Abstract
Pure bacterial cultures were isolated from a highly enriched denitrifying consortium previously shown to anaerobically biodegrade naphthalene. The isolates were screened for the ability to grow anaerobically in liquid culture with naphthalene as the sole source of carbon and energy in the presence of nitrate. Three naphthalene-degrading pure cultures were obtained, designated NAP-3-1, NAP-3-2, and NAP-4. Isolate NAP-3-1 tested positive for denitrification using a standard denitrification assay. Neither isolate NAP-3-2 nor isolate NAP-4 produced gas in the assay, but both consumed nitrate and NAP-4 produced significant amounts of nitrite. Isolates NAP-4 and NAP-3-1 transformed 70 to 90% of added naphthalene, and the transformation was nitrate dependent. No significant removal of naphthalene occurred under nitrate-limited conditions or in cell-free controls. Both cultures exhibited partial mineralization of naphthalene, representing 7 to 20% of the initial added 14C-labeled naphthalene. After 57 days of incubation, the largest fraction of the radiolabel in both cultures was recovered in the cell mass (30 to 50%), with minor amounts recovered as unknown soluble metabolites. Nitrate consumption, along with the results from the 14C radiolabel study, are consistent with the oxidation of naphthalene coupled to denitrification for NAP-3-1 and nitrate reduction to nitrite for NAP-4. Phylogenetic analyses based on 16S ribosomal DNA sequences of NAP-3-1 showed that it was closely related to Pseudomonas stutzeri and that NAP-4 was closely related to Vibrio pelagius. This is the first report we know of that demonstrates nitrate-dependent anaerobic degradation and mineralization of naphthalene by pure cultures.
Hydrocarbon contamination in sediments has been the subject of continuous environmental and human health concern over the last few decades. Polycyclic aromatic hydrocarbons (PAHs), components of petroleum waste, airborne combustion particulates, and creosote are particularly persistent in sedimentary subsurface environments due to their relatively low aqueous solubilities, low volatility, and high affinity for sediment particles. PAH-contaminated sediments are a concern to human health because of the potential for exposure through the consumption of contaminated seafood stocks (19, 23, 33, 37). Fish and shellfish frequently bioaccumulate PAHs to concentrations that are several orders of magnitude greater than their aqueous solubility (2). The human health effects of PAH exposure are well documented. Acute exposure effects range from skin and lung irritation to cyanosis (2). Exposure to some PAHs has been implicated as carcinogenic and tumorigenic to both humans and wildlife (2). Because of these human health concerns, accurate estimates of the persistence of PAHs in the environment are important in assessing the risk presented by PAH contamination.
A key parameter affecting the fate of PAHs in the environment is the extent of loss due to mechanisms of biodegradation. The ability of aerobic microorganisms to degrade naphthalene, phenanthrene, and other low-molecular-weight PAHs is well known (5). However, subsurface organic-impacted sediments are commonly anaerobic. Although PAHs typically had been thought to be recalcitrant to biodegradation without oxygen (e.g., references 3, 11, 14), recent studies have demonstrated PAH degradation under sulfate-reducing (7, 8, 30, 38) and nitrate-reducing (22, 24, 25, 30; K. J. Rockne and S. E. Strand, submitted for publication) conditions. Because PAHs can be biodegraded without oxygen, their persistence in anaerobic environments might be less than was previously thought. While it has been shown that PAHs are degraded anaerobically, little is known about the microorganisms responsible for this activity. With the exception of a recent report (22), pure cultures have not been reported thus far with the ability to degrade PAHs under anaerobic conditions. Furthermore, nitrate-dependent PAH transformation by microbial isolates has never been demonstrated. Virtually nothing is known about the types of bacteria responsible for anaerobic PAH biodegradation. Further knowledge of the biochemical reactions, enzymes, and genetics of anaerobic PAH biodegradation would be facilitated by the isolation and study of pure cultures. In this study we describe several anaerobic naphthalene-degrading pure cultures isolated from a highly enriched PAH-degrading, nitrate-reducing culture originating from a PAH-contaminated site.
MATERIALS AND METHODS
Isolation of pure cultures.
The source of inoculum for these isolations was a highly enriched consortium of microorganisms obtained from a fluidized bed reactor (FBR) originally seeded with PAH-contaminated marine sediment from Eagle Harbor in Puget Sound, Wash. (30; Rockne and Strand, submitted). The FBR was operated by continuous feedings of low concentrations of naphthalene, phenanthrene, and biphenyl in the presence of nitrate, as previously described (30). Samples from the FBR were transferred to serum bottles containing anaerobically prepared artificial seawater (ASW) (29), plus nitrate (2 mM), and biphenyl, naphthalene, or phenanthrene and sealed with Teflon-lined stoppers. The approximate concentrations of the substrates ranged from 3 to 4 μM phenanthrene, 20 to 23 μM biphenyl, and 30 to 80 μM naphthalene (Rockne and Strand, submitted). The cultures were incubated for 90 days (in the case of the naphthalene-fed cultures) with three refeedings of naphthalene as described elsewhere (Rockne and Strand, submitted). Results from the biphenyl- and phenanthrene-fed cultures were inconclusive, so we focus here solely on the naphthalene-fed cultures. Samples taken from the naphthalene-fed bottles were streaked onto modified anaerobic M-R2A agar plates (6) adjusted for sea salts concentrations and amended with KNO3 (5 mM). The plates were inoculated and incubated at room temperature (20 to 25°C) in an anaerobic glove box with a headspace consisting of 80% N2, 18% CO2, and 2% H2. Aseptic and strict anaerobic techniques were used throughout this study (17).
