Abstract
Xenorhabdus nematophilus is an insect pathogen that lives in a symbiotic association with a specific entomopathogenic nematode. During prolonged culturing, variant cells arise that are deficient in numerous properties. To understand the genetic mechanism underlying variant cell formation, a transposon mutagenesis approach was taken. Three phenotypically similar variant strains of X. nematophilus, each of which contained a single transposon insertion, were isolated. The insertions occurred at different locations in the chromosome. The variant strain, ANV2, was further characterized. It was deficient in several properties, including the ability to produce antibiotics and the stationary-phase-induced outer membrane protein, OpnB. Unlike wild-type cells, ANV2 produced lecithinase. The emergence of ANV2 from the nematode host was delayed relative to the emergence of the parental strain. The transposon in ANV2 had inserted in a gene designated var1, which encodes a novel protein composed of 121 amino acid residues. Complementation analysis confirmed that the pleiotropic phenotype of the ANV2 strain was produced by inactivation of var1. Other variant strains were not complemented by var1. These results indicate that inactivation of a single gene was sufficient to promote variant cell formation in X. nematophilus and that disruption of genetic loci other than var1 can result in the same pleiotropic phenotype.
Xenorhabdus nematophilus is harbored as a symbiont in an intestinal vesicle of the infective juvenile stage of the entomopathogenic nematode, Steinernema carpocapsae (19, 20, 24, 25, 31, 35, 36). The bacteria are carried into a susceptible insect larva by the nematode and are subsequently released into the insect hemolymph, where they participate in the killing of the insect host (3, 4, 11, 17). X. nematophilus proliferates within the hemolymph and eventually enters stationary phase. Under stationary-phase conditions, the bacterium produces numerous products that play roles in providing a nutrient base for the developing nematodes (25, 42, 45). During the final stages of nematode development, the bacteria and nematode reassociate, and the symbiotic pair leaves the insect cadaver in search of a new host (4, 25, 36). The symbiotic association presumably protects the bacterium so that it can exist outside of the insect host (3, 11).
The X. nematophilus cells obtained from the infective juvenile nematode are referred to as primary or phase I cells (1, 8–10, 12, 13, 43, 48). The primary cells of all strains of X. nematophilus studied to date possess the following characteristics: they are motile, are able to bind dye, can produce antibiotics, hemolysins, proteases, and crystal proteins, can stimulate hemagglutination, can elaborate fimbriae on agar surfaces, and are able to synthesize the outer membrane protein, OpnB, during post-exponential-phase growth (25, 26, 27, 28, 34, 44). These properties will be referred to as primary-specific traits. Other properties, such as lecithinase and lipase production, are more variable between strains. During prolonged culturing of the bacteria, variant cells arise spontaneously at a variable frequency. The variant cells have been called secondary or phase II cells (8, 9, 12, 13). In the secondary cells, the primary-specific traits mentioned above are either absent or greatly reduced. In some strains, lipase and lecithinase activity are increased in the secondary cells (8, 12). The membranes of the variant secondary forms are less fluid than their respective primary forms (21). The formation of secondary cells occurs in all species of Xenorhabdus (8) and also occurs in the closely related Photorhabdus spp. (24). Low osmolarity in Photorhabdus luminescens (32) and microaerophilic conditions in X. nematophilus (9, 25) have been shown to enhance formation of the secondary cells. Although the biological role of the secondary cells remains unsolved, this form has been shown to grow faster than the primary cells after experiencing starvation conditions (41). It was suggested that the secondary cells may be better adapted to life as a free-living organism.
The primary and secondary cells are equally pathogenic towards larvae of the greater wax moth, Galleria mellonella (17, 25, 31, 46). The secondary cells of Xenorhabdus support growth and development of the nematodes in vitro (18, 46). In contrast, they are defective in their ability to support growth of the nematode in the insect (1). These findings suggest that the products missing in the secondary cells are not essential for virulence but are required for normal in vivo growth of the nematode. A major difference between the secondary cells of Xenorhabdus and Photorhabdus is that the latter do not support in vitro growth of their symbiotic partner, the Heterorhabditidae nematodes (18).
The genetic mechanism underlying the formation of the secondary cell type is not known. DNA rearrangement (5) and loss of plasmids (33) do not appear to play a role in this process. In Photorhabdus, both posttranscriptional (25, 30) and posttranslational (16, 47) mechanisms have been proposed to be involved in the loss of primary-specific traits. Since numerous properties are altered in the secondary cells, it is conceivable that inactivation of a putative global regulatory gene could result in the coordinate loss of the primary-specific products. The question arises whether a unique genetic locus controls variant cell formation. Alternatively, since secondary cells form under prolonged incubation conditions, it is possible that the accumulation of multiple mutations is necessary for the formation of the variant form. In the present study, a transposon mutagenesis approach was taken to address these questions.
MATERIALS AND METHODS
Bacterial strains, plasmids, media, and growth conditions.
Bacterial strains and plasmids used in this study are shown in Table 1. X. nematophilus strain AN6/I (ATCC 19061/I) was maintained on Luria broth (LB) containing 50 μg of ampicillin per ml, 0.0025% bromothymol blue (BTB), and 0.004% triphenyl-tetrazonium chloride (LBTA) (26, 34). ANV1, ANV2, and ANV4 were maintained on LB containing 30 μg of kanamycin per ml. The rifampicin-resistant strain AN6/I was maintained on LB containing 100 μg of rifampicin per ml. Escherichia coli S17-1 λ(pir) was used to conjugally transfer plasmids to X. nematophilus (40). X. nematophilus strains were grown at 30°C, and E. coli was grown at 37°C.
TABLE 1.
