Abstract
Culture-independent molecular phylogenetic methods were used to explore the breadth of diversity and environmental distribution of members of the division-level “candidate” phylogenetic group WS6, recently discovered in a contaminated aquifer and with no cultivated representatives. A broad diversity of WS6-affiliated sequences were cloned from 7 of 12 environments investigated: mainly from anaerobic sediment environments. The number of sequences representing the WS6 candidate division was increased from 3 to 60 in this study. The extent of phylogenetic divergence (sequence difference) in this candidate division was found to be among the largest of any known bacterial division. This indicates that organisms representing the WS6 phylogenetic division offer a broad diversity of undiscovered biochemical and metabolic novelty. These results provide a framework for the further study of these evidently important kinds of organisms and tools, the sequences, with which to do so.
Perspective on the extent of bacterial diversity has expanded substantially in the past decade. In 1987, Woese could describe 12 main relatedness groups comprising the domain Bacteria, using 16S rRNA oligonucleotide catalogs and the few continuous 16S rRNA sequences then available (12). These relatedness groups have been termed “kingdoms,” “phyla,” or “divisions,”. We use the term “division” to describe a phylogenetic relatedness group of 16S rRNA sequences that are reproducibly monophyletic and unaffiliated with all other deepest branchings in the bacterial tree. Application of rapid sequencing techniques to cloned 16S rRNA genes from cultures, and especially to environmental samples, has revealed substantial additional diversity beyond the 12 divisions described by Woese (11). Currently, 36 to 38 phylogenetic divisions of Bacteria are indicated by analysis of ca. 15,000 rRNA sequences from cultured and environmental organisms (5). Thirteen of those phylogenetic divisions have only been encountered in sequence-based environmental surveys and currently have no cultivated representatives. Some sequence-defined phylogenetic divisions are represented by only a few (<10) sequences, so the extents of diversity within the clades are unknown.
Recent studies have found that representatives of bacterial divisions with few or no cultivated members are widely distributed in the environment and numerically seem to dominate many of the environments examined. Specifically, members of the bacterial divisions verrucomicrobia, acidobacteria, green nonsulfur, and “candidate” division OP11 occur abundantly in many different environments (3, 5, 6, 9). (The term “candidate” has been used to denote clades with no cultivated representative [5, 10] or with too few members [sequences] for reliable phylogenetic assessments [6].) It is still unclear how many division-level clades emerged during the evolution of the phylogenetic domain Bacteria and how widely these clades are distributed environmentally.
A recent survey of the microbial diversity in an anaerobic, hydrocarbon-contaminated aquifer described six novel, division-level clades of Bacteria (4). These new relatedness groups were indicated by only a few unique environmental rRNA sequences, however. One of the candidate divisions encountered in that contaminated aquifer study, WS6, was noteworthy for its abundance. WS6 sequences were present as a large percentage (up to 24%) of clone libraries in the study, indicating that organisms represented by the sequences are prevalent in the environment sampled (4) and may be important in other environments. Although the division was indicated by only three specific sequences in the original study, these sequences were greatly divergent in phylogenetic analyses from those of other known bacterial divisions. This extensive divergence has resulted in base changes in a region of the 16S rRNA gene (515F region) that in other bacteria is universally conserved. This sequence divergence facilitates the detection of WS6-related sequences. In order to substantiate the WS6 candidate division and document the breadth of phylogenetic diversity that it represents, as well as to explore its distribution in the environment, we analyzed 12 different environments for their content of WS6 organisms. The results indicate that representatives of the WS6 clade are widely distributed and in some environments are sufficiently abundant that they are probably important in biogeochemical processes.
MATERIALS AND METHODS
Sample collection and DNA extraction.
