Abstract
Background
Accelerated reproductive aging, in women indicated by early natural menopause, is associated with increased coronary heart disease (CHD) risk in observational studies. Conversely, an adverse CHD risk profile has been suggested to accelerate menopause.
Objectives
To study the direction and evidence for causality of the relationship between reproductive aging and (non-)fatal CHD and CHD risk factors in a bidirectional Mendelian randomization (MR) approach, using age at natural menopause (ANM) genetic variants as a measure for genetically determined reproductive aging in women. We also studied the association of these variants with CHD risk (factors) in men.
Design
Two-sample MR, using both cohort data as well as summary statistics, with 4 methods: simple and weighted median-based, standard inverse-variance weighted (IVW) regression, and MR-Egger regression.
Participants
Data from EPIC-CVD and summary statistics from UK Biobank and publicly available genome-wide association studies were pooled for the different analyses.
Main Outcome Measures
CHD, CHD risk factors, and ANM.
Results
Across different methods of MR, no association was found between genetically determined reproductive aging and CHD risk in women (relative risk estimateIVW = 0.99; 95% confidence interval (CI), 0.97-1.01), or any of the CHD risk factors. Similarly, no associations were found in men. Neither did the reversed analyses show evidence for an association between CHD (risk factors) and reproductive aging.
Conclusion
Genetically determined reproductive aging is not causally associated with CHD risk (factors) in women, nor were the genetic variants associated in men. We found no evidence for a reverse association in a combined sample of women and men.
Keywords: reproductive aging, Mendelian Randomization, coronary heart disease, risk factors
Coronary heart disease (CHD) is the leading cause of death in both men and women (1). Accelerated reproductive aging, as indicated by early menopause in women, has been associated with increased risk of CHD (2-6). The mechanisms underlying these associations are not fully understood yet; deterioration of traditional coronary heart disease risk factors, in particular cholesterol, has been suggested to play a role (7, 8). For example, women with an early menopause might be exposed to higher levels of these CHD risk factors longer, which might result in the association with CHD in observational studies.
In observational studies, it is difficult to disentangle the potential independent effect of accelerated reproductive aging on CHD risk from the effect of general aging because residual confounding can still be present. Furthermore, reversed causality can also play a role here because women with an unfavorable CHD risk profile have been reported to experience accelerated reproductive aging (9). Mendelian randomization (MR) designs, exploiting the principle of random independent segregation of alleles at meiosis, are a means to establish causality in situations where randomized clinical trials are impossible (10, 11). In MR studies, single nucleotide polymorphisms (SNPs) associated with the exposure as found in genome-wide association studies (GWAS) are used as instrumental variables.
A GWAS has been conducted by Day et al for the reproductive aging trait age at natural menopause (ANM) in 69 360 women of European decent with ANM between 40 and 60 years (12). This GWAS reported 56 SNPs, mainly implicated in genome stability (DNA repair), immune function, and mitochondrial biogenesis (12), which are biological processes not specific to women and known to be impaired upon aging. Furthermore, a recent CHD GWAS showed that only 10 of the 241 CHD variants were sex specific (13), which did not include any of the 56 ANM variants. Therefore, we hypothesize that the same 56 ANM variants, even though they may not be associated with reproductive aging in men, could still be associated with CHD in both women and men because of the biological processes they are involved in and their link to biological aging.
A recent study in 3 cohorts (14) showed that an earlier ANM, genetically determined by the 56 SNPs, was associated with an increased CHD risk in women (meta-analyzed hazards ratio [HR] = 1.12; 95% CI, 1.02-1.24), but not in men (meta-analyzed HR = 1.05; 95% CI, 0.94-1.16). However, although studying relationships between genetic risk scores and disease risk provides higher statistical power than formal MR analysis, it is also associated with substantial risk of false-positive findings resulting from horizontal pleiotropy (15). A formal MR analysis by the authors in publicly available data from the Coronary Artery Disease Genome wide Replication and Meta-analysis (CARDIoGRAM) consortium from 2011 did not confirm a causal role of ANM in CHD risk (14). In addition, the authors did not investigate cardiovascular risk factors as an outcome nor studied the reversed association.
The aims of the present study are to disentangle putative causal links between reproductive aging and fatal or nonfatal CHD and CHD risk factors in women. In addition, we also studied the reversed association (eg, between fatal or nonfatal CHD and CHD risk factors and reproductive aging) because hypotheses exist that an adverse CHD risk profile may cause advanced reproductive aging (9, 16). Finally, we aim to assess whether the genetic variants are associated with CHD and traditional CHD risk factors in men as well.
Methods
Ethics Approval
All procedures performed in studies involving human participants were in accordance with the ethical standards of the institutional and/or national research committee and with the 1964 Helsinki Declaration and its later amendments or comparable ethical standards. The studies were approved by local medical ethical committees as described in the Supplemental methods (17).
To study the different hypotheses as described in the introduction, we conducted 4 different MR analyses.
MR to study the association between ANM variants and CHD risk
MR to study the association between ANM variants and CHD risk factors
MR to study the association between CHD variants and ANM
MR to study the association between CHD risk factor variants and ANM
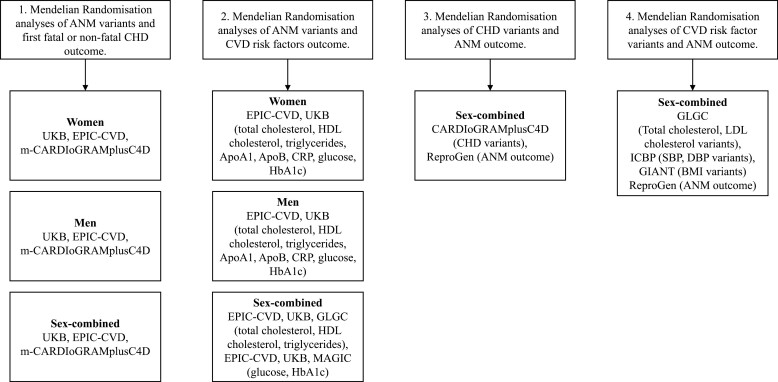
Next, we describe the methods for each MR. In addition, Fig. 1 shows which data sources were used for each MR.
Figure 1.
Visual of the data sources used for each Mendelian randomization.
Methods for MR to Study the Association Between ANM Variants and CHD Risk
Outcome
The main outcome was the first event of fatal or nonfatal CHD, defined by codes 410 through 414 of the International Classification of Diseases, Ninth Edition, and codes I20 through I25 of the 10th Edition.
Data sources
A genome-wide meta-analysis by Day et al identified 56 SNPs associated with ANM among 70 000 women of European descent, 54 common HapMap SNPs and 2 Exome chip SNPs (12), a list of SNPs can be found in Supplemental Table 1 (17). As described by Day et al, all SNPs passed the threshold of P < 5e-8 in the joint model. Pleiotropic effects were investigated by searching the GRASP database and NHGRI catalog for the SNPs or their proxies (R2 > 0.5) (12). We used the 56 ANM variants as instrument for genetically determined reproductive aging in women and investigated whether these variants were associated with CHD in men.
We studied these variants in relation to CHD outcome data from 417 579 participants of European descent (including 49 150 CHD cases) from 3 studies: the EPIC-CVD case-cohort study (18), the UK Biobank (19), and a modified version of the CARDIoGRAMplusC4D consortium (m-CARDIoGRAMplusC4D) because we could only include those studies that provided us with sex-specific summary data (Cardiogenics, Thiseas, AMC-PAS, Duke 2, CCGB 2, ITH 2, OHGS A2, OHGS B2, OHGS C2, Germifs I, Germifs II, Germifs III, Germifs IV, LIFE-Heart, and LURIC (20)). Details of the 3 studies (EPIC-CVD, UK Biobank, and m-CARDIoGRAMplusC4D), including ethical approval and definitions of fatal or nonfatal CHD in each study, can be found in the Supplemental methods (17).
