Abstract
The enzymes chlorocatechol-1,2-dioxygenase, chloromuconate cycloisomerase, dienelactone hydrolase, and maleylacetate reductase allow Ralstonia eutropha JMP134(pJP4) to degrade chlorocatechols formed during growth in 2,4-dichlorophenoxyacetate or 3-chlorobenzoate (3-CB). There are two gene modules located in plasmid pJP4, tfdCIDIEIFI (module I) and tfdDIICIIEIIFII (module II), putatively encoding these enzymes. To assess the role of both tfd modules in the degradation of chloroaromatics, each module was cloned into the medium-copy-number plasmid vector pBBR1MCS-2 under the control of the tfdR regulatory gene. These constructs were introduced into R. eutropha JMP222 (a JMP134 derivative lacking pJP4) and Pseudomonas putida KT2442, two strains able to transform 3-CB into chlorocatechols. Specific activities in cell extracts of chlorocatechol-1,2-dioxygenase (tfdC), chloromuconate cycloisomerase (tfdD), and dienelactone hydrolase (tfdE) were 2 to 50 times higher for microorganisms containing module I compared to those containing module II. In contrast, a significantly (50-fold) higher activity of maleylacetate reductase (tfdF) was observed in cell extracts of microorganisms containing module II compared to module I. The R. eutropha JMP222 derivative containing tfdR-tfdCIDIEIFI grew four times faster in liquid cultures with 3-CB as a sole carbon and energy source than in cultures containing tfdR-tfdDIICIIEIIFII. In the case of P. putida KT2442, only the derivative containing module I was able to grow in liquid cultures of 3-CB. These results indicate that efficient degradation of 3-CB by R. eutropha JMP134(pJP4) requires the two tfd modules such that TfdCDE is likely supplied primarily by module I, while TfdF is likely supplied by module II.
Ralstonia eutropha JMP134 is able to grow in media containing 2,4-dichlorophenoxyacetate (2,4-D) and 3-chlorobenzoate (3-CB), as well as other chloroaromatics (3, 5, 26). Most of its catabolic abilities are encoded in the plasmid pJP4 (5, 6). The enzymes for the catabolism of chloroaromatics in pJP4 have been intensively studied (19, 25–27, 32, 33, 35, 36). Catabolism of 2,4-D is started by the products of the 2,4-D/α-ketoglutarate dioxygenase (tfdA) (10, 11) and 2,4-dichlorophenol hydroxylase (tfdB) genes on pJP4, to form 3,5-dichlorocatechol (3,5-DCC). Metabolism of 3-CB is initiated by a chromosomally encoded, low-specificity benzoate dioxygenase and 1,2-dihydro-1,2-dihydroxybenzoate dehydrogenase to form 3-chlorocatechol (3-CC) and 4-chlorocatechol (4-CC), as has been reported for Alcaligenes eutrophus B9 (29). Chlorocatechol metabolism is supposed to be performed by the enzymes encoded in the tfdCIDIEIFI gene module present in the EcoRI-B fragment of pJP4. Genes tfdCI, tfdDI, and tfdEI encode for chlorocatechol-1,2-dioxygenase, chloromuconate cycloisomerase, and dienelactone hydrolase, respectively (7). Interruption of these genes by transposon mutagenesis resulted in mutants no longer able to grow in 2,4-D, supporting the notion that these gene products play a major role in the metabolism of this substrate. It has been proposed that the fourth gene of this module, tfdFI, encodes a functional maleylacetate reductase (14). An additional chromosomally encoded maleylacetate reductase was reported to be recruited for chloroaromatic degradation in R. eutropha (20). The presence in pJP4 of a second module of genes (tfdDIICIIEIIFII) possibly coding for enzymes for chlorocatechol metabolism has only recently been reported (9, 22). Leveau and coworkers have observed that transcription of the genes of both modules I and II takes place during adaptation to 2,4-D in cells of R. eutropha JMP134 growing on fructose (23). It is not known if the tfdII gene products are functional for chlorocatechol degradation.
The objective of this work was to investigate the function of the tfdCIDIEIFI and tfdDIICIIEIIFII gene modules in R. eutropha growing on 3-CB as sole carbon and energy source. Each of the two gene modules was cloned into the medium-copy-number plasmid vector pBBR1MCS-2 (17), under the control of the LysR-type transcriptional activator tfdR (21) and its corresponding putative promoter sequences. The tfdR/Ptfd-ItfdCIDIEIFI and tfdR/Ptfd-IItfdDIICIIEIIFII gene modules were independently introduced into two strains able to transform 3-CB into chlorocatechols, i.e., R. eutropha JMP222, a derivative of strain JMP134 cured of plasmid pJP4, and Pseudomonas putida KT2442. In the derivatives obtained, the expression of Tfd enzymes and the ability to grow with 3-CB were assessed.
MATERIALS AND METHODS
Bacterial strains, plasmids, and culture conditions.
Bacterial strains and plasmids used in this study are listed in Table 1. R. eutropha JMP222 and P. putida KT2442 were grown at 30°C in a chloride-free minimal medium (18) with 3 mM benzoate plus streptomycin (1,000 μg/ml) or rifampycin (50 μg/ml), respectively. R. eutropha JMP134 and 3-CB-mineralizing derivatives of JMP222 and KT2442 were grown in minimal medium with 3 mM 3-CB. Derivatives of strain KT2442 not capable of growing with 3-CB were grown in minimal medium with 3 mM benzoate plus kanamycin (50 μg/ml). P. putida KT2442(pJP4) was grown in minimal medium with 3 mM benzoate or in Luria-Bertani (LB) medium containing 0.5 mM merbromin. Escherichia coli strains were maintained on LB agar plates containing the appropriate antibiotic: 50 μg of ampicillin, kanamycin, or rifampycin per ml or 20 μg of tetracycline or cloramphenicol per ml. Growth in 3-CB was determined as increase in optical density at 660 nm (OD660). At least three replicate growth measurements were performed.
TABLE 1.
