consistent with previous studies, LPS stimulation caused a shift in TED-seq reads upon stimulation (0–1 h), indicating a PAL increase in TNF (Fig. 2C). However, our extended time-course data revealed that this increase is transient and followed by a rapid reduction in tail length after 1 h. Moreover, this increase was not due to any local PAS switching, since 3′-seq shows only one single dominant 3′-seq peak (98% of reads from the major isoform) in TNF throughout the time course (Fig. 2C; Proportion of a PAS isoform over all five isoforms [PPI] ≥0.98). TNF inhibition after a transient activation is known to be integral to the macrophage response (Carballo et al. 1998). Our TED-seq data, generated at multiple time-points, revealed more complex changes in TNF PAL than previously appreciated.
To characterize temporal PAL dynamics in the transcriptome, we applied k-means clustering to identify coordinated PAL change patterns across genes (Fig. 2D, left; n = 1520). The majority of genes are in clusters 1 to 3 (C1–C3), each of which show gradual PAL decreases. Other clusters exhibit PAL increases (C4–C6), revealing diverse and distinctive temporal patterns (Fig. 2D, left). C4 and C5 undergo early PAL increases (within 2 h), followed by rapid or gradual decreases in PAL, respectively. In contrast, C6 exhibits persistent PAL lengthening later in macrophage activation. Notably, C7–C9 show early decreases in PAL, which is reversed to initial or longer lengths. These data reveal that PAL regulation during macrophage activation is more widespread and complex than previously recognized.
To gain insights into the biological relevance of PAL changes, we examined gene ontology (GO) enrichments in each cluster (Fig. 2D, right). Genes in C1–C3, with gradual PAL decreases, are enriched in transmembrane proteins (C1), phagocytosis factors (C2), and oxidoreductases (C3). These GO terms may reflect reduced requirements for the corresponding products during the inflammatory response. More interestingly, genes in C4–C6, characterized by PAL increases upon activation (Fig. 2D–F), are enriched with immune-related terms such as cytokines, chemokines, and chemotaxis (Fig. 2D, right). These enrichments imply a role of PAL control during the inflammatory process. In particular, enrichment of proinflammatory genes in C4, characterized by early transient increases in PAL, is notable given that immediate expression of proinflammatory genes is integral to early macrophage activation (Carpenter et al. 2014; Corbett 2018). Within C4, we also observed enrichment of 3′-UTR binding proteins, including ZFP36 and ELAVL1, factors known to regulate TNF (Katsanou et al. 2005; Tiedje et al. 2012; Mukherjee et al. 2014; highlighted in red, Fig. 2E). Moreover, RNA binding proteins (RBPs) are also enriched in C8 and C9. Thus, mRNAs with extensive PAL changes during activation encode genes important to immune function and trans-factors related to the poly(A) tail itself.
3′-UTR sequence features associated with changes in PAL
Poly(A) tail length control is often mediated by interactions between 3′-UTR cis elements and trans-acting factors. AREs are well-characterized 3′-UTR cis-elements that mediate rapid decay of many short-lived mRNAs, in particular, of cytokine and chemokine transcripts during immune responses (Caput et al. 1986; Xu et al. 1997). ARE-binding RBPs such as ZFP36 recognize AREs and destabilize mRNAs by recruiting deadenylation and decay factors (Lai et al. 2003; Sanduja et al. 2011). To gain insights into possible relationships between AREs and PAL changes, we first examined the association between 3′-UTR A/U content and LPS-induced PAL changes. We classified transcript isoforms into three groups: those with increases in PAL (ΔPAL ≥ 10 nt), those with decreases (ΔPAL ≤ −10 nt), and those with little or no change (|ΔPAL| ≤ 5 nt). We then compared 3′-UTR AU content across the three ΔPAL groups (Fig. 3A, left). 3′ UTRs of PAL-decreased transcripts (0 h to 4 h) have lower AU content compared to PAL-unchanged and -increased transcripts (P < 10−8; K–S test). This association between low AU content and tail shortening was observed for all time intervals except for the 1 h to 2 h interval (Fig. 3A, right and Supplemental Fig. S3B,C). We also examined the association between ΔPAL and other potentially relevant features, such as 3′-UTR length and codon optimality, which revealed that 3′-UTR AU content is the most strongly correlated feature with ΔPAL (Fig. 3A, right and Supplemental Fig. S3D). In parallel, we performed 6-mer enrichment analysis to search for sequence motifs enriched in the 3′ UTRs of PAL-increased transcripts, which revealed that AU-rich 6-mers are enriched in the 3′ UTRs of PAL-increased genes (Student's t-test FDR < 0.1; Supplemental Fig. S3E).
FIGURE 3.
RNA features contributing to PAL control during the macrophage immune response. (A) Association of different transcript features with ΔPAL. (Left) CDF of 3′-UTR AU content (x-axis) in transcripts grouped by ΔPAL between 0 and 4 h (down, DN; no change, NC; up, UP). Indicated P-value denotes two-tailed K–S tests between ΔPAL UP and DN groups. (Right) Heatmap of Pearson correlation coefficients between transcript features (y-axis), and ΔPAL at indicated time intervals (x-axis). When calculating ΔPAL, average value of mean PALs from two biological replicates was compared between two different time points (e.g., PAL4 h–PAL0 h). (B) Heatmap of AU content across 3′ UTRs in transcripts with increased and decreased PAL (left and right panels, respectively). Rows are transcripts ordered by 3′-UTR lengths, and columns (x-axis) are consecutive nonoverlapping 50 nt windows of 3′ UTR for a given transcript isoform, aligned to the PAS. Each cell contains AU content calculated in a given window, where the value is colored from blue to orange (low to high). (C) Association between 3′-UTR motif location and ΔPAL during activation. Number of unique 6-mers associated (Student's t-test, FDR < 0.1) with ΔPAL (y-axis), partitioned by location (first and last 500 nt of 3′ UTR; x-axis), in six time intervals (subpanels). Six-mers grouped and color-coded by number of A or U bases [n(A|U)]. (D) Distribution of ΔΔPAL of the unique 6-mers identified in C, with respect to 3′-UTR locations. The comparison of 0 and 4 h is shown as a representative. Top and bottom subpanels indicate front and rear 3′-UTR positions, respectively. ΔΔPAL (x-axis) is the difference of ΔPAL (0 to 4 h) values between the highest quartile isoforms with a given 6-mer and the lowest quartile isoforms with the 6-mer. 6-mers color-coded as in C. (E) Pairs of 6-mers showing stronger association with ΔPAL (0 h vs. 1 h) in combination than individually (K–S test, FDR < 0.1). The 6-mers identified as associated with ΔPAL (0 h vs. 1 h) in the last 500 nt 3′-UTR windows were tested for their combinatorial effects. The top combinatorial 6-mers are shown as a representative and the other pairs of 6-mers for other time point comparisons are provided in Supplemental Table S6. See also Supplemental Figure S3.
There is growing evidence that 3′-UTR cis-elements are functionally sensitive to their location within the 3′ UTR (Grimson et al. 2007; Piqué et al. 2008; Geissler and Grimson 2016; Geissler et al. 2016; Dai et al. 2019). Therefore, we examined how the association of AU content with poly(A) tail length change depends on the relative location of AU content within the 3′ UTR, which revealed that AU content is enriched near the 3′ ends of 3′ UTRs of PAL-increased transcripts compared to the PAL-decreased set (Fig. 3B; Supplemental Fig. S3F). These results suggested an association between changes in poly(A) tail length and AREs located toward the 3′ terminus of the 3′ UTR. To test this interpretation, we examined the association between individual 6-mers (n = 4096) and ΔPAL in the first and the last 500 nt segments of 3′ UTRs, for 3′ UTRs longer than 1 kb. For each 6-mer, we binned transcripts into four quartiles by the frequency of each 6-mer in each 500 nt segment, and then compared ΔPAL values between the top and bottom quartiles (Student's t-test, FDR < 0.1). AU-rich 6-mers are more frequently associated with PAL changes when located in the last 500 nt compared to the first 500 nt of the 3′ UTRs (Fig. 3C). Moreover, for those 6-mers significantly associated with ΔPAL (Student's t-test, FDR < 0.1; Fig. 3C), we obtained ΔΔPAL, the difference in mean ΔPAL between the top and bottom quartiles. Positive ΔΔPALs indicate that the 6-mers promote PAL increases, and those with negative ΔΔPALs are associated with PAL decreases. The AU-rich 6-mers tend to have ΔΔPAL > 0 (Fig. 3D). In addition, we repeated the 6-mer analysis using alternative smaller terminal segments of 300 and 100 nt, and obtained consistent results using 300 nt segments (Supplemental Fig. S4A). The 100 nt segments showed less clear positional preferences, suggesting that cis elements that control PAL may be excluded somewhat from the 3′-most terminal region of 3′ UTRs. Collectively, these results demonstrate that 3′-UTR AU content is a major feature associated with LPS-induced PAL changes and this association manifests near the 3′ end of 3′ UTRs.
3′-UTR regulatory elements often work in combination to mediate poly(A) tail length control (Piqué et al. 2008; Dai et al. 2019). Therefore, we examined combinations of 6-mer elements associated with PAL changes during macrophage activation. We identified 6-mer pairs that may be functioning combinatorially by comparing the ΔPAL of transcripts harboring both 6-mers to transcripts containing two instances of either of the 6-mers. For every pair of 6-mers sampled from those identified as individually associated with ΔPAL in the last 500 nt regions, we assessed whether the ΔPAL of the mRNAs containing both 6-mers (once each) are significantly greater than ΔPALs of the mRNA containing the two instances of each 6-mers. This strategy controls for the total number of the tested 6-mers in one mRNA. We found a total of 138 6-mer pairs that may act in combination to mediate PAL control upon macrophage activation in any time-point comparison (Supplemental Tables S5, S6). The 6-mers were assigned to known RBP motifs based on position weight matrix scores (Ray et al. 2013). For example, mRNAs with the 3′ terminal colocalization of specific pairs of AU-rich motifs tend to undergo greater tail length increase during the early stage of macrophage activation (between 0 and 1 h, Fig. 3E). The RBPs corresponding to these 6-mer pairs include many previously established poly(A)-tail-associated proteins (e.g., ELAVL1, PABPC1, CREBs, TIA1, and ZFP36), but also some relatively uncharacterized proteins (e.g., IGF2B2, HNRNPs, SYNCRIP, SART3, U2AF2, and RALY). This result indicates the complexity of poly(A) tail length control, and implicates various proteins with poly(A) tail length control.
Poly(A) tail length correlates with post-transcriptional changes
In post-embryonic systems, due to the presence of active and dynamic transcriptional regulation, assessing the relative role of post-transcriptional events is challenging. In particular, multiple mechanisms could explain poly(A) tail length changes in somatic cells upon stimulation. Our observations of poly(A) tail length increases in 237 transcripts (Fig. 2A; Supplemental Fig. S2F) could result from rapid accumulation of new transcripts deriving from an LPS-induced transcriptional burst, increased cotranscriptional polyadenylation, decreases in deadenylation, and/or cytoplasmic readenylation (Supplemental Fig. S4B). These possible mechanisms may apply differently to different transcripts or transcript isoforms (Kondrashov et al. 2012). We first examined whether the poly(A) tail length increases we observed derived from increased synthesis of transcripts, which are expected to have longer tails initially. Thus, we measured transcriptional activity genome-wide using a nascent RNA profiling assay, Precision-Run-On sequencing (PRO-seq; Kwak et al. 2013). PRO-seq profiles transcription activity genome-wide by performing a nuclear-run-on reaction with biotin-labeled nucleotides (biotin-NTPs). Incorporation of biotin-NTPs occurs at the 3′ end of nascent RNAs, providing a molecular handle with which to selectively purify nascent RNAs and construct libraries for sequencing. We performed PRO-seq across the macrophage immune response (0, 1, 2, and 4 h upon LPS; two biological replicates) (Supplemental Fig. S4C; Supplemental Table S7), and found that transcriptionally up-regulated genes exhibited increases in poly(A) tail length compared to those with transcriptional down-regulation (Fig. 4A; Supplemental Fig. S4D,E). Thus, distinguishing the impact of de novo transcription from post-transcriptional events is essential to determine the degree of post-transcriptional poly(A) tail length control.
FIGURE 4.