Sediment samples were collected from the upper 6 in. of the Bolinas and Berkeley marine estuaries in the San Francisco Bay, Calif., as well as from nonmarine Lake Lemon and Fairfax Swamp in Indiana. Soil samples were obtained from the methanogenic zone in a hydrocarbon-contaminated aquifer in Alameda, Calif., and from the upper 2 in. of a landscaped topsoil on the campus of the University of California, Berkeley. DNA from human intestinal wall samples was provided by Dan Frank (University of Colorado, Boulder). DNA from human fecal matter was provided by Phil Hugenholtz (University of Queensland, Sydney, Australia). Three hot spring samples were analyzed, all from Yellowstone National Park. One sample was taken from the sediment of Obsidian Pool, a 75 to 95°C hot spring that is rich in reduced iron, sulfide, carbon dioxide, and probably hydrogen and that contains a broad microbial diversity (2, 6). A second Yellowstone sample was derived from a dark green microbial mat in a 72°C pool in the White Creek area. A third Yellowstone sample was from 70°C orange-colored sediment in the Queens Laundry pool, in the Sentinal Creek area. The final sample examined was derived from a microbialite (stromatolite-like carbonate deposits possibly mediated by microorganisms) located at a depth of 46 feet in Pavillion Lake, a mountain lake in central British Columbia, Canada. Collected samples were generally frozen immediately in liquid nitrogen and then stored at −80°C until processed for nucleic acid extraction. The microbialite sample was preserved in 70% ethanol until extracted. DNA was extracted from samples by use of a bead beating protocol. Generally, the following protocol was used. Sample (0.5 to 1.0 g) was resuspended in 0.5 ml of sodium dodecyl sulfate (SDS)-buffer solution (200 mM Tris [pH 8.0], 20 mM EDTA, 200 mM NaCl, 2% SDS) and incubated for 20 min at 70°C. Samples were reciprocated on a Mini-Beadbeater (Biospec) at low speed for 2 min in the presence of 0.3 g of acid-washed zirconium-silica beads (0.1-mm diameter) and phenol. Nucleic acids were precipitated from the supernatant with sodium acetate and isopropanol and purified by passage through a Chroma Spin+TE-1000 column (Clontech Laboratories, Inc.).
PCR and cloning.
Community ribosomal DNAs (rDNAs) were amplified by PCR from 1 to 50 ng of DNA in reaction mixtures containing (as final concentrations) 1× PCR buffer II (Perkin-Elmer), 2.5 mM MgCl2, 200 μM each deoxynucleoside triphosphate, 300 nM each forward and reverse primer, and 0.025 U of AmpliTaq Gold DNA polymerase (Perkin-Elmer) per ml. Reaction mixtures were incubated in a Mastercycler Gradient thermal cycler (Eppendorf) at 94°C for 12 min (for initial denaturation and activation of AmpliTaq Gold); followed by 25 to 35 cycles at 94°C for 30 s, 50°C for 45 s, and 72°C for 1.5 min; followed by a final extension period of 12 min at 72°C. For all clone libraries except BMS (Table 1), rDNAs were amplified with universal reverse oligonucleotide primer 1492R (5′-GGTTACCTTGTTACGACTT-3′) (7) and WS6-specific forward primer WS6514F (5′-CGTGCCAGAAGCATCGGTG-3′) for both primary and secondary amplifications. For clone library BMS, the initial round of PCR was performed with the forward primer 27F (specific for Bacteria) (5′-AGAGTTTGATCCTGGCTCAG-3′) (7) and 1492R, and the second round was performed with 27F and OP11-specific primer OP11-1090R (5′-TCGTTGTCCCACTTAA-3′). (The WS6 clones from the OP11-specific primer represent nontarget sequences obtained from the BMS library.) PCR products were cloned with a TOPO TA cloning kit in accordance with the manufacturer's instructions (Invitrogen Corp.). Plasmid DNAs containing inserts were analyzed by restriction fragment length polymorphism (RFLP) analysis and sequenced as reported previously (4).
TABLE 1.
Environments sampled for members of candidate division WS6
| Environmenta | Annealing temp attempted (°C) | PCR product obtainedb | Library annealing temp (°C) | Library name | No. of clones
|
|
|---|---|---|---|---|---|---|
| Analyzed | Unique | |||||
| Hot spring cyanobacterial mat (YNP) | 50, 55, 60 | ++ | 55 | 1A | 48 | 2 |
| Berkeley marina sediment (Calif.) | 45, 50, 55, 60, 65 | ++ | 60 | BMS1 | 96 | 11 |
| Berkeley marina sediment (Calif.) | 55c | + | 55 | BMS | 96 | 3 |
| Bolinas sediment (Calif.) | 45, 50, 55, 60, 65 | ++ | 55 | BOL | 128 | 20 |
| Hydrocarbon-contaminated aquifer (Calif.) | 45, 50, 55, 60, 65 | ++ | 55 | CA1 | 128 | 11 |
| Hydrocarbon-contaminated aquifer (Mich.)d | 50 | ++ | 50 | WCHB | 192 | 3 |
| Obsidian Pool hot spring sediment (YNP) | 50, 55, 60 | + | 55 | OP | 4 | 1 |
| Lake Lemon (Ind.) sediment | 45, 50, 55, 60, 65 | ++ | 60 | LL | 32 | 8 |
| Landscaped topsoil (Berkeley) | 50, 55, 60 | ++ | 55 | D1 | 96 | 1 |
| Pavilion Lake (Canada) microbialite | 55, 60 | − | ||||
| Fairfax Swamp (Ind.) sediment sample | 55, 60 | − | ||||
| 2 Human fecal samples | 50, 55, 60 | − | ||||
| 4 Human intestine samples | 50, 55, 60 | − | ||||
| Queen's Laundry hot spring orange sediment (YNP) | 50, 55, 60 | − | ||||
YNP, Yellowstone National Park.
