Abstract
Background
Last two decades, many association studies on genetic variants and chronic obstructive pulmonary disease (COPD) risk have been published. But results from different studies are inconsistent. Therefore, we performed this article to systematically evaluate results from previous meta-analyses and genome-wide association studies (GWASs). Material and Methods. Firstly, we retrieved meta-analyses in PubMed, Embase, and China National Knowledge Infrastructure and GWASs in PubMed and GWAS catalog on or before April 7th, 2022. Then, data were extracted and screened. Finally, two main methods—Venice criteria and false-positive report probability test—were used to evaluate significant associations.
Results
As a result, eighty-eight meta-analyses and 5 GWASs were deemed eligible for inclusion. Fifty variants in 26 genes obtained from meta-analyses were significantly associated with COPD risk. Cumulative epidemiological evidence of an association was graded as strong for 10 variants in 8 genes (GSTM1, CHRNA, ADAM33, SP-D, TNF-α, VDBP, HMOX1, and HHIP), moderate for 6 variants in 5 genes (PI, GSTM1, ADAM33, TNF-α, and VDBP), and weak for 40 variants in 23 genes. Five variants in 4 genes showed convincing evidence of no association with COPD risk in meta-analyses. Additionally, 29 SNPs identified in GWASs were proved to be noteworthy based on the FPRP test.
Conclusion
In summary, more than half (52.38%) of genetic variants reported in previous meta-analyses showed no association with COPD risk. However, 13 variants in 9 genes had moderate to strong evidence for an association. This article can serve as a useful reference for further studies.
1. Background
Chronic obstructive pulmonary disease (COPD) is a chronic inflammatory disease with progressive limitation of airflow [1]. According to the latest WHO prediction that COPD will become the third leading cause of death worldwide by 2030, COPD remains a huge health threat and high burden on health care resources to human beings [2]. Smoking and air pollution are widely believed to be the main risk factors of COPD, but only 15–20% of smokers develop this pathology and many COPD cases cannot be explained by environmental risk factors alone [3, 4], suggesting that other factors such as genetic variations may contribute to the development of COPD as well [5]. Moreover, different clinical presentation and severity of COPD between different races and the familial clustering pattern can be seen in COPD cases, indicating substantial evidence of genetic variations to COPD [6, 7]. As early as 2009, Smolonska et al. [8] conducted meta-analyses about 20 polymorphisms in 12 genes and demonstrated that three polymorphisms in TGF-β1 (rs2241712, rs1982073, and rs6957) were significant with COPD risk in the “diverse populations.” Another three polymorphisms were reported significantly only in Asians (IL1RN rs2234663, TNF-α rs1800629, and GSTP1 rs1695). One year later, Castaldi et al. [9] added three more loci (GSTM1 null variant, TGF-β1 rs1800470, and SOD3 rs1799896) related to COPD risk. Follow-up studies reported some other polymorphisms that mainly focused on IL, MMP, ADRB2, CHRNA, ADAM33, VDBP, SP-A/B/D, and COX2.
Last two decades, a surge of studies investigating genetic polymorphisms and COPD risk have been published, including genome-wide association studies (GWASs), candidate-gene association studies, and meta-analyses. Covering the whole genome, GWASs detect millions of single nucleotide polymorphisms (SNPs) and are a powerful and efficient tool for identifying the association between genetic variants and complex diseases [10]. On the contrary, most of the candidate-gene association studies are underpowered to detect moderate-sized genetic effects. In addition, systematically integrating data from individual studies, meta-analyses have an advantage of developing a single conclusion with greater statistical power [11]. Despite the prominence and growing number of results from GWASs, candidate-gene association studies are still one of the main methods to identify common COPD susceptibility alleles. Some of these associations reported in candidate-gene association studies may be true associations; however, inconsistent results from different studies suggest the possibility of false-positive associations. Even results from meta-analyses cannot be replicated in follow-up studies, indicating that these associations lack robustness. Guidelines for assessing the cumulative epidemiological evidence of genetic associations, known as the Venice criteria, were proposed by the Human Genome Epidemiology Network (HuGENet) in 2008 [12]. These guidelines help evaluate gene-disease associations [13]. However, as far as we know, there is still no such integrative and updated evaluation for all the reported polymorphisms associated with COPD in current literature.
Therefore, we collected evidence from meta-analyses and GWASs to conduct an integrative assessment of the gene-COPD associations based on the Venice criteria and false-positive report probability (FPRP) test, thus providing a synopsis of our current understanding of the genetic basis of COPD risk.
2. Materials and Methods
2.1. Study Eligibility and Literature Search
A systematic strategy was used to identify all relevant publications. Firstly, we searched PubMed, Embase, and China National Knowledge Infrastructure (CNKI) using the terms “COPD or chronic obstructive pulmonary disease,” “genetic or genetic association or single nucleotide polymorphism or SNP or polymorphism or genotype or variant or mutation or susceptibility,” and “meta-analysis or systematic review or literature review” for meta-analyses. We searched PubMed using the terms “COPD or chronic obstructive pulmonary disease” and “GWAS or genome-wide association study” as well as checking the GWAS catalog for GWASs on or before April 7th, 2022. Secondly, after manually screening the title and abstract, all references cited in relevant studies were also reviewed to identify additional studies. Inclusion criteria for meta-analyses were as follows: data were published in a peer-reviewed journal; studies used a cross-sectional, case-control, or cohort design; the association was about the etiology of COPD; and necessary information was provided for the FPRP test or/and Venice criteria. GWASs were eligible for inclusion if they met the following criteria: the association was directly about the etiology of COPD; P value <5 × 10−8; and both discovery and replication phases were reported. We removed duplicated and unrelated articles by screening the title and abstract or reading the full-text if necessary. Literature search and studies screening were done by Liu and Ran together. Any disagreement was resolved by consensus.
2.2. Data Extraction
Two reviewers, Liu and Ran, extracted data separately and then exchanged and cross-checked. Any disagreement was resolved by consensus. We collected the following data from meta-analyses: PMID, gene name, genetic variant, first author, publication year, comparison model, ethnicity, OR and 95% CI, adjustment for smoking (Yes/No), I2, the number of studies, cases and controls, minor allele frequency (MAF) in controls, P value of Egger's test and P value of the Q test, and the number of test alleles or genotypes. We collected the following data from GWASs: PMID, gene name, genetic variant, first author, publication year, risk allele, ethnicity, OR and 95% CI, risk allele frequency, P value, and the number of cases and controls. Caucasian and Asian were the two major ethnicities reported. We defined ethnicity as “diverse populations” if a combination of two or more ethnicities were reported. We extracted significant results of subgroup analysis stratified by ethnicity. Cigarette smoking is the major environmental risk factor for COPD and may influence the distribution of genetic polymorphisms [14]; therefore, results after adjusted for smoking were extracted as well. Except several variants (e.g., PIMZ and TGF-β1 rs2241718), all other were investigated exclusively by one meta-analysis, most of the variants were widely investigated. We extracted data from them all, but only one meta-analysis was chosen for further assessment considering publication year, comparison model, sample size, between-study heterogeneity, and study design altogether. Because there was a considerable overlap among these studies, to avoid the variant nomenclature confusion from different articles, we used the uniform identifiers (“rs” number) of variants in the dbSNP. For the variants without any “rs” number, we used the common nomenclature (e.g., GSTT1 null/present and SERPINA3 Ala9Thr). Associations were considered statistically significant if the reported P value was less than 0.05 or if the 95% CI excluded 1.0. Most of the meta-analyses provided multiple results under different genetic models; in order to avoid selection bias, we used the allelic model as the unified model. When data under allelic model were not available, then other models were used. Statistical analyses were performed with Stata, version 12.0, using the original data provided in meta-analyses if there was no sufficient information for the Venice criteria.
