Abstract
Cold stress is a major environmental factor that affects plant growth and development, as well as fruit postharvest life and quality. MicroRNAs (miRNAs) are a class of non-coding small RNAs that play crucial roles in various abiotic stresses. Peanuts (Arachis hypogaea L.), one of the most important grain legumes and source of edible oils and proteins, are cultivated in the semi-arid tropical and subtropical regions of the world. To date, there has been no report on the role of miRNAs in the response to cold stress in cultivated peanuts. In this study, we profiled cold-responsive miRNAs in peanuts using deep sequencing in cold-sensitive (WQL20) alongside a tolerant variety (WQL30). A total of 407 known miRNAs and 143 novel peanut-specific miRNAs were identified. The expression of selected known and novel miRNAs was validated by northern blotting and six known cold-responsive miRNAs were revealed. Degradome sequencing identified six cold-responsive miRNAs that regulate 12 target genes. The correlative expression patterns of several miRNAs and their target genes were further validated using qRT-PCR. Our data showed that miR160-ARF, miR482-WDRL, miR2118-DR, miR396-GRF, miR162-DCL, miR1511-SRF, and miR1511-SPIRAL1 modules may mediate cold stress responses. Transient expression analysis in Nicotiana benthamiana found that miR160, miR482, and miR2118 may play positive roles, and miR396, miR162, and miR1511 play negative roles in the regulation of peanut cold tolerance. Our results provide a foundation for understanding miRNA-dependent cold stress response in peanuts. The characterized correlations between miRNAs and their response to cold stress could serve as markers in breeding programs or tools for improving cold tolerance of peanuts.
Keywords: peanut, microRNA, cold stress, cold response, cold tolerance
Introduction
Cultivated peanuts (Arachis hypogaea L.) are vital oil plants and cash crops that globally play an essential role in consuming edible vegetable oil and leisure food. However, low temperatures limit the scope of crops to a large extent and cause yield reduction, quality decline, and occasionally no harvest in severe cases (Jeon and Kim, 2013; Barrero-Gil and Salinas, 2018). The annual loss of crops caused by low-temperature damage worldwide is as high as hundreds of billions of dollars (Xiao and Song, 2011; Abla et al., 2019). Therefore, the improvement of cold tolerance has essential application value for crops and other plants and is an urgent research topic.
As a thermophilic crop, peanuts require relatively high temperatures throughout their development. Cold injury causes different degrees of harm to the peanut at seedling emergence, flowering, maturity, and other key growth stages. Among them, the impact of the seedling stage is the most common, which could delay peanut germination and seedling growth, resulting in uneven seedling emergence, weak seedling potential, prolonged seedling emergence time, lack of seedlings, and ridge cutting. Research on cold stress in plants began in the early 1830s and today has a history of more than 190 years. Breeders have been trying to develop new varieties to resolve the problem of peanut chilling damage and have made progress. A few cold-tolerant and early maturing varieties have been cultivated (Upadhyaya et al., 2003; Zhang et al., 2019). However, the mechanism of cold tolerance is a complex quantitative trait that always appears in combination or continuously and is not controlled by a single regulatory pathway or gene, making traditional cold tolerance breeding methods challenging with longer breeding cycles (Park et al., 2015; Shi et al., 2018; Zhang et al., 2019). Therefore, understanding peanut cold defense mechanisms is necessary to accelerate the cultivation of peanut varieties with high cold tolerance. Under low non-freezing temperature conditions, plants reprogram their gene expression through transcriptional, post-transcriptional, and post-translational mechanisms and undergo physiological and biochemical adjustments to improve their tolerance to low temperatures. Many studies have shown that the molecular mechanisms of cold defense in many plants are evident. The plant’s cold defense molecular mechanism is a highly complex network composed of numerous positive and negative regulatory factors, which can form a center that integrates a variety of internal and external signals by binding to the C-repeat binding transcription factor (CBF)/dehydrate responsive element binding factor (DREB) promoter (Barrero-Gil and Salinas, 2018). The core mechanism is that cold stress activates the plant’s ICE-CBF-COR regulatory pathway. Under cold stress, the transcription factor CBF responds to cold signals with the participation of the transcription activator ICE (inducer of CBF expression), binds to the cis-acting element CRT (C-repeat)/DER (dehydration) of the cold-induced genes COR (cold-regulated) responsive element sequence, initiates the expression of a series of cold-induced genes and proteins, and then changes the physiology, metabolism, and growth of plants (Gilmour et al., 1998; Wang et al., 2017; Barrero-Gil and Salinas, 2018; Shi et al., 2018; Guo et al., 2019). The ICE-CBF-COR transcriptional cascade has been established as the main regulatory response toolkit for cold signaling and freezing tolerance. Peanuts are sensitive to low temperatures, but they have evolved a unique low-temperature survival mechanism (Zhu, 2016). Studies have reported that overexpression of AtDREB2A in peanuts can improve their tolerance to low temperature, drought, and salt stress (Pruthvi et al., 2014). AhPNDREB1 (a homologous gene of AtCBF1), which is intron-less and constitutively expressed, was isolated from the cultivar Luhua 14 (Zhang et al., 2009). The AP2 domain in AhPNDREB1 is specific to the CRT/DER element combining ability, and the gene can be vigorously and rapidly induced to be expressed under cold and drought stress. Chen et al. (2012) cloned peanut AhCBF2 and AhCBF15, analyzed their amino acid sequences, and found that they both contained the unique ERF/AP2 domain of CBF/DREB family genes. However, there are few reports on the mechanism of peanut cold tolerance, which requires further exploration.
Over the past few years, many studies have shown that small RNA (sRNA) play a pivotal role in regulating cold stress in various plants. sRNA, as a type–21-24 nucleotide long non-coding RNA molecule, is widespread in eukaryotes and has two main forms: microRNA (miRNA) and small interfering RNA (siRNA) (Jin, 2008). It can form an RNA-induced silencing complex by combining with the ribozyme complex containing the AGO protein to cleave target mRNA or inhibit its translation and is widely involved in the cold stress responses of various plants, such as Arabidopsis (Sunkar and Zhu, 2004; Zhou et al., 2008; Yan et al. 2016), poplar (Lu et al., 2008), wheat (Tang et al., 2012), rice (Yang et al., 2013), soybean (Zhang et al., 2014), tomato (Shi et al., 2019), and wild grape (Wang et al., 2019). By targeting stress-related transcription factors, such as MYB, WRKY, and bHLH (Zhang et al., 2014; Shi et al., 2019; Wang et al., 2019), genes related to the reactive oxygen scavenging system (Yang et al., 2013) or ABA signaling pathway genes (Yan et al., 2016), these miRNAs may regulate the cold tolerance of plants. Studies have shown that sRNA-mediated gene silencing, histone modification, phosphorylation, and ubiquitination can regulate CBF expression at post-transcriptional and post-translational levels (Barrero-Gil and Salinas, 2018; Lantzouni et al., 2020). Thus, we believe that exploring the peanut cold defense regulatory network from small RNA-mediated gene silencing will be helpful in revealing the peanut response and regulatory mechanisms of cold stress.
To understand the role of miRNAs in peanut cold tolerance, we used two representative peanut lines in this study: WQL20 (waiting for quiz line 20) and WQL30 were eighth-generation recombinant inbred lines (RILs) from a cross between cultivars of Yuanza 9102 (female) and Xuzhou 68–4 (male). The main difference between the two RILs is their ability to withstand low temperatures: WQL30 is resistant to low temperatures, whereas WQL20 is susceptible to low temperatures. These two peanut lines were investigated to understand peanut cold tolerance by analyzing plant miRNA expression patterns and their target genes.
Materials and Methods
Plant Materials
The RIL population from a cross between cultivars of Yuanza 9102 (female) and Xuzhou 68–4 (male) includes 188 F8 lines. Yuanza 9102, the female parent of the RIL population, belonging to A. hypogaea subsp. fastigiata var. vulgaris, was derived from interspecific hybridization between cultivated cultivar Baisha 1,016 and a diploid wild species A. diogoi. Xuzhou 68–4, the male parent, which belongs to A. hypogaea subsp. hypogaea var. hypogaea. Yuanza 9102 is a popular cultivar in China that is resistant to cold stress, while Xuzhou 68–4 is susceptible to cold stress.
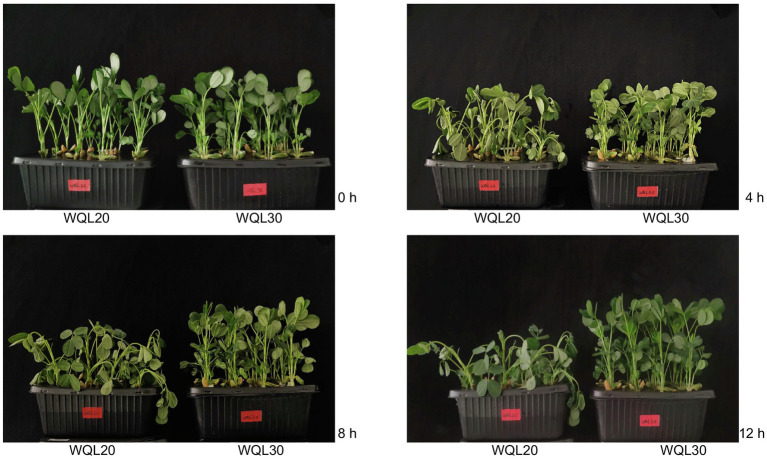
We conducted hydroponic culture of the two lines in Hoagland solution in a growth chamber, and then culture them in illumination incubator in the laboratory. The temperature was 27°C and the light–dark cycle was 14 h of light and 10 h of darkness. For low-temperature treatment, 6 four-leaf stage seedlings for each line were kept at 6°C, 70% RH, and a 14-h/10-h light/dark photoperiod. Lines WQL30 and WQL20 showed apparent difference resistance to cold. After 4 h of cold treatment (HCT), WQL20 began to wilt, while WQL30 did not change significantly. After 8 h of cold treatment (HCT), WQL20 wilted significantly, but WQL30 still had no noticeable change. After 12 h of cold treatment (HCT), WQL20 wilted and fell, while WQL30 showed slight wilting symptoms (Figure 1). Samples of four HCT (0 h, 4 h, 8 h, and 12 h) were collected from whole seedlings for miRNAome and degradome analysis.