Bacterial strains and plasmids used in the study
| Bacterial strain or plasmid | Relevant genotype | Source or reference |
|---|---|---|
| X. nematophilus strains | ||
| AN6/I (ATCC 19061/I) | Wild type, Ampr | R. J. Akhurst |
| AN6/II | Spontaneous variant type, Ampr | R. J. Akhurst |
| ATCC 19061/II | Spontaneous variant type, Ampr | R. Hurlbert |
| ANV2 | AN6/I::Tn10 Ampr Kmr | This study |
| ANV1 | AN6/I::Tn10 Ampr Kmr | This study |
| ANV4 | AN6/I::Tn10 Ampr Kmr | This study |
| Plasmids | ||
| pLOF-Kmr | Ampr, Tn10-based delivery plasmid with Kmr | 29 |
| pJQ200KS | sacB, R6Kori, Rp4mob, Gmr | 37 |
| pG52 | pGEM3Z containing 2.1-kb EcoRI/HindIII fragment from ANV2 | This study |
| pBK9 | pBK-CMV containing a 5.2-kb EcoRI insert from the wild-type X. nematophilus AN6/I | This study |
| pBK10 | pBK9 with 945-bp EcoRV deletion | This study |
| pBK12 | pBK9 with 1.5-kb deletion at Csp45I and ClaI sites | This study |
| pJV9 | pJQ200KS containing a 5.2-kb EcoRI insert from the wild-type X. nematophilus AN6/I | This study |
| pJV10 | pJV9 with 945-bp EcoRV deletion | This study |
| pJV12 | pJV9 with 1.5-kb deletion at Csp45I and ClaI sites | This study |
Transposon mutagenesis.
The mini-Tn10 transposon carried on pLOF-Kmr was introduced into X. nematophilus by conjugal transfer. Cultures used for transposon mutagenesis were grown to logarithmic phase, were centrifuged for 10 min at room temperature, and were resuspended in 0.6 ml of LB. One hundred microliters of E. coli 17-1/pLOF-Kmr and 400 μl of AN6/I were mixed and placed on the surface of an LB plate containing 0.1 mM isopropyl-β-d-thiogalactopyranoside (IPTG) and were incubated for 12 to 14 h at 30°C. Bacteria were resuspended in 1.5 ml of LB, were serially diluted, and were plated on LBTA (26) containing 30 μg of kanamycin per ml and 100 μg of rifampicin per ml. The kanamycin-resistant red colonies were screened for several in vitro phenotypes.
In vitro biochemical assays.
Motility on soft agar, outer membrane proteins, crystal proteins, antibiotic production, hemolysis, and egg yolk lecithinase reaction were analyzed as described previously (46).
Fimbria production and hemagglutination.
Fimbria production was examined by electron microscopy as described by Brehelin et al. (13). For the hemagglutination assay, cultures from nutrient broth agar plates incubated at 30°C for 3 days were used (34). Cells were serially diluted in Grace's medium (25 μl). Twenty-five microliters of sheep blood (4% in phosphate-buffered saline) was added to the cells, and the mixture was shaken gently to mix. The result was recorded after 2 h of incubation at room temperature.
Initial growth of bacteria released from the nematode.
Infective dauer juveniles raised on a lawn of either AN6 or ANV2 were harvested from water traps. There was no detectable difference in the yields of nematodes grown on the respective strains. The bacteria were subsequently surface sterilized by washing in sterile 0.9% NaCl, followed by brief vortexing in a mixture of 10% Clorox and 0.9% NaCl, and were subsequently washed three times with sterile 0.9% NaCl. The effectiveness of the sterilization procedure in removing bacteria from the nematode surface was assessed by using the BacLight vital stain (Molecular Probes, Inc., Eugene, Oreg.). Bacteria were not detected on the surface of the sterile nematodes. Five hundred nematodes were inoculated into 5 ml of either LB (wild-type strain) or LB containing 30 μg of kanamycin per ml (mutant strains). The nematodes were incubated at 28°C, and the bacterial growth was monitored by turbidity by using a Klett meter. Each experimental condition was performed in triplicate. The released bacteria were plated on LBTA containing either 50 μg of ampicillin per ml (wild-type strain) or 30 μg of kanamycin per ml (mutant strains). X. nematophilus cultures with plasmids were grown and plated on LBTA containing 10 μg of gentamicin per ml. The experiment was repeated three times with nearly identical results.
Natural infection of Manduca sexta.
Fourth-instar M. sexta larvae were naturally infected with 50 surface-sterilized dauer juveniles per caterpillar. The nematodes were raised on either the wild-type strain, secondary strains, mutant strains, or mutant strains carrying plasmids, as described above. Nematodes were placed on sterile filter paper soaked in 0.9% NaCl solution prior to infection of the caterpillars. The growth of the M. sexta larvae was monitored by weight gain. The time at which 50% of the insects had died (LT50) was determined by monitoring mortality by lack of movement and loss of body turgor. The pathogenicity experiments were carried out three times for each experimental condition (26).
Southern hybridization.
Southern hybridization was carried out as previously described (38). Briefly, DNA separated on a 1% agarose gel run in Tris-borate-EDTA buffer was transferred onto a nylon membrane by capillary transfer and was hybridized with 32P-labeled probes under stringent conditions (65°C). After hybridization, blots were washed at high stringency (65°C, 0.1× SSC [1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate], 0.1% sodium dodecyl sulfate [SDS]) (44).
Cloning var1 and construction of var1-orf2 plasmids.
Attempts to clone the 6.8-kb EcoRI fragment containing the inactivated var1 of ANV2 were unsuccessful (6, 38). To clone the inactivated var1, we took advantage of the single HindIII site that exists in the Kmr cassette. The chromosome of ANV2 was digested with HindIII and EcoRI, and the resulting fragments were cloned into pGEM3Z (Promega). Following transformation of E. coli, colonies containing the kanamycin resistance cassette were detected by colony hybridization. The 1.5-kb NotI fragment of pLOFKmr was used as the probe. The radiolabeled probe was hybridized to the filter at 65°C. Blots were washed at 65°C in a solution containing 0.1× SSC and 0.1% SDS. One positive clone containing a 1.2-kb flanking region adjacent to the kanamycin resistance cassette (pG52) was analyzed by sequencing. To clone the full-length EcoRI fragment that contained the wild-type var1, an EcoRI library of the AN6/1 was created in Lambda ZAPII EcoRI (Stratagene). Plaque hybridization was performed with nylon filters (MagnaGraph). The radiolabeled probe containing the 5′ region of var1 (1.2-kb EcoRI/NotI fragment of pG52) was used for plaque hybridization. After hybridization, blots were washed at 65°C with a solution containing 0.5× SSC and 0.5% SDS. The identified positive lambda clones were converted into plasmid (pBK-CMV) with the in vivo excision protocol, described by Stratagene, resulting in pBK9 (9.7 kb). A 5.2-kb insert was cut out from pBK9 with BamHI and XbaI and was subcloned into pJQ200KS, resulting in pJV9 (10.1 kb). In order to delete the open reading frames (ORFs) downstream of the ybhE-like gene, we used EcoRV, which has three restriction sites (see Fig. 4). The pBluescript vector (pBK9) containing the 5.2-kb wild-type fragment from X. nematophilus was digested with EcoRV, and the 8.7-kb var1-containing fragment was isolated from the gel. The new construction (pBK10, 8.7 kb in size) was digested with XbaI and SalI to clone var1 into pJQ200KS, resulting in pJV10 (9.1 kb in size). To construct pJV12, pBK9 was digested with Csp45I and ClaI. The larger (8.2-kb) fragment was isolated from the gel and religated (pBK12). The insert was cut out with XbaI and SalI and was cloned into identical restriction sites of pJQ200KS.
