Abstract
Mycoplasma pneumoniae and Mycoplasma genitalium are cell wall-less bacteria with strongly reduced genome content and close phylogenetic relatedness. In humans, the only known natural host, the microorganisms colonize the respiratory or genitourinary mucosa and may cause a broad range of clinical presentations. Besides fundamental differences in their tissue specificity, transmission route, and ability to cause prevalence peaks, both species share similarities such as the occurrence of asymptomatic carriers, preferred populations for infection, and problems with high rates of antimicrobial resistance. To further understand the epidemiology of these practically challenging bacteria, typing of strains is necessary. Since the cultivation of both pathogens is difficult and not performed outside of specialized laboratories, molecular typing methods with adequate discriminatory power, stability, and reproducibility have been developed. These include the characterization of genes containing repetitive sequences, of variable genome regions without the presence of repetitive sequences, determination of single and multi-locus variable-number tandem repeats, and detection of single nucleotide polymorphisms in different genes, respectively. The current repertoire of procedures allows reliable differentiation of strains circulating in different populations and in different time periods as well as comparison of strains occurring subsequently in individual patients. In this review, the methods for typing M. pneumoniae and M. genitalium, including the results of their application in different studies, are summarized and current knowledge regarding the association of typing data with the clinical characteristics of infections is presented.
Keywords: Mycoplasma pneumoniae, Mycoplasma genitalium, molecular typing, epidemiology, SNP, MLST, tandem repeats
Introduction
During the evolutionary interplay with their hosts, the genomes of species of the class Mollicutes (“mycoplasma”) have been greatly reduced. Besides limited metabolic capabilities, the lack of a classical bacterial cell wall is the most striking result of this interaction. In humans, different species can be found as commensals whereas the most clinically relevant Mycoplasma pneumoniae and Mycoplasma genitalium are host-specific pathogens that infect the respiratory and genitourinary mucosa. Mycoplasma pneumoniae is transmitted via contaminated aerosols and is a frequent cause of community-acquired respiratory tract infections including severe cases of interstitial pneumonia (Khoury et al., 2016). Infections can occur in all age groups but school-aged children are the preferred population. Besides small-scale outbreaks in settings with close person-to-person contacts, such as military camps or schools (Waites et al., 2017), epidemic peaks are registered every 3–7 years (Brown et al., 2016; Kenri et al., 2020). During these periods, which sometimes can be registered worldwide, M. pneumoniae may cause up to 50% of all community-acquired respiratory infections (Ho et al., 2015; Kogoj et al., 2015). In addition, a broad spectrum of extrapulmonary manifestations is described, mainly affecting the central nervous system and the skin (Narita, 2016). In contrast, M. genitalium is a sexually transmitted pathogen that causes non-gonococcal urethritis in men and is associated with urethritis, cervicitis, endometritis, and pelvic inflammatory disease in women (Jensen et al., 2022). The prevalence of infection ranges between 1.3 and 3.9% in the general population (Baumann et al., 2018) but can be significantly higher in risk groups such as men who have sex with men (MSM) and HIV-positive patients (Jansen et al., 2020; Latimer et al., 2020).
Treatment of infections with both species is challenging as mycoplasmas are intrinsically resistant to betalactams. Generally, tetracyclines and quinolones are effective but have side effects for relevant patient groups (treatment of M. pneumoniae in pediatrics) or are of limited clinical efficacy (M. genitalium and doxycycline). Hence, macrolides are the antibiotics recommended primarily for treatment of adults and children with severe clinical disease and in attempts at eradication in symptomatic or asymptomatic patients. Unfortunately, high rates of acquired resistance to macrolides (M. pneumoniae) and to macrolides and quinolones (M. genitalium) are described (Fernandez-Huerta et al., 2020a; Machalek et al., 2020; Pereyre and Tardy, 2021). Despite strong epidemiological differences, such as the location of colonized tissues and consequently the transmission route of infections, there are similarities in epidemiological characteristics. Besides the high rates of resistance, these include the occurrence of asymptomatic carriers probably able to transmit the pathogens (Spuesens et al., 2013; Gnanadurai and Fifer, 2020; de Groot et al., 2021).
As is typical for mycoplasmas, genomes of species are small but closely related phylogenetically, with around 580 kbp (M. genitalium) and 816 kbp (M. pneumoniae). Many orthologous proteins (~480) can be found in both pathogens and identity between them has been calculated as around 67% (Himmelreich et al., 1997). In the last few years, whole genome sequencing of isolates of different geographic origin and from varying time periods resulted in strong similarities between compared strains (>99%) and classification into two main lineages. Within these main genotypes, the genomes of both species were identical in >99.5%, which is remarkably high (Lluch-Senar et al., 2015; Xiao et al., 2015; Diaz et al., 2017; Fookes et al., 2017; Lee et al., 2019). Moreover, construction of phylogenetic trees of M. pneumoniae genomes resulted in different clades within the main types showing differences in the frequency of in vivo occurrence and in the number of recombination events (Kenri et al., 2020; Hsieh et al., 2022). Despite the overall conserved genomes, regions with higher heterogeneity were shown to be associated in most cases with the occurrence of repetitive elements distributed in similar but non-identical copies in the genomes. Four repetitive elements (RepMp1, 2/3, 4, and 5) were found in M. pneumoniae, whereas repeated sequences from M. genitalium cluster in nine discrete regions are known as MgPar. Genome parts with copies of repetitive elements can be exchanged by homologous recombination (Musatovova et al., 2008; Spuesens et al., 2009; Hakim et al., 2021). This also applies to genes coding for surface-localized and antigenic proteins with special importance for the infection process such as the adhesins P1 and P40/P90 from M. pneumoniae, and MgpB and MgpC from M. genitalium (also known as P140 and P110). Resulting modifications of these proteins has been postulated as immune escape mechanism of the bacteria (Rocha and Blanchard, 2002; Hakim et al., 2021).
To further understand the epidemiology of infections by M. pneumoniae and M. genitalium, typing of strains is important. Unfortunately, cultivation of both species is difficult and only realized in few specialized centers. Therefore, molecular detection is common in routine laboratories and positive DNA from clinical material is the most frequent specimen available for typing in practice.
Development of Typing Methods
The first molecular method for typing M. pneumoniae isolates was PCR-mediated DNA fingerprinting, which confirmed that there were two main strain types (Su et al., 1990; Ursi et al., 1994). Supported by data from early whole genome sequencing of M. pneumoniae (type 1 strain M129; Himmelreich et al., 1996), more targeted methods were introduced in the following years. These included the restriction fragment length polymorphism procedure, which uses primers that amplify both regions of the p1 gene (MPN141) containing copies of the repetitive elements RepMP2/3 and RepMP4 (Sasaki et al., 1996). This approach distinguished the two main p1 types and a limited number of additional genotypes (Kenri et al., 2020). To detect all sequence variations, the more laborious amplification of RepMp copies in the p1 gene followed by Sanger sequencing has been used (Dumke et al., 2006, 2015).
In 2009, multi-locus variable-number tandem-repeat analysis (MLVA) was introduced using five loci [HsdS (MPN089), intergenic, and hypothetical proteins (MPN501, MPN524, and MPN613)] to determine the number of repeats (Degrange et al., 2009). The results of typing of isolates showed an excellent discriminatory index (DI) of 0.92. The method was adapted for investigation of M. pneumoniae-positive DNA (Dumke and Jacobs, 2011). Unfortunately, the locus with the highest DI (Mpn1, coding for a subunit of type I restriction-modification enzyme, MPN089) was found to be instable (Benitez et al., 2012; Sun et al., 2013) and must be removed from the list of repeats. For the remaining four repeats, interpretation guidelines for use of MLVA were established to allow reliable interlaboratory comparison of results (Chalker et al., 2015). In some strains, differences in the length of distinct tandem repeat loci must be considered (Xue et al., 2018; Kenri et al., 2020). Further tandem repeats were tested and might be an alternative to Mpn1 to increase the discrimination of the method (Zhang et al., 2017). In addition, AGT repeats can be found in the region between the repetitive elements in the p1 gene. Using this single locus of tandem repeats, strains with identical MLV type were differentiated and the number of p1 repeats is associated with the main p1 types (Zhao et al., 2011; Tian et al., 2013; Xiao et al., 2020). Unfortunately, there is evidence that this marker is unstable and this needs further analysis (Spuesens et al., 2016; Dumke, unpubl.). Recently, a new target of VNTR analysis was reported using the tandem repeats in subunit S of the type I restriction-modification system (MPN085) of M. pneumoniae for differentiation of strains (Lee et al., 2022).
Based on comprehensive comparison of the increasing number of whole genome data, two methods that use determination of single nucleotide polymorphisms (SNPs) for typing were developed. SNPs in eight genes (MPN003, MPN185, MPN246, MPN307, MPN528, MPN576, MPN600, and MPN628) coding for house-keeping proteins (Brown et al., 2015; here called MLS typing) or for house-keeping proteins (MPN004, MPN050, MPN168, MPN246, and MPN516), hypothetical lipoproteins (MPN442, MPN582), and the P1 adhesin [(MPN141); Touati et al., 2015; SNP typing] were selected. Both methods can be used not only for characterization of isolates but also for investigation of strains in DNA-positive clinical samples (Dumke and Rodriguez, 2021) and result in a numerical code or a SNP profile, which can be easily exchanged between laboratories. For a high discriminatory power of differentiation if MLS and SNP typing is performed in parallel, use of same loci [gmk (MPN246)] is disadvantageous. In addition, SNP measured in the p1 gene (SNP typing) is located in the repetitive element RepMp2/3 and homologous recombination of this locus cannot be excluded. For the method of Brown et al., a database was established that comprises listing, consecutive numbering, and comparison of detected MLS types.1
In addition to the aforementioned methods, various other typing methods were developed but are not widely used. Real-time PCR with high-resolution melting point analysis is able to differentiate p1 type 1 and 2 strains (Schwartz et al., 2009). Moreover, pyrosequencing of two targets (MPN141 and MPN528a) resulted in correct classification of both main p1 types (Spuesens et al., 2010). The MPN142 gene (historically named ORF6) contains a copy of the repetitive element RepMp5 and amplification/sequencing can be used to distinguish the main types 1 and 2 as well as some, but not all, other p1 types (Ruland et al., 1994; Kenri et al., 2020). Investigation of the protein composition of bacteria by MALDI-ToF is not only suitable for reliable characterization of mycoplasmas on the species level but also for differentiation of the two main types of M. pneumoniae (Pereyre et al., 2013; Xiao et al., 2014). Of note, the procedure is described for investigation of isolates. Finally, nanorod array surface-enhanced Raman spectroscopy was used for detection of M. pneumoniae and typing of isolates and strains in clinical throat swabs (Hennigan et al., 2010; Henderson et al., 2015). Special equipment and experienced staff are required for this typing method which is, therefore, reserved for specialized laboratories.