Strains and plasmids used in this work
| Strain or plasmid | Relevant phenotype or genotypea | Source or reference |
|---|---|---|
| Bacteria | ||
| R. eutropha | ||
| JMP134 | 2,4-D+, 3-CB+, Hgr, pJP4 | DSMZb |
| JMP222 | R. eutropha JMP134 derivative, 2,4-D−, 3-CB−, Smr | H. Knackmuss |
| P. putida | ||
| KT2442 | P. putida mt-2 derivative, hsdR1 hsdM+, Rifr | 12 |
| E. coli | ||
| DH5α | 1 | |
| HB101 | 1 | |
| Plasmids | ||
| pRK600 | Cmr, IncPα, tra+ | 4 |
| pBluescript II KS | Apr | Stratagene |
| pUC18 | Apr | GIBCO-BRL |
| pUC18Not (Sfi) | Apr | 12 |
| pUCP19 | Apr | 31 |
| pBBR1MCS-2 | Kmr, broad host range | 17 |
| pVJE22 | Tcr, tfdCIDIEIFI, tfdB | 34 |
| pJRC105 | Apr, tfdCIDIEIFI, pUC18Not derivative | This work |
| pJRC67 | Apr, tfdCIDIEIFI, pBSKSII derivative | This work |
| pJRC48 | Apr, tfdCIDIEIFI, pUC18Not derivative | This work |
| pJRC42 | Apr, tfdCIDIEIFI, pUC18Not derivative | This work |
| pUCLG | Apr, Ptfd-ItfdCIDI′, pUC18Not derivative | This work |
| pUCLG1 | Apr, Ptfd-ItfdCIDIEIFI, pUC18Not derivative | This work |
| pUCLG2 | Apr, tfdR/Ptfd-ItfdCIDIEIFI, pUC18Not derivative | This work |
| pUCLG4 | Apr, tfdR/Ptfd-ItfdCIDI′, pUC18Not derivative | This work |
| pUCPM-I | Apr, tfdR/Ptfd-ItfdCIDIEIFI, pUCP19 derivative | This work |
| pUCDP1 | Apr, pJP4 EcoRI-E fragment, pUC18 derivative | This work |
| pUCDP2 | Apr, pJP4 EcoRI-G fragment, pUC18 derivative | This work |
| pBSDP3 | Apr, tfdR/Ptfd-IItfdDIICIIEII′, pBSKSII derivative | This work |
| pBSDP4 | Apr, tfdR/Ptfd-IItfdDIICIIEIIFII, pBSKSII derivative | This work |
| pBBR1M-I | Kmr, tfdR/Ptfd-ItfdCIDIEIFI, pBBR1MCS-2 derivative | This work |
| pBBR1M-II | Kmr, tfdR/Ptfd-IItfdDIICIIEIIFII, pBBR1MCS-2 derivative | This work |
2,4-D and 3-CB, able to grow in 2,4-D and 3-CB, respectively; IncPα tra, IncP transference functions; tfd, catabolic genes from pJP4; tfdR, regulatory gene of pJP4; Ptfd-I, putative promoter region for the tfdCIDIEIFI cluster; Ptfd-II, putative promoter region for the tfdDIICIIEIIFII cluster; Ap, ampicillin; Tc, tetracycline; Km, kanamycin; Rf, rifampycin; Cf, cloramphenicol; Sm, streptomycin; pBSKSII, pBluescript II KS.
DSMZ, Deutsche Sammlung von Mikroorganismen und Zellkulturen, Braunschweig, Germany.
DNA manipulation.
Restriction, ligation, and dephosphorylation reactions and purification and electroporation of DNA were performed by standard procedures (1). Derivatives of the broad-host-range plasmid vector pBBR1MCS-2 were mobilized from E. coli to R. eutropha JMP222 or P. putida KT2442 by using a triparental mating with E. coli HB101(pRK600) as the helper strain. A donor-to-helper-to-recipient ratio of 1:1:2 was used. After incubation, cells were resuspended, and the transconjugants were selected on agar plates containing minimal medium with 3 mM benzoate plus 50 μg of kanamycin per ml. Plasmid pJP4 was transferred to P. putida by biparental mating with R. eutropha JMP134 as the donor, and selection was performed on LB agar plates containing 0.5 mM merbromin plus 50 μg of rifampycin per ml. The presence of pJP4 in P. putida transconjugants was determined by plasmid extraction.
Construction of a tfdR/Ptfd-ItfdCIDIEIFI gene module.
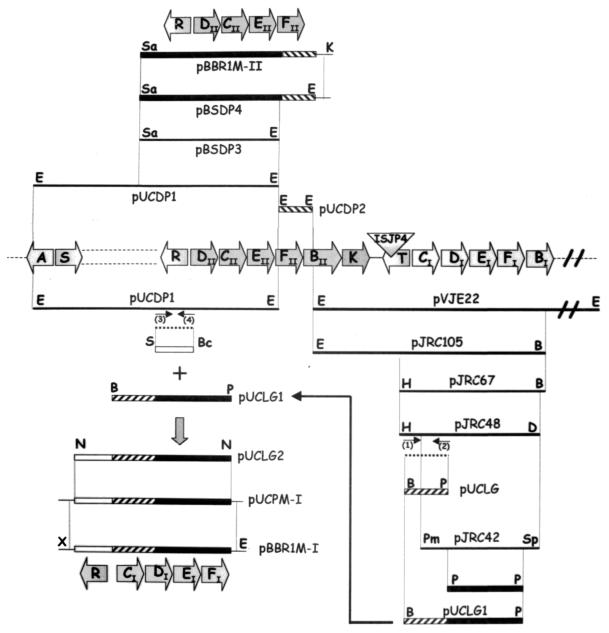
The tfdCIDIEIFI genes were cloned (Fig. 1) from the EcoRI-B pJP4 fragment previously inserted in plasmid pVJE22 (34). Plasmid pJRC105 was generated by cloning the 10.5-kb EcoRI/BamHI fragment of pVJE22 into pUC18Not. The 6.7-kb HpaI/BamHI fragment of pJRC105 was subcloned into pBluescript II KS digested with SmaI/BamHI to make pJRC67. An EcoRI/DraI digestion of pJRC67 produced a 4.8-kb fragment that was introduced into pUC18Not digested with EcoRI/HincII to form pJRC48. The insert in pJRC48 contains a 0.6-kb region upstream of the initiation codon of gene tfdIC. Finally, digestion of pJRC48 DNA with PmlI and SphI allowed cloning of tfdICDEF into pUC18Not digested with SmaI/SphI, to form pJRC42.
FIG. 1.
