Abstract
In this review, we discuss the central role of fibroblast growth factor (FGF) signalling in mammalian tooth development. The FGF family consists of 22 members, most of which bind to four different receptor tyrosine kinases, which in turn signal through a cascade of intracellular proteins. This signaling regulates a number of cellular processes, including proliferation, differentiation, cell adhesion and cell mobility. FGF signaling first becomes important in the presumptive dental epithelium at the initiation stage of tooth development, and subsequently, it controls the invagination of the dental epithelium into the underlying mesenchyme. Later, FGFs are critical in tooth shape formation and differentiation of ameloblasts and odontoblasts, as well as in the development and homeostasis of the stem cell niche that fuels the continuously growing mouse incisor. In addition, FGF signaling is critical in human teeth, as mutations in genes encoding FGF ligands or receptors result in several congenital syndromes characterized by alterations in tooth number, morphology or enamel structure. The parallel roles of FGF signaling in mouse and human tooth development demonstrate the conserved importance of FGF signaling in mammalian odontogenesis.
Keywords: Fibroblast growth factors, Tooth development, Adult stem cells, Mouse, Human
Introduction
Every species has a unique array of teeth. Humans develop two sets of dentition, one deciduous (milk teeth) and one permanent, with four types of teeth in a continuous row. Unlike humans, mice only have one set of teeth with two different tooth types: three molars proximally and one incisor distally in each quadrant, separated by a toothless region called the diastema. In contrast to the molars, mouse incisors grow continuously throughout the life of the animal, and this continuous growth is fueled by somatic stem cells that reside in the proximal portion of the incisor and give rise to the differentiated cell types of the tooth [1]. Therefore, the mouse provides an excellent model to study both the genetics and molecular mechanisms of tooth patterning and the function of stem cells in tooth development.
The stages of early tooth development are similar in all mammals. In the mouse, tooth formation is initiated by a signal from the oral epithelium at around embryonic day (E) 9.5 [2, 3], and the thickening of the prospective dental epithelium is the first visible sign of tooth development. The epithelium signals to the underlying mesenchyme to initiate odontogenesis, and the mesenchyme induces the localized thickening of the oral epithelium to form the dental lamina at the position of the future teeth. The dental lamina then grows into the underlying mesenchyme at the sites of tooth formation, and the odontogenic mesenchymal cells condense around the invaginating epithelium to form the tooth bud. The development of the tooth crown and acquisition of tooth shape occurs during the subsequent cap and bell stages.
During tooth development, two sets of transient signaling centers called enamel knots (EKs) form in the epithelium. The primary EK (pEK) appears at the bud stage at the tip of the dental epithelium, and it expresses several signaling molecules that regulate the bud to cap transition by controlling cell proliferation and apoptosis and later determine cusp morphogenesis [4, 5]. Molar teeth develop a secondary EK (sEK), which determines the multicuspid pattern of molar crowns. During the transition from bud to cap stage, the mesenchymal cells nearest to the tip of the epithelial bud give rise to the dental papilla. At the later bell stage, the odontoblasts differentiate from the dental papilla and produce the dentin matrix, while the ameloblasts arise from the epithelium and secrete the enamel matrix [6].
Fibroblast growth factor (FGF) signaling induces the growth and differentiation of many different cell types in the embryo [7–11]. The role of FGFs as inductive embryonic signals was first reported during mesoderm formation in Xenopus embryos [12], and further studies in Drosophila [13] and mammals [14] showed that FGFs are widely required for development in animals. Here, we focus on the roles of FGF signaling in mammalian tooth development and review how FGFs regulate dental positioning, initiation, invagination and differentiation during tooth formation. We also discuss how FGFs control the function of stem cells in the continuously growing incisor in mouse and how dysregulation of FGF signaling in humans affects dental development.
Fibroblast growth factors and their receptors
The FGF family is one of the largest growth factor families, consisting of 22 members that share 13–71 % sequence homology in mammals [15]. Most FGFs mediate their biological responses as extracellular proteins by binding to and activating cell surface tyrosine kinase FGF receptors (FGFRs) [15, 16]. FGFs can be subdivided into several subfamilies based on sequence similarities and functional properties such as receptor specificity and binding affinity [15–17] (Table 1). Among these FGFs, FGF11–14 function as intracellular proteins, called iFGFs, which act in an FGFR-independent manner [18]. A principal role of FGF signaling during embryonic tooth development, as discussed below, is the regulation of morphogenesis between the epithelial layer and underlying mesenchyme. Similar processes have also been observed in the development of lungs [19], salivary glands [20], mammary glands [21], limb buds [22], brain [23], and other organs.
Table 1.
FGF families and their members
| FGF family | FGF members |
|---|---|
|
| |
| FGF1 | FGF1, FGF2 |
| FGF4 | FGF4, FGF5, FGF6 |
| FGF7 | FGF3, FGF7, FGF10, FGF22 |
| FGF8 | FGF8, FGF17, FGF18 |
| FGF9 | FGF9, FGF16, FGF20 |
| FGF11 | FGF11, FGF12, FGF13, FGF14 |
| FGF19/15 | FGF19/15, FGF21, FGF23 |
FGFRs are four related transmembrane proteins consisting of an extracellular ligand binding domain, a single transmembrane domain, and an intracellular tyrosine kinase domain. Fgfr1–3 mRNA undergoes alternative splicing events that can result in three alternative versions of the Ig-like domain III of the extracellular component of FGFR; this, in turn, can alter ligand binding properties of the extracellular domain. Individual splice forms are called IIIa, IIIb, IIIc in FGFRs 1–3 [24–26]; Fgfr4 mRNA is not alternatively spliced in this region [27]. The FgfrIIIa splice form encodes a protein with a terminal Ig-like domain resulting in a soluble FGF binding protein without known signaling function [28]. The splice variants IIIb and IIIc influence specificity of ligand binding and appear to be regulated in a tissue-dependent manner. IIIb splice forms are predominantly expressed in epithelial lineages and transduce signals initiated by FGF ligands expressed in the mesenchyme. The IIIc splice variant expression is restricted to mesenchymal lineages and is responsible for transduction of signaling from FGF ligands expressed in the epithelium [29–33].
The dimerization of receptors results in trans-phos-phorylation and activation of FGFRs [34], which initiates signaling through multiple downstream intracellular pathways. The cytosolic domain of the activated receptor binds a range of adaptor proteins, including growth factor receptor-bound protein 2 (GRB2) and SHP2 [35–38]. These recruit the guanosine nucleotide exchange factors (GEFs) son-of-sevenless (SOS) 1 and 2, which convert the small GTPase Ras from inactive Ras-GDP to activated Ras-GTP. Once activated, Ras signals through multiple effector pathways, including RAF/MEK/ERK, phosphatidylinositol-3-kinase (PI3K)/AKT, T-cell lymphoma invasion and metastasis 1 (TIAM1)/Rac, and Ral guanine nucleotide dissociation stimulator (RALGDS)/Ral [39–42]. In the simplest terms, Ras activates RAF (RAF-1, ARAF, BRAF, and CRAF), which phosphorylates and activates MEK1/2, which in turn activates ERK1/2. Activated ERK1/2 phosphorylates several targets, including transcription factors of the ETS family, such as JUN and ELK1, which promote cell cycle progression and proliferation [43]. Activated Ras can also bind PI3K [44, 45], which activates phosphoinositide-dependent kinase 1 (PDK1) and AKT to promote cell cycle progression and cell survival [46]. The TIAM-1/Rac pathway is involved in cytoskeletal remodeling [47, 48], while RALGDS/Ral is involved in endocytosis, exocytosis, and actin skeleton organization [49]. The contribution of each downstream effector pathway to various biological processes is an active area of research, and complex interactions between the pathways have yet to be fully characterized.
Since the intensity and duration of FGF signaling are critical for controlling various cellular functions, the pathway has multiple regulators. One such group of regulators is encoded by the Sprouty genes, whose gene products negatively regulate FGF signaling. Although the biochemical functions of Sprouty proteins are still unclear, it is known that Ras signaling induces expression of Sprouty genes, and it is thought that Sprouty proteins bind GRB2, preventing SOS localization and activation of Ras [50], and RAF, interfering with its interaction with downstream MEK [51, 52].