Genotyping
EPIC-CVD participants were genotyped with the Human Core Exome array, Illumina 660 Quad array, and Omni Exome Express array (21). Genotyping in the UK Biobank was performed using the Affymetrix UK BiLEVE Axiom array and the Affymetrix UK Biobank Axiom Array (19, 22). The m-CARDIoGRAMplusC4D studies have used various genotyping methods as described previously (20).
Statistical analysis
We verified whether the ANM variants were a valid instrument for the MR analysis in women by calculating the F-statistic according to the method described previously (23), using the SD (5.8 years) for ANM from the imputed data in the EPIC-CVD subcohort and the betas for the ANM variants from the GWAS (12).
Regarding the outcome CHD, summary statistics (odds ratios and SEs for the SNP-CHD associations) were derived through the contact persons of UK Biobank and the included CARDIoGRAMplusC4D studies. For the EPIC-CVD case-cohort study, Prentice-weighted Cox proportional hazards regression adjusted for age, country, the first 2 principal components, and array was used to calculate HRs and SEs for the associations between reproductive aging SNPs and CHD outcome.
We performed a 2-sample MR using 4 separate methods to estimate causal effects for binary (CHD) outcomes: the simple median-based method, the weighted median-based method, the standard inverse-variance weighted (IVW) regression and the MR-Egger regression using the “Mendelian Randomization” package in R (24). The IVW provides a consistent estimate and assumes that all assumptions of the instrumental variable are met, the median-based and MR-Egger methods provide estimates under weaker assumptions, with the MR-Egger additionally providing an intercept that represents the average pleiotropic effect (25, 26). When unbalanced horizontal pleiotropy is absent, results of all methods are expected to be consistent (27). We first conducted sex-specific MR analyses for ANM on CHD in all 3 studies (UK Biobank, m-CARDIoGRAMplusC4D, EPIC-CVD) separately. Subsequently, we pooled the estimates with a fixed effect model as is standard in MR studies. All analyses were conducted with R version 3.2.0 (28).
Methods for MR to Study the Association Between ANM Variants and CHD Risk Factors
Outcomes
The outcomes used for this MR are traditional CHD risk factors: total cholesterol, high-density lipoprotein (HDL) cholesterol, triglycerides, HbA1c, and glucose. More risk factors are associated with CHD, but we could only use risk factors for which publicly available summary statistics were accessible.
Data sources
We again used the 56 variants from the study by Day et al (12) as instrument for the exposure ANM. We applied these variants in MR analyses using data from EPIC-CVD and UK Biobank combined with publicly available GWAS summary statistics from the Global Lipids Genetics Consortium (n = 188 577) (29) (total cholesterol, HDL cholesterol, triglycerides) and MAGIC (n = 122 744) (30, 31) (HbA1c, fasting glucose) for the CHD risk factor outcomes. Details on these consortia can be found in the Supplemental methods (17).
Genotyping
As described previously, EPIC-CVD participants were genotyped with the Human Core Exome array, Illumina 660 Quad array, and Omni Exome Express array (21). Genotyping in the UK Biobank was performed using the Affymetrix UK BiLEVE Axiom array and the Affymetrix UK Biobank Axiom Array (19, 22). The Global Lipids Genetics Consortium and MAGIC used different assays as described previously (29-31). Finally, the 56 SNPs were selected according to the study by Day et al as described previously (12).
Statistical analysis
For the CHD risk factor outcomes, we derived effect estimates and SEs for the reproductive aging SNPs with the cardiovascular risk factors total cholesterol, HDL cholesterol, and triglycerides in the Global Lipids Genetics Consortium (29), and in MAGIC (30, 31) for diabetes risk factors HbA1c and fasting glucose using Phenoscanner (32) or the original publication. In the random subcohort of EPIC-CVD, we first imputed the missing observational data of EPIC-CVD (nongenetic data only) using multiple imputation with the MICE package in R (33) with 10 imputations and 50 iterations, including the cardiovascular disease (CVD) risk factors, SNPs, and other baseline characteristics as predictors. Subsequently, we derived regression coefficients with linear regression in the subcohort only, separately in each imputation, using the same adjustments as for CHD. Thereafter, we pooled the results with Rubin’s rule (34), a method designed to pool parameter estimates of multiple imputed datasets, taking into account that the imputed datasets are drawn from the same source dataset. The estimates for the UK Biobank data were downloaded from the Nealelab (35).
We performed a 2-sample MR using 4 separate methods to estimate causal effects for continuous (total cholesterol, HDL cholesterol, triglycerides, apolipoprotein A, apolipoprotein B, C-reactive protein, glucose, HbA1c) outcomes: the simple median-based method, the weighted median-based method, the standard IVW regression, and the MR-Egger regression using the “Mendelian Randomization” package in R (24). MR analyses were performed for each cardiovascular risk factor in each study separately (EPIC-CVD, UK Biobank, Global Lipids Genetics Consortium, MAGIC) and then pooled using a fixed effects model. Sex-specific analyses were possible in EPIC-CVD and UK Biobank, and pooled results with both sexes conducted with Global Lipids Genetics Consortium and MAGIC. All analyses were conducted with R version 3.2.0 (28).
Methods for MR to Study the Association Between CHD Variants and ANM (Reversed Association)
Outcomes
For this MR, ANM was the outcome defined as the age at last naturally occurring menstrual period followed by at least 12 consecutive months of amenorrhea (12).
Data sources
To study causality of the reversed association, we used the genome-wide significant variants for CHD in the CARDIoGRAMplusC4D (20) (n = 185 000) data as the instrument for the exposure CHD. The ReproGen (n = 70 000) (12) data were used for ANM as the outcome. We used the sex-combined GWAS summary statistics for the exposure because sex-specific summary statistics were not available. The outcome ANM is only available in women, so the outcome variants are in women only.
Genotyping
The CARDIoGRAMplusC4D consortium consists of 40 different studies with all slightly different methods for genotyping. Details can be found in the paper by Nikpay et al (20). The 56 SNPs selected according to the study by Day et al from the ReproGen study were used as outcome variants (12).
Statistical analysis
We performed a 2-sample MR using 4 separate methods to estimate causal effects for the continuous ANM outcome: the simple median-based method, the weighted median-based method, the standard IVW regression, and the MR-Egger regression using the “Mendelian Randomization” package in R (24). The analyses were conducted for both sexes combined. All analyses were conducted with R version 3.2.0 (28).
Methods for MR to Study the Association Between CHD Risk Factor Variants and ANM (Reversed Association)
Outcomes
For this MR, ANM was the outcome defined as the age at last naturally occurring menstrual period followed by at least 12 consecutive months of amenorrhea (12).
Data sources
To study causality of the reversed association, we used genome-wide significant variants from publicly available GWAS summary statistics of the Global Lipids Genetics Consortium (n = 188 577) (29) (total cholesterol, low-density lipid [LDL] cholesterol), the International Consortium for Blood Pressure GWAS (n = 200 000) (36) (systolic blood pressure, diastolic blood pressure), and the GIANT consortium (n = 339 224) (37) (body mass index) as instruments for the exposures, and ReproGen (n = 70 000) (12) for ANM as the outcome. We used the sex-combined GWAS summary statistics for the exposure because sex-specific summary statistics were not available. The outcome ANM is only available in women, so the outcome variants are in women only.
Genotyping
We used the genetic variants as defined in the Global Lipids Genetics Consortium, the International Consortium for Blood Pressure GWAS, and the GIANT consortium. Details on genotyping and SNP selection can be found in references (29, 36, 37). The 56 SNPs selected according to the study by Day et al from the ReproGen study were used as outcome variants (12).
Statistical analyses
We performed a 2-sample MR using 4 separate methods to estimate causal effects for continuous (total cholesterol, LDL cholesterol, systolic blood pressure, diastolic blood pressure, and body mass index) outcomes: the simple median-based method, the weighted median-based method, the standard IVW regression, and the MR-Egger regression using the “Mendelian Randomization” package in R (24). These analyses with ANM as an outcome were conducted for both sexes combined. All analyses were conducted with R version 3.2.0 (28).