Association between changes in PAL and mRNA abundance regardless of transcriptional changes. (A) CDF of ΔPAL (x-axis) with respect to transcriptional changes (ΔTXN; PRO-seq, log2 fold change) between 0 and 4 h post-activation. Transcripts are binned into six groups based on ΔTXN (e.g., (−4, −2] denoting −4 < ΔTXN ≤ −2), and their ΔPAL values are plotted as a group. K–S test P-value compares ΔPAL values from lowest and highest ΔTXN bins. TXN values (RPKM) were averaged from two biological replicates of PRO-seq at each time point, and the fold change value between two different time points was log2-transformed for ΔTXN calculation. (B) Box plots of ΔPAL for transcript isoforms with PAL changes out of the genes with minimal transcriptional changes (|ΔTXN| < 0.5 in all pairwise time intervals) for 0 and 4 h comparison. The number of transcripts is denoted. PAL changes were defined as |ΔPAL| ≥ 10 nt and K–S test FDR < 0.01. (C) Analysis of genes with minimal ΔTXN between 0 h through 4 h (|ΔTXN| < 0.5 in all pairwise time intervals). CDF of ΔPAL (x-axis) among the sets of transcripts grouped based on RNA abundance changes (ΔRNA; DN, NC, and UP) defined in Supplemental Figure S4F. K–S test P-value between DN and UP groups indicated. 3′-seq read count values were averaged from two biological replicates at each time point, and then the fold change between two different time points was log2-transformed for ΔRNA calculation. (D) CDF of ΔPAL (x-axis) differentiated among the three ΔRNA groups (ΔRNA; DN, NC, and UP). Transcripts grouped by ΔRNA proceeds with RSS by ΔTXN levels (between 0 and 4 h post-activation). Inset violin plot shows ΔTXN per ΔRNA group. K–S test P-value for comparison between DN and UP groups indicated. (E) CDF of ΔRNA/ΔTXN (x-axis; log2 [3′-seq FC/PRO-seq FC]) comparing 0 to 2 h (left) and 2 to 4 h (right), for genes grouped by ΔPAL between 0 and 2 h post-LPS (DN; ΔPAL < −10, NC; |ΔPAL| < 5, UP; ΔPAL > 10). K–S test P-values between ΔPAL UP and DN groups indicated. (F) ΔRNA/ΔTXN with respect to ΔPAL in transcriptionally up-regulated genes. ΔRNA/ΔTXN between 0 to 2 h (left) and 2 to 4 h (right), for genes grouped by ΔPAL (between 0 to 2 h as defined in E). Student's t-test P-values are denoted by asterisk. (G) ΔRNA/ΔTXN with respect to ΔPAL in transcriptionally down-regulated genes; otherwise as in F. (Significant differences indicated as follows: [*] P < 0.05; [**] P < 0.01; [***] P < 0.001). Number of transcript isoforms or genes for the corresponding bins or groups are in parentheses. (FC) Fold change, (RSS) random stratified sampling, (DN) down, (NC) no change, (UP) up. See also Supplemental Figures S4 and S5.
To determine the extent of post-transcriptional poly(A) tail length changes and the influence such changes have on transcript abundance, we integrated our PRO-seq data with our mRNA tail and abundance profiling data (TED-seq and 3′-seq). First, we selected transcript isoforms with minimal transcriptional changes (PRO-seq, |log2 fold change (FC)| < 0.5), and from this set of genes, identified transcript isoforms with |ΔPAL| greater than 10 nt (FDR < 0.01; K–S test) between two time points. We included only genes with minimal transcriptional changes for all pairwise time intervals, and still identified transcripts that exhibited significant shifts in the poly(A) tail length distribution (Fig. 4B; Supplemental Fig. S4F), implying that their poly(A) tail length changes are post-transcriptional. Notably, the association between low AU content and tail shortening was also observed in the set of genes that exhibited minimal transcriptional changes (Supplemental Fig. S3C). Taken together, these results suggest that transcription alone cannot explain the observed poly(A) tail length dynamics and post-transcriptional events contribute to tail length regulation.
Poly(A) tail length regulation and its association with mRNA fate have been observed for several genes in various physiological contexts, including neuronal cells (Wells et al. 2001; Weill et al. 2012). More recently, a transcriptome-wide tail length study showed that PAL changes (ΔPAL) correlate with changes in transcript abundance (ΔRNA), stability and translation efficiency during the endoplasmic reticulum stress response (Woo et al. 2018). However, it is unclear whether these relationships apply in macrophage activation. To characterize the association of poly(A) tail length changes (ΔPAL) with changes in RNA abundance (ΔRNA) independent of transcriptional changes (ΔTXN), we made robust control sets of genes, only including those with minimal ΔTXN. We selected genes with minimal ΔTXN (PRO-seq, |log2 FC| < 0.5) throughout the time-course (0 h through 4 h), and split them into three groups (down, no change, up) based on ΔRNA (3′-seq, log2 FC [4 h/0 h] threshold = 1). We then applied stratified random sampling to the groups to normalize the ΔTXN distribution; we split the genes in each group into 10 bins based on ΔTXN (PRO-seq, log2 FC [4 h/0 h]), and sampled the same number of genes from each ΔTXN bin across the three ΔRNA groups (Supplemental Fig. S4G). This analysis revealed a significant relationship between ΔPAL and ΔRNA after precisely controlling for ΔTXN (Fig. 4C), which holds true for other time point comparisons (Supplemental Fig. S5A, 0 h and 1 h comparison shown as a representative). The association was also tested after grouping genes based on ΔPAL and examining ΔRNA (Supplemental Fig. S5B). The same relationship was observed when we examined genes across the entire range of transcriptional changes after stratified sampling (Fig. 4D; Supplemental Fig. S5C), and when genes were binned by ΔPAL and then ΔRNA were assessed (Supplemental Fig. S5D). Collectively, these results indicate that during macrophage activation, post-transcriptional events couple changes in tail length to RNA abundance under conditions of extensive changes in transcriptional regulation.
To further dissect the post-transcriptional relationships between poly(A) tail dynamics and RNA abundance, we used the ΔRNA/ΔTXN metric, which approximates mRNA stability (Woo et al. 2018; Patel et al. 2020; Blumberg et al. 2021), and explored the relationship between ΔPAL and ΔRNA/ΔTXN in our time-resolved data. This analysis revealed that ΔPAL has a strong positive association with ΔRNA/ΔTXN only when ΔPAL was from the preceding interval than ΔRNA/ΔTXN (e.g., 0 h vs. 2 h for ΔPAL and 2 h vs. 4 h for ΔRNA/ΔTXN; Fig. 4E, right), but not when both were from the same interval (e.g., 0 h vs. 2 h for ΔPAL and ΔRNA/ΔTXN both; Fig. 4E, left). These results suggest that during macrophage activation, changes in mRNA stability are coupled to changes in poly(A) tail lengths in a temporally delayed manner.
Given the evidence that poly(A) tail length might control RNA abundance during macrophage activation, we investigated if tail control can affect opposite influences of transcription on mRNA dynamics. We selected genes that are transcriptionally up-regulated (log2 FC ([1 h, 2 h, and 4 h]/0 h) > 1), and compared how the changes in poly(A) tail lengths (ΔPAL, 0 h to 2 h) are associated with the ΔRNA/ΔTXN metric (Fig. 4F; Supplemental Fig. S5E). The transcriptionally up-regulated transcripts did not exhibit changes in the mRNA stability metric between 0 and 2 h regardless of changes in tail length (Fig. 4F, left panel). However, when we considered changes in the mRNA stability metric during the 2 to 4 h interval, changes in stability (ΔRNA/ΔTXN) occurred in the same direction as the ΔPAL from the earlier time interval (Fig. 4F, right panel). In particular, mRNA levels at later time-points are reduced in genes with decreased tail length (0 h to 2 h) despite increases in transcript synthesis (Supplemental Fig. S5E, see mean (RNA) in PAL:DN). Collectively, these analyses demonstrate that changes in mRNA abundance coupled to PAL shortening may override the influence of increased transcription, and post-transcriptional control is evident even in genes under active and opposing transcriptional control. Additionally, we also examined genes that were transcriptionally repressed during the time-course (Fig. 4G; Supplemental Fig. S5F). Genes that exhibited reductions in tail length have greater decreases in RNA stability (ΔRNA/ΔTXN) than those with no PAL change, and their difference manifested at a later time interval than the same time interval (Fig. 4G; Supplemental Fig. S5F). Together, these results indicate that poly(A) length control mediates significant and widespread impacts on transcript abundance during macrophage activation.
Profiling readenylation during initiation of the macrophage immune response
TNF is thought to be regulated through cytoplasmic polyadenylation in mouse macrophages (Crawford et al. 1997), although this readenylation hypothesis has not been examined in human macrophages. Moreover, this phenomenon has not been examined at a transcriptome-wide level during macrophage activation. The added complexity of dynamic transcriptional regulation during macrophage activation further complicates this question. Therefore, we repeated our TED-seq profiling after inhibiting transcription with actinomycin D (ActD), which was performed prior to LPS stimulation (Fig. 5). The inhibition of transcription by ActD was confirmed by a bulk poly(A) tail length assay that revealed a global shortening of poly(A) tails after ActD treatment (Supplemental Fig. S6A; Kojima and Green 2015). In addition, for selected genes known to be induced by LPS stimulation, we verified by qRT PCR that ActD treatment was sufficient to negate induction (Supplemental Fig. S6B). Next, we generated TED-seq libraries from ActD-treated cells, at 0, 1, and 2 h post-LPS activation, constructing a pair of biological replicate libraries for each time point. The resulting poly(A) length profiles were well-correlated between replicates (Supplemental Fig. S6C). Quantitative analysis of the suppression of known LPS induced transcripts in the ActD TED-seq replicates also showed at least 95% suppression by ActD.
FIGURE 5.
Widespread readenylation during macrophage activation. (A) Genome browser tracks of TED-seq reads for TNF in the presence or absence of Actinomycin D (ActD) at different time-points post-activation (y-axis), together with 3′-seq (ActD-untreated) at the same time-points. Mean PAL is displayed at the corners of TED-seq tracks. 3′-seq peak indicates the position of PAS for the given gene. One of two biological replicates is shown on a genome browser for TED-seq and 3′-seq. (B) PAT assay (tail length, y-axis) for IL1B and TNF, during LPS activation time-course in the presence of ActD. (*) denotes nonspecific band. (C) CDF of ΔRNA (x-axis) compared between readenylated (UP) and control (CTRL) transcripts. K–S test P-value for the comparison between UP and CTRL is denoted. (D) CDF of ΔRNA/ΔTXN (x-axis) for readenylated and control transcripts defined as in C (orange and gray lines, respectively). K–S test P-value for the comparison between UP and CTRL is denoted. (E) Association of RBP motifs with readenylated transcripts. (Left) Statistical significance (y-axis) of RBP motifs tested for enrichment within the 3′-terminal 500 nt of 3′ UTRs of readenylated transcripts compared to control transcripts. (Right) Sequence logos of top ranked motifs. (F) Gene ontology terms enriched in transcripts undergoing readenylation (Fisher's exact test, FDR < 0.2). See also Supplemental Figures S6 and S7.
Upon ActD-treatment, we expect PAL increases only for mRNAs targeted by cytoplasmic polyadenylation. This readenylation is necessary and sufficient to explain transcription independent PAL increases without de novo synthesis of longer PAL transcripts. PAL changes were quantified for the PAS isoforms (n = 6876 derived from 5609 genes) that passed a cutoff of 50 TED-seq reads across all time points (0, 1, 2 h). Following ActD treatment, TED-seq identified tail length increases for TNF, with ΔPAL values of 8.6 nt (10.6 and 6.6 nt, respectively in each of the replicates) (Fig. 5A). This increase was validated with PAT assays under ActD treatment (Fig. 5B), in which we included in vitro deadenylated controls (Supplemental Fig. S6D). As an additional control for the PAT assay, we also included IL1B, which did not exhibit any change in poly(A) tail length in TED-seq (Fig. 5B; Supplemental Fig. S6D). In our normal (no ActD treatment) LPS time-course TED-seq data, we observed significant overlap of PAL increase genes between biological replicates using a smaller cutoff of ΔPAL > 5 (P < 10−8, Fisher's exact test), and therefore applied this threshold to the ActD-LPS data to identify readenylation targets with a higher sensitivity. Transcripts that exhibited ΔPAL > 5 across both biological replicates (FDR < 0.2; K–S test) include TNF and 266 potential readenylation targets (n = 61, 166 and 86 for 0 h vs. 1 h, 0 h vs. 2 h, and 1 h vs. 2 h comparisons, respectively) out of 6876 transcript isoforms considered upon macrophage activation, corresponding to 255 (of 5609) genes (Supplemental Fig. S6E; Supplemental Table S8). The majority of the potential readenylation targets have intermediate tail lengths, and only six transcripts had very short tails (<25 nt) that may be the targets of oligouridylation (Supplemental Fig. S6F; Chang et al. 2014; Lim et al. 2014). Moreover, the potential readenylation target transcripts exhibited tail length increases averaging 14 nt (Supplemental Fig. S6G), and there was no relationship apparent between tail increase and starting tail length (Supplemental Fig. S6H,I).
To examine the potential impact of readenylation, we investigated the association between readenylation and changes in RNA abundance (ΔRNA, log2 FC) upon LPS stimulation. Potential readenylation targets (ΔPAL UP, ActD-treated) exhibited greater ΔRNA (3′-seq, ActD-untreated), compared to nontarget genes (CTRL) at 0–1 h and 1–2 h, but not at 0–2 h (Fig. 5C; Supplemental Fig. S7A, right), after stratified random sampling to equalize transcription change (ΔTXN; PRO seq, log2 FC; Supplemental Figs. S6J, S7A, left). These results were also observed using the ΔRNA/ΔTXN metric (log2 transformed [3′-seq FC/PRO-seq FC], Fig. 5D). Additionally, we confirmed the association between readenylation and RNA abundance, even when restricting the analysis to the subset of genes with minimal changes in transcription (PRO-seq, |log2 FC| < 0.5, Supplemental Fig. S7B). Collectively, these results implicate readenylation as a process responsible for stabilizing transcripts during macrophage activation.