Figure 1.
The phenotype of peanut exposed to low temperature. For low-temperature treatment, four-leaf stage seedlings of WQL30 and WQL20 were kept in a growth chamber at 6°C, 70% RH, and a 14-h/10-h light/dark photoperiod. With the increase of treatment time, WQL20 began to wilt, while WQL30 did not change significantly after 4 h of cold treatment. After 8 h of cold treatment, WQL20 wilted significantly, but WQL30 still had no noticeable change. After 12 h of cold treatment, WQL20 wilted and fell, while WQL30 showed slight wilting symptoms as shown in this figure. The pictures were taken at 0 HCT, 4 HCT, 8 HCT, and 12 HCT and assembled in Adobe Photoshop CS6.
sRNA Library Construction and DNA Sequencing
RNA degradation and contamination was monitored on 1% agarose gels. RNA purity was checked using the NanoPhotometer® spectrophotometer (IMPLEN, CA, United States). RNA concentration was measured using Qubit® RNA Assay Kit in Qubit® 2.0 Fluorometer (Life Technologies, CA, United States). A total amount of 3 μg total RNA per sample was used as input material for the small RNA library. Sequencing libraries were generated using NEBNext®Multiplex Small RNA Library Prep Set for Illumina® (NEB, United States.) following manufacturer’s recommendations and index codes were added to attribute sequences to each sample. Briefly, Total RNA was ligated to the RNA 3′ and RNA 5′ adapters and reverse transcription, followed by PCR, was performed to make cDNA constructs of the small RNAs. PCR products were purified on a 8% polyacrylamide gel (100 V, 80 min). DNA fragments corresponding to 140 ~ 160 bp (the length of small non-coding RNA plus the 3′ and 5′ adaptors) were recovered and dissolved in 8 μl elution buffer. At last, library quality was assessed on the Agilent Bioanalyzer 2,100 system using DNA High Sensitivity Chips. We then performed single-end sequencing (50 bp) on an Illumina Hiseq2500 at the LC-BIO (Novogene Experimental Department, Beijing, China) following the vendor’s recommended protocol.
Degradome Library Construction and Sequencing
To identify the potential targets, equal amounts of RNAs from WQL20 (0 HCT, 4 HCT, 8 HCT, and 12 HCT) and WQL30 samples (0 HCT, 4 HCT, 8 HCT, and 12 HCT) were mixed together for degradome library construction and deep sequencing. The two libraries named as DS1 and DS2, respectively. Through preprocessing, clean tags are generated. Then, clean tags were classified by alignment with GenBank, Rfam database, and miRNA database. Next, the reads were mapped to the reference genome.1 The sense strand of peanut cDNA was used to predict miRNA cleavage sites using CLeaveL and pipeline (Addo-Quaye et al., 2009). Based on the signature number and abundance of cleaved position at each occupied transcript, the cleaved transcripts could be categorized into five categories (0, 1, 2, 3, and 4).
Northern Blotting Analysis
RNA blot analyses for miRNAs from total extracts were performed as described previously (Katiyar-Agarwal and Jin, 2007). Total RNA was extracted using TRIzol reagent (Takara, Japan) following the manufacturer’s instructions. RNA was resolved on a 14% denaturing 8 M urea-PAGE gel and then transferred and chemically crosslinked onto a Hybond N+ membrane (GE Healthcare Life Sciences) using N-(3-Dimethylaminopropyl)-N′-ethylcarbodiimide hydrochloride. miRNA probes were end-labeled with [r-32P] ATP by T4 polynucleotide kinase (New England Biolabs). For internal control experiments, blots were stripped for 20 min and 4 times at 80°C in stripping buffer (0.1 X SSC/0.5% SDS). After detecting no signal, the stripped blots were rehybridized with r-32P ATP-labeled U6 gene fragments to confirm loading amounts. All signals were normalized to the U6 signals obtained from each blot. Expression levels were quantified using Image J as instructed.
qRT-PCR
The RNA used for qRT-PCR was from the same set RNA for northern blotting analysis. The RNA was then reverse transcribed into cDNA using the PrimeScript RT reagent Kit (Takara, Japan). The qRT-PCR was performed in a 15 μl reaction mixture consisting of 1.5 μl 1 × SYBR Green (Invitrogen, United States), 1.5 μl PCR buffer, 0.3 μl 10 mM dNTPs (Takara, Japan), 0.3 μl Taq, 0.3 μl ROX DYE2 (Vazyme, China), 1.5 μl 2 mM each primer and 2 μl appropriate diluted cDNA. The conditions for qRT-PCR were as follow: 94°C for 3 min, then 40 cycles at 94°C for 30 s and 58°C for 30 s, followed by 72°C for 35 s for PCR amplification. Transcript levels of each gene were measured by the Applied Biosystems 7500 (Applied Biosystems, United States) according to the manufacturer’s instructions. 18S rRNA was used as a quantitative control in the qRT-PCR analysis. Primers used in this study are listed in Supplementary Table S4.
Transient Expression Analysis of miRNA in Nicotiana benthamiana
The precursor of miRNA, MIRNA, cloned from peanut was sub-cloned into the overexpression vector pCAMBIA1300 destination vector using LR clonase II (Invitrogen). Transient co-expression assays in N. benthamiana were performed by infiltrating 4-week-old N. benthamiana plants with Agrobacterium GV3101 [OD600 (optical density at 600 nm) = 1.0] harboring constructs containing the MIRNA (pCAMBIA1300). The Agrobacterium-mediated transformation was performed according to a previously described method (Niu et al., 2018; Zhang et al., 2018). After cultivation for 48 h under 16 h light/8 h dark at 23°C conditions, the injected tobacco plants were treated with 4°C stress for 24 h in an incubator (Dongqi, Ningbo, China). Then the injected tobacco leaves were collected for RNA extraction.
Results
High-Throughput Sequencing and Annotation of Peanut sRNAs
To identify endogenous cold stress-responsive miRNAs that are potentially involved in cold response, we treated the two Lines (WQL20 and WQL30) with low-temperature 6°C and sampled tissue from treated plants at four different time points after treatment [0, 4, 8, and 12 h after cold treatment (HCT)]. Total RNA from each sample was used for sRNA library construction. As shown in Table 1, 9,576,091, 8,391,121, 9,343,483, and 7,954,268 total reads were generated from each library representing the different time points in line WQL20, respectively. 9,376,386, 11,205,178, 10,219,016, and 8,626,070 total reads were in line WQL30, respectively. By mapping to the peanut genome (Cultivated peanut, A. hypogaea cv. Tifrunner),2 9,349,005, 8,234,001, 9,129,232 and 7,784,811 peanut sRNA reads were obtained, corresponding to 2,093,285, 2,057,134, 2,270,139 and 1,634,313 unique reads in WQL20, respectively. 9,242,332, 11,025,444, 9,875,961 and 8,492,492 peanut sRNA reads were obtained in WQL30, corresponding to 1,833,592, 2,107,138, 2,030,541 and 1,998,859 unique reads, respectively. These reads were further searched against the Rfam database3 to remove known sRNA, such as ribosomal RNA (rRNA), transfer RNA (tRNA), and small nucleolar RNAs (snoRNAs), the Repbase databases4 to remove repeats. In the end, 5,676,589, 5,327,270, 5,435,094 and 4,049,334 reads were retrieved from each library of WQL20, and 4,964,372, 5,982,719, 5,551,346 and 5,019,867 reads were retrieved from each library of WQL30. The reads were considered as sRNA originated.
Table 1.
Distribution of sRNAs among different categories in each library.
| Types | Normalized Reads | |||||||
|---|---|---|---|---|---|---|---|---|
| WQL20 | WQL30 | |||||||
| 0 h | 4 h | 8 h | 12 h | 0 h | 4 h | 8 h | 12 h | |
| Total sRNA | 9,576,091 | 8,391,121 | 9,343,483 | 7,954,268 | 9,376,386 | 11,205,178 | 10,219,016 | 8,626,070 |
| Mapped sRNA | 9,349,005 | 8,234,001 | 9,129,232 | 7,784,811 | 9,242,332 | 11,025,444 | 9,875,961 | 8,492,492 |
| Unique reads | 2,093,285 | 2,057,134 | 2,270,139 | 1,634,313 | 1,833,592 | 2,107,138 | 2,030,541 | 1,998,859 |
| Rfam | ||||||||
| rRNA | 3,024,554 | 2,104,932 | 2,902,873 | 2,898,576 | 3,674,164 | 4,202,996 | 3,637,792 | 2,867,982 |
| tRNA | 144,762 | 152,863 | 124,882 | 215,951 | 154,697 | 292,961 | 187,397 | 146,373 |
| snRNA | 918 | 844 | 1,586 | 873 | 861 | 1,584 | 847 | 829 |
| snoRNA | 13,669 | 15,491 | 38,404 | 16,492 | 16,673 | 24,703 | 13,390 | 13,998 |
| Repeat | 488,513 | 632,601 | 626,393 | 603,585 | 431,565 | 520,481 | 485,189 | 443,443 |
| Peanut small RNA | 5,676,589 | 5,327,270 | 5,435,094 | 4,049,334 | 4,964,372 | 5,982,719 | 5,551,346 | 5,019,867 |
| Conserved_miRNA (Total) | 1,089,490 | 942,979 | 359,409 | 555,137 | 1,004,326 | 1,207,299 | 1,090,170 | 881,849 |
| Conserved_miRNA (Unique) | 1,328 | 1,260 | 1,126 | 1,184 | 1,295 | 1,414 | 1,339 | 1,233 |
| Novel_miRNA (Total) | 58,424 | 70,267 | 34,750 | 42,150 | 65,449 | 103,589 | 84,218 | 73,171 |
| Novel_miRNA (Unique) | 1,243 | 1,439 | 1,398 | 1,166 | 1,370 | 1,546 | 1,536 | 1,386 |
| TAS (Total) | 3,094 | 6,961 | 4,133 | 2,944 | 3,508 | 5,193 | 5,104 | 4,408 |
| TAS (Unique) | 300 | 366 | 298 | 267 | 292 | 333 | 297 | 304 |
| NAT (Total) | 654,060 | 757,250 | 904,777 | 694,258 | 687,859 | 852,495 | 774,593 | 663,985 |
| NAT (Unique) | 157,239 | 181,283 | 199,877 | 147,463 | 153,205 | 180,847 | 169,278 | 161,026 |
| exon:+ | 206,065 | 230,953 | 266,696 | 164,444 | 183,927 | 223,993 | 201,463 | 170,121 |
| exon:− | 126,392 | 148,788 | 134,160 | 99,348 | 120,449 | 169,910 | 139,523 | 118,291 |
| intron:+ | 172,157 | 214,436 | 249,986 | 156,912 | 171,819 | 209,054 | 203,835 | 188,352 |
| intron:− | 103,208 | 127,995 | 154,537 | 96,073 | 109,887 | 123,634 | 124,446 | 120,489 |
| other | 3,263,699 | 2,827,641 | 3,326,646 | 2,238,068 | 2,617,148 | 3,087,552 | 2,927,994 | 2,799,201 |
A peanut sRNA deep sequencing summary is presented. The abundance in different libraries was normalized to that in the library at 0 h, and sRNAs were calculated as reads per million.
