Abstract
We describe a new PCR-based method for distinguishing human and cow fecal contamination in coastal waters without culturing indicator organisms, and we show that the method can be used to track bacterial marker sequences in complex environments. We identified two human-specific genetic markers and five cow-specific genetic markers in fecal samples by amplifying 16S ribosomal DNA (rDNA) fragments from members of the genus Bifidobacterium and the Bacteroides-Prevotella group and performing length heterogeneity PCR and terminal restriction fragment length polymorphism analyses. Host-specific patterns suggested that there are species composition differences in the Bifidobacterium and Bacteroides-Prevotella populations of human and cow feces. The patterns were highly reproducible among different hosts belonging to the same species. Additionally, all host-specific genetic markers were detected in water samples collected from areas frequently contaminated with fecal pollution. Ease of detection and longer survival in water made Bacteroides-Prevotella indicators better than Bifidobacterium indicators. Fecal 16S rDNA sequences corresponding to our Bacteroides-Prevotella markers comprised closely related gene clusters, none of which exactly matched previously published Bacteroides or Prevotella sequences. Our method detected host-specific markers in water at pollutant concentrations of 2.8 × 10−5 to 2.8 × 10−7 g (dry weight) of feces/liter and 6.8 × 10−7 g (dry weight) of sewage/liter. Although our aim was to identify nonpoint sources of fecal contamination, the method described here should be widely applicable for monitoring spatial and temporal fluctuations in specific bacterial groups in natural environments.
Fecal pollution is a serious environmental problem that affects many coastal regions in the United States and worldwide. Pathogens associated with fecal pollution can lead to human disease and economic losses in industries that depend on coastal waters, such as shell fisheries (13). Despite efforts to minimize fecal input into coastal waterways, the problem persists, partly due to an inability to reliably identify nonpoint sources. These sources may include inefficient sewage treatment plants, leaking septic systems, agricultural runoff, or wildlife (53). Knowing the source of the contamination is crucial to effective resource management and, ultimately, solution of the problem.
The most widely used method for measuring fecal pollution is to quantify viable fecal coliforms by culturing them (3). This method, however, does not identify the source of fecal contamination. In addition, the extent to which fecal coliforms settle, grow, and are resuspended after they are released into receiving waters remains controversial (14), leaving the method's accuracy in question. Thus, there is a need for a reliable method for identifying the source of fecal pollution that does not rely on measuring coliform concentrations.
Several researchers have suggested that members of the genera Bacteroides and Bifidobacterium could be used as indicator organisms (1, 20, 47). Members of both of these genera are strict anaerobes, are restricted to warm-blooded animals, and, unlike coliforms, make up a significant portion of fecal bacteria. Additionally, because they are strict anaerobes, they do not survive very long once they are released into receiving waters (5, 10, 35, 47). The use of these organisms as indicators, however, has been limited because strict anaerobes are often difficult to grow. The difficulty of growing strict anaerobes can be circumvented by using molecular methods rather than culture-based methods to detect them.
Molecular approaches have become popular and efficient methods for characterizing and tracking changes in the community structures of microbial populations (26, 43, 55). Our approach, however, was to identify and track spatial and temporal fluctuations in individual bacterial markers in a natural environment. We used recently developed techniques that distinguish members of mixtures of bacterial gene sequences by detecting differences in the number of base pairs in a particular gene fragment (4, 9). Length heterogeneity PCR (LH-PCR) (55) and terminal restriction fragment length polymorphism (T-RFLP) analyses (8, 12, 37) are methods which are used to analyze differences in the lengths of gene fragments due to insertions and deletions and to estimate the relative abundance of each fragment. To track a bacterial marker sequence, it first must be uniquely identified by using a combination of specific primers for PCR and appropriate restriction enzymes. Once a reliable identification has been made, the sequence can be monitored easily and quickly in many samples.
Our goals were (i) to develop 16S ribosomal DNA (rDNA) markers that were based on fecal anaerobes and distinguished human fecal pollution from cow fecal pollution and (ii) to show that these markers could be recovered from natural waters and identified, indicating that they could be used to measure and distinguish the source of fecal pollution.
Starting with cow and human fecal samples, we used LH-PCR and T-RFLP analyses to characterize the community profiles of members of the Bacteroides-Prevotella group and the genus Bifidobacterium and to look for unique host species-specific patterns or markers. We identified host-specific markers that were highly reproducible among hosts belonging to the same species, but the Bifidobacterium markers were more difficult to detect.
To show that the markers could also be recovered from natural waters, we analyzed water samples from Tillamook Bay, Oreg. Pollution in this bay, which is the site of a major shellfish industry, is thought to be mostly due to dairy cows, but the possibility that there is human fecal contamination from septic tanks and sewage treatment plants cannot be ruled out. We recovered our host-specific markers from water samples, and sequence analyses confirmed their identities.
MATERIALS AND METHODS
Sample collection. (i) Fecal samples.
Human fecal samples were donated by healthy adult and child volunteers from Corvallis, Oreg., including Caucasian, Asian, and Hispanic individuals. Samples were collected in sterile containers and stored at −80°C. Fresh cow fecal samples were collected from healthy Holstein dairy cows from two farms in Corvallis, Oreg., and three farms in Tillamook County, Oreg. We collected Corvallis cow fecal samples during three different seasons from 1996 to 1998 and Tillamook County cow fecal samples during the fall of 1996. Samples were collected with sterile utensils, placed in sterile 50-ml tubes, kept on ice for transport to the lab, and stored at −80°C.
(ii) Water samples.
Samples were collected from multiple bay and river sites in the Tillamook Bay watershed and from sewage treatment facilities in Corvallis and Tillamook, Oreg. We selected sites representing three rivers, the Tillamook River, the Trask River, and the Wilson River, that are frequently contaminated with fecal pollution, and four sites along a north-south transect starting near the confluence of the Tillamook and Trask rivers and ending at a site near the mouth of the estuary. Water samples were collected in sterile 1-liter containers from surface waters and were stored on ice during transport to the lab. Upon return to the lab, we filtered water samples through 0.2-μm-pore-size Supor-200 filters. The filters were placed in separate plastic bags or 50-ml disposable centrifuge tubes containing 5 ml of lysis buffer (20 mM EDTA, 400 mM NaCl, 750 mM sucrose, 50 mM Tris; pH 9) and stored at −80°C. We measured fecal coliform concentrations by using standard methods (3).
DNA extraction. (i) Fecal samples.
