Abstract
Rhizobacteria closely related to two recently described species of pseudomonads, Pseudomonas brassicacearum and Pseudomonas thivervalensis, were isolated from two geographically distinct wheat field soils in South Australia. Isolation was undertaken by either selective plating or immunotrapping utilizing a polyclonal antibody raised against P. brassicacearum. A subset of 42 isolates were characterized by amplified 16S ribosomal DNA restriction analysis (ARDRA), BIOLOG analysis, and gas chromatography-fatty acid methyl ester (GC-FAME) analysis and separated into closely related phenetic groups. More than 75% of isolates tested by ARDRA were found to have >95% similarity to either Pseudomonas corrugata or P. brassicacearum-P. thivervalensis type strains, and all isolates had >90% similarity to either type strain. BIOLOG and GC-FAME clustering showed a >70% match to ARDRA profiles. Strains representing different ARDRA groups were tested in two soil types for biological control activity against the soilborne plant pathogen Gaeumannomyces graminis var. tritici, the causative agent of take-all of wheat and barley. Three isolates out of 11 significantly reduced take-all-induced root lesions on wheat plants grown in a red-brown earth soil. Only one strain, K208, was consistent in reducing disease symptoms in both the acidic red-brown earth and a calcareous sandy loam. Results from this study indicate that P. brassicacearum and P. thivervalensis are present in Australian soils and that a level of genetic diversity exists within these two novel species but that this diversity does not appear to be related to geographic distribution. The result of the glasshouse pot trial suggests that some isolates of these species may have potential as biological control agents for plant disease.
The search for alternatives to chemical control of plant pathogens, such as biological control, has gained momentum in recent years. Emergence of fungicide-resistant pathogens, health concerns for producer and consumer, and the phasing out of chemicals such as methyl bromide have prompted research into viable alternative practices to achieve more sustainable levels of agricultural production. Since the 1980s rhizobacteria and other microorganisms have been investigated as possible replacements for chemicals used to control a broad range of plant diseases. Isolates from the genus Pseudomonas have been tested due to their widespread distribution in soil, ability to colonize the rhizospheres of host plants, and ability to produce a range of compounds antagonistic to a number of serious plant pathogens (3, 13, 20, 24, 30).
One of the difficulties in developing microorganisms as viable alternatives to chemical control is that many biological control agents are found to be active only in certain soil types. The physical and chemical nature of soils varies greatly between agricultural regions; factors such as soil texture, organic matter, pH, water and oxygen availability, and competition for nutrients with indigenous microflora may significantly dampen the biological activity of introduced inocula. It has previously been demonstrated that an effective biological control strain isolated from one region may not perform in other soil and/or climatic conditions (5, 10, 16, 31). For this reason, one of the important factors to be considered when screening new isolates is their activity in the range of environments in which they would be expected to be used, in particular different soil types. New species of plant-associated rhizobacteria may provide potentially new biological control agents with novel mechanisms of disease suppression active in a range of environments.
Two new species of Pseudomonas isolated from soil samples in France have recently been described in the literature (1). Pseudomonas brassicacearum was isolated from the rhizospheres of plants belonging to the family Brassicaceae, while Pseudomonas thivervalensis was isolated from the rhizosphere of Arabidopsis in the Thiverval-Grignon region (1). Isolation of these new species from soil samples was undertaken by immunotrapping utilizing a polyclonal antibody raised against P. brassicacearum or by plating on selective media (1). P. brassicacearum and P. thivervalensis are discriminated from other Pseudomonas species based on unique restriction profiles generated by amplified 16S ribosomal DNA (rDNA) restriction analysis (ARDRA). These two new species are subsequently distinguished from each other based on isoelectric focusing of pyoverdines (1). Pseudomonas corrugata, a species closely related to the newly described pseudomonads, may also be isolated by immunotrapping with the same polyclonal antibody (2). Despite this cross-reaction, previous results have demonstrated that ARDRA profiles generated by TaqI digestion were able to distinguish between P. corrugata and both P. brassicacearum and P. thivervalensis (8).
In vitro trials with these isolates have demonstrated that strains of the two new species were able to antagonize a range of pathogenic fungi (8), including Gaeumannomyces graminis (Sacc.) von Arx and Olivier var. tritici the causative agent of take-all, a serious disease of wheat and barley. These results suggested that these new species might be useful as biological control agents against take-all if in vitro antagonism can be matched with disease control in soil.
The purpose of this study was to isolate P. brassicacearum and P. thivervalensis from wheat field soils in South Australia using a polyclonal antibody raised against P. brassicacearum (1). Soils from two sites where wheat monoculture has been practiced for many years, including a calcareous take-all suppressive soil, were selected for sampling. A take-all suppressive soil was included in the sampling, as potential biological control agents are often isolated from soils where the pathogen is present but disease symptoms are not observed on susceptible crops (7). The phenomenon of take-all decline in wheat monoculture, where the incidence of disease diminishes over time, has been attributed principally to biological factors, predominantly indigenous microflora suppressing the pathogen and thus limiting the incidence of disease (23). Isolates obtained by immunotrapping and plating on a semiselective medium were taxonomically characterized by ARDRA and siderophore typing as well as BIOLOG and gas chromatography-fatty acid methyl ester (GC-FAME) analyses. These isolates were then tested for in vitro antagonism of G. graminis var. tritici, and selected strains were subsequently tested for their ability to suppress take-all symptoms on wheat in two contrasting soil types.
MATERIALS AND METHODS
Strains and culture conditions.