Scheme of cloning for tfdR/Ptfd-ItfdCIDIEIFI (bottom) and tfdR/Ptfd-IItfdDIICIIEIIFII (top). The genetic map of the 22-kb region of pJP4 containing the tfd genes is depicted. DNA fragments used in cloning steps are indicated by restriction enzyme sites and by the names of plasmids that contain them (Table 1). The drawing is not to scale. B, BamHI; Bc, BclI; D, DraI; E, EcoRI; H, HpaI; N, NotI; Pm, PmlI; P, PstI; S, SstI; Sa, SacI; Sp, SphI; X, XbaI. Dashed lines represent fragments obtained by PCR amplification. Small arrows indicate primer pairs: (1), PR-1; (2), VAL-2; (3), PR-2; (4), PR-3.
The tfdR/Ptfd-ItfdCIDIEIFI gene module was obtained as follows (Fig. 1). First, the PCR product with primer pairs PR-1 and VAL-2 (see below) from pJRC48 was digested with BamHI and PstI and was inserted into pUC18Not to give pUCLG. This 1.9-kb BamHI/PstI fragment includes a 219-base region upstream of the first nucleotide of tfdIC, and, therefore, contains the putative cis regulatory region (Ptfd-I), plus the tfdCIDI′ genes, but without the last 72 bases of tfdDI (i.e., tfdDI′). Then, a 2.2-kb PstI fragment from pJRC42 containing the last 72 bases of tfdDI plus tfdEIFI was cloned in pUCLG to give pUCLG1. The PCR product with primers PR-2 and PR-3 (see below) of the tfdR-containing EcoRI-E pJP4 fragment, previously cloned in pUC18 to give pUCDP1, was digested with SstI and BclI and was introduced into pUCLG1 to give pUCLG2.
Plasmid pUCLG2 containing tfdR/Ptfd-ItfdCIDIEIFI was digested with NotI, and 5′ overhangs of NotI fragment were filled in by Klenow DNA polymerase I (Gibco BRL) and were ligated to pUCP19 digested with SmaI to form pUCPM-I. An EcoRI/XbaI digestion of pUCPM-I produced a 5.1-kb fragment, containing the tfdR/Ptfd-ItfdCIDIEIFI gene module, that was introduced into pBBR1MCS-2 to generate pBBR1M-I.
To amplify Ptfd-ItfdCIDI′, primer pairs PR-1 (5′-CTGTCTTATTTCCAGGATCCGTCCCG-3′ [bp 127 to 152 of GenBank accession no. M35097/XO7754, modified to introduce a BamHI recognition sequence]) and VAL-2 (5′-GCCGTGGAATTCGCCAGTGGGAACCTGCAG-3′ [bp 2136 to 2165, modified to introduce an EcoRI recognition sequence]) were used. To amplify tfdR, primer pairs PR-2 (5′-CCACCAGGAGTGATCAATGGAGTTTCG-3′ [bp 40 to 66 of GenBank accession no. S80112, modified to create a BclI recognition sequence]) and PR-3 (5′-ACGTAGCCGAGCTCGCTATTTCTGTCCTTTCCCG-3′ [bp 984 to 1017, modified to create a SstI recognition sequence]) were used. Conditions for PCR were 95°C for 2 min; 35 cycles of 95°C for 45 s, 55°C for 30 s, and 72°C for 3 min; and then 72°C for 10 min.
Construction of a tfdR/Ptfd-IItfdDIICIIEIIFII gene module.
The tfdR and tfdDIICIIEIIFII genes with their intergenic region were cloned (Fig. 1) from the EcoRI-E and EcoRI-G pJP4 fragments previously inserted in pUC18 to give pUCDP1 and pUCDP2, respectively. pUCDP1 was digested with SacI and EcoRI to yield a 4.2-kb fragment containing tfdR, an intergenic region, and tfdDIICIIEII. This fragment was subcloned into pBluescript II KS digested with SacI/EcoRI, generating pBSDP3. The EcoRI-G fragment containing the tfdFII gene was obtained from pUCDP2 and was inserted in the EcoRI site of pBSDP3 to give pBSDP4. The plasmid pBSDP4 was digested with SacI/KpnI, and the 5.9-kb fragment containing the tfdR/Ptfd-IItfdDIICIIEIIFII gene module was introduced in pBBR1MCS-2 to give pBBR1M-II.
Enzyme assays.
For enzyme assays, cells were grown in minimal medium containing 3 mM 3-CB. Additionally, strains not able to grow in 3-CB were grown in minimal medium containing 3 mM benzoate plus the corresponding antibiotic and were induced at late exponential growth phase with 1 mM 3-CB for 3 h. About 100 ml of each culture was harvested at the end of the exponential phase and was centrifuged, washed twice, and resuspended in 5 ml of a solution containing 50 mM Tris-acetate (pH 7.5) and 1 mM MnSO4. Cells were disrupted by sonication (Vibracell; Sonics & Materials, Inc.). The soluble protein fraction was obtained after 1 h of centrifugation at 130,000 × g in a Beckman L-80 ultracentrifuge. Cell extracts (0.1 to 5.0 mg of protein per ml) were used without further purification. Assays contained 50 μM substrate, 33 mM Tris-acetate buffer (pH 7.5), 1 mM MnSO4, and a volume of crude extract corresponding to 1 to 100 μg of protein (0.002 to 0.02 enzyme units). One unit of enzyme activity was the amount of crude extract that forms or consumes 1 μmol of product or substrate, respectively, per min. Protein determinations were performed as previously described (2). Enzyme activities were determined in assays performed in a diode-array Hewlett Packard HP 8452-A UV/Vis spectrophotometer.
(i) Chlorocatechol-1,2-dioxygenase.
Chlorocatechol-1,2-dioxygenase activity was measured with 3,5-dichlorocatechol (3,5-DCC), 4-chlorocatechol (4-CC), or 3-chlorocatechol (3-CC) (Helix Biotechnology, Inc., Vancouver, British Columbia, Canada) as substrate by following product formation as indicated by OD260. The molar absorption coefficients were 2,4-dichloromuconate (2,4-DCM), ɛ260 = 12,000 M−1cm−1; 3-chloromuconate (3-CM), ɛ260 = 12,400 M−1cm−1; and 2-chloromuconate (2-CM), ɛ260 = 17,100 M−1cm−1, respectively (8).