Results
Results of MR to Study the Association Between ANM Variants and CHD Risk
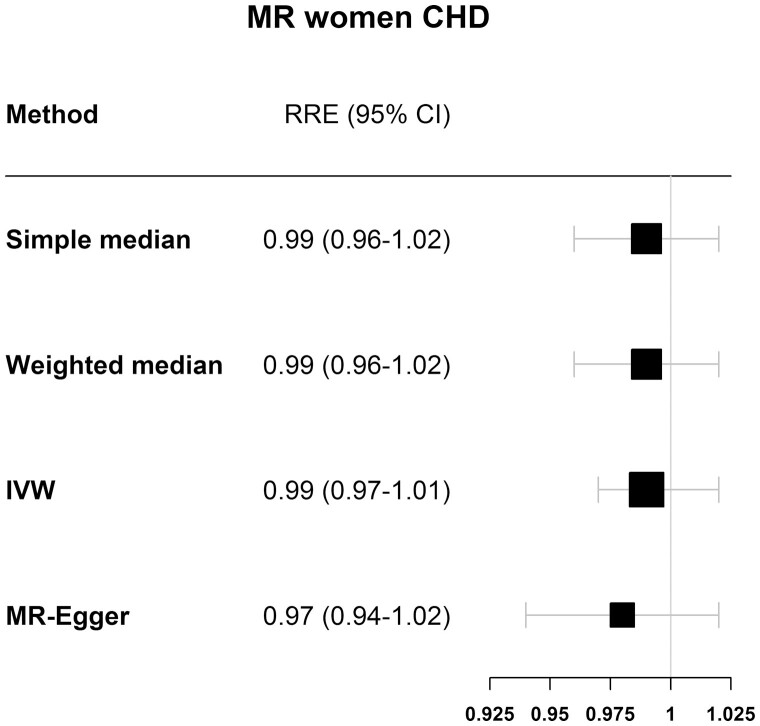
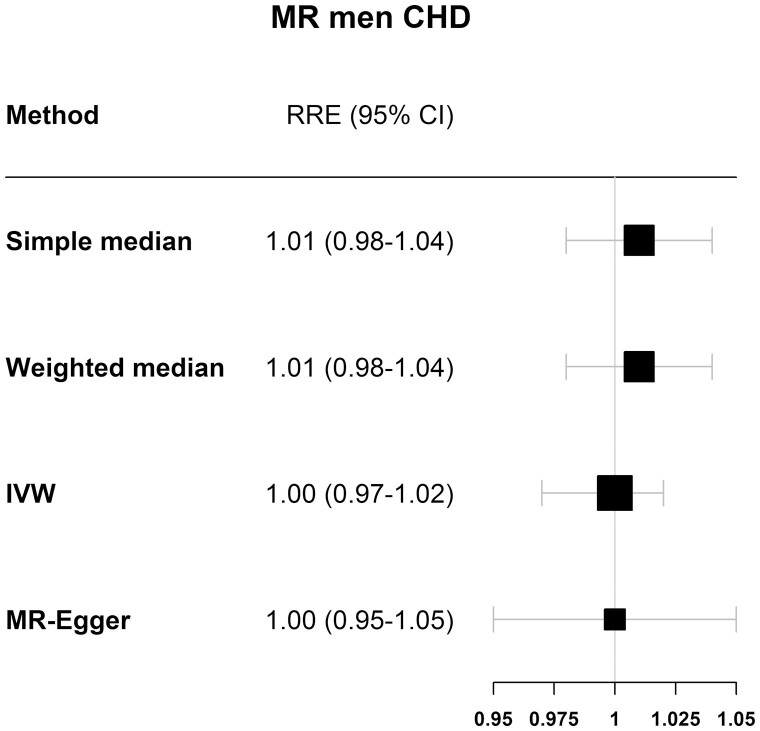
The F-statistic for genetically determined reproductive aging in women in EPIC-CVD was 93.7, indicating that our instrument was strong in women. Table 1 shows the results for the analysis of the association between genetically determined reproductive aging and CHD risk, stratified by sex and by study (UKBiobank, m-CARDIoGRAMplusC4D, and EPIC-CVD). In women (Fig. 2 and Table 1), the IVW analyses in each study separately showed no causal association between genetically determined reproductive aging and CHD, nor when studies were pooled together (relative risk estimate [RRE]IVW = 0.99; 95% CI, 0.97-1.01). The MR-Egger method indicated no pleiotropic effects (intercept = 0.004, P = 0.318) and resulted in an RREMR-Egger of 0.97 (95% CI, 0.94-1.02) in the pooled data. Similar results were found for men (Fig. 3 and Table 2) with a pooled RREIVW of 1.00 (95% CI, 0.97-1.02), also indicating no pleiotropic effects (RREMR-Egger = 1.00 (95% CI, 0.95-1.05), intercept = 0.000, P = 0.948).
Table 1.
Relative risk estimates of the association between genetically determined reproductive aging and coronary heart disease for individual studies and the pooled cohorts
| UK Biobank OR (95% CI) |
m-CARDIoGRAM plusC4D OR (95% CI) |
EPIC-CVD HR (95% CI) |
Pooled relative risk estimates | |
|---|---|---|---|---|
| Women | ||||
| Simple median | 0.99 (0.96-1.02) | 0.97 (0.89-1.06) | 0.97 (0.78-1.21) | 0.99 (0.96-1.02) |
| Weighted median | 0.99 (0.96-1.02) | 0.97 (0.89-1.06) | 1.08 (0.88-1.33) | 0.99 (0.96-1.02) |
| IVW | 0.99 (0.97-1.02) | 0.98 (0.91-1.05) | 1.02 (0.89-1.17) | 0.99 (0.97-1.01) |
| MR-Egger | 0.98 (0.94-1.02) | 0.88 (0.76-1.02) | 1.29 (0.91-1.83) | 0.97 (0.94-1.02) |
| Men | ||||
| Simple median | 0.99 (0.95-1.02) | 1.05 (0.99-1.12) | 0.98 (0.81-1.18) | 1.01 (0.98-1.04) |
| Weighted median | 0.99 (0.96-1.03) | 1.05 (0.99-1.12) | 0.84 (0.71-1.01) | 1.01 (0.98-1.04) |
| IVW | 0.99 (0.96-1.01) | 1.03 (0.99-1.08) | 0.93 (0.82-1.05) | 1.00 (0.97-1.02) |
| MR-Egger | 0.98 (0.93-1.03) | 1.02 (0.92-1.12) | 0.85 (0.62-1.16) | 1.00 (0.95-1.05) |
Abbreviations: IVW, inverse-variance weighted; MR, Mendelian randomization; OR, odds ratio.
Figure 2.
Results for the MR of ANM variants and coronary heart disease in women of the 4 different Mendelian randomization methods used.
Figure 3.
Results for the MR of ANM variants and coronary heart disease in men of the 4 different Mendelian randomization methods used.
Table 2.
Estimates of the association between CHD and CHD risk factors and genetically determined reproductive aging
| CHD beta (95% CI) |
Total cholesterol beta (95% CI) |
LDL cholesterol beta (95% CI) |
Systolic blood pressure beta (95% CI) |
Diastolic blood pressure beta (95% CI) |
Body mass index beta (95% CI) |
|
|---|---|---|---|---|---|---|
| Simple median | 0.064 (-0.104 to 0.232) |
0.000 (-0.005 to 0.005) |
0.004 (-0.002 to 0.009) |
-0.006 (-0.030 to 0.019) |
0.018 (-0.022 to 0.058) |
-0.385 (-0.658 to -0.112) |
| Weighted median | 0.057 (-0.100 to 0.231) |
-0.005 (-0.009 to 0.001) |
0.000 (-0.004 to 0.004) |
-0.013 (-0.036 to 0.009) |
-0.008 (-0.043 to 0.028) |
-0.114 (-0.379 to 0.151) |
| IVW | 0.063 (-0.050 to 0.176) |
-0.002 (-0.006 to 0.002) |
0.003 (-0.001 to 0.006) |
0.015 (-0.018 to 0.049) |
0.023 (-0.031 to 0.077) |
-0.069 (-0.294 to 0.156) |
| MR-Egger | -0.005 (-0.260 to 0.251) |
-0.007 (-0.013 to 0.000) |
0.001 (-0.005 to 0.008) |
-0.004 (-0.112 to 0.104) |
-0.060 (-0.237 to 0.117) |
0.418 (-0.116 to 0.953) |
We used the sex-combined genome-wide association study summary statistics for the exposure because sex-specific summary statistics were not available. The outcome at natural menopause is only available in women, so the outcome variants are in women only.