2.3. Evaluation of Cumulative Evidence
For statistically significant associations from meta-analyses, we applied the Venice criteria to grade the credibility of cumulative epidemiological evidence. Three aspects were assigned to the Venice criteria: the amount of evidence, replication of association, and protection from bias. For each of the three aspects, three levels (A, B, or C) were assigned, based on which credibility was defined as strong, moderate, or weak. Amount of evidence was determined by the sum of test alleles or genotypes among cases and controls: A (n > 1000), B (100 < n < 1000), and C (n < 100). We did not apply this criterion for rare variants with frequency less than 1% since an A grade was unlikely to obtain [15]. The replication of association was determined depending on between-study heterogeneity: A (I2 < 25%), B (25% < I2 < 50%), and C (I2 > 50%). Protection from bias should take various potential sources of bias into consideration. A grade of A was assigned when there was no demonstrable bias or the bias would unlikely invalidate the association. A grade of B was assigned when there was no sufficient information for identifying evidence although there was no obvious bias. A grade of C was assigned when the bias was evident or/and was likely to explain the presence of association. Furthermore, if any of the following situations occurred, a grade of C was assigned: the sensitivity analysis indicated that the significant results can be substantially changed; the magnitude of the association was low (e.g., OR < 1.15 and OR > 0.87 for risk effect and protective effect, respectively) unless the association had been replicated prospectively by GWASs or several studies with no evidence of publication bias; and obvious publication bias (Egger's test P value <0.05). Cumulative epidemiological evidence was categorized as strong if all grades were A and weak if any grade was C. All of the rest combinations were categorized as moderate. If the data were insufficient, then associations were not evaluated (n = 6).
An approach developed by Wacholder et al. [16] was also used to calculate the FPRP for all the significant associations. The FPRP was determined by three parameters: the observed P value, the statistical power, and the prior probability. We used the prior probability of 0.01 and preset the FPRP noteworthiness value at 0.2 in our study. FPRP values and statistical power were calculated by the excel spreadsheet offered by Wacholder et al. We considered the association noteworthy if the calculated FPRP value was less than 0.2, indicating a true association. FPRP < 0.05, 0.05 < FPRP < 0.2, and FPRP > 0.2 were considered strong, moderate, and weak evidence of true association, respectively. We upgraded cumulative epidemiological evidence from moderate to strong, from weak to moderate, if evidence based on the FPRP was strong. We downgraded cumulative epidemiological evidence from strong to moderate, from moderate to weak, if evidence based on the FPRP was weak [17].
3. Results
3.1. Overall Characteristics
A total of 88 meta-analyses met the eligibility criteria (Figure 1), reporting 86 variants in 40 genes. One hundred and fourteen associations (66 significant associations and 48 nonsignificant associations) were addressed between these variants and the risk of COPD. Except 9 associations, results were obtained based on at least three original studies. The included meta-analyses had a mean of 1822 cases (range 120–10466) and 4848 controls (range 100–95336). Publication time ranged from 2004 to 2019, and most of them (n = 77, 92.8%) were published since 2010. More detailed information was presented in the tables.
Figure 1.
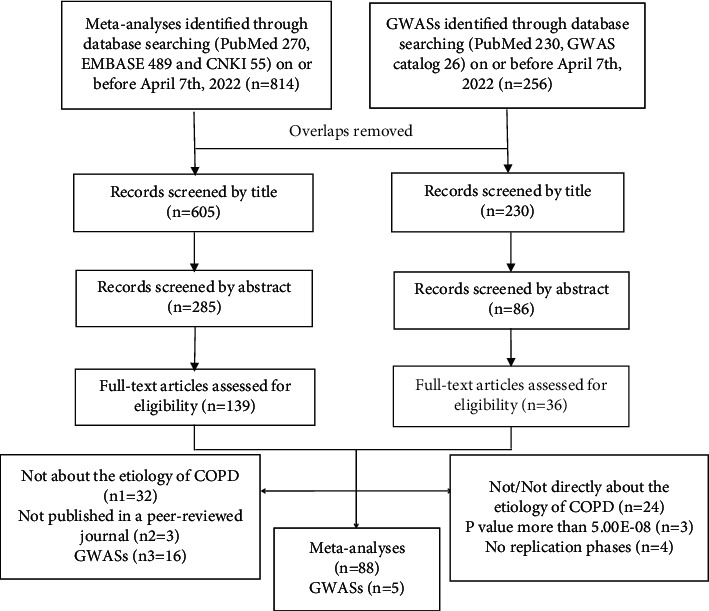
Selection of studies.
A total of 36 GWASs were included for full-text assessment (Figure 1), but most of them were excluded for not/not directly about the etiology of COPD. The P value of 3 GWASs did not meet the threshold of 5 × 10−8 [18–20], and in another 4 GWASs there was a lack of replication phases [21–24]. At last, 5 GWASs met the criteria for inclusion and reported 32 SNPs in or near 25 genes [25–29].
3.2. Significant Associations in Meta-Analyses
As mentioned above, 66 significant associations were identified, including 38 associations obtained from the main meta-analyses and 28 associations obtained from the subgroup meta-analyses [30–57]. These associations involved 50 variants in 26 genes. In the main meta-analyses, 43 variants reported ORs higher than 1.0 with a mean value of 1.61 (range 1.14–3.26, median 1.43), while protective effect was found for the other 7 variants with a mean value of 0.70 (range 0.51–0.86, median 0.76). In the subgroup meta-analyses, 16 variants reported ORs higher than 1.0 with a mean value of 1.66 (range 1.14–2.64, median 1.53), while protective effect was found for the other 7 variants, with a mean value of 0.65 (range 0.46–0.79, median 0.63). The average number of included studies in the main meta-analyses was 9, while the median was 6 (range 1–38). The average number of total sample size was 7818, while the median number was 4168 (range 220–49520). Most of the meta-analyses contained diverse ethnicities, while only three reported one specific ethnicity, including one for Caucasians and two for Asians.