Expression of FGFs, FGFRs, and Sprouty genes during tooth development
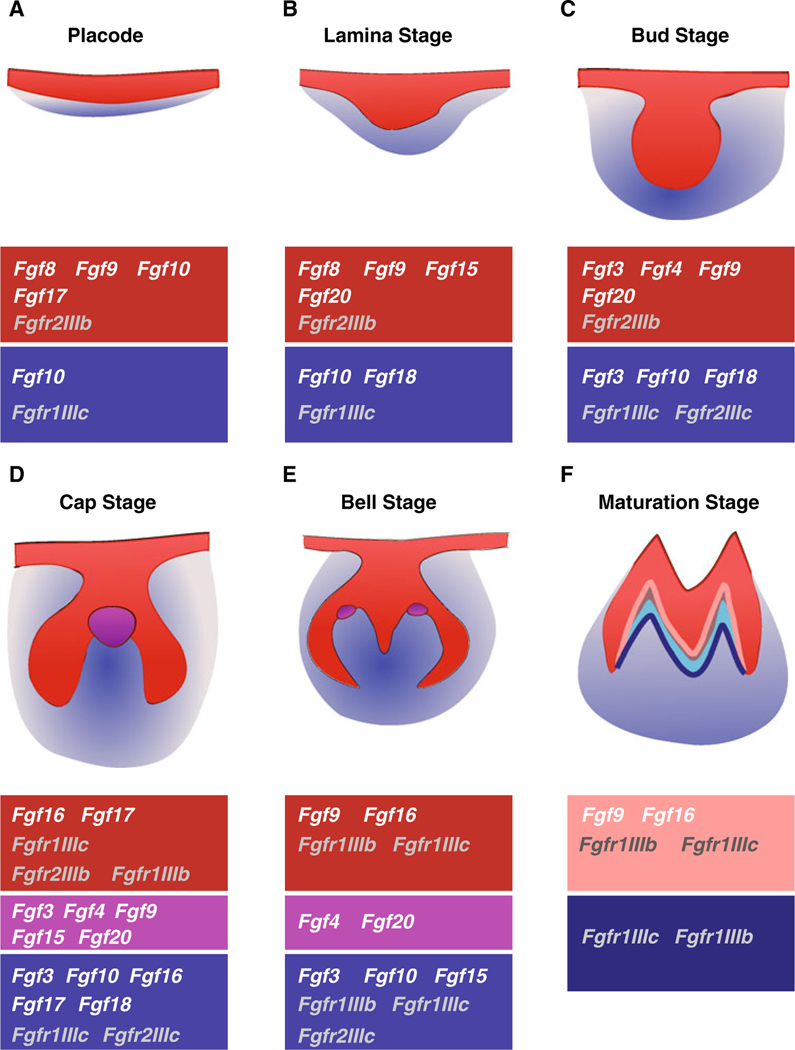
To date, the expression of twelve FGF ligands has been reported at different stages of tooth development (Fig. 1). Four FGFs, Fgf8, Fgf9, Fgf10 and Fgf17, and two FGF receptors, Fgfr2IIIb and Fgfr1IIIc, are expressed early in the prospective tooth region (Fig. 1a) [53–55]. The expression of Fgfr2IIIb and Fgfr1IIIc is maintained throughout tooth development. When the epithelium in the prospective tooth region becomes thickened to form the dental lamina (Fig. 1b), Fgf10 expression in the epithelium is diminished [55], while six other FGFs are expressed in the prospective tooth area: Fgf8, Fgf9, Fgf15 and Fgf20 in the epithelium, Fgf10 and Fgf18 in the mesenchyme. As the epithelium within the dental lamina continues to form the epithelial bud (Fig. 1c), the expression of Fgf9 and Fgf20 is maintained in the epithelium, and expression of Fgf4 is initiated.Fgf10 and Fgf18 continue to be expressed in the mesenchyme, and Fgf3 is expressed in both epithelium and mesenchyme. In addition to Fgfr1IIIc, Fgfr2IIIc expression is established in the mesenchyme at this stage. After formation of the pEK (Fig. 1d), Fgf3, Fgf4, Fgf9, Fgf15 and Fgf20 are restricted to this structure. Other FGFs are expressed as follows: Fgf3, Fgf10 and Fgf18 in the mesenchyme, and Fgf16 and Fgf17 in both epithelium and mesenchyme of the cervical loop. Fgfr2IIIb, Fgfr1IIIb and Fgfr1IIIc are expressed in the enamel epithelium, and Fgfr1IIIc and Fgfr2IIIc in the buccal mesenchyme.
Fig. 1.
Expression of FGFs and their receptors in molar development. a At the placode (initiation) stage, Fgf8, Fgf9, Fgf10 and Fgf17 are expressed in epithelium (red) together with Fgfr2IIIb. Fgf10 is also expressed in mesenchyme (blue) with Fgfr1IIIc. b At the dental lamina stage, expression of Fgf8, Fgf9, Fgf15, Fgf20 and Fgfr2IIIb is localized in epithelium and Fgf10, Fgf18 and Fgfr1IIIc in mesenchyme. c At bud stage, Fgf3, Fgf4, Fgf9, Fgf20 and Fgfr2IIIb are expressed in epithelium and Fgf3, Fgf10, Fgf18, Fgfr1IIIc and Fgfr2IIIc are expressed in mesenchyme. d At cap stage, Fgf16, Fgf17 and Fgfr1IIIb, Fgfr1IIIc and Fgfr2IIIb are expressed in dental epithelium (red). Expression of Fgf3, Fgf4, Fgf9, Fgf15 and Fgf20 is restricted to the enamel knot (violet). Mesenchyme forms the dental papilla (blue) at this stage which expresses Fgf3, Fgf10, Fgf16, Fgf17, Fgf18, Fgfr1IIIc and Fgfr2IIIc. e At bell stage, Fgf9, Fgf16, Fgfr1IIIb and Fgfr1IIIc are expressed in dental epithelium. Fgf4 and Fgf20 are expressed only in the secondary enamel knots (violet). Fgf3, Fgf10, Fgf15, Fgfr1IIIb, Fgfr1IIIc and Fgfr2IIIc are expressed in the mesenchyme of the dental papilla. f During maturation stage, the expression of Fgf9 and Fgf16 is localized to ameloblasts (pink) derived from the epithelium (red) together with Fgfr1IIIb and Fgfr1IIIc. Only odontoblasts (dark blue) from mesenchymal cells (light blue) retain expression of Fgfr1IIIb and Fgfr1IIIc. At this stage, ameloblasts secrete enamel matrix (grey) and odontoblast secrete dentin (cyan)
At the late bell stage (Fig. 1e), Fgf4 and Fgf20 are expressed at the tip of the forming cusps in the secondary enamel knot (sEK), and Fgf9 and Fgf16 in the differentiating ameloblasts (Fig. 1f). In the mesenchyme, Fgf3 is restricted to the dental papilla, Fgf10 to the differentiating odontoblasts, and Fgf15 to the mesenchyme underlying the sEK. Fgfr1IIIb and Fgfr1IIIc are expressed in the differentiating ameloblasts, and Fgfr1IIIb and Fgfr1IIIc are expressed in the odontoblasts [55]. In the incisor at the bell stage, Fgf1, Fgf9, Fgf16 and Fgf17 are expressed in the epithelium of the cervical loop, and in the mesenchyme, six FGFs are detected at this point: Fgf3, Fgf7, Fgf10, Fgf16, Fgf18 and Fgf21 [54, 56].
Sprouty (Spry) genes are expressed in different tissue compartments during tooth development [57]. At the cap stage, Spry1 is expressed at low levels in the diastema buds and at higher levels in the first molar (M1) tooth germs. Spry2 is abundantly expressed in the epithelium adjacent to dental mesenchyme, including the diastema and M1 tooth germ. Spry4 is almost exclusively expressed in the dental mesenchyme in both diastema and M1 tooth germs. Spry3, which is expressed in adult brain and testis, is not expressed in the tooth germ.
FGFs are critical in the determination of the presumptive tooth epithelium and formation of the dental lamina
Vertebrate organogenesis is often initiated at sites that are histologically indistinguishable from the surrounding tissues. In murine tooth formation, the first signals are derived from the presumptive tooth epithelium at E9.5 [58]. The oral ectoderm thickens in the prospective tooth-forming regions, and the expression of Fgf8, Fgf9, and Fgf17 in the epithelium at this stage suggests that these FGFs may signal to initiate tooth formation [54, 56]. Early work showed that FGF8 protein is sufficient to induce Pax9 expression at E9.5, which marks sites of prospective odontogenesis in mice and is required for tooth development beyond the bud stage [59]. Conditional deletion of Fgf8 with Nestin-Cre in the ectoderm of the first branchial arch (BA1) resulted in decreased expression of Pax9 in the presumptive molar region, and molar tooth formation was arrested at the initiation stage. The expression of Pax9 was not affected in the presumptive incisor region, and the incisor developed normally in this mutant. FGF8 is essential for mesenchymal cell survival, since without FGF8, the mesenchymal cells undergo apoptosis in the proximal region of BA1 [60]. These data suggested that FGF8 is a key inductive signal in the mesenchyme during initiation of molar odontogenesis. However, deletion of Fgf8 resulted in agenesis of the entire posterior portion of BA1, and therefore, conditional deletion of Fgf8 after proper formation of the mandible and before the initiation of molar odontogenesis will be needed to provide a clearer picture of FGF8 function during tooth development.
Another FGF family member, Fgf10, is expressed in both epithelium and mesenchyme at this early stage [55]. Fgf10 null mice develop teeth, although the formation of the stem cell compartment in the apical incisor bud is disrupted [61], and deletion of another gene expressed at early stages, Fgf9, does not affect tooth formation in mice [62, 63]. These data indicate that neither FGF9 nor FGF10 is involved in positioning of the tooth sites, or that there is redundancy between these FGF ligands during tooth initiation. Fgf17, a more recently studied gene, is expressed at the initiation stage and belongs to the FGF8 subfamily. Fgf17 is expressed in the prospective molar but not incisor epithelial region, suggesting that like FGF8, FGF17 plays a role in the positioning of the presumptive molar sites [54]. FGF8 is thought to play a critical role in tooth type determination [59, 64], and FGF17 may participate in this process as well. Bmp2 and Bmp4 antagonize the inductive effects of Fgf8 on Pax9 expression at E10, prior to thickening of the dental ectoderm, and it has been suggested that odontogenesis is initiated only in regions in which the inducer FGF is present, its antagonists (BMPs) are absent, and the mesenchyme is competent to respond to the inducer.
At the prospective tooth-forming region, the epithelium thickens to form a multilayered epithelium that later forms the dental lamina. At this stage, the expression of Fgf10 is downregulated [55]. Expression of Fgf8 and Fgf9 in the epithelium persists, expression of Fgf15 is initiated on the lingual side of the dental lamina, and Fgf20 is expressed at the tip of the dental lamina, suggesting that these genes play a role during the thickening of the epithelium [54].