(ii) Chloromuconate cycloisomerase.
Chloromuconate cycloisomerase activity was measured by substrate consumption (as indicated by OD260) with 2,4-DCM, 3-CM, or 2-CM. With 2,4-DCM, a reaction coefficient of ɛ260 = 5,800 M−1cm−1 was used (for explanation, see reference 19). These muconates were prepared by incubation of the corresponding chlorocatechols with a cell suspension of E. coli DH5α harboring plasmid pUCLG4. This pUC18Not derivative contains the tfdR/Ptfd-ItfdCIDI′ genes but lacks the last 72 bases of the gene coding for the chloromuconate cycloisomerase and, therefore, expresses an active chlorocatechol-1,2-dioxygenase but an inactive chloromuconate cycloisomerase. E. coli DH5α(pUCLG4) was grown in 50 ml of LB broth containing 100 μg of ampicillin per ml and 0.5 mM isopropyl-β-d-thiogalactopyranoside (IPTG). After overnight growth, 1 mM IPTG was added to the culture, and cells were incubated for an additional 2 h. Cells were pelleted, washed twice, and resuspended in 5 ml of 10 mM Tris-HCl, pH 8.5. The suspension was incubated in a shaker at 30°C for 120 min with three additions of a 0.5 mM solution of the respective chlorocatechol, performed at 0, 15, and 60 min. After 120 min, the cell suspension was centrifuged, and the supernatant was stored at −20°C. Quantitative transformation of chlorocatechols and no formation of dienelactones were verified by high-pressure liquid chromatography analysis. This was carried out with an LC-10AD Shimadzu liquid chromatograph equipped with an SC 125- by 4.6-cm Lichrospher 100 RP8 5.0-μm-particle-size column (Bischoff, Leonberg, Germany) and by using as elution solvent an aqueous system containing 36% methanol plus 0.1% phosphoric acid.
(iii) Dienelactone hydrolase.
Dienelactone hydrolase activity was measured by consumption of cis-dienelactone as indicated by OD280 (ɛ280 = 17,000 M−1cm−1) (30). The assay contained 10 mM histidine-HCl buffer, pH 6.5, and 0.08 μM cis-dienelactone, in addition to crude extract. cis-Dienelactone was a gift of W. Reineke and S. Kaschabek (15).
(iv) Maleylacetate reductase.
Maleylacetate reductase was measured by consumption of NADH as indicated by OD340 (ɛ340 = 6,300 M−1cm−1). The substrates were generated in situ with a crude extract of R. eutropha JMP134 pregrown in 2 mM 2,4-D. The diluted crude extract was incubated at room temperature with 100 μM of 4-CC or 3,5-DCC until complete conversion (approximately 120 min) to maleylacetate or chloromaleylacetate, respectively, had occurred. During the incubation, UV spectral changes corresponding to the complete removal of the chlorocatechol, along with formation of the maleylacetate (maximum OD at 245 to 253 nm) were observed. No signals for aromatic compounds were detected by gas chromatography-mass spectrometry analysis at the end of incubation. Only one peak, corresponding to 2-chloromaleylacetate or maleylacetate was observed by high-pressure liquid chromatography analysis (L. Padilla, V. Matus, P. Zenteno, and B. González, unpublished data). The formation of maleylacetates was also supported by the fact that no product was detected in incubations without chlorocatechol or in incubations with chlorocatechol plus NADH (because the maleylacetate was converted to β-ketoadipate). The final concentration of maleylacetate was 0.1 mM. These compounds were used immediately. After the incubation of the crude extract of R. eutropha JMP134, the maleylacetate reductase lost its activity. That was confirmed by the addition of NADH at the end of incubation. These control assays did not show spectral changes at 340 nm. Contribution of the NADH oxidation due to components other than maleylacetate was determined by adding fresh crude extract plus NADH to incubations performed without chlorocatechol, and this contribution was subtracted from reported measurements. The assay was started after the addition of 200 μM NADH and fresh crude extract.
RESULTS
Growth in 3-CB of derivatives of R. eutropha JMP222 and P. putida KT2442 containing pBBR1M-I or pBBR1M-II.
R. eutropha JMP222 derivatives containing pBBR1M-I were able to grow in liquid cultures with 3-CB as the sole carbon and energy source after 1 to 2 days of incubation. Surprisingly, R. eutropha JMP222 derivatives containing pBBR1M-II were also able to grow in liquid cultures with 3-CB after 4 to 5 days of incubation. The generation time for these derivatives growing in 3 mM 3-CB are shown in Table 2. R. eutropha JMP222(pBBR1M-I) grew about four times faster than strain JMP222(pBBR1M-II) and even faster than wild-type strain JMP134(pJP4). Of all the P. putida KT2442 derivatives, only the derivative containing pBBR1M-I was able to grow in liquid cultures of 3-CB, with a generation time about twice that of the corresponding R. eutropha strain (Table 2). P. putida derivatives containing pJP4 or pBBR1M-II were unable to grow in liquid cultures with 3-CB, even after 15 days of incubation. However, P. putida KT2442(pJP4) and strain KT2442(pBBR1M-II) formed small colonies on 3-CB agar plates. P. putida KT2442(pJP4) was also able to form small colonies on 2,4-D agar plates. These small colonies were clearly different from those occasionally seen in plates without a carbon source or those inoculated with strains without plasmids.
TABLE 2.
Growth in 3-CB of R. eutropha JMP134 and JMP222 and P. putida KT2442 derivatives containing tfdR/Ptfd-ItfdCIDIEIFI and/or tfdR/Ptfd-IItfdDIICIIEIIFII
| Strain/derivative | Growth in 3-CBa | Generation time (h) |
|---|---|---|
| R. eutropha JMP134(pJP4) | + | 5.0 |
| R. eutropha JMP222(pBBR1M-I) | + | 3.4 |
| R. eutropha JMP222(pBBR1M-II) | + | 13.4 |
| P. putida KT2442(pJP4) | − | Nonapplicable |
| P. putida KT2442(pBBR1M-I) | + | 6.0 |
| P. putida KT2442(pBBR1M-II) | − | Nonapplicable |
Growth was tested in liquid cultures containing 3 mM 3-CB incubated for up to 15 days. Growth was observed after 1 to 2 days of incubation for the wild-type strain and derivatives with pBBR1M-I and after 4 to 5 days of incubation for the derivative with pBBR1M-II.