We extracted DNA from fecal samples by the bead-beating method of Gray and Herwig (27), with the following modifications: 0.5 g of 0.1-mm-diameter glass beads (acid washed and baked) was used, polyvinylpolypyrrolidone was omitted from the lysis buffer, crude extracts were ethanol precipitated, and the resulting pellets dried under a vacuum and resuspended in InstaGene Matrix (Bio-Rad, Hercules, Calif.) or TE (10 mM Tris, 1 mM EDTA; pH 8). The DNA extracts were purified by phenol-chloroform extraction followed by ethanol precipitation and resuspension in TE.
(ii) Water samples.
The method used to extract DNA from water samples was based on the method of Giovannoni (25), except that the cesium trifluoroacetate purification steps were omitted. Instead, samples were cleaned by using one of the following methods: (i) 1 volume of 20% polyethylene glycol 8000 in 2.5 M NaCl was added, the samples were incubated for 15 min at 37°C and centrifuged for 10 min at 14,000 × g, and the resulting pellets were washed twice with ice-cold 80% ethanol; (ii) guanidine thiocyanate (Fluka, Buchs, Switzerland) purification based on the method of Pitcher and colleagues (46) was used; or (iii) polyvinylpolypyrrolidone (Aldrich, Milwaukee, Wis.) spin columns (6) were used.
PCR.
Approximately 2 to 4 ng of fecal DNAs from individual humans and cows was amplified by PCR. In addition to analyzing individual samples, we also analyzed pooled PCR products from multiple individuals obtained from each host species. DNAs from 14 human samples were amplified with both Bacteroides-Prevotella and Bifidobacterium primers (Table 1). DNAs from eight Corvallis cows and eight Tillamook cows were amplified with Bacteroides-Prevotella primers, but DNAs from only four Corvallis cows and four Tillamook cows were amplified with the Bifidobacterium primers. Each 50-μl PCR mixture contained 1× Taq polymerase buffer, each primer at a concentration of 10 μM, each deoxynucleoside triphosphate at a concentration of 200 μM, 1.25 U of Taq polymerase, 640 ng of bovine serum albumin per μl (34), and 1.5 mM MgCl2. Bif601R and Bac32F were labeled with the fluorophore 6-FAM (GenSet, La Jolla, Calif.). Non-fluorophore-labeled primers Bac303R, Bac708R, and Bif164F were synthesized by Gibco BRL (Gaithersburg, Md.). New primers Bif601R, Bac32F, and Bac708R were designed by using the Probe Design function of ARB (54), and their specificities were confirmed by using CHECK_PROBE of the Ribosomal Database Project (39) and Probe Match of ARB. We established primer conditions by using DNAs from cultured Bacteroides and Bifidobacterium strains. A thermal minicycler (MJ Research, Watertown, Mass.) was used for all reactions with the following conditions: 35 cycles consisting of 94°C for 30 s, 53°C for 1 min, and 72°C for 2 min, followed by a final 6-min extension at 72°C. We quantified the products in a 1% agarose gel by comparing the band intensities to the intensity of a low-molecular-weight DNA mass ladder (Gibco BRL).
TABLE 1.
Primers used in this study
| Primera | Sequence (5′-3′) | Target | Reference |
|---|---|---|---|
| Bac32F | AACGCTAGCTACAGGCTT | Bacteroides-Prevotella | This study |
| Bac303R | CCAATGTGGGGGACCTTC | Bacteroides-Prevotella | —b |
| Bac708R | CAATCGGAGTTCTTCGTG | Bacteroides-Prevotella | This study |
| Bif164F | GGGTGGTAATGCCGGATG | Bifidobacterium | 36 |
| Bif601R | TAAGCGATGGACTTTCACACC | Bifidobacterium | This study |
Bac, Bacteroides-Prevotella; Bif, Bifidobacterium. The numbers correspond to numbers in the E. coli 16S rRNA gene.
Modified from the study of Manz et al. (40).
Restriction endonuclease digestion.
We chose restriction enzymes based on an analysis of previously published sequences in the GenBank database by using Mapsort (Genetics Computer Group, Madison, Wis.). Enzymes that produced the greatest number of terminal restriction fragments with different lengths from the Bacteroides-Prevotella or Bifidobacterium 16S rDNA sequences were tested empirically. Enzymes were purchased from New England Biolabs (Beverly, Mass.). PCR products amplified with Bac32F and Bac708R were digested overnight at 37°C with either AciI or HaeIII. PCR products amplified with Bif164F and Bif601R were digested overnight with HaeIII (at 37°C) or TaqI (at 65°C). Each 20-μl digestion mixture contained 20 to 40 ng of PCR products, 10 U of enzyme, 1× enzyme buffer, and 100 μg of bovine serum albumin per μl (only for TaqI).
GeneScan analysis.
All analyses were performed with individual samples, as well as host-specific community samples. Approximately 25-fmol portions of PCR products or restriction digest products were resolved on a Long Ranger polyacrylamide gel (FMC, Rockland, Maine) by using a model ABI 377 automated DNA sequencer and GeneScan software (Applied Biosystems Inc., Fremont, Calif.) (ABI). An internal size standard, GENESCAN2500-ROX (ABI), was loaded into each lane. Fragment sizes were estimated by using the Local Southern Method in GeneScan, version 2.1 (ABI).
Clone library construction and analysis.
Fecal DNAs from individual cows or humans were amplified with Bac32F and Bac708R, and amplicons from 10 individuals belonging to each host species were pooled. The PCR products were gel purified with a Qiaquick gel extraction kit (Qiagen, Valencia, Calif.) and were cloned by using a pGEM-T Easy cloning kit (Promega, Madison, Wis.) as recommended by the manufacturer. A total of 192 transformants were randomly selected from each library and inoculated into 100-μl portions of Luria-Bertani broth supplemented with 100 μg of ampicillin per ml in 96-well microtiter plates. After incubation for 6 h, two replica plates were made from each original microtiter plate. All of the plates were incubated overnight at 37°C. The following day, clones from each row in a microtiter replica plate and clones from each column in another microtiter replica plate were pooled.
DNAs from the pooled rows and columns were amplified with Bac32F and either Bac303R or Bac708R. Bac32F was labeled with the fluorophore 6-FAM. PCR products amplified with Bac32F and Bac303R were analyzed by LH-PCR. PCR products amplified with Bac32F and Bac708R were digested with restriction enzyme HaeIII or AciI as described above and analyzed by the T-RFLP technique. The clones corresponding to cow and human genetic markers were identified by locating the intersection of a positive result in a row with a positive result in a column.
Sequencing of marker clones.
Plasmid DNAs from overnight cultures were prepared by using a Qiaprep spin column purification kit (Qiagen) as recommended by the manufacturer. DNA was quantified spectrophotometrically with a Shimadzu UV-visible light spectrophotometer. Bidirectional sequences were obtained by using T7 and SP6 priming sites on either side of the insert. Sequences were determined with a model ABI 377 DNA sequencer using dye terminator chemistry.