The Venice criteria and FPRP test were used to evaluate the epidemiological credibility of the 66 significant associations (Table 1). Firstly, the Venice criteria was applied, as a result grade A was given to 36, 21, and 53 associations for amount of evidence, replication of association, and protection from bias, respectively. Grade B was given to 25, 14, and 2 associations for these three aspects, respectively. Grade C was given to 3, 30, and 5 associations for these three aspects, respectively. It is worth nothing that the amount of evidence was not applied for one variant, PISZ, because of its low frequency (0.12%). As a result, 10, 17, and 33 associations were categorized as strong, moderate, and weak, respectively. We did not evaluate the other 6 associations because of the insufficient information. Secondly, FPRP was applied to test the evidence of true association. As a result, 17, 7, and 42 associations were categorized as strong, moderate, and weak, respectively. Finally, we upgraded cumulative evidence from moderate to strong for GSTM1 null/present in Caucasians, CHRNA rs1051730 in non-Asians, ADAM33 rs612709, SP-D rs721917 in Asians, TNF-α rs1800629 in Asians, and HMOX1 L allele in Asians and from weak to moderate for GSTM1 null/present, GSTM1 null/present in Asians, and TNF-α rs1800629. We downgraded cumulative evidence from strong to moderate for PIMS, ADAM33 rs3918396, and ADAM33 rs3918396 after adjusted for smoking and from moderate to weak for IL-13 rs1800925 in Caucasians, ADAM33 rs2280091 and rs511898, CYP1A1 rs4646903, TNF-α rs1800630, VDBP 1S, COX2 rs20417, IREB2 rs2568494, and ADRB2 rs1042714 in Asians. Altogether, cumulative epidemiological evidence of 13 associations involved 10 variants in 8 genes were considered to be strong, including GSTM1 null/present in Caucasians, CHRNA rs16969968, rs8034191, and rs1051730, CHRNA rs1051730 in non-Asians, ADAM33 rs612709, ADAM33 rs612709 in Caucasians, SP-D rs721917, SP-D rs721917 in Asians, TNF-α rs1800629 in Asians, VDBP 1F, HHIP rs13118928, and HMOX1 L allele in Asians. Eight associations involved 6 variants in 5 genes (PI, GSTM1, ADAM33, TNF-α, and VDBP) and 39 associations involved 40 variants in 23 genes were considered to be moderate and weak evidence, respectively.
Table 1.
Statistically significant variants from meta-analyses, false-positive report probabilities (FPRP), and cumulative epidemiological evidence.
| Gene | Variants | Year | Comparison | Ethnicity | OR (95% CI) | Adjustment for smoking | Publication bias/heterogeneity∗∗ | I 2 (%) | No. of studies | Cases/control | Maf (%)† | Number of test alleles or genotypes | Venice criteria | Power OR of 1.5 | FPRP values at prior probability of 0.01 | Cumulative epidemiological evidence | Reference |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| PI | PIMZ | 2004 | PIMZ vs PIMM | Diverse | 2.31 (1.60–3.35) | No | 0.15/0.002 | 58.6∗ | 16 | 3653/12340 | 4.43 | 822 | BCC‡ | 0.011 | 0.081 | Weak | [30] |
| PIMZ vs PIMM | Diverse | 1.61 (0.92–2.81) | Yes | No/0.15 | NA | 5 | 1600/7645 | — | — | — | — | — | — | [30] | |||
| PI | PISZ | 2005 | PISZ vs PIMM | Diverse | 3.26 (1.24–8.57) | No | 0.98/0.23 | 27.2∗ | 6 | 2404/8999 | 0.12 | 42 | NBC‡ |
0.058 | 0.966 | Weak | [31] |
| PI | PIMS | 2005 | PIMS vs PIMM | Diverse | 1.19 (1.02–1.38) | No | 1.0/0.54 | 0.0∗ | 17 | 3630/13030 | 7.26 | 1169 | AAA | 0.999 | 0.679 | Moderate | [31] |
| PIMS vs PIMM | Diverse | 1.02 (0.81–1.28) | Yes | NA/NA | NA | 5 | 1526/7859 | — | — | — | — | — | — | [31] | |||
| GSTM1 | Null/present | 2018 | Null vs present | Diverse | 1.52 (1.31–1.77) | No | No/<0.00001 | 64.0 | 37 | 4669/4914 | 46.99 | 5018 | ACA | 0.432 | 0.000 | Moderate | [32] |
| Null vs present | Asian | 1.59 (1.29–1.96) | No | No/<0.0001 | 64.0 | 23 | 2374/2630 | 44.79 | 2531 | ACA | 0.293 | 0.005 | Moderate | [32] | |||
| Null vs present | Caucasian | 1.27 (1.11–1.44) | No | No/0.03 | 49.0 | 11 | 1881/1938 | 49.85 | 2045 | ABA | 0.995 | 0.019 | Strong | [32] | |||
| GSTT1 | Null/present | 2018 | Null vs present | Diverse | 1.28 (1.09–1.50) | No | No/0.001 | 51.0 | 29 | 3273/3578 | 32.36 | 2338 | ACA | 0.975 | 0.188 | Weak | [32] |
| Null vs present | Asian | 1.30 (1.05–1.61) | No | No/0.002 | 57.0 | 18 | 2025/2253 | 39.61 | 1697 | ACA | 0.905 | 0.639 | Weak | [32] | |||
| TGF-b1 | rs1800470 | 2011 | C vs T | Diverse | 0.82 (0.70–0.96) | No | 0.326/0.01 | 57.6 | 10 | 1507/2542 | 43.14 | 3533 | ACA | 0.995 | 0.575 | Weak | [33] |
| C vs T | Caucasian | 0.77 (0.60–0.98) | No | No/0.02 | 65.9 | 5 | 840/1788 | 41.11 | 2061 | ACA | 0.879 | 0.791 | Weak | [33] | |||
| TGF-β1 | rs1982073※ | 2017 | C vs T | Caucasian | 0.79 (0.64–0.99) | No | No/0.006 | 75.7 | 4 | 1656/2564 | 40.72 | 3348 | ACA | 0.930 | 0.812 | Weak | [34] |
| IL1RN | rs2234663 | 2019 | L vs 2 | Diverse | 0.56 (0.39–0.80) | No | 0.02/0.25 | 30.3 | 5 | 322/365 | 10.11 | 171 | BBC‡ | 0.169 | 0.458 | Weak | [35] |
| IL-6 | rs1800796 | 2019 | CC + GC vs GG | Diverse | 2.23 (1.46–3.40) | No | 0.05/0.38 | 19.3 | 3 | 574/862 | 26.33 | 511 | BAC‡ | 0.033 | 0.370 | Weak | [35] |
