Introduction
Herpes simplex virus type-1 (HSV-1) is an alpha herpesvirus that infects over 60% of the human population [1]. Infection results in a variety of disease manifestations, including cold sores, encephalitis, and keratitis. The HSV-1 genome contains approximately 152 kb of double-stranded DNA and includes over 80 genes [2,3], which are sequentially transcribed by cellular RNA polymerase II (Pol II) [4,5]. Recently, studies using direct RNA sequencing, long-read sequencing, and ribosome profiling have revealed the transcriptional complexity of the HSV-1 genome and demonstrate that the genome actually contains over 200 open reading frames [6–8]. Viral genes are classified into 4 groups depending on their expression kinetics, including immediate early (α), early (β), leaky-late (γ1), and true late (γ2) genes. HSV-1 DNA replication is a key point in the infectious cycle, as it enables γ2 and amplifies γ1 transcription [9,10]. Below, we discuss the studies that defined the 4 HSV-1 gene classes and examine how viral DNA replication may facilitate a switch to regulate γ (γ1/γ2) gene transcription. Although the HSV-1 gene expression cascade was identified over 40 years ago, high-throughput sequencing approaches continue to reveal new insight into how each gene class is regulated [11–15].
Temporal and replication-dependent expression of HSV-1 genes
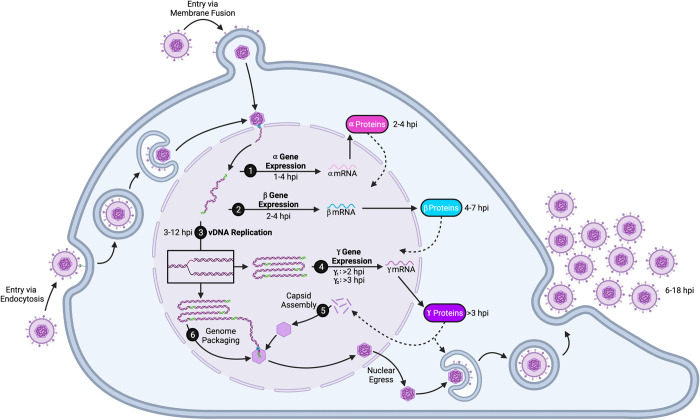
HSV-1 genes are temporally expressed during lytic infection. Taking advantage of synchronous infection during high multiplicity infection, the temporal expression of viral genes was previously characterized (Fig 1). Early studies demonstrated that α and β polypeptide levels peak between 3 to 4 and 5 to 7 hours post infection (hpi), respectively [16]. γ polypeptides are detected after 3 hpi, and levels continue to increase up to 12 hpi. More recently, RNA sequencing (RNA-seq) was used to measure HSV-1 mRNA levels throughout lytic infection [11]. α mRNAs are detected by 1 hpi and, in general, peak around 4 hpi. β mRNAs are detected by 2 hpi and decline after the onset of viral DNA replication (between 3 to 4 hpi). γ1 mRNA is expressed around 2 hpi, and expression is amplified by viral DNA replication. Additionally, γ2 mRNA is detected following the onset of DNA replication, with increasing levels observed through 16 hpi. The exact timing of the virus life cycle may vary depending on the cell type.
Fig 1. The HSV-1 infectious cycle.
Virions enter the cell via fusion with the host-cell membrane or endocytosis. The capsid docks at the nuclear pore and viral DNA enters the nucleus through the portal in the capsid (drawn in teal above). The HSV-1 genome is then expressed: (1) α genes are expressed upon entry; (2) α gene products drive β transcription; (3) after β protein expression, vDNA replication occurs; (4) γ1 gene expression is amplified, and γ2 gene expression is enabled by DNA replication; (5) capsid proteins, which are γ gene products, assemble; and then (6) replicated genomes are packaged into capsids through the portal. Packaged capsids leave the nucleus through nuclear egress and move through the secretory pathway, obtain an envelope spiked with viral glycoproteins, and nascent virions are released from the cell by exocytosis. Transcript and protein timing pictured above are approximated based on previous studies [11,16]. Note that the viral genome forms concatemers during replication as indicated above, and unit length genomes (flanked by green) are packaged into capsids. Also note that some models indicate that the viral genome circularizes after entry into the nucleus. Tegument proteins were omitted for simplicity. Figure created using www.biorender.com. hpi, hours post infection; HSV-1, herpes simplex virus type-1.