Phylogenetic analysis.
Sequences were analyzed with BLAST, version 2.0, to determine preliminary closest phylogenetic neighbors. We aligned the sequences manually with sequences of members of the Cytophaga-Flavobacter-Bacteroides group obtained from GenBank by using the DNA sequence editor in GCG, version 10 (Genetics Computer Group). Sequences and alignments were verified by comparing them to the 16S rRNA secondary structure of Bacteroides fragilis and to Bacteroides signature sequences (23). Evolutionary distances were calculated using the DNADIST program with the Kimura two-parameter model for nucleotide change and a transition/transversion ratio of 2.0 (31). Phylogenetic trees were inferred with the neighbor-joining algorithm (49) using the NEIGHBOR program in PHYLIP 3.5c (18). Regions where the alignment was ambiguous were not included in the analyses. To check the consistency of the resulting tree, we randomly resampled the sequences 100 times (bootstrapping) and obtained a consensus tree (17). Similarities were calculated by using Similarity Matrix, version 1.1, obtained from the Ribosomal Database Project.
Sensitivity analysis.
Serial dilutions of fresh cow feces or raw sewage were added to 1-liter samples of filter-sterilized bay water. The final concentrations in the 1-liter samples ranged from 2 × 10−7 to 2.0 mg (wet weight)/liter. Samples were filtered onto a 0.2-μm-pore-size Supor filter and stored in lysis buffer at −80°C as described above. The percentages of solids in the fecal samples were estimated by weighing replicate samples of wet feces and drying the samples with heat until no more weight was lost. To estimate the percentage of solids in raw sewage, we collected the solids by centrifugation, decanted the supernatant, and dried the samples overnight with heat. DNAs extracted from the filters were amplified with Bac32F and Bac708R as described above, and the PCR products were visualized in a 1% agarose gel. The products were digested as described above. We analyzed all of the samples by performing a T-RFLP analysis with 25 fmol of the most concentrated dilution (2.0 mg/liter) and equivalent volumes of all other dilutions.
GenBank accession numbers. The GenBank accession numbers are as follows: AF233400, AF233401, AF233402, AF233403, AF233404, AF233405, AF233406, AF233407, AF233408, AF233409, AF233410, AF233411, AF233412, and AF233413.
RESULTS
We amplified human and cow fecal DNAs with primers specific for the fecal anaerobes Bacteroides-Prevotella spp. and Bifidobacterium spp. We separated the amplified fragments by size with an ABI DNA sequencer using GeneScan software, which allowed us to identify DNA fragment lengths that were unique to either humans or cows. From these analyses, we identified seven potential host-specific 16S rDNA genetic markers in human and cow fecal DNAs (Table 2). To be considered a host-specific genetic marker, a gene fragment had to be present in all of the samples obtained from the host and absent in all of the samples obtained from the other host.
TABLE 2.
Potential host-specific genetic markers identified by LH-PCR or T-RFLP analysis of human and cow fecal DNAsa
| Host specificity | Primer pair | Enzyme used | Size of marker fragment (bp) |
|---|---|---|---|
| Human | Bac32F-Bac708R | HaeIII | 119 |
| Cow | Bac32F-Bac708R | HaeIII | 222 |
| Cow | Bac32F-Bac708R | AciI | 227 |
| Cow | Bac32F-Bac303R | None | 276 |
| Cow | Bif164F-Bif601R | HaeIII | 142–152 |
| Human | Bif164F-Bif601R | TaqI | 313 |
| Cow | Bif164F-Bif601R | None | 453 |
The markers are located in the Bacteroides or Bifidobacterium 16S rRNA genes.
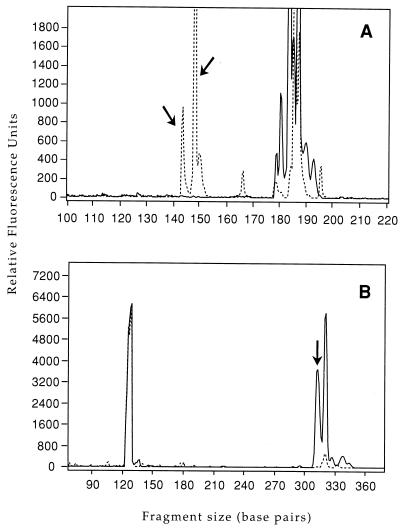
The LH-PCR analysis (55), which detected length differences in PCR amplicons, revealed cow-specific Bacteroides-Prevotella and Bifidobacterium genetic markers (Fig. 1). On the basis of the LH-PCR analysis of 16S rDNA amplicons amplified with Bac32F and Bac303R from human and cow feces we identified a peak at 276 bp as a potential cow-specific gene fragment, but no human-specific genetic markers were detected. LH-PCR analysis of 16S rDNA amplicons amplified with Bif164F and Bif601R revealed a cow-specific genetic marker at 453 bp.
FIG. 1.
LH-PCR analysis of 16S rDNA gene fragments amplified with Bac32F-FAM and Bac303R (A) and with Bif164F and Bif601R-FAM (B). The solid lines are human fecal DNA; the dotted lines are cow fecal DNA. The samples were mixtures of DNAs from seven or eight individuals. The arrows indicate cow-specific gene fragments.
We identified five additional host-specific genetic markers by cutting Bacteroides-Prevotella or Bifidobacterium PCR amplicons with restriction endonucleases and looking for unique size fragments among the fluorescently labeled terminal end fragments (T-RFLP) (37). Host-specific peaks, corresponding to terminal end fragments, were identified in T-RFLP analyses of human and cow fecal DNAs (Fig. 2 and 3). When PCR products amplified with Bac32F and Bac708R were digested with AciI, one cow-specific peak was found at 227 bp (Fig. 2A). There were additional host-specific peaks, which, upon sequence analysis, were found to be artifacts produced by partial digestion. Analysis of 16S rDNA amplicons from cow and human feces and sewage amplified with Bac32F and Bac708R and digested with HaeIII revealed a 119-bp human-specific peak and a 222-bp cow-specific peak (Fig. 2B).
FIG. 2.
T-RFLP analysis of 16S rDNA gene fragments amplified with Bac32F-FAM and Bac708R and cut with AciI (A) or HaeIII (B). The solid lines are human fecal DNA; the dotted lines are cow fecal DNA. The samples were mixtures of DNAs from seven or eight individuals. The arrows indicate host-specific genetic markers.
FIG. 3.