Abbreviations: CHD, coronary heart disease; IVW, inverse-variance weighted; MR, Mendelian randomization; LDL, low-density lipoprotein.
Results of MR to Study the Association Between ANM Variants and CHD Risk Factors
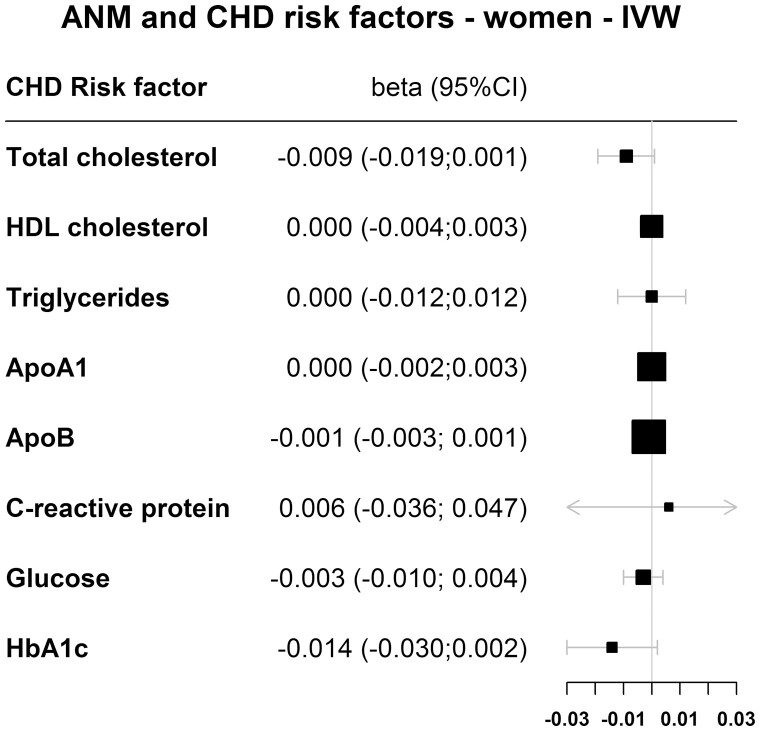
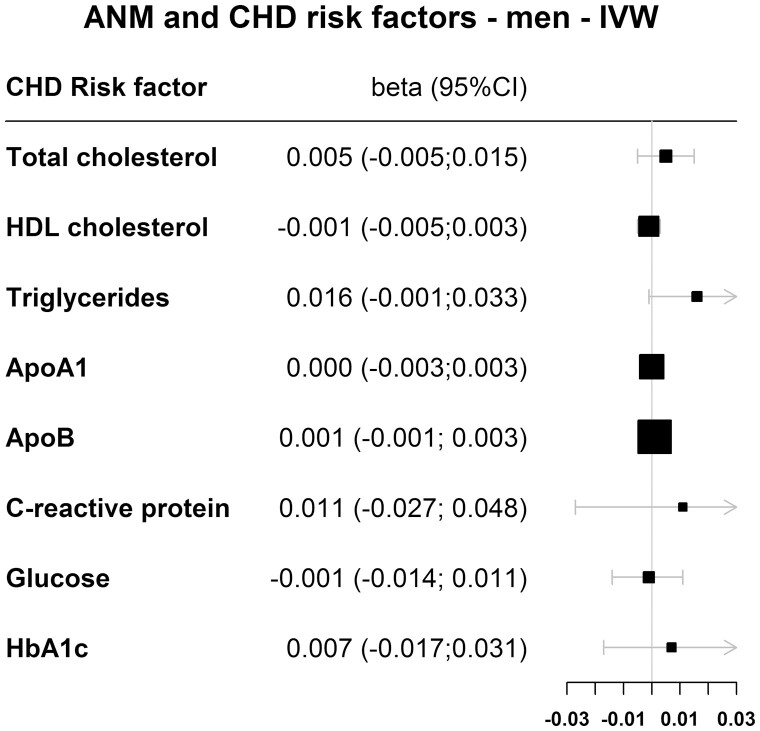
Figure 4 (women) and Fig. 5 (men) show the IVW results for the association between genetically determined reproductive aging and cardiovascular risk factors. Supplemental table 2 (17) shows the results for all 4 MR methods. For each 1-year decrease in genetically determined reproductive aging, total cholesterol levels decreased by 0.009 mmol/L in women in the IVW analysis; however, this was not statistically significant (95% CI, -0.019 to 0.001). Similarly, genetically determined reproductive aging was not causally associated with total cholesterol in men (betaIVW = 0.005 mmol/L; 95% CI, -0.005 to 0.015). Furthermore, no causal association was found for HDL cholesterol, triglycerides, apolipoprotein A, apolipoprotein B, C-reactive protein, glucose, or HbA1c in both women as well as men (Fig. 4, Fig. 5, Supplemental Table 2 (17)).
Figure 4.
Results for the MR of ANM variants and coronary heart disease risk factors in women for the inversed variance-weighted (IVW) method.
Figure 5.
Results for the MR of ANM variants and coronary heart disease risk factors in men for the inversed variance-weighted (IVW) method.
Results of MR to Study the Association Between CHD Variants and ANM
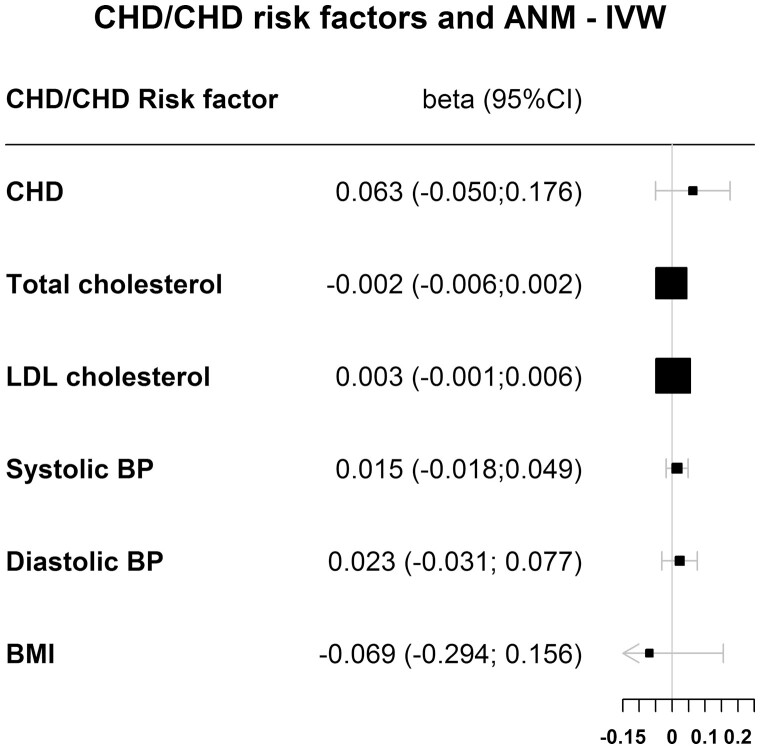
Figure 6 and Table 2 show the results for the association between CHD genetically determined reproductive aging for all 4 MR methods, investigating causality of the reversed association. The IVW analysis shows that CHD was not causally associated with earlier genetically determined reproductive aging (betaIVW = 0.063; 95% CI, -0.050 to 0.176).
Figure 6.
Results for the MR of CHD and CHD risk factors and ANM for the inversed variance-weighted (IVW) method. We used the sex-combined GWAS summary statistics for the exposure because sex-specific summary statistics were not available. The outcome ANM is only available in women, so the outcome variants are in women only.