In comparison with M. pneumoniae, the number of procedures for typing M. genitalium strains is relatively small. The first method was investigation of a variable part of the MG_191 gene, which codes for the adhesin MgpB (Hjorth et al., 2006). This gene contains repetitive elements but the typing region near the 5′ end (nt 180–460 in type strain G37) was found to be stable during in vivo and in vitro passage of isolates and is not influenced by homologous recombination. The results of the study by Hjorth et al. (2006) confirmed not only the sexual transmission of M. genitalium between couples but also the usefulness of culture-independent mgpB typing to investigate the circulation of genotypes in different populations and to characterize strains in cases of treatment failure. Furthermore, analysis of different short tandem repeats in the gene MG_309, which codes for a surface-localized lipoprotein, is suitable for typing (Ma and Martin, 2004; Ma et al., 2008; McGowin et al., 2009). Combining the number of these repeats (AGT/AAT) with mgpB typing increases the discriminatory power. Further targets of strain discrimination as well as MLVA testing were found to be not stable, not discriminatory enough, too discriminatory (Cazanave et al., 2012) or were used in a very limited number of studies (Ma and Martin, 2004; Pineiro et al., 2019) to date.
Evaluation of Common Typing Methods
A sufficiently large number of data for comparison of the results of different methods are available for p1, MLV, MLS, and SNP typing in M. pneumoniae and for mgpB and MG_309 typing in M. genitalium (Table 1). Using p1 typing, calculated DI’s (Hunter and Gaston, 1988) from selected studies ranged between 0.42 and 0.68. To date, 15 p1 types have been described (Kenri et al., 2020; Xiao et al., 2020). The main types 1 and 2 contain specific repetitive sequences in their genomes, suggesting early differentiation of M. pneumoniae strains into these two lineages (Spuesens et al., 2009; Diaz and Winchell, 2016). In contrast, the 13 known variants can be assigned to the main genotypes but differ in one or both repetitive copies in the p1 gene. Criteria for recognizing a strain as variant should be set in the future (e.g., length of sequence which must be different from known p1 types to define a new type, deposition of full-length sequences of both RepMP2/3, and four elements of p1 gene of the strain in databases). Interestingly, by contrast with p1 type 1, more variant strains have been characterized among type 2. This might be attributed to type-specific differences in the functionality of proteins putatively involved in DNA recombination and repair in M. pneumoniae and M. genitalium (Sluijter et al., 2010; Hakim et al., 2021).
Table 1.
Characteristics of frequently used molecular approaches for typing Mycoplasma pneumoniae and Mycoplasma genitalium (results of studies with >25 patients are included; if not presented in the study, HGDI’s are calculated according to the data).
p1, sequence differences in p1 gene (MPN141); MLVA, multilocus variable number of tandem-repeat analysis; MLST, multilocus sequence typing; SNP, determination of single polynucleotide polymorphisms; mgpB, sequence differences in mgpB gene (MG_191); VNTR MG_309, variable number of tandem repeat in gene MG_309; HGDI, discriminatory index (Hunter and Gaston, 1988); and n.d., not determined.
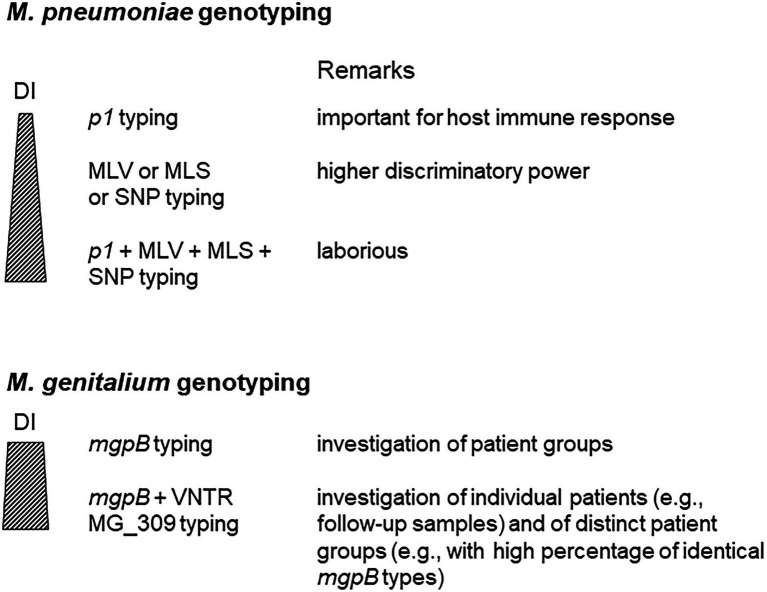
As an example for the occurrence of MLV types in various populations, the results of selected studies (characterization of >50 strains) in different countries are summarized in Table 1 and Figure 1A. With at least 24 known types, MLVA demonstrated a greater number of genotypes in comparison with p1 typing. However, calculated DI’s in these reports are not substantially higher due to the strong dominance (94%) of the three MLV types 4,572, 3,562, and 3,662, respectively. After review of further studies, additional MLV types were found in a small number of strains, underlining the need to summarize the typing results in an appropriate manner (preferably in a database). In contrast, 46 MLS types have been registered in the corresponding database and recent studies using MLST resulted in DI’s between 0.66 and 0.78, respectively. Higher DI’s up to 0.84 were calculated for SNP typing despite the fact that the overall number of SNP types is relatively low. However, it should be mentioned that this method was used to a lesser extent in comparison with the other typing methods up to now. Hence, data on the discriminatory power of this method might be preliminary.
Figure 1.
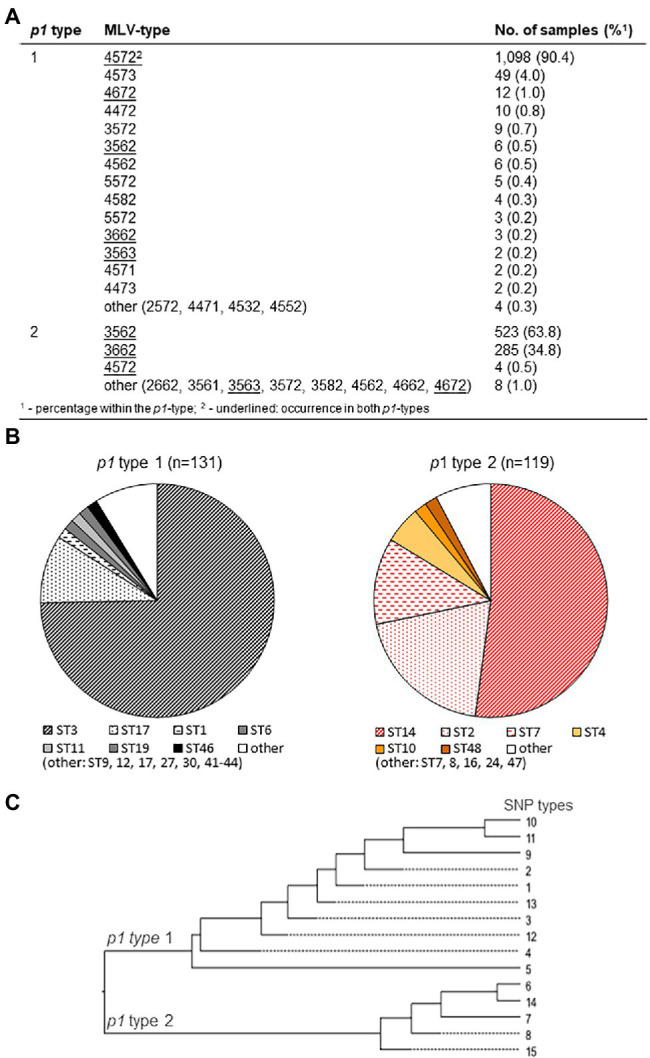
Association of main p1 types 1 and 2 (including variants) with multi-locus variable-number (MLV; A; using data of Benitez et al., 2012; Brown et al., 2015; Dumke et al., 2015; Touati et al., 2015; Kogoj et al., 2018; Zhao et al., 2019; Kenri et al., 2020; Xiao et al., 2020; Dumke and Rodriguez, 2021; Meyer Sauteur et al., 2021), MLS (B; data of Brown et al., 2015; Kenri et al., 2020; Meyer Sauteur et al., 2021; and Dumke and Rodriguez, 2021), and single nucleotide polymorphism (SNP) types (C; data of Touati et al., 2015; Kenri et al., 2020; Dumke and Rodriguez, 2021), respectively.
According to the calculated DI’s, adequate discrimination of M. pneumoniae strains (DI ≥ 0.9) can only be achieved if different typing methods are used. For selected examples (reports in which typing results for all strains are listed), the discrimination power of method combinations was calculated in Table 1. In practice, such an approach is laborious and the volume of DNA obtained after use of automated systems for DNA preparation from clinical specimens could be too low to perform three or four typing procedures in parallel.