To identify regulatory sequences involved in readenylation, we examined the enrichment of any putative RBP motifs in the 3′ terminal regions (500 nt) of 3′ UTRs of the readenylation targets. To identify RBPs that mediate PAL increases during macrophage activation, we used position weight matrices (PWM; n = 202) from the Cis-RBP database (Ray et al. 2013). For each RBP expressed in THP-1 cells (n = 86), we calculated PWM scores in the last 500 nt of 3′ UTRs of PAL increased transcripts, and assessed the occurrences over background. The top significant motifs include the sequences bound by RALY, ZFP36, HNRNPC, CPEB2, ELAVL1, and U2AF2, all of which are characterized by poly(U) sequences (Fig. 5E). These enrichments were also observed when we repeated the analysis examining the 3′ terminal 300 and 100 nt of 3′ UTRs (Supplemental Fig. S7C,D).
Additionally, to understand the role of post-transcriptional poly(A) tail length elongation (post-TXN ΔPAL UP), we performed gene ontology analysis (Huang et al. 2009) of potential readenylation targets, using genes expressed at all time points as the background set (Fig. 5F). These readenylation targets were enriched with RNA binding terms, including AU-rich element binding, RNA binding, poly(A) RNA binding, and post-translational modification (PTM) targets such as phosphoproteins, acetylation, and ubiquitin conjugation (Fisher's exact test, FDR < 0.1). To a lesser significance (Fisher's exact test, 0.1 ≤ FDR < 0.2), immune-related terms such as viral process, NF-κB signaling pathway as well as cellular localization and intracellular transport were detected (Fig. 5F). These observations suggest that RBP and PTM target proteins are prevalently regulated at a post-transcriptional level through cytoplasmic polyadenylation at the early stage of macrophage activation, which is expected to exert broader secondary effects on their downstream targets later in macrophage activation.
Concomitant readenylation of ZFP36 and its target mRNAs upon early activation
ZFP36, also known as tristetraprolin (TTP), is an RBP that regulates mRNAs of proinflammatory genes to attenuate inflammation during macrophage activation (Brooks and Blackshear 2013). ZFP36 binding to AREs in target mRNAs, such as TNF, results in the recruitment and activation of deadenylase complexes and translational repression. However, the mechanisms underlying regulation of ZFP36 activity in early macrophage activation are not well understood, in particular at the post-transcriptional level. Intriguingly, in our TED-seq data, ZFP36 itself was identified as one of the potentially readenylated targets upon macrophage activation (Fig. 6A). We validated ZFP36 readenylation by performing PAT assay in the ActD-treated condition (Fig. 6B). Of note, comparing PAT assay results with and without RNase H (lane 1, 2, and 4) indicates that ZFP36 transcripts exist in very short poly(A) tail forms (near A0). Next, we tested whether ZFP36 tail length increases are affected by mutating poly(U) sequences in the 3′ UTR of ZFP36 mRNAs. We constructed reporter genes fused with human ZFP36 3′-UTR sequences, either wild-type (WT) or with poly(U) regions disrupted, and expressed in the THP-1 cells (Fig. 6C). The poly(U)-containing motifs in the 3′ UTR of ZFP36 mRNA were either deleted (MUT-DEL) or substituted with G and C nucleotides (MUT-GC). We used lentiviral transduction to introduce these constructs into THP-1 cells, confirming that delivery did not elicit immune activation (Supplemental Fig. S7E). Next, we differentiated and stimulated the cells, as described previously (Fig. 1A). We performed PAT assays on the 4 THP-1 RNAs (endogenous ZFP36 mRNAs, and the three reporter mRNAs) with and without LPS stimulation using a primer that can detect both endogenous and transgenic ZFP36 poly(A) tails and focused on the tail length changes for short-tailed mRNAs. Tail length increased only in the endogenous and WT 3′-UTR reporter mRNAs, but in none of the mutant 3′-UTR reporter mRNAs upon activation (Fig. 6D; Supplemental Fig. S7F). Notably, the near-completely deadenylated products (A0, red asterisk band in Fig. 6D) disappear in both endogenous and WT, but not in MUT samples. This indicates that even if the PAT assay is detecting both the endogenous and transgenic ZFP36 poly(A) tail populations in the WT sample, the very short poly(A) tail forms from both populations disappeared and were converted to longer tail forms. The greater degree of the length increases in WT transgene compared to the endogenous sample in the gel quantification analysis also supports this finding (Fig. 6D, right panel). Taken together, these results demonstrate that ZFP36 mRNAs undergo readenylation mediated by poly(U) readenylation control sequences in the 3′ UTR during macrophage activation.
FIGURE 6.
ZFP36 mRNAs are readenylated through poly(U)-containing 3′-UTR elements upon macrophage activation. (A) Genome browser tracks of TED-seq reads for ZFP36 in the presence or absence of Actinomycin D (ActD) at different time-points post-activation (y-axis), together with 3′-seq at the same time-points. One of two biological replicates is shown on a genome browser for TED-seq and 3′-seq. (B) PAT assay (tail length, y-axis) for ZFP36, during LPS activation time-course in the presence or absence of ActD (one of three replicates shown as a representative). Red asterisk indicates the PCR product for completely deadenylated mRNAs, derived from RNase H treatment in the presence of oligo dT. (C) Schematic of GFP reporters with either wild-type (WT) or mutant versions (MUT-DEL, MUT-GC) of human ZFP36 3′ UTR. Three distinct THP-1 stable cell lines were generated with each expressing one of the GFP-ZFP36 reporters by lentiviral transduction. Known RBP motifs were searched in the 3′-UTR region. For the motifs containing consecutive Us (≥3 Us) and with at least half of the motif length composed of Us (annotated in red), the consecutive Us were modified to have a deletion or GC substitution (annotated in green). The 3′-UTR length is indicated under each construct name in brackets. The PAT assay forward primer is represented as a red line for each construct with their distances from the cleavage site in brackets. (D) PAT assay on the human ZFP36 3′-UTR reporter mRNAs of WT, MUT-DEL, and MUT-GC versions expressed in the differentiated THP-1 cells before and after LPS treatment (1 h). The gel image of PAT assay (left panel) was digitally quantified by image J (right panel). Red asterisk denotes the PCR product of completely deadenylated mRNA (A0). See also Supplemental Figure S7.
Intriguingly, both TNF and ZFP36 were identified to undergo readenylation early upon activation in our data. These observations suggest that readenylation rapidly induces TNF expression, but this induction is transient, as ZFP36 readenylation and the resulting induction of ZFP36 protein results in negative regulation of TNF. In support of this model, we found that levels of ZFP36 exhibited a robust increase of both total protein level and dephosphorylated isoforms at 1 h upon LPS stimulation compared to the nontreated condition (Fig. 7A). Dephosphorylated ZFP36 isoforms are known to act preferentially on their target mRNAs (Chrestensen et al. 2004; Stoecklin et al. 2004). Therefore, we examined whether this readenylation model explains early transient expression of a larger set of ZFP36-targeted mRNAs including TNF.
FIGURE 7.
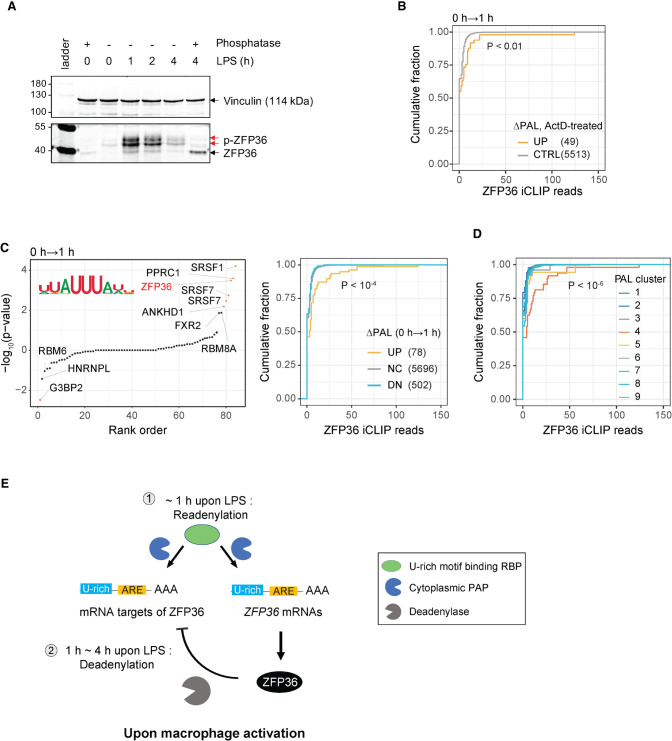
Concomitant readenylation of ZFP36 and its target mRNAs upon macrophage activation. (A) LPS-induced changes in overall protein expression and phosphorylation level for ZFP36 protein was measured by western blot, together with staining for Vinculin as a loading control. One of three replicates is displayed as a representative. (B) CDF of ZFP36 iCLIP binding (x-axis) in the 3′ UTRs of readenylated and control transcripts. The number of transcripts in each group is displayed in the legend. K–S test P-value between UP and CTRL groups is denoted. (C) Association of RBP motifs with ΔPAL (0 to 1 h). (Left) Motif enrichment P-values (Fisher's exact test) in the last 500 nt of 3′ UTRs of transcripts with increased PAL compared to decreased PAL. (Right) CDF of ZFP36 iCLIP binding (x-axis) in the 3′ UTRs of transcripts grouped by PAL changes (DN, NC, and UP). Indicated P-value denotes K–S test P-value between ΔPAL UP and DN groups. The number of transcripts in each group is labeled in the legend. (D) ZFP36 binding across PAL clusters defined in Figure 2D. CDF of ZFP36 binding (x-axis) in 3′ UTRs of transcripts in different PAL-clusters. Wilcoxon test P-values between cluster 4 and the rest of clusters indicated. In B–D, ZFP36 iCLIP data from bone marrow derived macrophage (BMDM) post-activation was used. (E) Model of post-transcriptional feedback loop via ZFP36 mRNA readenylation during macrophage activation. See also Supplemental Figure S7.
First, we asked whether there are other ZFP36-targeted mRNAs in our readenylation candidates. Our RBP binding motif enrichment analysis revealed enrichment of ZFP36 motifs in the 3′ UTRs of readenylation target mRNAs (Fig. 5E). To confirm this association, we turned to existing ZFP36 iCLIP data in bone marrow derived macrophage (BMDM) cells upon 1 h LPS treatment (Tiedje et al. 2016). Transcripts undergoing rapid (0 to 1 h) PAL increases in our data are associated with in vivo ZFP36 binding in BMDM cells, compared to transcripts with no changes in PAL (K–S test P < 0.01, Fig. 7B). In addition, putative ZFP36 binding motifs (Fisher's exact test, FDR < 0.2, Fig. 7C, left) and in vivo ZFP36 binding sites (K–S test P < 10−4, Fig. 7C, right) are strongly enriched in the set of transcripts with LPS-induced PAL increases in our normal (no ActD treatment) LPS time-course TED-seq data, implying that poly(A) tails of ZFP36 targeted mRNAs are elongated. In addition, transcripts with increased PAL tend to have higher ZFP36 motif density (Supplemental Fig. S7G). Collectively, these analyses suggest that ZFP36 mRNA itself and ZFP36-targeted mRNAs undergo readenylation together at the early stage of macrophage activation.
Next, we investigated tail length dynamics of the ZFP36-targeted mRNAs. Our readenylation model predicts that ZFP36-targeted mRNAs exhibit early and transient tail lengthening during macrophage activation, due to early readenylation followed by ZFP36 mediated deadenylation. Indeed, using the ZFP36 iCLIP data from BMDM cells (Tiedje et al. 2016), we found that transcripts bound by ZFP36 were specifically enriched in a set of transcripts undergoing early transient increases in PAL and characterized by GO terms associated with proinflammation (Cluster 4 in Fig. 2D, Wilcoxon test P < 10−6; Fig. 7D). This finding was corroborated using ZFP36 bound mRNAs identified using HITS-CLIP data from activated CD4+ T cells (Wilcoxon test P < 10−14, Supplemental Fig. S7H; Stoecklin et al. 2004; Moore et al. 2018). These observations suggest that ZFP36 readenylation upon macrophage activation plays a key role in shaping the transient expression of proinflammatory genes in macrophages (Fig. 7E).
DISCUSSION
In this study, we set out to examine the prevalence and consequences of poly(A) tail regulation in a somatic-cell context. We selected a model of macrophage activation, in which we examined transcription, RNA abundance and poly(A) tail length in unstimulated cells and across a time-course following LPS stimulation, enabling us to study tail dynamics in a complex regulatory environment. Importantly, our approach enabled us to profile the tail with 3′-UTR isoform resolution. We found extensive regulation of transcript abundance associated with poly(A) tail control. In response to activation, many transcripts exhibited tail lengthening, associated with increased transcript abundance. These transcripts preferentially encoded proteins associated with immune function and trans-acting factors that function in post-transcriptional regulation.
Poly(A) tail length dynamics upon macrophage activation
Prior to our work, TNF has been the sole example of a transcript known to be regulated by poly(A) tail control during macrophage activation (Crawford et al. 1997). Our study revealed that in addition to TNF, more than a thousand transcripts undergo poly(A) tail length changes during the macrophage immune response. Many are likely to be regulated by deadenylation, a well-established mode of control. However, several hundred transcripts appear to be readenylated, greatly expanding the scope of such regulation from TNF alone, and implicating readenylation as a major mode of control during macrophage activation.