The type strain for P. corrugata, ATCC 29736 (27), was utilized in this study. Two novel pseudomonads, P. brassicacearum CFBP 11699 and P. thivervalensis CFBP 11261, were described previously (1). P. corrugata strain 2140 was isolated from wheat field soil in New South Wales and has previously been demonstrated to be a biological control agent against take-all of wheat (25). Pseudomonas fluorescens Pf5 was obtained from C. Howell, College Station, Tex. Bacterial field isolates obtained in this study that were subsequently selected for the pot trial are described in Table 1. G. graminis var. tritici isolate 8 was collected by H. McDonald from Avon, South Australia, in 1979.
TABLE 1.
ARDRA groups and phenotypic characteristics of Pseudomonas field isolates from Avon and Kapunda soils
| Strain | Source | ARDRA groupa | BIOLOG groupb | GC-FAME groupb | Fluorescence on King's medium Bc | Inhibition of G. graminis var. tritici (mm)d | Reference or source |
|---|---|---|---|---|---|---|---|
| Type strains | |||||||
| P. brassicacearum CFBP11699 | Rape rhizosphere | 8 | M1 | R | +/− | 4.8 | 1 |
| P. thivervalensis CFBP11261 | Arabidopsis rhizosphere | 8 | M1 | P2 | + | 3.3 | 1 |
| P. corrugata ATCC29736 | Tomato | 7 | NTe | P1 | − | 7.2 | 27 |
| Field isolates | |||||||
| A101 | Wheat rhizosphere | 1 | J | Q2 | + | 0.6 | This study |
| A113 | Wheat rhizosphere | 1 | J | Q2 | + | 2.0 | This study |
| A115 | Wheat rhizosphere | 1 | L | Q2 | ++ | 1.3 | This study |
| A211 | Wheat rhizoplane | 1 | K | Q2 | + | 2.3 | This study |
| A214 | Wheat rhizoplane | 1 | NT | NT | NT | NT | This study |
| A234 | Wheat rhizoplane | 1 | M1 | R | + | NDf | This study |
| K100 | Wheat rhizosphere | 1 | J | Q3 | + | 1.7 | This study |
| K253 | Wheat rhizoplane | 1 | K | Q3 | + | 2.8 | This study |
| K254 | Wheat rhizoplane | 1 | K | Q2 | + | 1.8 | This study |
| K255 | Wheat rhizoplane | 1 | K | Q2 | + | 2.8 | This study |
| A300 | Bulk soil | 2 | NT | NT | ++ | ND | This study |
| A303 | Bulk soil | 2 | K | Q2 | ++ | ND | This study |
| A307 | Bulk soil | 2 | NT | P1 | ++ | 4.0 | This study |
| K400 | Wheat rhizosphere | 3 | K | Q2 | ++ | 3.7 | This study |
| K404 | Wheat rhizosphere | 3 | K | Q2 | ++ | 3.7 | This study |
| K218 | Wheat rhizoplane | 4 | J | NT | + | ND | This study |
| A508 | Wheat rhizoplane | 4 | NT | P2 | ± | 0.7 | This study |
| K279 | Wheat rhizoplane | 4 | M1 | P2 | + | 1.3 | This study |
| A203 | Wheat rhizoplane | 5 | K | Q2 | ++ | 2.0 | This study |
| A418 | Wheat rhizosphere | 5 | NT | NT | + | 2.0 | This study |
| A427 | Wheat rhizosphere | 5 | K | Q2 | + | 0.2 | This study |
| A451 | Wheat rhizosphere | 5 | K | Q2 | ++ | ND | This study |
| A514 | Wheat rhizoplane | 5 | N | Q3 | + | 2.5 | This study |
| A515 | Wheat rhizoplane | 5 | NT | NT | ++ | 1.0 | This study |
| K208 | Wheat rhizoplane | 5 | M2 | P2 | + | 0.8 | This study |
| K240 | Wheat rhizoplane | 6 | J | NT | + | 1.5 | This study |
| A129 | Wheat rhizosphere | 8 | M1 | Q1 | + | 1.2 | This study |
| A132 | Wheat rhizosphere | 8 | L | Q1 | ++ | 0.2 | This study |
| A134 | Wheat rhizosphere | 8 | NT | P1 | − | 9.3 | This study |
| A138 | Wheat rhizosphere | 8 | M1 | Q1 | ++ | 1.8 | This study |
| A229 | Wheat rhizoplane | 8 | M1 | P2 | + | 1.0 | This study |
| A413 | Wheat rhizosphere | 8 | M2 | P2 | + | 0.7 | This study |
| A424 | Wheat rhizosphere | 8 | M2 | P2 | + | ND | This study |
| A433 | Wheat rhizosphere | 8 | M2 | P2 | + | 2.2 | This study |
| A436 | Wheat rhizosphere | 8 | M2 | P2 | + | 0.7 | This study |
| A526 | Wheat rhizoplane | 8 | M1 | P2 | + | 6.7 | This study |
| K108 | Wheat rhizosphere | 8 | M2 | P2 | + | 0.8 | This study |
| K214 | Wheat rhizoplane | 8 | M1 | R | + | 1.8 | This study |
| K241 | Wheat rhizoplane | 8 | M2 | P2 | + | 0.7 | This study |
| K265 | Wheat rhizoplane | 8 | M2 | P2 | + | 1.0 | This study |
| K276 | Wheat rhizoplane | 8 | M2 | P2 | − | 2.2 | This study |
| A119 | Wheat rhizosphere | 10 | K | Q1 | + | 3.8 | This study |
| Other strains | |||||||
| P. corrugata 2140 | Wheat rhizosphere | 9 | NT | P1 | − | 7.0 | 25 |
| P. fluorescens Pf5 | NAg | NA | NA | NA | ++ | NA | 15 |
Groups are based on the dendrograms in Fig. 3.
Fluorescence compared to that of P. fluorescens Pf5. ++, similar fluorescence; +, fluorescence detected; +/−, poor fluorescence; −, no fluorescence detected.
Zone of inhibition between colony edge and hyphal mat. Values are averages from three independent replicates.
NT, not tested.