Mouse molecular genetic approaches targeting individual FGF genes during tooth development have provided some understanding of the role of FGF signaling in dental lamina formation. Deletion of Fgf9, Fgf10 or Fgf20 does not appear to affect the thickening of oral epithelium or dental lamina formation [63, 65]. These results may be due to compensatory effects among the FGFs, and conditional deletion of FGFs in combination at this stage is needed to determine their effects on lamina formation. FGF18 is a newly identified FGF that belongs to the FGF8 subfamily. Unlike other FGF8 family members, which are expressed in the epithelium, Fgf18 mRNA is found at the buccal side of the mesenchyme at the lamina stage. Its function in tooth development is still unknown, and further study is required to determine whether FGF18 plays a role during odontogenesis. During these early stages of tooth development, Fgf2rIIIb is expressed in the odontogenic epithelium, and Fgf1rIIIc is expressed in the underlying mesenchyme [56].
FGF signaling regulates invagination of the dental epithelium
The dental lamina invaginates into the mesenchyme and induces mesenchymal condensation around the epithelium, which forms a tooth bud and later cap. The expression pattern of FGFs (Fig. 1) indicates that, at the invagination and bud stages, FGF signaling is activated in the epithelium by FGF3 and FGF10 binding to Fgfr2IIIb. In the mesenchyme, FGF4, FGF8 and FGF20 likely bind FGFR1IIIc, and FGF4, FGF8, FGF9, FGF16, FGF18 and FGF20 bind FGFR2IIIc [53, 54]. In Fgfr2 mutant mice, tooth development is arrested after epithelial thickening, and mesenchymal condensation is not observed in the Fgfr2−/− tooth germ. Although mesenchymal Fgf3 and Fgf10 expression is detectable, the epithelial expression of Fgf3 in Fgfr2 mutants is diminished [66].
FGF3 and FGF10 signal through FGFR2IIIb, suggesting that these FGFs are critical in the transition to the bud stage [67, 68]. However, mice with single mutations in Fgf3 or Fgf10 do not show any defect in early tooth development, and tooth germs proceed to cap stage normally. Further study in the Fgf3−/−;Fgf10−/− double null mutants revealed that, in the embryo, molar development is arrested prior to the bud stage, indicating that Fgf3 and Fgf10 can compensate for each other during dental epithelium invagination [69, 70].
Fgf9 is strongly expressed in the tip of the bud epithelium (Fig. 1). Fgf9 null mice do not show any defect in tooth bud invagination, although the differentiation of progenitor cells in the incisor is affected [62, 63]. Interestingly, exogenous FGF9 protein rescues the epithelium invagination defect in Runx2−/− tooth germs [62, 71], which suggests that FGF9 is a required downstream target of RUNX2 in tooth invagination. These findings highlight the compensatory effects that occur between FGF9 and other FGFs expressed in the epithelium. FGF9 also positively regulates the homeobox-containing transcription factor Msx1, which is an essential molecule for bud invagination [56, 72].
PITX2 is an important transcription factor that is regulated by FGF signaling during tooth bud invagination. Two molecules, BMP4 and FGF8, initially control the expression of Pitx2 in the oral epithelium; FGF8 positively regulates Pitx2 expression and BMP4 represses it [73]. In the absence of Pitx2, the expression of Fgf8 in the oral epithelium is diminished [74, 75]. FGF8 and BMP4 may act as positive–negative feedback regulators to control Pitx2 expression and regulate invagination of the tooth bud.
At E13.5, FGF4 expression initiates at the tip of the epithelium. Fgf4 expression is diminished in Lef1 null tooth germs at E13, resulting in a defect in mesenchyme condensation [76], and exogenous FGF4 protein rapidly induces the expression of Fgf3 in dental mesenchyme and fully rescues the developmental arrest of Lef1−/− tooth germs [77]. These data indicate that Fgf4 is a transcriptional target of WNT signaling. FGF18 is present in the mesenchyme, except underneath the bud epithelium, at this stage, and further investigation is required to understand the function of this protein in odontogenesis [54]. FGF20 expression is restricted in the epithelium to the tip of the tooth bud. Like Fgf9, deletion of Fgf20 in teeth does not disrupt early tooth development, and mutant mice form teeth normally [63]. These redundant roles will make it necessary to analyze double or triple deletion of FGFs to clearly understand gene function at this stage.
FGF signaling regulates tooth shape and cusp formation
Tooth shape characteristics are determined during embryonic development. The pEK, a signaling center that regulates tooth size and shape, is composed of non-proliferating cells [4] that express signaling molecules and their antagonists, including FGFs, Sprouty genes, Shh, several WNTs, BMPs and Follistatin [78]. The pEK cells themselves do not express FGF receptors and thus are unable to respond to the mitogenic stimuli of FGFs [56]. The lack of proliferation in the pEK combined with extensive proliferation around it may regulate the epithelial folding and the bud to cap transition [4, 79]. In multicuspid teeth, the pEK induces the formation of the sEKs. A network of activators and inhibitors has been suggested to determine the spatial arrangement of the sEKs [80, 81]. The secreted molecules from the sEKs regulate the proliferation and differentiation of the epithelium, which specifies the position and shape of the cusps and thus determines the shape of the tooth crown.
The size of the pEK in the molar is responsible for shaping the invaginated dental epithelium. If the pEK is too small, the folding of the dental epithelium and formation of cervical loop and sEKs are affected, which results in reduction of tooth size and cusp number; thisoccurs,for example, in mice that are null for ectodysplasin (Eda) or Traf6, members of the TNF-α family that regulate tooth development [82, 83]. Compromising the signaling from the pEK by altering its size orshape leads to changes in the arrangement of the sEKs in the molar, resulting in cusp defects. Experimentally, manipulating the level of gene expression in the EKs also results in variation of molar morphologies. For example, modulation of SHH, BMP and WNT signaling results in altered molar shapes and cusp patterns [84–88].
In terms of the FGF family, Fgf4 and Fgf9 are strongly expressed in the pEK and sEKs, and these proteins maintain Fgf3 expression in the dental mesenchyme. FGF4 in the EK may stimulate proliferation and thereby regulate the growth of tooth cusps [4]. In addition to stimulating cell division, FGF4 prevents apoptosis in the dental mesenchyme and epithelium [5,89]. However, inactivating either Fgf4 or Fgf9 individually has no effect on tooth number or shape [62, 63].
Another FGF family member, Fgf20, is expressed in the anterior bud of the dental lamina as well as in the EK, where Fgf3, Fgf4, Fgf9 and Fgf15 are also expressed [54, 56, 90]. FGF20 lies downstream of EDA during tooth development, as the expression of Fgf20 is significantly decreased in Eda−/− molars, whereas the K14-Eda mice show increased expression of Fgf20 [63]. Deletion of Fgf20 in mice results in smaller molars with a mildly altered anterior cusp morphology, but the overall cusp pattern of the Fgf20 mutants appears normal, and thus, FGF20 is involved in the regulation of tooth size and fine-tuning of anterior cusp patterning. Deletion of both Fgf9 and Fgf20 has a significant additive effect, shortening the EK length significantly compared to the length of either single mutant, demonstrating the redundant functions of the two FGFs [63].
Mesenchymal FGFs also affect tooth shape during development. In Fgf3−/−;Fgf10+/− mice, the molars are small, similar to Fgf20 null molars [63, 70], and FGF10 protein partially rescues the Eda−/− molar phenotype in vitro [82]. Thus, reduction of either epithelial or mesenchymal FGF signaling can cause similar effects on tooth formation.
Besides the regulation of individual tooth size and shape, FGF signaling also tightly regulates tooth number and arrangement within the dentition. Adult mice have a reduced dentition of three molars and one incisor in each quadrant, and rudimentary tooth buds have been described in mouse embryos in the incisor and cheek regions [91]. These have their own signaling centers, which resemble EKs of functional teeth [92]. The rudiments arrest at the bud stage or possibly fuse with the first molar primordium to give rise to the anterior extension of the crown of the lower M1, called the anteroconid [93]. Supernumerary teeth have been reported in several mutant mice, and these are mostly located at the putative site of the premolar. These teeth are thought to represent revitalization of evolutionarily suppressed tooth rudiments. The first transgenic mouse line discovered with ectopic teeth was the Eda overexpressor (K14-Eda) [94]. Later experiments showed that deletion of Fgf20 in this genetic background increased the frequency of extra tooth formation, but deletion of Fgf20 alone was not sufficient to induce extra molar formation [63]. Supernumerary teeth anterior to the first molar as well as extra incisors are also present in knockouts of Sprouty genes, presumably from rescued vestigial buds [57, 95]. These results demonstrate a role for FGFs as stimulators of tooth formation and for Sprouty genes as important endogenous inhibitors of FGF activity in tooth formation.
FGF signaling regulates ameloblast and odontoblast differentiation
At the later bell stage, dental papilla cells differentiate into odontoblasts that produce a dentin matrix. This matrix induces the epithelium to differentiate into ameloblasts that secrete enamel matrix, forming the hard tissues of the tooth crown, dentin and enamel [6]. It is thought that FGFs from the EK induce the differentiation of odontoblasts [96, 97]. At this stage, FGF3 and FGF10 are expressed in the mesenchyme. When dental papilla cells differentiate into odontoblasts, the expression of Fgf3 and Fgf10 is downregulated [55].
FGF signaling also plays an important role in the differentiation of ameloblasts. Fgf4 and Fgf9 are expressed in the inner enamel epithelium (IEE) [56], Fgf2 is expressed in supporting cells called the stellate reticulum and Fgfr1 and Fgfr2IIIb are expressed in the ameloblasts at the bell stage. Inactivation of Fgfr1 in the epithelium resulted in dysfunctional ameloblasts that produced disorganized enamel [98]. Overexpression of Fgf2 in cultured embryonic molars resulted in decreased expression of amelogenin, whereas inhibition of FGF2 increased amelogenin expression and enamel formation [99]. Tbx1, which encodes a transcription factor, is expressed in the dental epithelium; addition of FGF2 and FGF4 in tooth cultures induces Tbx1 expression, and Tbx1 expression is decreased in Fgfr2−/− mice [100]. Furthermore, Tbx1 is necessary for ameloblast differentiation, as incisors from Tbx1−/− mice cultured in vitro lack ameloblasts and do not form enamel [101]. Interestingly, Ras superfamily members that are downstream of FGFs play a role in amelogenesis, including Rac, a GTPase involved in cytoskeletal remodeling. Conditional inactivation of Rac1 in the epithelium in mouse results in ameloblasts that express decreased levels of amelogenin and have loose attachment to the secreted enamel matrix, resulting in hypo-mineralized enamel [102].