| IL-13 | rs1800925 | 2017 | C vs T | Diverse | 1.57 (1.21–2.04) | No | No/0.015 | 57.9 | 9 | 1477/891 | 16.27 | 963 | BCA | 0.366 | 0.166 | Weak | [36] |
| C vs T | Asian | 1.88 (1.23–2.87) | No | 0.268∗/0.016 | 67.1 | 5 | 555/544 | 14.71 | 425 | BCA | 0.148 | 0.698 | Weak | [36] | |||
| C vs T | Caucasian | 1.30 (1.01–1.67) | No | 0.644∗/0.314 | 15.6 | 4 | 922/347 | 18.73 | 538 | BAA | 0.869 | 0.820 | Weak | [36] | |||
| IL-27 | rs153109 | 2016 | G vs A | Asian | 0.51 (0.34–0.74) | No | NA/NA | NA | 1 | 120/100 | 45.50 | NA | XXX | 0.079 | 0.329 | NA | [37] |
| MMP-9 | rs3918242 | 2017 | T vs C | Diverse | 1.32 (1.02–1.71) | No | 0.12/<0.001 | 77.0 | 13 | 2295/2539 | 11.17 | 1408 | ACA | 0.833 | 0.808 | Weak | [38] |
| CT + TT vs CC | Asian | 1.80 (1.04–3.11) | No | No/<0.001 | 85.0 | 8 | 1101/1147 | 14.34 | 645 | BCA | 0.257 | 0.931 | Weak | [38] | |||
| MMP-12 | rs652438 | 2014 | G vs A | Diverse | 1.62 (1.08–2.42) | No | >0.05/0.070 | 57.5 | 4 | 710/1652 | 10.57 | 328 | BCA | 0.354 | 0.838 | Weak | [39] |
| CHRNA | rs16969968 | 2014 | C vs T | Diverse | 1.27 (1.17–1.39) | No | No/0.655 | 0.0 | 3 | 1996/6463 | 29.42 | 4979 | AAA | 1.000 | 0.000 | Strong | [40] |
| CHRNA | rs1051730 | 2014 | T vs C | Diverse | 1.14 (1.10–1.18) | No | No/0.318 | 14.0 | 9 | 10466/39054 | 30.80 | 30264 | AAA§ | 1.000 | 0.000 | Strong | [40] |
| T vs C | Non-Asian | 1.14 (1.10–1.18) | No | No/0.124 | 42.2 | 6 | 8620/37099 | 32.32 | 30088 | ABA§ | 1.000 | 0.000 | Strong | [40] | |||
| CHRNA | rs8034191 | 2014 | C vs T | Diverse | 1.29 (1.18–1.41) | No | No/0.644 | 0.0 | 5 | 2652/2565 | 24.09 | 2933 | AAA | 1.000 | 0.000 | Strong | [40] |
| CHRNA | rs6495309 | 2014 | C vs T | Diverse | 1.26 (1.09–1.45) | No | Pre/0.109 | 50.4 | 4 | 1977/2131 | 48.12 | 3756 | ACC | 0.993 | 0.112 | Weak | [40] |
| ADAM33 | rs3918396 | 2014 | GG + AG vs AA | Diverse | 1.42 (1.12–1.79) | No | 0.168/0.89 | 0.0 | 6 | 1422/3034 | 25.61 | 3806 | AAA | 0.679 | 0.304 | Moderate | [41] |
| GG + AG vs AA | Asian | 1.45 (1.14–1.85) | No | NA/0.665∗ | 0.0 | 3 | 642/630 | 67.38 | 645 | BAX | 0.607 | 0.313 | NA | [41] | |||
| GG + AG vs AA | Diverse | 1.45 (1.14–1.84) | Yes | No/0.91 | 0.0 | 3 | 836/848 | 54.07 | 1052 | AAA | 0.610 | 0.266 | Moderate | [41] | |||
| ADAM33 | rs597980 | 2014 | A vs G | Caucasian | 1.25 (1.05–1.48) | No | NA/0.158 | 49.9 | 2 | 296/2294 | 40.87 | 2148 | ABX | 0.983 | 0.492 | NA | [42] |
| ADAM33 | rs2280091 | 2014 | GG vs AA + AG | Diverse | 1.76 (1.27–2.43) | No | 0.483/0.348 | 10.5 | 8 | 2112/3952 | 13.18 | 197 | BAA | 0.166 | 0.262 | Weak | [42] |
| G vs A | Asian | 2.03 (1.40–2.94) | No | 0.857/0.006 | 76.0 | 4 | 1130/1135 | 13.04 | 798 | BCA | 0.055 | 0.245 | Weak | [42] | |||
| ADAM33 | rs511898 | 2019 | A vs G | Diverse | 1.16 (1.04–1.30) | No | 0.726/0.048 | 44.5 | 12 | 2454/4481 | 33.17 | 4763 | ABA | 1.000 | 0.514 | Weak | [51] |
| ADAM33 | rs612709 | 2014 | A vs G | Diverse | 0.60 (0.52–0.68) | No | 0.070/0.107 | 44.7 | 6 | 1341/3359 | 67.61 | 5345 | ABA | 0.050 | 0.000 | Strong | [42] |
| A vs G | Asian | 0.61 (0.42–0.89) | No | 0.256/0.021 | 74.1 | 3 | 767/763 | 26.54 | 664 | BCA | 0.322 | 0.760 | Weak | [42] | |||
| A vs G | Caucasian | 0.64 (0.53–0.77) | No | 0.324/0.798 | 0.0 | 3 | 574/2596 | 79.68 | 4681 | AAA | 0.333 | 0.001 | Strong | [42] | |||
| ADAM33 | rs2280090※ | 2014 | A vs G | Asian | 0.46 (0.33–0.66) | No | 0.581/0.021 | 74.1 | 3 | 767/763 | 32.31 | 763 | BCA | 0.022 | 0.101 | Weak | [42] |
| CYP1A1 | rs4646903 | 2015 | CC vs TT | Diverse | 1.73 (1.18–2.55) | No | No/0.34 | 11.0 | 5 | 532/591 | 23.43 | 141 | BAA | 0.236 | 0.703 | Weak | [43] |
| CC vs TT | Asian | 1.84 (1.11–3.06) | No | NA/0.65 | 0.0 | 2 | 158/154 | 35.27 | 88 | CAX | 0.216 | 0.896 | NA | [43] | |||
| CYP1A1 | rs1048943 | 2015 | GG vs AA | Diverse | 3.23 (1.50–6.93) | No | No/0.16 | 45.0 | 3 | 510/553 | 9.01 | 41 | CBA | 0.024 | 0.914 | Weak | [43] |
| SP-A | Common polymorphisms | 2013 | W vs M | Diverse | 1.53 (1.14–2.05) | No | >0.05/0.043 | 56.5 | 3 | 383/369 | 23.83 | 828 | BCA | 0.447 | 0.493 | Weak | [44] |
| SP-D | rs721917 | 2019 | T vs C | Diverse | 1.39 (1.18–1.65) | No | 0.375/0.901 | 0.0 | 5 | 561/550 | 45.36 | 1106 | AAA | 0.808 | 0.020 | Strong | [45] |
| T vs C | Asian | 1.44 (1.20–1.73) | No | 0.457/0.978 | 0.0 | 4 | 499/435 | 43.56 | 917 | BAA | 0.669 | 0.014 | Strong | [45] | |||
| SP-A/B/D | Common polymorphisms | 2013 | W vs M | Asian | 1.43 (1.15–1.78) | No | >0.05/0.001∗ | 71.3∗ | 4 | 485/488 | 27.28 | 1080 | ACA | 0.666 | 0.169 | Weak | [44] |
| TNF-α | rs1800629 | 2016 | A vs G | Diverse | 1.56 (1.29–1.89) | No | No/0.000 | 70.8 | 38 | 3951/5110 | 10.96 | 2326 | ACA | 0.344 | 0.002 | Moderate | [46] |
| A vs G | Asian | 2.40 (1.98–2.90) | No | No/0.093 | 29.8 | 22 | 2067/2167 | 7.63 | 1011 | ABA | 0.000 | 0.000 | Strong | [46] | |||
| A vs G | Diverse | 1.13 (0.95–1.35) | Yes | No/0.089 | 34.9 | 15 | 1500/1239 | — | — | — | — | — | — | [46] | |||