Further experiments were conducted to define the expression requirements of each gene class. It was found that transcription of α genes does not require de novo viral protein synthesis [16], indicating that cellular proteins and proteins brought into the cell with the infecting virion are sufficient to stimulate α gene expression. On the other hand, both β and γ protein synthesis depend on α protein expression [9,16], including the major viral transcription factor ICP4 [17]. Treatment of infected cells with viral DNA synthesis inhibitors results in the selective inhibition of γ viral gene expression [18–20]. γ1 transcripts are expressed in the absence of viral DNA replication and levels increase in a replication-dependent manner [12]. It is possible that this occurs because an increased number of viral genomes within the cell are available to serve as additional templates for transcription. γ2 transcription, however, is dependent on viral DNA replication, and viral transcripts are not detectable in the absence of viral DNA replication. This implies that replication results in a switch to enable their transcription. Taken together, these observations indicate that the HSV-1 gene expression cascade is highly coordinated during lytic infection.
Classification of HSV-1 late genes
RNA-seq was used to globally classify which viral mRNAs are expressed in a replication-dependent manner by comparing expression of viral genes between HSV-1 lab strain KOS and a UL30 (HSV-1 DNA polymerase) mutant that is defective for viral DNA synthesis [12]. Twenty-two γ1 mRNAs and 16 γ2 mRNAs were identified as either having decreased (γ1) or negligible (γ2) expression during infection with the UL30 mutant compared to the lab strain (Table 1). These results are consistent with previous reports, reviewed in [21], with the exception that UL42 was more recently classified as a γ1, rather than a β gene [12]. In general, α genes encode proteins involved in viral transcription regulation, β genes encode proteins that facilitate viral genome replication, and γ1 and γ2 genes encode factors involved in virion assembly and exit. Taken together, the regulatory cascade synthesizes viral proteins in an as needed manner during the HSV-1 infectious cycle.
Table 1. Classification of leaky late and late genes, along with their general functions [12].
| Gene class | Gene name | Protein identity | Function |
|---|---|---|---|
|
Leaky late (γ1) |
UL18 | VP23 | Capsid proteins |
| UL19 | VP5/ICP5 | ||
| UL35 | VP26 | ||
| UL26.5 | UL26.5 | Capsid scaffold protein | |
| UL32 | UL32 | Packaging protein | |
| UL21 | UL21 | Tegument proteins | |
| UL36 | UL36 | ||
| UL41 | VHS | ||
| UL46 | VP11/12 | ||
| UL48 | VP16 | ||
| UL49 | VP22 | ||
| US9 | US9 | ||
| US10 | US10 | ||
| UL27 | gB | Membrane glycoproteins | |
| US4 | gG | ||
| US6 | gD | ||
| US7 | gI | ||
| US8 | gE | ||
| UL45 | UL45 | Integral membrane protein | |
| UL11 | UL11 | Egress proteins | |
| UL34 | UL34 | ||
| UL42 | UL42 | DNA polymerase processivity factor | |
|
Late (γ2) |
UL38 | VP19c | Capsid protein |
| UL25 | UL25 | Packaging protein | |
| UL37 | UL37 | Capsid assembly proteins | |
| UL3 | UL3 | Tegument proteins |
|
| UL16 | UL16 | ||
| UL47 | VP13/14 | ||
| UL51 | UL51 | ||
| US2 | US2 | ||
| US11 | Vmw21 | ||
| UL1 | gL | Membrane glycoproteins | |
| UL10 | gM | ||
| UL44 | gC | ||
| UL49A | gN | ||
| US5 | gJ | ||
| UL31 | UL31 | Egress protein |
Initial rounds of HSV-1 DNA replication are sufficient to license late viral gene expression
An important question is whether ongoing DNA replication is continuously required for γ genes to be expressed. To address this, viral DNA replication was inhibited with acyclovir at 0, 2, 3, 4, or 6 hpi, and the effects on viral gene expression were determined by RNA-seq at 12 hpi [22]. If acyclovir was added before the onset of viral DNA replication (0 or 2 hpi), genome replication did not occur and γ2 mRNAs were not expressed. However, if acyclovir was added at 3 or 4 hpi, allowing for 1 to 2 rounds of viral DNA replication as measured by real-time PCR to quantify viral genome number, γ2 mRNA levels observed at 12 hpi were similar to an untreated control. These data may indicate that DNA replication is not continuously required for γ2 gene expression, and that the alterations that enable their expression occur during initial rounds of replication. It is possible that alterations in viral chromatin or the architecture of the viral genome enable this switch. However, it was recently demonstrated that although chromatin dynamics dictate the transcriptional competency during the onset of lytic infection, it does not appear to play a role in regulating the temporal expression of individual classes of viral genes [13,14].