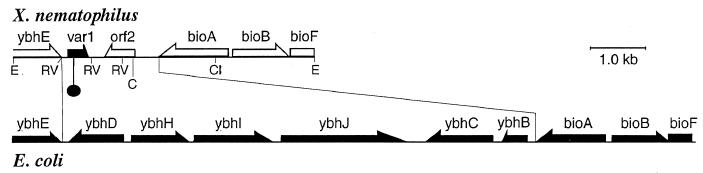
FIG. 4.
Comparison of the ybh-bio regions in X. nematophilus and E. coli. The site where the Tn10 inserted is marked with the circle. E, EcoRI; RV, EcoRV; C, Csp45I; Cl, ClaI.
Mobilization of the plasmids and complementation.
pG52 was transformed into SB221 as previously described (22). The plasmids pBK10, pBK12, pJV9, pJV10, and pJV12 were electroporated into S17-1 in disposable microelectroporation chambers (Life Technologies) as described in the manufacturer's procedure. The plasmids pJV9, pJV10, and pJV12 were conjugated into ANV2.
Nucleotide sequence accession number.
The sequences reported herein have been deposited in the GenBank database under accession no. AF191556.
RESULTS
Transposon mutagenesis.
The genetic mechanism by which secondary cells of X. nematophilus are formed is not known. To determine whether inactivation of a single gene could produce the pleiotropic phenotype associated with the secondary cell, a transposon mutagenesis approach was taken. The strain AN6, which is equivalent to the type strain ATCC 19061 (Table 1), was mutagenized by using a mini-Tn10 carrying a kanamycin resistance gene. Exconjugants were selected for kanamycin resistance and simultaneously screened for lack of BTB binding. Out of 2,600 transposon mutant strains isolated, 50 lacked the ability to bind BTB. These strains were subsequently tested for numerous phenotypic properties associated with the secondary cell (Table 2). Eight mutant strains which displayed the secondary cell phenotype were isolated. Southern hybridization and restriction enzyme analysis showed that each of the eight strains contained a single transposon insertion and that the chromosomal position of the insertion was different in the individual strains.
TABLE 2.
Phenotypic characterization of ANV2
| Phenotypic trait | Primary | Variant type
|
|
|---|---|---|---|
| ANV2 | Secondarya | ||
| BTB absorption | + | − | − |
| Antibiotic production | + | − | − |
| Crystal protein production | + | − | − |
| Lecithinase | − | + | + |
| OpnB | + | − | − |
| OpnS | + | +++ | + |
| Swimming motility | + | + | + |
| Swarming motility | + | − | − |
| Fimbriae | + | −b | −b |
| Hemagglutination | + | − | ± |
| Hemolysis | + | − | − |
| Cell shape in stationary phase | Short rods | Long rods | Short rods |
19061/II.
Fimbriae were dramatically reduced but not absent on the variant cells.
During the course of this study, we found that three of the eight mutant strains did not revert during storage at −80°C or during storage on agar plates. In contrast, several phenotypic characteristics reverted to the wild-type state in the remaining five strains. The phenotypes of the three stable strains, ANV1, ANV2, and ANV4, were similar but not identical. They differed in the amount of lecithinase, lipase, and the stationary-phase-induced outer membrane protein (OpnS) produced. ANV1 and ANV4 produced higher levels of lecithinase (Table 3) and lipase (data not shown) than ANV2. In addition, OpnS production in ANV2 (Fig. 1, lane 2) and ANV1 (data not shown) was elevated while ANV4 (data not shown) produced wild-type levels of this protein. Finally, ANV2 and ANV4 retained a long rod shape under stationary-phase conditions while ANV1 exhibited a shorter rod shape (Table 3). ANV2 was chosen for further study.
TABLE 3.
Variable traits of the secondary and Tn10 variant forms of X. nematophilus
| Phenotypic trait | Secondary variants
|
Transposon variants
|
||||||
|---|---|---|---|---|---|---|---|---|
| AN6 | 19061 | F1 | A24 | NC116 | ANV2 | ANV1 | ANV4 | |
| Lecithinase | + | + | − | − | − | + | ++ | ++ |
| Swimming motility | − | + | − | − | − | + | + | + |
| Pathogenicity | − | + | + | NAa | NA | + | + | + |
| Shape in stationary phase | Short, thin, rods | Short rods | Short rods | NA | NA | Long rods | Short rods | Long rods |
NA, not available.
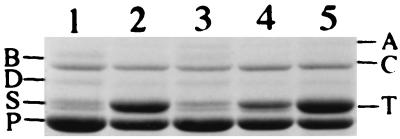
FIG. 1.
Outer membrane proteins of strains grown under stationary-phase conditions in LB. Lane 1, AN6; lane 2, ANV2; lane 3, ANV2 with pJV9; lane 4, ANV2 with pJV12; lane 5, ANV2 with pJV10.
In vitro characterization of ANV2.