As the genomes of strains belonging to one of the two main lineages of M. pneumoniae are strongly related, an association of p1 types 1 and 2 with genotypes identified by other typing methods is probable. As summarized in Figure 1A, many MLV types (79%) can be assigned exclusively to one of the main genotypes. In contrast, five MLV types occurred in both main lineages but in most cases (like 4,572, 3,562, and 3,662) with a clear numerical preference for a p1 type. As errors in counting the tandem repeats cannot be excluded, consideration of guide line for VNTR typing of M. pneumoniae (Chalker et al., 2015) is strongly recommended. Although the number of strains with a MLV type not belonging to the preferred p1 type is low (0.9%), MLV typing cannot be used to replace p1 typing. In contrast, the known MLS and SNP types are in complete agreement with p1 types (Figures 1B,C).
Regarding M. genitalium, mgpB typing has resulted in a large number of types, confirming the variability of the region of mgpB gene used. Nearly 250 mgpB types have now been described (Table 1). To answer epidemiological questions regarding the distribution of strains in different human populations, the stability and strong heterogeneity of the typing region allow reliable characterization of isolates. The corresponding DI’s for mgpB sequencing in different studies varied between 0.82 and 0.99 whereas MG_309 typing resulted in discrimination between 0.84 and 0.95. Lower DI’s seem to be associated with the investigation of more local than national populations (Dumke et al., 2020; Dumke and Spornraft-Ragaller, 2021), indicating a lower number of circulating genotypes. DI’s of ≥0.95 were obtained after the mgpB and MG_309 methods were combined. With these values, further optimization of typing for M. genitalium does not seem necessary. For standardized comparison of mgpB sequences, the defined part of the gene (position 221,749–222,029 in the genome of strain G37, GenBank no. NC_000908.2) should be fully sequenced. For example, when analyzing several deposited sequences, differentiation between type 2 and 74 is not possible due to their short lengths. Additionally, determination of the number of tandem repeats at locus MG_309 can be complicated in practice by the presence of mixed sequences in some samples. Careful inspection of the results after sequencing in both directions can help to solve this problem (Ma et al., 2008).
Association of Typing Results With Epidemiological and Clinical Parameters of Infections
Besides the use of typing to elucidate the epidemiological correlations of infections due to M. pneumoniae and M. genitalium, relating typing results to clinically relevant aspects is of practical importance for clinicians. These include associations between genotypes and severity of clinical disease, distinct symptoms of infection, site of infection, or antibiotic resistance. Regarding M. pneumoniae, the association between typing results and clinical aspects has been investigated in only a few studies. In comparison with other MLVA types, a statistically significantly higher pneumonia severity index, longer duration of cough, and older age of patients were demonstrated after infection with the mainly p1 1-specific type 4/5/7/2 (Qu et al., 2013). In a further study, a higher rate of severe pneumonia was demonstrated if children were infected with p1 type 1 vs. type 2 (Fan et al., 2017). In contrast, Yan et al. (2019) reported a higher rate of pediatric patients with pleural effusion as a severe complication of M. pneumoniae pneumonia after infection with MLVA type 3/5/6/2 (mainly p1 type 2) vs. infection with type 4/5/7/2. In a study among Slovenian children, infection with p1 type 2 strains resulted in an elevated C-reactive protein level and a higher rate of hospital admissions in comparison with p1 type 1 infections (Rodman Berlot et al., 2021). Regarding extrapulmonary manifestations of infections, only very few reports have dealt with the possible influence of the genotype on this complex of diseases. In the case of M. pneumoniae-induced mucocutaneous disease (Steven-Johnson syndrome), a rare but severe complication of infection, genotype does not seem to be a determinant of clinical symptoms (Olson et al., 2015; Watkins et al., 2017; Meyer Sauteur et al., 2021). In conclusion, the limited and partly heterogeneous results underline the need for further clinical studies with well-characterized patient populations of different age to confirm or exclude correlations of genotypes with clinical manifestations and outcomes of M. pneumoniae infections. This should include investigation of the pathogen in the lower and upper respiratory tract of patients, which might influence the ratio of p1 genotypes (Xiao et al., 2020). Furthermore, typing studies among symptomatic patients and asymptomatic carriers would be helpful to clarify if genotypes play a role in the differences in the clinical manifestation of M. pneumoniae infections between the groups (Spuesens et al., 2013; de Groot et al., 2021). In addition, further genetic, proteomic, and phenotypic investigations will help to understand differences between genotypes, which might explain clinical aspects of infections. To date, the main p1 types vary with regard to the development of biofilms (Simmons et al., 2013) and expression of the CARDS toxin (Lluch-Senar et al., 2015), which is a virulence factor with special relevance in pathogenesis (Su et al., 2021).
Circulation of different p1 types in the human population has been suggested to explain the typical epidemiology of infections as the immunodominant P1 protein is crucial for adhesion of bacteria to the cells of the respiratory epithelium as the first and essential stage in clinical manifestation. Regions of this adhesin, which contains repetitive sequences, were characterized as surface-located and it can be assumed that type-specific antibodies will be produced during host colonization (Dumke et al., 2008; Schurwanz et al., 2009; Nakane et al., 2011; Vizarraga et al., 2020). If these antibodies play a role in the adherence process, their quantitative occurrence in a host population could influence the distribution of type 1 or 2 strains (Figure 2). Based on the time-dependent level of herd immunity, the reported type shifts of p1 types in combination with an increase in the prevalence of infection might be explained by the presence of and change in type-specific antibodies. Studies have confirmed the polyclonality of strains in nation-wide investigations as a precondition for changes of genotypes and a varying dominance of type 1 or 2 in different regions (Kogoj et al., 2018; Lee et al., 2018) as well as a time period of 5–10 years for type change in Japan (Kenri et al., 2020). In some cases, the temporary dominance of a p1 type reaches more than 90% (Sun et al., 2017; Kenri et al., 2020). In contrast, if levels of type-specific antibodies do not differ greatly, the occurrence of both p1 types without dominance of one lineage is demonstrated (Jacobs et al., 2015; Xiao et al., 2020; Guo et al., 2022). Further studies are needed to confirm the association between type and type-specific antibodies experimentally (Dumke et al., 2010). However, results of mathematical models support the hypothesis of co-circulation of both p1 types and the importance of herd immunity for the ratio of genotypes (Omori et al., 2015; Zhang et al., 2019). This pattern seems primarily independent of the rate of resistance in the corresponding population. Despite the regional emergence of types with high rates of macrolide resistance (Ho et al., 2015; Lee et al., 2018; Hung et al., 2021), a clear association of resistance with distinct genotypes of M. pneumoniae was not found. Recently, an association between the number of tandem repeats in subunit S of the type I restriction-modification system (MPN085) and macrolide resistance in MLST-3 strains was reported (Lee et al., 2022). This interesting aspect should be investigated in future studies including further types. It can be assumed that the regional/national use of macrolides and the treatment regime in particular patients will be crucial for the development of resistant strains. Up to now, there is no in vivo or in vitro evidence for a greater rate of resistance among the described genotypes. Thus, antibiotic pressure in a population will lead to an increasing rate of resistance in the dominating type(s), which is primarily determined by the circulation of p1 types and, due to the association with the main p1 types, secondarily by the regional occurrence of MLV, MLS, or SNP types (Figure 1). Consequently, unsubstantiated use of macrolides in combination with the lack of resistance-guided therapy regimes in many settings worldwide will result in the selection of resistant strains. This hypothesis is supported by the results of studies reporting a decrease in the resistance rate after genotype change (Nakamura et al., 2021), followed by a subsequent increase among strains of the previously nondominant genotype (Wang et al., 2021). In small-scale outbreaks, time-dependent emergence of resistance among strains of the same genotype can be found (Hubert et al., 2021), emphasizing the need for mutation analysis in cases of treatment failure.
Figure 2.
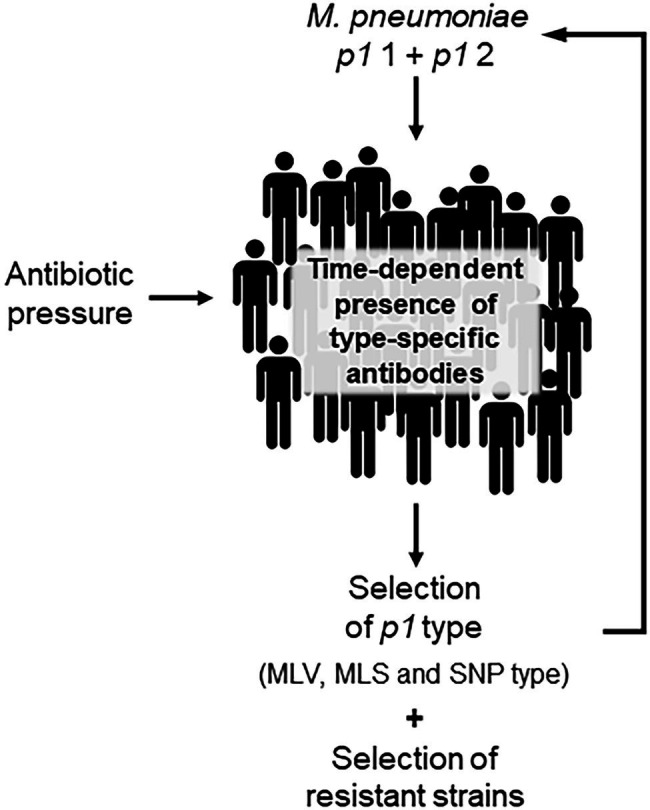
Schematic illustration of time-dependent shift of Mycoplasma pneumoniae genotypes.