Colonies from the plates were removed with an inoculating loop scraped once over one agar surface and transferred to Balch tubes containing ASW (20 ml) plus 1 mM nitrate. The headspace for these and all further anaerobic bottle incubations consisted of oxygen-free nitrogen and were incubated at room temperature (20 to 25°C). Naphthalene was added to the medium using small sections (ca. 0.1 cm3) of sterile glass fiber filters containing naphthalene (1% [wt/vol] in methylene chloride) and allowing the methylene chloride to evaporate. The final naphthalene concentration added was 15.6 μM, which resulted in an aqueous equilibrium concentration of 12 to 12.5 μM. The same naphthalene delivery method was used in liquid cultures throughout the remainder of this study unless otherwise stated. Controls consisted of uninoculated medium. All liquid cultures were incubated quiescently in an anaerobic glove box in an inverted position.
Liquid cultures were monitored for loss of naphthalene by gas chromatography (GC) and nitrate depletion by high-pressure liquid chromatography (HPLC; see chemical analyses section, below). Naphthalene and nitrate were added to the cultures as needed. After incubation in liquid cultures (6 to 8 weeks), serial dilutions were prepared in anaerobic ASW and used to inoculate agar shake tubes. Agar shakes consisted of ASW (20 ml) plus nitrate (1 mM) and low-melting-point agarose (2%). Tubes were inoculated with diluted culture (1 ml) in cooled but molten agar and then mixed and solidified. Naphthalene was added in hexadecane (0.5 ml, 1% [vol/vol]) as an overlay to the agar. Four different colonies were removed from the naphthalene-containing agar shakes (10−6 dilution) using a syringe while under a stream of oxygen-free nitrogen. Individual colonies were transferred to ASW (20 ml) plus 1 mM nitrate and naphthalene, incubated in an anaerobic glove box, and monitored for decreases in naphthalene and nitrate concentrations.
Culture characterization.
Cultures that demonstrated naphthalene transformation and nitrate reduction were restreaked for purity onto sea salts M-R2A plates and transferred back to ASW plus nitrate (1 mM) and naphthalene. Purified cultures were also inoculated into ASW containing nitrate (5 mM), peptone, yeast extract, casamino acids (0.5 g liter−1 each), and pyruvate (3.4 mM) to assay for nitrate reduction activity. A Durham tube was added to each culture to capture evolved gas. Denitrification activity was determined by growth (turbidity) and by visual observation of gas following 6 days of incubation. Nitrate and nitrite concentrations were analyzed by using HPLC with UV detection. The pure cultures were assayed for Gram stain, oxidase, and catalase reactions using standard methods (34). Salinity requirements of the isolates were assayed on M-R2A liquid at 0, 0.35, 0.5, 3.5, 10, 17.5, 35, and 70 ppt (total sea salts concentration). The presence or absence of visual turbidity was reported after 10 days of incubation. The cultures were also tested for growth under aerobic and anaerobic growth conditions (in the presence or absence of 5 mM nitrate) on sea salts M-R2A broth. Turbidity was recorded after 7 days of incubation. Pure cultures were maintained on either ASW plus naphthalene medium or sea salts M-R2A plates. All assays were performed in duplicate.
Naphthalene mineralization and nitrate dependence.
Pure cultures were tested for the ability to mineralize naphthalene and the requirement of nitrate for naphthalene transformation. Prior to the experiment, cells which had previously been growing on naphthalene were grown on anaerobic ASW (20 ml) with either acetate (1 mM, NAP-3-1) or pyruvate (1 mM) plus yeast extract (100 mg l−1, NAP-4) in order to provide higher cell concentrations for use in this experiment. Nitrate was added to provide sufficient amounts of electron acceptor to oxidize all of the added carbon source, 1.6 or 2 mM NO3− for the acetate- or pyruvate-fed cultures, respectively (nitrate limiting), or to leave a 25% excess nitrate residual for use by cells during the oxidation of naphthalene in the mineralization experiment. In addition, naphthalene (15.6 μM) was added to all of the cultures. After 3 weeks of growth, concentrations of nitrate were measured in each culture to confirm that it was completely utilized in the nitrate-limiting cultures.
Radiolabeled [U-14C]naphthalene (49.8 μCi μmol−1; Sigma Chemical Co., St. Louis, Mo.) was added along with nonradiolabeled naphthalene to clean, empty serum bottles via a methylene chloride solution to give a final activity of 1.6 μCi per bottle (the total added naphthalene concentration was 12.5 μM). The methylene chloride was allowed to evaporate prior to addition of the cells. The cultures growing on acetate or pyruvate were then transferred to the 14C-naphthalene-coated serum bottles under a stream of oxygen-free N2 and sparged for 10 to 15 min. The initial naphthalene concentrations were measured because of the potential for residual naphthalene carryover in the culture fluid during preincubation with naphthalene. All cultures were incubated in duplicate with control bottles consisting of [U-14C]naphthalene-containing sterile ASW. Subsamples were removed to determine initial nitrate and naphthalene concentrations and the 14C specific activity. The test bottles were placed in an anaerobic glove box for incubation. At time points during incubation, aliquots (2 ml) of the cultures were taken for quantification of 14CO2 using the method of Rockne and Strand (submitted). Naphthalene, nitrate, soluble 14C metabolites, and incorporation of 14C into biomass were measured at the end of the experiment.
Chemical analyses.