Increasing FGF signaling by decreasing Sprouty gene expression results not only in formation of supernumerary teeth but also in ectopic enamel formation [57]. In Spry2+/−;Spry4−/− mice, ameloblasts differentiate on the lingual aspect of the incisor and form ectopic enamel [103, 104]. Mice with increased signaling of HRas, which lies downstream of FGFs, had disorganized, hypo-mineralized enamel, and inhibiting the MAPK pathway rescued this phenotype [105].
FGFs regulate adult stem cells in the continuously growing mouse incisor
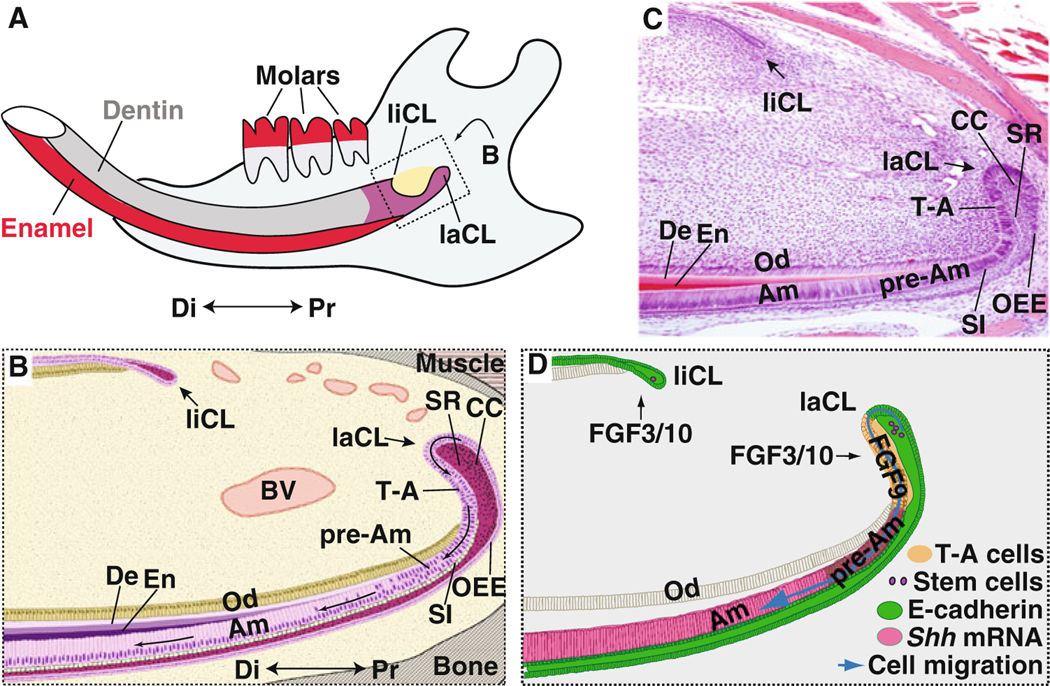
Rodent incisors grow continuously throughout the life of the animal, and the cervical loop, located in the proximal end of the incisor, is the niche that houses the dental stem cells [1]. Continuous incisor growth is counterbalanced by abrasion, which in rodent incisors is facilitated by absence of enamel on the lingual surface. This lack of lingual enamel is due to the absence of ameloblasts on that side [106]. Asymmetric abrasion not only maintains the incisor length but also generates a sharp tip (Fig. 2a), and a number of signaling factors and adhesion proteins have been implicated in the maintenance of the continuous growth of the incisor. Histologically, the cervical loop contains several cell types: inner enamel epithelium (IEE), outer enamel epithelium (OEE), stellate reticulum (SR), stratum intermedium (SI), transit-amplifying (T-A) and differentiated ameloblasts. There is an additional group of more tightly condensed cells located between the SR and OEE [107] (Fig. 2b, c), and the function of this cell type is not clear.
Fig. 2.
FGF signaling regulates the behavior of stem cells in the mouse incisor. a Schematic diagram of an adult mouse hemimandible with three molars and an incisor containing the lingual (liCL) and labial (laCL) cervical loops at the proximal end. Enamel, secreted by ameloblasts, is present only on the labial surface of the incisor. Dentin, produced by odontoblasts, is deposited on both the labial and lingual surfaces. Schematic diagram (b) and image of H&E stained (c) sagittal sections of the adult mouse incisor showing the various cell types present. The dental epithelial stem cells reside in the stellate reticulum (SR), condensed cell (CC) or outer enamel epithelium (OEE) regions of the labial cervical loop (laCL). The epithelial stem cells differentiate to form proliferating progenitors, called transit-amplifying (T-A) cells. T-A cells give rise to the pre-ameloblasts (pAm) that differentiate into ameloblasts (Am). The stratum intermedium (SI), which is a single layer of cells that subtends the ameloblasts, also arises from stem cells. BV, blood vessel; Di, distal; De, dentin; En, enamel; liCL, lingual cervical loop; Od, odontoblasts; Pr, proximal. d FGF signaling regulates dental stem cell maintenance, proliferation and differentiation. FGF3 and FGF10 signals regulate the expression of E-cadherin protein and cell proliferation in the CLs. FGF9 is present in the T-A region to maintain a low level of Shh expression in the progenitor cells and avoid their premature cell differentiation in the laCL
Fgf10 is expressed in the mesenchyme that surrounds the epithelium of the apical part of the cervical loop, as well as in the mesenchyme underlying the inner enamel epithelium. Fgf10 deletion leads to morphologically abnormal formation of the cervical loop and hypoplasia of the incisor [108]. Deletion of Fgf10 causes decreased proliferation of ameloblast progenitor cells in the cervical loop, suggesting that Fgf10 is essential for the maintenance and proliferation of the progenitor cells in the niche. Fgf3 is asymmetrically expressed in the mesenchyme, with higher levels in the labial cervical loop, where Fgf3 expression underlies the T-A cells. In the absence of Fgf3, the lower incisor lacks enamel, and the incisors are thin and frequently break. In Fgf3−/−;Fgf10+/− mice, the enamel layer is either very thin or missing in the incisors, and the labial cervical loop is not fully formed [70].
In order for stem cells to proliferate and differentiate in the cervical loop, FGFs must downregulate E-cadherin expression in the stem cells so that they can move out of the niche, proliferate and become T-A cells, which differentiate into more mature ameloblasts. E-cadherin is not downregulated in the T-A region in Fgf3−/−;Fgf10+/− mice, and cell proliferation is dramatically decreased in this region [107]. In contrast, Spry2+/−;Spry4−/− have ectopic Fgf3 expression in the lingual mesenchyme, which is associated with the formation of lingual E-cadherin negative T-A cells and ameloblasts [103, 107].
Unlike Fgf3 and Fgf10, which are expressed in the mesenchyme, Fgf9 expression persists in the epithelium of the cervical loop in mouse [56]. Deletion of Fgf9 results in a smaller labial cervical loop, and the region of Shh expression extends toward a more posterior position in the labial cervical loop [62]. Ectopic FGF9 significantly downregulates the expression of Shh mRNA in incisor explants [62]. Since Shh expression in the T-A region is required for ameloblast differentiation [109], Fgf9 may protect progenitors from exposure to the Shh signal and keep them in an undifferentiated state in the cervical loop.
Signaling by FGF10 and FGF9 is mediated by FGFR2b. Conditional deletion of or decreased signaling through Fgfr2b results in lack of ameloblasts and enamel, suppressed Shh expression and decreased cellular proliferation [110, 111], which further supports the hypothesis that FGF9 regulates the proliferation and differentiation of the progenitor cells in the cervical loop (Fig. 2d). Recently, expression of additional FGF molecules was discovered in the mouse incisor, including Fgf1, Fgf7, Fgf16, Fgf17, Fgf18 and Fgf21 [54]. However, the influence of these FGFs on stem cell behavior is not yet known.