ND, not detected.
NA, not applicable.
All bacterial strains were routinely grown in the laboratory in Luria-Bertani (LB) broth (26) or on TZCA agar medium modified as described by Kelman (17) and comprising, per liter, 10 g of Proteose Peptone no. 3 (Difco), 1 g of Casamino Acids (Difco), 5 g of glucose, 50 mg of 2,3,5-triphenyl tetrazolium chloride, and 15 g of Bacto agar. G. graminis var. tritici was cultured on half-strength potato dextrose agar (Difco). All cultures were incubated at 25°C.
Isolation of field strains.
Wheat field soils collected from Avon (34°14′S,138°19′E) and Kapunda (34°21′S,138°54′E) were selected for isolating Pseudomonas strains from both bulk soil and wheat rhizosphere. Avon soil is a calcareous sandy loam classified as a fine mixed thermic Calcic Palexeralf (29), pH 8.4 (CaCl2). Kapunda soil is a red-brown earth classified as a fine mixed thermic Natrixeralf (29), pH 5.5 (CaCl2). Soil samples were obtained from random sites at both field sites. Wheat seeds (cv. Stiletto) were surface sterilized sequentially with 2% calcium hypochlorite and 10% H2O2 and sown into the soil samples in 300-ml pots. After 4 weeks of incubation at 15°C, plants were harvested and the roots were separated from shoots. Bulk soil and rhizosphere soil (soil loosely clinging to roots) samples were serially diluted in sterile distilled water. Rhizoplane isolates were obtained by macerating washed roots with a mortar and pestle prior to serial dilution as described above. Bacterial isolates were obtained either by plating samples on RCS semiselective agar medium (2) or by immunotrapping in 96-well microtiter plates, utilizing a polyclonal antibody against P. brassicacearum by methods previously described (2).
ARDRA.
16S rDNA primers rD1 and fD1 (32) were used to amplify a 1.6-kb internal region of the 16S rRNA gene. Genomic DNA was obtained by freezing 1.5 ml of an overnight culture grown in LB broth in liquid nitrogen and then rapid thawing in a 50°C water bath prior to phenol-chloroform extraction and washing in ice-cold 70% ethanol, drying, and subsequent suspension in 50 μl of TE8 (26). The PCR solution comprised, per 50-μl reaction mixture, 1 U of Taq polymerase (Promega), 5.0 μl of reaction buffer (Promega), 0.3 μg each of primers rD1 and fD1, 1.0 μl of genomic DNA, final concentrations of 1.25 mM MgCl2 (Promega), and 0.25 mM deoxynucleoside triphosphates (Promega); the volume was taken to 50 μl with autoclaved ultrapure water. The cycles used were as follows: 1 cycle at 94°C for 4 min; 35 cycles at 94°C for 1 min, 55°C for 2 min and 72°C for 2 min; and one cycle at 72°C for 3 min. Amplification was undertaken in a PTC-100 Thermal Controller (M.J. Research, Watertown, Mass.). This amplified 16S rDNA was digested with six different restriction endonucleases, AluI, CfoI, HaeIII, HinfI, RsaI, and TaqI (Boehringer Mannheim), as per the manufacturer's instructions. Profiles of digested 16S rDNA were separated by gel electrophoresis using a 3.0% agarose gel (Nusieve; FMC BioProducts) in 0.5× Tris-borate-EDTA buffer at 5.0 V cm−1 (26).
Restriction fragment profiles were used to determine phenetic relationships among the field isolates, the three type strains, and P. corrugata strain 2140, using Nei's genetic similarity statistic (22). Pairwise comparisons were made between all isolates and the values were used to generate a similarity matrix. Hierarchical cluster analysis, using the unweighted pair-group method of arithmetic averages (UPGMA), was then used to identify genetically similar groups. These results were used to generate a dendrogram displaying the hierarchical associations between all isolates. GENSTAT 5, release 3.2 (Lawes Agricultural Trust, Rothamsted Experimental Station, 1995), was used for phenetic data analysis.
BIOLOG analysis.
Field isolates were assessed for their ability to metabolize 95 carbon substrates using the BIOLOG GN Microtiter system. Isolates grown in LB broth were washed and resuspended in 0.85% (wt/vol) NaCl solution. The A550 was adjusted to 0.25, 150 μl was added to each microtiter well, and the plates were incubated at 28°C for 24 h. Color development was assessed using the associated Microlog 2 computer software (BIOLOG Inc., Hayward, Calif.).
GC-FAME.
Bacterial isolates were subcultured twice on tryptic soy agar (Difco) for 24 h at 28°C prior to fatty acid extraction and methylation according to the Microbial Identification System procedure (MIDI Inc, Newark, Del.). Extracted samples were analyzed with a Hewlett-Packard S5890 Series II gas chromatograph. Fatty acid peaks were identified by the Microbial Identification System version 4, and profiles were compared to the Sherlock TSBA Library version 3.80 (Microbial ID; MIDI Inc.).
Fluorescence on King's B agar medium.
The ability of isolates to produce fluorescent siderophores was tested by plating bacteria on King's medium B (18) and incubating for 2 days at 25°C. Plates were then inspected under 366-nm UV light, and fluorescence was compared visually to those of P. fluorescens Pf5 (15) and P. corrugata strain 2140 as positive and negative controls respectively.
Isoelectric focusing of pyoverdines.
Strains were grown in CAA medium, comprising, per liter, Bacto Casamino Acids (Difco) (5 g), K2HPO4 · 3H2O (1.54 g), and MgSO4 · 7H2O (0.25 g) at 25°C for 48 h. After centrifugation to pellet cells, the cell-free supernatants were then concentrated 20-fold by lyophilization. Isoelectric focusing of pyoverdines was undertaken by the method of Koedam et al. (19) with 1.5 μl of concentrated supernatant. Bands corresponding to specific pyoverdines were visualized under UV light (1).