For naphthalene analysis, liquid samples (2 ml) were placed into 4-ml HPLC vials with Teflon-lined septa and extracted into hexane (2 ml) with vigorous shaking for 90 min on a vortex mixer. The hexane was transferred into GC vials and analyzed on a Hewlett-Packard gas chromatograph (model 5890) fitted with a DB-5 column and flame ionization detector as described previously (30). Nitrate and nitrite was determined by the HPLC-UV method of Shroeder (32) as described previously (Rockne and Strand, submitted). Confirmation of nitrate and nitrite concentrations was made after injection onto an HPLC Partisil 10 SAX column (Waters Corp., Milford, Mass.), eluted with 50 mM phosphate buffer (pH 3.0) at 1.2 ml min−1, and detected by UV absorption at 210 nm. Total nonvolatile 14C metabolites were determined after gradient-HPLC fraction collection. The HPLC (Waters model 501) had a flow rate of 1.5 ml min−1 with the following gradient: 95% H3PO4 (0.1%)–5% methanol, proceeding to 95% methanol–5% H3PO4 (0.1%) with a linear gradient over 18 min, hold for 7 min, and return to initial conditions. Fractions at 1-min intervals (1.5 ml) were collected from the HPLC effluent in scintillation vials (7 ml) with scintillation cocktail (5 ml; Ecolume, Los Angeles, Calif.) and counted on a Packard TriCarb 1600 TR liquid scintillation counter (LSC; Packard, Downers Grove, Ill.) for 20 min.
Samples of the culture (5 ml) were filtered through 0.2-μm (pore-size) polypropylene syringe filters (Gelman Sciences, Ann Arbor, Mich.) to determine 14C radiolabel incorporation into biomass. The filter was washed with sequential washes of deionized water (10 ml) and methanol (10 ml) to remove radiolabel in the aqueous phase and nonpolar radiolabel (e.g., naphthalene) sorbed to the cells. The extracted filter was then placed in an acid fumer for removal of any absorbed 14CO2 (1), transferred to a scintillation vial (20 ml) with scintillation cocktail (20 ml; Ecolume) and counted by using the LSC. The total activity was counted and 14C radiolabeled fractions were determined as percentages of the total [U-14C]naphthalene added following quench corrections by the standard addition technique as described previously (29). Total radiolabel recoveries ranged from 60 to 90%.
Reaction stoichiometry.
Incomplete mineralization and electron donor incorporation into cell mass (as was observed in these cultures) would affect the stoichiometry. To take this into account, we calculated the expected nitrate-to-naphthalene stoichiometry by incorporating the fraction of electron donor coupled to cell synthesis (fs) as described by Rockne and Strand (submitted). This parameter was used to calculate the nitrate-to-naphthalene stoichiometry using the following cell energetics equations:
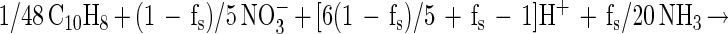 |
1 |
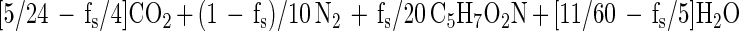 |
with dinitrogen as the terminal nitrate reduction product and
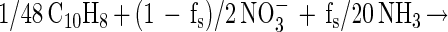 |
2 |
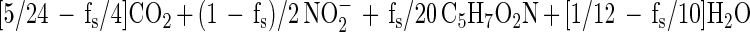 |
with nitrite as the terminal nitrate reduction product.
We calculated fs from the naphthalene consumption and cell mass incorporation data (using either equation 1 or equation 2) as follows: fs = (10/48)(5/20)−1(naphthalene-carbon/cell mass carbon)−1. The expected nitrate-to-naphthalene stoichiometric ratio was calculated using fs by either [(1 − fs)/5](1/48)−1 from equation 1 or [(1 − fs)/2](1/48)−1 from equation 2.
16S rDNA sequencing and phylogenetic analysis.
For phylogenetic analysis, colonies of isolates NAP-3-1 and NAP-4 grown anaerobically on seawater M-R2A plates were harvested and genomic DNA was isolated using the Instagene Kit (Bio-Rad, Hercules, Calif.). 16S ribosomal DNA (rDNA) was PCR amplified from the genomic DNA preparations using universal primers (28) and the following parameters: 32 cycles of 1.5 min at 94°C, 1 min at 42°C, and 4 min at 72°C with the last step of the last cycle continuing for 10 min. PCR products were purified using Ultrafree-MC cellulose filter units (Millipore, Bedford, Mass.), digested with NotI, and cloned into pBluescript II KS(+) (Stratagene, La Jolla, Calif.) using standard techniques (31).
A single clone representing each strain was sequenced using the Taq DyeDeoxy Terminator Cycle Sequencing kit (Applied Biosystems, Foster City, Calif.) and 16S rDNA-specific forward and reverse primers (10). Contiguous 16S rDNA sequences were assembled using the SeqApp program (D. G. Gilbert, Seq-App 1.9a169, 1992) and were initially aligned with similar sequences by the Ribosomal Database Project (RDP) version 5.0 ALIGN_SEQUENCE program (21). Manual corrections were made to the alignment. Additional sequences used in the phylogenetic analyses were accessed from the RDP (21).
The data sets were analyzed using several phylogenetic methods. Maximum-likelihood and parsimony analyses were performed by fastDNAml (26) and PAUP 3.0 (35), respectively. Programs used in the neighbor-joining analysis were obtained from the PHYLIP package, version 3.2 (12).
Nucleotide accession numbers.
The nearly complete 16S rDNA sequences for isolates NAP-3-1 and NAP-4 have been deposited in GenBank with the following accession numbers: AF064636 and AF064637. The accession numbers of all the microbial cultures used in the construction of the phylogenetic tree are as follows (reading clockwise in the tree from Haemophilus influenzae): M35019, J01695, X74702, AF064637, X74726, X67024, AF150806, L34955, X67022, M22365, L28676, AF064636, M34133, M34139, and M34131.
RESULTS
General characteristics of the pure cultures.