The lengths of the unique, valid reads ranged from 18 to 30 nucleotides (nt), and the 21–24 nt sequences were predominant in each library, with the 24 (nt) sequences being the most common (Supplementary Figure S1).
Identification of Known, Conserved, and Novel miRNAs
To identify known, conserved, and novel miRNAs in peanut, all the total peanut sRNAs were classified into three categories according to their origination and structural features: (1) conserved miRNAs with a hairpin structure of their precursor sequences were aligned to plant miRNAs in miRBase5; (2) novel peanut miRNAs have a hairpin structure of their precursor sequences but are not listed in the microRNA database; and (3) peanut siRNAs that do not have a hairpin structure of their precursor sequences. According to this criteria, we retrieved 1,328, 1,260, 1,126, and 1,184 unique conserved peanut miRNAs in each library of WQL20 and 1,295, 1,414, 1,339, and 1,233 unique conserved peanut miRNAs in each library of WQL30, respectively (Table 1; Supplementary Table S1). In addition, we obtained 1,243, 1,439, 1,398, and 1,166 novel peanut miRNAs in each library of WQL20 and 1,370, 1,546, 1,536, and 1,386 novel peanut miRNAs in each library of WQL30, respectively.
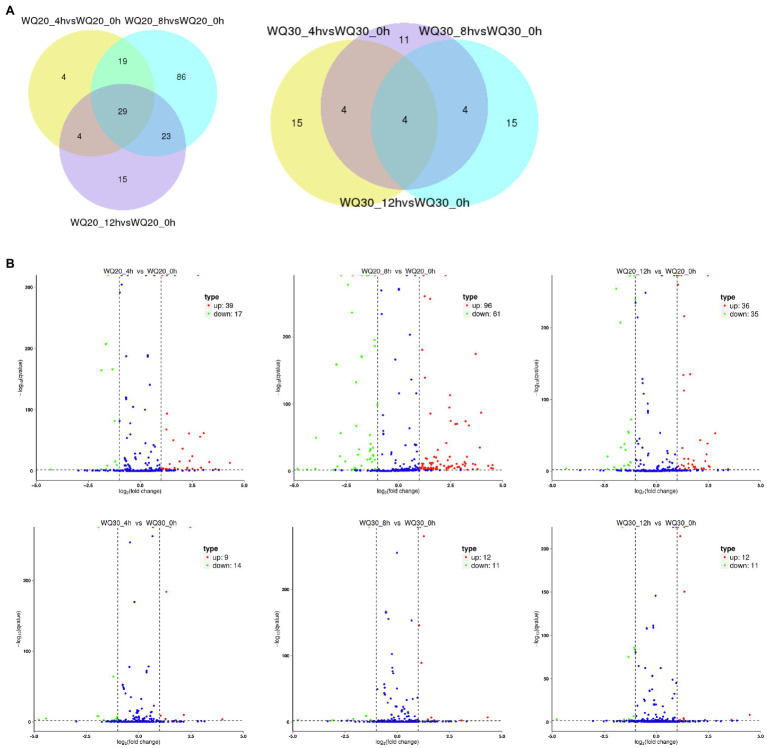
Comparison of Differentially Expressed miRNAs Between the Two Peanut RILs
To identify the differentially expressed miRNAs, we compared the frequencies of occurrence of differentially expressed miRNAs at the 0 HCT, 4 HCT, 8 HCT, and 12 HCT stages between the two lines after the miRNA reads were normalized to transcripts per million (TPM). We identified 56, 71, and 157 differentially expressed miRNAs in the WQL20_4 h vs. WQL20_0 h, WQL20_8 h vs. WQL20_0 h, and WQL20_12 h vs. WQL20_0 h comparisons, and 23, 23, and 23 differentially expressed miRNAs in the WQL30_4 h vs. WQL30_0 h, WQL30_8 h vs. WQL30_0 h and WQL30_12 h vs. WQL30_0 h comparisons, respectively (Figure 2A). Among these, 39, 96, and 36 miRNAs were upregulated expression, and 17, 61, and 35 miRNAs were downregulated at 4 HDT, 8 HDT, and 12 HDT compared to 0 HDT in RIL WQL20, respectively. 9, 12, and 12 miRNAs were upregulated expression, and 14, 11, and 11 miRNAs downregulated expression in RIL WQL30, respectively (Figure 2B). To identify the miRNAs responsive to cold stress, the miRNAs with opposite changes between the two peanut RILs both abundantly and significantly differentially expressed were identified by employing the following criteria: (1) total reads ≥3,000; (2) [treated/mock] ≥ 2 or [treated/mock] ≤ 0.5 in at least one stage. In the end, we found 12 significantly differentially expressed conserved peanut miRNAs (DE-miRNAs; Table 2).
Figure 2.
Differentially expressed miRNAs in two peanut RILs. (A) Venn diagrams showing the number of common and specific miRNAs in comparisons of the eight libraries. (B) Volcanic diagrams showing the number of differentially expressed miRNAs in each comparison. The red dots indicate miRNAs with significant differences, and the blue dots indicate that the difference was not significant for miRNA expression.
Table 2.
List of some candidate miRNAs involved in low-temperature response in each library.
| miRNA Identifier | Normalized reads (reads/mgs) | Fold change | |||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| WQL20 | WQL30 | WQL20 | WQL30 | ||||||||||||
| 0 h | 4 h | 8 h | 12 h | 0 h | 4 h | 8 h | 12 h | 4 h/0 h | 8 h/0 h | 12 h/0 h | 4 h/0 h | 8 h/0 h | 12 h/0 h | ||
| ahy-miR396 | ath-miR396a-5p | 12078.55 | 17044.83 | 18074.27 | 27523.55 | 16443.92 | 14907.85 | 14650.86 | 13752.88 | 1.41 | 1.50 | 2.28 | 0.91 | 0.89 | 0.84 |
| gma-miR396h | 12091.90 | 17038.83 | 18086.71 | 27498.87 | 16440.42 | 14907.85 | 14641.29 | 13748.15 | 1.41 | 1.50 | 2.27 | 0.91 | 0.89 | 0.84 | |
| zma-miR396g-3p | 12058.79 | 17032.24 | 18064.93 | 27494.12 | 16426.41 | 14890.62 | 14634.91 | 13732.61 | 1.41 | 1.50 | 2.28 | 0.91 | 0.89 | 0.84 | |
| vvi-miR396a | 4968.34 | 6500.29 | 7497.62 | 10354.21 | 3998.15 | 2943.47 | 2942.41 | 2629.60 | 1.31 | 1.51 | 2.08 | 0.74 | 0.74 | 0.66 | |
| ahy-miR167 | sly-miR167b-5p | 7017.83 | 16039.02 | 18027.59 | 18560.48 | 17306.84 | 16602.84 | 19235.74 | 16172.98 | 2.29 | 2.57 | 2.64 | 0.96 | 1.11 | 0.93 |
| ahy-miR1511 | gma-miR1511 | 2425.43 | 8460.94 | 17159.43 | 15457.25 | 10430.93 | 10406.25 | 8992.64 | 9098.53 | 3.49 | 7.07 | 6.37 | 1.00 | 0.86 | 0.87 |
| ahy-miR482 | gso-miR482a | 10571.60 | 3956.68 | 7737.22 | 6511.26 | 8098.48 | 10688.83 | 9343.68 | 8679.52 | 0.37 | 0.73 | 0.62 | 1.32 | 1.15 | 1.07 |
| ahy-miR2118 | pvu-miR2118 | 1911.19 | 576.38 | 692.35 | 950.53 | 1144.33 | 6018.87 | 2322.76 | 2762.74 | 0.30 | 0.36 | 0.50 | 5.26 | 2.03 | 2.41 |
| gma-miR2118a-3p | 1911.19 | 576.38 | 692.35 | 950.53 | 1143.75 | 6018.87 | 2322.76 | 2762.74 | 0.30 | 0.36 | 0.50 | 5.26 | 2.03 | 2.42 | |
| ahy-miR156 | mdm-miR156ad | 978.82 | 758.11 | 356.29 | 997.06 | 743.82 | 1090.94 | 1330.25 | 1290.14 | 0.77 | 0.36 | 1.02 | 1.47 | 1.79 | 1.73 |
| ahy-miR162 | bdi-miR162 | 656.29 | 915.25 | 1754.99 | 1099.61 | 629.97 | 554.33 | 477.10 | 408.19 | 1.39 | 2.67 | 1.68 | 0.88 | 0.76 | 0.65 |
| ahy-miR160 | ath-miR160a-5p | 300.11 | 267.50 | 231.82 | 147.18 | 280.24 | 338.21 | 379.24 | 339.26 | 0.89 | 0.77 | 0.49 | 1.21 | 1.35 | 1.21 |
Variations in expression were analyzed. Peanut miRNAs with opposite changes in WQL20 and WQL30 and significantly differentially expressed were identified using the following criteria: [1] total reads ≥ 3,000; [2] [treated/mock] ≥ 2 or [treated/mock] ≤ 0.5 in at least one stage.