The growth rate of ANV2 at 30°C in different growth media was identical to that of the parent cell. ANV2 did not produce antibiotics, crystal proteins, or OpnB (Table 2). These products have been previously shown to be induced during the post-exponential-growth phase (23, 24, 25, 42). ANV2 also possessed dramatically reduced levels of fimbriae, was unable to stimulate hemagglutination, lacked hemolytic activity, and did not swarm on agar plates. In contrast, ANV2 was as motile in broth culture as the parent strain and continued to be motile in stationary phase, while motility of the parent was reduced. In this regard, ANV2 and the secondary form of 19061 (19061/II) differed from secondary cells characterized previously in that the latter were shown to be nonmotile in liquid culture (Table 3).
Certain products were made at higher levels in ANV2 than in the parent strain. The production of lecithinase was increased in ANV2 and was not detectable in the parent cells (Table 2). In other strains of X. nematophilus, lecithinase activity was detected in the parent strain and was reduced in the secondary strains (Table 3). ANV2 also produced a higher level of OpnS (Fig. 1, lane 2) than did the parent strain (lane 1). Overall, the phenotype of ANV2 closely resembled that of 19061/II (Table 2). However, ANV2 produced more OpnS and retained its long rod shape under stationary-phase conditions while 19061/II became shorter in stationary phase.
In vivo characterization of ANV2.
To assess the pathogenic properties of ANV2, approximately 300 bacterial cells were injected into either fourth- or fifth-instar M. sexta. The LT50s of the injected animals were determined. The LT50 was 24 h when the bacteria were injected into fourth-instar caterpillars and was 27 h when cells were injected into fifth-instar M. sexta. These values were identical to those found for the parent strain. These findings indicated that ANV2 was fully pathogenic towards the larval insect host and that the properties missing in the variant strain were not essential for virulence.
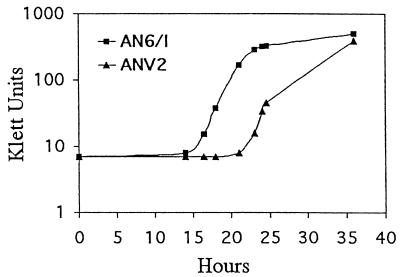
We next addressed the question of whether ANV2 was defective in its interaction with the nematode host. It was previously shown that S. carpocapsae was able to mature and reproduce on secondary cells of X. nematophilus (18, 46). The ability of the nematode to grow on, retain, and subsequently release the bacteria was evaluated as described in Materials and Methods. Figure 2 shows a representative experiment. The release and subsequent growth of AN6 were initially detected 14.7 h after inoculation. In contrast, the initial appearance of ANV2 in the culture did not occur until 20.7 h after inoculation. These findings suggest that ANV2 was defective in some aspect of its survival within, or in its release from, the nematode.
FIG. 2.
Nematode-bacteria assay. Five hundred surface-sterilized dauer juvenile nematodes were inoculated into 5 ml of LB, and the bacterial growth was monitored.
Cloning and sequence analysis of var1 and orf2.
Since a single Tn10 insertion was found in ANV2, it was possible that inactivation of a single gene was responsible for the pleiotropic phenotype of this variant cell. To confirm this possibility, the location of the Tn10 insertion in ANV2 was determined. A 2.1-kb fragment containing the upstream sequence flanking the transposon was cloned and sequenced. A partial ORF that encoded 279 amino acid residues was found to share 43% identity with the ybhE gene of E. coli. The function of ybhE in E. coli is not known. The final nine codons of the ybhE-like gene of X. nematophilus are shown in Fig. 3. The transposon was found to be inserted 320 base pairs downstream of the stop codon of the ybhE-like gene. An ORF encoding 84 residues to the point of the transposon insertion was identified. This ORF was named var1 since its inactivation appeared to be involved in the formation of the variant type cell. To obtain the entire nucleotide sequence of var1, a var1-containing clone was obtained from the chromosomal library of the wild type. Sequence analysis of the DNA region containing var1 is shown in Fig. 3. The var1 gene encodes a protein consisting of 121 amino acid residues. The transposon had inserted into the codon for Ala84. A consensus ribosome binding site was found 10 bp upstream of the predicted start codon. Var1 showed no sequence similarity to any protein in the GenBank database.
FIG. 3.
Nucleotide sequences of var1 and orf2 and their deduced amino acid sequences. The putative ribosome binding sites are underlined. The putative promoter consensus sequences are double underlined. The site of transposon insertion is marked with an arrowhead. Restriction sites used for complementation analysis are indicated with small letters and are labeled above the sequence.
The ORF located downstream of var1 was named orf2. The orf2 gene encoded a protein of 192 amino acid residues and showed no sequence similarity to any known gene. The orf2 gene, which is transcribed in the opposite direction of var1, contained an identifiable ribosome binding site and consensus −10 and −35 ς70 promoter sequences. The nucleotide sequence of the region upstream of orf2 was also determined (Fig. 4). Three genes which shared amino acid identity with the E. coli genes bioA, bioB, and bioF were identified. The level of sequence identity was 72, 69, and 58%, for bioA, bioB, and bioF, respectively. These genes are involved in biotin biosynthesis in E. coli and other enteric bacteria. The level of identity for bioA was based on the complete sequence of this gene while that for bioB and bioF was based on a partial sequence. This sequence analysis revealed that in X. nematophilus the six ybh genes found in this region of the E. coli chromosome have been replaced with the var1-orf2 sequence in X. nematophilus. Interestingly, the guanine-plus-cytosine content of var1 and orf2 was 41 and 36%, respectively, while the average guanine-plus-cytosine content of the X. nematophilus genes so far sequenced, including those presented in this study, is 46%.
Complementation of in vitro phenotypes.