++, product after one round of PCR; +, product after two rounds of PCR; −, no PCR product detected.
WS6-specific primer not used in amplification.
Original study.
Phylogenetic analyses and chimera detection.
Sequences were compared to those in available databases by use of the BLAST (Basic Local Alignment Search Tool) network service (1) to determine their approximate phylogenetic affiliations. Partial sequences were compiled in AutoAssembler 2.1 (PE Applied Biosystems); compiled sequences were aligned by use of the ARB database (O. Strunk and W. Ludwig, ARB: a software environment for sequence data, 1999 [http://www.mikro.biologie.tu-muenchen.de]). Chimeric sequences were identified by secondary-structure anomalies and by branching-order discrepancies of independently inferred regions of the alignment as previously described (6). Sequence alignments used for phylogenetic inference were minimized by use of the Lane mask (8), which removes hypervariable regions of the SSU-rRNA alignment from the analysis, for bacterial data sets. (The alignment is available at http://crab3.colorado.edu/publications.html.) The dendrogram (Fig. 1) was constructed by use of the ARB database with evolutionary distance analysis (neighbor-joining algorithms with Olsen correction). The robustness of inferred topologies was tested by bootstrap resampling of trees calculated by evolutionary distance (test version 4.0b2 of PAUP*, a neighbor-joining algorithm with either Kimura two-parameter correction or maximum-likelihood correction with an empirically determined gamma distribution model of site-to-site rate variation and empirically determined base frequencies), parsimony (test version 4.0b2 of PAUP*; heuristic search), and maximum likelihood (fastDNAml) analyses [D. L. Swofford, PAUP*. Phylogenetic Analysis Using Parsimony (* and Other Methods). Version 4. Sinauer Associates, Sunderland, Mass., 1998.]
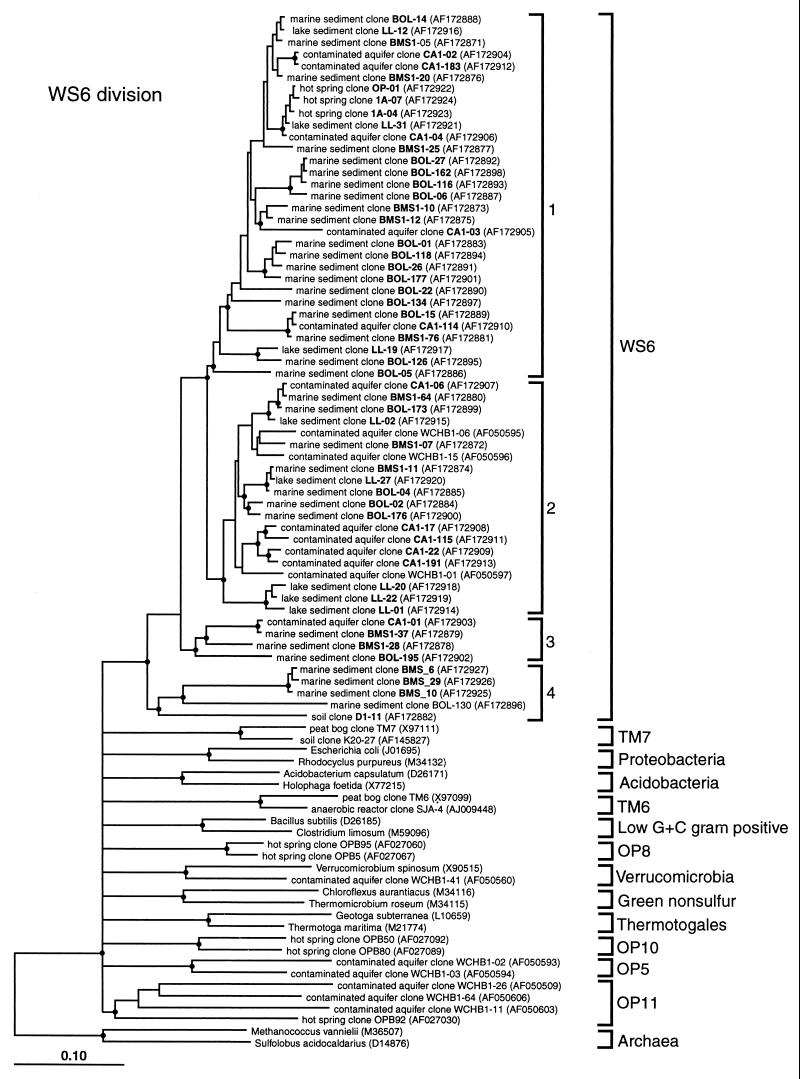
FIG. 1.