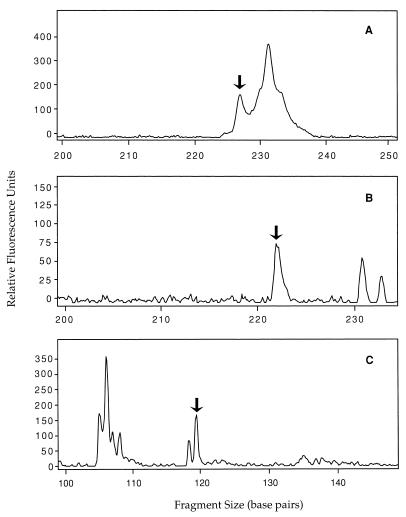
T-RFLP analysis of 16S rDNA gene fragments amplified with Bif164F and Bif601R-FAM and cut with HaeIII (A) or TaqI (B). The solid lines are community profiles obtained with human fecal DNA; the dotted lines are community profiles obtained with cow fecal DNA. The arrows indicate host-specific genetic markers.
T-RFLP analysis of 16S rDNA genes amplified from cow feces with Bif164F and Bif601R and cut with HaeIII revealed a cow-specific cluster of peaks at 142 to 152 bp (Fig. 3A). Analysis of 16S rDNA amplicons from human feces and sewage amplified with Bif164F and Bif601R and digested with TaqI revealed a human-specific peak at 313 bp, but no cow-specific peaks were detected in the amplicons obtained from cow feces (Fig. 3B).
A comparison of the Bacteroides-Prevotella and Bifidobacterium communities in sewage samples obtained from two Oregon cities, Corvallis and Tillamook, and in feces obtained from 14 humans revealed that both the Bacteroides-Prevotella and Bifidobacterium community profiles were very similar, although sometimes there were differences in the proportions of the LH-PCR and T-RFLP peaks present (data not shown). Similarly, DNAs obtained from cow feces collected at different times of the year, from different farms, and from different towns produced very similar patterns. These results suggest that although there may be slight intraspecies variation, at the level of variability detected by our markers the host-specific patterns are the same.
To verify the identities of the Bacteroides-Prevotella genetic markers, we constructed 16S rDNA clone libraries from cow and human fecal DNAs with primers Bac32F and Bac708R. We screened 192 clones from each host and sequenced the clones that had the LH-PCR or T-RFLP pattern of interest. Because the Bacteroides-Prevotella group is a more promising indicator (see below), we cloned 16S rDNA genes from this group but not from members of the genus Bifidobacterium. In the library obtained from human feces, we found six different clones that corresponded to the 119-bp human-specific marker (Fig. 4). Further analysis of these sequences revealed that the fragment size estimated by the T-RFLP method was 1 bp smaller than the actual size determined from the sequences (120 bp). Four of these sequences (HF8, HF102, HF117, and HF145), although not identical, were more than 98.9% similar to each other and were 97.5 to 98.0% similar to the Bacteroides vulgatus sequence. These sequences formed the closely related HF8 gene cluster (Fig. 4) but did not exactly correspond to any previously described sequence. HF74 was 93.9 to 94.9% similar to the clones in the HF8 cluster and 93.2% similar to the B. vulgatus sequence. One other human fecal clone, HF10, was 97.7% similar to the Bacteroides uniformis sequence.
FIG. 4.
Phylogenetic relationships among partial 16S rDNA sequences (558 positions) of human (HF) and cow (CF) host-specific genetic markers identified from fecal clone libraries. The tree was inferred by using the neighbor-joining method. The numbers above the branch points are the percentages of bootstrap replicates that support the branching order. Bootstrap values less than 50% are not shown. Cytophaga fermentans was used to root the tree.
None of the cow-specific clones was closely related to the sequence of any previously characterized microorganism. These clones formed two distinct gene clusters within the Bacteroides-Prevotella group (Fig. 4). We recovered seven clones from cow feces that produced the 227-bp fragment when the DNAs were amplified with Bac32F and Bac708R and cut with AciI. Partial 16S rDNA sequencing revealed five different sequences, each with the same T-RFLP profile, which formed the CF123 gene cluster. The fragment sizes estimated by the T-RFLP analysis were about 2 bases larger than the fragment sizes determined from the sequences (225 bp). The levels of similarity within this cluster ranged from 91.6 to 95.2%. Sequence analysis of the clones corresponding to the 222-bp cow-specific marker (T-RFLP analysis with HaeIII) and the 276-bp cow-specific marker (LH-PCR analysis) revealed that these markers represented the same sequences. We found four clones that represented three different sequences corresponding to these two markers. These three sequences were 92 to 94.4% similar and were all included in the CF151 gene cluster (Fig. 4). Again, the sizes of the fragments estimated by the T-RFLP and LH-PCR methods were 1 to 2 bases different from the sizes predicted from the sequences.
We tested a variety of river and estuarine water samples to determine whether they contained Bacteroides-Prevotella and Bifidobacterium DNAs and also the marker genes. The fecal coliform levels in these samples ranged from 0 to 120 CFU/100 ml (Table 3). Bacteroides-Prevotella DNAs were detected in all eight samples tested, but Bifidobacterium DNAs were detected in only two of these samples. Additionally, the product yields of Bifidobacterium amplicons detected in these water samples were considerably less than the product yields obtained from the same samples when Bacteroides primers were used. All seven host-specific genetic markers were detected in at least one water sample (Table 3 and Fig. 5). In subsequent experiments, we used sequence data to validate the identities of the Bacteroides-Prevotella markers obtained from water samples. We recovered sequences of Bacteroides-Prevotella markers belonging to the HF8, CF123, and CF151 gene clusters (data not shown).
TABLE 3.
Fecal coliform concentrations and presence of Bifidobacterium and Bacteroides-Prevotella host-specific markers in water samples collected from Tillamook Bay, Oreg., and three of its tributaries
| Sample type | Date collected (mo/day/yr) | Fecal coliform concn (CFU/100 ml) | Presence ofa:
|
||||||
|---|---|---|---|---|---|---|---|---|---|
|
Bifidobacterium markers
|
Bacteroides-Prevotella markers
|
||||||||
| 142-152 bp | 313 bp | 453 bp | 119 bp | 222 bp | 276 bp | 227 bp | |||
| River | 12/12/97 | 2 | + | − | + | ND | ND | + | + |
| River | 12/12/97 | 6 | − | − | − | − | − | − | + |
| River | 5/17/98 | 120 | − | − | − | ND | ND | − | + |
| River | 10/30/98 | 36 | + | + | − | + | − | − | + |
| River mouth | 10/30/98 | 112 | − | − | − | + | + | − | + |
| Estuary | 10/30/98 | 35 | − | − | − | − | − | − | + |
| Estuary | 10/30/98 | 57 | − | − | − | + | + | − | + |
| Estuary mouth | 10/30/98 | 0 | − | − | − | − | − | + | − |
+, peak detected in the LH-PCR or T-RFLP analysis; −, no peak detected; ND, no data.