Biological control of take-all in planta.
Field isolates were tested for in vitro antagonism of G. graminis var. tritici on half-strength potato dextrose agar at 25°C. Measurements of zones of inhibition between the edge of the bacterial colony and a growing hyphal mat of G. graminis var. tritici were taken after 3 to 4 days of incubation, and the average was obtained from three replicates per isolate.
Antagonistic isolates representing different 16S rDNA groups were selected to test their capacity to suppress take-all symptoms on wheat (cv. Excalibur) in both Avon and Kapunda soils. Soils were inoculated with G. graminis var. tritici grown on autoclaved rye grass seed (28) at a rate of 1.0 g per kg of soil. The pot trial was set up as previously described (25). Approximately 30 g of polyethylene beads (3 to 4 mm in diameter) was placed on top of the soil to prevent water loss by evaporation. Eight replicate pots per treatment, each with five plants per pot, were prepared and watered to a predetermined weight on a regular basis with 10% Hoagland's solution (14).
After 4 weeks of incubation, the wheat plants were harvested and examined for take-all-induced lesions on the seminal roots. Lesions on seminal roots were measured and expressed as a percentage of seminal root length. Shoot lengths were measured from the crown to the tip of the longest leaf of each plant. Root and shoot dry weights were measured after drying plant material at 60°C for 3 days. Water uptake by the wheat plants over the last 2 weeks of the pot trial was also monitored by weighing pots prior to replenishing pots at each watering time.
Means of treatment results were subjected to analysis of variance (Statistix version 3.5; Analytical Software, Tallahassee, Fla.), and results are presented with Fisher's protected least significant difference. Correlations between means of measured parameters were calculated using product moment correlation coefficients with obtained r values compared to product moment correlation values at the 0.05 and 0.01 levels of significance (11).
RESULTS
Isolation of pseudomonads from field soils.
Two hundred isolates were obtained from Avon soil and 118 isolates were obtained from Kapunda soil by immunotrapping or semiselective plating. Approximately 50% of the isolates were obtained by each method. Isolates were obtained from bulk soil as well as the rhizoplanes and rhizospheres of wheat seedlings grown in soil taken from both sites. More than 95% of the field strains obtained were isolated from either the rhizospheres or rhizoplanes of the wheat plants. Isolates were designated with a strain number and an alphabetical prefix denoting the site from which it was obtained (A for Avon and K for Kapunda).
Phenetic characterization.
Forty-two field isolates selected at random but representing each field site and environmental niche (source location) were analyzed by ARDRA (Table 1). Digestion of amplified 16S rDNA with six endonucleases revealed eight different groups of ARDRA patterns after hierarchical cluster analysis by Nei's genetic similarity statistic (22) (Fig. 1; Table 2). Six of these groups contained more than one representative isolate. P. corrugata type strain ATCC 29736 was distinguished from the type strains of the other two species by digesting the 16S rDNA PCR product with TaqI, as reported by Degraeve (8). P. corrugata strain 2140 was found to be different from the P. corrugata type strain based on DNA fragment profiles obtained from HaeIII- and TaqI-digested 16S rDNA.
FIG. 1.
(A) ARDRA banding patterns of amplified 16S rDNA. A total of 47 strains, including 42 isolates obtained from Avon and Kapunda soils, were analyzed. Molecular marker X (Boehringer Mannheim) was used to determine fragment lengths. Different banding patterns generated by single enzyme digestion are designated A, B, and C, except for RsaI, where only one pattern was observed for all isolates except A119 (see Table 2). (B) Results of cluster analysis of the ARDRA patterns.
TABLE 2.
ARDRA banding patterns of Pseudomonas field isolates using six restriction enzymesa
| Group | ARDRA banding pattern
|
|||||
|---|---|---|---|---|---|---|
| AluI | RsaI | HaeIII | TaqI | HinfI | CfoI | |
| 1 | A | A | A | A | A | A |
| 2 | A | A | A | A | A | B |
| 3 | A | A | A | A | B | A |
| 4 | A | A | A | B | A | A |
| 5 | A | A | A | C | A | A |
| 6 | A | A | A | C | A | B |
| 7b | B | A | A | A | A | A |
| 8c | B | A | A | B | A | A |
| 9d | B | A | B | B | A | A |
| 10 | B | Be | A | B | A | A |
Based on ARDRA profiles shown in Fig. 1. Eight groups (excluding groups 7 and 9) contained isolates from Avon and/or Kapunda soils.
P. corrugata ATCC 29736.
Includes P. brassicacearum and P. thivervalensis type strains.
P. corrugata 2140.
Isolate A119 exhibited a band of 750 bp instead of 800 bp (not shown in Fig. 1) but otherwise had a restriction pattern identical to pattern A when digested with RsaI.
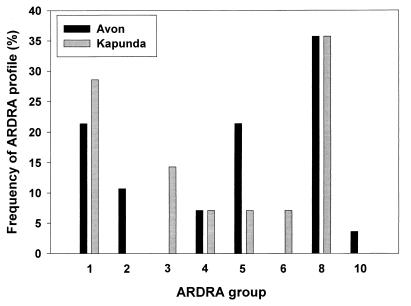
Most of the eight ARDRA groups contained isolates occurring in both Avon and Kapunda soils (Fig. 2) and had a high genetic similarity to either P. brassicacearum and P. thivervalensis (group 8) or P. corrugata ATCC 29736 (group 7) (Fig. 1). Fifteen isolates (10 from Avon soil and 5 from Kapunda soil) exhibited ARDRA patterns identical to those of the P. brassicacearum and P. thivervalensis type strains (group 8), while a further 13 isolates (9 from Avon soil and 4 from Kapunda soil) were closely related to the P. corrugata type strain. ARDRA groups 2 and 3 contained isolates found exclusively in a single environmental niche, i.e., bulk soil collected from Avon and the rhizosphere of wheat grown in Kapunda soil, respectively.