The above results suggested that the variant cell phenotype of ANV2 was caused by inactivation of var1. Because orf2 was transcribed in the opposite direction, it was not likely that the Tn10 insertion had a polar effect on this gene. To ensure that it was inactivation of var1 alone that produced the ANV2 phenotype, complementation experiments were carried out. The plasmid pJV9, carrying a 5.3-kb fragment containing var1, orf2, and the bio genes, was constructed. Introduction of pJV9 into ANV2 completely complemented the variant type cell (Table 4) and restored the OpnS production to wild-type levels (Fig. 1, lane 3). These results supported the idea that var1 function was required for the expression of the numerous traits that were altered in ANV2. To further analyze whether var1 alone was sufficient for the complementation of ANV2, the DNA sequence spanning the upstream regulatory region and the first 10 amino acid residues of orf2 was deleted, generating pJV12. This plasmid contained the var1 and bio genes but lacked the orf2 gene. ANV2 containing pJV12 was almost completely complemented (Table 4). Thus, var1 function alone was sufficient to restore almost all of the phenotypic traits altered in ANV2. Interestingly, pJV12 only partially restored the production of OpnB and OpnS to wild-type levels (Fig. 1, lane 4), suggesting that orf2 may play a role in the production and/or processing of these stationary-phase outer membrane proteins. To be certain that the bio genes were not involved in the complementation of ANV2, the plasmid pJV10, which lacked var1 and orf2 but retained the bio genes, was constructed. As expected, pJV10 was unable to complement ANV2 (Table 4 and Fig. 1, lane 5). These results confirm that the var1 gene itself is required for the production of the numerous phenotypic traits altered in ANV2 and that the inactivation of this gene resulted in the formation of the variant type cell.
TABLE 4.
Complementation analysis of ANV2
| Phenotypic trait | Plasmids
|
||
|---|---|---|---|
| pJV9 (var1+ orf2+) | pJV12 (var1+ orf2 mutant) | pJV10 (var1 mutant orf2 mutant) | |
| BTB absorption | + | + | − |
| Antibiotic production | + | + | − |
| Crystal protein production | + | + | − |
| Lecithinase | − | − | + |
| OpnB | + | ± | − |
| OpnS | + | ++ | +++ |
| Swimming motility | + | + | ++ |
| Hemolysis | + | + | − |
| Cell shape in stationary phase | Short rods | Short rods | Long rods |
To address the question whether var1 was inactivated in other variant cells, pJV9 was conjugated into the secondary cells of both 19061 and AN6. These strains were not complemented by pJV9, indicating that genes other than var1 were altered in these variant cells. Finally, while restriction analysis suggested that the transposon had inserted into different chromosomal locations in ANV1, ANV2, and ANV4, it was possible that the transposon had actually inserted into different positions of var1. This was not the case, since pJV9 did not complement ANV1 and ANV4, indicating that genes other than var1 were affected in these variant strains.
Complementation of in vivo phenotypes.
Since plasmids containing var1 were able to complement ANV2 in vitro, we addressed whether the defect in its interaction with, or survival within, the nematode could be complemented. Dauer juveniles grown on ANV2 carrying either pJV9, pJV10, or pJV12 were surface sterilized and were inoculated in LB. The initial time of bacterial growth was monitored as shown in Table 5. The initial growth of ANV2 was previously shown to be delayed for 6 h relative to that of AN6 (Fig. 2 and Table 5). When ANV2 carried plasmids containing var1 (pJV9 or pJV12), the time of initial bacterial growth was comparable to that of AN6, indicating that var1 was able to correct the defect in the interaction with the nematode. In contrast, this defect was not corrected in ANV2 carrying pJV10, further supporting the conclusion that var1 was required for normal interaction with, or survival within the nematode.
TABLE 5.
Initial growth of bacteria after inoculating dauer juveniles into LBa
| Bacterial strain | Time of initial growth (h) ± SE |
|---|---|
| AN6/I | 14.7 ± 0.4 |
| ANV2 | 20.7 ± 0.9 |
| ANV2/pJV9b | 15.6 ± 0.2 |
| ANV2/pJV10b | 20.4 ± 0.5 |
| ANV2/pJV12b | 15.8 ± 0.2 |
Five hundred dauer juveniles were used in the experiments.
LB containing 10 μg of gentamicin per ml was used.
The question of whether a delay in the initial time of bacterial growth would be reflected in a delay in killing of the insect host during natural infection was next addressed. Fourth-instar M. sexta larvae were naturally infected with nematodes carrying the different bacterial strains described above. As shown in Table 6, the LT50 for nematodes containing ANV2/pJV10 was 45.8 h, which was 7 to 8 h longer than that for nematodes carrying the parent strain. In contrast, the LT50s of ANV2 carrying either pJV9 or pJV12 were comparable to that of AN6. These findings indicated that the delay in the initial growth of ANV2 correlated with a delay in the killing of the insect when the bacteria were introduced via the nematode. When ANV2 contained var1-bearing plasmids, both the delay in initial growth and the delay in insect killing were corrected.
TABLE 6.
LT50s from natural infection of fourth-instar M. sexta larvae
| Bacterial strain | LT50 (h) ± SE |
|---|---|
| AN6/I/pJQ200KS | 38.0 ± 0.8 |
| ANV2/pJV9 | 37.8 ± 0.5 |
| ANV2/pJV10 | 45.8 ± 0.7 |
| ANV2/pJV12 | 38.0 ± 1.0 |
DISCUSSION
During prolonged culturing of Xenorhabdus spp., secondary cells that lack numerous primary-specific traits arise spontaneously at an unpredictable frequency. Secondary variants have not been isolated during exponential growth. At least eight diverse primary-specific characteristics are consistently affected in the secondary cells of all strains of X. nematophilus studied to date (Table 2). Other characteristics, such as lecithinase production, swimming motility, and cell shape, were more variable. The mechanism underlying secondary cell formation is not known. We show that disruption of a single gene, var1, produced a secondary cell that was very similar to the spontaneously formed 19061/II strain. This result suggests that spontaneous formation of the secondary variant cell could result from a mutation in a single gene. This event appears to occur under prolonged culturing conditions but not during exponential growth. Our results show that var1 is not the only gene involved in secondary cell formation since plasmids containing the var1 gene did not complement the 19061/II, AN6/II, ANV1, and ANV4 strains. Furthermore, Tn10 insertions in the ANV1, ANV2, and ANV4 strains were located in different chromosomal positions. Thus, the expression of primary-specific traits is complex and may involve multiple levels of regulation.