One challenge in studying poly(A) tails in transcriptionally active, non-steady state systems is the difficulty in discriminating tail changes mediated post-transcriptionally from those derived from nascent transcription upon cellular activation. Our approach was to use PRO-seq to quantify transcriptional changes, and thus discriminate between transcriptional and post-transcriptional inputs on tail length. This approach was accomplished by stratifying genes by their transcriptional state: first, we examined genes with stable transcription, and second, we examined if genes with changes in transcriptional status also exhibit changes in RNA abundance as a function of poly(A) tail status. Thus, by normalizing transcriptional inputs, our integrative analysis demonstrated that transcriptional change alone does not explain changes in poly(A) tail status. Most importantly, changes in tail length and RNA abundance are correlated regardless of transcriptional change, indicating that post-transcriptional regulation is a major component of overall gene regulatory changes during macrophage activation.
This study was designed to examine the impact of poly(A) tail dynamics during a rapid cellular response in a differentiated cell, here, macrophage activation; the temporal relationships between changes in transcription, transcript abundance and the status of the poly(A) tail revealed a global preference for tail length changes that preceded changes in RNA abundance. This observation implies that post-transcriptional tail changes influence mRNA stability, and most mRNAs do not decay immediately upon deadenylation, but undergo intervening rate-limiting steps. We further reveal that tail length control can override transcriptional influences on RNA abundance. For many transcripts, their abundance is correlated primarily to rapid and transient changes in tail length. These results support a view of post-transcriptional control as a major component of gene regulation, even for genes under extensive transcriptional control, a potentially important phenomenon during rapid transitions.
Basis for rapid and transient poly(A) tail lengthening
Notably, we report widespread transcription-independent poly(A) tail lengthening, including for TNF. The simplest and most likely explanation is that such transcripts undergo readenylation. This interpretation relies on our transcription inhibition experiments, yet we acknowledge that such experiments may have limitations deriving from secondary effects, and from technical limitations inherent to the complexity of the experiment, that is LPS stimulation concomitant with transcriptional inhibition. Future work using approaches such as RNA metabolic labeling will be needed to confirm these striking results. Nevertheless, the rapid and transient nature of macrophage activation and the timing of poly(A) tail changes allow us to reason that they are not the secondary effects of transcription inhibition. Moreover, the common 3′-UTR features in these target transcripts suggest that the rapid and transient transcript readenylation has a post-transcriptional mechanistic basis. This observation is particularly meaningful because the extent of post-transcriptional readenylation has been less understood in somatic cells, and our transcription-independent experiments addressed a major challenge in the field by distinguishing polyadenylation from reduced deadenylation of de novo transcripts.
Interestingly, transcripts encoding RNA binding proteins (RBPs) often exhibited changes in poly(A) tail length; indeed, many encode proteins involved in post-transcriptional regulation by 3′-UTR cis elements. Thus, a regulatory network linked through post-transcriptional control may play a significant role in macrophage activation. We identified factors associated with changes in the poly(A) tail using 3′-UTR RBP binding inferences and CLIP-seq, converging to AU and U rich sequences, and their corresponding trans-factors. We elucidated widespread readenylation during macrophage activation in ZFP36 bound transcripts. We also found pronounced enrichment of poly(U)-containing RBP motifs in the 3′ UTRs of readenylated mRNAs, which we tested in GFP-3′-UTR reporter assays. In yeast, 3′-UTR poly(U) sequences are known to protect mRNAs from deadenylation (Muhlrad and Parker 2005), which is functionally relevant to our findings in human cells. In humans, CPEB1 mediates cytoplasmic polyadenylation by binding to poly(U)-containing cis elements called cytoplasmic polyadenylation elements (CPE) during oocyte maturation (Hake and Richter 1994), and host-viral mRNA readenylation in cytomegalovirus infection (Batra et al. 2016). However, CPEB1 is not expressed in THP-1 cells. Thus, other noncanonical factors likely engage in readenylation during macrophage activation. Notably, ELAVL1 is expressed in THP-1 cells, and its binding motif is among the most enriched elements in transcripts undergoing tail lengthening. ELAVL1 stabilizes mRNAs, but whether this is mediated through readenylation is unknown (Charlesworth et al. 2013). Further efforts will be needed to test this and the relative contribution of ZFP36 and ELAVL1 or other factors to poly(A) tail dynamics during macrophage activation.
The identity of the polymerase responsible for readenylation during macrophage activation is an important question arising herein. In humans, multiple noncanonical poly(A) polymerases (TENT1-TENT6) exist (Warkocki et al. 2018; Liudkovska and Dziembowski 2021), all relatively uncharacterized but for TENT2, a well-known poly(A) polymerase responsible for maternal mRNA polyadenylation. Based on our RNA-seq data, only TENT2, TENT4A/B, and TENT5A transcripts are expressed in THP-1 cells. As TENT4 recruitment is mediated by 3′-UTR structure rather than U-rich elements (Kuchta et al. 2016; Warkocki et al. 2018), we propose TENT5A as a candidate responsible for readenylation, recruited by a probable interaction with ELAVL1 (bioGRID interactome; Stark et al. 2006). ELAVL1-mediated recruitment of TENT5A during macrophage activation is a testable model to explain the widespread changes in post-transcriptional poly(A) tail lengthening and ensuing consequences on transcript abundance. Future studies elucidating these complex mechanistic models that underlie the widespread poly(A) tail control would be essential to understand the role of post-transcriptional regulation in macrophage activation.
MATERIALS AND METHODS
Cell lines, cell culture, and compound treatment
THP-1 cells used in this study are an authenticated cell line purchased from ATCC (TIB-202; human male). THP-1 cells were cultured at 37°C in 5% CO2 in RPMI1640 (Gibco, 11875093) supplemented with 10% FBS (VWR) and 1% antibiotics (Gibco, 15240062). THP-1 cells were differentiated to macrophage-like cells by incubating them overnight in complete media containing 200 ng/mL PMA (Sigma-Aldrich, P1585-1MG), followed by 3 d incubation in fresh media without PMA. The resulting differentiated cells were stimulated with 200 ng/mL LPS (Sigma-Aldrich, L4391) and collected at four time points: 0 h post-stimulation (no stimulation), and 1, 2, and 4 h post-stimulation. Total RNA was extracted with TRIzol (Invitrogen, 15596018). To inhibit transcription, THP-1 cells were incubated in media with 10 µg/mL actinomycin D (Sigma Aldrich, A9415) for 15 min prior to stimulation with LPS.
3′-sequencing library preparation
Total RNA was extracted using TRIzol from differentiated THP-1 cells throughout the LPS stimulation time-course (0, 1, 2, and 4 h). For each sample, poly(A) RNA was isolated from 10 µg of total RNA (Dynabeads mRNA Purification Kit; Invitrogen, 61006) followed by RNA fragmentation with 0.1 N NaOH, 5′ RNA phosphorylation (NEB, M0201S), and 5′ RNA ligation (NEB, M0204L) to VRA5 (5′-CCUUGGCACCCGAGAAUUCCA-3′). After heat denaturation at 65°C for 2 min, 5′ adapter-ligated poly(A)-containing RNA fragments were reverse transcribed by superscript II enzyme (Invitrogen,18064-014) using RT primer (CPS_RTP: 5′-GTTCAGAGTTCTACAGTCCGACGATCNNNNNNNNT8VN-3′) at 50°C for 1 h. The 3′-terminal ten nucleotides of CPS_RTP were designed to anneal to the junction between the poly(A) tail and the site of cleavage and polyadenylation within the transcript, and also contain an eight-nucleotide (nt) unique molecular index (UMI) barcode for PCR deduplication, with the remaining sequence designed for PCR amplification (NEB, M0530L). The resulting cDNA molecules were amplified by PCR for 14 cycles with RP-1 primer (See Supplemental Table S9) and RPI-X primers (See Supplemental Table S9) using Phusion High-Fidelity DNA Polymerase (NEB, M0530), followed by gel purification of 200 to 500 bp products on a 6% PAGE gel in TBE buffer. PCR products were eluted from the excised gel in TE-TW buffer overnight at 37°C, and then filtered through a DNase-free spin X column (Costar Spin-X centrifuge tube filters; Corning, CLS8160) and purified using Ampure XP beads (Beckman Coulter, A63881). The purified, barcoded libraries were quantified and pooled prior to Illumina sequencing on a Next500 platform (75 bp single-end reads). Unless otherwise stated, enzymatic reactions were performed as described in the manufacturer's protocols. The 3′-sequencing libraries were prepared in two independent biological replicates.
3′-seq data preprocessing and mapping
5′ RNA adaptor sequence was removed from the 3′ end of sequencing reads using Cutadapt (Martin 2011) with option -e 0.10, –overlap 2, –minimum-length = 10, –nextseq-trim 20. After adaptor removal, low quality reads were removed (those with quality scores <20, at any position). The first 30 nt, containing the 8 nt UMI, were used to deduplicate the reads (FASTX-Toolkit v0.0.11, http://hannonlab.cshl.edu/fastx_toolkit/, 2018; PRINSEQ v0.20.4 [Schmieder and Edwards 2011]; Seqtk v1.3-r106, https://github.com/lh3/seqtk, 2012). After trimming 16 nt (8 nt UMI and 8 nt corresponding to the dT8 portion of the adapter oligonucleotide) from the 5′ end of each read, reads with at least 10 nt remaining were mapped to the human genome (hg38; UCSC), using STAR aligner (Dobin et al. 2013) with the option –sjdbGTFfile “gencode.v26.annotation.gtf” –alignSJDBoverhangmin 3, –outFilterMultimapNmax 1. The aligned reads were represented by their 5′ end mapping coordinate on the opposite strand, and converted to BedGraph format, where the mapping position and the corresponding read counts of a 3′-seq peak were used to determine the cleavage and polyadenylation site and mRNA abundance of a transcript isoform.
Determination of cleavage and polyadenylation sites (PAS)
To avoid potential contamination of 3′-seq reads by the annealing of oligo-dT primers to internal A-rich sequences, we filtered the 3′-seq reads to remove internally primed reads from A-rich internal regions, as previously described (Fu et al. 2011; Li et al. 2012). Briefly, we searched for consecutive A sequences (>5 consecutive A nt) downstream from 3′-seq peaks, filtering out these reads from our 3′-seq reads. Then the 3′-seq read counts were normalized by counts per million mapped reads (CPM). The 3′-seq peaks were collapsed across all samples (0, 1, 2, and 4 h) with the read count per position totaled. Next, each 3′-seq peak position was converted to a 10 nt-wide window, and the overlapping windows of 3′-seq peaks within the window merged, totaling the merged 3′-seq peaks, retaining the midpoint of the merged window as the PAS coordinate. Merged windows with five or more normalized reads (final PAS window) were retained. For each 3′-seq time point data, the read counts of the 3′-seq peaks mapped in a final PAS window were summed to represent mRNA abundance of the PAS isoform expressed in the given sample. All final PAS located in the reference (GENCODE V26)-annotated 3′ UTR(s) + 1 kb downstream region, were considered as distinct 3′-UTR isoforms expressed in THP-1 cells. Finally, a custom transcript isoform annotation (bed12) file was built by modifying the reference transcript isoforms to terminate at our experimentally determined 3′-seq PAS sites. Poly(A) tail lengths were estimated for this comprehensive set of experimentally determined PAS isoforms. Additional PAS analysis was performed as follows. The identified PAS sites (n = 47,986) were tested for their locations (within gene or intergenic) within annotated genes (n = 12,336) using bedtools intersect function. Similarly, we then tested the positions of the PAS sites (n = 44,791) located within genes against the genomic coordinates of annotated 3′ UTRs or within 1000 nt downstream from the 3′ terminus (n = 10,589), CDS, intron, or 5′ UTR. Multiple PAS were defined based on the genes with at least one mapped PAS site (n = 12,336), containing multiple PAS sites (n = 7760), or a single site (n = 4576). Discrepant PAS were defined by the distance between the mapped PAS and the annotated PAS, and binned by the following distance: 10 to 300 nt (33%) or greater than 300 nt (35%). Based on this classification, we generated a customized reference transcript annotation file representing 30,141 3′-UTR isoforms from 10,589 genes for downstream usage (Fig. 1; Supplemental Fig. S1).
TED-seq library preparation
Tail end displacement sequencing (TED-seq) was applied to total RNA samples (5–10 µg) collected at multiple time points (0, 1, 2, and 4 h) after LPS treatment. TRIzol-purified RNA was subjected to poly(A) RNA purification using the manufacturer's protocol (Dynabeads mRNA Purification Kit, Invitrogen), and ligated with the adaptor molecule (RA3; Supplemental Table S9) to their 3′ terminus. The products of the ligation reaction were purified using TRIzol, and then fragmented with 0.1 N NaOH. Fragmented RNAs were purified with a P-30 column (Bio-Rad, 732-6251), and poly(A)-containing fragments enriched using Dynabeads mRNA Purification Kit. T4 polynucleotide kinase (PNK; NEB, M0201S) was used to phosphorylate the 5′ terminus of RNA fragments, enabling ligation of the 5′ terminus to the adaptor oligonucleotide containing UMIs (RA5; Supplemental Table S9). The resulting RNA libraries were reverse transcribed and PCR amplified using KAPA HiFi HotStart ReadyMix PCR Kit (Kapa Biosystems, KR0370), using no more than eight cycles of amplification. Prior to sequencing, 350–360 bp DNA molecules were purified using PAGE. Following PCR amplification, PAGE-mediated size selection was repeated on the amplified DNA. The resulting size-selected libraries were pooled and sequenced on an Illumina NextSeq500 (75 bp single-end reads). The TED-seq libraries were prepared in two independent biological replicates.