FIG. 5.
T-RFLP analyses of 16S rDNA gene fragments amplified from DNA extracted from Tillamook Bay water samples. DNA was amplified with Bac32F and Bac708R and digested with AciI (A) or HaeIII (B and C). The arrows indicate host-specific markers. (A and B) Cow-specific markers (227 and 222 bp, respectively). (C) Human-specific marker (119 bp).
We evaluated the sensitivity of host-specific DNA detection in water samples by performing assays with filter-sterilized bay water amended with fresh feces or raw sewage. We used feces and sewage rather than cultured organisms since fecal organisms were the intended targets. The limits of detection of host-specific markers differed. The 222-bp cow-specific marker was the least sensitive (2.8 × 10−5 g [dry weight] of feces/liter), followed by the 119-bp human-specific marker (6.8 × 10−7 g [dry weight] of sewage/liter); the 227-bp cow-specific marker was the most sensitive (2.8 × 10−8 g [dry weight] of feces/liter).
DISCUSSION
We identified species composition differences in the Bacteroides-Prevotella and Bifidobacterium populations when we compared human and cow feces. These differences could be useful for identifying fecal pollution sources in coastal waters. The human-specific genetic markers for the Bacteroides-Prevotella group were closely related, but not identical, to sequences of Bacteroides species commonly found in human intestines and feces, B. uniformis and B. vulgatus (50). The cow-specific genetic markers formed new gene clusters in the Bacteroides-Prevotella group, which is in the Cytophaga-Flavobacter-Bacteroides phylum, a group whose members are physiologically diverse but phylogenetically similar (23, 45). Gene clusters are sets of gene sequences that are more closely related to each other than to the sequences of any previously characterized species; they have been found in many diverse natural bacterial populations (19, 24, 28, 44).
The discovery of new gene clusters for members of the Bacteroides-Prevotella group in cows reflects the lack of characterization of the diversity in this habitat. Conversely, the human intestine is a better characterized habitat due to the clinical significance of the bacteria in this region. The microbial diversity of human fecal and colonic bacteria has been the subject of many culture-based studies, but only since molecular techniques have become available have researchers had tools which can be used to assess the diversity more accurately. Although culture bias may be less of a problem in enriched, highly selective environments, such as feces, it is still likely to occur, especially for anaerobic bacteria that may be difficult to grow (2). Comparisons of 16S rDNA diversity with the diversity assessed by culture methods in human feces (58, 59) and bovine rumens (32) suggest that diversity is underestimated by culturing alone.
Most of the clones comprising the HF8 cluster were more than 99% similar; the only exception was HF102. The other three clones (HF8, HF117, and HF145) varied by only 1 to 2 nucleotides over a 700-base sequence, which falls within the range of predicted Taq polymerase error rates (48). Three of the six deviant nucleotides were consistent with common Taq errors (16, 56), and two others were incompatible with secondary structure, suggesting that PCR or sequencing errors may have occurred. We think that it is possible that these three sequences are actually the same. Although HF102 was in the same gene cluster, it differed from the other three sequences in the cluster at 9 to 11 nucleotide positions. The differences, however, were in a hypervariable region of the gene that was not included in the phylogenetic analysis because of ambiguous alignment.
The LH-PCR and T-RFLP methods proved to be highly reproducible, although the estimated peak size often differed by 1 to 2 bp from the size predicted from the sequence. The differences between the peak sizes and the predicted sizes may have several explanations. First, there may have been slight differences in the electrophoretic mobilities of the ROX-labeled standard and the FAM-labeled samples. Second, our observations based on analyses of single clones suggest that addition of an adenine to the end of PCR products by Taq polymerase is variable, which leads to products that are exactly 1 bp different. Third, differences in sequences may cause products to migrate anomalously compared to a standard fragment of the same size. All of these variables may have contributed to the 1- to 2-bp differences which we observed between the peak sizes and the sizes predicted from the sequences. We also observed that the size of the difference appeared to increase as the size of the fragment increased. Despite these differences, the methods were reproducible, and the variances were ±0.3 bp for fragments up to 350 bp long.
Our comparisons of the Bacteroides-Prevotella and Bifidobacterium gene profiles for 14 humans and 16 cows suggested that intraspecies variation was insignificant and that most of the differences were differences in proportions rather than differences in the species present. Human feces were collected from coworkers and their families, so it is possible that these individuals share intestinal flora (11, 42). However, sewage samples from Tillamook and Corvallis (cities separated by 100 miles and a mountain range) also produced nearly identical patterns, suggesting that the host-specific patterns were widely distributed. This does not mean that the commensal bacterial communities in individuals from geographically distinct populations are identical. Instead, it demonstrates that our method does not reveal variability at the level of the individual but does reveal variability between host species.
Previous analyses of human fecal flora in which culture techniques were used did not reveal major differences in bacterial species composition even when populations with different diets were compared (21), although the relative frequencies in individuals varied (29). Other studies, however, have suggested that there are major differences in the compositions of Bifidobacterium and Lactobacillus communities in humans (30, 41). Our data suggested that low levels of intraspecific variation occur in bacterial populations. The differences described above may be explained by the differences in the methods used. Culturing bacteria from samples distinguishes organisms at the species level or even the strain level. Methods based on sizes and compositions of gene fragments, such as the LH-PCR and T-RFLP methods, may distinguish organisms only at the phylogenetic group level or the gene cluster level. It is possible that individuals harbor different species or strains of bacteria within a particular gene cluster; this would not necessarily be detected by the methods described in this paper.
We concluded that species belonging to the Bacteroides-Prevotella group were better indicators than Bifidobacterium species for coastal waters. Although we detected host species differences in the Bifidobacterium populations, the Bifidobacterium genetic markers proved to be less robust than the Bacteroides-Prevotella genetic markers. First, we were sometimes unable to detect Bifidobacterium spp. in cow fecal samples. It has been shown that large numbers of members of the genus Bifidobacterium are present in the rumen (7), but the prevalence of these organisms in feces may be affected by acidic conditions in the stomach or by the actions of certain antibiotics (15, 51). We collected samples only from cows that were not being given antibiotics, but the antibiotic history of the individual cows was not considered at the time of collection.