Transcription factors associate with nascent viral DNA
Experiments conducted to identify proteins that interact with the HSV-1 genome during viral replication reveal insight into the potential mechanism through which γ gene expression is regulated. Using an adaptation of iPOND (isolation of proteins on nascent DNA), replicating viral DNA was pulse labeled with the nucleoside analog 5-ethynyl-2′-deoxycytidine (EdC) to enable subsequent tagging and purification of replicated viral DNA, followed by the identification of associated proteins by mass spectrometry [22]. In addition to the viral replication machinery, cellular Pol II and transcription regulatory proteins were enriched on EdC-labeled viral replication forks.
iPOND was also used to compare the relative enrichment of individual proteins at viral replication forks with that of nascent viral DNA post replication [22]. For this experiment, an EdC pulse followed by a chase with 2′-deoxycytidine was conducted. This enabled purification of EdC-labeled viral DNA during (pulse) or post (chase) replication. The Mediator complex and the basal transcription factors TFIID and TATA binding protein (TBP) were more enriched on pulse-labeled viral DNA than chased. Pol II and ICP4 were equally enriched on both populations of nascent viral DNA. Additionally, factors involved in cotranscriptional RNA processing were more enriched on chased DNA. The relative enrichment of promoter-binding factors including Mediator, TBP, and TFIID on pulse-labeled DNA; ICP4 and Pol II on pulse-labeled and chased DNA; and RNA processing factors on chased DNA may indicate that the act of DNA replication enables transcription factor binding to promoters, resulting in transcription initiation.
Late gene promoters and transcription factor binding
Transcription reporters have been used to map key regions of HSV-1 γ2 gene promoters that are necessary for replication-dependent transcription (Fig 2). The region immediately upstream from the transcription start site and another region downstream in the 5′ untranslated region of the gene are responsible for replication-dependent expression of representative γ2 genes [23–26]. This region contains a TATA box, initiator (Inr) element, and downstream activation sequence (DAS), which help to facilitate TBP and TFIID binding to γ2 gene promoters in an ICP4-dependent manner [12,13,27–29]. The structure of γ1 genes are less well understood, but representative promoters contain SP1 binding sites, a TATA box, and an Inr element [30]. It is likely that the difference between the cis elements of γ1 and γ2 promoters contributes to the differences in their dependence on DNA replication.
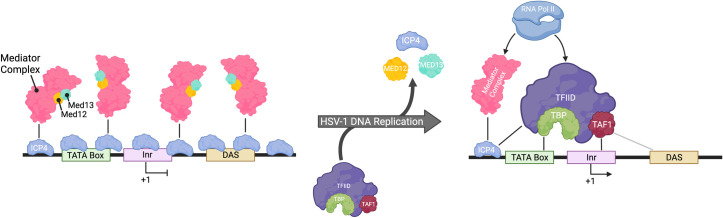
Fig 2. Model of HSV-1 γ2 gene promoter interactions before and after DNA replication.