Consequences of FGF signaling dysregulation in human tooth development
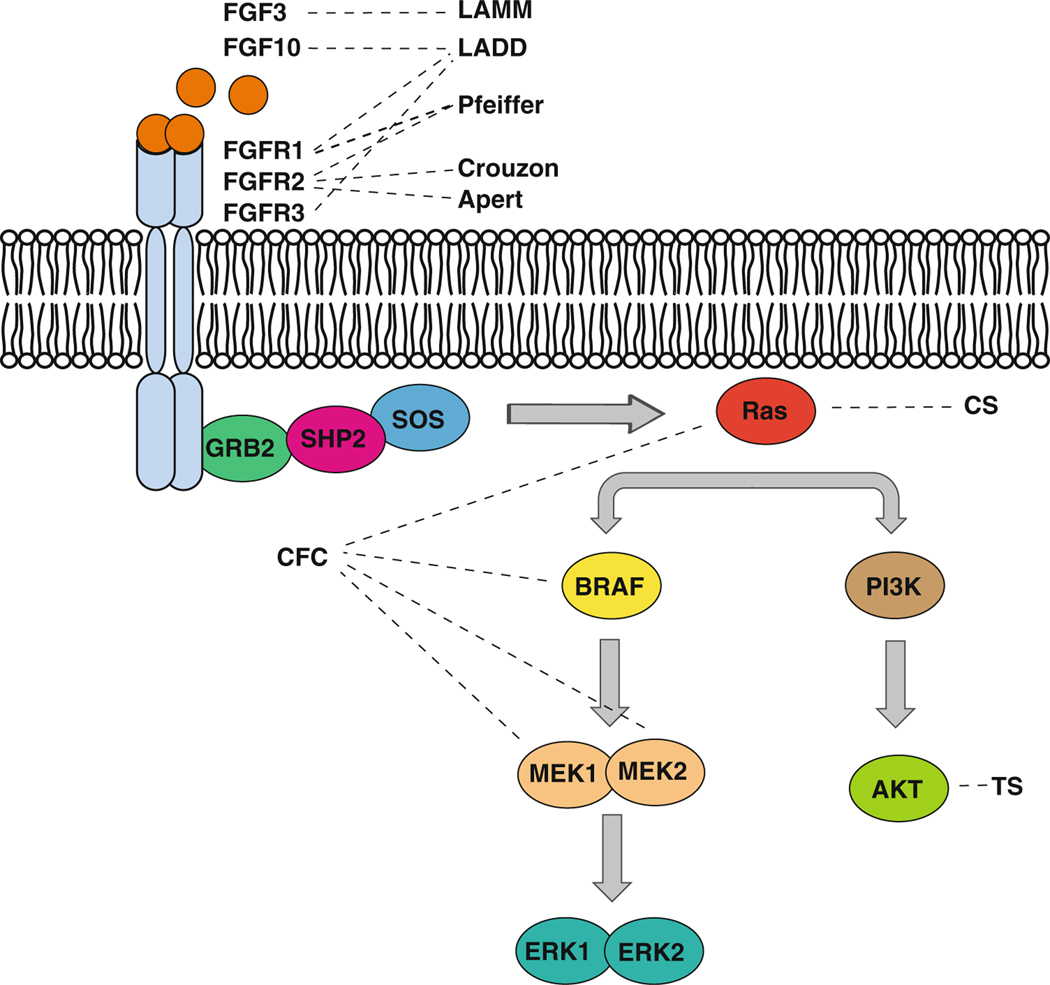
Dysregulation of FGF signaling can have profound consequences on human tooth development, including tooth agenesis and enamel defects (Fig. 3). Several syndromes are caused by mutations in FGFRs, including the Apert, Crouzon, and Pfeiffer cranio-synostosis syndromes. Apert syndrome (OMIM #101200) is characterized by craniosynostosis, midface hypoplasia, and syndactyly of the hands and feet, commonly with bony fusion, and it is caused by gain of function mutations in FGFR2 [112]. Similarly, Crouzon syndrome (OMIM #123500) is caused by mutations in FGFR2 and characterized by craniosyn-ostosis and secondary effects on craniofacial structures including hypertelorism, parrot-beaked nose, short upper lip, maxillary hypoplasia, and mandible prognathism [113]. Pfeiffer syndrome (OMIM #101600) is caused by mutations in both FGFR1 and FGFR2 and characterized by craniosynostosis, resulting in hypertelorism, short nose, and midface hypoplasia, cutaneous syndactyly of the hands and feet, and short and broad fingers and toes [114, 115]. Little is known about the dental phenotype of Pfeiffer syndrome, but Apert and Crouzon syndrome patients have hypodontia, most commonly of the third molar, maxillary lateral incisor, and mandibular second premolar [116, 117].
Fig. 3.
Syndromes caused by mutations in genes encoding FGFs, FGFRs and their downstream effectors have distinct dental characteristics. Diagram of the FGF signalling pathway with dashed lines connecting syndromes to the protein encoded by the causative gene. Although these syndromes are caused by mutations in the same pathway, they have distinct dental characteristics, described in the text. (LADD, Lacrimo-auriculodento-digital; LAMM, Autosomal recessive congenital deafness with labyrinthine aplasia, microtia, and microdontia; CFC, Cardio-facio-cutaneous; CS, Costello; TS, Tuberous sclerosis)
Lacrimo-auriculo-dento-digital (LADD; OMIM #149730) syndrome is an autosomal dominant congenital disorder characterized by aplasia, atresia or hypoplasia of lacrimal and salivary glands, cup-shaped ears, hearing loss, and digital anomalies. LADD is caused by heterozygous missense mutations in FGFR2, FGFR3, and FGF10 that are thought to result in loss of function [118–120]. Individuals with LADD present with varying dental anomalies, including missing teeth, peg shaped teeth, and enamel hypoplasia [121]. Autosomal recessive congenital deafness with labyrinthine aplasia, microtia, and microdontia (LAMM; OMIM #610706) is similarly characterized by malformed or missing inner ear structures, malformed external ear, and small, peg shaped teeth. This condition is caused by homozygous or compound heterozygous mutations in FGF3 [122–125].
Increasing signaling downstream of FGFs also causes disruption in enamel formation in humans. A group of syndromes termed the RASopathies are caused by activating mutations in the Ras pathway. Two of these syndromes are Cardio-facio-cutaneous syndrome (CFC; OMIM #115150) and Costello syndrome (CS; OMIM #218040), and both CS and CFC are characterized by craniofacial dysmorphia, ectodermal abnormalities, congenital heart defects, growth delay, and neurocognitive deficits[126,127].Inaddition,CS individuals present with musculoskeletal anomalies [128, 129]. Nearly all CS patients have a heterozygous, de novo germline mutation in HRAS that results in a constitutively active Ras protein [130], whereas CFC is caused by activating mutations in genes encoding proteins downstream of Ras: BRAF, MAP2K1, MAP2K2, and KRAS [131, 132]. Interestingly, even though both CS and CFC individuals have activated Ras signaling, their tooth number, shape, and morphology are normal. However, whereas the enamel of CFC individuals appeared normal [133], CS individuals had hypo-mineralized enamel. Likewise, mice with increased HRas signaling had disorganized, hypo-mineralized enamel [105].Tuberoussclerosis (TS; OMIM #191100)iscausedby mutations in TSC1 or TSC2, which encode proteins that function downstream of AKT, and gingival fibromas and enamel pitting of the maxillary incisors and canines have been reported in this syndrome [134]. Thus, dysregulation of FGF signaling can dramatically affect tooth formation, resulting in tooth agenesis or anomalies in tooth morphology and enamel structure. It is intriguing that mutations in the same pathway can have different manifestations in tooth development, highlighting the complexity of Ras signaling and its effectors downstream of FGFs.
Conclusion
Here, we have summarized some of the important roles of FGF signaling in tooth development. To date, expression of 12 of the 22 FGF family members has been reported in teeth, and in many cases these ligands play important roles from initiation of tooth development through formation of mineralized tissues. Specifically in rodents, FGFs are important for maintenance of stem cell pools fueling the continuously growing incisor throughout the life of the animal. The tooth provides an excellent model to gain further insight into the transduction and regulation of FGF signaling in developmental and stem cell biology. Insights from studies in the tooth will be applicable to other organ systems and may help to lay the foundation for development of pharmaceuticals that treat dysregulation of FGF signaling in patients.
Acknowledgments
The authors thank their colleagues in the Klein laboratory for helpful discussions. The authors were funded in part by NIH R01-DE021420.
Contributor Information
Chun-Ying Li, Zhongshan Hospital of Dalian University, Dalian 116001, China; Department of Orofacial Sciences and Program in Craniofacial and Mesenchymal Biology, University of California San Francisco, 513 Parnassus ave, HSE1508, San Francisco, CA 94143, USA.
Jan Prochazka, Department of Orofacial Sciences and Program in Craniofacial and Mesenchymal Biology, University of California San Francisco, 513 Parnassus ave, HSE1508, San Francisco, CA 94143, USA.
Alice F. Goodwin, Department of Orofacial Sciences and Program in Craniofacial and Mesenchymal Biology, University of California San Francisco, 513 Parnassus ave, HSE1508, San Francisco, CA 94143, USA
Ophir D. Klein, Department of Orofacial Sciences and Program in Craniofacial and Mesenchymal Biology, University of California San Francisco, 513 Parnassus ave, HSE1508, San Francisco, CA 94143, USA Department of Pediatrics and Institute for Human Genetics, University of California San Francisco, 513 Parnassus ave, HSE1508, San Francisco, CA 94143, USA.