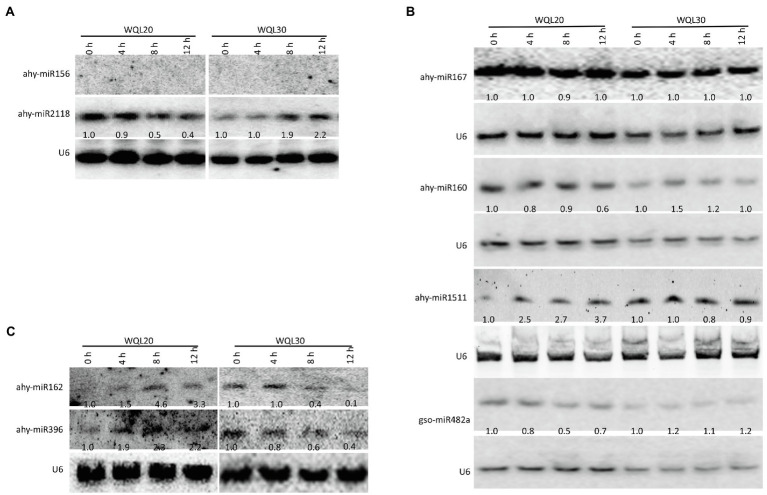
Validation of Cold-Responsive miRNAs by Northern Blotting
By using reverse-cDNA fragments as probes, we validated our bioinformatics prediction by northern blotting assay (there is only one nucleotide difference between ath-miR396a-5p and gma-miR396h, between ath-miR396a-5p and vvi-miR396a, between pvu-miR2118 and gma-miR2118a-3p; therefore, detection using probes reverse complementary to the aforementioned sRNAs represents the expression of the family). The eight northern blottings that were used to detect the candidate miRNAs. ahy-miR2118 and ahy-miR160 showed discernible downregulation in line WQL20 and upregulation in line WQL30 (Figures 3A,B). ahy-miR482 showed discernible downregulation in line WQL20 and weak upregulation in line WQL30 (Figure 3B). ahy-miR1511 showed discernible upregulation in line WQL20 and weak downregulation in line WQL30 (Figure 3B). ahy-miR396 and ahy-miR162 showed discernible upregulation in line WQL20 and downregulation in line WQL30 (Figure 3C). Surprisingly, ahy-miR167 showed strong expression with no noticeable expression variation, which is different to the bioinformatic prediction (Figure 3B). ahy-miR156 expressed under the detectable level (Figure 3A).
Figure 3.
Expression patterns of miRNAs upon low-temperature treatment. Four-leaf stage seedlings were treated with low temperature (6°C), and total RNA was extracted from leaves at the indicated time points. Detection of the peanut miRNAs using Northern blotting. A total of 100 μg of total RNA was loaded. RNA blots were hybridized with DNA oligonucleotide probes complementary to the indicated miRNAs. U6 was used as a loading control. Values below each section represent the relative abundance of miRNA normalized to U6.
Identification of Target Genes of miRNAs by Degradome Analysis
To further understand the biological functions of cold-responsive miRNAs, the miRNA target genes were identified by Degradome Analysis. In this study, we constructed two degradome libraries, DS1 (including WQL20_0h, WQL20_4h, WQL20_8h, and WQL20_12h) and DS2 (including WQL30_0h, WQL30_4h, WQL30_8h, and WQL30_12h). In total, we obtained 34,299,289 and 43,309,050 raw reads from DS1 and DS2, respectively. After removing the reads without the 3′ adaptor sequence, we obtained 10,139,640 and 9,850,766 unique raw reads from the DS1 and DS2 libraries, respectively. The unique reads were aligned to the peanut genome database, and 25,396,751 (74.04%) and 30,399,096 (70.19%) reads were mapped to the genome, respectively. The mapped reads represented 7,005,430 (69.09%) and 6,538,802 (66.38%) annotated peanut genes in the DS1 and DS2 libraries, respectively (Supplementary Table S2). We identified cleaved targets for miRNAs based on a method in the Cleaveland pipeline (Addo-Quaye et al., 2009), in which a host gene with an alignment score of 7 or less was considered to be a potential target. According to the signature number and abundance of putative cleaved position at each occupied transcript, these cleaved transcripts could be categorized into five classes (0, 1, 2, 3, and 4). In total, 7,561 and 7,296 targets were identified from the DS1 and DS2 libraries, respectively (Supplementary Table S3). For these targets, 291, 70, 2,421, 991, and 3,788 were classified to categories 0, 1, 2, 3, and 4 in the DS1 library, and 365, 41, 2,358, 1,188, and 3,344 belonged to a category <4 in the DS2 library (Supplementary Table S3). There were 11,615 differentially expressed target genes between the DS1 and DS2 libraries. 6,317 genes were upregulated expression for these targets, and 5,298 genes were downregulated expression (Supplementary Table S3).
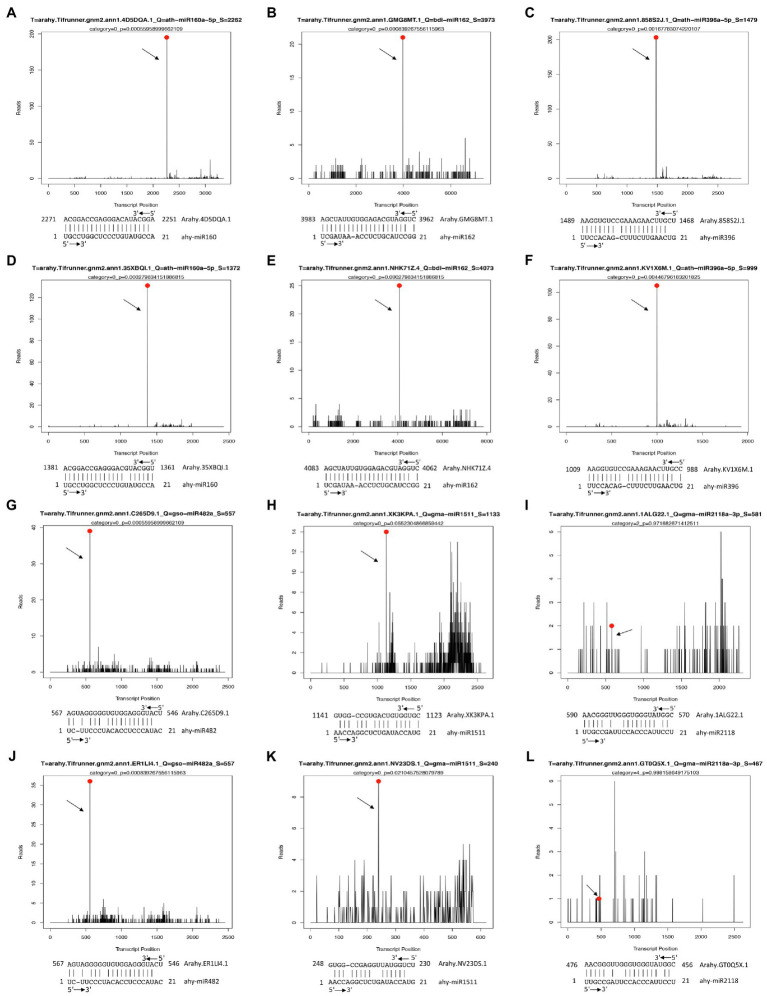
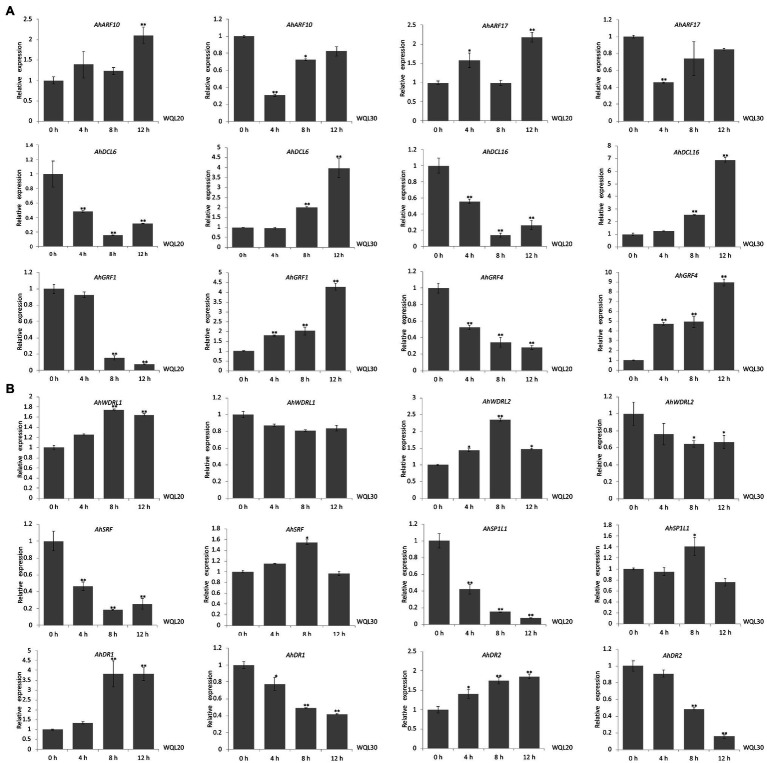
For the cold-responsive miRNAs, we found that ahy-miR160 targets auxin response factor genes AhARF10 (Arahy.4D5DQA) and AhARF17 (Arahy.35XBQI) (Figures 4A,D); ahy-miR162 targets DEAD-box ATP-dependent RNA helicase genes AhDCL6 (Arahy.GMG8MT) and AhDCL16 (Arahy.NHK71Z) (Figures 4B,E); ahy-miR396 targets the growth-regulating factor 1 genes AhGRF1 (Arahy.858S2J) and AhGRF4 (Arahy.KV1X6M) (Figures 4C,F); ahy-miR482 targets WD repeat-containing protein 26-like genes AhWDRL1 (Arahy.C265D9) and AhWDRL2 (Arahy.ER1LI4) (Figures 4G,J); ahy-miR1511 targets protein kinase superfamily protein gene AhSRF (Arahy.XK3KPA) and SPIRAL1-like 1 gene AhSP1L1 (Arahy.NV23DS) (Figures 4H,K); ahy-miR2118 targets disease resistance protein genes AhDR1 (Arahy.1ALG22) and AhDR2 (Arahy.GT0Q5X) (Figures 4I,L).