Three pure cultures were obtained in this study: NAP-4, NAP-3-1, and NAP-3-2. These cultures were all gram-negative rods (Table 1). NAP-3-1 and NAP-3-2 were catalase and oxidase positive, whereas NAP-4 was oxidase negative and only weakly catalase positive. All cultures were salt tolerant, and NAP-4 had an apparent requirement for greater than 3.5 ppt salinity for growth on M-R2A under the conditions of the assay. Gas production was observed for NAP-3-1 after 6 days of incubation in the denitrification assay. Isolate NAP-3-1, which degrades naphthalene, was confirmed to be a denitrifier by measuring N2 production in the denitrification assay. Isolate NAP-3-2 grew on the denitrification medium but did not produce measurable quantities of gas. Similarly, isolate NAP-4 did not produce gas in the denitrification medium, but unlike NAP-3-2, NAP-4 grew poorly on the medium. Significant accumulation of nitrite (2 mM) occurred with isolate NAP-4. No nitrate remained in any of the cultures, and no nitrite was detected for isolates NAP-3-1 and NAP-3-2. All cultures grew on M-R2A both aerobically and anaerobically with nitrate, but only NAP-4 grew on M-R2A anaerobically without nitrate present.
TABLE 1.
Summary of general characteristics of the pure cultures
| Isolate | Gram staina | Catalasea | Oxidasea | Salinity toleranceb at:
|
Sea salt M-R2Ac
|
Denitrification assayd
|
||||
|---|---|---|---|---|---|---|---|---|---|---|
| 0.35–3.5 ppt | 10–70 ppt | Aerobic | Anaerobic (+NO3−) | Anaerobic (−NO3−) | Growth | Gas | ||||
| NAP-3-1 | − | + | + | + | + | + | + | − | ++ | ++ |
| NAP-3-2 | − | + | + | + | + | + | + | − | + | − |
| NAP-4 | − | + (weak) | − | − | + | + | + | + | + (weak) | − |
Tested after 24 h of anaerobic growth on sea salts M-R2A (medium is defined in Materials and Methods).
Visible turbidity after 10 days of anaerobic growth on M-R2A with corresponding salinity.
Visible turbidity after 7 days of growth.
Visible turbidity and gas production in Durham tube test after 6 days of growth.
Naphthalene transformation by pure cultures.
The three strains were assayed for naphthalene and nitrate transformation in liquid cultures. After 18 days, the naphthalene-fed isolates demonstrated a loss in naphthalene concentration along with nitrate removal (data not shown). Isolate NAP-4 removed more than 33% of the naphthalene and 30% of the nitrate relative to the uninoculated control. Similarly, both NAP-3-1 and NAP-3-2 removed 22% of the naphthalene, with 22 and 25% of the nitrate reduced, respectively, relative to the uninoculated control. All three cultures contained small amounts of nitrite (data not shown). No significant losses of nitrate or aqueous-phase naphthalene were observed in the uninoculated naphthalene-containing control. The remainder of this study was focused on isolates NAP-3-1 and NAP-4 since NAP-3-2 demonstrated a rate of naphthalene degradation similar to that of NAP-3-1.
Nitrate-dependent naphthalene transformation and mineralization.
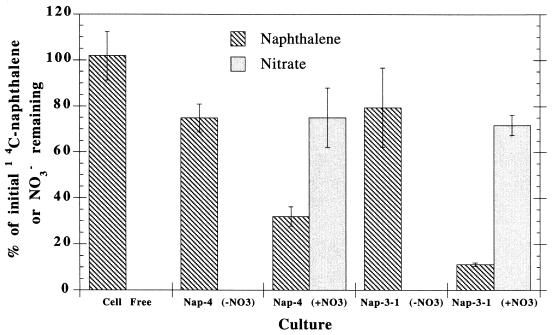
Isolates NAP-4 and NAP-3-1 both exhibited nitrate-dependent naphthalene transformation (Fig. 1) after pregrowth on pyruvate or acetate, respectively. No statistically significant (P = 0.05) loss in naphthalene was measured in the NAP-3-1 culture under nitrate-limiting conditions compared to cell-free controls after 57 days of incubation. NAP-4 had a small but significant loss of naphthalene in the absence of nitrate. In the presence of nitrate, isolates NAP-4 and NAP-3-1 degraded 70 to 90% of the labeled naphthalene with significant removal of nitrate (Fig. 1).
FIG. 1.
Nitrate-dependent anaerobic transformation of 14C-naphthalene by pure cultures after 57 days of incubation. The percentages of initial values are relative to the initial nitrate concentration and the 14C scintillation counts. The control consisted of uninoculated ASW plus naphthalene after 57 days. The average of duplicates ± the standard error of the mean (SEM), indicated by error bars, is shown.
Isolate NAP-4 mineralized naphthalene without a lag period, with one of the replicates reaching a maximum of 18% mineralization after 25 days (Fig. 2). After this period naphthalene mineralization appeared to cease in both replicates. Mineralization of naphthalene by isolate NAP-3-1 increased steadily in both replicates throughout the experiment, reaching maxima of 10 to 20% mineralization after 57 days.
FIG. 2.
Mineralization of naphthalene by nitrate-reducing naphthalene-degrading pure cultures of NAP-4 (A) and NAP-3-1 (B). The percentages of the initial 14C-naphthalene carbon recovered as 14C-CO2 are given as the average of duplicates ± the SEM.
Carbon mass balance and reaction stoichiometry.
Minor amounts (≤4%) of radiolabel were recovered in the nonvolatile, aqueous fraction at the end of the experiment, suggesting that water-soluble metabolites did not accumulate significantly outside the cell (Table 2). The recovery of radiolabel at the end of the experiment was 57% for NAP-3-1 and 91% for NAP-4, comparing well with recoveries reported in other studies using radiolabeled PAH (4, 15, 16, 29; Rockne and Strand, submitted). The largest fraction of radiolabel was associated with cell mass comprising 30 to 50% of the initial 14C from naphthalene.