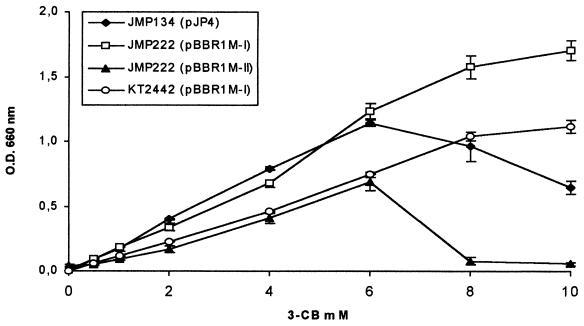
The effect of substrate concentration on 3-CB growth was studied with the four strains that grew in liquid cultures. R. eutropha JMP222 containing module I exhibited a better growth performance than the wild type at high 3-CB concentrations (Fig. 2). In contrast, the slow-growing derivative containing module II reached lower growth yields at concentrations up to 6 mM, then declined in yield at 8 and 10 mM 3-CB. P. putida (pBBR1M-I) exhibited a behavior similar to R. eutropha JMP222(pBBR1M-I), but with lower yields (Fig. 2).
FIG. 2.
Effect of substrate concentration in growth of R. eutropha and P. putida strains in 3-CB. OD was measured at stationary phase, i.e., 1 to 2 days of incubation for the wild type and derivatives with pBBR1M-I and 4 to 5 days of incubation for the derivative with pBBR1M-II. Values correspond to means ± deviation of triplicates. ⧫, R. eutropha JMP134(pJP4); □, R. eutropha JMP222(pBBR1M-I); ▴, R. eutropha(pBBR1M-II); and ○, P. putida KT2442(pBBR1M-I).
Expression of Tfd gene products in derivatives of R. eutropha JMP222 containing pBBR1M-I or pBBR1M-II.
To further study the role of tfd modules I and II in the degradation of 3-CB, the activity of Tfd enzymes was determined in derivatives of strain JMP222 containing only one of these modules. The activities of chlorocatechol-1,2-dioxygenase, chloromuconate cycloisomerase, dienelactone hydrolase, and maleylacetate reductase in crude extracts of strains JMP222(pBBR1M-I), JMP222(pBBR1M-II), and JMP134(pJP4) grown in 3 mM 3-CB are shown in Table 3. The activity for chlorocatechol-1,2-dioxygenase in strain JMP222(pBBR1M-I) was two times higher than the activity of the wild-type strain JMP134(pJP4) and was two to three times higher than the activity encoded in module II. A very low activity against chlorocatechols was found in the crude extract of the recipient strain JMP222 (Table 3), supporting the notion that both modules encode an active chlorocatechol-1,2-dioxygenase. The highest activity for chloromuconate cycloisomerase (Table 3) was also found in the crude extract of strain JMP222(pBBR1M-I). This activity was two to three times higher than that observed in the wild type and was more than four times higher than that observed in derivatives containing module II. No chloromuconate cycloisomerase activity was found in the recipient strain JMP222. Therefore, the differences in amount and specificity (see below) of chloromuconate cycloisomerase in both crude extracts are completely due to the tfd genes. Significant differences in substrate specificity of chloromuconate cycloisomerase were observed in cell extracts of R. eutropha derivatives containing module I or II. The chloromuconate cycloisomerase of module II showed higher ratios of 2,4-DCM to 3-CM and 2,4-DCM to 2-CM utilization (5.2 and 52, respectively [Table 3]) than the corresponding enzyme in module I (2.8 and 36, respectively [Table 3]). Another important difference was found for the dienelactone hydrolase activity (Table 3). The dienelactone hydrolase activity of module I was 50 times higher than that of module II. In contrast, the highest activity of maleylacetate reductase was detected in strain JMP222(pBBR1M-II). It was approximately 3 times higher than in the wild-type JMP134(pJP4). Maleylacetate reductase activity was observed at low rates in both JMP222 grown in 3 mM benzoate and in strain JMP222(pBBR1M-I) grown in 3-CB (Table 3). tfdFI was cloned under the control of the Ptac promoter in pVLT35, a medium-copy-number plasmid that replicates in R. eutropha (4), and was introduced in R. eutropha JMP222. No significant differences in maleylacetate reductase activity were found in IPTG-induced cells with respect to noninduced cells or cells of strain JMP222. It can therefore be concluded that a functional maleylacetate reductase was not produced from tfdFI at all, or it has a very low activity. The activity of maleylacetate reductase observed in JMP222 cells grown on benzoate and induced with 3-CB (Table 3) may correspond to the chromosomal activity involved in the ability of R. eutropha JMP222 to grow in 2,4,6-trichlorophenol (3; L. Padilla, V. Matus, P. Zenteno, and B. González, unpublished data).
TABLE 3.
Specific activities of enzymes encoded in modules tfdR/Ptfd-ItfdCIDIEIFI and tfdR/Ptfd-IItfdDIICIIEIIFII in crude extracts of R. eutropha JMP134 and R. eutropha JMP222 derivatives grown in 3 mM 3-CBa
| Enzyme | Substrate | Sp act of enzymes in R. eutropha strain:
|
|||
|---|---|---|---|---|---|
| JMP134(pJP4)a | JMP222b | JMP222(pBBR1M-I)a | JMP222(pBBR1M-II)a | ||
| CC-1,2-DO | 3,5-DCC | 0.46 ± 0.02 | <0.02 | 0.95 ± 0.23 | 0.32 ± 0.02 |
| 4-CC | 0.26 ± 0.03 | <0.02 | 0.47 ± 0.22 | 0.19 ± 0.02 | |
| 3-CC | 0.27 ± 0.04 | <0.01 | 0.52 ± 0.21 | 0.28 ± 0.02 | |
| CMCI | 2,4-DCM | 0.53 ± 0.03 | No activity | 1.08 ± 0.13 | 0.26 ± 0.008 |
| 3-CM | 0.13 ± 0.01 | No activity | 0.39 ± 0.02 | 0.05 ± 0.002 | |
| 2-CM | 0.01 ± 0.001 | No activity | 0.03 ± 0.002 | <0.005 | |
| DLH | DL | 0.61 ± 0.11 | No activity | 1.96 ± 0.25 | 0.04 ± 0.008 |
| MAR | MA | 0.43 ± 0.02 | 0.02 ± 0.01 | 0.03 ± 0.01 | 1.57 ± 0.03 |
| 2-CMA | 0.32 ± 0.01 | 0.02 ± 0.01 | 0.02 ± 0.01 | 1.12 ± 0.02 | |
Cells grown in 3 mM 3-CB.