Evolutionary distance dendrograms of the candidate division WS6 and selected bacterial divisions. Division designations are listed outside the brackets. Branch points supported (bootstrap values of >74%) by rate-corrected maximum likelihood, parsimony, and distance analyses are indicated by solid circles. Branch points without circles were not resolved (bootstrap values of <75%) as specific groups in different analyses, and at the division level, they were collapsed back to the next significant node. The bar represents 10% sequence divergence.
Nucleotide sequence accession numbers.
The sequences of the rDNA clones have GenBank accession no. AF172871 to AF172927.
RESULTS
Twelve different environments, as summarized in Table 1 and described in Materials and Methods, were analyzed for the presence of members of the candidate phylogenetic division WS6. DNA purified from environmental samples was used as a template for PCR, with a universal reverse primer and generally a WS6-specific forward primer. For all environments that showed a negative result in the first PCR, the reaction mixture of the first PCR was used as a template for a second PCR amplification with the same primers.
Five of the 12 environmental DNA samples showed an rDNA-sized product after 25 to 35 cycles of PCR, while two others showed a product only after the initial reaction mixture was used as a template in a second round of PCR. Five environments yielded no PCR product after two rounds of PCR. These relative responses to the PCR round provide a rough gauge of the abundances of WS6 sequences in the environments sampled. Qualitatively, the product after one round of PCR indicates a relatively high concentration of WS6-related sequences compared to that in samples that required two rounds to obtain product; no PCR product after two PCR amplification series indicates the absence or rarity of WS6 sequences in the sample analyzed.
PCR products were cloned and screened by RFLP analysis for different sequences, which were then determined (Materials and Methods). Fifty-seven new sequences (typically 1 kb in length), representing a broad diversity of the WS6 relatedness group, were determined.
WS6 sequences were encountered in both marine sediment samples, the hot spring cyanobacterial mat, one of two hot spring sediment samples, one of two freshwater sediment samples, the topsoil sample, and the contaminated aquifer sample. WS6 sequences were not obtained from the very-low-biomass lake microbialite or from any of the human samples. (PCRs from both the lake microbialite and human samples were positive with more general primers.) Clone libraries derived from DNA extracted from marine sediments from Bolinas and Berkeley Marina, Lake Lemon sediment, and contaminated aquifer soil had the broadest diversity of unique WS6 clones, while libraries derived from DNA extracted from the landscaped topsoil and hot springs had small amounts of diversity compared to, e.g., Bolinas sediment (Table 1).
The WS6 sequences obtained were reproducibly monophyletic and distinct from all other known bacterial sequences in phylogenetic analyses (see Fig. 1 and Discussion). The depth of phylogenetic divergence within the WS6 group, which indicates the extent of diversity represented by members of the group, exceeds that of well-known divisions of Bacteria such as the Proteobacteria (see Table 2 and Discussion). To date, the WS6 division consists of four reproducible subgroupings. The subclade termed group 4 in Fig. 1 is notably deeply divergent from the other three groups, but is still robustly monophyletic with the other WS6 sequences. We discuss other phylogenetic aspects of this division in the following section.
TABLE 2.