Second, detecting members of the genus Bifidobacterium by PCR in water samples was also difficult. It is possible that the signal was simply too weak to be detected by PCR. Resnick and Levin (47) found that members of the genus Bifidobacterium could not be cultured after 5 h in freshwater or 10 h salt water. These short survival times make it difficult to detect Bifidobacterium spp. in water in which fecal pollution is much more than 10 h old. Carrillo and colleagues (10) also observed very low levels of survival of Bifidobacterium adolescentis in a tropical environment and suggested that Bifidobacterium spp. could only be used to detect very recent pollution.
Conversely, host-specific markers for the Bacteroides-Prevotella group were detected in all water samples from Tillamook Bay and its tributaries and in all fecal samples. Additionally, we did not detect any fecal markers in water samples collected from the Sargasso Sea or Crater Lake, Oreg., neither of which would be expected to be polluted with human or cow feces. Our assay was not designed to quantify fecal pollution; rather, it was designed to determine the presence or absence of a particular source. Therefore, direct comparisons to fecal coliform results are inappropriate. The presence of fecal markers in samples that contained no detectable coliforms (Table 3) was probably the result of differences in the sensitivity of the assays or the viability of the coliforms. The sample containing no detectable coliforms was obtained from the mouth of the estuary, where salinity is highest and coliforms would probably be stressed or dead. We did not use methods for resuscitating stressed organisms, so it is unlikely that we would have detected stressed organisms by culturing them. DNA, however, has been detected after several days to 2 weeks if conditions are optimal (35).
The results of our sensitivity assays are comparable to the results of other studies in which the workers used PCR to detect single Bacteroides species in feces (34, 35, 57). The 119-bp human-specific marker and the 227-bp cow-specific marker appear to be more sensitive, as determined by the T-RFLP method with general Bacteroides-Prevotella PCR primers. It is possible that by designing primers specific for these markers, the sensitivity may be increased. The 222-bp cow-specific marker, which represents the same sequences as the 276-bp marker, was the least sensitive marker by 3 to 4 orders of magnitude. We also observed some samples that were positive for one marker but not the other marker (Table 3). Since the sensitivity for these markers was much lower, it is possible that the source contamination was at or near the limit of detection; therefore, inconsistent detection of these two markers in the same water sample is not surprising.
Bacteroides account for as much as 30% of fecal isolates (29) and 62% of the eubacterial fecal rDNA (59) and are found in both humans and cows (38). Franks and colleagues (22) found that the Bacteroides population from one human fluctuated less over time than Bifidobacterium populations fluctuated. Moreover, Bacteroides cells have been isolated from environmental water samples for at least several days after they were dispersed in the water (5, 35, 52). Survival of Bacteroides cells depends primarily on temperature and predation (35), and these organisms can survive for up to 6 days under oxygen stress conditions (5).
Thus, the Bacteroides-Prevotella group is a promising indicator that could be used to identify the source of fecal contamination in water samples. We identified one human-specific and two cow-specific gene clusters of fecal markers from Bacteroides-Prevotella species and demonstrated that these markers can be recovered from natural fresh- and saltwater samples. We also identified these marker genes phylogenetically as genes from members of the Bacteroides-Prevotella group, but they represented uncharacterized species. Armed with the sequence data obtained in this study, we are currently designing new primers specific for each cluster of genetic markers. These primers will then be used to identify the most likely sources of fecal contamination in natural water samples. Additionally, since real-time quantitative PCR methods are now available, we may now be able to develop a quantitative assay.
In this study we focused on two host species, and thus, we can only determine the absence of a particular pollution source with certainty. We have not investigated the distribution of the genetic markers in other animals yet. It is possible that some of these markers may not be limited to humans or cows. In future studies we will test feces from other potential pollution sources, such as swine, waterfowl, and other common wildlife.
We demonstrated that the LH-PCR and T-RFLP methods can be used to identify and track bacterial markers in complex natural environments. These methods have the advantage of being specific, rapid, and sensitive to changes as subtle as 1 bp (12, 37). Potential applications of these methods include tracking environmentally important species, genetically engineered species released into the environment, and pathogens in clinical specimens.
ACKNOWLEDGMENTS
This work was supported in part by grant NA76RG0476 (project R/ECO-04) from the National Oceanic and Atmospheric Administration to the Oregon State University Sea Grant College Program and by appropriations made by the Oregon state legislature. This work was also supported by the Research Council of Oregon State University.
We are grateful for the assistance provided by Amanda Barry, Debbie Colbert, James McManus, James Moore, Mike Rappé, and Kevin Vergin. We also thank Abigail Salyers for providing DNAs from Bacteroides cultures and Caprice Rosato and Nanci Adair for performing the GeneScan analyses and sequencing.
REFERENCES
- 1.Allsop K, Stickler J D. An assessment of Bacteroides fragilis group organisms as indicators of human faecal pollution. J Appl Bacteriol. 1985;58:95–99. doi: 10.1111/j.1365-2672.1985.tb01433.x. [DOI] [PubMed] [Google Scholar]
- 2.Amann R I, Ludwig W, Schleifer K-H. Phylogenetic identification and in situ detection of individual microbial cells without cultivation. Microbiol Rev. 1995;59:143–169. doi: 10.1128/mr.59.1.143-169.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.American Public Health Association. Standard methods for the examination of water and wastewater. 18th ed. Washington, D.C.: American Public Health Association; 1992. [Google Scholar]
- 4.Avaniss-Aghajani E A, Jones K, Chapman D, Brunk C. A molecular technique for identification of bacteria using small subunit ribosomal RNA sequences. BioTechniques. 1994;17:144–149. [PubMed] [Google Scholar]