Cells grown in 3 mM benzoate and induced with 3-CB. Values for R. eutropha JMP134 grown on 3 mM benzoate and induced with 3-CB were approximately 20 to 30% (T. Ledger, C. Varela, R. Céspedes, D. Pérez-Pantoja, L. Guzmán, D. Pieper, and B. González, unpublished data) of those found in cell extracts grown on 3-CB CC-1,2-DO, chlorocatechol-1,2-dioxygenase; CMCI, chloromuconate cycloisomerase; DLH, dienelactone hydrolase; MAR, maleylacetate reductase: DL, cis-dienelactone; MA, maleylacetate; and 2-CMA, 2-chloromaleylacetate. Values are expressed as units per milligram and correspond to averages of two or three independent experiments.
P. putida derivatives were studied for expression of chlorocatechol dioxygenase. Crude extracts of cells grown in 3-CB or in benzoate and induced with 3-CB showed specific activities that were 25 to 50% lower than those of the corresponding R. eutropha derivatives.
DISCUSSION
This work showed that both tfd modules found in R. eutropha JMP134(pJP4) encode functional enzymes for chlorocatechol metabolism. High levels of activity were found for enzymes encoded by genes tfdCI, tfdCII, tfdDI, tfdDII, tfdEI, and tfdFII. Low activity levels were found for the enzyme encoded by tfdEII. No, or very low, activity was observed for the enzyme encoded by tfdFI. This work also showed that cloning of each module in the medium-copy-number plasmid vector pBBR1MCS-2 and introduction of each module into strains that accumulate chlorocatechols from 3-CB allow such derivatives to grow in it. In order to study the role in chlorocatechol degradation of the two tfd modules harbored in pJP4, we chose to clone each of these modules into a medium-copy-number plasmid vector that replicates in R. eutropha and P. putida. Two reasons explain that choice. First, preliminary work performed with tfdR/Ptfd-ItfdCIDIEIFI and tfdR/Ptfd-IItfdDIICIIEIIFII in E. coli showed very little expression of Tfd enzymes. Second, introduction of a single copy of these tfd gene modules into the R. eutropha chromosome, by using miniTn5-derived vectors, also gave very low expression of the Tfd enzymes (T. Ledger, C. Varela, R. Céspedes, D. Pérez-Pantoja, L. Guzmán, D. Pieper, and B. González, unpublished data). These R. eutropha derivatives have 2 to 10% of the enzyme activity levels found in the wild-type strain or in derivatives containing module II or I in pBBRMCS-2 (this work) and were not able to grow in 3-CB (T. Ledger, C. Varela, R. Céspedes, D. Pérez-Pantoja, L. Guzmán, D. Pieper, and B. González, unpublished data). Both chlorocatechol metabolism gene modules were cloned under the control of tfdR. The role of tfdR, located upstream of tfdDIICIIEIIFII, as a regulator of the tfdA gene and tfdCIDIEIFI and tfdDIICIIEIIFII gene modules has been proposed (13, 21, 24). Leveau and van der Meer (21) showed that expression of tfdCI is activated in R. eutropha by tfdR, which was previously thought to encode a repressor protein, whereas tfdT, located upstream of tfdCIDIEIFI, does not encode a functional regulatory protein, as it is inactivated by insertion of the ISJP4 element. The presence of tfdR ensures proper recognition of Ptfd-I, and Ptfd-II, the putative promoter sequences for modules I and II, respectively.
The introduction of each tfdR-regulated module into bacterial strains that accumulate chlorocatechols from 3-CB allowed us to assess the role of these modules for the degradation of chloroaromatics. Two observations arose from this part of the work. First, the introduction of module I resulted in a more-efficient 3-CB-degrading phenotype than introduction of module II. Second, the growth properties related to tfd genes were better expressed in the R. eutropha strain than in the P. putida strain. The last observation can be explained by the expected differences in gene expression and gene background between a homologous and a heterologous system. The first observation, however, may be explained in several ways. One of them is that transcriptional activation of module I is higher than that of module II. However, steady-state mRNA levels of transcripts from modules I and II show no obvious difference (23). Another possibility is that the very low activity of TfdDII with 2-CM or TfdEII becomes the rate-limiting step in catabolism of 3-CB by strains containing module II.
The expression of chlorocatechol metabolism enzymes was studied in R. eutropha JMP222 derivatives containing module II or I that were able to grow in 3-CB. Both modules expressed a chlorocatechol-1,2-dioxygenase activity whose substrate specificity resembled that of the wild-type strain. It has been recently reported that both genes are transcribed during growth of strain JMP134 in a chemostat fed with 2,4-D (23). Therefore, it is highly probable that both genes play a role in catabolism of 3-CB. Both modules express the second enzyme of the pathway, chloromuconate cycloisomerase. Although sequence comparisons indicate that the tfdDII gene is clustered apart from the other chloromuconate cycloisomerase genes reported in gram-negative bacteria (9), both chloromuconate cycloisomerase activities possess the higher dichlorinated-to-monochlorinated substrate activity ratio of all known chloromuconate cycloisomerases (36). However, the enzyme encoded in module II has a more pronounced preference for 2,4-DCM with very little activity toward 2-CM. In this context, it is worth mentioning that 2-CM is the main muconate formed during catabolism of 3-CB (28). This may be the main reason for the poor growth on 3-CB observed with module II. The activities of dienelactone hydrolases were also tested in this work. There was a significant (50-fold) difference of this activity in both modules. However, as accumulation of dienelactones was never observed during transformation of 2,4-DCM, 3-CM, or 2-CM, it can be assumed that the activity of each dienelactone hydrolase is not rate limiting. Another important difference between enzyme expression from both tfd modules was observed in the activity of maleylacetate reductase. Maleylacetate reductase encoded by tfdFII was, by far, more active than tfdFI. Although similar steady-state levels of mRNAs corresponding to both tfdFI and tfdFII are found in R. eutropha JMP134 growing in 2,4-D (23), it should be noted that the NH2-terminal sequence of the maleylacetate reductase purified from a culture of R. eutropha JMP134 grown in 2,4-D (32) perfectly matched the amino acid sequence deduced for tfdFII (GenBank accession no. U16782). Therefore, the expression and substrate specificity of the enzyme encoded by tfdFII makes it highly probable that this maleylacetate reductase plays the main role in 3-CB, as well as 2,4-D, metabolism. Our observations may explain early reports of no phenotype associated with the tfdFI gene (7). It should be noted that derivatives containing module I, and therefore not having a maleylacetate reductase encoded in the plasmid, grow similarly to the wild type. The expression, at low levels, of a chromosomally encoded maleylacetate reductase (Table 3) (20, 32; L. Padilla, V. Matus, P. Zenteno, and B. González, unpublished data) should be enough to support growth of such derivatives in 3-CB. The higher level of activity of the maleylacetate reductase encoded in module II may be needed for growth in 2,4-D (but not for 3-CB), since this carbon source produces 2-chloromaleylacetate which is not as good a substrate as maleylacetate and requires two steps catalyzed by this enzyme (16).