TABLE 2.
Recovery of 14C after 57 days of incubation by pure cultures grown on naphthalene and nitratea
| Culture | 14CO2 (%) | 14C recovered in biomass (%) | Soluble 14C metabolites (%) | Hexane extractable 14C (includes naphthalene) (%) | Total 14C recovered (%) |
|---|---|---|---|---|---|
| NAP-3-1 | 15 ± 5 | 29 ± 2 | 3 ± 0.3 | 11 ± 1 | 57 ± 8 |
| NAP-4 | 12 ± 4 | 43 ± 8 | 4 ± 0.1 | 32 ± 4 | 91 ± 16 |
Average of duplicates ± the SEM. The percentage of the 14C activity was measured at the start of the experiment.
At the end of the mineralization experiment, the calculated ratios of nitrate-to-naphthalene consumption were 5.3 ± 0.1 and 12.4 ± 2.7 (moles of NO3− per mole of naphthalene) for isolates NAP-3-1 and NAP-4, respectively (Table 3). The expected nitrate-to-naphthalene ratio using the 14C incorporation data and assumed nitrate reduction products were calculated using equations 1 and 2. If NAP-3-1 coupled naphthalene mineralization to the reduction of nitrate to dinitrogen gas, the expected nitrate-to-naphthalene stoichiometry would be 7.0 mol of NO3− per mol of naphthalene, a value higher than the observed ratio of 5.3 (Table 3). Assuming NAP-4 reduced nitrate to nitrite (based on the previous data), the nitrate-to-naphthalene stoichiometry would be 11 mol of NO3− per mol of naphthalene, within the range of the observed ratio (Table 3).
TABLE 3.
Nitrate consumption and the ratio (mole per mole) of nitrate to naphthalene consumption by nitrate-reducing naphthalene-degrading pure cultures after 57 days of incubation
| Isolate | Nitrate consumeda (μM) | Naphthalene consumeda (μM) | Ratio of nitrate to naphthalene consumedb (mol/mol) | With incorporation of radiolabel resultsc
|
||
|---|---|---|---|---|---|---|
| Assumed nitrate reduction product | Fraction of naphthalene utilized for cell synthesis (fs) | Expected ratio of nitrate to naphthalene consumed (mol/mol) | ||||
| NAP-3-1 | 48 ± 7 | 9.1 ± 1.2 | 5.3 ± 0.1 | N2 | 0.27 | 7.0 |
| NAP-4 | 196 ± 90 | 15.4 ± 2.8 | 12.4 ± 2.7 | NO2− | 0.53 | 11 |
Average of duplicates ± the SEM. The initial nitrate concentrations were 123 ± 8.4 and 795 ± 34 μM for NAP-3-1 and NAP-4, respectively. The initial naphthalene concentrations were 10.2 ± 2 and 22.73 ± 3 μM for NAP-3-1 and NAP-4, respectively.
Average of ratio for each replicate.
The naphthalene consumed and the amount of 14C-naphthalene incorporated into biomass (from Table 2) were used to calculate the fraction of electron donor coupled to production of cell mass (fs) using equations 1 and 2.
Phylogenetic analysis.
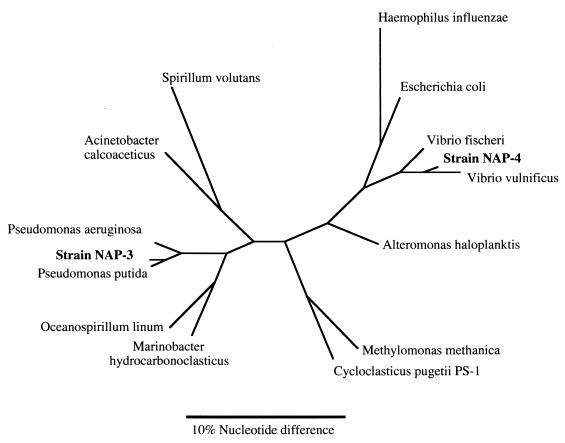
Comparison of the complete 16S rDNA sequences of NAP-3-1 and NAP-4 indicated that they were both phylogenetically similar to bacteria in the gamma proteobacteria group but were distant from each other within the group (Fig. 3). NAP-4 was phylogenetically similar to Vibrio pelagius (99.2% homology), which reduces nitrate to nitrite, and other Vibrio spp. NAP-3-1 was also in the gamma proteobacteria and was most closely related phylogenetically with Pseudomonas stutzeri (99.7% homology), as well as with P. putida and P. aeruginosa. These bacteria are in rRNA group I of the Pseudomonadaceae and are well known aerobic aromatic compound degraders (9, 36). P. aeruginosa is a denitrifier able to use nitrate, nitrite, or nitrous oxide as terminal electron acceptors (9). P. putida is not a denitrifier and does not degrade toluene under oxygen-limited conditions (2 mg of O2 liter−1), either with or without nitrate (naphthalene degradation was not assayed in that study [20]).
FIG. 3.
Phylogenetic tree based on 16S rDNA sequences showing isolates NAP-4 and NAP-3-1 and other members of the gamma proteobacteria. The tree is the most parsimonious tree based on a branch and bound analysis using PAUP.
DISCUSSION
Recent studies have clearly demonstrated that highly enriched cultures can degrade unsubstituted aromatic compounds such as naphthalene without oxygen. Further understanding of the mechanisms of anaerobic naphthalene transformation and the bacteria responsible for the degradation activity, however, has been hampered by the lack of isolates. This is the first study presenting evidence of pure cultures capable of nitrate-dependent, anaerobic naphthalene degradation and mineralization.