As regard M. genitalium, data about an association between genotypes and clinical aspects of infection are not currently available. After establishing appropriate and comparable methods, more than 20 typing studies have been published and have found many interesting aspects of the epidemiology of the pathogen (Table 2). This aggregation of data includes not only aspects of the transmission of infections among patients with different sexual preferences but also investigated treatment approaches in individual patients that are of great importance for comprehensive evaluation of the clinical management of infections. Overall, these reports confirmed the remarkable genetic diversity of the typing region of the mgpB gene among patients of different populations. However, great differences in type frequency have been found. Especially in the risk group of MSM, a striking dominance of type 4 strains in different geographic regions was noted (Figure 3A). It remains unclear whether this is a result of the widespread occurrence of this type in a relatively self-contained sexual network (Fernandez-Huerta et al., 2020b) and/or of a selection advantage in the rectal microenvironment (Guiraud et al., 2021). Despite the high discriminatory power of mgpB typing, future spread of distinct types in particular populations might require an increased use of the MG_309 method for successful differentiation (e.g., for comparison of first samples and a positive test of cure; Pineiro et al., 2019; Dumke et al., 2020; Dumke and Spornraft-Ragaller, 2021). In addition, type 4 strains have been found to be macrolide resistant in many cases. However, high rates of macrolide resistance are also observed in other genotypes that occur frequently among the different MSM populations but appear to be lower in rarer types (Figure 3B). In contrast, a clear distribution pattern of quinolone resistance among these genotypes is lacking. Independent mutation events can be assumed, suggesting that the development of resistance is multiclonal, which might explain the lack of a correlation between genotypes and resistance. Besides transmission of resistant strains, acquired resistance after drug exposure is important for the spread of unsusceptible types (Pineiro et al., 2019). As discussed for M. pneumoniae, acquisition of resistance can be expected for any genotype of M. genitalium, and regional differences in prescriptions and consumption of macrolides and quinolones might play an important role in the resistance rate among circulating strains (Kenyon et al., 2021). This is especially the case in risk groups for sexually transmitted infections, who often receive antibiotic therapy against infections with pathogens other than M. genitalium. Currently, little is known about the consequences for the fitness of mycoplasma strains of acquisition of antibiotic resistance and this might be an object for future studies (Guiraud et al., 2021). In practice, clinicians should be aware that types with high rates of resistance commonly circulate in risk populations. Analogously to the P1 adhesin of M. pneumoniae, MgpB of M. genitalium contains repetitive elements and is an immunodominant protein. Different studies have confirmed changes of the gene sequence in the course of infection, which are related to distinct regions of the gene (Iverson-Cabral et al., 2006; Ma et al., 2010; Burgos et al., 2018; Wood et al., 2020). These recombination processes are of importance for the interaction with the host immune system but do not involve the typing region. This part of the protein was found to be antigenic as well as surface-localized but corresponding antibodies were not associated with the inhibition of hemadsorption (Iverson-Cabral et al., 2015; Aparicio et al., 2020). Nevertheless, sequence differences between mgpB types result in amino acid changes (Musatovova and Baseman, 2009; Ma et al., 2010; Dumke et al., 2020; Fernandez-Huerta et al., 2020b). Further studies should analyze if these differences or potential conformation changes of MgpB after recombination events in strains with constant mgpB type will have an influence on pathogenesis (Wood et al., 2020). Examples of long-term colonization of patients with the same type (Hjorth et al., 2006; Dumke et al., 2020; Dumke and Spornraft-Ragaller, 2021) and the non-reactivity of the conserved N-terminus with antibodies from infected animals (Iverson-Cabral et al., 2015) may suggest that the typing region of MgpB is of limited importance for interaction with the host immune system.
Table 2.
Results of Mycoplasma genitalium typing studies.
| Main aspect | Typing method | Patients/country | No. of patients or samples | Main result(s) of the study | Reference |
|---|---|---|---|---|---|
| Methology/epidemiology | mgpB | Not specified + couples/worldwide | 267 | First description of mgpB typing; usefulness for investigation of sexual networks and treatment failures | Hjorth et al., 2006 |
| MgpB recombination | mgpB | Women/Kenya | 9 | Intrastrain mgpB heterogeneity due to recombination | Iverson-Cabral et al., 2006 |
| Methology/epidemiology | mgpB + VNTR MG_309 | Not specified + couples/worldwide | 105 | Description of MG_309 typing; usefulness of mgpB + VNTR MG_309 typing for general epidemiological studies | Ma et al., 2008 |
| Methology/epidemiology | mgpB + VNTR MG_309 | Men + women with and without symptoms/France + Tunisia | 76 | Comparison of methods; mgpB typing for general epidemiological studies; mgpB + VNTR MG_309 typing for sexual-network studies; and MLVA not suitable | Cazanave et al., 2012 |
| Epidemiology | mgpB | Women with previous STD and partners/United States | 80 | Evaluation of sequence variability between strains from partners and occurrence of reinfections | Musatovova and Baseman, 2009 |
| Resistance | mgpB | Not specified/France | 136 | Evaluation of sequence variability, selection for mutation during treatment; and polyclonality of macrolide resistance | Chrisment et al., 2012 |
| Epidemiology | mgpB | Women in sexual health and family planning clinics/Guinea-Bissau | 30 | Diversity of circulating strains | Olsen et al., 2012 |
| Methology | mgpB + VNTR MG_309 |
Not specified/Cuba |
12 | Importance of typing for documentation of absence of cross-contamination | Mondeja et al., 2013 |
| Resistance/epidemiology | mgpB + VNTR MG_309 | Men with and without urethritis/United Kingdom | 22 | Two major clusters of genotypes with macrolide resistance in both clusters | Pond et al., 2014 |
| Resistance/epidemiology | mgpB + VNTR MG_309 | Men with NGU/Japan | 20 | Evaluation of genotype variability | Kikuchi et al., 2014 |
| Resistance/epidemiology | mgpB + VNTR MG_309 | MSM/Germany | 19 | Evaluation of genotype variability; comparison of first and follow-up samples | Dumke et al., 2016 |
| Resistance/follow-up | mgpB + VNTR MG_309 | Men (mainly MSM)/Germany | 163 | Evaluation of genotype variability; comparison of first and follow-up samples | Dumke et al., 2020 |
| Resistance/epidemiology | mgpB + VNTR MG_309 | Women in antenatal clinics/Solomon Islands | 41 | Two major clusters of genotypes, strain replacement after mass drug administration for trachoma elimination | Harrison et al., 2019 |
| Resistance/follow-up | mgpB + VNTR MG_309 | Patients with suspected STD/Spain | 79 | Differentiation of persistent and recurrent infections | Pineiro et al., 2019 |
| Resistance/epidemiology | mgpB + VNTR MG_309 | Heterosexual couples/US | 33 | Concordance of strains in couples | Xiao et al., 2019 |
| Resistance/epidemiology | mgpB + VNTR MG_309 | Mainly MSM/Spain | 54 | Two major clusters of genotypes with correlation to sexual networks and to macrolide resistance | Fernandez-Huerta et al., 2020b |
| Methology/epidemiology | mgpB | Patients of a sexual health center/Australia | 52 | Establishment of a custom amplicon sequencing approach for mgpB typing | Plummer et al., 2020 |
| Resistance/epidemiology | mgpB | Not specified/Australia | 89 | Genotype variability correlated with de novo acquisition of resistance | Sweeney et al., 2020 |
| Resistance/epidemiology | mgpB | Men in STD clinics/France | 78 | Lower diversity of types among macrolide-resistant strains | Guiraud et al., 2021 |
| Epidemiology | mgpB + VNTR MG_309 | Men and women with and without symptoms/South Africa | 38 | Circulation of different genotypes without geographic clustering | Laumen et al., 2021 |
| Resistance/follow-up | mgpB + VNTR MG_309 | Mainly MSM/Germany | 54 | Evaluation of first and follow-up samples during a resistance-guided treatment regime; two major clusters of genotypes with correlation to MSM and macrolide resistance | Dumke and Spornraft-Ragaller, 2021 |
| Resistance/epidemiology | mgpB | Asymptomatic MSM/Australia | 94 | Resistance not restricted to specific genotypes | Chua et al., 2021 |
STD, sexually transmitted disease; VNTR MG_309, variable number of tandem repeat in gene MG_309; NGU, non-gonococcal urethritis; and MSM, men who have sex with men.
Figure 3.
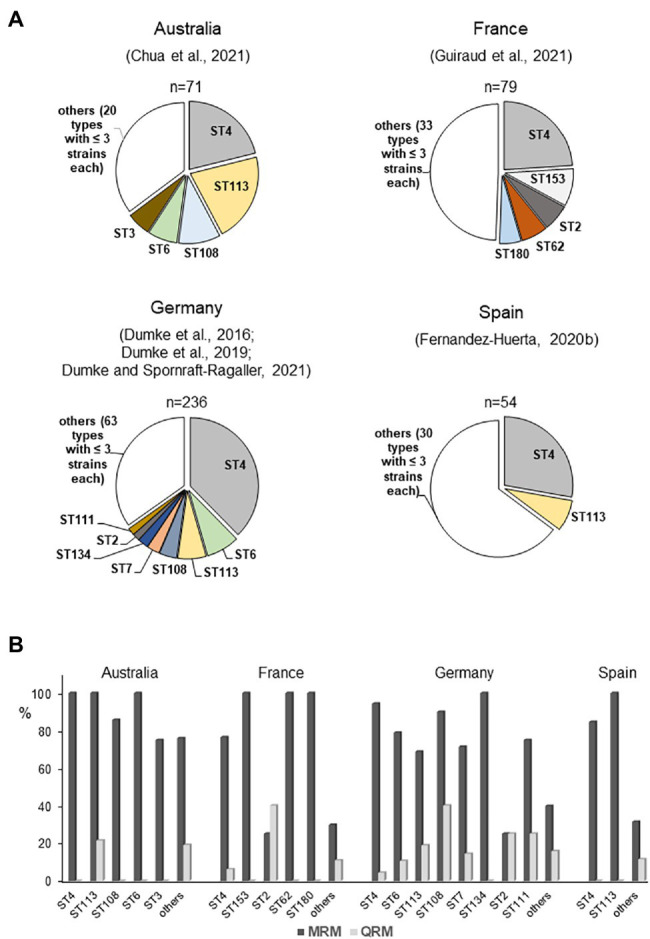
Regional distribution of mgpB types (A) and association between mgpB type and resistance (B) among strains in selected studies included mainly men who have sex with men (MSM; Dumke et al., 2016, 2020; Fernandez-Huerta et al., 2020b; Chua et al., 2021; Dumke and Spornraft-Ragaller, 2021; Guiraud et al., 2021). Strains with S83N change of ParC are not included. ST, mgpB type; MRM, macrolide resistance-associated mutation; and QRM, quinolone resistance-associated mutation.