The expression profile of Tfd enzymes reported here may explain the presence of both tfd modules in pJP4 (a very unusual feature in genes encoding chloroaromatic metabolism) with both modules complementing each other (e.g., tfdCIDIEI plus tfdFII).
ACKNOWLEDGMENTS
M. Schlömann and H.-J. Knackmuss kindly provided pVJE22 and R. eutropha JMP222, respectively. We thank M. Klemba for helpful discussion.
This work was supported by grants 1960262 and 8990004 from FONDECYT-Chile and by the collaborative grant 95005 from FUNDACION ANDES/CONICYT, Chile, and BMBF-FZK/Karlsruhe, Germany. L. Guzmán was supported by postdoctoral FONDECYT grant 3970030.
REFERENCES
- 1.Ausubel F, Brent R, Kingston R, Moore D, Seidman J, Smith J, Struhl K, editors. Short protocols in molecular biology. 2nd ed. New York, N.Y: Greene Publishing Associates; 1992. [Google Scholar]
- 2.Bradford M M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1976;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- 3.Clément P, Matus V, Cárdenas L, González B. Degradation of trichlorophenols by Alcaligenes eutrophus JMP134. FEMS Microbiol Lett. 1995;127:51–55. doi: 10.1111/j.1574-6968.1995.tb07449.x. [DOI] [PubMed] [Google Scholar]
- 4.de Lorenzo V, Eltis L, Kessler B, Timmis K N. Analysis of Pseudomonas gene products using lacIq/Ptrp-lac plasmids and transposons that confer conditional phenotypes. Gene. 1993;123:17–24. doi: 10.1016/0378-1119(93)90533-9. [DOI] [PubMed] [Google Scholar]
- 5.Don R H, Pemberton J M. Properties of six pesticide degradation plasmids isolated from Alcaligenes paradoxus and Alcaligenes eutrophus. J Bacteriol. 1981;145:681–686. doi: 10.1128/jb.145.2.681-686.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Don R H, Pemberton J M. Genetic and physical map of the 2,4-dichlorophenoxyacetic acid-degradative plasmid pJP4. J Bacteriol. 1985;161:466–468. doi: 10.1128/jb.161.1.466-468.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Don R H, Weightman A J, Knackmuss H-J, Timmis K N. Transposon mutagenesis and cloning analyses of the pathways for degradation of 2,4-dichlorophenoxyacetic acid and 3-chlorobenzoate in Alcaligenes eutrophus JMP134 (pJP4) J Bacteriol. 1985;161:85–90. doi: 10.1128/jb.161.1.85-90.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dorn E, Knackmuss H-J. Chemical structure and biodegradability of halogenated aromatic compounds. Substituent effects on 1,2-dioxygenation of catechol. Biochem J. 1978;174:85–94. doi: 10.1042/bj1740085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Eulberg D, Kourbatova E M, Golovleva L A, Schlömann M. Evolutionary relationship between chlorocatechol catabolic enzymes from Rhodococcus opacus 1CP and their counterparts in proteobacteria: sequence divergence and functional convergence. J Bacteriol. 1998;180:1082–1094. doi: 10.1128/jb.180.5.1082-1094.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fukumuri F, Hausinger R P. Alcaligenes eutrophus JMP134 “2,4-dichlorophenoxyacetate monooxygenase” is an α-ketoglutarate-dependent dioxygenase. J Bacteriol. 1993;175:2083–2086. doi: 10.1128/jb.175.7.2083-2086.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fukumuri F, Hausinger R P. Purification and characterization of 2,4-dichlorophenoxyacetate/α-ketoglutarate dioxygenase. J Biol Chem. 1993;268:24311–24317. [PubMed] [Google Scholar]
- 12.Herrero M, de Lorenzo V, Timmis K N. Transposon vectors containing non-antibiotic selection markers for cloning and stable chromosomal insertion of foreign DNA in gram-negative bacteria. J Bacteriol. 1990;172:6557–6567. doi: 10.1128/jb.172.11.6557-6567.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kaphammer B, Kukor J J, Olsen R H. Regulation of tfdCDEF by tfdR of the 2,4-dichlorophenoxyacetate acid degradation plasmid pJP4. J Bacteriol. 1990;172:2280–2286. doi: 10.1128/jb.172.5.2280-2286.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kasberg T, Daubaras D L, Chakrabarty A M, Kinzelt D, Reineke W. Evidence that operons tcb, tfd, and clc encode maleylacetate reductase, the fourth enzyme of the modified ortho pathway. J Bacteriol. 1995;177:3885–3889. doi: 10.1128/jb.177.13.3885-3889.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kaschabek S R, Reineke W. Synthesis of bacterial metabolites from haloaromatic degradation. 1. Fe(III)-catalyzed peracetic oxidation of halocatechols, a facile entry to cis,cis-2-halo-2,4-hexadienedioic acids and 3-halo-5-oxo-2(5H)-furanylideneacetic acids. J Org Chem. 1994;59:4001–4003. [Google Scholar]
- 16.Kaschabek S R, Reineke W. Maleylacetate reductase of Pseudomonas sp. strain B13: specificity of substrate conversion and halide elimination. J Bacteriol. 1995;177:320–325. doi: 10.1128/jb.177.2.320-325.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kovach M E, Elzer P H, Hill D S, Robertson G T, Farris M A, Roop II R M, Peterson K M. Four new derivatives of the broad-host-range cloning vector pBBR1MCS, carrying different antibiotic-resistance cassettes. Gene. 1995;166:175–176. doi: 10.1016/0378-1119(95)00584-1. [DOI] [PubMed] [Google Scholar]