References
- 1.Harada H, Kettunen P, Jung HS, et al. Localization of putative stem cells in dental epithelium and their association with Notch and FGF signaling. J Cell Biol. 1999;147:105–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Mina M, Kollar EJ. The induction of odontogenesis in nondental mesenchyme combined with early murine mandibular arch epithelium. Arch Oral Biol. 1987;32:123–7. [DOI] [PubMed] [Google Scholar]
- 3.Lumsden AG. Spatial organization of the epithelium and the roleof neural crest cells in the initiation of the mammalian tooth germ. Development. 1988;103(Suppl):155–69. [DOI] [PubMed] [Google Scholar]
- 4.Jernvall J, Kettunen P, Karavanova I, Martin LB, Thesleff I. Evidence for the role of the enamel knot as a control center in mammalian tooth cusp formation: non-dividing cells express growth stimulating Fgf-4 gene. Int J Dev Biol. 1994;38:463–9. [PubMed] [Google Scholar]
- 5.Vaahtokari A, Aberg T, Thesleff I. Apoptosis in the developing tooth: association with an embryonic signaling center and suppression by EGF and FGF-4. Development. 1996;122:121–9. [DOI] [PubMed] [Google Scholar]
- 6.Thesleff I, Hurmerinta K. Tissue interactions in tooth development. Differentiation. 1981;18:75–88. [DOI] [PubMed] [Google Scholar]
- 7.Crossley PH, Martinez S, Martin GR. Midbrain developmentinduced by FGF8 in the chick embryo. Nature. 1996;380:66–8. [DOI] [PubMed] [Google Scholar]
- 8.Crossley PH, Minowada G, MacArthur CA, Martin GR. Rolesfor FGF8 in the induction, initiation, and maintenance of chick limb development. Cell. 1996;84:127–36. [DOI] [PubMed] [Google Scholar]
- 9.Christen B, Slack JM. FGF-8 is associated with anteroposteriorpatterning and limb regeneration in Xenopus. Dev Biol. 1997;192:455–66. [DOI] [PubMed] [Google Scholar]
- 10.Vogel A, Rodriguez C, Izpisua-Belmonte JC. Involvement ofFGF-8 in initiation, outgrowth and patterning of the vertebrate limb. Development. 1996;122:1737–50. [DOI] [PubMed] [Google Scholar]
- 11.Phillips BT, Bolding K, Riley BB. Zebrafish fgf3 and fgf8 encode redundant functions required for otic placode induction. Dev Biol. 2001;235:351–65. [DOI] [PubMed] [Google Scholar]
- 12.Slack JM, Darlington BG, Heath JK, Godsave SF. Mesoderminduction in early Xenopus embryos by heparin-binding growth factors. Nature. 1987;326:197–200. [DOI] [PubMed] [Google Scholar]
- 13.Sutherland D, Samakovlis C, Krasnow MA. Branchless encodesa Drosophila FGF homolog that controls tracheal cell migration and the pattern of branching. Cell. 1996;87:1091–101. [DOI] [PubMed] [Google Scholar]
- 14.Feldman B, Poueymirou W, Papaioannou VE, DeChiara TM,Goldfarb M. Requirement of FGF-4 for postimplantation mouse development. Science. 1995;267:246–9. [DOI] [PubMed] [Google Scholar]
- 15.Ornitz DM, Itoh N. Fibroblast growth factors. Genome Biol.2001;2(REVIEWS300):5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Itoh N, Ornitz DM. Evolution of the Fgf and Fgfr gene families.Trends Genet. 2004;20:563–9. [DOI] [PubMed] [Google Scholar]
- 17.Popovici C, Roubin R, Coulier F, Birnbaum D. An evolutionaryhistory of the FGF superfamily. BioEssays. 2005;27:849–57. [DOI] [PubMed] [Google Scholar]
- 18.Goldfarb M Fibroblast growth factor homologous factors: evolution, structure, and function. Cytokine Growth Factor Rev. 2005;16:215–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Metzger RJ, Krasnow MA. Genetic control of branching morphogenesis. Science. 1999;284:1635–9. [DOI] [PubMed] [Google Scholar]
- 20.Hoffman MP, Kidder BL, Steinberg ZL, et al. Gene expressionprofiles of mouse submandibular gland development: FGFR1 regulates branching morphogenesis in vitro through BMP- and FGF-dependent mechanisms. Development. 2002;129:5767–78. [DOI] [PubMed] [Google Scholar]
- 21.Mailleux AA, Spencer-Dene B, Dillon C, et al. Role of FGF10/FGFR2b signaling during mammary gland development in the mouse embryo. Development. 2002;129:53–60. [DOI] [PubMed] [Google Scholar]
- 22.Mariani FV, Ahn CP, Martin GR. Genetic evidence that FGFshave an instructive role in limb proximal-distal patterning. Nature. 2008;453:401–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jukkola T, Lahti L, Naserke T, Wurst W, Partanen J. FGF regulated gene-expression and neuronal differentiation in the developing midbrain-hindbrain region. Dev Biol. 2006;297:141–57. [DOI] [PubMed] [Google Scholar]
- 24.Werner S, Duan DS, de Vries C, et al. Differential splicing in theextracellular region of fibroblast growth factor receptor 1 generates receptor variants with different ligand-binding specificities. Mol Cell Biol. 1992;12:82–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chellaiah AT, McEwen DG, Werner S, Xu J, Ornitz DM.Fibroblast growth factor receptor (FGFR) 3. Alternative splicing in immunoglobulin-like domain III creates a receptor highly specific for acidic FGF/FGF-1. J Biol Chem. 1994;269:11620–7. [PubMed] [Google Scholar]
- 26.Johnson DE, Lu J, Chen H, Werner S, Williams LT. The humanfibroblast growth factor receptor genes: a common structural arrangement underlies the mechanisms for generating receptor forms that differ in their third immunoglobulin domain. Mol Cell Biol. 1991;11:4627–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Vainikka S, Partanen J, Bellosta P, et al. Fibroblast growthfactor receptor-4 shows novel features in genomic structure, ligand binding and signal transduction. EMBO J. 1992;11: 4273–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Duan DS, Werner S, Williams LT. A naturally occurringsecreted form of fibroblast growth factor (FGF) receptor 1 binds basic FGF in preference over acidic FGF. J Biol Chem.1992;267:16076–80. [PubMed] [Google Scholar]
- 29.Avivi A, Yayon A, Givol D. A novel form of FGF receptor-3 using an alternative exon in the immunoglobulin domain III. FEBS Lett. 1993;330:249–52. [DOI] [PubMed] [Google Scholar]
- 30.Orr-Urtreger A, Bedford MT, Burakova T, et al. Developmentallocalization of the splicing alternatives of fibroblast growth factor receptor-2 (FGFR2). Dev Biol. 1993;158:475–86. [DOI] [PubMed] [Google Scholar]
- 31.Alarid ET, Rubin JS, Young P, et al. Keratinocyte growth factorfunctions in epithelial induction during seminal vesicle development. Proc Natl Acad Sci USA. 1994;91:1074–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yan G, Fukabori Y, McBride G, Nikolaropolous S, McKeehanWL. Exon switching and activation of stromal and embryonic fibroblast growth factor (FGF)-FGF receptor genes in prostate epithelial cells accompany stromal independence and malignancy. Mol Cell Biol. 1993;13:4513–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gilbert E, Del Gatto F, Champion-Arnaud P, Gesnel MC,Breathnach R. Control of BEK and K-SAM splice sites in alternative splicing of the fibroblast growth factor receptor 2 pre-mRNA. Mol Cell Biol. 1993;13:5461–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Eswarakumar VP, Lax I, Schlessinger J. Cellular signaling byfibroblast growth factor receptors. Cytokine Growth Factor Rev. 2005;16:139–49. [DOI] [PubMed] [Google Scholar]
- 35.Clark SG, Stern MJ, Horvitz HR. C. elegans cell-signalling gene sem-5 encodes a protein with SH2 and SH3 domains. Nature. 1992;356:340–4. [DOI] [PubMed] [Google Scholar]
- 36.Li W, Nishimura R, Kashishian A, et al. A new function for aphosphotyrosine phosphatase: linking GRB2-Sos to a receptor tyrosine kinase. Mol Cell Biol. 1994;14:509–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Saxton TM, Henkemeyer M, Gasca S, et al. Abnormal mesoderm patterning in mouse embryos mutant for the SH2 tyrosine phosphatase Shp-2. EMBO J. 1997;16:2352–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hadari YR, Kouhara H, Lax I, Schlessinger J. Binding of Shp2 tyrosine phosphatase to FRS2 is essential for fibroblast growth factor-induced PC12 cell differentiation. Mol Cell Biol. 1998;18:3966–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kouhara H, Hadari YR, Spivak-Kroizman T, et al. A lipidanchored Grb2-binding protein that links FGF-receptor activation to the Ras/MAPK signaling pathway. Cell.1997;89:693–702. [DOI] [PubMed] [Google Scholar]
- 40.Lowenstein EJ, Daly RJ, Batzer AG, et al. The SH2 and SH3 domain-containing protein GRB2 links receptor tyrosine kinases to ras signaling. Cell. 1992;70:431–42. [DOI] [PubMed] [Google Scholar]
- 41.Lebreton S, Boissel L, Moreau J. Control of embryonic Xenopusmorphogenesis by a Ral-GDS/Xral branch of the Ras signalling pathway. J Cell Sci. 2003;116:4651–62. [DOI] [PubMed] [Google Scholar]
- 42.Wilson R, Vogelsang E, Leptin M. FGF signalling and themechanism of mesoderm spreading in Drosophila embryos. Development. 2005;132:491–501. [DOI] [PubMed] [Google Scholar]
- 43.Chambard JC, Lefloch R, Pouyssegur J, Lenormand P. ERKimplication in cell cycle regulation. Biochim Biophys Acta. 2007;1773:1299–310. [DOI] [PubMed] [Google Scholar]
- 44.Pacold ME, Suire S, Perisic O, et al. Crystal structure andfunctional analysis of Ras binding to its effector phosphoinositide 3-kinase gamma. Cell. 2000;103:931–43. [DOI] [PubMed] [Google Scholar]