Figure 4.
Examples of T-plots of miRNA targets in two peanut RILs confirmed by degradome sequencing. The T-plots show the distribution of the degradome tags along the full length of the target mRNA sequence. The vertical red line indicates the cleavage site of each transcript and is also shown by an arrow. (A,D) The cleavage features in AhARF10 (Arahy.4D5DQA.1) and ARF17 (Arahy.35XBQI.1) mRNA by ahy-miR160 in DS1. (B,E) The cleavage features in AhDCL6 (Arahy.GMG8MT.1) and AhDCL16 (Arahy.NHK71Z.4) mRNA by ahy-miR162 in DS1. (C,F) The cleavage features in AhGRF1 (Arahy.858S2J.1) and AhGRF4 (Arahy.KV1X6M.1) mRNA by ahy-miR396 in DS1. (G,J) The cleavage features in AhWDRL1 (Arahy.C265D9.1) and AhWDRL2 (Arahy.ER1LI4.1) mRNA by ahy-miR482 in DS1. (H,K) The cleavage features in AhSRF (Arahy.XK3KPA.1) and AhSP1L1 (Arahy.NV23DS.1) mRNA by ahy-miR1511 in DS1. (I,L) The cleavage features in AhDR1 (Arahy.1ALG22.1) and AhDR2 (Arahy.GT0Q5X.1) mRNA by ahy-miR2118 in DS1.
We further examined the in vivo expression of each predicted target gene upon low-temperature treatment. The RNA used for qRT-PCR was from the same set RNA for northern blot analyses. Both of the transcriptions of ARF10 and ARF17 (targeted by ahy-miR160) were significantly upregulated expression in line WQL20, but significantly downregulated expression in line WQL30, respectively. Same as AhGRF1 and AhGRF4 (targeted by ahy-miR396), AhDCL6 and AhDCL16 (targeted by ahy-miR162) showed similar expression profiles between the two RIL lines. In line WQL20, they were significantly downregulated expression, but significantly upregulated expression in line WQL30, respectively (Figure 5A). AhDR1 and AhDR2, target genes of ahy-miR2118, showed sustained increase with the low-temperature treatment in line WQL20, while sustained decrease in line WQL30, respectively. The transcriptions of AhSRF and AhSP1L1 showed a noticeable reduction since 4 h and reached less than 40% of the original level at 8 h in line WQL20, but significantly increase in WQL30 with low-temperature treatment, which corresponds to the overall expression tendency of ahy-miR1511. The expressions of AhWDRL1 and AhWDRL2 are cognately inverse to the expression pattern of ahy-miR482, further suggesting a regulatory relationship between these two parties (Figure 5B). In summary, our research indicates that these genes are authentic target genes of their cognate miRNAs. These miRNAs may be involved in the cold-responsive process in peanut by negatively regulating their target genes.
Figure 5.
Comparison of the relative expression levels of target genes in two RILs. (A) qRT-PCR analyses of target genes of ahy-miR160, ahy-miR162, and ahy-miR396 expression levels in two peanut RILs upon low-temperature (6°C) treatment at the indicated time points, respectively. (B) qRT-PCR analyses of target genes of ahy-miR482, ahy-miR1511, and ahy-miR2118 expression levels in two peanut RILs upon low-temperature (6°C) treatment at the indicated time points, respectively. Data are shown as means ± SD (n = 3). Student’s t-test was used to determine the significance of differences between 0 HCT and the indicated time points. Asterisks indicate significant differences (*p < 0.05 and **p < 0.01). Similar results were obtained from three biological replicates.
Functional Analysis of the Six Cold-Responsive miRNAs in Tobacco
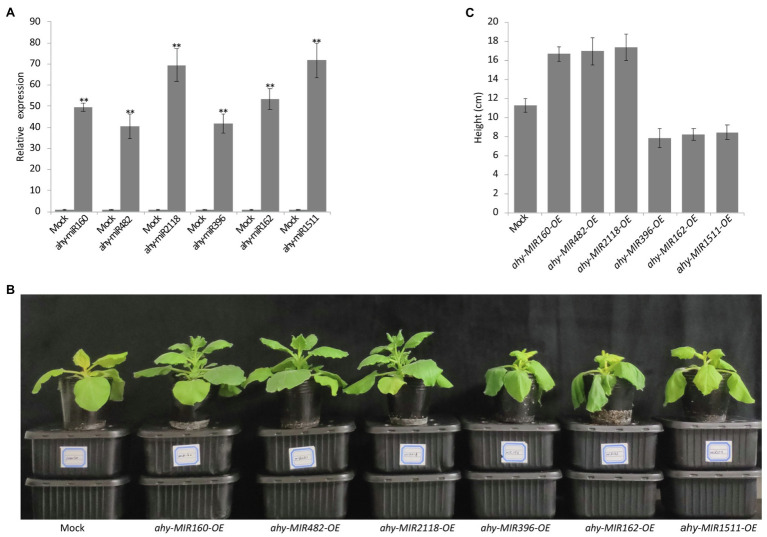
To further analyze the function of cold-responsive miRNAs, we transiently overexpressed the six cold-responsive miRNAs in tobacco leaves using Agrobacterium expressing pCAMBIA1300-miRNAs (Figure 6A). After 24 h cold treatment, the tobacco plants showed visual phenotypes. Compared with the control, the leaves overexpressing ahy-miR160, ahy-miR482, and ahy-miR2118 exhibited better growth status, but the leaves overexpressing ahy-miR396, ahy-miR162, and ahy-miR1511 showed poor growth status (Figures 6B,C). These results suggest that tobacco leaves overexpressing ahy-miR160, ahy-miR482, and ahy-miR2118 have enhanced cold tolerance. On the contrary, tobacco leaves overexpressing ahy-miR396, ahy-miR162, and ahy-miR1511 have weakened cold tolerance.
Figure 6.
Functional analysis of the six cold-responsive miRNAs in tobacco infiltrated by Agrobacterium tumefaciens strain GV3101. (A) The transcript analysis of peanut miRNAs in infiltrated tobacco leaves. Student’s t-test was used to determine the significance of differences between mock and the indicated lines. Asterisks indicate significant differences (**p < 0.01). Similar results were obtained from three biological replicates. (B) The phenotype observed in infiltrated tobacco leaves overexpressing pCAMBIA1300 and pCAMBIA1300-MIRNAs with 4°C treatment for 24 h. (C) The different height of infiltrated tobacco overexpressing pCAMBIA1300 and pCAMBIA1300-MIRNAs with 4°C treatment for 24 h.
Discussion
The Cold-Responsive miRNAs in Peanut
Cold stress is the main limiting factor for the distribution, yield, and quality of crops (Zhang et al., 2019). The cold signaling pathway has been well studied (Jeon and Kim, 2013; Barrero-Gil and Salinas, 2018; Shi et al., 2018), and many studies have shown that miRNAs play critical roles in cold stress in plants. For example, osa-miR319 can silence OsPCF5 and OsPCF8, and positively regulate cold tolerance in rice (Yang et al., 2013); osa-miR156k targets the cold stress-related gene SPL and negatively regulates cold tolerance in rice (Cui et al., 2015); sha-miR319 can silence Gamyb-like1 genes that positively regulate cold tolerance in tomatoes (Shi et al., 2019). As a typical semi-arid tropical and subtropical plant, peanuts are highly sensitive to cold stress and may have evolved differential regulatory mechanisms to respond to cold stress. However, the function of small RNA-mediated gene silencing in peanut cold defense has not been reported. In this study, the transcription levels of miRNAs were analyzed between tolerant and sensitive lines with cold treatment in peanuts, and 407 known miRNAs and 143 novel peanut-specific miRNAs in total were identified. Our data showed that known miRNAs and novel miRNAs were involved in cold responses from eight libraries (Supplementary Table S1). Some of these miRNAs have been reported to be responsive to low temperatures in other plants. For example, specific members of the miR171 and miR396 families were also found to respond to cold treatment in peanuts, indicating that they are also conserved in dicotyledons. Likewise, some previously identified cold-induced conserved miRNAs, such as miR393 and miR402 in Arabidopsis (Sunkar and Zhu, 2004), were not found in our data, suggesting that they were unaffected by cold or that induction levels of certain miRNAs may be too low to be detected as significant in peanuts. Finally, through experimental verification, we found that six miRNAs might play key roles in peanut cold defense.
Based on miRNA expression profiling and degradome analysis combined with northern blotting and qRT-PCR validation, we found that several miRNA target pairs (miR160-ARF, miR482-WDRL, miR2118-DR, miR162-DCL, miR1511-SRF, miR1511-SPIRAL1, and miR396-GRF) might play key roles in the regulation of cold tolerance in peanuts. Specially, miR160-ARF, miR482-WDRL, and miR2118-DR showed opposite expression profiles to miR162-DCL, miR1511-SRF, miR1511-SPIRAL1, and miR396-GRF in tolerant and sensitive lines, indicating that they may be key factors leading to differences in cold tolerance.
mirnas Mediated Positive Regulation Pathway in Peanut Cold Response
ahy-miR160, ahy-miR482, and ahy-miR2118 were upregulated in cold-tolerant line WQL30 but downregulated in cold-sensitive line WQL20 (Figure 3). According to previous reports, highly conserved miR160 with ARF targets responds to cold tolerance in maize, wheat, Populus trichocarpa, Populus simonii × P. nigra, and wild banana (Lu et al., 2005; Liu et al., 2018; Zhou et al., 2019; Aydinoglu, 2020). Likewise, the target gene auxin response factor (ARF) not only regulates plant growth but also participates in abiotic stress responses, especially cold stress. For example, in Arabidopsis, AtARF genes are upregulated during cold acclimation (Hannah et al., 2005). Many ARF genes are repressed or induced in bananas (Musa acuminata L.) following cold, salt, and osmotic stress (Hu et al., 2015). In maize, 13 ARFs were upregulated in seedling leaves after cold stress (Aydinoglu, 2020). SLR/IAA14, a transcriptional repressor of auxin signaling, plays a crucial role in the integration of miRNAs in auxin and cold responses in Arabidopsis roots (Aslam et al., 2020). In peanuts, the target genes of ahy-miR160 (AhARF10 and AhARF17) also responded to cold stress (Figures 4, 5), and overexpression of ahy-miR160 enhanced cold tolerance in tobacco (Figure 6). These studies indicate that under cold stress conditions, upregulated ahy-miR160 may reduce ARF transcript levels, suppressing ARF-mediated auxin-responsive gene expression, leading to peanut growth and development attenuation, eventually enhancing cold stress tolerance in peanuts.