FIG. 2.
Frequency of ARDRA groups at each sampling site. Frequencies were obtained by dividing the number of isolates from each site represented in a particular group by the total number of isolates tested from that site. Most isolates analyzed were found to be either identical to P. brassicacearum and P. thivervalensis (group 8) or closely related to P. corrugata (group 1). Four groups contained isolates from a single site only; the total number of isolates in each of these groups ranged from one (groups 6 and 10) to three (group 2) (see Table 1 for details of isolate numbers).
Analysis of carbon utilization on BIOLOG GN plates resulted in field isolates exhibiting >90% homology with the P. brassicacearum and P. thivervalensis type strains from France. Differences in utilization of carbon compounds between the Australian field isolates and the French type strains were generally similar for all isolates tested, with the Australian isolates being able to grow on glycogen, α-d-glucose, d-gluconic acid, and uridine but unable to metabolize N-acetyl-d-glucosamine, γ-hydroxybutyric acid, and sebacic acid.
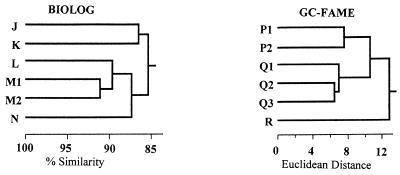
Cluster analyses of BIOLOG and GC-FAME data revealed distinct groups for the field isolates (Fig. 3; Table 1). Most isolates (82%) fell within two distinct BIOLOG groups, with one group further divided into two subgroups containing a similar numbers of isolates in each. Two distinct GC-FAME groups accounted for 73% of all isolates tested. When BIOLOG and GC-FAME clusters were compared to each other, it was found that 81% of the isolates analyzed remained within the same group. For example, 80% of BIOLOG group K isolates exhibited similar GC-FAME profiles (group Q2), while all BIOLOG group M2 isolates shared the same GC-FAME profile (group P2) (Table 1). Comparison of ARDRA profiles to BIOLOG and GC-FAME clusters revealed that isolates from each ARDRA group fell into distinct BIOLOG or GC-FAME clusters (91 and 85%, respectively), suggesting that results from the three separate analyses were consistent with each other in assessing phenetic relationships among bacterial isolates.
FIG. 3.
BIOLOG and GC-FAME dendrograms of Pseudomonas isolates from Avon and Kapunda soils. BIOLOG analysis revealed that most isolates were clustered in group K (32% of total isolates tested) or groups M1 and M2 (24 and 26%, respectively). When compared to GC-FAME data, 82% of BIOLOG group K isolates were clustered in GC-FAME group Q2, while 75% of BIOLOG group M1 and M2 isolates were clustered in GC-FAME group P2. The relationship of these groups to the ARDRA groups is included in Table 1.
Isolates were tested for the in vitro production of fluorescent siderophores. When fluorescence was compared to that of strain Pf5 under UV light, it was found that >95% of isolates tested produced fluorescent siderophores after 48 h of incubation on King's B agar at 25°C (Table 1).
Results of the isoelectric focusing of pyoverdine siderophores indicated that 85% of isolates tested were P. brassicacearum. Although P. thivervalensis isolates made up only 15% of the strains tested, representative isolates were obtained from both the Avon and Kapunda field sites for this species.
Biological control.
In vitro antagonism experiments with G. graminis var. tritici revealed that 86% of the 42 isolates tested demonstrated detectable antifungal activity. Eleven isolates, six from Avon and five from Kapunda, consistently induced reproducible zones of fungal inhibition on half-strength potato dextrose agar medium (Table 1); these were selected for the pot trial. Two ARDRA group 8 isolates from Avon, A134 and A526, were the most antagonistic against the take-all fungus, while the other isolates displayed various degrees of antifungal activity.
Results from the pot trial showed large variations between bacterial isolates in biological control activity against take-all (Tables 3 and 4). Most isolates showed greater biocontrol activity in the Kapunda soil than in the Avon soil. P. thivervalensis isolate K208 performed best in suppressing take-all lesions on wheat roots in both soils; in Kapunda soil this strain performed marginally better than the biological control strain P. corrugata 2140. Ability to antagonize G. graminis var. tritici in vitro did not necessarily correlate with significant disease suppression in planta; strains A134 and A526 had no significant effect on disease suppression in either soil type despite producing the largest in vitro inhibition zones (Table 1). Significant negative correlations were observed between incidence of lesions (disease rating) and the other plant growth parameters measured (Table 1). The highest negative correlations were observed between disease ratings and water consumption (r = −0.85 and −0.93 for Avon and Kapunda soils, respectively).
TABLE 3.
Biological control of take-all and health of wheat plants inoculated with Pseudomonas strains and grown in Avon soila
| Inoculant | Root lesions (%)b | H2O uptake (g)c | Root dry wt (mg/pot)d | Shoot dry wt (mg/pot)d | Shoot length (mm) |
|---|---|---|---|---|---|
| Control, no added G. graminis var. tritici | 0.17 | 45* | 197* | 194 | 250* |
| Control: added G. graminis var. tritici | 15.74 | 32 | 150 | 171 | 225 |
| P. corrugata 2140 | 15.09 | 30 | 168 | 170 | 228 |
| A119 | 14.41 | 29 | 149 | 173 | 230 |
| A134 | 15.15 | 32 | 157 | 177 | 227 |
| A221 | 12.30 | 31 | 143 | 162 | 230 |
| A307 | 13.28 | 31 | 153 | 175 | 234 |
| A514 | 16.34 | 28 | 148 | 166 | 227 |
| A526 | 14.83 | 34 | 164 | 185 | 228 |
| K208 | 11.05† | 30 | 156 | 168 | 227 |
| K255 | 15.06 | 31 | 158 | 173 | 234 |
| K276 | 14.65 | 34 | 160 | 184 | 235 |
| K400 | 14.49 | 30 | 158 | 170 | 228 |
| K404 | 18.53 | 30 | 141 | 170 | 214 |
| P (treatment)e | 0.061 | <0.01 | <0.01 | >0.1 | <0.01 |
| Correlation (r) to percent root lesions | Not applicable | −0.85 | −0.80 | −0.54 | −0.54 |
∗ and † indicate treatments with results that were significantly different from those of the control treatment with an added inoculum of the take-all fungus (G. graminis var. tritici). Use of different symbols indicates that treatments were significantly different from each other. Control treatments contained no bacterial inoculant. All data are means from eight replicate pots.