Results of MR to Study the Association Between CHD Risk Factor Variants and ANM
Figure 6 and Table 2 show the results for the association between CHD risk factors and genetically determined reproductive aging for all 4 MR methods, investigating causality of the reversed association. The IVW analysis shows that total cholesterol (betaIVW = -0.002; 95% CI, -0.006-0.002), LDL cholesterol (betaIVW = 0.003; 95% CI, -0.001 to 0.006), systolic blood pressure (betaIVW = 0.015; 95% CI, -0.018 to 0.049), diastolic blood pressure (betaIVW = 0.023; 95% CI, -0.031 to 0.077), and body mass index (betaIVW = -0.069; 95% CI, -0.0294 to 0.0156) were not causally associated with earlier genetically determined reproductive aging.
Discussion
This study did not find a causal association between reproductive aging and CHD risk or CHD risk factors, including cholesterol levels, in women. This study does not provide evidence for an association between genetic variants for female reproductive aging and CHD risk or CHD risk factors in men. Furthermore, this study does not provide evidence for causality of the reversed association because we did not find a causal association between CHD and CHD risk factors and genetically determined reproductive aging.
Our findings regarding CHD are partly in contrast with 1 previous study investigating the association between ANM SNPs and CHD death, which reported a significantly increased risk of CHD death with a weighted genetic risk score (wGRS) in women, but not in men (14). However, our findings are in line with those of the MR analysis in women, presented in the same paper, using CARDIoGRAMplusC4D data only, which was also null. The discrepancy between the wGRS and MR findings is potentially because the wGRS analysis was adjusted for several known CVD risk factors (current smoking, body mass index, hypertension, type 2 diabetes, total cholesterol, and lipid treatment). This might induce a biased association between the genetic variant and the outcome through confounder(s), also known as collider bias (38, 39). In addition, the number of cases used for the wGRS analyses was small (only 541 CHD deaths in women), so a chance finding cannot be ruled out either. However, the discrepancy between studies might also be caused by the heterogeneity of outcome definitions. These definitions slightly differ between this study (CHD) and the previously published study (CVD), the composite CVD also includes stroke and congestive heart failure. This should be considered when interpreting the results.
Our MR study suggests that the association between genetically determined reproductive aging and CHD is not causal. However, most observational studies do find an association between early age at menopause and CHD in women. A possible explanation is that observational studies are susceptible to residual confounding. Postmenopausal women are older than premenopausal women, making it challenging to separate the effects of biological aging from the various phases of the reproductive aging process. Hence, residual confounding due to age may still be present in observational studies. Another possible explanation might be survivor bias in the GWAS we used. It is possible that women, who died of a CHD event before they went through menopause, although this is very rare (16), were not included in the GWAS. Therefore, variants associated with both ANM and CHD might not have been found. Furthermore, reverse causation could be a potential problem in observational studies. Although most studies assume that an early ANM increases CHD risk, it might be possible that an unfavorable cardiovascular risk profile, or accelerated vascular aging, causes an early ANM. One previous observational study showed that higher cholesterol levels before menopause were associated with earlier menopause (9). One other observational study found no association between premenopausal CVD and subsequent age at menopause (40); if anything, women who developed CVD before menopause had a lower risk of becoming postmenopausal than women without premenopausal CVD (HR = 0.98 for CVD and HR = 0.90 for myocardial infarction), indicating that menopause occurred later in these women (40), but none of these results was statistically significant the result of the small number of premenopausal cases. However, another study did find an accelerated menopause for women with CVD before the age of 35 (16). Our reversed MR analysis does not support evidence for a reversed association where CHD increases the risk of early menopause.
MR uses SNPs, which are randomly assigned by birth, as instrumental variables, and as such provides a method to assess causality (41). However, an MR study makes several assumptions that have to be taken into account (42). The first assumption is that the genetic marker is associated with the exposure. The SNPs used in our study were all associated with ANM at a P value < 5e-6 in the latest and largest GWAS (13). As discussed previously, this may not be true in men. The second and third assumptions are that the association between the genetic marker and the outcome is explained exclusively through the exposure of interest and is unconfounded. This is often referred to as the absence of pleiotropy, which means that the genetic variant is not associated with other phenotypes. Although our Phenoscanner search showed that a few of the SNPs are associated with age at menarche or sex hormone levels, and thus that some pleiotropy may be present, our MR-Egger analysis showed no indication of pleiotropy because all intercepts were 0 or very close to 0 and nonsignificant (26). We therefore assume that our results are not biased by pleiotropy.
Strengths and Limitations
Strengths of this study are that, to the best of our knowledge, this is the largest MR study of associations between reproductive aging and CHD to date with 20 169 CHD events in women and 27 397 in men. We used several methods for MR analyses all yielding consistent results for the tested hypotheses, and in women the instrument we used was strong (F-statistic 93.7). Some limitations need to be acknowledged. First, we cannot establish whether the ANM risk score is a valid instrument for reproductive aging in men. The F-statistic is calculated using observed menopausal age in women, but men do not have a similar trait with an abrupt stop in reproductive potential. Because the SNPs we used are mainly implicated in mechanisms that are not specific for women, and the SNPs were not sex-specific, we hypothesized that there are common mechanisms of reproductive aging for women and men, and that, therefore, the same variants can be used as marker for genetically determined reproductive aging in men. However, corresponding phenotypic traits in men need to be further investigated. Second, the GWAS on ANM included women with an ANM between 40 and 60 years only and therefore did not include women with an extremely early menopause (< 40) or premature ovary insufficiency (POI). Most of the observational studies did include women with an extremely early menopause or POI, and two recent systematic reviews and meta-analyses of observational studies showed that POI is associated with both fatal and non-fatal CHD and CVD (43, 44). Although we could not study an effect of extremely early menopause in our MR study, a recent GWAS on early menopause revealed no new genetic variants for early menopause and showed that the genetic etiology of early menopause overlaps with that of ANM; thus, early menopause is at least partly explained by the same polygenic variants as ANM (45). The GWAS also excluded women using hormone replacement therapy because long-term hormone replacement therapy use might be associated with lower CHD risk. However, this only induces confounding in observational studies and not in MR studies. Third, we cannot fully rule out reversed causation because the analyses are conducted in a sample of men and women combined, a consequence of using publicly accessible data, whereas the outcome (ANM) is estimated in women only. To rule out reversed causation, these analyses should be rerun in women and men separately. Fourth, our analyses with glucose were based on both fasting (MAGIC) and nonfasting estimates (EPIC-CVD). Although both are associated with an increased CVD risk (46, 47), it might not be appropriate to combine them because different SNPs might drive the association and underlying mechanisms could be different. Fifth, some prevalent cases might have been present at the start of the study, which can be problematic in observational studies. However, in an MR, one can argue that study entry is not “time zero,” but the allocation of genetic variants at conception is; therefore, all events are incident events. Furthermore, there were no women that had an event before they became postmenopausal.
Conclusion
In summary, we found no convincing evidence that reproductive aging is causally associated with CHD and CHD risk factors in women, nor are the SNPs related to CHD and CHD risk factors in men. We also found no evidence for causality of the reverse association in a combined sample of women and men. However, there is a discrepancy between the definition of CHD in the studies used and we could only analyze men and women together in the reversed association. Still, our results suggest that the association between early menopause and CHD risk in observational studies might be the result of residual confounding, reversed causation, or reflect a shared etiology that results in both earlier menopause and higher CHD risk.
Acknowledgments
This research has been conducted using the UK Biobank Resource under Application Number 29916. Data from m-CARDIoGRAMplusC4D have been contributed by CARDIoGRAMplusC4D investigators of the respective studies included. Statistics Netherlands is acknowledged for providing causes of death for the Dutch contribution to EPIC-CVD. We thank all EPIC participants and staff for their contribution to the study. We also thank staff from the EPIC-CVD and EPIC-InterAct Coordinating Centres for sample preparation and data handling, particularly Sarah Spackman (EPIC-CVD Data Manager) and Nicola Kerrison (EPIC-InterAct Data Manager, MRC Epidemiology Unit).