- 45.Rodriguez-Viciana P, Warne PH, Dhand R, et al. Phosphatidylinositol-3-OH kinase as a direct target of Ras. Nature. 1994;370:527–32. [DOI] [PubMed] [Google Scholar]
- 46.Castellano E,Downward J. RAS interactionwithPI3K: more thanjust another effector pathway. Genes Cancer. 2011;2:261–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Habets GG, Scholtes EH, Zuydgeest D, et al. Identification of aninvasion-inducing gene, Tiam-1, that encodes a protein with homology to GDP-GTP exchangers for Rho-like proteins. Cell. 1994;77:537–49. [DOI] [PubMed] [Google Scholar]
- 48.Michiels F, Habets GG, Stam JC, van der Kammen RA, CollardJG. A role for Rac in Tiam1-induced membrane ruffling and invasion. Nature. 1995;375:338–40. [DOI] [PubMed] [Google Scholar]
- 49.Ferro E, Trabalzini L. RalGDS family members couple Ras toRal signalling and that’s not all. Cell Signal. 2010;22:1804–10. [DOI] [PubMed] [Google Scholar]
- 50.Hanafusa H, Torii S, Yasunaga T, Nishida E. Sprouty1 andSprouty2 provide a control mechanism for the Ras/MAPK signalling pathway. Nat Cell Biol. 2002;4:850–8. [DOI] [PubMed] [Google Scholar]
- 51.Kim HJ, Bar-Sagi D. Modulation of signalling by Sprouty: adeveloping story. Nat Rev Mol Cell Biol. 2004;5:441–50. [DOI] [PubMed] [Google Scholar]
- 52.Sasaki A, Taketomi T, Kato R, et al. Mammalian Sprouty4 suppresses Ras-independent ERK activation by binding to Raf1. Nat Cell Biol. 2003;5:427–32. [DOI] [PubMed] [Google Scholar]
- 53.Kettunen P, Karavanova I, Thesleff I. Responsiveness ofdeveloping dental tissues to fibroblast growth factors: expression of splicing alternatives of FGFR1, −2, −3, and of FGFR4; and stimulation of cell proliferation by FGF-2, −4, −8, and −9. Dev Genet. 1998;22:374–85. [DOI] [PubMed] [Google Scholar]
- 54.Porntaveetus T, Otsuka-Tanaka Y, Basson MA, et al. Expressionof fibroblast growth factors (Fgfs) in murine tooth development. J Anat. 2011;218:534–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Kettunen P, Laurikkala J, Itaranta P, et al. Associations of FGF3 and FGF-10 with signaling networks regulating tooth morphogenesis. Dev Dyn. 2000;219:322–32. [DOI] [PubMed] [Google Scholar]
- 56.Kettunen P, Thesleff I. Expression and function of FGFs-4, −8,and −9 suggest functional redundancy and repetitive use as epithelial signals during tooth morphogenesis. Dev Dyn. 1998;211:256–68. [DOI] [PubMed] [Google Scholar]
- 57.Klein OD, Minowada G, Peterkova R, et al. Sprouty genescontrol diastema tooth development via bidirectional antagonism of epithelial-mesenchymal FGF signaling. Dev Cell. 2006;11:181–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Ferguson CA, Tucker AS, Sharpe PT. Temporospatial cellinteractions regulating mandibular and maxillary arch patterning. Development. 2000;127:403–12. [DOI] [PubMed] [Google Scholar]
- 59.Neubuser A, Peters H, Balling R, Martin GR. Antagonisticinteractions between FGF and BMP signaling pathways: a mechanism for positioning the sites of tooth formation. Cell. 1997;90:247–55. [DOI] [PubMed] [Google Scholar]
- 60.Trumpp A, Depew MJ, Rubenstein JL, Bishop JM, Martin GR.Cre-mediated gene inactivation demonstrates that FGF8 is required for cell survival and patterning of the first branchial arch. Genes Dev. 1999;13:3136–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Yokohama-Tamaki T, Ohshima H, Fujiwara N, et al. Cessationof Fgf10 signaling, resulting in a defective dental epithelial stem cell compartment, leads to the transition from crown to root formation. Development. 2006;133:1359–66. [DOI] [PubMed] [Google Scholar]
- 62.Kurosaka H, Islam MN, Kuremoto K, et al. Core binding factorbeta functions in the maintenance of stem cells and orchestrates continuous proliferation and differentiation in mouse incisors. Stem Cells. 2011;29:1792–803. [DOI] [PubMed] [Google Scholar]
- 63.Haara O, Harjunmaa E, Lindfors PH, et al. Ectodysplasin regulates activator-inhibitor balance in murine tooth development through Fgf20 signaling. Development. 2012;139:3189–99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Tucker AS, Yamada G, Grigoriou M, Pachnis V, Sharpe PT.Fgf-8 determines rostral-caudal polarity in the first branchial arch. Development. 1999;126:51–61. [DOI] [PubMed] [Google Scholar]
- 65.Ohuchi H, Hori Y, Yamasaki M, et al. FGF10 acts as a majorligand for FGF receptor 2 IIIb in mouse multi-organ development. Biochem Biophys Res Commun. 2000;277:643–9. [DOI] [PubMed] [Google Scholar]
- 66.Kettunen P, Spencer-Dene B, Furmanek T, et al. Fgfr2b mediated epithelial-mesenchymal interactions coordinate tooth morphogenesis and dental trigeminal axon patterning. Mech Dev. 2007;124:868–83. [DOI] [PubMed] [Google Scholar]
- 67.De Moerlooze L, Spencer-Dene B, Revest JM, et al. Animportant role for the IIIb isoform of fibroblast growth factor receptor 2 (FGFR2) in mesenchymal-epithelial signalling during mouse organogenesis. Development. 2000;127:483–92. [DOI] [PubMed] [Google Scholar]
- 68.Hosokawa R, Deng X, Takamori K, et al. Epithelial-specificrequirement of FGFR2 signaling during tooth and palate development. J Exp Zool B Mol Dev Evol. 2009;312B:343–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Harada H, Toyono T, Toyoshima K, et al. FGF10 maintainsstem cell compartment in developing mouse incisors. Development. 2002;129:1533–41. [DOI] [PubMed] [Google Scholar]
- 70.Wang XP, Suomalainen M, Felszeghy S, et al. An integratedgene regulatory network controls stem cell proliferation in teeth. PLoS Biol. 2007;5:e159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.D’Souza RN, Aberg T, Gaikwad J, et al. Cbfa1 is required forepithelial-mesenchymal interactions regulating tooth development in mice. Development. 1999;126:2911–20. [DOI] [PubMed] [Google Scholar]
- 72.Satokata I, Maas R. Msx1 deficient mice exhibit cleft palate andabnormalities of craniofacial and tooth development. Nat Genet. 1994;6:348–56. [DOI] [PubMed] [Google Scholar]
- 73.St Amand TR, Zhang Y, Semina EV, et al. Antagonistic signalsbetween BMP4 andFGF8 definethe expression of Pitx1 andPitx2 in mouse tooth-forming anlage. Dev Biol. 2000;217:323–32. [DOI] [PubMed] [Google Scholar]
- 74.Lu MF, Pressman C, Dyer R, Johnson RL, Martin JF. Functionof Rieger syndrome gene in left-right asymmetry and craniofacial development. Nature. 1999;401:276–8. [DOI] [PubMed] [Google Scholar]
- 75.Lin CR, Kioussi C, O’Connell S, et al. Pitx2 regulates lungasymmetry, cardiac positioning and pituitary and tooth morphogenesis. Nature. 1999;401:279–82. [DOI] [PubMed] [Google Scholar]
- 76.van Genderen C, Okamura RM, Farinas I, et al. Development ofseveral organs that require inductive epithelial-mesenchymal interactions is impaired in LEF-1-deficient mice. Genes Dev. 1994;8:2691–703. [DOI] [PubMed] [Google Scholar]
- 77.Kratochwil K, Galceran J, Tontsch S, Roth W, Grosschedl R.FGF4, a direct target of LEF1 and Wnt signaling, can rescue the arrest of tooth organogenesis in Lef1(−/−) mice. Genes Dev. 2002;16:3173–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Tummers M, Thesleff I. The importance of signal pathwaymodulation in all aspects of tooth development. J Exp Zool B Mol Dev Evol. 2009;312B:309–19. [DOI] [PubMed] [Google Scholar]
- 79.Jernvall J, Thesleff I. Reiterative signaling and patterning duringmammalian tooth morphogenesis. Mech Dev. 2000;92:19–29. [DOI] [PubMed] [Google Scholar]
- 80.Salazar-Ciudad I, Jernvall J. A gene network model accountingfor development and evolution of mammalian teeth. Proc Natl Acad Sci USA. 2002;99:8116–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Salazar-Ciudad I, Jernvall J. A computational model of teeth andthe developmental origins of morphological variation. Nature. 2010;464:583–6. [DOI] [PubMed] [Google Scholar]
- 82.Pispa J, Jung HS, Jernvall J, et al. Cusp patterning defect inTabby mouse teeth and its partial rescue by FGF. Dev Biol. 1999;216:521–34. [DOI] [PubMed] [Google Scholar]
- 83.Ohazama A, Courtney JM, Tucker AS, et al. Traf6 is essentialfor murine tooth cusp morphogenesis. Dev Dyn. 2004;229: 131–5. [DOI] [PubMed] [Google Scholar]
- 84.Dassule HR, Lewis P, Bei M, Maas R, McMahon AP. Sonichedgehog regulates growth and morphogenesis of the tooth. Development. 2000;127:4775–85. [DOI] [PubMed] [Google Scholar]
- 85.Harjunmaa E, Kallonen A, Voutilainen M, et al. On the difficulty of increasing dental complexity. Nature. 2012;483:324–7. [DOI] [PubMed] [Google Scholar]
- 86.Jarvinen E, Salazar-Ciudad I, Birchmeier W, et al. Continuoustooth generation in mouse is induced by activated epithelial Wnt/beta-catenin signaling. Proc Natl Acad Sci USA. 2006;103:18627–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Liu F, Chu EY, Watt B, et al. Wnt/beta-catenin signaling directsmultiple stages of tooth morphogenesis. Dev Biol. 2008;313: 210–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Wang XP, Suomalainen M, Jorgez CJ, et al. Modulation ofactivin/bone morphogenetic protein signaling by follistatin is required for the morphogenesis of mouse molar teeth. Dev Dyn. 2004;231:98–108. [DOI] [PubMed] [Google Scholar]
- 89.Jernvall J, Aberg T, Kettunen P, Keranen S, Thesleff I. The lifehistory of an embryonic signaling center: BMP-4 induces p21 and is associated with apoptosis in the mouse tooth enamel knot. Development. 1998;125:161–9. [DOI] [PubMed] [Google Scholar]