Synthesis of spike-in poly(A) standards
Poly(A) spike-in RNAs of 40, 80, 120, and 160 nt were generated by in vitro transcription of a PCR amplified double-stranded DNA template composed of a T7 promoter sequence, unique sequences for alignment from plasmid vector backbones (pmRFP-C1 [Addgene, 54764] for A40; pEGFP-C1 [Clontech] at EGFP ORF for A80; pEGFP-C1 at NeoR/KanR ORF for A120; pGL4.23 [Promega, E8411] for A160), and poly(A) repeats of desired lengths (Supplemental Table S9). To generate four distinct 700-bp backbone sequences, different coding sequences were targeted and PCR-amplified with the set of gene-specific primers (Supplemental Table S9) using Phusion High-Fidelity DNA Polymerase (25 cycles of 10 sec at 98°C, 30 sec at 60°C, and 30 sec at 72°C, followed by 5 min extension at 72°C in a final volume 100 µL; 70 µL H2O, 20 µL 5× buffer, ∼2.5 ng template plasmid, 300 nM forward and reverse primers, 250 µM dNTP and 1 µL Phusion polymerse). After PAGE purification, oligo(Dt) tails of distinct sizes were added to the 3′ end of the corresponding backbone template, and amplified with the specific primers (Supplemental Table S9) using Phusion Polymerase (two PCR cycles of 10 sec at 98°C and 50 sec at 68°C, followed by 5 min extension at 72°C). After PAGE purification, the resulting four distinct 700 bp templates with different coding sequences and tail sizes were in vitro transcribed using MAXIscript T7 Transcription Kit (Invitrogen, AM1314). The RNA products were purified by denaturing polyacrylamide electrophoresis (Urea; Thermo Scientific, U15-500) and quantified using a nanodrop. The spike-in RNAs were added to purified mRNAs from samples used for TED-seq library generation (1 ng of each spike-in RNA species per 100 ng of poly[A]-selected RNA). The entire sequences of individual spike-in poly(A) standards are provided in Supplemental Table S9.
TED-seq data preprocessing and mapping
For sequencing reads ending with >10 A residues, consecutive (A) sequences were trimmed from the 3′ end (PRINSEQ v0.20.4; 33). After poly(A) tail trimming, reads with a length ≥ 15 nt and mean quality score > 20 were retained for further analysis. PCR duplicates were removed using the first 15 nt of the trimmed reads, which includes an 8 nt UMI (FASTX-Toolkit v0.0.11, PRINSEQ v0.20.4, Seqtk v1.3-r106). Nucleotides corresponding to the UMI were then trimmed from the 5′ end of the deduplicated reads, followed by the exclusion of trimmed reads shorter than 15 nt. The resulting reads were mapped to the human genome (hg38) using STAR (2.4.2a; 26) with the option –sjdbGTFfile “gencode.v26.annotation.gtf” –alignSJDBoverhangmin 3, –outFilterMultimapNmax 1. BWA (Li and Durbin 2009) was used to align the reads corresponding to the poly(A) spike-in standards. The relationship between the mapped TED-seq reads, the cleavage and polyadenylation site, and the library insert size enables the 5′ terminus mapping coordinate of the aligned TED-seq reads to shift the 3′ tail ends upstream into the 3′ UTR by the insert size of the library (300 nt): for transcripts with longer poly(A) tails, TED-seq reads map closer to the PAS, whereas those with shorter tails map further from the PAS and into the 3′ UTR. Accordingly, 3′-UTR isoform-specific poly(A) tail length distribution is reproduced immediately upstream of the corresponding PAS by 5′ termini of the mapped TED-seq reads, and visualized on a genome browser track with the IGV genome browser (Robinson et al. 2011) (Fig. 1; Supplemental Fig. S1).
Poly(A) tail length estimation and identifying significant changes in poly(A) tail length
From the library insert size (I) and the distance from 5′ termini of TED-seq reads to PAS (D), poly(A) tail length (L) is derived as L = I – D. GENCODE V26 annotation (Harrow et al. 2012; Frankish et al. 2019) of the human transcriptome (bed12 format) was amended to reflect experimentally determined THP-1 cleavage and polyadenylation sites (PAS) identified using 3′-seq. Finally, a frequency table of TED-seq read 5′ termini located within the 3′ terminal 500 nt of 3′ exons within the custom transcriptome annotation was constructed. Transcript isoforms with ≥50 mapped reads in the terminal 500 nt region were used to calculate the mean value of poly(A) lengths and to represent the distribution of poly(A) tail reads for that region. Due to the dependency of reliable poly(A) tail length calculations on precise PAS annotations, any shifts in APA isoform preferences occurring within the 300 nt window (referred to as local PAS switch hereafter), limit the ability of TED-seq to determine poly(A) tail length changes. To avoid any errors in poly(A) tail inferences caused by a shift in APA usage, we removed PAS isoforms subject to potential bias from APA by processing our 3′-seq data as follows. First, we defined tandem PAS clusters of size = 300 nt by clustering experimentally determined PAS isoforms within ≤300 nt (9658 PAS in 5128 genes clustered to 6745 tandem clusters) and testing whether APA usage in a cluster was significantly altered upon LPS stimulation for all isoforms. Switching in the tandem poly(A) sites in a tandem PAS cluster was determined based on previously described approaches (Fu et al. 2011; Jia et al. 2017). The PPI (proportion of an individual PAS isoform in each tandem cluster) index was calculated for each time point across the activation time course. If the PPI within each cluster significantly differs between any pair of time points of the macrophage LPS activation time-course (χ2 test with FDR < 0.1), we considered those as locally shifted (124 tandem clusters, 410 PAS), and removed these isoforms from further analysis. We only considered PAS clusters with consistent APA isoform usage across the time-course for further analysis. Additionally, polychoric correlation coefficients, a version of Pearson correlation coefficient with discrete UTR indices, were calculated to estimate the switching direction, and defined as USI, UTR switching index (described as TSI; Harrow et al. 2012). A positive USI value (USI > 0.1) indicated a switch to the longer tandem 3′ UTRs (distal), while a shift to short tandem 3′ UTR (proximal) has a negative USI value (USI < −0.1). Next, if a PAS cluster contained multiple PAS, the PAS with the most read counts was defined as the major PAS isoform, whereas PAS isoforms with fewer read counts were considered as minor PAS isoforms (n = 2979). To avoid redundant use of a collection of TED-seq reads for the PAS in the same cluster, minor PAS isoforms were removed from the tail length analysis, leaving 6269 isoforms (5079 genes). For this set of transcript isoforms, differences in poly(A) tail length for a given transcript isoform between two biological conditions were compared using the Kolmogorov–Smirnov test and the P-value adjusted by FDR with the criteria of FDR < 0.1, and |ΔPAL| (difference in mean poly[A] tail length) ≥ 10 nt (Fig. 2; Supplemental Fig. S2D–F).
Identification of significant changes in tandem 3′-UTR isoform expression
By analyzing our custom transcript isoform annotation, each gene was scrutinized for tandem 3′-UTR expression. If a gene had multiple transcript isoforms that shared the same 3′ terminal exon (5′ splice site, thereof), but were cleaved and polyadenylated at different positions, these transcript isoforms were considered as tandem 3′-UTR isoforms. For analytic stringency, we only considered transcript isoforms with ≥50 TED-seq reads within the 300 nt region upstream of the PAS for further analysis. This approach allowed us to remove any potential decay intermediates and other artifacts derived from internal priming from our collection of PAS isoforms, and focus on a more confident set of transcript isoforms. In cases with multiple transcript isoforms located within ≤300 nt, we determined the isoform with the highest read counts as the major isoform, and the rest as minor isoforms. Finally, for the final set of major transcript isoforms, each transcript isoform was indexed based on their genomic location, as follows. In the case of a 3′ terminal exon expressing multiple PAS isoforms, the PAS closest to the stop codon was indexed as 1, incrementing the index with increasing distance from the stop codon. For a tandem 3′-UTR isoform, mRNA abundance was calculated as the read counts of the given isoform. As described in “Poly(A) tail length estimation,” we determined the switching direction using USI, and a χ2 test was performed to determine the statistical significance of the 3′-UTR switch. Under the criteria of the adjusted P-value (FDR) <0.1, a positive USI value (USI > 0.1) indicated a switch to the longer tandem 3′ UTRs (distal APA isoforms), while a shift to short tandem 3′ UTRs (proximal APA isoforms) has a negative USI value (USI < −0.1) (Supplemental Fig. S2A–C).
PRO-seq library preparation
PRO-seq libraries for differentiated THP-1 cells were produced essentially as previously described (Mahat et al. 2016). A total of 1 × 107 cells were permeabilized in 100 µL buffer D (50 mM Tris-Cl pH 8.0, 25% glycerol, 5 mM MgAc2, 0.1 mM EDTA, 5 mM DTT) for each PRO-seq sample. 2× concentrated nuclear-run-on buffer (20 mM Tris-HCl pH 8.0, 10 mM MgCl2, 2 mM DTT, 600 mM KCl, 500 µM ATP [Thermo Fisher, R0481], 500 µM GTP [Thermo Fisher, R0481], 50 µM biotin-11-CTP [PerkinElmer, NEL542001EA], 50 µM biotin-11-UTP [PerkinElmer; NEL543001EA], 1% sarkosyl [Fisher Scientific; AC612075000], 1 µL SUPERase In [Thermo Fisher, AM2694]) was added to the permeabilized cell, followed by incubation for 3 min at 37°C, followed by 3× volume addition of TRIzol LS (Thermo Fisher, 10296028) to stop the reaction. GlycoBlue (Thermo Fisher, AM9515) was added to the run-on products and subjected to ethanol precipitation. The resulting pellet was dissolved in DEPC-treated water, denatured at 65°C, and subject to base hydrolysis in 0.2 N NaOH on ice for 15 min, stopped by the addition of the same volume of 1 M Tris-HCl (pH 6.8). Biotinylated RNAs were isolated using Streptavidin-conjugated magnetic beads (Thermo Fisher, #11205D). Subsequently, 3′RNA adaptor ligation was performed (NEB; M0204L), followed by another round of bead binding, 5′ decapping using RppH (NEB, M0356S), 5′ end phosphorylation (NEB, M0201S), and then 5′ adaptor ligation. After the 5′ adaptor ligation, a final round of bead binding was performed; the purified products were reverse transcribed and PCR amplified, followed by Ampure XP-mediated size selection. Adaptor dimers were removed from the sequencing library by PAGE-mediated size selection, retaining fragments ≥175 nt, and the resulting libraries sequenced from their 3′ ends using an Illumina NextSeq500 platform. PRO-seq libraries were prepared in two biological replicates.
PRO-seq data preprocessing, mapping, and quantifying transcription activity
PRO-seq reads were trimmed by removing the adaptor sequence TGGAATTCTCGGGTGCCAAGG using Cutadapt (Martin 2011) with the parameter of –e 0.10 –overlap 2. From the trimmed reads, PCR duplicates were removed by collapsing reads with the same UMI barcodes, followed by UMI trimming. The de-duplicated reads with lengths ≥15 nt were aligned to the human genome (hg38) using BWA (Li and Durbin 2009). The resulting uniquely mapped reads were used for further analysis. The 5′ ends of the aligned reads represent the 3′ termini of the nascent RNA, but are located on the opposite strand. Therefore, the bam file of uniquely mapped reads was sorted, converted to bed format (BedTools; Quinlan and Hall 2010), retaining the 5′ position of the reads, and switched to the opposite strand to correct transcript orientation. Finally, the number of reads mapped to the gene body region, which spans from 500 nt downstream from the transcription start site (TSS + 500 bp) to 500 nt upstream of the poly(A) site (PAS—500 bp) for transcript isoforms longer than 1 kb, was calculated based on hg38 GENCODE (V26) transcript annotation, and normalized to reads per kilobase per million mapped (RPKM).
Combined analysis of PRO-seq, 3′-seq, and TED-seq
Stratified random sampling (SRS) was performed as follows. Genes were grouped by ΔRNA [log2 fold change (3′-seq RPM)] or ΔPAL [Δ(mean poly(A) length)] gene groups (down, no change, up) then randomly sampled to maintain the same distribution of ΔTXN [log2 fold change (PRO-seq)] across each group. Genes in each ΔRNA (or ΔPAL) group were split into bins based on ΔTXN, with the same number of genes sampled from each ΔTXN bin across the three ΔRNA (or ΔPAL) groups. For the analysis of genes with minimal transcriptional changes, we selected genes with minimal ΔTXN [log2 fold change (PRO-seq) < 0.5] throughout the time-course (0 h through 4 h), splitting such genes into three groups (down, no change, up) based on ΔRNA (or ΔPAL), followed by SRS based on ΔTXN (4 h/0 h) for more stringent ΔTXN equalization. These analyses were used to test the relationship between ΔPAL and ΔRNA after controlling for ΔTXN (Figs. 4–6; Supplemental Figs. S4, S5, S7).
qRT-PCR
To quantify RNA abundance, total RNA was reverse transcribed using Maxima H Minus reverse transcriptase (Thermo Scientific, EP0751) with random hexamers at 50°C for 1 h, followed by qPCR using SYBR green master mix (Bio-Rad, 172-5270), as described in the manufacturer's guide. The following primers were used to amplify individual target genes: human GAPDH (forward, 5′-CAGCAAGAGCACAAGAGGAA-3′; reverse, 5′-TGGTTGAGCACAGGGTACTTT-3′); GAPDH signal was used to normalize signals from other genes. The entire qPCR primer sets are detailed in Supplemental Table S9. Reactions were performed in 10 µL volume per reaction in four technical replicates, which was repeated twice (two biological replicates). The data are presented as mean ± SEM, and two different conditions were compared using a two-tailed Student's t-test.