The fact that a single transposon insertion eliminated the production of the primary-specific traits, and that these traits are also affected in the secondary variants of all strains of X. nematophilus examined, suggests that the primary-specific genes are coordinately regulated. Since none of the primary-specific genes have been cloned, we can only speculate about how inactivation of var1 affects the production of the primary-specific genes. It is conceivable that the primary-specific genes are coordinately regulated at the level of transcription. Although BLAST search analysis indicated that the Var1 protein was not similar to any known regulatory protein, it may function as a small DNA binding protein, such as integration host factor, that globally regulates gene expression. Primary-specific genes may require this type of regulatory protein for their expression. It is also possible that the production of individual gene products is controlled posttranscriptionally or at the level of protein processing and/or protein secretion. For example, Var1 may function as a specific chaperone that is necessary for the proper folding or secretion of primary-specific products. The pleiotropic phenotype of ANV2 may also be due to an alteration in protein transport and secretion pathways or altered membrane properties. The primary-specific products may be secreted via the specific transport system which would be absent or nonfunctional in secondary cells. It is conceivable that OpnB participates in these transport processes. Since crystal protein production and stationary-phase cell shape were also altered in ANV2, it appears that var1 affects other cellular functions as well. Understanding the level of regulation of the individual primary-specific traits should help to elucidate whether, and how, these genes are coordinately regulated.
ANV2 and ANV4 were longer rods under stationary-phase conditions while the parent strain bacteria were shorter rods under these conditions. E. coli cells also become short rods under stationary-phase conditions. The alternative sigma factor, rpoS, is involved in controlling genes that regulate this stationary-phase cell shape phenotype (39). It remains to be determined whether var1 affects rpoS function in X. nematophilus.
While the primary-specific properties are thought to be important for the growth of the nematode in the insect cadaver, it was not known whether they were directly involved in the interaction between the bacterium and the nematode. We show that when nematodes carrying either ANV2 or 19061/II were placed in broth culture, initial bacterial growth was delayed relative to that seen with the parent strain (Fig. 2). The LT50 for natural infection with ANV2-bearing nematodes was also increased (Table 5). The delay in initial bacterial growth may indicate that the ANV2 and secondary strains survive less well in the nematode than does the parent strain. Fewer bacterial cells in the intestinal sac would result in a longer lag phase for initial bacterial growth. One intriguing possibility is that the crystal proteins play a role in survival of the bacterium in the nematode. Another possible explanation for the delay in initial growth is that X. nematophilus may require an active process such as swarming motility to leave the nematode. Since ANV2 and secondary cells do not swarm, they may leave the nematode via a slower, passive process. To elucidate the role of primary-specific traits in the interaction between the nematode and the bacteria, it will be necessary to create mutant strains in which individual primary-specific genes are inactivated.
Secondary (phase II) variants have also been isolated during prolonged culturing of Photorhabdus spp. (1, 3, 24). The secondary variants of Photorhabdus are pathogenic but do not support growth of the Heterorhabditidae nematodes in vitro (1, 8). It was recently shown that an inactivation of either one of the two different crystal genes, cipA and cipB, resulted in a pleiotropic phenotype that resembled secondary cells (7). The cipA mutant and cipB mutant strains grew normally and were fully pathogenic, but did not support growth of nematodes in vitro. Taken together, these findings support the idea that primary-specific traits are involved in the symbiotic interaction between the bacteria and the nematode. Several primary-specific products, such as antibiotic and crystal protein production, are shared by both Xenorhabdus and Photorhabdus. However, the biochemical properties of these products are very different. The antibiotics produced by Xenorhabdus are either indole derivatives or belong to the xenocoumacin or xenorhabdin family of compounds, while the antibiotics of Photorhabdus are hydroxystilbenes (2, 24, 42). The biochemical properties of the crystal proteins of the respective bacteria are also very different (7, 14). It is therefore probable that the genes responsible for these phenotypic traits were acquired laterally and have been retained to participate in the symbiotic function of the bacterium. The acquired genes presumably came under the control of a preexisting regulatory network in the cell.
While primary-specific traits were altered in all secondary cells so far studied, several traits, such as lecithinase and OpnS production, stationary-phase cell shape, and swimming motility, varied among the various strains. For example, ANV2 overproduced OpnS and grew as long rods during stationary phase, whereas OpnS was not overproduced in 19061/II and cells were short rods under stationary-phase conditions. There were also significant differences among the secondary cells of different strains of X. nematophilus. 19061/II was able to swim and produced lecithinase while the secondary cells of other strains (Table 3) were nonmotile and lacked lecithinase activity. These findings suggest that the regulation of the primary-specific and variable traits is complex. Identifying the genes that have been altered in the various secondary and ANV strains would help to elucidate the genetic network involved in the formation of secondary cells of X. nematophilus. With this information, it may be possible to understand the role of primary-specific and variable traits in the symbiotic interaction between the bacterium and the nematode.
ACKNOWLEDGMENTS
We thank H. Owen for her assistance in the electron microscopy analysis performed in this study and J. Witten for kindly providing M. sexta larvae. We are also grateful to B. Jester for helping with figure design and editing the manuscript. We thank B. Boylan and A. Givaudan for their critical reading of the manuscript.