- 18.Kröckel L, Focht D. Construction of chlorobenzene-utilizing recombinants by progenitive manifestation of a rare event. Appl Environ Microbiol. 1987;53:2470–2475. doi: 10.1128/aem.53.10.2470-2475.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kuhm A E, Schlömann M, Knackmuss H-J, Pieper D. Purification and characterization of dichloromuconate cycloisomerase from Alcaligenes eutrophus JMP134. Biochem J. 1990;266:877–883. [PMC free article] [PubMed] [Google Scholar]
- 20.Kukor J, Olsen R, Siak J-S. Recruitment of a chromosomally encoded maleylacetate reductase for degradation of 2,4-dichlorophenoxyacetic acid by the plasmid pJP4. J Bacteriol. 1989;171:3385–3390. doi: 10.1128/jb.171.6.3385-3390.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Leveau J H, van der Meer J R. The tfdR gene product can successfully take over the role of the insertion element-inactivated tfdT protein as a transcriptional activator of the tfdCDEF gene cluster, which encodes chlorocatechol degradation in Ralstonia eutropha JMP134 (pJP4) J Bacteriol. 1996;178:6824–6832. doi: 10.1128/jb.178.23.6824-6832.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Leveau J H, Zehnder A J, van der Meer J R. The tfdK gene product facilitates uptake of 2,4-dichlorophenoxyacetate by Ralstonia eutropha JMP134 (pJP4) J Bacteriol. 1998;180:2237–2243. doi: 10.1128/jb.180.8.2237-2243.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Leveau J H, Konig F, Fuchslin H, Werlen C, van der Meer J R. Dynamics of multigene expression during catabolic adaptation of Ralstonia eutropha JMP134 (pJP4) to the herbicide 2,4-dichlorophenoxyacetate. Mol Microbiol. 1999;33:396–406. doi: 10.1046/j.1365-2958.1999.01483.x. [DOI] [PubMed] [Google Scholar]
- 24.Matrubutham U, Harker A R. Analysis of duplicated gene sequences associated with tfdR and tfdS in Alcaligenes eutrophus JMP134. J Bacteriol. 1994;176:2348–2353. doi: 10.1128/jb.176.8.2348-2353.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Perkins E J, Gordon M P, Cáceres O, Lurquin P F. Organization and sequence analysis of the 2,4-dichlorophenol hydroxylase and dichlorocatechol oxidative operons of plasmid pJP4. J Bacteriol. 1990;172:2352–2359. doi: 10.1128/jb.172.5.2351-2359.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pieper D H, Reineke W, Engesser K H, Knackmuss H-J. Metabolism of 2,4-dichlorophenoxyacetic acid, 4-chloro-2-methylphenoxyacetic acid and 2-methylphenoxyacetic acid by Alcaligenes eutrophus JMP134. Arch Microbiol. 1988;150:95–102. [Google Scholar]
- 27.Pieper D H, Engesser K H, Knackmuss H-J. Regulation of catabolic pathways of phenoxyacetic acids and phenols in Alcaligenes eutrophus JMP134. Arch Microbiol. 1989;151:356–371. [Google Scholar]
- 28.Pieper D H, Knackmuss H-J, Timmis K N. Accumulation of 2-chloromuconate during metabolism of 3-chlorobenzoate by Alcaligenes eutrophus JMP134. Appl Microbiol Biotechnol. 1993;39:563–567. [Google Scholar]
- 29.Reineke W, Knackmuss H-J. Chemical structure and biodegradability of halogenated aromatic compounds. Substituent effects on 1,2-dioxygenation of benzoic acid. Biochim Biophys Acta. 1978;542:412–433. doi: 10.1016/0304-4165(78)90372-0. [DOI] [PubMed] [Google Scholar]
- 30.Schlömann M, Schmidt E, Knackmuss H-J. Different types of dienelactone hydrolase in 4-fluorobenzoate-utilizing bacteria. J Bacteriol. 1990;172:5112–5118. doi: 10.1128/jb.172.9.5112-5118.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Schweizer H P. Escherichia-Pseudomonas shuttle vectors derived from pUC18/19. Gene. 1991;97:109–112. doi: 10.1016/0378-1119(91)90016-5. [DOI] [PubMed] [Google Scholar]
- 32.Seibert V, Stadler-Fritzsche K, Schlömann M. Purification and characterization of maleylacetate reductase from Alcaligenes eutrophus JMP134 (pJP4) J Bacteriol. 1993;175:6745–6754. doi: 10.1128/jb.175.21.6745-6754.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Streber W, Timmis K N, Zenk M H. Analysis, cloning and high-level expression of 2,4-dichlorophenoxyacetate monooxygenase gene tfdA of Alcaligenes eutrophus JMP134. J Bacteriol. 1987;169:2950–2955. doi: 10.1128/jb.169.7.2950-2955.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Streber W. Der bakterielle Metabolismus des herbizids 2,4-D: Struktur und Expression des Gens tfdA aus Alcaligenes eutrophus JMP134. M.Sc. thesis. Munich, Germany: Ludwig-Maximilians- Universität; 1987. [Google Scholar]
- 35.Vollmer M D, Schlömann M. Conversion of 2-chloro-cis,cis-muconate and its metabolites 2-chloro- and 5-chloromuconolactone by chloromuconate cycloisomerases of pJP4 and pAC27. J Bacteriol. 1995;177:2938–2941. doi: 10.1128/jb.177.10.2938-2941.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Vollmer M D, Schell U, Seibert V, Lakner S, Schlömann M. Substrate specificities of the chloromuconate cycloisomerases from Pseudomonas sp. B13, Ralstonia eutropha JMP134 and Pseudomonas sp. P51. Appl Microbiol Biotechnol. 1999;51:598–605. doi: 10.1007/s002530051438. [DOI] [PubMed] [Google Scholar]