The results obtained with nitrate-limited cultures demonstrated that nitrate was required for degradation of naphthalene by NAP-3-1 and NAP-4. There was no significant loss of nitrate or naphthalene in cell-free controls. Nitrate was required by both isolates NAP-3-1 and NAP-4 for significant transformation and partial mineralization of naphthalene. The minor loss of naphthalene in the nitrate-limited culture of NAP-4 (Fig. 1) may have been due to the presence of a small unquantified amount of nitrate left in the culture at the start of the experiment. In addition to demonstrating nitrate-dependent naphthalene degradation, the use of stringent anaerobic techniques in this experiment also precluded the possibility of any involvement of oxygen during naphthalene degradation.
The nitrate-to-naphthalene stoichiometric ratio would be expected to be lower than the theoretical ratio of 9.6 if naphthalene mineralization were incomplete, as was the case here. Using the 14C incorporation into cell mass results and the likely nitrate reduction product, calculated stoichiometric ratios of nitrate-to-naphthalene utilization (using equations 1 and 2) were close to what would be expected from theory. For isolate NAP-3-1, the ratio was 30% higher than the expected ratio assuming denitrification to dinitrogen. Isolate NAP-4, which was not a denitrifier, demonstrated a higher molar ratio of nitrate utilization to the amount of naphthalene transformed than would be expected assuming denitrification. This result is not unexpected if we assume nitrite is the product of nitrate reduction coupled to naphthalene oxidation by NAP-4. With this assumption, the cell mass incorporation data give a calculated nitrate-to-naphthalene ratio that was not significantly different than the observed amount.
The preceding cell energetics analysis assumes that nitrite is the terminal nitrate reduction product by NAP-4. Alternatively, isolate NAP-4 may have reduced nitrate to ammonia. With nitrate reduction to ammonia, the corresponding nitrate-to-naphthalene ratio for NAP-4 would be 2.8 mol/mol (with cell mass incorporation), a level much lower than the observed ratio of 12.4 or the predicted ratio of 11 with nitrate reduction to nitrite. Further, NAP-4 did not produce nitrous oxide or dinitrogen gas from nitrate. Although significant production of nitrite was observed in this culture during the denitrification assay, further studies are required to determine the nitrate reduction pathway employed by isolate NAP-4 during degradation of naphthalene.
The phylogenetic analysis was consistent with standard physiological and biochemical characterizations of the isolates. Isolate NAP-3-1 is a denitrifier related to Pseudomonas spp. in the gamma proteobacteria. The Pseudomonas family comprises many aerobic naphthalene-degrading bacteria. McNally et al. (22) reported biodegradation of naphthalene, acenaphthene, anthracene, phenanthrene, and pyrene in the absence of detectable oxygen by isolates putatively identified as P. stutzeri, P. putida, and P. fluorescens. Although the study by McNally et al. (22) did not prove nitrate-dependent naphthalene degradation, the results do suggest that other members of the Pseudomonadaceae may possess anaerobic PAH biodegradation ability, which is consistent with the results of this study.
Isolate NAP-4 also was in the gamma proteobacteria, but not closely related to NAP-3-1. NAP-4 was closely related phylogenetically to Vibrio spp. Phenotypically, Vibrio spp. are characterized by a lack of gas production (18), a finding also consistent with the results from the denitrification assay for the NAP-4 culture. Although some known marine Vibrio can degrade PAHs under aerobic conditions (13), none are known to degrade aromatic compounds under anaerobic conditions (27). Together with the NAP-3-1 results, these data suggest that phylogenetically diverse bacteria within the gamma proteobacteria can degrade naphthalene without oxygen.
The isolation of the pure cultures obtained in this study was facilitated by the use of a highly enriched source of inoculum. The subcultures obtained from the FBR studies that were used as the source of inoculum in this study had been developed over a period of 2 years. Even with this extended enrichment period, the rate of naphthalene degradation was slow, necessitating the need for more readily utilizable carbon sources to assist in growth and isolation. Nitrate in marine sediment ecosystems is not thought to be a significant electron sink and, consequently, nitrate reducers are generally not considered important anaerobic organisms for significant removal of organic contaminants. The limited presence of these microorganisms in marine sediment may be one reason they have been overlooked as important contaminant degraders in the past.
Further studies are needed to determine what the biochemical mechanisms are for naphthalene degradation activity. The slow growth rates of our isolates on naphthalene and the resultant low cell mass production will make this task difficult. However, our technique of growing the cultures on alternate substrates (such as acetate) in the presence of naphthalene resulted in much higher cell mass production than when grown on naphthalene alone. This method could be used to facilitate experiments by increasing the cell mass. It is not possible to determine from the present data whether anaerobic naphthalene degradation is an intrinsically slow process or whether it is due to unknown growth-limiting factors. McNally et al. (22) reported anoxic PAH degradation rates comparable to or only slightly slower than aerobic degradation rates, including complete degradation of 24 μM naphthalene in 6 to 8 h. In contrast, Rockne and Strand (30) reported anaerobic specific PAH biodegradation rates that were an order of magnitude slower than other published aerobic rates. Elucidating the initial enzymatic attack on the ring structure may aid in determining the rate-limiting step of anaerobic PAH biodegradation. Recent work has shown that ring carboxylation is an early reaction of the anaerobic naphthalene and phenanthrene biodegradation pathways in a sulfate-reducing enrichment (38). It is unknown whether this pathway is also used by the cultures in this study to degrade naphthalene.