| A vs G | Asian | 1.26 (0.69–2.30) | Yes | No/0.063 | 52.2 | 6 | 570/616 | — | — | — | — | — | — | [46] | |||
| TNF-α | rs1800630 | 2013 | A vs C | Diverse | 0.76 (0.60–0.97) | No | No/0.158 | 42.3 | 4 | 606/622 | 16.64 | 155 | BBA | 0.854 | 0.761 | Weak | [47] |
| A vs C | Asian | 0.56 (0.37–0.85) | No | NA/0.640 | 0.0 | 2 | 202/203 | 16.50 | 40 | CAX | 0.206 | 0.756 | NA | [47] | |||
| EPHX1 | rs1051740 | 2012 | CC vs TT | Diverse | 1.33 (1.06–1.69) | No | 0.158/<0.0001 | 73.0 | 25 | 4808/24888 | 52.59 | 5186 | ACA | 0.837 | 0.699 | Weak | [48] |
| CC vs TT | Caucasian | 1.61 (1.12–2.31) | No | No/<0.0001 | 81.0 | 12 | 3930/23907 | 49.91 | 4343 | ACA | 0.350 | 0.733 | Weak | [48] | |||
| EPHX1 | rs2234922 | 2012 | Extremely slow vs normal | Diverse | 1.77 (1.23–2.55) | No | No/<0.0001 | 76.0 | 15 | 3146/21391 | NA | 4180 | ACA | 0.187 | 0.535 | Weak | [48] |
| Slow vs normal | Diverse | 1.44 (1.13–1.85) | No | No/<0.00001 | 74.0 | 15 | 4248/28597 | NA | 12488 | ACA | 0.625 | 0.407 | Weak | [48] | |||
| Extremely slow vs normal | Caucasian | 2.64 (1.30–5.38) | No | No/<0.0001 | 85.0 | 8 | 2685/20884 | NA | 3813 | ACA | 0.060 | 0.926 | Weak | [48] | |||
| Slow vs normal | Caucasian | 1.31 (1.01–1.71) | No | No/0.01 | 62.0 | 8 | 3563/27956 | NA | 11763 | ACA | 0.840 | 0.847 | Weak | [48] | |||
| VDBP | 1F | 2015 | 1F vs 1S | Diverse | 1.47 (1.22–1.77) | No | No/0.447 | 0.0 | 8 | 809/1407 | 20.47 | 1125 | AAA | 0.584 | 0.008 | Strong | [49] |
| 1F vs 1S | Asian | 1.53 (1.22–1.92) | No | 0.133/0.935 | 0.0 | 5 | 510/460 | 30.84 | 319 | BAA | 0.432 | 0.052 | Moderate | [49] | |||
| VDBP | 1S | 2015 | 1S vs non 1S | Diverse | 0.86 (0.77–0.96) | No | No/0.12 | 35.0 | 11 | 1410/1595 | 46.14 | 2578 | ABA | 1.000 | 0.416 | Weak | [50] |
| 1S vs non 1S | Asian | 0.76 (0.64–0.89) | No | No/0.23 | 26.0 | 7 | 785/719 | 31.71 | 869 | BBA | 0.948 | 0.064 | Moderate | [50] | |||
| HHIP | rs13118928 | 2019 | G vs A | Diverse | 1.14 (1.08–1.20) | No | No/0.930 | 0.0 | 7 | 5157/9768 | 36.39 | 10250 | AAA§ | 1.000 | 0.000 | Strong | [57] |
| COX2 | rs20417 | 2014 | C vs G | Diverse | 1.33 (1.06–1.67) | No | >0.05/0.209 | 30.2 | 6 | 930/719 | 17.04 | 597 | BBA | 0.850 | 0.621 | Weak | [39] |
| IREB2 | rs2568494 | 2016 | AA vs GG + AG | Diverse | 1.38 (1.09–1.76) | No | No/0.171 | 40.1 | 4 | 2001/2167 | 22.70 | 322 | BBA | 0.749 | 0.555 | Weak | [52] |
| HMOX1 | L allele | 2017 | L vs S + M | Diverse | 2.02 (1.31–3.11) | No | 0.79/0.08 | 52.0 | 5 | 601/547 | 8.78 | 287 | BCB | 0.088 | 0.612 | Weak | [53] |
| L vs S + M | Asian | 2.23 (1.68–2.95) | No | No/0.46 | 0.0 | 4 | 471/452 | 9.29 | 262 | BAB | 0.003 | 0.001 | Strong | [53] | |||
| ACE | I/D※ | 2018 | D vs I | Asian | 1.63 (1.17–2.27) | No | No/0.065 | 54.7 | 5 | 380/206 | 39.86 | 685 | BCA | 0.311 | 0.549 | Weak | [54] |
| ADRB2 | rs1042714※ | 2018 | G vs C | Asian | 1.29 (1.02–1.65) | No | 0.970∗/0.34 | 10.0 | 4 | 613/587 | 11.46 | 314 | BAA | 0.885 | 0.826 | Weak | [55] |
| FAM13A | rs7671167 | 2017 | CC + CT vs TT | Diverse | 0.76 (0.62–0.94) | No | No/0.002 | 71.0 | 7 | 3571/4344 | 53.96 | 5892 | ACA | 0.887 | 0.560 | Weak | [56] |
| CC + CT vs TT | Caucasian | 0.63 (0.43–0.93) | No | NA/0.12 | 59.0 | 2 | 607/1009 | 52.73 | 1216 | ACX | 0.388 | 0.837 | NA | [56] |
NA: not available; No: significant publication bias was not found; present: significant publication bias was found; W vs M: wild allele vs mutant allele. †Frequency of minor allele in controls. ‡The grade of C is assigned to protection from bias because the sensitivity analysis indicated that the significant summary OR can be substantially changed. §The grade of A is assigned to protection from bias though the OR is less than 1.15, and the association is replicated by GWAS. ¶Frequency less than 1%, the amount of evidence is not applied considering an A grade is unlikely to obtain. ※Reported nonsignificantly in the diverse populations, but significant results were reported in the stratified analysis by ethnicity. ∗P value or I2 was calculated according to the original data provided in the article with Stata 12.0 since the article did not present these values. ∗∗P value of Egger's test/test for between-study heterogeneity.
3.3. Significant Associations in GWASs
Thirty-two SNPs were reported in GWASs (Table 2), among which 25 SNPs showed an association with increased COPD risk with a mean OR value of 1.17 (range 1.08–1.36, median 1.15), while protective effect was found for 4 SNPs with a mean OR value of 0.73 (range 0.73–0.74, median 0.73). There were no sufficient data in GWASs for the rest 3 SNPs. Unsurprisingly, based on the FPRP test, all the 29 SNPs were proved to be noteworthy. Given that the Venice criteria was not applicable to GWASs, we did not further evaluate these results. Additionally, we extracted data of another 77 SNPs from the two GWASs excluded for lacking of replication phases. Because the study of Sakornsakolpat et al. [21] is one of the largest COPD GWASs to date (Supplementary Table 1), these data might provide an overview of known and well-established COPD SNPs from GWASs.