Obtained by measuring take-all-induced lesions of seminal roots as a percentage of total seminal root length.
Total water lost per pot over the last 14 days of a 28-day experiment.
Each pot contained five plants.
P value for root lesion analysis excluded control treatment with no added G. graminis var. tritici.
TABLE 4.
Biological control of take-all and health of wheat plants inoculated with Pseudomonas strains and grown in Kapunda soila
| Inoculant | Root lesions (%)b | H2O uptake (g)c | Root dry wt (mg/pot)d | Shoot dry wt (mg/pot)d | Shoot length (mm) |
|---|---|---|---|---|---|
| Control, no added G. graminis var. tritici | 0.23 | 74* | 263* | 280* | 287* |
| Control: added G. graminis var. tritici | 22.44 | 46 | 184 | 230 | 258 |
| P. corrugata 2140 | 12.92† | 56† | 202 | 252† | 269 |
| A119 | 19.72 | 46 | 175 | 216 | 257 |
| A134 | 18.45 | 50 | 187 | 236 | 265 |
| A221 | 17.60 | 45 | 180 | 231 | 265 |
| A307 | 16.77 | 51 | 210 | 248 | 262 |
| A514 | 15.89 | 51 | 193 | 237 | 270 |
| A526 | 17.94 | 47 | 169 | 231 | 260 |
| K208 | 10.70† | 56† | 216† | 261† | 276* |
| K255 | 17.31 | 48 | 192 | 238 | 253 |
| K276 | 14.99† | 48 | 176 | 231 | 259 |
| K400 | 14.51† | 47 | 176 | 221 | 271 |
| K404 | 15.51 | 48 | 184 | 235 | 258 |
| P (treatment)e | <0.01 | <0.001 | <0.001 | <0.001 | <0.01 |
| Correlation (r) to percent root lesions | Not applicable | −0.93 | −0.85 | −0.83 | −0.86 |
∗ and † indicate treatments with results that were significantly different from those of the control treatment with an added inoculum of the take-all fungus (G. graminis var. tritici). Use of different symbols indicates that treatments were significantly different from each other. Control treatments contained no bacterial inoculant. All data are means from eight replicate pots.
Obtained by measuring take-all-induced lesions of seminal roots as a percentage of total seminal root length.
Total water lost per pot over the last 14 days of a 28-day experiment.
Each pot contained five plants.
P value for root lesion analysis excluded control treatment with no added G. graminis var. tritici.
DISCUSSION
As P. corrugata is closely related to P. brassicacearum and a polyclonal antibody raised against the new species will cross-react with P. corrugata, it was expected that a proportion of the field isolates obtained by immunotrapping would be shown to be more closely related to the P. corrugata type strain than to P. brassicacearum or P. thivervalensis. In this study, almost 35% of the field isolates characterized by ARDRA were closely related to P. corrugata. Overall, more than 80% of the 42 isolates examined in this study showed >90% homology to either P. corrugata or P. brassicacearum-P. thivervalensis, and all 42 isolates showed >85% homology to the type strains.
In the original study describing P. brassicacearum and P. thivervalensis, it was found that these species were regularly isolated from the rhizospheres of Brassica napus and Arabidopsis thaliana in a range of soil types in France (1). This report shows that P. brassicacearum and P. thivervalensis are present in at least two distinct geographic locations and in dissimilar soils, implying that these new species may be widespread in Australia. Furthermore, the presence of these new species in soils subjected to wheat monoculture and their ability to be isolated from the rhizospheres of wheat seedlings suggest that they may be able to colonize a broad range of host plants, increasing the likelihood that they will be found in a range of environments.
P. brassicacearum and P. thivervalensis were originally isolated from the rhizospheres of rape and Arabidopsis (1). Taken together with the data from this study of wheat field soil isolates, the results suggest a close interaction between these new species of rhizobacteria and host plants. The high proportion of isolates obtained from the rhizospheres and rhizoplanes of wheat roots compared to bulk soil (95 and 97% for Avon and Kapunda soils, respectively) observed in this study provides further evidence of a possible plant-microbe relationship. It is yet to be established what the overall nature of these relationships is, i.e., whether these species are beneficial or benign or if there are plant-deleterious strains of these species. Studies with these new species have yet to establish whether they have evolved with and play a role in the growth of commercially important crops.
Comparison of BIOLOG results indicated a >90% homology between the P. brassicacearum and P. thivervalensis type strains from France and the field isolates obtained from Australian soils. It was interesting that the differences in carbon utilization between the French and Australian isolates were identical. This may be an evolutionary factor whereby geographically separate strains of the same species isolated from the rhizospheres of different host plants have acquired the capacity to metabolize different carbon compounds. It has been shown that root exudates and lysates from different host plants can influence the structure of rhizobacterial populations in the short term (12). Over a longer period, however, the influence of different root-derived carbon compounds may influence the genetic characteristics of individual species. Another explanation for this observation is that different root exudates from distinct host plants exert a selective pressure in the rhizosphere, favoring isolates with specific carbon utilization pathways that are able to exploit the differing ranges of exudates and lysates.