Glossary
Abbreviations
- ANM
at natural menopause
- CHD
coronary heart disease
- CARDIoGRAM
Coronary Artery Disease Genome wide Replication and Meta-analysis
- CVD
cardiovascular disease
- GWAS
genome-wide association study
- HbA1c
glycated hemoglobin
- HDL
high-density lipoprotein
- HR
hazard ratio
- IVW
inverse-variance weighted
- LDL
low-density lipoprotein
- MR
Mendelian randomization
- POI
premature ovary insufficiency
- RRE
relative risk estimate
- SNP
single-nucleotide polymorphism
- wGRS
weighted genetic risk score
Contributor Information
Veerle Dam, Julius Center for Health Sciences and Primary Care, University Medical Center Utrecht, Utrecht University, GA 3508 Utrecht, the Netherlands; Netherlands Heart Institute, DG 3501 Utrecht, the Netherlands.
N Charlotte Onland-Moret, Julius Center for Health Sciences and Primary Care, University Medical Center Utrecht, Utrecht University, GA 3508 Utrecht, the Netherlands.
Stephen Burgess, MRC Biostatistics Unit, University of Cambridge, Cambridge CB2 0SR, UK; MRC/BHF Cardiovascular Epidemiology Unit, Department of Public Health and Primary Care, University of Cambridge, Cambridge, UK; Homerton College, Cambridge, UK.
Maria-Dolores Chirlaque, Department of Epidemiology, Regional Health Authority, IMIB-Arrixaca, Murcia University, 30001 Murcia, Spain; Department of Public Health and Clinical Medicine, Umea University, 901 87 Umea, Sweden.
Sanne A E Peters, Julius Center for Health Sciences and Primary Care, University Medical Center Utrecht, Utrecht University, GA 3508 Utrecht, the Netherlands; The George Institute for Global Health, Imperial College London, London W12 0BZ, UK.
Ewoud Schuit, Julius Center for Health Sciences and Primary Care, University Medical Center Utrecht, Utrecht University, GA 3508 Utrecht, the Netherlands.
Kaja Tikk, Division of Clinical Epidemiology and Aging Research, German Cancer Research Centre (DKFZ), 69120 Heidelberg, Germany; German Cancer Consortium, DKFZ, 69120 Heidelberg, Germany.
Elisabete Weiderpass, International Agency for Research on Cancer, World Health Organization, 69372 Lyon, France.
Clare Oliver-Williams, MRC/BHF Cardiovascular Epidemiology Unit, Department of Public Health and Primary Care, University of Cambridge, Cambridge, UK; Homerton College, Cambridge, UK.
Angela M Wood, MRC/BHF Cardiovascular Epidemiology Unit, Department of Public Health and Primary Care, University of Cambridge, Cambridge, UK.
Anne Tjønneland, Danish Cancer Society Research Center, DK-2100 Copenhagen, Denmark; Department of Public Health, Faculty of Health and Medical Sciences, University of Copenhagen, DK-2200 Copenhagen, Denmark.
Christina C Dahm, Department of Public Health, Aarhus University, 8000 Aarhus, Denmark.
Kim Overvad, Department of Public Health, Aarhus University, 8000 Aarhus, Denmark; Department of Cardiology, Aalborg University Hospital, 9000 Aalborg, Denmark.
Marie-Christine Boutron-Ruault, INSERM, Centre for Research in Epidemiology and Population Health, U1018, Nutrition, Hormones, and Women’s Health Team, Institut Gustave Roussy, 94 805 Villejuif, France.
Matthias B Schulze, Department of Epidemiology, German Institute of Human Nutrition, Potsdam-Rehbruecke, Nuthetal, Germany; Institute of Nutritional Science, University of Potsdam, 14558 Nuthetal, Germany.
Antonia Trichopoulou, Hellenic Health Foundation, 115 27 Athens, Greece; WHO Collaborating Center for Nutrition and Health, Unit of Nutritional Epidemiology and Nutrition in Public Health, Department of Hygiene, Epidemiology and Medical Statistics, School of Medicine, National and Kapodistrian University of Athens, Athens 115 27, Greece.
Pietro Ferrari, International Agency for Research on Cancer, World Health Organization, 69372 Lyon, France.
Giovanna Masala, Cancer Risk Factors and Life-Style Epidemiology Unit, Institute for Cancer Research, Prevention and Clinical Network - ISPRO, 50139 Florence, Italy.
Vittorio Krogh, Epidemiology and Prevention Unit, Fondazione IRCCS Istituto Nazionale dei Tumori, 20133 Milan, Italy.
Rosario Tumino, Cancer Registry and Histopathology Department, “Civic - M.P. Arezzo” hospital, ASP Ragusa, 97100 Ragusa, Italy.
Giuseppe Matullo, Department of Medical Sciences, University of Torino, 10124 Torino, Italy; Italian Institute for Genomic Medicine–IIGM/HuGeF, 10126 Torino, Italy.
Salvatore Panico, Dipartimento di medicina clinica e chirurgia, Federico II University, 80126 Naples, Italy.
Jolanda M A Boer, National Institute for Public Health and the Environment, 3720 BA Bilthoven, the Netherlands.
W M Monique Verschuren, Julius Center for Health Sciences and Primary Care, University Medical Center Utrecht, Utrecht University, GA 3508 Utrecht, the Netherlands; National Institute for Public Health and the Environment, 3720 BA Bilthoven, the Netherlands.
Marit Waaseth, Department of Pharmacy, Faculty of Health Sciences, UiT the Arctic University of Norway, N-9037 Tromsø, Norway.
Maria José Sánchez Pérez, Escuela Andaluza de Salud Pública. Instituto de Investigación Biosanitaria ibs.GRANADA, Universidad de Granada, 18011 Granada, Spain; CIBER de Epidemiología y Salud Pública (CIBERESP), 28029 Madrid, Spain.
Pilar Amiano, CIBER de Epidemiología y Salud Pública (CIBERESP), 28029 Madrid, Spain; Public Health Division of Gipuzkoa, Biodonostia Research Institute, 20014 San Sebastian, Spain.
Liher Imaz, CIBER de Epidemiología y Salud Pública (CIBERESP), 28029 Madrid, Spain; Public Health Division of Gipuzkoa, Biodonostia Research Institute, 20014 San Sebastian, Spain.
Conchi Moreno-Iribas, Instituto de Salud Pública de Navarra, IdiSNA, Navarre Institute for Health Research, REDISSEC, 31008, Pamplona, Spain.
Olle Melander, Department of Clinical Sciences, Lund University, SE-221 00 Malmö, Sweden.
Sophia Harlid, Department of Radiation Sciences, Oncology, Umea University, 901 87 Umea, Sweden.
Maria Nordendahl, Department of Public Health and Clinical Medicine, Umea University, 901 87 Umea, Sweden.
Patrik Wennberg, Department of Public Health and Clinical Medicine, Umea University, 901 87 Umea, Sweden.
Timothy J Key, Nuffield Department of Population Health, University of Oxford, OX3 7LF Oxford, England.
Elio Riboli, Department of Epidemiology and Biostatistics, School of Public Health, Imperial College London, SW7 2AZ London, UK.
Carmen Santiuste, CIBER de Epidemiología y Salud Pública (CIBERESP), 28029 Madrid, Spain; Department of Epidemiology, Murcia Regional Health Authority, IMIB-Arrixaca, 30001 Murcia, Spain.
Rudolf Kaaks, Division of Cancer Epidemiology, DKFZ, Foundation under Public Law, D-69120 Heidelberg, Germany.
Verena Katzke, Division of Cancer Epidemiology, DKFZ, Foundation under Public Law, D-69120 Heidelberg, Germany.
Claudia Langenberg, MRC Epidemiology Unit, University of Cambridge School of Clinical Medicine, CB2 0SL Cambridge, UK.
Nicholas J Wareham, MRC Epidemiology Unit, University of Cambridge School of Clinical Medicine, CB2 0SL Cambridge, UK.
Heribert Schunkert, Deutsches Herzzentrum München, Technische Universität München, 80636 Munich, Germany; DZHK (German Center for Cardiovascular Research), Partner Site Munich Heart Alliance, 80636 Munich, Germany.
Jeanette Erdmann, Institute for Cardiogenetics, University of Lübeck, 23562 Lübeck, Germany.