Table 2.
Statistically significant variants from GWASs.
| Gene | Variants | Risk allele | Year | Ethnicity | OR (95% CI) | RAF† | P value‡ | Cases/controls | Power OR of 1.5 | FPRP values at prior probability of 0.01/0.001 | Ref. | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| CHRNA 3/5 | rs8034191 | C | 2009 | Caucasian | NA | 0.26† | 1.48E − 10 | 1818 | 2567 | — | — | [25] |
| CHRNA 3/5 | rs1051730 | T | 2009 | Caucasian | NA | 0.27† | 5.74E − 10 | 1818 | 2567 | — | — | [25] |
| FAM13A | rs1903003 | C | 2010 | Caucasian | NA | 0.43† | 9.47E − 11 | 4552 | 4582 | — | — | [26] |
| RAB4B-EGLN2 | rs7937 | C | 2012 | Caucasian | 0.73 (0.63–0.83) | 0.44 | 2.88E − 09 | 3499 | 1922 | 0.917 | 0.000/0.002 | [27] |
| MIA-RAB4B | rs2604894 | A | 2012 | Caucasian | 0.74 (0.65–0.84) | 0.44 | 3.41E − 08 | 3499 | 1922 | 0.947 | 0.000/0.003 | [27] |
| FAM13A | rs7671167 | C | 2012 | Caucasian | 0.73 (0.66–0.81) | 0.49 | 2.22E − 09 | 3499 | 1922 | 0.956 | 0.000/0.000 | [27] |
| FAM13A | rs1964516 | C | 2012 | Caucasian | 0.73 (0.66–0.81) | 0.49 | 1.88E − 09 | 3499 | 1922 | 0.956 | 0.000/0.000 | [27] |
| CHRNA3 | rs12914385 | T | 2014 | Diverse | 1.28 (1.20–1.36) | 0.31† | 6.38E − 14 | 6633 | 5704 | 1.000 | 0.000/0.000 | [28] |
| FAM13A | rs4416442 | C | 2014 | Diverse | 1.28 (1.20–1.36) | 0.45† | 1.12E − 14 | 6633 | 5704 | 1.000 | 0.000/0.000 | [28] |
| MMP12 | rs626750 | G | 2014 | Diverse | 1.36 (1.23–1.51) | 0.80† | 5.35E − 09 | 3497 | 5704 | 0.967 | 0.000/0.000 | [28] |
| HHIP | rs13141641 | T | 2017 | Diverse | 1.22 (1.19–1.26) | 0.59 | 9.10E − 41 | 24754 | 57684 | 1.000 | 0.000/0.000 | [29] |
| CHRNA5 | rs17486278 | C | 2017 | Diverse | 1.18 (1.15–1.22) | 0.35 | 1.77E − 28 | 24754 | 57684 | 1.000 | 0.000/0.000 | [29] |
| HTR4 | rs7733088 | G | 2017 | Diverse | 1.18 (1.14–1.21) | 0.60 | 5.33E − 26 | 24754 | 57684 | 1.000 | 0.000/0.000 | [29] |
| ADGRG6 | rs9399401 | T | 2017 | Diverse | 1.15 (1.12–1.19) | 0.72 | 1.81E − 19 | 24754 | 57684 | 1.000 | 0.000/0.000 | [29] |
| THSD4 | rs1441358 | G | 2017 | Diverse | 1.13 (1.10–1.16) | 0.33 | 8.22E − 16 | 24754 | 57684 | 1.000 | 0.000/0.000 | [29] |
| FAM13A | rs6837671 | G | 2017 | Diverse | 1.12 (1.09–1.15) | 0.41 | 7.48E − 15 | 24754 | 57684 | 1.000 | 0.000/0.000 | [29] |
| GSTCD | rs11727735 | A | 2017 | Diverse | 1.26 (1.18–1.33) | 0.94 | 3.84E − 14 | 24754 | 57684 | 1.000 | 0.000/0.000 | [29] |
| RIN3 | rs754388 | C | 2017 | Diverse | 1.15 (1.11–1.20) | 0.82 | 4.96E − 14 | 24754 | 57684 | 1.000 | 0.000/0.000 | [29] |
| ADAM19 | rs113897301 | A | 2017 | Diverse | 1.16 (1.12–1.21) | 0.17 | 1.58E − 13 | 24754 | 57684 | 1.000 | 0.000/0.000 | [29] |
| TET2 | rs2047409 | A | 2017 | Diverse | 1.12 (1.08–1.15) | 0.62 | 2.46E − 13 | 24754 | 57684 | 1.000 | 0.000/0.000 | [29] |
| EEFSEC | rs2955083 | A | 2017 | Diverse | 1.18 (1.13–1.24) | 0.88 | 4.16E − 13 | 24754 | 57684 | 1.000 | 0.000/0.000 | [29] |
| CFDP1 | rs7186831 | A | 2017 | Diverse | 1.12 (1.08–1.16) | 0.43 | 1.12E − 11 | 24754 | 57684 | 1.000 | 0.000/0.000 | [29] |
| TGFB2 | rs10429950 | T | 2017 | Diverse | 1.11 (1.07–1.14) | 0.73 | 1.66E − 10 | 24754 | 57684 | 1.000 | 0.000/0.000 | [29] |
| AGER | rs2070600 | C | 2017 | Diverse | 1.24 (1.16–1.32) | 0.95 | 5.94E − 10 | 24754 | 57684 | 1.000 | 0.000/0.000 | [29] |
| ARMC2 | rs2806356 | C | 2017 | Diverse | 1.12 (1.08–1.16) | 0.18 | 8.34E − 10 | 24754 | 57684 | 1.000 | 0.000/0.000 | [29] |
| PID1 | rs16825267 | C | 2017 | Diverse | 1.19 (1.12–1.25) | 0.93 | 1.68E − 09 | 24754 | 57684 | 1.000 | 0.000/0.000 | [29] |
| DSP | rs2076295 | T | 2017 | Diverse | 1.09 (1.06–1.12) | 0.55 | 3.97E − 09 | 24754 | 57684 | 1.000 | 0.000/0.000 | [29] |
| MTCL1 | rs647097 | C | 2017 | Diverse | 1.10 (1.06–1.13) | 0.27 | 6.14E − 09 | 24754 | 57684 | 1.000 | 0.000/0.000 | [29] |
| RARB | rs1529672 | C | 2017 | Diverse | 1.11 (1.07–1.15) | 0.83 | 2.47E − 08 | 24754 | 57684 | 1.000 | 0.003/0.026 | [29] |
| SFTPD | rs721917 | G | 2017 | Diverse | 1.08 (1.05–1.11) | 0.42 | 2.49E − 08 | 24754 | 57684 | 1.000 | 0.000/0.000 | [29] |
| CYP2A6 | rs12459249 | C | 2017 | Diverse | 1.10 (1.06–1.14) | 0.66 | 3.42E − 08 | 24754 | 57684 | 1.000 | 0.000/0.000 | [29] |
| CCDC101 | rs17707300 | C | 2017 | Diverse | 1.10 (1.06–1.13) | 0.37 | NA | 24754 | 57684 | 1.000 | 0.000/0.000 | [29] |
NA: not available. †Risk allele frequency from the dbSNP. ‡The P values are all less than 5.00E − 08.