PAT assay
Poly(A) tails on specific mRNAs were analyzed with a Poly(A) Tail-Length Assay Kit (Thermo Fisher, 764551KT), as follows. Total RNA was isolated and denatured at 65°C for 3 min. 3′ termini were extended by poly(A) polymerase with guanosine and inosine nucleotides. The G/I tailed poly(A) RNAs were reverse transcribed using a universal primer provided by the manufacturer, which was annealed to the G/I tail region, and then amplified by PCR with the following gene-specific custom forward primer and universal reverse primer; TNF F primer 5′-TGACCAACTGTCACTCATT-3′, IL1B F primer 5′-GTGCTCTCTTTAAATCAAGTCCT-3′. Other gene-specific primers and oligos used in this study are listed in Supplemental Table S9. To confirm the size of the deadenylated mRNAs for each mRNA species, poly(A) tails were digested by RNase H, as follows. Two micrograms of total RNA was denatured in the presence of oligo d(T)18 at 65°C for 5 min, followed by slowing cooling to 30°C. The poly(A) tails, hybridized to oligo dTs, were digested with 5U of RNase H (NEB, M0297) at 37°C for 1 h, followed by heat inactivation at 65°C for 20 min, and TRIzol extraction. The purified RNAs were subjected to 3′ RNA ligation with an RNA adaptor sequence, followed by reverse transcription (Thermo Scientific, EP0441) as described in the manufacturer's protocol. PAT PCR was performed as described by the manufacturer. Finally, the size of the PAT PCR product was assessed by running half of the PCR reaction through 6% PAGE gel and staining the gel with SYBR Gold (Invitrogen, S11494). PAT assay was repeated at least twice for each gene of interest, and one of the replicates was shown to be representative.
Association of AU content in the 3′ UTR with poly(A) tail length changes
For each transcript isoform, AU content in the 3′ UTR was calculated by the number of A and U nucleotides divided by the 3′-UTR length, using the hg38 reference sequence of the target transcript's 3′ UTR. For ΔPAL association tests, we classified transcript isoforms with ΔPAL ≥ 10 nt as PAL-increased, those with ΔPAL ≤ -10 nt as PAL-decreased, and with |ΔPAL| ≤ 5 nt as not undergoing changes in tail length. 3′-UTR length was calculated by integrating our experimentally determined PAS into the reference 3′-UTR annotations (GENCODE v26). For a given transcript isoform, codon optimality was calculated as the mean of the codon stabilization coefficients (CSC) of all codons constituting the corresponding coding sequence using CSC values previously determined in four human cell lines, HEK293T, HeLa, RPE, and K562 (Wu et al. 2019) and is related to Figure 3A,B and Supplemental Figure S3.
Association of 6-mer frequency with poly(A) tail length changes
A matrix of 6-mer counts for 3′ UTRs corresponding to our experimentally determined isoforms was created by counting the number of instances of each 6-mer (n = 4096) in each 3′ UTR, which were normalized by the 3′-UTR length. Alternatively, the first and last 500 nt of 3′-UTR regions were used in genes with 3′-UTR lengths longer than 1 kb. The associations of each 6-mer count with changes in poly(A) tail length were tested by binning genes into four quartile groups based on the given 6-mer content: 0%–25%, 25%–50%, 50%–75%, and 75%–100%; comparing ΔPAL between the top and bottom quartiles; and assessing by Student's t-test. After FDR correction, 6-mers were considered to be associated with poly(A) tail length changes with FDR < 0.1. The identified 6-mers (significant 6-mers) were grouped into seven groups based on the number of A and U residues; 0, 1, 2, 3, 4, 5, and 6. To examine the effects of the 6-mer on poly(A) tail length change, the value ΔΔPAL was defined for each of the 6-mers significantly associated with ΔPAL, as follows:
For 6-mers with ΔΔPAL > 0, we considered the corresponding 6-mer as associated with poly(A) tail lengthening in response to LPS stimulation, and vice versa for 6-mers with ΔΔPAL < 0, indicating association with a decrease in tail length. This is related to Figure 3C,D.
Combinatorial codes analysis in the 3′ UTR
To identify two different cis-elements whose co-presence is more associated with ΔPAL than that of a single type, we examined the combinatorial effects of two different 6-mers (A and B, hereafter) in the association with ΔPAL using a set of the 6-mers whose frequency was individually identified as associated with ΔPAL in the last 500 nt 3′-UTR region. Transcript isoforms were categorized into four groups based on the frequency of A and B in the last 500 nt 3′-UTR regions; (1) none of A and B, (2) A only (n = 2), (3) B only (n = 2), and (4) both A and B (n = 1 for each; total n = 2), where the total number of the tested 6-mers were fixed to avoid the potential bias from the number of motifs. This analysis was iterated for all different time point comparisons. To predict RBPs that are likely to bind to a given 6-mer, RBP motif scores were calculated for the 6-mer using a corresponding position weight matrix (pwm) of a given RBP, which was iterated across all RBPs (n = 202) using the CISBP-RNA database (Ray et al. 2013). If a RBP has a pwm with more than 6 nt positions, the given 6-mer was tested for a match to the pwm in a nucleotide increment; for a RBP motif pwm containing seven positions, two motif scores were generated for a given 6-mer, for example. Any RBP showing the RBP motif score ≥6 was considered as a RBP whose motif aligns to a given 6-mer.
RBP enrichment in the 3′ UTR
To scan for sites corresponding to RBP (RNA binding proteins), we used the CISBP-RNA database, which consists of 202 position weight matrixes for human RBP recognition motifs (Ray et al. 2013). Only the RBPs that are expressed in THP-1 cells, as defined by the 3′-seq data, were included (n = 86). Here, 3′-UTR sequences of experimentally determined PAS isoforms were used in the analysis. To count instances of an RBP binding motif enriched in the 3′ UTRs of interest (PAL increase) relative to the background (no change in PAL) or counterpart (PAL decrease), each 3′-UTR sequence was searched for the given RBP binding site with a score ≥6, calculated as the natural log transformation of the odds ratio of the occurrence of the given motif in the examined 3′-UTR sequence versus a two-order Markov model background. Fisher's exact test was performed to calculate the statistical significance, and the corresponding P-value was adjusted by FDR with a threshold for significant RBP motifs set as FDR < 0.2 (Figs. 5E, 6E; Supplemental Fig. S7D).
Analysis of ActD-pretreated LPS activation
Poly(A) tail length changes were quantified for the PAS isoforms that passed a cutoff of 50 TED-seq reads across all time points (0, 1, 2 h) in ActD pretreated samples (n = 7771 derived from 6069 genes). The genes with a mean PAL increase greater than five (ΔPAL ≥ 5 and FDR < 0.2) in both biological replicates were defined as cytoplasmic/post-transcriptional polyadenylation targets and those with no change or decrease (ΔPAL < 1) defined as controls. Otherwise, data was processed as described in the section “Poly(A) tail length estimation and identifying significant changes in poly(A) tail length” (Figs. 5, 6; Supplemental Figs. S6, S7).
Western blot
At different indicated time points upon LPS stimulation, cells were lysed in ice-cold lysis buffer (10 mM Tris, 10 mM NaCl [pH 8.0], 1% NP-40, 2 mM MnCl2) containing 1x Protease Inhibitor (Roche, 11873580001) on ice. After scraping the cells into a tube, samples were sonicated until the lysate solution cleared. For the phosphatase-untreated samples, SDS was added to a final concentration of 1%, followed by 10 min incubation at 95°C. For the control sample (phosphatase-treated), a 30 min incubation with Lambda phosphatase (NEB, P0753S) was performed at room temperature, followed by a 10 min incubation with Alkaline phosphatase (NEB, M0290). After addition of SDS to a final concentration of 1%, phosphatase-treated samples were boiled for 10 min at 95°C to inactivate the phosphatase enzymes. Protein concentration was determined by BCA assay, then equal amounts of proteins (50 µg) were prepared, mixed with 2× Laemmli buffer containing 2.5% β-mercaptoethanol, and then boiled again at 95°C for 5 min before loading. After electrophoresis in 7% SDS-PAGE gel and transfer to a nitrocellulose membrane, the blotted membrane was incubated with 3% BSA containing TBS-T, followed by overnight incubation with primary antibodies for ZFP36 (Cell Signaling, #71632), and vinculin (Sigma-Aldrich, V9131). After incubation with infrared fluorescent dye-conjugated secondary antibodies (IRDye 800CW goat anti-rabbit for ZFP36, IRDye 680RD goat anti-mouse for vinculin), proteins were detected on a LI-COR Odyssey. Phosphorylated and unphosphorylated ZFP36 were discriminated from one another by their difference in molecular weight and sensitivity to phosphatase treatment (Wang et al. 2015). This experiment was repeated three times, and one of the replicates is shown as a representative in the main figure.
Bulk poly(A) assay
A total of 3 µg of total RNA (extracted by TRIzol) was labeled with 33.3 µM pCp-Biotin (Jena Bioscience, NU-1706-BIO) with 20 U T4 ssRNA ligase (NEB, M0204S) in a 50 µL reaction overnight at 16°C, followed by RNA purification by RNA Clean and Concentrator (Zymo Research, R1013). The labeled RNAs were digested by RNase A/T1 mixture at a 1:1000 dilution (Thermo Scientific, EN0551) for 30 min at 37°C. The resulting poly(A) tails, which are insensitive to RNase A/T1 enzymes, were purified by TRIzol, followed by electrophoresis on a denaturing 7% 7M Urea-PAGE gel, together with a labeled RNA ladder (Invitrogen, AM7145). For northern blotting, the RNAs were transferred to a nitrocellulose membrane on a Semi-Dry Transfer Blotting System (Bio-Rad, 1703940). After membrane blocking and subsequent wash steps, the 3′ biotin-labeled poly(A) tails were detected with streptavidin-conjugated horseradish peroxidase (HRP) and a 1-min incubation with the chemiluminescent substrate (Luminol/Enhancer Solution), followed by brief exposure to X-ray film. All the materials used post transfer, including blocking buffer, wash buffer, streptavidin-HRP, and Luminol/Enhancer Solution, were components of the North2South Chemiluminescent Hybridization and Detection Kit (Thermo Scientific, #17097).
Cloning of human ZFP36 3′-UTR reporter constructs
For transfection into RAW 264.7 cells, an EGFP coding region and the human ZFP36 3′-UTR sequences, either wild-type (WT) or mutant versions (MUT-DEL, MUT-GC), were cloned downstream from the CMV promoter of the vector pCMV-7.1 (Addgene #47948) by Gibson assembly. For transduction into THP-1 cells, CMV-EGFP-ZFP36 3′-UTR fragments, with either wild-type or mutant (MUT-DEL, MUT-GC) versions of the human ZFP36 3′ UTR, were cloned into a modified lentiviral pGFP-3′-UTR plasmid (Clontech) by Gibson assembly. The human ZFP36 3′-UTR sequences (WT, MUT-DEL, MUT-GC) were ordered as gBlocks (IDT) with vector-overlapping sequences at the fragment ends to be compatible with Gibson assembly.
Designing mutant versions of human ZFP36 3′-UTR sequence
Wild-type 3′-UTR sequence of human ZFP36 was scanned for known RBP motifs using CISBP-RNA database. Motif score was computed by summing up weights (PWM) at corresponding positions. Out of the RBP motifs with a binding score ≥5, those which have (1) number of consecutive Us ≥3 (e.g., UUU and UUUU), and (2) the portion of U in the given motif >0.5, were regarded as poly(U)-containing RBP motifs, subject to sequence modification. In all chosen poly(U) motifs, the consecutive poly(U) sequences were deleted to build a MUT-DEL version of ZFP36 3′ UTR, or replaced with G and C of the same length as the U stretch. The 3′-UTR sequences of WT, MUT-DEL, and MUT-GC versions are provided in Supplemental Table S9.
Viral packaging and transduction
Lentiviral packaging and lentivirus infection with the 3′-UTR reporter constructs were performed following the protocols from Broad Institute GPP (The Genetic Perturbation Platform) web portal (https://portals.broadinstitute.org/gpp/public/resources/protocols). For lentiviral packaging, HEK293T cells were seeded to a density of 5 × 105 cells in 10 cm culture dishes. Twenty-four hours later, transfection was carried out using TransIT-LT1 transfection reagent (Mirus Bio, #MIR2304) to introduce packaging and lentiviral plasmids into HEK293T cells. After harvesting the media containing lentivirus, the virus supernatant was filtered using 0.45 µM syringe filters (VWR; #514–4133) and stored at −80°C until the lentiviral transduction. For transduction of THP-1 cells, 20,000 cells were plated in six-well plates, followed by spinfection with lentivirus at 300g at 30°C for 1.5 h. After 24 h of incubation, 10 mL fresh medium was added to the cells and transferred to a T25 flask. Transduction status of the 3′-UTR reporter constructs was monitored by checking GFP intensity on a microscope on a daily basis. Three days post-infection, G418 (Gibco, #10131027) was added to the cells at a final concentration of 1 mg/mL, followed by a 2-wk selection for the virus-integrated cells with a media change twice/week. After antibiotic selection, cells were differentiated with 200 ng/mL PMA overnight, followed by media change with regular fresh media (without PMA). Three days later, the differentiated cells were treated with or without 200 ng/mL LPS for 1 h before total RNA extraction for PAT assay.