Christina Willenborg, Institute for Cardiogenetics, University of Lübeck, 23562 Lübeck, Germany.
Christian Hengstenberg, Department of Internal Medicine II, Division of Cardiology, Medical University of Vienna, 1090 Vienna, Austria.
Marcus E Kleber, Vth Department of Medicine, Medical Faculty Mannheim, Heidelberg University, Mannheim, Germany.
Graciela Delgado, Vth Department of Medicine, Medical Faculty Mannheim, Heidelberg University, Mannheim, Germany.
Winfried März, Vth Department of Medicine, Medical Faculty Mannheim, Heidelberg University, Mannheim, Germany; Synlab Academy, Synlab Holding Deutschland GmbH, 68167 Mannheim, Germany; Clinical Institute of Medical and Chemical Laboratory Diagnostics, Medical University of Graz, 8036 Graz, Austria.
Stavroula Kanoni, William Harvey Research Institute, Barts and The London School of Medicine and Dentistry, Queen Mary University of London, London E1 4NS, UK.
George Dedoussis, Department of Nutrition-Dietetics/Harokopio University, 17671 Athens, Greece.
Panos Deloukas, William Harvey Research Institute, Barts and The London School of Medicine and Dentistry, Queen Mary University of London, London E1 4NS, UK; Centre for Genomic Health, Queen Mary University of London, London E1 4NS, UK; Princess Al-Jawhara Al-Brahim Centre of Excellence in Research of Hereditary Disorders, King Abdulaziz University, Jeddah 21589, Saudi Arabia.
Majid Nikpay, Ruddy Canadian Cardiovascular Genetics Centre, University of Ottawa Heart Institute, Ottawa, Ontario K1Y 4W7, Canada.
Ruth McPherson, Ruddy Canadian Cardiovascular Genetics Centre, University of Ottawa Heart Institute, Ottawa, Ontario K1Y 4W7, Canada.
Markus Scholz, Institute for Medical Informatics, Statistics and Epidemiology, University of Leipzig, 04107 Leipzig, Germany; LIFE Research Center for Civilization Diseases, University of Leipzig, 04103 Leipzig, Germany.
Andrej Teren, LIFE Research Center for Civilization Diseases, University of Leipzig, 04103 Leipzig, Germany; Heart Center Leipzig, 04289 Leipzig, Germany.
Adam S Butterworth, MRC/BHF Cardiovascular Epidemiology Unit, Department of Public Health and Primary Care, University of Cambridge, Cambridge, UK.
Yvonne T van der Schouw, Julius Center for Health Sciences and Primary Care, University Medical Center Utrecht, Utrecht University, GA 3508 Utrecht, the Netherlands.
Funding
The EPIC-CVD project was supported by the European Union Framework 7 (HEALTH-F2-2012-279233), the European Research Council (268834), the UK Medical Research Council (G0800270, MR/L003120/1), the British Heart Foundation (SP/09/002, RG/08/014, RG13/13/30194), and the UK National Institute of Health Research (to EPIC-CVD). The establishment of the subcohort was supported by the EU Sixth Framework Programme (FP6) (grant LSHM_CT_2006_037197 to the InterAct project) and the Medical Research Council Epidemiology Unit (grants MC_UU_12015/1 and MC_UU_12015/5). The national EPIC cohorts are supported by Danish Cancer Society (Denmark); Ligue Contre le Cancer, Institut Gustave Roussy, Mutuelle Générale de l’Education Nationale, Institut National de la Santé et de la Recherche Médicale (INSERM) (France); Deutsche Krebshilfe, Deutsches Krebsforschungszentrum and Federal Ministry of Education and Research (Germany); Ministry of Health and Social Solidarity, Stavros Niarchos Foundation and Hellenic Health Foundation (Greece); Italian Association for Research on Cancer (AIRC) and National Research Council (Italy) and MIUR “Dipartimenti di Eccellenza”(Project D15D18000410001) to the Department of Medical Sciences; Dutch Ministry of Public Health, Welfare and Sports (VWS), LK Research Funds, Dutch Prevention Funds, Dutch ZON (Zorg Onderzoek Nederland), World Cancer Research Fund (WCRF); ERC-2009-AdG 232997 and Nordforsk, Nordic Centre of Excellence programme on Food, Nutrition and Health (Norway); Health Research Fund (FIS), Regional Governments of Andalucía, Asturias, Basque Country, Murcia and Navarra, ISCIII RETIC (RD06/0020) (Spain); Swedish Cancer Society, Swedish Scientific Council and Regional Government of Skåne and Västerbotten (Sweden); and Cancer Research UK, Medical Research Council (United Kingdom). LIFE-Heart is funded by the Leipzig Research Center for Civilization Diseases (LIFE). LIFE is an organizational unit affiliated to the Medical Faculty of the University of Leipzig. LIFE is funded by means of the European Union, by the European Regional Development Fund (ERDF), and by funds of the Free State of Saxony within the framework of the excellence initiative. This work is supported by the Dutch Heart Foundation (2013T083 to V.D.), by a UK Medical Research Council Skills Development Fellowship (MR/P014550/1 to S.A.E.P.), and by a Sir Henry Dale Fellowship jointly funded by the Welcome Trust and the Royal Society (204623/Z/16/Z to S.B.). None of the funding sources had a role in the collection, analysis, and interpretation of the data, nor in the decision to submit the article for publication.
Conflict of Interest
Dr. Clare Oliver-Williams received £1000 in prize money from Novartis. None of the other authors report any potential conflict of interest. Where authors are identified as personnel of the International Agency for Research on Cancer/World Health Organization, the authors alone are responsible for the views expressed in this article and they do not necessarily represent the decisions, policy or views of the International Agency for Research on Cancer/World Health Organization.
Data Availability
Restrictions apply to the availability of some or all data generated or analyzed during this study to preserve patient confidentiality or because they were used under license. The corresponding author will on request detail the restrictions and any conditions under which access to some data may be provided.