- 90.Aberg T, Wang XP, Kim JH, et al. Runx2 mediates FGF signaling from epithelium to mesenchyme during tooth morphogenesis. Dev Biol. 2004;270:76–93. [DOI] [PubMed] [Google Scholar]
- 91.Peterkova R, Peterka M, Viriot L, Lesot H. Development of thevestigial tooth primordia as part of mouse odontogenesis. Connect Tissue Res. 2002;43:120–8. [DOI] [PubMed] [Google Scholar]
- 92.Prochazka J, Pantalacci S, Churava S, et al. Patterning by heritage in mouse molar row development. Proc Natl Acad Sci USA. 2010;107:15497–502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Tureckova J, Lesot H, Vonesch JL, et al. Apoptosis is involvedin the disappearance of the diastemal dental primordia in mouse embryo. Int J Dev Biol. 1996;40:483–9. [PubMed] [Google Scholar]
- 94.Mustonen T, Pispa J, Mikkola ML, et al. Stimulation of ectodermal organ development by Ectodysplasin-A1. Dev Biol. 2003;259:123–36. [DOI] [PubMed] [Google Scholar]
- 95.Charles C, Hovorakova M, Ahn Y, et al. Regulation of toothnumber by fine-tuning levels of receptor-tyrosine kinase signaling. Development. 2011;138:4063–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Thesleff I, Keranen S, Jernvall J. Enamel knots as signalling centers linking tooth morphogenesis and odontoblast differentiation. Adv Dent Res. 2001;15:14–8. [DOI] [PubMed] [Google Scholar]
- 97.Tompkins K Molecular mechanisms of cytodifferentiation in mammalian tooth development. Connect Tissue Res. 2006;47:111–8. [DOI] [PubMed] [Google Scholar]
- 98.Takamori K, Hosokawa R, Xu X, et al. Epithelial fibroblastgrowth factor receptor 1 regulates enamel formation. J Dent Res. 2008;87:238–43. [DOI] [PubMed] [Google Scholar]
- 99.Tsuboi T, Mizutani S, Nakano M, Hirukawa K, Togari A. Fgf-2 regulates enamel and dentine formation in mouse tooth germ. Calcif Tissue Int. 2003;73:496–501. [DOI] [PubMed] [Google Scholar]
- 100.Mitsiadis TA, Tucker AS, De Bari C, Cobourne MT, Rice DP. Aregulatory relationship between Tbx1 and FGF signaling during tooth morphogenesis and ameloblast lineage determination. Dev Biol. 2008;320:39–48. [DOI] [PubMed] [Google Scholar]
- 101.Caton J, Luder HU, Zoupa M, et al. Enamel-free teeth: Tbx1 deletion affects amelogenesis in rodent incisors. Dev Biol. 2009;328:493–505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Huang Z, Kim J, Lacruz RS, et al. Epithelial-specific knockoutof the Rac1 gene leads to enamel defects. Eur J Oral Sci. 2011;119(Suppl 1):168–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Klein OD, Lyons DB, Balooch G, et al. An FGF signaling loopsustains the generation of differentiated progeny from stem cells in mouse incisors. Development. 2008;135:377–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Boran T, Peterkova R, Lesot H, et al. Temporal analysis ofectopic enamel production in incisors from sprouty mutant mice. J Exp Zool B Mol Dev Evol. 2009;312B:473–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Goodwin AF, Tidyman WE, Jheon AH, et al. Abnormal Rassignaling in costello syndrome (CS) negatively regulates enamel formation. Hum Mol Genet. 2013. [Epub ahead of print]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Smith CE, Warshawsky H. Histological and three dimensional organization of the odontogenic organ in the lower incisor of 100 gram rats. Am J Anat. 1975;142:403–29. [DOI] [PubMed] [Google Scholar]
- 107.Li CY, Cha W, Luder HU, et al. E-cadherin regulates thebehavior and fate of epithelial stem cells and their progeny in the mouse incisor. Dev Biol. 2012;366:357–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Harada H, Toyono T, Toyoshima K, Ohuchi H. FGF10 maintains stem cell population during mouse incisor development. Connect Tissue Res. 2002;43:201–4. [DOI] [PubMed] [Google Scholar]
- 109.Seidel K, Ahn CP, Lyons D, et al. Hedgehog signaling regulatesthe generation of ameloblast progenitors in the continuously growing mouse incisor. Development. 2010;137:3753–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Parsa S, Kuremoto K, Seidel K, et al. Signaling by FGFR2bcontrols the regenerative capacity of adult mouse incisors. Development. 2010;137:3743–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Lin Y, Cheng YS, Qin C, et al. FGFR2 in the dental epitheliumis essential for development and maintenance of the maxillary cervical loop, a stem cell niche in mouse incisors. Dev Dyn. 2009;238:324–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Wilkie AO, Slaney SF, Oldridge M, et al. Apert syndromeresults from localized mutations of FGFR2 and is allelic with Crouzon syndrome. Nat Genet. 1995;9:165–72. [DOI] [PubMed] [Google Scholar]
- 113.Reardon W, Winter RM, Rutland P, et al. Mutations in thefibroblast growth factor receptor 2 gene cause Crouzon syndrome. Nat Genet. 1994;8:98–103. [DOI] [PubMed] [Google Scholar]
- 114.Muenke M, Schell U, Hehr A, et al. A common mutation in thefibroblast growth factor receptor 1 gene in Pfeiffer syndrome. Nat Genet. 1994;8:269–74. [DOI] [PubMed] [Google Scholar]
- 115.Lajeunie E, Ma HW, Bonaventure J, et al. FGFR2 mutations inPfeiffer syndrome. Nat Genet. 1995;9:108. [DOI] [PubMed] [Google Scholar]
- 116.Stavropoulos D, Bartzela T, Bronkhorst E, Mohlin B, HagbergC. Dental agenesis patterns of permanent teeth in Apert syndrome. Eur J Oral Sci. 2011;119:198–203. [DOI] [PubMed] [Google Scholar]
- 117.Reitsma JH, Ongkosuwito EM, van Wijk AJ, Prahl-Andersen B.Patterns of tooth agenesis in patients with the syndrome of crouzon or apert. Cleft Palate Craniofac J. 2012. [Epub ahead of print]. [DOI] [PubMed] [Google Scholar]
- 118.Rohmann E, Brunner HG, Kayserili H, et al. Mutations in different components of FGF signaling in LADD syndrome. Nat Genet. 2006;38:414–7. [DOI] [PubMed] [Google Scholar]
- 119.Milunsky JM, Zhao G, Maher TA, Colby R, Everman DB.LADD syndrome is caused by FGF10 mutations. Clin Genet. 2006;69:349–54. [DOI] [PubMed] [Google Scholar]
- 120.Shams I, Rohmann E, Eswarakumar VP, et al. Lacrimo-auriculo-dento-digital syndrome is caused by reduced activity of the fibroblast growth factor 10 (FGF10)-FGF receptor 2 signaling pathway. Mol Cell Biol. 2007;27:6903–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Guven Y, Rosti RO, Tuna EB, Kayserili H, Aktoren O. Orodental findings of a family with lacrimo-auriculo-dento digital (LADD) syndrome. Oral Surg Oral Med Oral Pathol Oral Radiol Endod. 2008;106:e33–44. [DOI] [PubMed] [Google Scholar]
- 122.Tekin M, Hismi BO, Fitoz S, et al. Homozygous mutations infibroblast growth factor 3 are associated with a new form of syndromic deafness characterized by inner ear agenesis, microtia, and microdontia. Am J Hum Genet. 2007;80:338–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Tekin M, Ozturkmen Akay H, Fitoz S, et al. Homozygous FGF3 mutations result in congenital deafness with inner ear agenesis, microtia, and microdontia. Clin Genet. 2008;73:554–65. [DOI] [PubMed] [Google Scholar]
- 124.Alsmadi O, Meyer BF, Alkuraya F, et al. Syndromic congenital sensorineural deafness, microtia and microdontia resulting from a novel homoallelic mutation in fibroblast growth factor 3 (FGF3). Eur J Hum Genet. 2009;17:14–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Sensi A, Ceruti S, Trevisi P, et al. LAMM syndrome withmiddle ear dysplasia associated with compound heterozygosity for FGF3 mutations. Am J Med Genet A. 2011;155A:1096–101. [DOI] [PubMed] [Google Scholar]
- 126.Roberts A, Allanson J, Jadico SK, et al. The cardiofaciocutaneous syndrome. J Med Genet. 2006;43:833–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Rauen KA. HRAS and the Costello syndrome. Clin Genet. 2007;71:101–8. [DOI] [PubMed] [Google Scholar]
- 128.Tidyman WE, Lee HS, Rauen KA. Skeletal muscle pathology inCostello and cardio-facio-cutaneous syndromes: developmental consequences of germline Ras/MAPK activation on myogenesis. Am J Med Genet C Semin Med Genet. 2011;157C:104–14. [DOI] [PubMed] [Google Scholar]
- 129.Stevenson DA, Allen S, Tidyman WE, et al. Peripheral muscle weakness in RASopathies. Muscle Nerve. 2012;46:394–9. [DOI] [PubMed] [Google Scholar]
- 130.Aoki Y, Niihori T, Kawame H, et al. Germline mutations inHRAS proto-oncogene cause Costello syndrome. Nat Genet. 2005;37:1038–40. [DOI] [PubMed] [Google Scholar]
- 131.Niihori T, Aoki Y, Narumi Y, et al. Germline KRAS and BRAFmutations in cardio-facio-cutaneous syndrome. Nat Genet. 2006;38:294–6. [DOI] [PubMed] [Google Scholar]
- 132.Rodriguez-Viciana P, Tetsu O, Tidyman WE, et al. Germline mutations in genes within the MAPK pathway cause cardio-facio-cutaneous syndrome. Science. 2006;311:1287–90. [DOI] [PubMed] [Google Scholar]
- 133.Goodwin AF, Oberoi S, Landan M, et al. Craniofacial and dental development in cardio-facio-cutaneous syndrome: the importance of Ras signaling homeostasis. Clin Genet. 2013;83:539–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Schwartz RA, Fernandez G, Kotulska K, Jozwiak S. Tuberous sclerosis complex: advances in diagnosis, genetics, and management. J Am Acad Dermatol. 2007;57:189–202. [DOI] [PubMed] [Google Scholar]