HSV-1 γ2 promoter regions contain a TATA box, Inr element, and DAS. The TATA box is within the ‒34 to ‒24 region in the promoter depending on the gene. The Inr flanks the +1 site and the DAS is in the 5′ untranslated region. Prior to DNA replication, ICP4 coats γ2 genes in a sequence-independent manner. ICP4 recruits the Mediator complex containing Med12 and Med13, which are members of the Mediator kinase domain that can inhibit transcription. Following the onset of DNA replication, ICP4 binding to γ2 genes decreases. This may expose the promoter, enabling ICP4 to recruit TFIID containing TBP and TAF1 to the promoter region. It is possible that loss of the Mediator kinase domain following replication licenses γ2 transcription. Collectively, Mediator and TFIID may recruit Pol II to γ2 genes. Note that this is a proposed model based on current data and other factors likely play a role in this process. Figure created using www.biorender.com. DAS, downstream activation sequence; HSV-1, herpes simplex virus type-1, hpi, hours post infection; Pol II, RNA polymerase II; TBP, TATA binding protein.
To better understand the mechanisms behind HSV-1 gene expression, chromatin immunoprecipitation followed by sequencing (ChIP-seq) was used to map TFIID, TBP, and Pol II binding to viral DNA before and after replication [12]. Acyclovir was also used to investigate transcription factor binding in the absence of viral DNA replication. The binding of TBP and TFIID subunit TAF1, 2 general transcription factors that recruit Pol II to γ2 promoters, was enabled by viral DNA replication. TBP binds to the TATA box sequence and TAF1 was enriched near the Inr element of γ2 viral genes after the onset of viral DNA replication. In the absence of DNA replication, Pol II was deficient on the promoters and bodies of γ2 genes [12,15]. A single genome duplication event was sufficient to recruit TBP, TAF1, and Pol II to γ2 genes[12], consistent with previous findings [22]. Additionally, continuous ICP4 expression is required for γ gene expression [13]. These data suggest that DNA replication induces a change to the viral genome architecture, thus increasing the availability of silent γ2 gene promoters to general transcription factors resulting in robust transcription.
Using ChIP-Seq, it was found that prior to DNA replication, ICP4 coats the HSV-1 genome at a high density [13]. Following replication, ICP4 is less abundant on the genome, as an increasing number of genomes may compete for the limited amount of ICP4 within the cell. How changes to ICP4 binding to viral DNA before and after the onset of replication facilate γ2 gene transcription is not completely understood. One possibility is that ICP4 recruits Mediator to γ2 genes before the onset of viral DNA replication in a form that may inhibit transcription (Fig 2). In support of this model, Mediator containing the kinase domain has been found to copurify with ICP4 [31]. In addition, in the EdC pulse-chase studies described above, Med12 and Med13 subunits of the Mediator complex associated with replicated viral DNA decreased substantially with increasing time after EdC labeling [22]. These subunits are part of the kinase domain, which blocks Mediator interaction with Pol II to inhibit transcription initiation. Replication may induce a change in the form of Mediator associated with viral DNA enabling a transcriptional switch.
Remaining questions
The coupling of HSV-1 γ2 gene expression to DNA replication is an intriguing concept with several remaining questions. Interestingly, other viruses such as the T4 bacteriophage also couple the expression of late genes to DNA replication [32]. Although unlike HSV-1, continuous DNA replication is necessary. It is currently unknown why initial rounds of DNA replication enable the expression of HSV-1 γ2 genes and what specific alterations to the viral genome allow this to occur. As mentioned above, there is likely an alteration to the genome architecture following viral DNA replication that enables a distinct change in the transcriptional competence of the genome. Understaninding the details of this change will further define how γ2 gene expression is enabled. It remains to be determined how the abundance of ICP4 or other forms of viral chromatin on the genome contribute to this switch or if other modifications of the genome, such as the repair of nicks and gaps or the formation of recombination intermediates, are involved. While Fig 2 is a proposed model of how γ2 genes are regulated, it does not role out the involvement of additional factors or alternate mechanisms. Another interesting observation is that during reactivation from latency, γ2 genes can be expressed independent of both prerequisite viral proteins and DNA replication [33,34]. It is unknown what contributes to these differences, but understanding the regulation of HSV-1 gene expression during latency and reactivation could potentially provide insight into factors that contribute to the regulation of γ2 genes. While there is significant progress made toward understanding the coupling of HSV-1 γ gene expression with viral DNA replication, there are several remaining paths for future investigation.