Conclusion
Although whole-genome sequencing (WGS) is likely to replace current methods for molecular typing of M. pneumoniae and M. genitalium, the simplicity of these assays suggests that they may still have considerable value for epidemiological investigations. This is especially the case if isolates are not available as expected outside of reference laboratories. Whereas the discriminatory power of mgpB/MG_309 characterization of M. genitalium is high enough for successful differentiation of strains in sexual networks as well as in individual patients, several methods must be combined to reach a DI ≥ 0.9 for typing of M. pneumoniae strains (Figure 4). In contrast to the variability of the typing region in the MgpB adhesin of M. genitalium, the occurrence of p1 types in M. pneumoniae seems naturally limited by the formation of a functional adhesion complex. For this pathogen, optimization of alternative typing approaches, such as an increase of loci for MLVA, has the potential to enhance their actual discriminatory power. Further studies are needed to evaluate the potential of combinations of current approaches to set internationally accepted recommendations for DI requirement of M. pneumoniae typing. At present, only limited knowledge is available regarding the correlation of genotypes with clinical aspects of infections caused by M. pneumoniae and M. genitalium. These include an association with high regional or population-specific rates of resistance.
Figure 4.
Use of different approaches for Mycoplasma pneumoniae and Mycoplasma genitalium typing. DI, discriminatory index.
Author Contributions
The author confirms being the sole contributor of this work and has approved it for publication.
Conflict of Interest
The author declares that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Footnotes
References
- Aparicio D., Scheffer M. P., Marcos-Silva M., Vizarraga D., Sprankel L., Ratera M., et al. (2020). Structure and mechanism of the nap adhesion complex from the human pathogen Mycoplasma genitalium. Nat. Commun. 11:2877. doi: 10.1038/s41467-020-16511-2, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baumann L., Cina M., Egli-Gany D., Goutaki M., Halbeisen F. S., Lohrer G. R., et al. (2018). Prevalence of Mycoplasma genitalium in different population groups: systematic review and meta-analysis. Sex. Transm. Infect. 94, 255–262. doi: 10.1136/sextrans-2017-053384, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benitez A. J., Diaz M. H., Wolff B. J., Pimentel G., Njenga M. K., Estevez A., et al. (2012). Multilocus variable-number tandem-repeat analysis of Mycoplasma pneumoniae clinical isolates from 1962 to the present: a retrospective study. J. Clin. Microbiol. 50, 3620–3626. doi: 10.1128/JCM.01755-12, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown R. J., Holden M. T., Spiller O. B., Chalker V. J. (2015). Development of a multilocus sequence typing scheme for molecular typing of Mycoplasma pneumoniae. J. Clin. Microbiol. 53, 3195–3203. doi: 10.1128/JCM.01301-15, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown R. J., Nguipdop-Djomo P., Zhao H., Stanford E., Spiller O. B., Chalker V. J. (2016). Mycoplasma pneumoniae epidemiology in England and Wales: a national perspective. Front. Microbiol. 7:157. doi: 10.3389/fmicb.2016.00157, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burgos R., Wood G. E., Iverson-Cabral S. L., Totten P. A. (2018). Mycoplasma genitalium nonadherent phase variants arise by multiple mechanisms and escape antibody-dependent growth inhibition. Infect. Immun. 86, e00866–e00817. doi: 10.1128/IAI.00866-17, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cazanave C., Charron A., Renaudin H., Bebear C. (2012). Method comparison for molecular typing of French and Tunisian Mycoplasma genitalium-positive specimens. J. Med. Microbiol. 61, 500–506. doi: 10.1099/jmm.0.037721-0, PMID: [DOI] [PubMed] [Google Scholar]
- Chalker V. J., Pereyre S., Dumke R., Winchell J., Khosla P., Sun H., et al. (2015). International Mycoplasma pneumoniae typing study: interpretation of M. pneumoniae multilocus variable-number tandem-repeat analysis. New Microb. New Infect. 7, 37–40. doi: 10.1016/j.nmni.2015.05.005, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chrisment D., Charron A., Cazanave C., Pereyre S., Bebear C. (2012). Detection of macrolide resistance in Mycoplasma genitalium in France. J. Antimicrob. Chemother. 67, 2598–2601. doi: 10.1093/jac/dks263, PMID: [DOI] [PubMed] [Google Scholar]
- Chua T. P., Bodiyabadu K., Machalek D. A., Garland S. M., Bradshaw C. S., Plummer E. L., et al. (2021). Prevalence of Mycoplasma genitalium fluoroquinolone-resistance markers, and dual-class-resistance markers, in asymptomatic men who have sex with men. J. Med. Microbiol. 70:001429. doi: 10.1099/jmm.0.001429, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Groot R. C. A., Cristina Estevão S., Meyer Sauteur P. M., Perkasa A., Hoogenboezem T., Spuesens E. B. M., et al. (2021). Mycoplasma pneumoniae carriage evades induction of protective mucosal antibodies. Eur. Respir. J. 59:2100129. doi: 10.1183/13993003.00129-2021, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Degrange S., Cazanave C., Charron A., Renaudin H., Bebear C., Bebear C. M. (2009). Development of multiple-locus variable-number tandem-repeat analysis for molecular typing of Mycoplasma pneumoniae. J. Clin. Microbiol. 47, 914–923. doi: 10.1128/JCM.01935-08, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diaz M. H., Desai H. P., Morrison S. S., Benitez A. J., Wolff B. J., Caravas J., et al. (2017). Comprehensive bioinformatics analysis of Mycoplasma pneumoniae genomes to investigate underlying population structure and type-specific determinants. PLoS One 12:e0174701. doi: 10.1371/journal.pone.0174701, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diaz M. H., Winchell J. M. (2016). The evolution of advanced molecular diagnostics for the detection and characterization of Mycoplasma pneumoniae. Front. Microbiol. 7:232. doi: 10.3389/fmicb.2016.00232, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dumke R., Jacobs E. (2011). Culture-independent multi-locus variable-number tandem-repeat analysis (MLVA) of Mycoplasma pneumoniae. J. Microbiol. Methods 86, 393–396. doi: 10.1016/j.mimet.2011.06.008, PMID: [DOI] [PubMed] [Google Scholar]
- Dumke R., Lück P. C., Noppen C., Schaefer C., von Baum H., Marre R., et al. (2006). Culture-independent molecular subtyping of Mycoplasma pneumoniae in clinical samples. J. Clin. Microbiol. 44, 2567–2570. doi: 10.1128/JCM.00495-06, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dumke R., Rodriguez N. (2021). Use of different approaches for the culture-independent typing of Mycoplasma pneumoniae from two geographically distinct regions. J. Microbiol. Methods 186:106239. doi: 10.1016/j.mimet.2021.106239, PMID: [DOI] [PubMed] [Google Scholar]
- Dumke R., Rust M., Glaunsinger T. (2020). MgpB types among Mycoplasma genitalium strains from men who have sex with men in Berlin, Germany, 2016-2018. Pathogens 9:12. doi: 10.3390/pathogens9010012, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dumke R., Schnee C., Pletz M. W., Rupp J., Jacobs E., Sachse K., et al. (2015). Mycoplasma pneumoniae and Chlamydia spp. infection in community-acquired pneumonia, Germany, 2011-2012. Emerg. Infect. Dis. 21, 426–434. doi: 10.3201/eid2103.140927, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dumke R., Schurwanz N., Jacobs E. (2008). Characterisation of subtype- and variant-specific antigen regions of the P1 adhesin of Mycoplasma pneumoniae. Int. J. Med. Microbiol. 298, 483–491. doi: 10.1016/j.ijmm.2007.06.002, PMID: [DOI] [PubMed] [Google Scholar]
- Dumke R., Spornraft-Ragaller P. (2021). Antibiotic resistance and genotypes of Mycoplasma genitalium during a resistance-guided treatment regime in a German university hospital. Antibiotics 10:962. doi: 10.3390/antibiotics10080962, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dumke R., Thürmer A., Jacobs E. (2016). Emergence of Mycoplasma genitalium strains showing mutations associated with macrolide and fluoroquinolone resistance in the region Dresden, Germany. Diagn. Microbiol. Infect. Dis. 86, 221–223. doi: 10.1016/j.diagmicrobio.2016.07.005, PMID: [DOI] [PubMed] [Google Scholar]
- Dumke R., von Baum H., Lück P. C., Jacobs E. (2010). Subtypes and variants of Mycoplasma pneumoniae: local and temporal changes in Germany 2003-2006 and absence of a correlation between the genotype in the respiratory tract and the occurrence of genotype-specific antibodies in the sera of infected patients. Epidemiol. Infect. 138, 1829–1837. doi: 10.1017/S0950268810000622, PMID: [DOI] [PubMed] [Google Scholar]
- Fan L., Li D., Zhang L., Hao C., Sun H., Shao X., et al. (2017). Pediatric clinical features of Mycoplasma pneumoniae infection are associated with bacterial P1 genotype. Exp. Ther. Med. 14, 1892–1898. doi: 10.3892/etm.2017.4721, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernandez-Huerta M., Barbera M. J., Serra-Pladevall J., Esperalba J., Martínez-Gomez X., Centeno C., et al. (2020a). Mycoplasma genitalium and antimicrobial resistance in Europe: a comprehensive review. Int. J. STD AIDS 31, 190–197. doi: 10.1177/0956462419890737, PMID: [DOI] [PubMed] [Google Scholar]