Results of in vitro antagonism assays have demonstrated that, like other biological control agents, P. brassicacearum and P. thivervalensis are capable of producing antifungal metabolites (reference 8 and this study). This study has further shown that some isolates of the new species are useful in controlling take-all in planta. A P. thivervalensis isolate obtained from Kapunda soil, K208, exhibited disease suppression in both Kapunda and Avon soils. This may be due to a number of factors, including in planta antifungal metabolite production or possible systemic induced resistance. Other mechanisms, such as rhizosphere exclusion or competition for nutrients, may also play a role in biological control of take-all.
An important factor to emerge from the pot trial was the detection of biological control rhizobacteria active in the two contrasting soil types. Take-all is a serious problem in alkaline soils (6), and the disease is still a threat to productivity in these soil types throughout southeastern Australia (4, 21). Previous attempts to isolate bacterial strains that can control take-all on wheat in calcareous sandy soils have been unsuccessful. A number of soil factors could contribute to this, including high pH, causing low survival and persistence of the inocula, and lack of expression of certain biocontrol-related genes in calcareous soil. In this study, strain K208 showed promise as a biological control agent. This strain reduced symptoms of take-all in both soil types, an important finding for the development of commercially viable biological control strains. Further tests in other soils, either as a single inoculant or in combination with other biocontrol isolates such as Bacillus and Trichoderma, need to be undertaken to ascertain the full potential of this isolate.
During the final 2 weeks of the pot trial, water consumption (as measured by changes in pot weight) was monitored. G. graminis var. tritici infests wheat through infection of the seminal roots, where thin-walled fungal microhyphae penetrate root cell walls through the production of cellulase and pectinase and subsequent colonization of vascular tissue (9, 33). Obstruction of the stele results in decreased nutrient flow in the plant, eventually leading to premature ripening (white heads with shriveled grain). Although measuring and rating roots and lesions still constitute the primary means for assessing disease severity, this is a time-consuming and generally destructive procedure that can be undertaken only after harvesting plants. Our results have shown a significant negative correlation between take-all-induced root lesions and the ability of plants to take up water (and associated soil nutrients). Measurement of water uptake by plants in a nondraining system may be a viable method for nondestructive monitoring of the state of the plants in an experiment. This can be used to indicate disease severity and plant health at any time point in the trial.
In conclusion, this study has shown the presence of two newly described species of pseudomonads, P. brassicacearum and P. thivervalensis, in Australian soils. ARDRA shows them to be readily identifiable and that although they are closely related to P. corrugata, they can be distinguished from this species. Further analysis with BIOLOG and GC-FAME indicated a degree of intraspecific variation, but this was not great enough to separate isolates into species-specific groups. Some isolates of these new species demonstrated a level of biological control of take-all of wheat comparable to those of previously described strains, suggesting that they have potential for use in the field and in a greater range of soil types. The full potential can be assessed only after field trials. Further studies to elucidate the genetic variability within this species, levels of host plant adaptation, whether particular genetic groups are more efficacious as biological control agents, and the mechanisms of biological control of these species may enable further progress towards commercial biological control of take-all, particularly in calcareous soils.
ACKNOWLEDGMENTS
This study was partially funded through the PICS program of CNRS, France.
We acknowledge the contribution of Bruce Hawke to the GC-FAME analysis. We also thank Clive Pankhurst and Suha Hare for critical review of the manuscript.
REFERENCES
- 1.Achouak W, Sutra L, Heulin T, Meyer J-M, Fromin N, Degraeve S, Christen R, Gardan L. Pseudomonas brassicacearum sp. nov. and Pseudomonas thivervalensis sp. nov, two root-associated bacteria from Brassica napus and Arabidopsis thaliana. Int J Syst Bacteriol Evol Microbiol. 2000;50:9–18. doi: 10.1099/00207713-50-1-9. [DOI] [PubMed] [Google Scholar]
- 2.Achouak W, Thiéry J M, Roubaud P, Heulin T. Impact of crop management on intraspecific diversity of Pseudomonas corrugata in bulk soil. FEMS Microbiol Ecol. 2000;31:11–19. doi: 10.1111/j.1574-6941.2000.tb00666.x. [DOI] [PubMed] [Google Scholar]
- 3.Anjaiah V, Koedam N, Nowak Thompson B, Loper J E, Höfte M, Tambong J T, Cornelis P. Involvement of phenazines and anthranilate in the antagonism of Pseudomonas aeruginosa PNA1 and Tn5 derivatives towards Fusarium spp. and Pythium spp. Mol Plant-Microbe Interact. 1998;11:847–854. [Google Scholar]
- 4.Brennan J P, Murray G M. Australian wheat diseases: assessing their economic importance. Agric Sci. 1988;2:26–35. [Google Scholar]
- 5.Capper A L, Higgins K P. Application of Pseudomonas fluorescens isolates to wheat as potential biological control agents against take-all. Plant Pathol. 1993;42:560–567. [Google Scholar]
- 6.Cook R J. The effect of soil reaction and physical conditions. In: Asher M J C, Shipton P J, editors. Biology and control of take-all. London, United Kingdom: Academic Press; 1981. pp. 343–352. [Google Scholar]