Our isolates may represent individual members of a consortium able to efficiently degrade naphthalene when present in coculture. Although NAP-4 is capable of naphthalene mineralization, it may degrade naphthalene or naphthalene intermediates more efficiently as a member of a consortium. For example, bacteria such as NAP-4, a nondenitrifying nitrate reducer, may transform naphthalene to utilizable substrates or may facilitate nutrient cross-feeding to other members of the microbial population. Consortium synergy may explain the difference between the slow degradation rates of our isolates and the rates observed in the FBR source of the isolate (30). In addition, there were similarities in the extent of naphthalene mineralization by the pure cultures in this study and those observed in the mixed enrichment subculture (the source of these isolates). Mineralization ranged from greater than 90% (for phenanthrene-fed subcultures) to 20% in the naphthalene-fed subcultures (Rockne and Strand, submitted); the latter result was similar to what was observed with both isolates NAP-4 and NAP-3-1. Interestingly, there was a large amount of naphthalene incorporated into the cell mass in the naphthalene-fed subcultures (Rockne and Strand, submitted), as was the case with NAP-4 but not NAP-3-1. The fraction of electron donor utilized for cell synthesis in the mixed culture was also in closer agreement with NAP-4 rather than NAP-3-1.
In summary, the isolates in this study are the first pure cultures shown to degrade naphthalene in the strict absence of oxygen using Hungate's technique, resazurin, and chemical reductants. Naphthalene was mineralized in the absence of oxygen, and degradation was nitrate dependent. The bacteria capable of this metabolism were phylogenetically diverse within the gamma proteobacteria, suggesting that the ability to degrade naphthalene anaerobically may be widely distributed within this group. Further studies are required to determine the metabolic pathways of anaerobic naphthalene mineralization. This knowledge may help to identify potential cometabolites and/or cofactors required for rapid growth by the isolates and potential requirements for anaerobic naphthalene utilization in the environment.
REFERENCES
- 1.American Public Health Association. Standard methods for the examination of water and wastewater. 19th ed. Washington, D.C.: American Public Health Association; 1995. [Google Scholar]
- 2.Agency for Toxic Substances and Disease Registry. Public health statement: creosote. Washington, D.C.: Agency for Toxic Substances and Disease Registry–Division of Toxicology; 1990. [Google Scholar]
- 3.Bauer J E, Capone D G. Degradation and mineralization of the polycyclic aromatic hydrocarbons anthracene and naphthalene in intertidal marine sediments. Appl Environ Microbiol. 1985;50:81–90. doi: 10.1128/aem.50.1.81-90.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Brodkorb T S, Legge R L. Enhanced biodegradation of phenanthrene in oil-tar-contaminated soils supplemented with Phanaerochaete chrysosporium. Appl Environ Microbiol. 1992;58:3117–3121. doi: 10.1128/aem.58.9.3117-3121.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cerniglia C E. Microbial metabolism of polycyclic aromatic hydrocarbons. Adv Appl Microbiol. 1984;30:31–76. doi: 10.1016/s0065-2164(08)70052-2. [DOI] [PubMed] [Google Scholar]
- 6.Chee-Sanford J, Frost J W, Tiedje J. Evidence for acetyl-CoA and cinnamoyl-CoA in the anaerobic mineralization pathway in Azoarcus tolulyticus Tol-4. Appl Environ Microbiol. 1996;62:964–973. doi: 10.1128/aem.62.3.964-973.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Coates J D, Anderson R T, Lovley D L. Oxidation of polycyclic aromatic hydrocarbons under sulfate-reducing conditions. Appl Environ Microbiol. 1996;62:1099–1101. doi: 10.1128/aem.62.3.1099-1101.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Coates J D, Woodward J, Allen J, Philp P, Lovley D L. Anaerobic degradation of polycyclic aromatic hydrocarbons and alkanes in petroleum-contaminated marine harbor sediments. Appl Environ Microbiol. 1997;63:3589–3593. doi: 10.1128/aem.63.9.3589-3593.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dagher F, D'eziel E, Lirette P, Paquette G, Bisaillon J G, Villemur R. Comparative study of five polycyclic aromatic hydrocarbon degrading bacterial strains isolated from contaminated soils. Can J Microbiol. 1997;43:368–377. doi: 10.1139/m97-051. [DOI] [PubMed] [Google Scholar]
- 10.Dyksterhouse S E, Gray J P, Herwig R P, Lara J C, Staley J T. Cycloclasticus pugetii gen. nov., sp. nov., an aromatic hydrocarbon-degrading bacterium from marine sediments. Int J Syst Bact. 1995;45:116–123. doi: 10.1099/00207713-45-1-116. [DOI] [PubMed] [Google Scholar]
- 11.Evans W C, Fuchs G. Anaerobic degradation of aromatic compounds. Annu Rev Microbiol. 1988;42:289–317. doi: 10.1146/annurev.mi.42.100188.001445. [DOI] [PubMed] [Google Scholar]
- 12.Felsenstein J. PHYLIP—Phylogeny Inference Package (version 3.2) Cladistics. 1989;5:164–166. [Google Scholar]
- 13.Geiselbrecht A D, Herwig R P, Deming J W, Staley J T. Enumeration and phylogenetic analysis of polycyclic aromatic hydrocarbon-degrading marine bacteria from Puget Sound sediments. Appl Environ Microbiol. 1996;62:3344–3349. doi: 10.1128/aem.62.9.3344-3349.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Genthner B R S, Townsend G T, Lantz S E, Mueller J G. Persistence of polycyclic aromatic hydrocarbon components of creosote under anaerobic enrichment conditions. Arch Environ Contam Toxicol. 1997;32:99–105. doi: 10.1007/s002449900160. [DOI] [PubMed] [Google Scholar]