- Fernandez-Huerta M., Serra-Pladevall J., Esperalba J., Moreno-Mingorance A., Fernandez-Naval C., Barbera M. J., et al. (2020b). Single-locus-sequence-based typing of the mgpB gene reveals transmission dynamics in Mycoplasma genitalium. J. Clin. Microbiol. 58, e01886–e01819. doi: 10.1128/JCM.01886-19, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fookes M. C., Hadfield J., Harris S., Parmar S., Unemo M., Jensen J. S., et al. (2017). Mycoplasma genitalium: whole genome sequence analysis, recombination and population structure. BMC Genomics 18:993. doi: 10.1186/s12864-017-4399-6, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gnanadurai R., Fifer H. (2020). Mycoplasma genitalium: A review. Microbiology 166, 21–29. doi: 10.1099/mic.0.000830 [DOI] [PubMed] [Google Scholar]
- Guiraud J., Lounnas M., Boissiere A., Le Roy C., Elguero E., Banuls A. L., et al. (2021). Lower mgpB diversity in macrolide-resistant Mycoplasma genitalium infecting men visiting two sexually transmitted infection clinics in Montpellier, France. J. Antimicrob. Chemother. 76, 43–47. doi: 10.1093/jac/dkaa410, PMID: [DOI] [PubMed] [Google Scholar]
- Guo Z., Liu L., Gong J., Han N., He L., Wang W., et al. (2022). Molecular features and antimicrobial susceptibility of Mycoplasma pneumoniae isolates from pediatric inpatients in Weihai, China. J. Glob. Antimicrob. Resist. 28, 180–184. doi: 10.1016/j.jgar.2022.01.002, PMID: [DOI] [PubMed] [Google Scholar]
- Hakim M. S., Annisa L., Jariah R. O. A., Vink C. (2021). The mechanisms underlying antigenic variation and maintenance of genomic integrity in Mycoplasma pneumoniae and Mycoplasma genitalium. Arch. Microbiol. 203, 413–429. doi: 10.1007/s00203-020-02041-4, PMID: [DOI] [PubMed] [Google Scholar]
- Harrison M. A., Harding-Esch E. M., Marks M., Pond M. J., Butcher R., Solomon A. W., et al. (2019). Impact of mass drug administration of azithromycin for trachoma elimination on prevalence and azithromycin resistance of genital Mycoplasma genitalium infection. Sex. Transm. Infect. 95, 522–528. doi: 10.1136/sextrans-2018-053938, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henderson K. C., Benitez A. J., Ratliff A. E., Crabb D. M., Sheppard E. S., Winchell J. M., et al. (2015). Specificity and strain-typing capabilities of nanorod array-surface enhanced Raman spectroscopy for Mycoplasma pneumoniae detection. PLoS One 10:e0131831. doi: 10.1371/journal.pone.0131831, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hennigan S. L., Driskell J. D., Dluhy R. A., Zhao Y., Tripp R. A., Waites K. B., et al. (2010). Detection of Mycoplasma pneumoniae in simulated and true clinical throat swab specimens by nanorod array-surface-enhanced Raman spectroscopy. PLoS One 5:e13633. doi: 10.1371/journal.pone.0013633, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Himmelreich R., Hilbert H., Plagens H., Pirkl E., Li B. C., Herrmann R. (1996). Complete sequence analysis of the genome of the bacterium Mycoplasma pneumoniae. Nucleic Acids Res. 24, 4420–4449. doi: 10.1093/nar/24.22.4420, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Himmelreich R., Plagens H., Hilbert H., Reiner B., Herrmann R. (1997). Comparative analysis of the genomes of the bacteria Mycoplasma pneumoniae and Mycoplasma genitalium. Nucleic Acids Res. 25, 701–712. doi: 10.1093/nar/25.4.701, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hjorth S. V., Björnelius E., Lidbrink P., Falk L., Dohn B., Berthelsen L., et al. (2006). Sequence-based typing of Mycoplasma genitalium reveals sexual transmission. J. Clin. Microbiol. 44, 2078–2083. doi: 10.1128/JCM.00003-06, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ho P. L., Law P. Y., Chan B. W., Wong C. W., To K. K., Chiu S. S., et al. (2015). Emergence of macrolide-resistant Mycoplasma pneumoniae in Hong Kong is linked to increasing macrolide resistance in multilocus variable-number tandem-repeat analysis type 4-5-7-2. J. Clin. Microbiol. 53, 3560–3564. doi: 10.1128/JCM.01983-15, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsieh Y. C., Li S. W., Chen Y. Y., Kuo C. C., Chen Y. C., Chang I., et al. (2022). Global genome diversity and recombination in Mycoplasma pneumoniae. Emerg. Infect. Dis. 28, 111–117. doi: 10.3201/eid2801.210497, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hubert D., Dumke R., Weichert S., Welker S., Tenenbaum T., Schroten H. (2021). Emergence of macrolide-resistant Mycoplasma pneumoniae during an outbreak in a primary school: clinical characterization of hospitalized children. Pathogens 10:328. doi: 10.3390/pathogens10030328, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hung H. M., Chuang C. H., Chen Y. Y., Liao W. C., Li S. W., Chang I. Y., et al. (2021). Clonal spread of macrolide-resistant Mycoplasma pneumoniae sequence type-3 and type-17 with recombination on non-P1 adhesin among children in Taiwan. Clin. Microbiol. Infect. 27, 1169.e1–1169.e6. doi: 10.1016/j.cmi.2020.09.035, PMID: [DOI] [PubMed] [Google Scholar]
- Hunter P. R., Gaston M. A. (1988). Numerical index of the discriminatory ability of typing systems: an application of Simpson’s index of diversity. J. Clin. Microbiol. 26, 2465–2466. doi: 10.1128/jcm.26.11.2465-2466.1988, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iverson-Cabral S. L., Astete S. G., Cohen C. R., Rocha E. P., Totten P. A. (2006). Intrastrain heterogeneity of the mgpB gene in Mycoplasma genitalium is extensive in vitro and in vivo and suggests that variation is generated via recombination with repetitive chromosomal sequences. Infect. Immun. 74, 3715–3726. doi: 10.1128/IAI.00239-06, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iverson-Cabral S. L., Wood G. E., Totten P. A. (2015). Analysis of the Mycoplasma genitalium MgpB adhesin to predict membrane topology, investigate antibody accessibility, characterize amino acid diversity, and identify functional and immunogenic epitopes. PLoS One 10:e0138244. doi: 10.1371/journal.pone.0138244, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacobs E., Ehrhardt I., Dumke R. (2015). New insights in the outbreak pattern of Mycoplasma pneumoniae. Int. J. Med. Microbiol. 305, 705–708. doi: 10.1016/j.ijmm.2015.08.021, PMID: [DOI] [PubMed] [Google Scholar]
- Jansen K., Steffen G., Potthoff A., Schuppe A. K., Beer D., Jessen H., et al. (2020). STI in times of PrEP: high prevalence of chlamydia, gonorrhea, and mycoplasma at different anatomic sites in men who have sex with men in Germany. BMC Infect. Dis. 20:110. doi: 10.1186/s12879-020-4831-4, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jensen J. S., Cusini M., Gomberg M., Moi H., Wilson J., Unemo M. (2022). 2021 European guideline on the management of Mycoplasma genitalium infections. J. Eur. Acad. Dermatol. Venereol. 36, 641–650. doi: 10.1111/jdv.17972, PMID: [DOI] [PubMed] [Google Scholar]
- Kenri T., Suzuki M., Sekizuka T., Ohya H., Oda Y., Yamazaki T., et al. (2020). Periodic genotype shifts in clinically prevalent Mycoplasma pneumoniae strains in Japan. Front. Cell. Infect. Microbiol. 10:385. doi: 10.3389/fcimb.2020.00385, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kenyon C., Manoharan-Basil S. S., van Dijck C. (2021). Is there a resistance threshold for macrolide consumption? Positive evidence from an ecological analysis of resistance data from Streptococcus pneumoniae, Treponema pallidum, and Mycoplasma genitalium. Microb. Drug Resist. 27, 1079–1086. doi: 10.1089/mdr.2020.0490, PMID: [DOI] [PubMed] [Google Scholar]
- Khoury T., Sviri S., Rmeileh A. A., Nubani A., Abutbul A., Hoss S., et al. (2016). Increased rates of intensive care unit admission in patients with Mycoplasma pneumoniae: a retrospective study. Clin. Microbiol. Infect. 22, 711–714. doi: 10.1016/j.cmi.2016.05.028, PMID: [DOI] [PubMed] [Google Scholar]
- Kikuchi M., Ito S., Yasuda M., Tsuchiya T., Hatazaki K., Takanashi M., et al. (2014). Remarkable increase in fluoroquinolone-resistant Mycoplasma genitalium in Japan. J. Antimicrob. Chemother. 69, 2376–2382. doi: 10.1093/jac/dku164, PMID: [DOI] [PubMed] [Google Scholar]
- Kogoj R., Mrvic T., Praprotnik M., Kese D. (2015). Prevalence, genotyping and macrolide resistance of Mycoplasma pneumoniae among isolates of patients with respiratory tract infections, Central Slovenia, 2006 to 2014. Euro Surveill. 20. doi: 10.2807/1560-7917.ES.2015.20.37.30018, PMID: [DOI] [PubMed] [Google Scholar]
- Kogoj R., Praprotnik M., Mrvic T., Korva M., Kese D. (2018). Genetic diversity and macrolide resistance of Mycoplasma pneumoniae isolates from two consecutive epidemics in Slovenia. Eur. J. Clin. Microbiol. Infect. Dis. 37, 99–107. doi: 10.1007/s10096-017-3106-5, PMID: [DOI] [PubMed] [Google Scholar]
- Latimer R. L., Shilling H. S., Vodstrcil L. A., Machalek D. A., Fairley C. K., Chow E. P. F., et al. (2020). Prevalence of Mycoplasma genitalium by anatomical site in men who have sex with men: a systematic review and meta-analysis. Sex. Transm. Infect. 96, 563–570. doi: 10.1136/sextrans-2019-054310, PMID: [DOI] [PubMed] [Google Scholar]