REFERENCES
- 1.Akhurst R J. Morphological and functional dimorphisms in Xenorhabdus spp., bacteria symbiotically associated with the insect pathogenic nematodes Neoaplectana and Heterorhabditis. J Gen Microbiol. 1980;121:303–309. [Google Scholar]
- 2.Akhurst R J. Antibiotic activity of Xenorhabdus spp., bacteria symbiotically associated with insect pathogenic nematodes of the families Heterorhabditidae and Steinernematidae. J Gen Microbiol. 1982;128:3061–3065. doi: 10.1099/00221287-128-12-3061. [DOI] [PubMed] [Google Scholar]
- 3.Akhurst R J, Boemare N. Biology and taxonomy of Xenorhabdus. In: Gaugler R, Kaya H K, editors. Entomopathogenic nematodes in biological control. Boca Raton, Fla: CRC Press; 1990. pp. 75–90. [Google Scholar]
- 4.Akhurst R J, Dunphy G B. Tripartite interactions between symbiotically associated entomopathogenic bacteria, nematodes, and their insect hosts. In: Beckage N, Thompson S, Federici B, editors. Parasites and pathogens of insects. Vol. 2. New York, N.Y: Academic Press; 1993. pp. 1–23. [Google Scholar]
- 5.Akhurst R J, Smigielski A J, Mari J, Boemare N, Mourant R G. Restriction analysis of phase variation in Xenorhabdus spp. (Enterobacteriaceae), entomopathogenic bacteria associated with nematodes. Syst Appl Microbiol. 1992;15:469–473. [Google Scholar]
- 6.Ausubel F M, Brent R, Kingston R E, Moore D D, Smith J A, Seidman J G, Struhl K, editors. Current protocols in molecular biology. Harvard Medical School, Massachusetts General Hospital. New York, N.Y: Wiley Interscience; 1987. Preparation of genomic DNA from bacteria; pp. 2.4.1–2.4.2. [Google Scholar]
- 7.Bintrim S B, Ensign J C. Insertional inactivation of genes encoding the crystalline inclusion proteins of Photorhabdus luminescens results in mutants with pleiotropic phenotypes. J Bacteriol. 1998;180:1261–1269. doi: 10.1128/jb.180.5.1261-1269.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Boemare N E, Akhurst R J. Biochemical and physiological characterization of colony form variants in Xenorhabdus spp. (Enterobacteriaceae) J Gen Microbiol. 1988;134:751–761. doi: 10.1099/00221287-134-7-1835. [DOI] [PubMed] [Google Scholar]
- 9.Boemare N E, Akhurst R J. Physiology of phase variation in Xenorhabdus nematophilus. In: Cooper D J, Drummond J, Pinnock D E, editors. Proceedings of the 5th International Colloquium on Invertebrate Pathology and Microbial Control. 1990. pp. 208–212. Adelaide, Australia. [Google Scholar]
- 10.Boemare N, Akhurst R J, Mourant R G. DNA relatedness between Xenorhabdus spp. (Enterobacteriaceae), symbiotic bacteria of entomopathogenic nematodes, and a proposal to transfer Xenorhabdus luminescens to a new genus, Photorhabdus gen. nov. Int J Syst Bacteriol. 1993;43:249–255. [Google Scholar]
- 11.Boemare N, Givaudan A, Brehelin M, Laumond C. Symbiosis and pathogenicity of nematode-bacterium complexes. Symbiosis. 1997;22:21–45. [Google Scholar]
- 12.Boemare N, Thaler J-O, Lanois A. Simple bacteriological tests for phenotypic characterization of Xenorhabdus and Photorhabdus phase variants. Symbiosis. 1997;22:167–175. [Google Scholar]
- 13.Brehelin M A, Cherqui L, Drif L, Luciani L, Akhurst R J, Boemare N E. Ultrastructure study of surface components of Xenorhabdus sp. in different cell phases and culture conditions. J Invertebr Pathol. 1993;61:188–191. [Google Scholar]
- 14.Couche G A, Gregson R P. Protein inclusions produced by the entomopathogenic bacterium Xenorhabdus nematophilus subsp. nematophilus. J Bacteriol. 1987;169:5279–5288. doi: 10.1128/jb.169.11.5279-5288.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.de Lorenzo V, Timmis K N. Analysis and construction of stable phenotypes in Gram-negative bacteria with Tn5 and Tn10-derived mini transposons. Methods Enzymol. 1994;235:386–405. doi: 10.1016/0076-6879(94)35157-0. [DOI] [PubMed] [Google Scholar]
- 16.Dowds B C A. Photorhabdus and Xenorhabdus—gene structure and expression, and genetic manipulation. Symbiosis. 1997;22:67–83. [Google Scholar]
- 17.Dunphy G B, Webster J M. Interaction of Xenorhabdus nematophilus subsp. nematophilus with the haemolymph of Galleria mellonella. Int J Parasitol. 1988;30:883–889. [Google Scholar]
- 18.Ehlers R-U, Stoessel S, Wyss U. The influence of phase variants of Xenorhabdus spp. and Escherichia coli (Enterobacteriaceae) on the propagation of entomopathogenic nematodes of the genera Steinernema and Heterorhabditis. Rev Nematol. 1990;13:417–424. [Google Scholar]
- 19.Fodor A. Genetic analysis of Photorhabdus and Xenorhabdus. In: Boemare N, Ehlers R-U, Fodor A, Szentirmai A, editors. Symbiosis and pathogenicity of nematode-bacteria complexes. COST 819 entomopathogenic nematodes report EUR 167727 EN, ECSC-EC-EAEC. Brussels, Belgium: European Commission; 1996. pp. 93–108. [Google Scholar]
- 20.Fodor A, Vecseri G, Farkas T. Caenorhabditis elegans as a model for the study of entomopathogenic nematodes. In: Gaugler R, Kaya H, editors. Entomopathogenic nematodes in biological control. Boca Raton, Fla: CRC Press; 1990. pp. 249–284. [Google Scholar]
- 21.Fodor E, Szallas E, Kiss Z, Fodor A, Horvath L I, Chitwood D J, Farkas T. Composition and biophysical properties of lipids in Xenorhabdus nematophilus and Photorhabdus luminescens, symbiotic bacteria associated with entomopathogenic nematodes. Appl Environ Microbiol. 1997;63:2826–2831. doi: 10.1128/aem.63.7.2826-2831.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Forst S, Comeau D, Norioka S, Inouye M. Localization and membrane topology of EnvZ, a protein involved in osmoregulation of OmpF and OmpC in Escherichia coli. J Biol Chem. 1987;262:16433–16438. [PubMed] [Google Scholar]
- 23.Forst S, Waukau J, Leisman G, Exner M, Hancock R. Functional and regulatory analysis of the OmpF-like porin, OpnP, of the symbiotic bacterium Xenorhabdus nematophilus. Mol Microbiol. 1995;18:779–789. doi: 10.1111/j.1365-2958.1995.mmi_18040779.x. [DOI] [PubMed] [Google Scholar]