Quantification and statistical analysis
Data are presented as mean ± SD or mean ± 95% CI (as indicated). Statistical significance was calculated with two-tailed Student's t-test, Kolmogorov–Smirnov test, or Wilcoxon signed-rank test with the significance denoted as follows: P < 0.05(*), P < 0.01(**), and P < 0.001 (***), unless noted otherwise. The type of statistical test, the value of n, and the statistical significance (P-value or FDR) are described in the figures, legends and/or “Results” section. All the experiments and the sequencing libraries were performed and prepared in two biological replicates, unless noted otherwise. For the screen shot of sequencing reads or immunoblot/PAT assay results, a single library/experiment was shown as a representative of ≥2 biological replicates confirmed to show consistent results. All the downstream sequencing data analyses were performed using the values averaged from two biological replicates. Stratified random samplings were used to determine the sample size. All graphs and statistical tests were performed using R.
DATA DEPOSITION
The code for processing TED-seq, PRO-seq, 3′-seq and the analyses reported in this paper is available at https://github.com/YeonuiKwak/Project_MacActivation. Raw and processed data of TED-seq, PRO-seq, and 3′-seq are available at GEO accession number GSE161188.
SUPPLEMENTAL MATERIAL
Supplemental material is available for this article.
Supplementary Material
ACKNOWLEDGMENTS
We thank Y. Woo in the Kwak laboratory for providing synthetic poly(A) RNA standards, the members of the Grimson and the Kwak laboratories for constructive discussion, and the Cornell Institute of Biotechnology, Genomics facility for next-generation sequencing. We also thank the Graduate Field of Genetics, Genomics and Development, and the Department of Molecular Biology at Cornell University for student and research support. This work was supported by discretionary funds provided to A.G. and H.K.
Author contributions: Y.K. and H.K. conceptualized the study; all authors contributed to the study design and methodological approaches. Experiments were performed by Y.K., and analysis of TED-seq and PRO-seq data was performed by Y.K. and H.K. The TED-seq, PRO-seq, 3′-seq and transcription inhibition data were analyzed by Y.K. under the supervision of A.G. and H.K. Y.K. and E.A.F performed 3′-UTR reporter assays with contributions from C.D. The manuscript was written by Y.K., A.G., and H.K.
Footnotes
Article is online at http://www.rnajournal.org/cgi/doi/10.1261/rna.078918.121.
Freely available online through the RNA Open Access option.
MEET THE FIRST AUTHOR
Yeonui Kwak.

Meet the First Author(s) is a new editorial feature within RNA, in which the first author(s) of research-based papers in each issue have the opportunity to introduce themselves and their work to readers of RNA and the RNA research community. Yeonui Kwak is the first author of this paper, “Dynamic and widespread control of poly(A) tail length during macrophage activation.” Yeonui is a graduate student co-advised by Dr. Andrew Grimson and Dr. Hojoong Kwak at Cornell University.
What are the major results described in your paper and how do they impact this branch of the field?
Most systematic, genome-wide investigations of poly(A) tail length control have been limited to some specific biological contexts, such as oocyte maturation. Most examples of poly(A) tail length regulation in nondevelopmental systems have only been shown with a handful of genes. By examining mRNA abundance, nascent transcripts, and poly(A) tail length across a time course of macrophage activation, we found some evidence that many genes are going through readenylation in this system. This result suggests that perhaps readenylation could be more common in post-embryonic systems.
What led you to study RNA or this aspect of RNA science?
RNA is an intermediate molecule that translates genetic information in DNA to protein. Gene expression is significantly modulated through a plethora of regulations acting on RNA. Therefore, gaining a full appreciation of gene regulation at the RNA level is crucial for understanding normal cellular physiology to human pathologies.
During the course of these experiments, were there any surprising results or particular difficulties that altered your thinking and subsequent focus?
It was surprising to us that many mRNAs that underwent tail extension encode proteins necessary for post-transcriptional regulation, including ZFP36. Particularly, it is intriguing that many mRNAs undergoing tail lengthening are, in turn, down-regulated by elevated levels of ZFP36. This result implies that macrophage activation involves a complex post-transcriptional feedback loop to ensure gene expression integral to macrophage activation.
If you were able to give one piece of advice to your younger self, what would that be?
Before carrying out an experiment, take enough time to think about what, why, and how you should do it. It is your solid answers to the “what, why, and how” that could drive your research in an effective way.
REFERENCES
- Batra R, Stark TJ, Clark E, Belzile J-P, Wheeler EC, Yee BA, Huang H, Gelboin-Burkhart C, Huelga SC, Aigner S, et al. 2016. RNA-binding protein CPEB1 remodels host and viral RNA landscapes. Nat Struct Mol Biol 23: 1101–1110. 10.1038/nsmb.3310 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blumberg A, Zhao Y, Huang Y-F, Dukler N, Rice EJ, Chivu AG, Krumholz K, Danko CG, Siepel A. 2021. Characterizing RNA stability genome-wide through combined analysis of PRO-seq and RNA-seq data. BMC Biol 19: 30. 10.1186/s12915-021-00949-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braun JE, Huntzinger E, Fauser M, Izaurralde E. 2011. GW182 proteins directly recruit cytoplasmic deadenylase complexes to miRNA targets. Mol Cell 44: 120–133. 10.1016/j.molcel.2011.09.007 [DOI] [PubMed] [Google Scholar]
- Brooks SA, Blackshear PJ. 2013. Tristetraprolin (TTP): interactions with mRNA and proteins, and current thoughts on mechanisms of action. Biochim Biophys Acta 1829: 666–679. 10.1016/j.bbagrm.2013.02.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caput D, Beutler B, Hartog K, Thayer R, Brown-Shimer S, Cerami A. 1986. Identification of a common nucleotide sequence in the 3′-untranslated region of mRNA molecules specifying inflammatory mediators. Proc Natl Acad Sci 83: 1670–1674. 10.1073/pnas.83.6.1670 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carballo E, Lai WS, Blackshear PJ. 1998. Feedback inhibition of macrophage tumor necrosis factor-α production by tristetraprolin. Science 281: 1001–1005. 10.1126/science.281.5379.1001 [DOI] [PubMed] [Google Scholar]
- Carpenter S, Ricci EP, Mercier BC, Moore MJ, Fitzgerald KA. 2014. Post-transcriptional regulation of gene expression in innate immunity. Nat Rev Immunol 14: 361–376. 10.1038/nri3682 [DOI] [PubMed] [Google Scholar]
- Chang H, Lim J, Ha M, Kim VN. 2014. TAIL-seq: genome-wide determination of poly(A) tail length and 3′ end modifications. Mol Cell 53: 1044–1052. 10.1016/j.molcel.2014.02.007 [DOI] [PubMed] [Google Scholar]
- Charlesworth A, Meijer HA, de Moor CH. 2013. Specificity factors in cytoplasmic polyadenylation. Wiley Interdiscip Rev RNA 4: 437–461. 10.1002/wrna.1171 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chrestensen CA, Schroeder MJ, Shabanowitz J, Hunt DF, Pelo JW, Worthington MT, Sturgill TW. 2004. MAPKAP kinase 2 phosphorylates tristetraprolin on in vivo sites including Ser178, a site required for 14-3-3 binding. J Biol Chem 279: 10176–10184. 10.1074/jbc.M310486200 [DOI] [PubMed] [Google Scholar]
- Corbett AH. 2018. Post-transcriptional regulation of gene expression and human disease. Curr Opin Cell Biol 52: 96–104. 10.1016/j.ceb.2018.02.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crawford EK, Ensor JE, Kalvakolanu I, Hasday JD. 1997. The role of 3′ poly(A) tail metabolism in tumor necrosis factor-α regulation. J Biol Chem 272: 21120–21127. 10.1074/jbc.272.34.21120 [DOI] [PubMed] [Google Scholar]
- Dai X-X, Jiang J-C, Sha Q-Q, Jiang Y, Ou X-H, Fan H-Y. 2019. A combinatorial code for mRNA 3′-UTR-mediated translational control in the mouse oocyte. Nucleic Acids Res 47: 328–340. 10.1093/nar/gky971 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dobin A, Davis CA, Schlesinger F, Drenkow J, Zaleski C, Jha S, Batut P, Chaisson M, Gingeras TR. 2013. STAR: ultrafast universal RNA-seq aligner. Bioinformatics 29: 15–21. 10.1093/bioinformatics/bts635 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eichhorn SW, Subtelny AO, Kronja I, Kwasnieski JC, Orr-Weaver TL, Bartel DP. 2016. mRNA poly(A)-tail changes specified by deadenylation broadly reshape translation in Drosophila oocytes and early embryos. Elife 5: e16955. 10.7554/eLife.16955 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eisen TJ, Eichhorn SW, Subtelny AO, Bartel DP. 2020a. MicroRNAs cause accelerated decay of short-tailed target mRNAs. Mol Cell 77: 775–785. 10.1016/j.molcel.2019.12.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eisen TJ, Eichhorn SW, Subtelny AO, Lin KS, McGeary SE, Gupta S, Bartel DP. 2020b. The dynamics of cytoplasmic mRNA metabolism. Mol Cell 77: 786–799. 10.1016/j.molcel.2019.12.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frankish A, Diekhans M, Ferreira A-M, Johnson R, Jungreis I, Loveland J, Mudge JM, Sisu C, Wright J, Armstrong J, et al. 2019. GENCODE reference annotation for the human and mouse genomes. Nucleic Acids Res 47: 766–773. 10.1093/nar/gky955 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu Y, Sun Y, Li Y, Li J, Rao X, Chen C, Xu A. 2011. Differential genome-wide profiling of tandem 3′ UTRs among human breast cancer and normal cells by high-throughput sequencing. Genome Res 21: 741–747. 10.1101/gr.115295.110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallie DR. 1991. The cap and poly(A) tail function synergistically to regulate mRNA translational efficiency. Genes Dev 5: 2108–2116. 10.1101/gad.5.11.2108 [DOI] [PubMed] [Google Scholar]
- Geissler R, Grimson A. 2016. A position-specific 3′UTR sequence that accelerates mRNA decay. RNA Biol 13: 1075–1077. 10.1080/15476286.2016.1225645 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geissler R, Simkin A, Floss D, Patel R, Fogarty EA, Scheller J, Grimson A. 2016. A widespread sequence-specific mRNA decay pathway mediated by hnRNPs A1 and A2/B1. Genes Dev 30: 1070–1085. 10.1101/gad.277392.116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giraldez AJ, Mishima Y, Rihel J, Grocock RJ, Van Dongen S, Inoue K, Enright AJ, Schier AF. 2006. Zebrafish miR-430 promotes deadenylation and clearance of maternal mRNAs. Science 312: 75–79. 10.1126/science.1122689 [DOI] [PubMed] [Google Scholar]
- Grimson A, Farh KK-H, Johnston WK, Garrett-Engele P, Lim LP, Bartel DP. 2007. MicroRNA targeting specificity in mammals: determinants beyond seed pairing. Mol Cell 27: 91–105. 10.1016/j.molcel.2007.06.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hake LE, Richter JD. 1994. CPEB is a specificity factor that mediates cytoplasmic polyadenylation during Xenopus oocyte maturation. Cell 79: 617–627. 10.1016/0092-8674(94)90547-9 [DOI] [PubMed] [Google Scholar]
- Harrow J, Frankish A, Gonzalez JM, Tapanari E, Diekhans M, Kokocinski F, Aken BL, Barrell D, Zadissa A, Searle S, et al. 2012. GENCODE: the reference human genome annotation for The ENCODE Project. Genome Res 22: 1760–1774. 10.1101/gr.135350.111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang DW, Sherman BT, Lempicki RA. 2009. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc 4: 44–57. 10.1038/nprot.2008.211 [DOI] [PubMed] [Google Scholar]
- Jalkanen AL, Coleman SJ, Wilusz J. 2014. Determinants and implications of mRNA poly(A) tail size: does this protein make my tail look big? Semin Cell Dev Biol 34: 24–32. 10.1016/j.semcdb.2014.05.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jia X, Yuan S, Wang Y, Fu Y, Ge Y, Ge Y, Lan X, Feng Y, Qiu F, Li P, et al. 2017. The role of alternative polyadenylation in the antiviral innate immune response. Nat Commun 8: 14605. 10.1038/ncomms14605 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katsanou V, Papadaki O, Milatos S, Blackshear PJ, Anderson P, Kollias G, Kontoyiannis DL. 2005. HuR as a negative posttranscriptional modulator in inflammation. Mol Cell 19: 777–789. 10.1016/j.molcel.2005.08.007 [DOI] [PubMed] [Google Scholar]
- Kim D, Lee Y-S, Jung S-J, Yeo J, Seo JJ, Lee Y-Y, Lim J, Chang H, Song J, Yang J, et al. 2020. Viral hijacking of the TENT4-ZCCHC14 complex protects viral RNAs via mixed tailing. Nat Struct Mol Biol 27: 581–588. 10.1038/s41594-020-0427-3 [DOI] [PubMed] [Google Scholar]