- Laumen J. G. E., van Alphen L. B., Maduna L. D., Hoffman C. M., Klausner J. D., Medina-Marino A., et al. (2021). Molecular epidemiological analysis of Mycoplasma genitalium shows low prevalence of azithromycin resistance and a well-established epidemic in South Africa. Sex. Transm. Infect. 97, 152–156. doi: 10.1136/sextrans-2019-054371, PMID: [DOI] [PubMed] [Google Scholar]
- Lee J. K., Lee J. H., Lee H., Ahn Y. M., Eun B. W., Cho E. Y., et al. (2018). Clonal expansion of macrolide-resistant sequence type 3 Mycoplasma pneumoniae, South Korea. Emerg. Infect. Dis. 24, 1465–1471. doi: 10.3201/eid2408.180081, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee J. K., Seong M. W., Shin D., Kim J. I., Han M. S., Yeon Y., et al. (2019). Comparative genomics of Mycoplasma pneumoniae isolated from children with pneumonia: South Korea, 2010-2016. BMC Genomics 20:910. doi: 10.1186/s12864-019-6306-9, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee J. K., Seong M. W., Yun K. W., Choi E. H. (2022). Association of tandem repeat number variabilities in subunit S of the type I restriction-modification system with macrolide resistance in Mycoplasma pneumoniae. J. Clin. Med. 11:715. doi: 10.3390/jcm11030715, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lluch-Senar M., Cozzuto L., Cano J., Delgado J., Llórens-Rico V., Pereyre S., et al. (2015). Comparative “-omics” in Mycoplasma pneumoniae clinical isolates reveals key virulence factors. PLoS One 10:e0137354. doi: 10.1371/journal.pone.0137354, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma L., Jensen J. S., Mancuso M., Hamasuna R., Jia Q., McGowin C. L., et al. (2010). Genetic variation in the complete MgPa operon and its repetitive chromosomal elements in clinical strains of Mycoplasma genitalium. PLoS One 5:e15660. doi: 10.1371/journal.pone.0015660, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma L., Martin D. H. (2004). Single-nucleotide polymorphisms in the rRNA operon and variable numbers of tandem repeats in the lipoprotein gene among Mycoplasma genitalium strains from clinical specimens. J. Clin. Microbiol. 42, 4876–4878. doi: 10.1128/JCM.42.10.4876-4878.2004, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma L., Taylor S., Jensen J. S., Myers L., Lillis R., Martin D. H. (2008). Short tandem repeat sequences in the Mycoplasma genitalium genome and their use in a multilocus genotyping system. BMC Microbiol. 8:130. doi: 10.1186/1471-2180-8-130, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Machalek D. A., Tao Y., Shilling H., Jensen J. S., Unemo M., Murray G., et al. (2020). Prevalence of mutations associated with resistance to macrolides and fluoroquinolones in Mycoplasma genitalium: a systematic review and meta-analysis. Lancet Infect. Dis. 20, 1302–1314. doi: 10.1016/S1473-3099(20)30154-7, PMID: [DOI] [PubMed] [Google Scholar]
- McGowin C. L., Ma L., Martin D. H., Pyles R. B. (2009). Mycoplasma genitalium-encoded MG309 activates NF-kappaB via toll-like receptors 2 and 6 to elicit proinflammatory cytokine secretion from human genital epithelial cells. Infect. Immun. 77, 1175–1181. doi: 10.1128/IAI.00845-08, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer Sauteur P. M., Panisova E., Seiler M., Theiler M., Berger C., Dumke R. (2021). Mycoplasma pneumoniae genotypes and clinical outcome in children. J. Clin. Microbiol. 59:e0074821. doi: 10.1128/JCM.00748-21, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mondeja B. A., Jensen J. S., Rodríguez I., Morier L. F., Kouri V., Rodríguez N. M., et al. (2013). Isolation of Mycoplasma genitalium from patients with urogenital infections: first report from the Latin-American region. New Microb. New Infect. 1, 22–26. doi: 10.1002/2052-2975.20, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Musatovova O., Baseman J. B. (2009). Analysis identifying common and distinct sequences among Texas clinical strains of Mycoplasma genitalium. J. Clin. Microbiol. 47, 1469–1475. doi: 10.1128/JCM.01602-08, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Musatovova O., Kannan T. R., Baseman J. B. (2008). Genomic analysis reveals Mycoplasma pneumoniae repetitive element 1-mediated recombination in a clinical isolate. Infect. Immun. 76, 1639–1648. doi: 10.1128/IAI.01621-07, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamura Y., Oishi T., Kaneko K., Kenri T., Tanaka T., Wakabayashi S., et al. (2021). Recent acute reduction in macrolide-resistant Mycoplasma pneumoniae infections among Japanese children. J. Infect. Chemother. 27, 271–276. doi: 10.1016/j.jiac.2020.10.007, PMID: [DOI] [PubMed] [Google Scholar]
- Nakane D., Adan-Kubo J., Kenri T., Miyata M. (2011). Isolation and characterization of P1 adhesin, a leg protein of the gliding bacterium Mycoplasma pneumoniae. J. Bacteriol. 193, 715–722. doi: 10.1128/JB.00796-10, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Narita M. (2016). Classification of extrapulmonary manifestations due to Mycoplasma pneumoniae infection on the basis of possible pathogenesis. Front. Microbiol. 7:23. doi: 10.3389/fmicb.2016.00023, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olsen B., Mansson F., Camara C., Monteiro M., Biai A., Alves A., et al. (2012). Phenotypic and genetic characterisation of bacterial sexually transmitted infections in Bissau, Guinea-Bissau, West Africa: a prospective cohort study. BMJ Open 2:e000636. doi: 10.1136/bmjopen-2011-000636, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olson D., Watkins L. K. F., Demirjian A., Lin X., Robinson C. C., Pretty K., et al. (2015). Outbreak of Mycoplasma pneumoniae-associated Stevens-Johnson syndrome. Pediatrics 136, e386–e394. doi: 10.1542/peds.2015-0278, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Omori R., Nakata Y., Tessmer H. L., Suzuki S., Shibayama K. (2015). The determinant of periodicity in Mycoplasma pneumoniae incidence: an insight from mathematical modelling. Sci. Rep. 5:14473. doi: 10.1038/srep14473, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pereyre S., Tardy F. (2021). Integrating the human and animal sides of mycoplasmas resistance to antimicrobials. Antibiotics 10:1216. doi: 10.3390/antibiotics10101216, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pereyre S., Tardy F., Renaudin H., Cauvin E., Del Pra Netto Machado L., Tricot A., et al. (2013). Identification and subtyping of clinically relevant human and ruminant mycoplasmas by use of matrix-assisted laser desorption ionization-time of flight mass spectrometry. J. Clin. Microbiol. 51, 3314–3323. doi: 10.1128/JCM.01573-13, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pineiro L., Idigoras P., Cilla G. (2019). Molecular typing of Mycoplasma genitalium-positive specimens discriminates between persistent and recurrent infections in cases of treatment failure and supports contact tracing. Microorganisms 7:609. doi: 10.3390/microorganisms7120609, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plummer E. L., Murray G. L., Bodiyabadu K., Su J., Garland S. M., Bradshaw C. S., et al. (2020). A custom amplicon sequencing approach to detect resistance associated mutations and sequence types in Mycoplasma genitalium. J. Microbiol. Methods 179:106089. doi: 10.1016/j.mimet.2020.106089, PMID: [DOI] [PubMed] [Google Scholar]
- Pond M. J., Nori A. V., Witney A. A., Lopeman R. C., Butcher P. D., Sadiq S. T. (2014). High prevalence of antibiotic-resistant Mycoplasma genitalium in nongonococcal urethritis: the need for routine testing and the inadequacy of current treatment options. Clin. Infect. Dis. 58, 631–637. doi: 10.1093/cid/cit752, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qu J., Yu X., Liu Y., Yin Y., Gu L., Cao B., et al. (2013). Specific multilocus variable-number tandem-repeat analysis genotypes of Mycoplasma pneumoniae are associated with diseases severity and macrolide susceptibility. PLoS One 8:e82174. doi: 10.1371/journal.pone.0082174, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rocha E. P., Blanchard A. (2002). Genomic repeats, genome plasticity and the dynamics of Mycoplasma evolution. Nucleic Acids Res. 30, 2031–2042. doi: 10.1093/nar/30.9.2031, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodman Berlot J., Krivec U., Mrvic T., Kogoj R., Kese D. (2021). Mycoplasma pneumoniae p1 genotype indicates severity of lower respiratory tract infections in children. J. Clin. Microbiol. 59:e0022021. doi: 10.1128/JCM.00220-21, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruland K., Himmelreich R., Herrmann R. (1994). Sequence divergence in the ORF6 gene of Mycoplasma pneumoniae. J. Bacteriol. 176, 5202–5209. doi: 10.1128/jb.176.17.5202-5209.1994, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sasaki T., Kenri T., Okazaki N., Iseki M., Yamashita R., Shintani M., et al. (1996). Epidemiological study of Mycoplasma pneumoniae infections in Japan based on PCR-restriction fragment length polymorphism of the P1 cytadhesin gene. J. Clin. Microbiol. 34, 447–449. doi: 10.1128/jcm.34.2.447-449.1996, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schurwanz N., Jacobs E., Dumke R. (2009). Strategy to create chimeric proteins derived from functional adhesin regions of Mycoplasma pneumoniae for vaccine development. Infect. Immun. 77, 5007–5015. doi: 10.1128/IAI.00268-09, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwartz S. B., Mitchell S. L., Thurman K. A., Wolff B. J., Winchell J. M. (2009). Identification of P1 variants of Mycoplasma pneumoniae by use of high-resolution melt analysis. J. Clin. Microbiol. 47, 4117–4120. doi: 10.1128/JCM.01696-09, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simmons W. L., Daubenspeck J. M., Osborne J. D., Balish M. F., Waites K. B., Dybvig K. (2013). Type 1 and type 2 strains of Mycoplasma pneumoniae form different biofilms. Microbiology 159, 737–747. doi: 10.1099/mic.0.064782-0, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sluijter M., Kaptein E., Spuesens E. B., Hoogenboezem T., Hartwig N. G., van Rossum A. M., et al. (2010). The Mycoplasma genitalium MG352-encoded protein is a Holliday junction resolvase that has a non-functional orthologue in Mycoplasma pneumoniae. Mol. Microbiol. 77, 1261–1277. doi: 10.1111/j.1365-2958.2010.07288.x, PMID: [DOI] [PubMed] [Google Scholar]