3.4. Nonsignificant Associations in Meta-Analyses
Forty-eight statistically nonsignificant associations were reported for 55 variants in 27 genes (Supplementary Table 2) [8, 9, 34–37, 39, 42, 44, 45, 47, 54, 55, 58–69]. These meta-analyses included a mean of 6 studies (range 1–15) and 5587 participants (range 220–100777). Except 48 statistically nonsignificant associations, 9 significant associations were identified in the stratified analysis by ethnicity [8, 36, 42, 54, 55, 59, 65, 70]. Only 4 associations met the following criteria for further assessment: genetic model must be an allelic model, and significant association was congruously confirmed across studies. However, all of them were graded as weak evidence of association with COPD risk. Additionally, 5 variants (TGF-β1 rs1800469, MMP-1 rs1799750, SERPINE2 rs3795879, and CHRNA rs578776 and rs588765) in 4 genes showed convincing evidence of no association with COPD risk in meta-analyses that included a minimum of 2400 cases and 3000 controls [34, 62, 67, 68].
3.5. Inconsistency among Meta-Analyses
Controversial results were reported for 23 variants in the meta-analyses (Supplementary Table 3) [8, 9, 32, 34–36, 38, 41, 42, 44–51, 57, 58, 61–64, 69–97]. Considering all of the factors we mentioned in “Data Extraction” comprehensively, 15 variants were deemed to have a significant association with COPD risk: TNF-α rs1800629 and rs1800630, GSTT1 null/present, MMP9 rs3918242, EPHX1 rs1051740 and rs2234922, IL-13 rs1800925, IL-6 rs1800796, ADAM33 rs2280091, rs511898, and rs612709, SP-D rs721917, VDBP 1F and 1S, and HHIP rs13118928. Eight variants were deemed to have a nonsignificant association with COPD risk: TNF-α rs1800610, TGF-β1 rs2241712, rs1982073, and rs6957, IL1β rs1143627, SOD3 rs1799895, ADAM33 rs528557, and VDBP GC-2. Still, results of seven variants should be prudently interpreted since studies with the similar sample size yielded opposing results, including TNF-α rs1800630, TGF-β1 rs2241712, ADAM33 rs2280091, rs612709, and rs511898, IL-6 rs1800796, and VDBP 1S.
4. Discussion
As far as we know, this study is the only one which aims to give a comprehensive assessment on all the reported genetic variants and COPD susceptibility with systematic methods. We retrieved relevant meta-analyses and GWASs, extracting useful data for further evaluation. The Venice criteria and FPRP test were the two major tools. As a result, 10 variants in 8 genes were graded as strong evidence of association with COPD risk, including: GSTM1 null/present, CHRNA rs16969968, rs8034191, and rs1051730, ADAM33 rs612709, SP-D rs721917, TNF-α rs1800629, VDBP 1F, HMOX1 L allele, and HHIP rs13118928. Six variants in 5 genes and 40 variants in 23 genes were graded as moderate and weak evidence, respectively. In addition, 29 SNPs identified in GWASs were proved to be noteworthy. There were overlaps between the two groups of SNP. Four SNPs (CHRNA rs1051730 and rs8034191, SP-D rs721917, and FAM13A rs7671167) reported in GWASs were also investigated by meta-analyses. Except FAM13A rs7671167, all the other three SNPs were graded as strong evidence, suggesting that methods we used do have the ability to pick out potential SNPs as long as a high-quality meta-analysis was included.
GSTM is a kind of the glutathione S-transferase (GST) cytoplasmic enzymes that metabolize various toxic substances [98]. GSTM1, located on chromosome 1p13.3, is highly expressed in the lung tissue. GSTM1 homozygous deletion leads to the absence of protein expression and production, preventing the detoxification. Previous studies have proven that the null genotype of GSTM1 was related to the increased risk of inflammatory lung diseases, thus taking an important part in COPD development [99]. Our study demonstrated that GSTM1 null genotype showed strong cumulative evidence for an association with COPD risk in Caucasians and moderate cumulative evidence was found in “diverse populations” and Asians. In addition, highly consistent positive results were reported in recent meta-analyses [8, 9, 32, 78, 79]. The association between GSTM1 null genotype and COPD risk was well established.
CHRNA3/5, located on chromosome 15q25, encodes the subunits of alpha-nicotinic acetylcholine receptor (nAChR). Gwilt et al. [100] reported that nAChR might play a role in modifying the inflammatory response to smoking. Moreover, variants in CHRNA3/5 were involved in altering mRNA levels of CHRNA5 in the lung tissue and influencing receptor response to nicotine agonists [101, 102]. Therefore, we can draw a conclusion that these variants may contribute to the development of COPD. Four variants (rs16969968, rs8034191, rs1051730, and rs6495309) in CHRNA3/5 were reported significantly in our study. Except rs6495309, all of them showed strong cumulative evidence for an association with COPD risk. Interestingly, two variants were also identified as susceptibility loci for COPD by GWASs [25]. However, these strong associations were limited to non-Asians because distribution of the minor allele varies extensively between different ethnicities. Hence, we recommend additional studies to confirm the association between these polymorphisms and COPD risk in Asians.
ADAM33, located on chromosome 20p13, is a member of the “a disintegrin and metalloprotease” (ADAM) family and is intricately related to airway hyper-responsiveness and obstruction [103]. Rs612709 in ADAM33 showed protective effect and was rated as strong evidence of association with COPD in both “diverse populations” (OR = 0.60, 95% CI = 0.52−0.68) and Caucasians (OR = 0.64, 95% CI = 0.53−0.77). Another meta-analysis with the similar publication year and data source yet yielded an opposing result (OR = 1.46, 95% CI = 1.14−1.87) [41]. A cognitive error in the minor allele may lead to this conflict (rs612709G>A).
SP-D, also known as SFTPD, is a kind of alveolar surfactant-associated protein (SP) and plays a prominent role in maintenance of lung function and immune regulation [104]. Our study revealed strong evidence for an association between the T allele of rs721917 and COPD risk in both “diverse populations” and Asians based on a total of 1111 and 934 subjects, respectively. Although sample size is relatively small, the high frequency (>40%) of the T allele makes it possible to achieve sufficient statistical power. However, no significant association was reported in Caucasians because data from Caucasians were scarce. Hence, further studies that focus on Caucasians are warranted.
TNF-α is a pivotal cytokine in inflammation of the lung [105]. Rs1800629 in TNF-α contains a G to A variation, carriers of the A allele are more likely to activate the TNF-α promoter region and lead to overexpression of TNF-α [106]. Highly consistent positive results of association between rs1800629 A allele and COPD risk in Asians but not in Caucasians were reported [8, 46, 71–75]. Our study also graded rs1800629 as having strong evidence of association with COPD risk in Asians. Nevertheless, no correlation was found between rs1800629 and COPD risk after adjustment for smoking (OR = 1.13, 95% CI = 0.95−1.35). It seems that other factors may contribute to the risk of COPD in smokers. Expanding studies with a sufficient number of smokers are needed.