- Kojima S, Green CB. 2015. Analysis of circadian regulation of poly(A) tail length. Methods Enzymol 551: 387–403. 10.1016/bs.mie.2014.10.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kondrashov A, Meijer HA, Barthet-Barateig A, Parker HN, Khurshid A, Tessier S, Sicard M, Knox AJ, Pang L, De Moor CH. 2012. Inhibition of polyadenylation reduces inflammatory gene induction. RNA 18: 2236–2250. 10.1261/rna.032391.112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kontoyiannis D, Pasparakis M, Pizarro TT, Cominelli F, Kollias G. 1999. Impaired on/off regulation of TNF biosynthesis in mice lacking TNF AU-rich elements: implications for joint and gut-associated immunopathologies. Immunity 10: 387–398. 10.1016/S1074-7613(00)80038-2 [DOI] [PubMed] [Google Scholar]
- Kuchta K, Muszewska A, Knizewski L, Steczkiewicz K, Wyrwicz LS, Pawlowski K, Rychlewski L, Ginalski K. 2016. FAM46 proteins are novel eukaryotic non-canonical poly(A) polymerases. Nucleic Acids Res 44: 3534–3548. 10.1093/nar/gkw222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwak H, Fuda NJ, Core LJ, Lis JT. 2013. Precise maps of RNA polymerase reveal how promoters direct initiation and pausing. Science 339: 950–953. 10.1126/science.1229386 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai WS, Kennington EA, Blackshear PJ. 2003. Tristetraprolin and its family members can promote the cell-free deadenylation of AU-rich element-containing mRNAs by poly(A) ribonuclease. Mol Cell Biol 23: 3798–3812. 10.1128/MCB.23.11.3798-3812.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Legnini I, Alles J, Karaiskos N, Ayoub S, Rajewsky N. 2019. FLAM-seq: full-length mRNA sequencing reveals principles of poly(A) tail length control. Nat Methods 16: 879–886. 10.1038/s41592-019-0503-y [DOI] [PubMed] [Google Scholar]
- Li H, Durbin R. 2009. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 25: 1754–1760. 10.1093/bioinformatics/btp324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, Sun Y, Fu Y, Li M, Huang G, Zhang C, Liang J, Huang S, Shen G, Yuan S, et al. 2012. Dynamic landscape of tandem 3′ UTRs during zebrafish development. Genome Res 22: 1899–1906. 10.1101/gr.128488.111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim J, Ha M, Chang H, Kwon SC, Simanshu DK, Patel DJ, Kim VN. 2014. Uridylation by TUT4 and TUT7 marks mRNA for degradation. Cell 159: 1365–1376. 10.1016/j.cell.2014.10.055 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim J, Lee M, Son A, Chang H, Kim VN. 2016. mTAIL-seq reveals dynamic poly(A) tail regulation in oocyte-to-embryo development. Genes Dev 30: 1671–1682. 10.1101/gad.284802.116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lima SA, Chipman LB, Nicholson AL, Chen Y-H, Yee BA, Yeo GW, Coller J, Pasquinelli AE. 2017. Short poly(A) tails are a conserved feature of highly expressed genes. Nat Struct Mol Biol 24: 1057–1063. 10.1038/nsmb.3499 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liudkovska V, Dziembowski A. 2021. Functions and mechanisms of RNA tailing by metazoan terminal nucleotidyltransferases. Wiley Interdiscip Rev RNA 12: e1622. 10.1002/wrna.1622 [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacDonald CC, McMahon KW. 2010. Tissue-specific mechanisms of alternative polyadenylation: testis, brain, and beyond. Wiley Interdiscip Rev RNA 1: 494–501. 10.1002/wrna.29 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahat DB, Kwak H, Booth GT, Jonkers IH, Danko CG, Patel RK, Waters CT, Munson K, Core LJ, Lis JT. 2016. Base-pair resolution genome-wide mapping of active RNA polymerases using precision nuclear run-on (PRO-seq). Nat Protoc 11: 1455–1476. 10.1038/nprot.2016.086 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin M. 2011. Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet.journal 17: 10–12. 10.14806/ej.17.1.200 [DOI] [Google Scholar]
- Mayr C. 2016. Evolution and biological roles of alternative 3′UTRs. Trends Cell Biol 26: 227–237. 10.1016/j.tcb.2015.10.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayr C, Bartel DP. 2009. Widespread shortening of 3′UTRs by alternative cleavage and polyadenylation activates oncogenes in cancer cells. Cell 138: 673–684. 10.1016/j.cell.2009.06.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Millevoi S, Vagner S. 2010. Molecular mechanisms of eukaryotic pre-mRNA 3′ end processing regulation. Nucleic Acids Res 38: 2757–2774. 10.1093/nar/gkp1176 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore MJ, Blachere NE, Fak JJ, Park CY, Sawicka K, Parveen S, Zucker-Scharff I, Moltedo B, Rudensky AY, Darnell RB. 2018. ZFP36 RNA-binding proteins restrain T cell activation and anti-viral immunity. Elife 7: e33057. 10.7554/eLife.33057 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muhlrad D, Parker R. 2005. The yeast EDC1 mRNA undergoes deadenylation-independent decapping stimulated by Not2p, Not4p, and Not5p. EMBO J 24: 1033–1045. 10.1038/sj.emboj.7600560 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mukherjee N, Jacobs NC, Hafner M, Kennington EA, Nusbaum JD, Tuschl T, Blackshear PJ, Ohler U. 2014. Global target mRNA specification and regulation by the RNA-binding protein ZFP36. Genome Biol 15: R12. 10.1186/gb-2014-15-1-r12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parameswaran N, Patial S. 2010. Tumor necrosis factor-α signaling in macrophages. Crit Rev Eukaryot Gene Expr 20: 87–103. 10.1615/CritRevEukarGeneExpr.v20.i2.10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park J-E, Yi H, Kim Y, Chang H, Kim VN. 2016. Regulation of poly(A) tail and translation during the somatic cell cycle. Mol Cell 62: 462–471. 10.1016/j.molcel.2016.04.007 [DOI] [PubMed] [Google Scholar]
- Patel RK, West JD, Jiang Y, Fogarty EA, Grimson A. 2020. Robust partitioning of microRNA targets from downstream regulatory changes. Nucleic Acids Res 48: 9724–9746. 10.1093/nar/gkaa687 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piqué M, López JM, Foissac S, Guigó R, Méndez R. 2008. A combinatorial code for CPE-mediated translational control. Cell 132: 434–448. 10.1016/j.cell.2007.12.038 [DOI] [PubMed] [Google Scholar]
- Quinlan AR, Hall IM. 2010. BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics 26: 841–842. 10.1093/bioinformatics/btq033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ray D, Kazan H, Cook KB, Weirauch MT, Najafabadi HS, Li X, Gueroussov S, Albu M, Zheng H, Yang A, et al. 2013. A compendium of RNA-binding motifs for decoding gene regulation. Nature 499: 172–177. 10.1038/nature12311 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rissland OS, Subtelny AO, Wang M, Lugowski A, Nicholson B, Laver JD, Sidhu SS, Smibert CA, Lipshitz HD, Bartel DP. 2017. The influence of microRNAs and poly(A) tail length on endogenous mRNA–protein complexes. Genome Biol 18: 211. 10.1186/s13059-017-1330-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson JT, Thorvaldsdóttir H, Winckler W, Guttman M, Lander ES, Getz G, Mesirov JP. 2011. Integrative genomics viewer. Nat Biotechnol 29: 24–26. 10.1038/nbt.1754 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sandler H, Kreth J, Timmers HTM, Stoecklin G. 2011. Not1 mediates recruitment of the deadenylase Caf1 to mRNAs targeted for degradation by tristetraprolin. Nucleic Acids Res 39: 4373–4386. 10.1093/nar/gkr011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanduja S, Blanco FF, Dixon DA. 2011. The roles of TTP and BRF proteins in regulated mRNA decay. Wiley Interdiscip Rev RNA 2: 42–57. 10.1002/wrna.28 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmieder R, Edwards R. 2011. Quality control and preprocessing of metagenomic datasets. Bioinformatics 27: 863–864. 10.1093/bioinformatics/btr026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smibert P, Miura P, Westholm JO, Shenker S, May G, Duff MO, Zhang D, Eads BD, Carlson J, Brown JB, et al. 2012. Global patterns of tissue-specific alternative polyadenylation in Drosophila. Cell Rep 1: 277–289. 10.1016/j.celrep.2012.01.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stark C, Breitkreutz B-J, Reguly T, Boucher L, Breitkreutz A, Tyers M. 2006. BioGRID: a general repository for interaction datasets. Nucleic Acids Res 34: D535–D539. 10.1093/nar/gkj109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stoecklin G, Stubbs T, Kedersha N, Wax S, Rigby WF, Blackwell TK, Anderson P. 2004. MK2-induced tristetraprolin:14-3-3 complexes prevent stress granule association and ARE-mRNA decay. EMBO J 23: 1313–1324. 10.1038/sj.emboj.7600163 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subtelny AO, Eichhorn SW, Chen GR, Sive H, Bartel DP. 2014. Poly(A)-tail profiling reveals an embryonic switch in translational control. Nature 508: 66–71. 10.1038/nature13007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian B, Pan Z, Lee JY. 2007. Widespread mRNA polyadenylation events in introns indicate dynamic interplay between polyadenylation and splicing. Genome Res 17: 156–165. 10.1101/gr.5532707 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tiedje C, Ronkina N, Tehrani M, Dhamija S, Laass K, Holtmann H, Kotlyarov A, Gaestel M. 2012. The p38/MK2-driven exchange between tristetraprolin and HuR regulates AU–rich element–dependent translation. PLoS Genet 8: e1002977. 10.1371/journal.pgen.1002977 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tiedje C, Diaz-Muñoz MD, Trulley P, Ahlfors H, Laaß K, Blackshear PJ, Turner M, Gaestel M. 2016. The RNA-binding protein TTP is a global post-transcriptional regulator of feedback control in inflammation. Nucleic Acids Res 44: 7418–7440. 10.1093/nar/gkw474 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang K-T, Wang H-H, Wu Y-Y, Su Y-L, Chiang P-Y, Lin N-Y, Wang S-C, Chang G-D, Chang C-J. 2015. Functional regulation of Zfp36l1 and Zfp36l2 in response to lipopolysaccharide in mouse RAW264.7 macrophages. J Inflamm (Lond) 12: 42. 10.1186/s12950-015-0088-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warkocki Z, Liudkovska V, Gewartowska O, Mroczek S, Dziembowski A. 2018. Terminal nucleotidyl transferases (TENTs) in mammalian RNA metabolism. Philos Trans R Soc Lond B Biol Sci 373: 20180162. 10.1098/rstb.2018.0162 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weill L, Belloc E, Bava F-A, Méndez R. 2012. Translational control by changes in poly(A) tail length: recycling mRNAs. Nat Struct Mol Biol 19: 577–585. 10.1038/nsmb.2311 [DOI] [PubMed] [Google Scholar]
- Wells DG, Dong X, Quinlan EM, Huang YS, Bear MF, Richter JD, Fallon JR. 2001. A role for the cytoplasmic polyadenylation element in NMDA receptor-regulated mRNA translation in neurons. J Neurosci 21: 9541–9548. 10.1523/JNEUROSCI.21-24-09541.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woo YM, Kwak Y, Namkoong S, Kristjánsdóttir K, Lee SH, Lee JH, Kwak H. 2018. TED-seq identifies the dynamics of poly(A) length during ER stress. Cell Rep 24: 3630–3641.e7. 10.1016/j.celrep.2018.08.084 [DOI] [PubMed] [Google Scholar]
- Wu L, Wells D, Tay J, Mendis D, Abbott MA, Barnitt A, Quinlan E, Heynen A, Fallon JR, Richter JD. 1998. CPEB-mediated cytoplasmic polyadenylation and the regulation of experience-dependent translation of α-CaMKII mRNA at synapses. Neuron 21: 1129–1139. 10.1016/S0896-6273(00)80630-3 [DOI] [PubMed] [Google Scholar]
- Wu L, Fan J, Belasco JG. 2006. MicroRNAs direct rapid deadenylation of mRNA. Proc Natl Acad Sci 103: 4034–4039. 10.1073/pnas.0510928103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu Q, Medina SG, Kushawah G, DeVore ML, Castellano LA, Hand JM, Wright M, Bazzini AA. 2019. Translation affects mRNA stability in a codon-dependent manner in human cells. Elife 8: e45396. 10.7554/eLife.45396 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu N, Chen CY, Shyu AB. 1997. Modulation of the fate of cytoplasmic mRNA by AU-rich elements: key sequence features controlling mRNA deadenylation and decay. Mol Cell Biol 17: 4611–4621. 10.1128/MCB.17.8.4611 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang D, Guelfi S, Garcia-Ruiz S, Costa B, Reynolds RH, D'Sa K, Liu W, Courtin T, Peterson A, Jaffe AE, et al. 2020. Incomplete annotation has a disproportionate impact on our understanding of Mendelian and complex neurogenetic disorders. Sci Adv 6: eaay8299. 10.1126/sciadv.aay8299 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng D, Ezzeddine N, Chen C-YA, Zhu W, He X, Shyu A-B. 2008. Deadenylation is prerequisite for P-body formation and mRNA decay in mammalian cells. J Cell Biol 182: 89. 10.1083/jcb.200801196 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.