- 5.Avelar K E S, Moraes S R, Pinto L J F, Silva e Souza W d G, Domingues R M C P, de Souza Ferreira M C. Influence of stress conditions on Bacteroides fragilis survival and protein profiles. Zentralbl Bakteriol. 1998;287:399–409. [PubMed] [Google Scholar]
- 6.Berthelet M, Whyte L G, Greer C W. Rapid, direct extraction of DNA from soils for PCR analysis using polyvinylpolypyrrolidone spin columns. FEMS Microbiol Lett. 1996;138:17–22. doi: 10.1111/j.1574-6968.1996.tb08128.x. [DOI] [PubMed] [Google Scholar]
- 7.Biavati B, Mattarelli P. Bifidobacterium ruminantium sp. nov. and Bifidobacterium merycicum sp. nov. from the rumens of cattle. Int J Syst Bacteriol. 1991;41:163–168. doi: 10.1099/00207713-41-1-163. [DOI] [PubMed] [Google Scholar]
- 8.Bruce K D. Analysis of mer gene subclasses within bacterial communities in soils and sediments resolved by fluorescent-PCR–restriction fragment length polymorphism profiling. Appl Environ Microbiol. 1997;63:4914–4919. doi: 10.1128/aem.63.12.4914-4919.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Brunk C F, Avaniss-Aghajani E, Brunk C A. A computer analysis of primer and probe hybridization potential with bacterial small-subunit rRNA sequences. Appl Environ Microbiol. 1996;62:872–879. doi: 10.1128/aem.62.3.872-879.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Carrillo M, Estrada E, Hazen T C. Survival and enumeration of the fecal indicators Bifidobacterium adolescentis and Escherichia coli in a tropical rain forest watershed. Appl Environ Microbiol. 1985;50:468–476. doi: 10.1128/aem.50.2.468-476.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Caugant D A, Levin B R, Selander R K. Distribution of multilocus genotypes of Escherichia coli within and between host families. J Hyg. 1984;92:377–384. doi: 10.1017/s0022172400064597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Clement B G, Kehl L E, DeBord K L, Kitts C L. Terminal restriction fragment patterns (TRFPs), a rapid, PCR-based method for the comparison of complex bacterial communities. J Microbiol Methods. 1998;31:135–142. [Google Scholar]
- 13.Crane S R, Moore J A. A management strategy to reduce bacterial pollution in shellfish areas: a case study. Environ Manag. 1986;10:41–51. [Google Scholar]
- 14.Davies C M, Long J H, Donald M, Ashbolt N J. Survival of fecal microorganisms in marine and freshwater sediments. Appl Environ Microbiol. 1995;61:1888–1896. doi: 10.1128/aem.61.5.1888-1896.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dennis S M, Nagaraja T G, Bartley E E. Effects of lasalocid or monensin on lactate-producing or -using rumen bacteria. J Anim Sci. 1981;52:418–426. doi: 10.2527/jas1981.522418x. [DOI] [PubMed] [Google Scholar]
- 16.Dunning A M, Talmud P, Humphries S E. Errors in the polymerase chain reaction. Nucleic Acids Res. 1988;16:10393. doi: 10.1093/nar/16.21.10393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Felsenstein J. Confidence limits on phylogenies: an approach using the bootstrap. Evolution. 1985;39:783–791. doi: 10.1111/j.1558-5646.1985.tb00420.x. [DOI] [PubMed] [Google Scholar]
- 18.Felsenstein J. PHYLIP—phylogeny inference package (v3.5) Cladistics. 1989;5:164–166. [Google Scholar]
- 19.Field K G, Gordon D, Rappé M, Urbach E, Vergin K, Giovannoni S J. Diversity and depth-specific distribution of SAR11 cluster rRNA genes from marine planktonic bacteria. Appl Environ Microbiol. 1997;63:63–70. doi: 10.1128/aem.63.1.63-70.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Fiksdal L, Make J S, LaCroix S J, Staley J T. Survival and detection of Bacteroides spp., prospective indicator bacteria. Appl Environ Microbiol. 1985;49:148–150. doi: 10.1128/aem.49.1.148-150.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Finegold S M, Sutter V L, Mathieson G E. Normal indigenous intestinal flora. In: Hentges D G, editor. Human intestinal microflora in health and disease. New York, N.Y: Academic Press; 1983. p. 568. [Google Scholar]
- 22.Franks A H, Harmsen H J M, Raangs G C, Jansen G J, Schut F, Welling G W. Variations of bacterial populations in human feces measured by fluorescent in situ hybridization with group-specific 16S rRNA-targeted oligonucleotide probes. Appl Environ Microbiol. 1998;64:3336–3345. doi: 10.1128/aem.64.9.3336-3345.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gherna R, Woese C R. A partial phylogenetic analysis of the “Flavobacter-Bacteroides” phylum: basis for taxonomic review. Syst Appl Microbiol. 1992;15:513–521. doi: 10.1016/S0723-2020(11)80110-4. [DOI] [PubMed] [Google Scholar]
- 24.Giovannoni S J, Britschgi T B, Moyer C L, Field K G. Genetic diversity in Sargasso Sea bacterioplankton. Nature. 1990;345:60–63. doi: 10.1038/345060a0. [DOI] [PubMed] [Google Scholar]
- 25.Giovannoni S J, DeLong E F, Schmidt T M, Pace N R. Tangential flow filtration and preliminary phylogenetic analysis of marine picoplankton. Appl Environ Microbiol. 1990;56:2572–2575. doi: 10.1128/aem.56.8.2572-2575.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Glockner F O, Fuchs B M, Amann R. Bacterioplankton compositions of lakes and oceans: a first comparison based on fluorescence in situ hybridization. Appl Environ Microbiol. 1999;65:3721–3726. doi: 10.1128/aem.65.8.3721-3726.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gray J P, Herwig R P. Phylogenetic analysis of the bacterial communities in marine sediments. Appl Environ Microbiol. 1996;62:4049–4059. doi: 10.1128/aem.62.11.4049-4059.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hales B A, Edwards C, Ritchie D A, Hall G, Pickup R W, Saunders J R. Isolation and identification of methanogen-specific DNA from blanket bog peat by PCR amplification and sequence analysis. Appl Environ Microbiol. 1996;62:668–675. doi: 10.1128/aem.62.2.668-675.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Holdeman L V, Good I J, Moore W E C. Human fecal flora: variation in bacterial composition within individuals and a possible effect of emotional stress. Appl Environ Microbiol. 1976;31:359–375. doi: 10.1128/aem.31.3.359-375.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kimura K, McCartney A L, McConnell M A, Tannock G W. Analysis of fecal populations of bifidobacteria and lactobacilli and investigation of the immunological responses of their human hosts to the predominant strains. Appl Environ Microbiol. 1997;63:3394–3398. doi: 10.1128/aem.63.9.3394-3398.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kimura M. A simple method for estimating evolutionary rates of base substitutions through comparative studies of nucleotide sequences. J Mol Evol. 1980;16:111–120. doi: 10.1007/BF01731581. [DOI] [PubMed] [Google Scholar]