Acknowledgments
We would like to thank Dr. Neal DeLuca (University of Pittsburgh School of Medicine) and Jessica Packard (Duquesne University) for their feedback on this article.
Funding Statement
JAD and JRH were supported by grant R21AI166879 and R01AI158361 from the National Institutes of Health. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Looker KJ, Magaret AS, May MT, Turner KM, Vickerman P, Gottlieb SL, et al. Global and Regional Estimates of Prevalent and Incident Herpes Simplex Virus Type 1 Infections in 2012. PLoS ONE. 2015;10(10):e0140765. Epub 20151028. doi: 10.1371/journal.pone.0140765 ; PubMed Central PMCID: PMC4624804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.McGeoch DJ, Dolan A, Donald S, Rixon FJ. Sequence determination and genetic content of the short unique region in the genome of herpes simplex virus type 1. J Mol Biol. 1985;181(1):1–13. doi: 10.1016/0022-2836(85)90320-1 . [DOI] [PubMed] [Google Scholar]
- 3.McGeoch DJ, Dalrymple MA, Davison AJ, Dolan A, Frame MC, McNab D, et al. The complete DNA sequence of the long unique region in the genome of herpes simplex virus type 1. J Gen Virol. 1988;69(Pt 7):1531–74. doi: 10.1099/0022-1317-69-7-1531 . [DOI] [PubMed] [Google Scholar]
- 4.Alwine JC, Steinhart WL, Hill CW. Transcription of herpes simplex type 1 DNA in nuclei isolated from infected HEp-2 and KB cells. Virology. 1974;60(1):302–7. doi: 10.1016/0042-6822(74)90390-0 . [DOI] [PubMed] [Google Scholar]
- 5.Ben-Zeev A, Becker Y. Requirement of host cell RNA polymerase II in the replication of herpes simplex virus in alpha-amanitin-sensitive and -resistant cell lines. Virology. 1977;76(1):246–53. doi: 10.1016/0042-6822(77)90300-2 . [DOI] [PubMed] [Google Scholar]
- 6.Depledge DP, Srinivas KP, Sadaoka T, Bready D, Mori Y, Placantonakis DG, et al. Direct RNA sequencing on nanopore arrays redefines the transcriptional complexity of a viral pathogen. Nat Commun. 2019;10(1):754. Epub 20190214. doi: 10.1038/s41467-019-08734-9 ; PubMed Central PMCID: PMC6376126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Whisnant AW, Jurges CS, Hennig T, Wyler E, Prusty B, Rutkowski AJ, et al. Integrative functional genomics decodes herpes simplex virus 1. Nat Commun. 2020;11(1):2038. Epub 20200427. doi: 10.1038/s41467-020-15992-5 ; PubMed Central PMCID: PMC7184758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tombácz D, Csabai Z, Szűcs A, Balázs Z, Moldován N, Sharon D, et al. Long-Read Isoform Sequencing Reveals a Hidden Complexity of the Transcriptional Landscape of Herpes Simplex Virus Type 1. Front Microbiol. 2017;8:1079. Epub 20170620. doi: 10.3389/fmicb.2017.01079 ; PubMed Central PMCID: PMC5476775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zhang YW, Wagner EK. The Kinetics of Expression of Individual Herpes Simplex Virus Type 1 Transcripts. Virus Genes. 1987;1(1):49–60. doi: 10.1007/BF00125685 [DOI] [PubMed] [Google Scholar]
- 10.Packard JE, Dembowski JA. HSV-1 DNA Replication-Coordinated Regulation by Viral and Cellular Factors. Viruses. 2021;13(10). Epub 20211007. doi: 10.3390/v13102015 ; PubMed Central PMCID: PMC8539067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Harkness JM, Kader M, DeLuca NA. Transcription of the herpes simplex virus 1 genome during productive and quiescent infection of neuronal and nonneuronal cells. J Virol. 