Studies have reported that miR2118 targets the protein family associated with disease resistance in response to biotic and abiotic stresses in many plants (de Vries et al., 2015; Wu et al., 2015; Cakir et al., 2016; Gao et al., 2016; Yang et al., 2020; Shi et al., 2021). In Caragana intermedia, cin-miR2118, a drought-resistant miRNA, positively affects drought stress tolerance. Overexpression of cin-miR2118 enhances drought tolerance in tobacco (Wu et al., 2015). miR2118 is a cold-responsive sRNA in wheat, banana, Astragalus membranaceus, and cassava (Xia et al., 2014; Song et al., 2017; Abla et al., 2019; Zhu et al., 2019). Similarly, our data confirmed that ahy-miR2118 was also a cold-responsive miRNA (Figure 3), and DR genes could also be targeted by ahy-miR2118 via the anti-correlation between miR2118 and the corresponding target AhDR genes in peanuts (Figures 4, 5). Moreover, enhanced cold tolerance was also observed in tobacco plants overexpressing ahy-miR2118 (Figure 6), indicating that miR2118 might play an important role in plant resistance to abiotic stress. Under cold treatment, pdu-miR482 was differentially expressed between H (a cold-tolerant genotype) and Sh12 (a cold-sensitive genotype) in almonds (Karimi et al., 2016), respectively. Csn-miR482 was also significantly different between two tea plant cultivars, “Yingshuang” (YS, a cold-tolerant tea plant cultivar) and “Baiye 1” (BY, a cold-sensitive tea plant cultivar; Zhang et al., 2014). In peanuts, the repression in sensitive lines and weak induction in tolerant lines for ahy-miR482 was consistent with the findings of the ovary tissues of H genotype and Sh12 under 0°C treatment in almonds (Figure 3). ahy-miR482 targeted AhWDRLs (Figure 4). AhWDRLs belong to the WD40 protein family, and the large gene family of WD40 proteins is involved in a broad spectrum of crucial plant stress resistance processes. HOS15, a WD40-repeat protein, plays a role in gene activation/repression via histone modification during plant acclimation to low-temperature conditions (Sharma and Pandey, 2015). RNA-seq data analysis also showed that numerous TaWD40s are involved in responses to cold stress in wheat (Hu et al., 2018). Similarly, we found that AhWDRL genes are also involved in responses to cold stress in peanuts (Figure 5). Moreover, the phenomenon of enhanced cold tolerance was observed in tobacco by overexpressing ahy-miR482 (Figure 6). In brief, miR160-ARF, miR482-WDRL, and miR2118-DR modules might mediate cold stress responses, and these three miRNAs may play roles in the positive regulation of peanut cold tolerance through related target genes in peanuts. Therefore, our data sheds light on the possibility of manipulating miRNAs to improve the tolerance of peanuts to cold stress.
miRNAs Mediated Negative Regulation Pathway in Peanut Cold Response
In addition to the above-mentioned miRNAs, we found three other miRNAs with differential responses to cold stress in tolerant and sensitive lines: ahy-miR396, ahy-miR162, and ahy-miR1511, which were upregulated in the cold-sensitive line WQL20 but downregulated in the cold-tolerant line WQL30 (Figure 3). Overexpressing the three miRNAs weakened cold tolerance in tobacco, which means that they may play roles as negative regulators of cold tolerance in peanuts.
Previous studies have shown that miR396 mediates the silencing of target gene GRFs and participates in the regulation of the cold signal response in many plants, such as Arabidopsis thaliana, rice, soybean, tomato, poplar, wheat, and wild grapes (Lu et al., 2008; Tang et al., 2012; Zhang et al., 2014; Yan et al., 2016; Chen et al., 2019; Shi et al., 2019; Wang et al., 2019; Lantzouni et al., 2020). GRFs are a family of transcription factors that mediate the development of seeds, leaves, flowers, root growth, and other essential life processes in plants, such as Arabidopsis thaliana, Brassica napus, Glycine max (soybean), Solanum tuberosum (potato), Oryza sativa (rice), and Zea mays (maize) (Kim et al., 2003; Omidbakhshfard et al., 2015). Additionally, GRFs are also the critical factors of cold signal transduction in plants, which can directly bind to the promoter region of the CBF gene to inhibit its transcription. For example, Arabidopsis AtGRF7 can bind to the tgtcagg cis element on the AtCBF3 promoter to inhibit the expression of AtCBF3 at the transcriptional level (Kim et al., 2012). Lantzouni et al. (2020) found that AtCBF genes (AtCBF1, AtCBF2, and AtCBF3) and the downstream AtCOR genes (AtCOR15a, AtCOR15b, and AtCOR29d) were significantly downregulated in vivo after overexpression of GRF5, and miR396/GRF module is a vital regulation mode for plants to balance growth, development, and cold response under cold stress. In peanuts, we also observed that ahy-miR396 can target AhGRFs and that overexpression of ahy-miR396 weakened cold tolerance in tobacco (Figures 4, 6), suggesting that it might help alleviate cold tolerance in peanuts through the target GRF gene. However, there is no direct evidence for a relationship between AhGRFs and the CBF pathway in peanuts.
Studies have shown that miR1511also is a cold-responsive sRNA in many plants. For example, pdu-miR1511 was differentially expressed between H and Sh12 in almond under cold treatment (Karimi et al., 2016). csi-miR1511 was also significantly different between YS and BY in tea plant (Zhang et al., 2014). mit-miR1511 showed remarkably higher levels at the cold stress temperatures (0°C) in the wild bananas (Liu et al., 2018). In agreement with previous reports, ahy-miR1511 showed significant differences between the tolerant and sensitive lines (Figure 3). It targets the AhSRF and AhSPIRAL1 genes (Figures 4, 5). In Arabidopsis, AtSPIRAL1 maintains microtubules (MT) stability and is closely associated with stress. For example, salt stress can lead to the UPS-dependent degradation of the MT-associated protein AtSPIRAL1. The degradation of AtSPIRAL1 results in the depolymerization of MTs, followed by the formation of new MTs that are better adapted to osmotic stress (Wang et al., 2011). For another target gene AhSRF, recent evidence has shown that homologous gene AtSRF6 is an identified regulator for COR in Arabidopsis (Wei et al., 2021). These reports suggest that the miR1511-SRF and miR1511-SPIRAL1 modules may attenuate cold tolerance by mediating MT stability or by regulating the ICE-CBF-COR pathway. Another anti-correlated module in peanuts is ahy-miR162, which targets AhDCL. It has been reported that miR162 not only regulates the key factors of plant growth and development but also responds to low-temperature stress. In almonds, pdu-miR162 is upregulated in H and downregulated in Sh12 under cold stress (Karimi et al., 2016). This difference in expression in tolerant and sensitive varieties agreed with our data in peanuts (Figure 3), suggesting that miR162 is conserved and functional in plant cold tolerance. Pdu-miR162 directly targets PdDCL1 for cleavage (Karimi et al., 2016). DCL proteins are key regulators of small RNA biogenesis (RNA interference, RNAi), and participate in the response to cold stress in plants. For example, rice, like peanuts, are low temperature-sensitive plants. Vyse et al. (2020) found that OsDCL1 is induced by cold acclimation in rice. We also observed significant differences in the expression of AhDCL6 and AhDCL16 targeted by ahy-miR162 in tolerant and sensitive lines (Figures 4, 5), suggesting that the miR162-DCL module probably weakens cold stress tolerance in peanuts by feedback regulation of small RNA biogenesis.
Conclusion
In summary, cold-responsive miRNAs and candidate target genes were identified through integrated sRNA and degradome analysis during cold treatment of tolerant and sensitive lines. ahy-miR160, ahy-miR162, ahy-miR396, ahy-miR482, ahy-miR1511, and ahy-miR2118, as well as transcriptional factors, including GRF, WDRL, and ARF, or genes, such as SRF, DCL, and SPIRAL, were differentially expressed. They might be the main contributors to the CBF and ARF pathways, protein kinase, small RNA biogenesis, and MT stability involved in cold tolerance of peanuts. These miRNAs may downregulate the expression of their target genes, which encode the regulatory and functional proteins involved in cold tolerance. These results increase our knowledge of sRNAs involved in the post-transcriptional regulation of cold tolerance and provide candidate genes for future functional analyses of cold tolerance-related signaling pathways in peanuts.
Data Availability Statement
The raw data of the sRNA and degradome libraries are available from the NCBI Sequence Read Archive (SRA) under accession numbers SRR19241918, SRR19241919, SRR19241920, SRR19241921, SRR19241922, SRR19241923, SRR19241924, SRR19241925 and SRR19262409, SRR19262410, respectively.
Author Contributions
DB and HJ designed the experiments. XZ, CR, YX, YT, HZ, NL, and CS performed the experiments. XZ, DB, and HJ performed the data analysis. XZ wrote the manuscript. All authors contributed to the article and approved the submitted version.
Funding
This work was supported by the National Natural Science Foundation of China (31871662), the Open Project of Key Laboratory of Biology and Genetic Improvement of Oil Crops, Ministry of Agriculture and Rural Affairs, P. R. China (KF2021004), the China Agricultural Research System (CARS-13), the Cultivation Project of National Natural Science Foundation (YGJPY1901), the Doctoral Research Fund Project, Technology Innovation Research Project of the Shanxi Academy of Agricultural Sciences (YBSJJ2014), the Research Program Sponsored by State Key Laboratory of Sustainable Dryland Agriculture (in preparation), Shanxi Agricultural University (NO. 202105D121008), and the earmarked fund for Modern Agro-industry Technology Research System (2022-05).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Acknowledgments
We are grateful to Huifang Jiang for kindly providing WQL20 and WQL30 seeds, and Editage (www.editage.cn) for English language editing.
Footnotes
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2022.920195/full#supplementary-material
Length distribution and abundance of the sequences. The lengths of the unique, valid reads ranged from 18 to 30 nucleotides (nt), and the 21-24 nt sequences were predominant in all libraries, with the 24 (nt) sequences being the most common.