- 15.Grosser R J, Warshawsky D, Vestal J R. Indigenous and enhanced mineralization of pyrene, benzo[A]pyrene, and carbazole in soils. Appl Environ Microbiol. 1991;57:3462–3469. doi: 10.1128/aem.57.12.3462-3469.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Heitkamp M A, Cerniglia C E. Polycyclic aromatic hydrocarbon degradation by a Mycobacterium sp. in microcosms containing sediment and water from a pristine ecosystem. Appl Environ Microbiol. 1989;55:1968–1973. doi: 10.1128/aem.55.8.1968-1973.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Holdeman L V, Cato E P, Moore W E C. Anaerobe laboratory manual. Blacksburg, Va: Anaerobe Laboratory, Virginia Polytechnic Institute and State University; 1977. [Google Scholar]
- 18.Holt J G. Bergey's manual of determinative bacteriology. 9th ed. Baltimore, Md: The Williams & Wilkins Co.; 1994. [Google Scholar]
- 19.Johnston N, Sadler R, Shaw G R, Connell D W. Environmental modification of PAH composition in coal tar containing samples. Chemosphere. 1993;27:1151–1158. [Google Scholar]
- 20.Kukor J J, Olsen R H. Catechol 2,3-dioxygenases functional in oxygen-limited (hypoxic) environments. Appl Environ Microbiol. 1996;62:1728–1740. doi: 10.1128/aem.62.5.1728-1740.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Larsen N, Olsen G J, Maidak B L, McCaughey M J, Overbeek R, Macke T J, Marsh T L, Woese C R. The ribosomal database project. Nucleic Acids Res. 1993;21:3021–3023. doi: 10.1093/nar/21.13.3021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.McNally D L, Mihelcic J R, Lueking D R. Biodegradation of three- and four-ring polycyclic aromatic hydrocarbons under aerobic and denitrifying conditions. Environ Sci Technol. 1998;32:2633–2639. [Google Scholar]
- 23.Menzie C A, Potocki B B, Santodonato J. Exposure to carcinogenic PAHs in the environment. Environ Sci Technol. 1992;26:1278–1284. [Google Scholar]
- 24.Mihelcic J R. Ph.D. thesis. Pittsburgh, Pa: Carnegie Mellon University; 1988. [Google Scholar]
- 25.Mihelcic J R, Luthy R G. Degradation of polycyclic aromatic hydrocarbons compounds under various redox conditions in soil-water systems. Appl Environ Microbiol. 1988;54:1182–1187. doi: 10.1128/aem.54.5.1182-1187.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Olsen G J, Matsuda H, Hagstrom R, Overbeek R. fastDNAml: a tool for construction of phylogenetic trees of DNA sequences using maximum likelihood. Comput Appl Biosci. 1994;10:41–43. doi: 10.1093/bioinformatics/10.1.41. [DOI] [PubMed] [Google Scholar]
- 27.Rabus R, Fukui M, Wilkes H, Widdel F. Degradative capacities and 16s rRNA-targeted hybridization of sulfate-reducing bacteria in an anaerobic enrichment culture utilizing alkylbenzenes. Appl Environ Microbiol. 1996;62:3605–3613. doi: 10.1128/aem.62.10.3605-3613.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Reysenbach A L, Wickham G S, Pace N R. Phylogenetic analysis of the hyperthermophilic pink filament community in Octopus Spring Yellowstone National Park. Appl Environ Microbiol. 1994;60:2113–2119. doi: 10.1128/aem.60.6.2113-2119.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Rockne K J, Stensel H D, Herwig R P, Strand S E. PAH degradation and bioaugmentation by a marine methanotrophic enrichment. Bioremed J. 1998;1:209–222. [Google Scholar]
- 30.Rockne K J, Strand S E. Biodegradation of bicyclic and polycyclic aromatic hydrocarbons in anaerobic enrichments. Environ Sci Technol. 1998;32:2962–3967. [Google Scholar]
- 31.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Plainview, N.Y: Cold Spring Harbor Press; 1989. [Google Scholar]
- 32.Schroeder D C. The analysis of nitrate in environmental samples by reversed-phase HPLC. J Chromatogr Sci. 1987;25:405–408. doi: 10.1093/chromsci/25.9.405. [DOI] [PubMed] [Google Scholar]
- 33.Shiaris M P. Seasonal biotransformation of naphthalene, phenanthrene, and benzo[a]pyrene in surficial estuarine sediments. Appl Environ Microbiol. 1989;55:1391–1399. doi: 10.1128/aem.55.6.1391-1399.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Smibert R M, Krieg N R. General characterization. In: Gerhardt P, Murray R G E, Costilow R N, editors. Manual of methods for general bacteriology. Washington, D.C.: American Society for Microbiology; 1981. pp. 409–433. [Google Scholar]
- 35.Swafford D L. PAUP: phylogenetic analysis using parsimony, 3.0 ed. Champaign, Ill: Illinois Natural History Survey; 1991. [Google Scholar]
- 36.Trzesicka-Mlynarz D, Ward O P. Degradation of polycyclic aromatic hydrocarbons (PAHs) by a mixed culture and its component pure cultures, obtained from PAH-contaminated soil. Can J Microbiol. 1995;41:470–476. doi: 10.1139/m95-063. [DOI] [PubMed] [Google Scholar]
- 37.Turney G L, Goerlitz D F. Organic contamination of ground water at gas-works-park, Seattle, Washington. Ground Water Monit Rev. 1990;10:187–198. [Google Scholar]
- 38.Zhang X, Young L Y. Carboxylation as an initial reaction in the anaerobic metabolism of naphthalene and phenanthrene by sulfidogenic consortia. Appl Environ Microbiol. 1997;63:4759–4764. doi: 10.1128/aem.63.12.4759-4764.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]