2014;88(12):6847–61. Epub 20140409. doi: 10.1128/JVI.00516-14 ; PubMed Central PMCID: PMC4054390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dremel SE, DeLuca NA. Genome replication affects transcription factor binding mediating the cascade of herpes simplex virus transcription. Proc Natl Acad Sci U S A. 2019;116(9):3734–9. Epub 2019/02/11. doi: 10.1073/pnas.1818463116 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dremel SE, DeLuca NA. Herpes simplex viral nucleoprotein creates a competitive transcriptional environment facilitating robust viral transcription and host shut off. Elife. 2019;8. Epub 20191022. doi: 10.7554/eLife.51109 ; PubMed Central PMCID: PMC6805162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hu M, Depledge DP, Flores Cortes E, Breuer J, Schang LM. Chromatin dynamics and the transcriptional competence of HSV-1 genomes during lytic infections. PLoS Pathog. 2019;15(11):e1008076. Epub 20191114. doi: 10.1371/journal.ppat.1008076 ; PubMed Central PMCID: PMC6855408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Birkenheuer CH, Baines JD. RNA Polymerase II Promoter-Proximal Pausing and Release to Elongation Are Key Steps Regulating Herpes Simplex Virus 1 Transcription. J Virol. 2020;94(5). Epub 20200214. doi: 10.1128/JVI.02035-19 ; PubMed Central PMCID: PMC7022368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Honess RR, Roizman B. Regulation of Herpesvirus Macromolecular Synthesis I. Cascade Regulation of the Synthesis of Three Groups of Viral Proteins. J Virol. 1974;14(1):8–19. doi: 10.1128/JVI.14.1.8-19.1974 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dixon RA, Schaffer PA. Fine-structure mapping and functional analysis of temperature-sensitive mutants in the gene encoding the herpes simplex virus type 1 immediate early protein VP175. J Virol. 1980;36(1):189–203. doi: 10.1128/JVI.36.1.189-203.1980 ; PubMed Central PMCID: PMC353630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jones PR, Roizman B. Regulation of Herpesvirus Macromolecular Synthesis: The Transcription Program Consists of Three Phases During Which Both Extent of Transcription and Accumulation of RNA in the Cytoplasm are Regulated. J Virol. 1979;31(2):299–314. doi: 10.1128/JVI.31.2.299-314.1979 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Powell KL, Purifoy DJ, Courtney RJ. The synthesis of herpes simplex virus proteins in the absence of virus DNA synthesis. Biochem Biophys Res Commun. 1975;66(1):262–71. doi: 10.1016/s0006-291x(75)80323-8 . [DOI] [PubMed] [Google Scholar]
- 20.Holland LE, Anderson KP, Shipman C Jr, Wagner EK. Viral DNA synthesis is required for the efficient expression of specific herpes simplex virus type 1 mRNA species. Virology. 1980;101(1):10–24. doi: 10.1016/0042-6822(80)90479-1 . [DOI] [PubMed] [Google Scholar]
- 21.Roizman B, Campadelli-Fiume G. Alphaherpes viral genes and their functions. In: Arvin A, Campadelli-Fiume G, Mocarski E, Moore PS, Roizman B, Whitley R, et al., editors. Human Herpesviruses: Biology, Therapy, and Immunoprophylaxis. Cambridge: Cambridge University Press; 2007. [PubMed] [Google Scholar]