- 32.Krause D O, Russell J B. How many ruminal bacteria are there? J Dairy Sci. 1996;79:1467–1475. doi: 10.3168/jds.S0022-0302(96)76506-2. [DOI] [PubMed] [Google Scholar]
- 33.Kreader C A. Design and evaluation of Bacteroides DNA probes for the specific detection of human fecal pollution. Appl Environ Microbiol. 1995;61:1171–1179. doi: 10.1128/aem.61.4.1171-1179.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kreader C A. Relief of amplification inhibition in PCR with bovine serum albumin or T4 gene 32 protein. Appl Environ Microbiol. 1996;62:1102–1106. doi: 10.1128/aem.62.3.1102-1106.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kreader C A. Persistence of PCR-detectable Bacteroides distasonis from human feces in river water. Appl Environ Microbiol. 1998;64:4103–4105. doi: 10.1128/aem.64.10.4103-4105.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Langendijk P S, Schut F, Jansen G J, Raangs G C, Kamphuis G R, Wilkinson M H F, Welling G W. Quantitative fluorescence in situ hybridization of Bifidobacterium spp. with genus-specific 16S rRNA-targeted probes and its application in fecal samples. Appl Environ Microbiol. 1995;61:3069–3075. doi: 10.1128/aem.61.8.3069-3075.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Liu W-T, Marsh T L, Cheng H, Forney L J. Characterization of microbial diversity by determining terminal restriction fragment length polymorphism of genes encoding 16S rRNA. Appl Environ Microbiol. 1997;63:4516–4522. doi: 10.1128/aem.63.11.4516-4522.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Macy J M, Probst I. The biology of the gastrointestinal Bacteroides. Annu Rev Microbiol. 1979;33:561–594. doi: 10.1146/annurev.mi.33.100179.003021. [DOI] [PubMed] [Google Scholar]
- 39.Maidak B L, Larsen N, McCaughey M J, Overbeek R, Olsen G J, Fogel K, Blandy J, Woese C R. The Ribosomal Database Project. Nucleic Acids Res. 1994;22:3485–3487. doi: 10.1093/nar/22.17.3485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Manz W, Amann R, Ludwig W, Vancanneyt M, Schleifer K-H. Application of a suite of 16S rRNA-specific oligonucleotide probes designed to investigate bacteria of the phylum cytophaga-flavobacter-bacteroides in the natural environment. Microbiology. 1996;142:1097–1106. doi: 10.1099/13500872-142-5-1097. [DOI] [PubMed] [Google Scholar]
- 41.McCartney A L, Wenzhi W, Tannock G W. Molecular analysis of the composition of the bifidobacterial and lactobacillus microflora of humans. Appl Environ Microbiol. 1996;62:4608–4613. doi: 10.1128/aem.62.12.4608-4613.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Mehta S K, Stevens D A, Mishra S K, Feroze F, Pierson D L. Distribution of Candida albicans genotypes among family members. Diagn Microbiol Infect Dis. 1999;34:19–25. doi: 10.1016/s0732-8893(98)00100-x. [DOI] [PubMed] [Google Scholar]
- 43.Murray A E, Hollibaugh J T, Orrego C. Phylogenetic compositions of bacterioplankton from two California estuaries compared by denaturing gradient gel electrophoresis of 16S rDNA fragments. Appl Environ Microbiol. 1996;62:2676–2680. doi: 10.1128/aem.62.7.2676-2680.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ohkuma M, Kudo T. Phylogenetic diversity of the intestinal bacterial community in the termite Reticulitermes speratus. Appl Environ Microbiol. 1996;62:461–468. doi: 10.1128/aem.62.2.461-468.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Paster B J, Ludwig W, Weisburg W G, Stackebrandt E, Hespell R B, Hahn C M, Reichenbach H, Stetter K O, Woese C R. A phylogenetic grouping of the Bacteroides, Cytophagas, and certain flavobacteria. Syst Appl Microbiol. 1985;6:34–42. [Google Scholar]
- 46.Pitcher D G, Saunders N A, Owen R J. Rapid extraction of bacterial genomic DNA with guanidium thiocyanate. Lett Appl Microbiol. 1989;8:151–156. [Google Scholar]
- 47.Resnick I G, Levin M A. Assessment of bifidobacteria as indicators of human fecal pollution. Appl Environ Microbiol. 1981;42:433–438. doi: 10.1128/aem.42.3.433-438.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Saiki R K, Gelfand D H, Stoffel S, Scharf S J, Higuchi R, Horn G T, Mullis K B, Erlich H A. Primer-directed enzymatic amplification of DNA with a thermostable DNA polymerase. Science. 1988;239:487–491. doi: 10.1126/science.2448875. [DOI] [PubMed] [Google Scholar]
- 49.Saitou N, Nei M. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol. 1987;4:406–425. doi: 10.1093/oxfordjournals.molbev.a040454. [DOI] [PubMed] [Google Scholar]
- 50.Salyers A A. Bacteroides of the human lower intestinal tract. Annu Rev Microbiol. 1984;38:293–313. doi: 10.1146/annurev.mi.38.100184.001453. [DOI] [PubMed] [Google Scholar]
- 51.Stewart C S, Bryant M P. The rumen bacteria. In: Hobson P N, editor. The rumen microbial ecosystem. New York, N.Y: Elsevier Applied Science; 1988. p. 527. [Google Scholar]
- 52.Straub D V, Dixon B. 38th Annual Western Fish Disease Workshop. Bodega Bay, Calif: Bodega Bay Marine Laboratory; 1997. Bacteroides vulgatus, an alternative indicator of the assessment of fecal contamination of shellfish and estuarine waters. [Google Scholar]
- 53.Strittholt J R, Garono R J, Frost P A. Spatial patterns in land use and water quality in the Tillamook Bay watershed: a GIS mapping project. Corvallis, Oreg: Earth Designs Consultants, Inc.; 1998. [Google Scholar]
- 54.Strunk O, Ludwig W. ARB, a software environment for sequence data. Munich, Germany: Technische Universitat Munchen; 1996. [Google Scholar]
- 55.Suzuki M T, Rappé M S, Giovannoni S J. Kinetic bias in estimation of coastal picoplankton community structure obtained by measurements of small subunit rRNA gene PCR amplicon length heterogeneity. Appl Environ Microbiol. 1998;64:4522–4529. doi: 10.1128/aem.64.11.4522-4529.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Tindall K R, Kunkel T A. Fidelity of DNA synthesis by the Thermus aquaticus DNA polymerase. Biochemistry. 1988;27:6008–6013. doi: 10.1021/bi00416a027. [DOI] [PubMed] [Google Scholar]
- 57.Wang R-F, Cao W-W, Cerniglia C E. PCR detection and quantitation of predominant anaerobic bacteria in human and animal fecal samples. Appl Environ Microbiol. 1996;62:1242–1247. doi: 10.1128/aem.62.4.1242-1247.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Wilson K H, Blitchington R B. Human colonic biota studied by ribosomal DNA sequence analysis. Appl Environ Microbiol. 1996;62:2273–2278. doi: 10.1128/aem.62.7.2273-2278.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Wood J, Scott K P, Avgustin G, Newbold C J, Flint H J. Estimation of the relative abundance of different Bacteroides and Prevotella ribotypes in gut samples by restriction enzyme profiling of PCR-amplified 16S rRNA gene sequences. Appl Environ Microbiol. 1998;64:3638–3689. doi: 10.1128/aem.64.10.3683-3689.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]