- 7.Cook R J. Problems and progress in the biological control of wheat take-all. Plant Pathol. 1994;43:429–437. [Google Scholar]
- 8.Degraeve S. Les peuplements et populations de bacteries associes aux racines de colza transformé per l'introduction d'un gene de chitinase. Ph.D. thesis. Nancy, France: Université Henri Poincaré; 1994. [Google Scholar]
- 9.Dori S, Hershenhorn J, Solel Z, Barash I. Characterisation of an endopolygalacturonase associated with take-all disease of wheat. Physiol Mol Plant Pathol. 1992;40:203–210. [Google Scholar]
- 10.Duffy B K, Ownley B H, Weller D M. Soil chemical and physical properties associated with suppression of take-all of wheat by Trichoderma koningii. Phytopathology. 1997;87:1118–1124. doi: 10.1094/PHYTO.1997.87.11.1118. [DOI] [PubMed] [Google Scholar]
- 11.Fowler J, Cohen L, Jarvis P. Practical statistics for field biology. 2nd ed. Chichester, England: John Wiley & Sons; 1998. [Google Scholar]
- 12.Grayston S J, Wang S, Campbell C D, Edwards A C. Selective influence of plant species on microbial diversity in the rhizosphere. Soil Biol Biochem. 1998;30:369–378. [Google Scholar]
- 13.Hill D S, Stein R I, Torkewitz N R, Morse A M, Howell C R, Pachlatko J P, Becker J O, Ligon J M. Cloning of genes involved in the synthesis of pyrrolnitrin from Pseudomonas fluorescens and role of pyrrolnitrin synthesis in biological control of plant disease. Appl Environ Microbiol. 1994;60:78–85. doi: 10.1128/aem.60.1.78-85.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hoagland D R, Arnon D I. The water culture method of growing plants without soil. University of California Agricultural Experiment Station circular no; 1938. p. 347. [Google Scholar]
- 15.Howell C R, Stipanovic R D. Suppression of Pythium ultimum-induced damping-off of cotton seedlings by Pseudomonas fluorescens and its antibiotic pyoluteorin. Phytopathology. 1980;70:712–715. [Google Scholar]
- 16.Johnsson L, Hokeberg M, Gerhardson B. Performance of the Pseudomonas chlororaphis biocontrol agent MA 342 against cereal seed-borne diseases in field experiments. Eur J Plant Pathol. 1998;104:701–711. [Google Scholar]
- 17.Kelman A. The relationship of pathogenicity in Pseudomonas solanacearum to colony appearance on a tetrazolium medium. Phytopathology. 1954;44:693–695. [Google Scholar]
- 18.King E O, Ward M K, Raney D E. Two simple media for the demonstration of pyocyanin and fluorescin. J Lab Clin Med. 1954;44:301–307. [PubMed] [Google Scholar]
- 19.Koedam N, Wittouck E, Gaballa A, Höfte M, Cornelis P. Detection and differentiation of microbial siderophores by isoelectric focussing and chrome azurol S overlay. BioMetals. 1994;7:287–291. doi: 10.1007/BF00144123. [DOI] [PubMed] [Google Scholar]
- 20.Maurhofer M, Keel C, Schnider U, Voisard C, Haas D, Défago G. Influence of enhanced antibiotic production in Pseudomonas fluorescens CHA0 on its disease suppressive capacity. Phytopathology. 1991;82:190–195. [Google Scholar]
- 21.Murray G M, Heenan D P, Taylor A C. The effect of rainfall and crop management on take-all and eyespot of wheat in the field. Aust J Exp Agric. 1991;31:645–651. [Google Scholar]
- 22.Nei M. Analysis of gene diversity in subdivided populations. Proc Natl Acad Sci USA. 1973;70:3321–3323. doi: 10.1073/pnas.70.12.3321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Raaijmakers J M, Weller D M. Natural plant protection by 2,4-diacetylphloroglucinol-producing Pseudomonas spp. in take-all decline soils. Mol Plant-Microbe Interact. 1998;11:144–152. [Google Scholar]
- 24.Rodriguez F, Pfender W F. Antibiosis and antagonism of Sclerotinia homoeocarpa and Drechslera poae by Pseudomonas fluorescens PF-5 in vitro and in planta. Phytopathology. 1997;87:614–621. doi: 10.1094/PHYTO.1997.87.6.614. [DOI] [PubMed] [Google Scholar]
- 25.Ryder M H, Rovira A D. Biological control of take-all of glasshouse-grown wheat using strains of Pseudomonas corrugata isolated from wheat field soil. Soil Biol Biochem. 1993;25:311–320. [Google Scholar]
- 26.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 27.Scarlett C M, Fletcher J T, Roberts P, Lelliott R A. Tomato pith necrosis caused by Pseudomonas corrugata n. sp. Ann Appl Biol. 1978;88:105–114. [Google Scholar]
- 28.Simon A, Rovira A D, Foster R C. Inocula of Gaeumannomyces graminis var. tritici for field and glasshouse studies. Soil Biol Biochem. 1987;19:363–370. [Google Scholar]
- 29.Soil Survey Staff. Keys to soil taxonomy, 4th ed. Soil Management Support Services technical monograph no. 6. Blacksburg: Virginia Polytechnic Institute and State University; 1990. [Google Scholar]
- 30.Thomashow L S, Bonsall R F, Weller D M. Antibiotic production by soil and rhizosphere microbes in situ. In: Hurst C J, Knudsen G R, McInerney M J, Stetzenbach L D, Walter M V, editors. Manual of environmental microbiology. Washington, D.C.: American Society for Microbiology; 1997. pp. 493–499. [Google Scholar]
- 31.Troxler J, Zala M, Moenne-Loccoz Y, Keel C, Défago G. Predominance of non-culturable cells of the biocontrol strain Pseudomonas fluorescens CHA0 in the surface horizon of large outdoor lysimeters. Appl Environ Microbiol. 1997;63:3776–3782. doi: 10.1128/aem.63.10.3776-3782.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Weisburg W G, Barns S M, Pelletier D A, Lane D J. 16S ribosomal DNA amplification for phylogenetic study. J Bacteriol. 1991;173:697–703. doi: 10.1128/jb.173.2.697-703.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Weste G. Extra-cellular enzyme production by various isolates of Ophiobolus graminis and O. graminis var. avenae. Phytopathol Z. 1970;67:189–204. [Google Scholar]