- 24.Forst S, Nealson K H. Molecular biology of the symbiotic-pathogenic bacteria Xenorhabdus spp. and Photorhabdus spp. Microbiol Rev. 1996;60:21–43. doi: 10.1128/mr.60.1.21-43.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Forst S, Dowds B, Boemare N, Stackebrandt E. Xenorhabdus spp. and Photorhabdus spp.: bugs that kill bugs. Annu Rev Microbiol. 1997;51:47–72. doi: 10.1146/annurev.micro.51.1.47. [DOI] [PubMed] [Google Scholar]
- 26.Forst S, Tabatabai N. Role of the histidine kinase, EnvZ, in the production of outer membrane proteins in the symbiotic-pathogenic bacterium, Xenorhabdus nematophilus. Appl Environ Microbiol. 1997;63:962–968. doi: 10.1128/aem.63.3.962-968.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Givaudan A, Baghdiguian S, Lanois A, Boemare N E. Swarming and swimming changes concomitant with phase variation in Xenorhabdus nematophilus. Appl Environ Microbiol. 1995;61:1408–1413. doi: 10.1128/aem.61.4.1408-1413.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Givaudan A, Lanois A, Boemare N E. Cloning and nucleotide sequence of a flagellin encoding genetic locus from Xenorhabdus nematophilus: phase variation leads to differential transcription of two flagellar genes (fliCD) Gene. 1996;183:243–253. doi: 10.1016/s0378-1119(96)00452-0. [DOI] [PubMed] [Google Scholar]
- 29.Herrero M, de Lorenzo V, Timmis K N. Transposon vectors containing non-antibiotic resistance selection markers for cloning and stable chromosomal insertion of foreign genes in gram-negative bacteria. J Bacteriol. 1990;172:6557–6567. doi: 10.1128/jb.172.11.6557-6567.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hosseini P K, Nealson K H. Symbiotic luminous soil bacteria: unusual regulation for an unusual niche. Photochem Photobiol. 1995;62:633–640. [Google Scholar]
- 31.Hurlbert R E. Investigations into the pathogenic mechanisms of the bacterium-nematode complex: the search for virulence determinants of Xenorhabdus nematophilus ATTC 19061 could lead to agriculturally useful products. ASM News. 1994;60:473–489. [Google Scholar]
- 32.Krasomil-Osterfeld K C. Influence of osmolarity on phase shift in Photorhabdus luminescens. Appl Environ Microbiol. 1995;61:3748–3749. doi: 10.1128/aem.61.10.3748-3749.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Leclerc M-C, Boemare N E. Plasmids and phase variation in Xenorhabdus spp. Appl Environ Microbiol. 1991;57:2597–2601. doi: 10.1128/aem.57.9.2597-2601.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Leisman G B, Waukau J, Forst S A. Characterization and environmental regulation of outer membrane proteins in Xenorhabdus nematophilus. Appl Environ Microbiol. 1995;61:200–204. doi: 10.1128/aem.61.1.200-204.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Nealson K H, Smith T M, Bleakley B. Biochemistry and physiology of Xenorhabdus. In: Gaugler R, Kaya H K, editors. Entomopathogenic nematodes in biological control. Boca Raton, Fla: CRC Press; 1990. pp. 271–284. [Google Scholar]
- 36.Poinar G O., Jr . Biology and taxonomy of Steinernematidae and Heterorhabditidae and physiology of Xenorhabdus. In: Gaugler R, Kaya H K, editors. Entomopathogenic nematodes in biological control. Boca Raton, Fla: CRC Press; 1990. pp. 271–284. [Google Scholar]
- 37.Quandt J, Hynes M F. Versatile suicide vectors which allow direct selection for gene replacement in Gram-negative bacteria. Gene. 1993;127:15–21. doi: 10.1016/0378-1119(93)90611-6. [DOI] [PubMed] [Google Scholar]
- 38.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1989. [Google Scholar]
- 39.Siegele D A, Kolter R. Life after log. J Bacteriol. 1992;174:345–348. doi: 10.1128/jb.174.2.345-348.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Simon R, Priefer U, Puhler A. A broad range host mobilization system for in vivo genetic engineering: transposon mutagenesis in gram negative bacteria. Bio/Technology. 1983;1:784–791. [Google Scholar]
- 41.Smigielski A, Akhurst R J, Boemare N E. Phase variation in Xenorhabdus nematophilus and Photorhabdus luminescens: differences in respiratory activity and membrane energization. Appl Environ Microbiol. 1994;60:120–125. doi: 10.1128/aem.60.1.120-125.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sundar L, Chang F N. Antimicrobial activity and biosynthesis of indole antibiotics produced by Xenorhabdus nematophilus. J Gen Microbiol. 1993;139:3139–3148. doi: 10.1099/00221287-139-12-3139. [DOI] [PubMed] [Google Scholar]
- 43.Szallas E, Koch C, Fodor A, Burghardt J, Buss O, Szentirmai A, Nealson K H, Stackebrandt E. Phylogenetic evidence for the taxonomic heterogeneity of Photorhabdus luminescens. Int J Syst Bacteriol. 1997;47:402–407. doi: 10.1099/00207713-47-2-402. [DOI] [PubMed] [Google Scholar]
- 44.Tabatabai N, Forst S. Molecular analysis of the two-component genes, ompR and envZ, in the symbiotic bacterium, Xenorhabdus nematophilus. Mol Microbiol. 1995;17:643–652. doi: 10.1111/j.1365-2958.1995.mmi_17040643.x. [DOI] [PubMed] [Google Scholar]
- 45.Thaler J-O, Duvic B, Givaudan A, Boemare N. Isolation and entomotoxic properties of the Xenorhabdus nematophilus F1 lecithinase. Appl Environ Microbiol. 1998;64:2367–2373. doi: 10.1128/aem.64.7.2367-2373.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Volgyi A, Fodor A, Szentirmai A, Forst S. Phase variation in Xenorhabdus nematophilus. Appl Environ Microbiol. 1998;64:1188–1193. doi: 10.1128/aem.64.4.1188-1193.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wang H, Dowds B C A. Phase variation in Xenorhabdus luminescens: cloning and sequencing of the lipase gene and analysis of its expression in primary and secondary phases of the bacterium. J Bacteriol. 1993;175:1665–1673. doi: 10.1128/jb.175.6.1665-1673.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Yamanaka S, Hagiwara A, Nishimura Y, Tanabe H, Ishibashi N. Biochemical and physiological characteristics of Xenorhabdus species, symbiotically associated with entomopathogenic nematodes including Steinernema kushidai and their pathogenicity against Spodoptera litura (Lepidoptera: Noctuidae) Arch Microbiol. 1992;158:387–393. [Google Scholar]