Examples of T-plots of miRNA targets in two peanut RILs confirmed by degradome sequencing. The T-plots show the distribution of the degradome tags along the full length of the target mRNA sequence. The vertical red line indicates the cleavage site of each transcript and is also shown by an arrow. (A,D) The cleavage features in AhARF10 (Arahy.4D5DQA.1) and AhARF17 (Arahy.35XBQI.1) mRNA by ahy-miR160 in DS2. (B,E) The cleavage features in AhDCL6 (Arahy.GMG8MT.1) and AhDCL16 (Arahy.NHK71Z.4) mRNA by ahy-miR162 in DS2. (C,F) The cleavage features in AhGRF1 (Arahy.858S2J.1) and AhGRF4 (Arahy.KV1X6M.1) mRNA by ahy-miR396 in DS2.
Examples of T-plots of miRNA targets in two peanut RILs confirmed by degradome sequencing. The T-plots show the distribution of the degradome tags along the full length of the target mRNA sequence. The vertical red line indicates the cleavage site of each transcript and is also shown by an arrow. (A,D) The cleavage features in AhWDRL1 (Arahy.C265D9.1) and AhWDRL2 (Arahy.ER1LI4.1) mRNA by ahy-miR482 in DS2. (B,E) The cleavage features in AhSRF (Arahy.XK3KPA.1) and AhSP1L1 (Arahy.NV23DS.1) mRNA by ahy-miR1511 in DS2. (C,F) The cleavage features in AhDR1 (Arahy.1ALG22.1) and AhDR2 (Arahy.GT0Q5X.1) mRNA by ahy-miR2118 in DS2.
List of identified miRNAs differentially expressed in peanut with cold treatment.
Statistics of degradome reads in DS1 and DS2.
Differentially-expressed target genes between the DS1 and DS2 libraries.
Primer information.
References
- Abla M., Sun H. G., Li Z. Y., Wei C. X., Gao F., Zhou Y. J., et al. (2019). Identification of miRNAs and their response to cold stress in Astragalus Membranaceus. Biomol. Ther. 9:182. doi: 10.3390/biom9050182, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Addo-Quaye C., Miller W., Axtell M. J. (2009). CleaveLand: a pipeline for using degradome data to find cleaved small RNA targets. Bioinformatics 25, 130–131. doi: 10.1093/bioinformatics/btn604, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aslam M., Sugita K., Qin Y., Rahman A. (2020). Aux/IAA14 regulates microRNA-mediated cold stress response in Arabidopsis roots. Int. J. Mol. Sci. 21:8441. doi: 10.3390/ijms21228441, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aydinoglu F. (2020). Elucidating the regulatory roles of microRNAs in maize (Zea mays L.) leaf growth response to chilling stress. Planta 251:38. doi: 10.1007/s00425-019-03331-y, PMID: [DOI] [PubMed] [Google Scholar]
- Barrero-Gil J., Salinas J. (2018). Gene regulatory networks mediating cold acclimation: the CBF pathway. Adv. Exp. Med. Biol. 1081, 3–22. doi: 10.1007/978-981-13-1244-1_1, PMID: [DOI] [PubMed] [Google Scholar]
- Cakir O., Candar-Cakir B., Zhang B. H. (2016). Small RNA and degradome sequencing reveals important microRNA function in Astragalus chrysochlorus response to selenium stimuli. Plant Biotechnol. J. 14, 543–556. doi: 10.1111/pbi.12397, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen N., Yang Q., Su M., Pan L. J., Chi X. Y., Chen M. N., et al. (2012). Cloning of six ERF family transcription factor genes from peanut and analysis of their expression during abiotic stress. Plant Mol. Biol. Report. 30, 1415–1425. doi: 10.1007/s11105-012-0456-0 [DOI] [Google Scholar]
- Chen X., Jiang L. R., Zheng J. S., Chen F. Y., Wang T. S., Wang M. L., et al. (2019). A missense mutation in Large Grain Size 1 increases grain size and enhances cold tolerance in rice. J. Exp. Bot. 70, 3851–3866. doi: 10.1093/jxb/erz192 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui N., Sun X., Sun M., Jia B., Duanmu H., Lv D., et al. (2015). Overexpression of osmiR156k leads to reduced tolerance to cold stress in rice (Oryza Sativa). Mol. Breed. 35:214. doi: 10.1007/s11032-015-0402-6 [DOI] [Google Scholar]
- de Vries S., Kloesges T., Rose L. E. (2015). Evolutionarily dynamic, but robust, targeting of resistance genes by the miR482/2118 gene family in the Solanaceae. Genome Biol. Evol. 7, 3307–3321. doi: 10.1093/gbe/evv225, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao F., Wang N., Li H. Y., Liu J. S., Fu C. X., Xiao Z. H., et al. (2016). Identification of drought-responsive microRNAs and their targets in Ammopiptanthus mongolicus by using high-throughput sequencing. Sci. Rep. 6:34601. doi: 10.1038/srep34601, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilmour S. J., Zarka D. G., Stockinger E. J., Salazar M. P., Houghton J. M., Thomashow M. F. (1998). Low temperature regulation of the Arabidopsis CBF family of AP2 transcriptional activators as an early step in cold-induced COR gene expression. Plant J. 16, 433–442. doi: 10.1046/j.1365-313x.1998.00310.x, PMID: [DOI] [PubMed] [Google Scholar]
- Guo J., Ren Y. K., Tang Z. H., Shi W. P., Zhou M. X. (2019). Characterization and expression profiling of the ICE-CBF-COR genes in wheat. PeerJ 7:e8190. doi: 10.7717/peerj.8190, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hannah M. A., Heyer A. G., Hincha D. K. (2005). A global survey of gene regulation during cold acclimation in Arabidopsis thaliana. PLoS Genet. 1:e26. doi: 10.1371/journal.pgen.0010026, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu R., Xiao J., Gu T., Yu X., Zhang Y., Chang J., et al. (2018). Genome-wide identification and analysis of WD40 proteins in wheat (Triticum aestivum L.). BMC Genomics 19:803. doi: 10.1186/s12864-018-5157-0, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu W., Zuo J., Hou X., Yan Y., Wei Y., Liu J., et al. (2015). The auxin response factor gene family in banana: genome-wide identification and expression analyses during development, ripening, and abiotic stress. Front. Plant Sci. 6:742. doi: 10.3389/fpls.2015.00742, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeon J., Kim J. (2013). Cold stress signaling networks in Arabidopsis. J. Plant. Biol. 56, 69–76. doi: 10.1007/s12374-013-0903-y [DOI] [Google Scholar]
- Jin H. L. (2008). Endogenous small RNAs and antibacterial immunity in plants. FEBS Lett. 582, 2679–2684. doi: 10.1016/j.febslet.2008.06.053, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karimi M., Ghazanfari F., Fadaei A., Ahmadi L., Shiran B., Rabei M., et al. (2016). The small-RNA profiles of almond (Prunus dulcis mill.) reproductive tissues in response to cold stress. PLoS One 11:e0156519. doi: 10.1371/journal.pone.0156519, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katiyar-Agarwal S., Jin H. (2007). Discovery of pathogen-regulated small RNAs in plants. Methods Enzymol. 427, 215–227. doi: 10.1016/S0076-6879(07)27012-0, PMID: [DOI] [PubMed] [Google Scholar]
- Kim J. H, Choi D., Kende H. (2003). The AtGRF family of putative transcription factors is involved in leaf and cotyledon growth in Arabidopsis. Plant J. 36, 94–104. doi: 10.1046/j.1365-313X.2003.01862.x, PMID: [DOI] [PubMed] [Google Scholar]
- Kim J. S., Mizoi J., Kidokoro S., Maruyama K., Nakajima J., Nakashima K., et al. (2012). Arabidopsis growth regulating factor7 functions as a transcriptional repressor of abscisic acid- and osmotic stress-responsive genes, including DREB2A. Plant Cell 24, 3393–3405. doi: 10.1105/tpc.112.100933, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lantzouni O., Alkofer A., Falter-Braun P., Schwechheimer C. (2020). GROWTH REGULATING FACTORS interact with DELLAs and regulate growth in cold stress. Plant Cell 32, 1018–1034. doi: 10.1105/tpc.19.00784, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu W. H., Cheng C. Z., Chen F. L., Ni S. S., Lin Y. L., Lai Z. X. (2018). High-throughput sequencing of small RNAs revealed the diversified cold-responsive pathways during cold stress in the wild banana (Musa itinerans). BMC Plant Biol. 18:308. doi: 10.1186/s12870-018-1483-2, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu S., Sun Y. H., Chiang V. L. (2008). Stress-responsive microRNAs in Populus. Plant J. 55, 131–151. doi: 10.1111/j.1365-313X.2008.03497.x, PMID: [DOI] [PubMed] [Google Scholar]
- Lu S., Sun Y. H., Rui S., Clark C., Chiang V. L. (2005). Novel and mechanical stress-responsive microRNAs in Populus trichocarpa that are absent from Arabidopsis. Plant Cell 17, 2186–2203. doi: 10.1105/tpc.105.033456, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niu D. D., Zhang X., Song X. O., Wang Z. H., Li Y. Q., Qiao L. L., et al. (2018). Deep sequencing uncovers rice long siRNAs and its involvement in immunity against Rhizoctonia solani. Phytopathology 108, 60–69. doi: 10.1094/Phyto-03-17-0119-R, PMID: [DOI] [PubMed] [Google Scholar]
- Omidbakhshfard M. A., Proost S., Fujikura U., Mueller-Roeber B. (2015). Growth-Regulating Factors (GRFs): A small transcription factorfamily with important functions in plant biology. Mol. Plant 8, 998–1010. doi: 10.1016/j.molp.2015.01.013, PMID: [DOI] [PubMed] [Google Scholar]
- Park S., Lee C. M., Doherty C. J., Gilmour S. J., Kim Y. S., Thomashow M. F. (2015). Regulation of the Arabidopsis CBF regulon by a complex low temperature regulatory network. Plant J. 82, 193–207. doi: 10.1111/tpj.12796, PMID: [DOI] [PubMed] [Google Scholar]