References
- 1. Timmis A, Townsend N, Gale C, et al. European Society of Cardiology: cardiovascular disease statistics 2017. Eur Heart J. 2018;39(7):508-579. [DOI] [PubMed] [Google Scholar]
- 2. van der Schouw YT, van der Graaf Y, Steyerberg EW, Eijkemans JC, Banga JD. Age at menopause as a risk factor for cardiovascular mortality. Lancet 1996;347(9003):714-718. [DOI] [PubMed] [Google Scholar]
- 3. Ossewaarde ME, Bots ML, Verbeek AL, et al. Age at menopause, cause-specific mortality and total life expectancy. Epidemiology 2005;16(4):556-562. [DOI] [PubMed] [Google Scholar]
- 4. Muka T, Oliver-Williams C, Kunutsor S, et al. Association of age at onset of menopause and time since onset of menopause with cardiovascular outcomes, intermediate vascular traits, and all-cause mortality: a systematic review and meta-analysis. JAMA Cardiol 2016;1(7):767-776. [DOI] [PubMed] [Google Scholar]
- 5. Zhu D, Chung H, Dobson AJ, et al. Age at natural menopause and risk of incident cardiovascular disease: a pooled analysis of individual patient data. Lancet Public Heal 2019;4(11):e553-e564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Dam V, Van Der Schouw YT, Onland-moret NC, et al. Association of menopausal characteristics and risk of coronary heart disease: a pan-European case-cohort analysis. Int J Epidemiol. 2019;48(4):1275-1285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Matthews KA, Crawford SL, Chae CU, et al. Are changes in cardiovascular disease risk factors in midlife women due to chronological aging or to the menopausal transition? J Am Coll Cardiol. 2009;54(25):2366-2373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. de Kat AC, Dam V, Onland-Moret NC, Eijkemans MJC, Broekmans FJM, van der Schouw YT. Unraveling the associations of age and menopause with cardiovascular risk factors in a large population-based study. BMC Med. 2017;15(1):2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Kok HS, van Asselt KM, van der Schouw YT, et al. Heart disease risk determines menopausal age rather than the reverse. J Am Coll Cardiol. 2006;47(10):1976-1983. [DOI] [PubMed] [Google Scholar]
- 10. Smith GD, Ebrahim S. “Mendelian randomization”: can genetic epidemiology contribute to understanding environmental determinants of disease? Int J Epidemiol. 2003;32(1):1-22. [DOI] [PubMed] [Google Scholar]
- 11. Davies NM, Holmes MV, Davey Smith G. Reading Mendelian randomisation studies: a guide, glossary, and checklist for clinicians. BMJ 2018;362:k601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Day FR. Europe PMC Funders Group. Large-scale genomic analyses link reproductive ageing to hypothalamic signaling. Breast cancer susceptibility and BRCA1-mediated DNA repair. Nat Genet. 2015;47(11):1294-1303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Aragam KG, Jiang T, Goel A, Kanoni S, Wolford BN.Discovery and systematic characterization of risk variants and genes for coronary artery disease in over a million participants. medRxiv. 2021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Sarnowski C, Kavousi M, Isaacs S, et al. Genetic variants associated with earlier age at menopause increase the risk of cardiovascular events in women. Menopause 2017;25(4):451-457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Richardson TG, Harrison S, Hemani G, Smith GD. An atlas of polygenic risk score associations to highlight putative causal relationships across the human phenome. eLife. 2019;8:e43657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Zhu D, Chung HF, Pandeya N, et al. Premenopausal cardiovascular disease and age at natural menopause: a pooled analysis of over 170,000 women. Eur J Epidemiol. 2019;34(3):235-246. [DOI] [PubMed] [Google Scholar]
- 17. Dam V. Supplemental material. 2022. doi: 10.5281/zenodo.5638886 [DOI] [Google Scholar]
- 18. Danesh J, Saracci R, Berglund G, et al. EPIC-Heart: the cardiovascular component of a prospective study of nutritional, lifestyle and biological factors in 520,000 middle-aged participants from 10 European countries. Eur J Epidemiol. 2007;22(2):129-141. [DOI] [PubMed] [Google Scholar]
- 19. Sudlow C, Gallacher J, Allen N, et al. UK Biobank: an open access resource for identifying the causes of a wide range of complex diseases of middle and old age. PLoS Med. 2015;12(3):e10017791-e10017710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Nikpay M, Goel A, Won H, et al. A comprehensive 1000 Genomes-based genome-wide association meta-analysis of coronary artery disease. Nat Genet. 2016;47(10):1121-1130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Zheng JS, Luan J, Sofianopoulou E, et al. Plasma vitamin C and type 2 diabetes: genome-wide association study and mendelian randomization analysis in European populations. Diabetes Care. 2021;44(1):98-106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Collins R. What makes UK Biobank special? Lancet 2012;379(9822):1173-1174. [DOI] [PubMed] [Google Scholar]
- 23. Burgess S, Davies NM, Thompson SG. Bias due to participant overlap in two-sample Mendelian randomization. Genet Epidemiol. 2016;40(7):597-608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Yavorska O, Burgess S.. Mendelian randomization: an R package for performing Mendelian randomization analyses using summarized data. Int J Epidemiol. 2017;46(6):1734-1739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Bowden J, Davey Smith G, Haycock PC, Burgess S. Consistent estimation in mendelian randomization with some invalid instruments using a weighted median estimator. Genet Epidemiol. 2016;40(4):304-314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Burgess S, Thompson SG. Erratum to: interpreting findings from Mendelian randomization using the MR-Egger method (Eur J Epidemiol, 10.1007/s10654-017-0255-x). Eur J Epidemiol. 2017;32(5):391-392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Dale CE, Fatemifar G, Palmer TM, et al. Causal associations of adiposity and body fat distribution with coronary heart disease, stroke subtypes, and type 2 diabetes mellitus: a Mendelian randomization analysis. Circulation. 2017;135(24):2373-2388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Team RC. R: A language and environment for statistical computing. 2016. Available at: https://www.r-project.org/
- 29. Willer CJ, Schmidt EM, Sengupta S, et al. ; Global Lipids Genetics Consortium. Discovery and refinement of loci associated with lipid levels. Nat Genet. 2013;45(11):1-24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Soranzo N, Sanna S, Wheeler E., et al. Common variants at 10 genomic loci influence hemoglobin A1c levels via glycemic and nonglycemic pathways. Diabetes. 2010;59(12):3229–3239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Dupuis J, Langenberg C, Prokopenko I, et al. New genetic loci implicated in fasting glucose homeostasis and their impact on type 2 diabetes risk. Nat Genet. 2010;42(Febrero):105-116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Staley JR, Blackshaw J, Kamat MA, et al. PhenoScanner: a database of human genotype-phenotype associations. Bioinformatics. 2016;32(20):3207-3209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Buuren S, Groothuis-Oudshoorn K. mice: multivariate imputation by chained equations in R. J Stat Softw. 2011;45(3). doi 10.18637/jss.v045.i03. [DOI] [Google Scholar]
- 34. Rubin DB. Multiple Imputation for Non Response in Surveys. John Wiley & Sons, Inc; 1987. [Google Scholar]
- 35. Biobank U. Rapid GWAS of thousands of phenotypes for 337,000 samples in the UK Biobank. Nealelab 2017. Available at: http://www.nealelab.is/blog/2017/7/19/rapid-gwas-of-thousands-of-phenotypes-for-337000-samples-in-the-uk-biobank.
- 36. Ehret G, Munroe P, Rice K, et al. Genetic variants in novel pathways influence blood pressure and cardiovascular disease risk. Nature. 2011;478(7367):103-109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Locke A, Kahali B, Berndt S, et al. Genetic studies of body mass index yield new insights for obesity biology. Nature. 2015;518:197-206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Haycock PC, Burgess S, Wade KH, Bowden J, Relton C, Smith GD. Statistical Commentary Best (but oft-forgotten) practices: the design, analysis, and interpretation of Mendelian randomization studies 1. Am J Clin Nutr. 2016;103(4):965-978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Day FR, Loh PR, Scott RA, Ong KK, Perry JRB. A robust example of collider bias in a genetic association study. Am J Hum Genet. 2016;98(2):392-393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Li J, Eriksson M, Czene K, Hall P, Rodriguez-Wallberg KA. Common diseases as determinants of menopausal age. Hum Reprod. 2016;31(12):2856-2864. [DOI] [PubMed] [Google Scholar]
- 41. Burgess S, Thompson SG.. Mendelian Randomization: Methods for Using Genetic Variants in Causal Estimation. CRC Press; 2015. [Google Scholar]
- 42. Vanderweele TJ, Tchetgen EJT, Kraft P. Methodological challenges in Mendelian randomization. Epidemiology. 2015;25(3):427-435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Tao XY, Zuo AZ, Wang JQ, Tao FB. Effect of primary ovarian insufficiency and early natural menopause on mortality: a meta-analysis. Climacteric. 2016;19(1):27-36. [DOI] [PubMed] [Google Scholar]
- 44. Roeters Van Lennep JE, Heida KY, Bots ML, Hoek A. Cardiovascular disease risk in women with premature ovarian insufficiency: a systematic review and meta-analysis. Eur. J. Prev. Cardiol. 2016;23(2):178-186. [DOI] [PubMed] [Google Scholar]
- 45. Perry JRB, Corre T, Esko T, et al. A genome-wide association study of early menopause and the combined impact of identified variants. Hum Mol Genet. 2013;22(7):1465-1472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Levitzky YS, Pencina MJ, D’Agostino RB, et al. Impact of impaired fasting glucose on cardiovascular disease. The Framingham Heart Study. J Am Coll Cardiol. 2008;51(3):264-270. [DOI] [PubMed] [Google Scholar]
- 47. Benn M, Tybjærg-Hansen A, McCarthy MI, Jensen GB, Grande P, Nordestgaard BG. Nonfasting glucose, ischemic heart disease, and myocardial infarction: a mendelian randomization study. J Am Coll Cardiol. 2012;59(25):2356-2365. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Restrictions apply to the availability of some or all data generated or analyzed during this study to preserve patient confidentiality or because they were used under license. The corresponding author will on request detail the restrictions and any conditions under which access to some data may be provided.