Typical SSU-rRNA sequence divergence in selected divisions
| Division | Typical % differencea |
|---|---|
| Cyanobacteria | 13 |
| Fusobacteria | 13 |
| Termite group 1 | 15 |
| TM7 | 16 |
| OP10 | 17 |
| Thermus/Deinococcus | 17 |
| Actinobacteria (high G+C gram positive) | 18 |
| Nitrospira | 19 |
| Thermotogales | 19 |
| Acidobacteria | 20 |
| Green sulfur | 20 |
| Verrucomicrobia | 20 |
| Cytophagales | 22 |
| Spirochetes | 22 |
| Green nonsulfur | 23 |
| Planctomycetes | 23 |
| Proteobacteria (α–ɛ) | 23 |
| Low G+C gram positive | 24 |
| WS6 | 26 |
| OP11 | 33 |
Taxa were selected from the ARB database to obtain approximately average sequence divergences within the divisions. The maximum percent differences in the sequence set (lane mask [8]) are listed. Chloroplast and mitochondrial sequences were excluded from this analysis.
In summary, these results collectively indicate that representatives of the WS6 division are widely distributed and apparently abundant in the environment. The sequence-based map (Fig. 1) of WS6 phylogenetic diversity is a guide to the further study of these evidently important organisms.
DISCUSSION
Since there are no cultivated representatives of the WS6 phylogenetic division, there is no perspective on what kinds of physiologies the members of the division might display. Only the properties that are general to representatives of the domain Bacteria—polymerase types, antibiotic patterns, many metabolic themes, etc.—can be inferred. The most diverse and abundant collections of WS6 sequences, as judged by the ability to obtain a PCR, were encountered in anaerobic sediments and in the anaerobic contaminated aquifer soil. None of the sequences was identical between the environments, although closely related sequences were detected (99% identity). Far fewer sequences were obtained from more aerobic settings. This is based on the observation that PCRs performed with DNA extracted from anaerobic environments produced relatively higher concentrations of WS6 DNA than DNA from the oxidized hot spring environment or the subaqueous microbialite (Table 1); we take this to indicate a higher initial concentration of WS6-related rDNA in the samples from anaerobic settings.
Attempts were made to determine if members of the WS6 division might occur in the human gut, an energy-rich, highly anaerobic ecosystem, but no PCR products were obtained from either fecal matter or intestinal tissue. This is obviously a very small subset of the microenvironments available to microorganisms in the human body, so the possibility that members of the WS6 division are human commensal organisms is not removed by this study. Similarly, it is not possible to say from the absence of PCR products that members of WS6 do not reside in aerobic environments. The overall evidence suggests, however, that members of the WS6 division are relatively abundant participants in organic-rich, anaerobic environmental communities. The recently described bacterial division OP11, also with no cultivated representatives, is similar to the WS6 group in the sense that its members appear to occur primarily in anaerobic ecosystems and are not detected in association with the human gut (unpublished observations). Additionally, members of the WS6 group, like members of the OP11 group, occur in low- and high-temperature environments, indicating a potentially wide range of metabolic capabilities in these groups.
Historically, most microbiological culture efforts have focused on aerobic organisms and used culture conditions such as nutrient broths that were devised originally for human pathogens. It is not surprising that major uncultured diversity lies in environments that are not aerobic and not associated with the human body. Moreover, traditional characterizations of microbes have demanded pure cultures of the organisms. Consequently, environmental organisms that are syntrophic, that rely on the activities of one or more other organisms, have seldom been studied or even detected by pure culture-based approaches. Since molecular techniques such as rRNA sequence analysis and single-cell hybridization probes can detect and identify specific organisms in the context of mixed communities, new avenues for studies of previously unculturable microbes are now available.
This study increases the number of sequences that represent the bacterial division WS6 from 3 to 60, establishing the clade as a significant phylogenetic entity, one of the main bacterial lines. The distribution of members of WS6 in the environment is extensive, and they are abundant, indicating their potential importance in biospheric processes. The WS6 clade has also been found to be a bacterial division that contains notably deep phylogenetic divergence. Table 2 lists the general extent of phylogenetic divergence (general sequence difference) within selected divisions, including the best-known and most deeply divergent of bacterial divisions. The extent of sequence variation within the clades is some measure of the extent of known diversity in the division. The WS6 division displays the second largest extent of rRNA sequence divergence in the Bacteria, a breadth of rRNA diversity exceeded only by the newly described division OP11. Because the number of sequences that define the WS6 clade is substantial and the diversity and environmental distribution of the sequences are extensive, we consider the WS6 group established as a phylogenetic entity, one of the 36 to 38 main clades of Bacteria that can be articulated at this time. We expect that some of these clades will coalesce as the base of the bacterial tree becomes better resolved and that new ones will be discovered.