- Pruthvi V., Narasimhan R., Nataraja K. N. (2014). Simultaneous expression of abiotic stress responsive transcription factors, AtDREB2A, AtHB7 and AtABF3 improves salinity and drought tolerance in peanut (Arachis hypogaea L.). PLoS One 9:e111152. doi: 10.1371/journal.pone.0111152, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma M., Pandey G. K. (2015). Expansion and function of repeat domain proteins during stress and development in plants. Front. Plant Sci. 6:1218. doi: 10.3389/fpls.2015.01218, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi Y., Ding Y., Yang S. (2018). Molecular regulation of CBF signaling in cold acclimation. Trends Plant Sci. 23, 623–637. doi: 10.1016/j.tplants.2018.04.002, PMID: [DOI] [PubMed] [Google Scholar]
- Shi X., Jiang F., Wen J., Wu Z. (2019). Overexpression of Solanum habrochaites microRNA319d (sha-miR319d) confers chilling and heat stress tolerance in tomato (S. lycopersicum). BMC Plant Biol. 19:214. doi: 10.1186/s12870-019-1823-x, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi R., Jiao W., Yun L., Zhang Z., Zhang X., Wang Q., et al. (2021). Utilization of transcriptome, small rna, and degradome sequencing to provide insights into drought stress and rewatering treatment in Medicago ruthenica. Front. Plant Sci. 12:675903. doi: 10.3389/fpls.2021.675903, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song G. Q., Zhang R. Z., Zhang S. J., Li Y. L., Gao J., Han X. D., et al. (2017). Response of microRNAs to cold treatment in the young spikes of common wheat. BMC Genomics 18:212. doi: 10.1186/s12864-017-3556-2, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sunkar R., Zhu J. K. (2004). Novel and stress-regulated microRNAs and other small RNAs from Arabidopsis. Plant Cell 16, 2001–2019. doi: 10.1105/tpc.104.022830, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang Z., Zhang L., Xu C., Yuan S., Zhang F., Zheng Y., et al. (2012). Uncovering small RNA-mediated responses to cold stress in a wheat thermosensitive genic male-sterile line by deep sequencing. Plant Physiol 159, 721–738. doi: 10.1104/pp.112.196048, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Upadhyaya H. D., Rodomiro O., Paula J. B., Sube S. (2003). Development of a groundnut core collection using taxonomical, geographical and morphological descriptors. Genet Resour. Crop. Ev. 50, 139–148. doi: 10.1007/s10722-003-6111-8 [DOI] [Google Scholar]
- Vyse K., Faivre L., Romich M., Pagter M., Schubert D., Hincha D. K., et al. (2020). Transcriptional and post-transcriptional regulation and transcriptional memory of chromatin regulators in response to low temperature. Front. Plant Sci. 11:39. doi: 10.3389/fpls.2020.00039, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang D. Z., Jin Y. N., Ding X. H., Wang W. J., Zhai S. S., Bai L. P., et al. (2017). Gene regulation and signal transduction in the ICE-CBF-COR signaling pathway during cold stress in plants. Biochemistry 82, 1103–1117. doi: 10.1134/S0006297917100030, PMID: [DOI] [PubMed] [Google Scholar]
- Wang P., Yang Y., Shi H., Wang Y., Ren F. (2019). Small RNA and degradome deep sequencing reveal respective roles of cold-related microRNAs across Chinese wild grapevine and cultivated grapevine. BMC Genomics 20:740. doi: 10.1186/s12864-019-6111-5, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang S., Kurepa J., Hashimoto T., Smalle J. A. (2011). Salt stress-induced disassembly of Arabidopsis cortical microtubule arrays involves 26S proteasome-dependent degradation of SPIRAL1. Plant Cell 23, 3412–3427. doi: 10.1105/tpc.111.089920, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei X. S., Liu S., Sun C., Xie G. S., Wang L. Q. (2021). Convergence and divergence: signal perception and transduction mechanisms of cold stress in Arabidopsis and rice. Plants-Basel 10:1864. doi: 10.3390/plants10091864, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu B. F., Li W. F., Xu H. Y., Qi L. W., Han S. Y. (2015). Role of cin-miR2118 in drought stress responses in Caragana intermedia and tobacco. Gene 574, 34–40. doi: 10.1016/j.gene.2015.07.072, PMID: [DOI] [PubMed] [Google Scholar]
- Xia J., Zeng C. Y., Chen Z., Zhang K. V., Chen X., Zhou Y. F., et al. (2014). Endogenous small-noncoding RNAs and their roles in chilling response and stress acclimation in cassava. BMC Genomics 15:634. doi: 10.1186/1471-2164-15-634, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao F. J., Song L. C. (2011). Analysis of extreme low-temperature events during the warm season in Northeast China. Na Hazards 58, 1333–1344. doi: 10.1007/s11069-011-9735-6 [DOI] [Google Scholar]
- Yan J., Zhao C., Zhou J., Yang Y., Wang P., Zhu X., et al. (2016). The miR165/166 mediated regulatory module plays critical roles in ABA homeostasis and response in Arabidopsis thaliana. PLoS Genet. 12:e1006416. doi: 10.1371/journal.pgen.1006416, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang C., Li D., Mao D., Liu X., Ji C., Li X., et al. (2013). Overexpression of microRNA319 impacts leaf morphogenesis and leads to enhanced cold tolerance in rice (Oryza sativa L.). Plant Cell Environ. 36, 2207–2218. doi: 10.1111/pce.12130, PMID: [DOI] [PubMed] [Google Scholar]
- Yang Z. M., Zhu P. P., Kang H., Liu L., Cao Q. H., Sun J., et al. (2020). High-throughput deep sequencing reveals the important role that microRNAs play in the salt response in sweet potato (Ipomoea batatas L.). BMC Genomics 21:164. doi: 10.1186/s12864-020-6567-3, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X., Bao Y. L., Shan D. Q., Wang Z. H., Song X. N., Wang Z. Y., et al. (2018). Magnaporthe oryzae induces the expression of a microRNA to suppress the immune response in rice. Plant Physiol. 177, 352–368. doi: 10.1104/pp.17.01665, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H., Dong J., Zhao X., Zhang Y., Ren J., Xing L., et al. (2019). Research progress in membrane lipid metabolism and molecular mechanism in peanut cold tolerance. Front. Plant Sci. 10:838. doi: 10.3389/fpls.2019.00838, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang M., Liu W., Bi Y. P., Wang Z. Z. (2009). Isolation and identification of PNDREB1: a new DREB transcription factor from peanut (Arachis hypogaea L.). Acta Agron. Sin. 35, 1973–1980. doi: 10.3724/SP.J.1006.2009.01973 [DOI] [Google Scholar]
- Zhang Y., Zhu X. J., Chen X., Song C. N. A., Zou Z. W., Wang Y. H., et al. (2014). Identification and characterization of cold-responsive microRNAs in tea plant (Camellia sinensis) and their targets using high-throughput sequencing and degradome analysis. BMC Plant Biol. 14:271. doi: 10.1186/s12870-014-0271-x, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou B., Kang Y., Leng J., Xu Q. (2019). Genome-wide analysis of the miRNA-mRNAs network involved in cold tolerance in Populus simonii x P. nigra. Genes-Basel 10:430. doi: 10.3390/genes10060430, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu J. K. (2016). Abiotic stress signaling and responses in plants. Cell 167, 313–324. doi: 10.1016/j.cell.2016.08.029, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu H., Zhang Y., Tang R., Qu H., Duan X., Jiang Y. (2019). Banana sRNAome and degradome identify microRNAs functioning in differential responses to temperature stress. BMC Genomics 20:33. doi: 10.1186/s12864-018-5395-1, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou X., Wang G., Sutoh K., Zhu J. K., Zhang W. (2008). Identification of cold-inducible microRNAs in plants by transcriptome analysis. BBA-Gene Regul. Mech. 1779, 780–788. doi: 10.1016/j.bbagrm.2008.04.005, PMID: [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Length distribution and abundance of the sequences. The lengths of the unique, valid reads ranged from 18 to 30 nucleotides (nt), and the 21-24 nt sequences were predominant in all libraries, with the 24 (nt) sequences being the most common.
Examples of T-plots of miRNA targets in two peanut RILs confirmed by degradome sequencing. The T-plots show the distribution of the degradome tags along the full length of the target mRNA sequence. The vertical red line indicates the cleavage site of each transcript and is also shown by an arrow. (A,D) The cleavage features in AhARF10 (Arahy.4D5DQA.1) and AhARF17 (Arahy.35XBQI.1) mRNA by ahy-miR160 in DS2. (B,E) The cleavage features in AhDCL6 (Arahy.GMG8MT.1) and AhDCL16 (Arahy.NHK71Z.4) mRNA by ahy-miR162 in DS2. (C,F) The cleavage features in AhGRF1 (Arahy.858S2J.1) and AhGRF4 (Arahy.KV1X6M.1) mRNA by ahy-miR396 in DS2.
Examples of T-plots of miRNA targets in two peanut RILs confirmed by degradome sequencing. The T-plots show the distribution of the degradome tags along the full length of the target mRNA sequence. The vertical red line indicates the cleavage site of each transcript and is also shown by an arrow. (A,D) The cleavage features in AhWDRL1 (Arahy.C265D9.1) and AhWDRL2 (Arahy.ER1LI4.1) mRNA by ahy-miR482 in DS2. (B,E) The cleavage features in AhSRF (Arahy.XK3KPA.1) and AhSP1L1 (Arahy.NV23DS.1) mRNA by ahy-miR1511 in DS2. (C,F) The cleavage features in AhDR1 (Arahy.1ALG22.1) and AhDR2 (Arahy.GT0Q5X.1) mRNA by ahy-miR2118 in DS2.
List of identified miRNAs differentially expressed in peanut with cold treatment.
Statistics of degradome reads in DS1 and DS2.
Differentially-expressed target genes between the DS1 and DS2 libraries.
Primer information.
Data Availability Statement
The raw data of the sRNA and degradome libraries are available from the NCBI Sequence Read Archive (SRA) under accession numbers SRR19241918, SRR19241919, SRR19241920, SRR19241921, SRR19241922, SRR19241923, SRR19241924, SRR19241925 and SRR19262409, SRR19262410, respectively.