- Spuesens E. B., Brouwer R. W., Mol K. H., Hoogenboezem T., Kockx C. E., Jansen R., et al. (2016). Comparison of Mycoplasma pneumoniae genome sequences from strains isolated from symptomatic and asymptomatic patients. Front. Microbiol. 7:1701. doi: 10.3389/fmicb.2016.01701, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spuesens E. B., Fraaij P. L., Visser E. G., Hoogenboezem T., Hop W. C., van Adrichem L. N., et al. (2013). Carriage of Mycoplasma pneumoniae in the upper respiratory tract of symptomatic and asymptomatic children: an observational study. PLoS Med. 10:e1001444. doi: 10.1371/journal.pmed.1001444, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spuesens E. B., Hoogenboezem T., Sluijter M., Hartwig N. G., van Rossum A. M., Vink C. (2010). Macrolide resistance determination and molecular typing of Mycoplasma pneumoniae by pyrosequencing. J. Microbiol. Methods 82, 214–222. doi: 10.1016/j.mimet.2010.06.004, PMID: [DOI] [PubMed] [Google Scholar]
- Spuesens E. B., Oduber M., Hoogenboezem T., Sluijter M., Hartwig N. G., van Rossum A. M. C., et al. (2009). Sequence variations in RepMP2/3 and RepMP4 elements reveal intragenomic homologous DNA recombination events in Mycoplasma pneumoniae. Microbiology 155, 2182–2196. doi: 10.1099/mic.0.028506-0, PMID: [DOI] [PubMed] [Google Scholar]
- Su C. J., Dallo S. F., Baseman J. B. (1990). Molecular distinctions among clinical isolates of Mycoplasma pneumoniae. J. Clin. Microbiol. 28, 1538–1540. doi: 10.1128/jcm.28.7.1538-1540.1990, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su X., You X., Luo H., Liang K., Chen L., Tian W., et al. (2021). Community-acquired respiratory distress syndrome toxin: unique exotoxin for M. pneumoniae. Front. Microbiol. 12:766591. doi: 10.3389/fmicb.2021.766591, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun H., Xue G., Yan C., Li S., Cao L., Yuan Y., et al. (2013). Multiple-locus variable-number tandem-repeat analysis of Mycoplasma pneumoniae clinical specimens and proposal for amendment of MLVA nomenclature. PLoS One 8:e64607. doi: 10.1371/journal.pone.0064607, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun H., Xue G., Yan C., Li S., Zhao H., Feng Y., et al. (2017). Changes in molecular characteristics of Mycoplasma pneumoniae in clinical specimens from children in Beijing between 2003 and 2015. PLoS One 12:e0170253. doi: 10.1371/journal.pone.0170253, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sweeney E. L., Tickner J., Bletchly C., Nimmo G. R., Whiley D. M. (2020). Genotyping of Mycoplasma genitalium suggests de novo acquisition of antimicrobial resistance in Queensland. Aust. J. Clin. Microbiol. 58, e00641–e00620. doi: 10.1128/JCM.00641-20, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian X. J., Dong Y. Q., Dong X. P., Li J. Y., Li D., Jiang Y., et al. (2013). P1 gene of Mycoplasma pneumoniae in clinical isolates collected in Beijing in 2010 and relationship between genotyping and macrolide resistance. Chin. Med. J. 126, 3944–3948. doi: 10.3760/cma.j.issn.0366-6999.20131643 [DOI] [PubMed] [Google Scholar]
- Touati A., Blouin Y., Sirand-Pugnet P., Renaudin H., Oishi T., Vergnaud G., et al. (2015). Molecular epidemiology of Mycoplasma pneumoniae: genotyping using single nucleotide polymorphisms and SNaPshot technology. J. Clin. Microbiol. 53, 3182–3194. doi: 10.1128/JCM.01156-15, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ursi D., Ieven M., van Bever H., Quint W., Niesters H. G., Goossens H. (1994). Typing of Mycoplasma pneumoniae by PCR-mediated DNA fingerprinting. J. Clin. Microbiol. 32, 2873–2875. doi: 10.1128/jcm.32.11.2873-2875.1994, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vizarraga D., Kawamoto A., Matsumoto U., Illanes R., Perez-Luque R., Martin J., et al. (2020). Immunodominant proteins P1 and P40/P90 from human pathogen Mycoplasma pneumoniae. Nat. Commun. 11:5188. doi: 10.1038/s41467-020-18777-y, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waites K. B., Xiao L., Liu Y., Balish M. F., Atkinson T. P. (2017). Mycoplasma pneumoniae from the respiratory tract and beyond. Clin. Microbiol. Rev. 30, 747–809. doi: 10.1128/CMR.00114-16, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y., Xu B., Wu X., Yin Q., Wang Y., Li J., et al. (2021). Increased macrolide resistance rate of M3562 Mycoplasma pneumoniae correlated with macrolide usage and genotype shifting. Front. Cell. Infect. Microbiol. 11:675466. doi: 10.3389/fcimb.2021.675466, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watkins L. K. F., Olson D., Diaz M. H., Lin X., Demirjian A., Benitez A. J., et al. (2017). Epidemiology and molecular characteristics of Mycoplasma pneumoniae during an outbreak of M. pneumoniae-associated Stevens-Johnson syndrome. Pediatr. Infect. Dis. J. 36, 564–571. doi: 10.1097/INF.0000000000001476, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wood G. E., Iverson-Cabral S. L., Gillespie C. W., Lowens M. S., Manhart L. E., Totten P. A. (2020). Sequence variation and immunogenicity of the Mycoplasma genitalium MgpB and MgpC adherence proteins during persistent infection of men with non-gonococcal urethritis. PLoS One 15:e0240626. doi: 10.1371/journal.pone.0240626, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao L., Ptacek T., Osborne J. D., Crabb D. M., Simmons W. L., Lefkowitz E. J., et al. (2015). Comparative genome analysis of Mycoplasma pneumoniae. BMC Genomics 16:610. doi: 10.1186/s12864-015-1801-0, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao L., Ratliff A. E., Crabb D. M., Mixon E., Qin X., Selvarangan R., et al. (2020). Molecular characterization of Mycoplasma pneumoniae isolates in the United States from 2012 to 2018. J. Clin. Microbiol. 58, e00710–e00720. doi: 10.1128/JCM.00710-20, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao L., Waites K. B., van der Pol B., Aaron K. J., Hook E. W., 3rd, Geisler W. M. (2019). Mycoplasma genitalium infections with macrolide and fluoroquinolone resistance-associated mutations in heterosexual African American couples in Alabama. Sex. Transm. Dis. 46, 18–24. doi: 10.1097/OLQ.0000000000000891, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao D., Zhao F., Zhang H., Meng F., Zhang J. (2014). Novel strategy for typing Mycoplasma pneumoniae isolates by use of matrix-assisted laser desorption ionization-time of flight mass spectrometry coupled with ClinProTools. J. Clin. Microbiol. 52, 3038–3043. doi: 10.1128/JCM.01265-14, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xue G., Li M., Wang N., Zhao J., Wang B., Ren Z., et al. (2018). Comparison of the molecular characteristics of Mycoplasma pneumoniae from children across different regions of China. PLoS One 13:e0198557. doi: 10.1371/journal.pone.0198557, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan C., Xue G., Zhao H., Feng Y., Li S., Cui J., et al. (2019). Molecular and clinical characteristics of severe Mycoplasma pneumoniae pneumonia in children. Pediatr. Pulmonol. 54, 1012–1021. doi: 10.1002/ppul.24327, PMID: [DOI] [PubMed] [Google Scholar]
- Zhang J., Song X., Ma M. J., Xiao L., Kenri T., Sun H., et al. (2017). Inter- and intra-strain variability of tandem repeats in Mycoplasma pneumoniae based on next-generation sequencing data. Future Microbiol. 12, 119–129. doi: 10.2217/fmb-2016-0111, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X. S., Zhao H., Vynnycky E., Chalker V. (2019). Positively interacting strains that co-circulate within a network structured population induce cycling epidemics of Mycoplasma pneumoniae. Sci. Rep. 9:541. doi: 10.1038/s41598-018-36325-z, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao F., Cao B., Li J., Song S., Tao X., Yin Y., et al. (2011). Sequence analysis of the p1 adhesin gene of Mycoplasma pneumoniae in clinical isolates collected in Beijing in 2008 to 2009. J. Clin. Microbiol. 49, 3000–3003. doi: 10.1128/JCM.00105-11, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao F., Li J., Liu J., Guan X., Gong J., Liu L., et al. (2019). Antimicrobial susceptibility and molecular characteristics of Mycoplasma pneumoniae isolates across different regions of China. Antimicrob. Resist. Infect. Control 8:143. doi: 10.1186/s13756-019-0576-5, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]