Since members of the bacterial division WS6 are so phylogenetically deeply divergent and widely distributed in the environment, their further characterization is warranted. Studies of pure cultures or simple consortia can provide biochemical information. Cultivation of WS6-representative microorganisms may be difficult; however, in situ methods such as in situ rRNA hybridization can be used to track uncultivated organisms in the environment and thereby to study their natural history. Members of divisions such as WS6, OP11, and a number of other bacterial divisions with few or no cultivated members have been found in recent years to constitute the majority of environmental biodiversity from the sequence perspective. The identification of such organisms opens many opportunities for environmental microbiologists to use classical and modern molecular techniques to determine their natures and the roles these microorganisms play in their ecosystems.
In summary, this survey of environmental sequences substantiates the division-level nature of the WS6 phylogenetic group, currently without any cultured representative. These results provide a framework for the further study of these apparently important kinds of organisms and the tools, the sequences, with which to do so in their environmental settings.
ACKNOWLEDGMENTS
We thank Scott Dawson, Dan Frank, Phil Hugenholtz, and Jose de la Torre for providing DNA samples and Galina Ishkanova for operating the ABI 373 sequencer.
This research was supported by grants to N.R.P. from the NIH (GM34527) and NSF (OCE-9870880).
REFERENCES
- 1.Altschul S F, Madden T L, Schäffer A A, Zhang J, Zhang Z, Miller W, Lipman D J. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Barns S M, Fundyga R E, Jeffries M W, Pace N R. Remarkable archaeal diversity detected in a Yellowstone National Park hot spring environment. Proc Natl Acad Sci USA. 1994;91:1609–1613. doi: 10.1073/pnas.91.5.1609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Barns S M, Takala S L, Kuske C L. Wide distribution and diversity of members of the bacterial kingdom Acidobacterium in the environment. Appl Environ Microbiol. 1999;65:1731–1737. doi: 10.1128/aem.65.4.1731-1737.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Dojka M A, Hugenholtz P, Haack S K, Pace N R. Microbial diversity in a hydrocarbon- and chlorinated-solvent-contaminated aquifer undergoing intrinsic bioremediation. Appl Environ Microbiol. 1998;64:3869–3877. doi: 10.1128/aem.64.10.3869-3877.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hugenholtz P, Goebel B M, Pace N R. Impact of culture-independent studies on the emerging phylogenetic view of bacterial diversity. J Bacteriol. 1998;180:4765–4774. doi: 10.1128/jb.180.18.4765-4774.1998. . (Erratum, 180:6793.) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hugenholtz P, Pitulle C, Hershberger K L, Pace N R. Novel division level bacterial diversity in a Yellowstone hot spring. J Bacteriol. 1998;180:366–376. doi: 10.1128/jb.180.2.366-376.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lane D J. 16S/23S rRNA sequencing. In: Stackebrandt E, Goodfellow M, editors. Nucleic acid techniques in bacterial systematics. New York, N.Y: John Wiley and Sons; 1991. pp. 115–175. [Google Scholar]
- 8.Lane D J, Pace B, Olsen G J, Stahl D A, Sogin M L, Pace N R. Rapid determination of 16S ribosomal RNA sequences for phylogenetic analyses. Proc Natl Acad Sci USA. 1985;82:6955–6959. doi: 10.1073/pnas.82.20.6955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ludwig W, Bauer S H, Bauer M, Held I, Kirchhof G, Schulze R, Huber I, Spring S, Hartmann A, Schleifer K-H. Detection and in situ identification of representatives of a widely distributed new bacterial phylum. FEMS Microbiol Lett. 1997;153:181–190. doi: 10.1111/j.1574-6968.1997.tb10480.x. [DOI] [PubMed] [Google Scholar]
- 10.Murray R G E, Schleifer K H. Taxonomic notes: a proposal for recording the properties of putative taxa of procaryotes. Int J Syst Bacteriol. 1994;44:174–176. doi: 10.1099/00207713-44-1-174. [DOI] [PubMed] [Google Scholar]
- 11.Pace N R. A molecular view of microbial diversity and the biosphere. Science. 1997;276:734–740. doi: 10.1126/science.276.5313.734. [DOI] [PubMed] [Google Scholar]
- 12.Woese C R. Bacterial evolution. Microbiol Rev. 1987;51:221–271. doi: 10.1128/mr.51.2.221-271.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]