- 22.Dembowski JA, Dremel SE, DeLuca NA. Replication-Coupled Recruitment of Viral and Cellular Factors to Herpes Simplex Virus Type 1 Replication Forks for the Maintenance and Expression of Viral Genomes. PLoS Pathog. 2017;13(1):e1006166. Epub 20170117. doi: 10.1371/journal.ppat.1006166 ; PubMed Central PMCID: PMC5271410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Homa FL, Otal TM, Glorioso JC, Levine M. Transcriptional control signals of a herpes simplex virus type 1 late (gamma 2) gene lie within bases -34 to +124 relative to the 5’ terminus of the mRNA. Mol Cell Biol. 1986;6(11):3652–66. doi: 10.1128/mcb.6.11.3652-3666.1986 PubMed Central PMCID: PMC367126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Guzowski JF, Wagner EK. Mutational analysis of the herpes simplex virus type 1 strict late UL38 promoter/leader reveals two regions critical in transcriptional regulation. J Virol. 1993;67(9):5098–108. doi: 10.1128/JVI.67.9.5098-5108.1993 ; PubMed Central PMCID: PMC237907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kibler PK, Duncan J, Keith BD, Hupel T, Smiley JR. Regulation of herpes simplex virus true late gene expression: sequences downstream from the US11 TATA box inhibit expression from an unreplicated template. J Virol. 1991;65(12):6749–60. doi: 10.1128/JVI.65.12.6749-6760.1991 ; PubMed Central PMCID: PMC250758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Steffy KR, Weir JP. Mutational analysis of two herpes simplex virus type 1 late promoters. J Virol. 1991;65(12):6454–60. doi: 10.1128/JVI.65.12.6454-6460.1991 ; PubMed Central PMCID: PMC250683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Petroski MD, Devi-Rao GB, Rice MK, Wagner EK. The downstream activation sequence of the strict late Herpes Simplex Virus Type 1 U(L)38 promoter interacts with hTAF(II)70, a component of TFIID. Virus Genes. 2001;22(3):299–310. doi: 10.1023/a:1011162106727 . [DOI] [PubMed] [Google Scholar]
- 28.Zabierowski SE, Deluca NA. Stabilized binding of TBP to the TATA box of herpes simplex virus type 1 early (tk) and late (gC) promoters by TFIIA and ICP4. J Virol. 2008;82(7):3546–54. Epub 20080123. doi: 10.1128/JVI.02560-07 ; PubMed Central PMCID: PMC2268492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sampath P, Deluca NA. Binding of ICP4, TATA-binding protein, and RNA polymerase II to herpes simplex virus type 1 immediate-early, early, and late promoters in virus-infected cells. J Virol. 2008;82(5):2339–49. Epub 20071219. doi: 10.1128/JVI.02459-07 ; PubMed Central PMCID: PMC2258917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lieu PT, Wagner EK. Two leaky-late HSV-1 promoters differ significantly in structural architecture. Virology. 2000;272(1):191–203. doi: 10.1006/viro.2000.0365 . [DOI] [PubMed] [Google Scholar]
- 31.Wagner LM, DeLuca NA. Temporal association of herpes simplex virus ICP4 with cellular complexes functioning at multiple steps in PolII transcription. PLoS ONE. 2013;8(10):e78242. Epub 20131011. doi: 10.1371/journal.pone.0078242 ; PubMed Central PMCID: PMC3795685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Geiduschek EP. Regulation of expression of the late genes of bacteriophage T4. Annu Rev Genet. 1991;25:437–60. doi: 10.1146/annurev.ge.25.120191.002253 . [DOI] [PubMed] [Google Scholar]
- 33.Kim JY, Mandarino A, Chao MV, Mohr I, Wilson AC. Transient reversal of episome silencing precedes VP16-dependent transcription during reactivation of latent HSV-1 in neurons. PLoS Pathog. 2012;8(2):e1002540. Epub 20120223. doi: 10.1371/journal.ppat.1002540 ; PubMed Central PMCID: PMC3285597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Du T, Zhou G, Roizman B. HSV-1 gene expression from reactivated ganglia is disordered and concurrent with suppression of latency-associated transcript and miRNAs. Proc Natl Acad Sci U S A. 2011;108(46):18820–4. Epub 20111107. doi: 10.1073/pnas.1117203108 ; PubMed Central PMCID: PMC3219146. [DOI] [PMC free article] [PubMed] [Google Scholar]