Heme oxygenase plays an important role in resisting damage caused by oxidative stress. HMOX1, belongs to the heme oxygenase isoforms, was reported to have the ability of protecting against airway inflammation and emphysema [107, 108]. The L allele (long GT repeat sequence) of HMOX1 was rated as strong evidence of association with 2.23-fold increased risk of COPD based on 923 subjects in Asians. The sample size was relatively small, and the frequency of the L allele was low (9.29%). Additionally, only one meta-analysis was retrieved in our study. Although we used the Venice criteria and FPRP test to evaluate the credibility of an association, a firm conclusion should not be drawn until studies with more subjects can be included in the meta-analysis.
HHIP gene, located on chromosome 4q31, encodes an inhibitory protein for sonic hedgehog. The hedgehog is known to be essential for branching morphogenesis in the developing lung [109]. Zhou et al. [110] demonstrated that HHIP expression at both mRNA and protein levels is reduced in COPD lung tissues. These studies showed that HHIP plays an important role in maintaining the normal lung function. HHIP rs13118928 was firstly reported by Pillai et al. [25] in GWASs but did not reach genome-wide significance levels. However, follow-up studies [98, 111] including ours have proved the association.
VDBP, located on chromosome 4q12-q13, is also known as GC and participates in binding substantial quantities of 25-hydroxyvitamin D and vitamin D [111]. Two mutations (C⟶T and G⟶A) of SNPs (rs4588 and rs7041) in VDBP result in 3 common isoforms (GC1S, GC1F, and GC2) and different protein products. We found strong and moderate evidence of association between GC1F and the risk of COPD in “diverse populations” and Asians, respectively. Among 8 meta-analyses we retrieved, only one meta-analysis reported a significant association with decreased risk of COPD in Caucasians [94]. The association between polymorphisms and COPD risk varied between ethnicities. Reasons may be summarized as follows: different genetic backgrounds, different degrees of environmental exposure, and small sample size results in poor statistical power.
Six variants in 5 genes and 40 variants in 23 genes showed moderate and weak evidence of association with COPD risk in our study, respectively. Among which, two variants (PIMS and ADAM33 rs3918396) and eight variants (IL-13 rs1800925, ADAM33 rs2280091, CYP1A1 rs4646903, TNF-α rs1800630, VDBP 1S, COX2 rs20417, IREB2 rs2568494, and ADRB2 rs1042714) were downgraded from strong to moderate and moderate to weak due to high FPRP values (>0.2), respectively. As we mentioned in “Materials and Methods,” three parameters may influence the calculated FPRP values. We used the prior probability of 0.01 for FPRP calculations; however, the calculated FPRP values might be smaller if a softer prior probability was used (e.g., 0.05) [17]. Therefore, we recommend further investigations on these variants since significant associations may have been excluded.
There was one more variant worth noting, FAM13A rs7671167; this showed weak evidence of association with COPD susceptibility because of the strong heterogeneity between studies. But it was identified by GWASs [27]. Results from GWASs, however, are more statistically significant and convincing. The Venice criteria, undoubtedly, is a useful tool to evaluate the cumulative epidemiological evidence of associations between SNPs and diseases. But sometimes omissions might occur because “weak evidence” was identified as long as a “C” was assigned to any of the three aspects. Results would be more credible if a more high-quality meta-analysis was included or different weights were assigned to the three aspects included in the Venice criteria.
Forty-eight statistically nonsignificant associations were reported for 55 variants in 27 genes. Five variants (TGF-β1 rs1800469, MMP-1 rs1799750, SERPINE2 rs3795879, and CHRNA rs578776 and rs588765) showed no association with COPD risk in meta-analyses including a minimum of 2400 patients and 3000 controls. The MAFs of these variants ranged from 20% to 50%. In other words, these meta-analyses provided greater than 88% power to detect an OR of 1.15 under different genetic models for these variants. In addition, no inconsistency was found among meta-analyses investigating these variants; therefore, we can safely draw a conclusion that these variants are not associated with the risk of COPD. Further studies are unlikely to yield significant results.
Limitations of our study must be addressed. Scopus and Web of Science were not retrieved. Non-English literature except literature in Chinese and gray literature were not included. Although a systematic literature search was done, and some articles were overlooked. Only one meta-analysis was chosen for further evaluation, which might introduce bias to some extent. The latest candidate-gene association studies might be missed. We extracted results from subgroup analyses to address the heterogeneity, but heterogeneity does exist. High heterogeneity influenced the evaluation directly.
5. Conclusions
In summary, combining Venice criteria and FPRP test, cumulative epidemiological evidence of significant associations between genetic variants and COPD risk was evaluated. As a result, 13 variants showed moderate to strong evidence. Our study can provide not only reliable evidence but also helpful clues for further investigations.
Acknowledgments
Part of the suggestion for the discussion and proofreading of English grammar were provided by Dr. Wenzhuo Zhao.
Data Availability
All data generated or analyzed during this study are included in this published article and its supplementary information files.
Conflicts of Interest
The authors declared that no conflicts of interest exist.
Authors' Contributions
Caiyang Liu and Ran Ran contributed equally to this work and thus are co-first authors.
Supplementary Materials
Supplementary Table 1: statistically significant variants from the other two GWASs. Supplementary Table 2: statistically nonsignificant gene-variant COPD overall associations. Supplementary Table 3: conflicting gene-variant COPD overall associations.
References
- 1.MacNee W. Pathogenesis of chronic obstructive pulmonary disease. Proceedings of the American Thoracic Society; 2005; New York, NY, USA. pp. 258–266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.World Health Organisation. Chronic Obstructive Pulmonary Disease (COPD) Geneva, Switzerland: World Health Organisation; https://www.who.int/respiratory/copd/en/ [Google Scholar]
- 3.Soares S., Costa I., Luísa Neves A., Couto L. Characterisation of a population at increased risk of COPD. Revista Portuguesa de Pneumologia . 2010;16(2):237–252. doi: 10.1016/s2173-5115(10)70033-6. [DOI] [PubMed] [Google Scholar]
- 4.Eisner M. D., Anthonisen N., Coultas D., et al. An official American Thoracic Society public policy statement: novel risk factors and the global burden of chronic obstructive pulmonary disease. American Journal of Respiratory and Critical Care Medicine . 2010;182(5):693–718. doi: 10.1164/rccm.200811-1757ST. [DOI] [PubMed] [Google Scholar]
- 5.Kim W. J. Genetics of COPD. Annual Review of Physiology . 2017;82:413–431. doi: 10.1146/annurev-physiol-021317-121224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hersh C. P., Hokanson J. E., Lynch D. A., et al. Family history is a risk factor for COPD. Chest . 2011;140(2):343–350. doi: 10.1378/chest.10-2761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dransfield M. T., Bailey W. C. COPD: racial disparities in susceptibility, treatment, and outcomes. Clinics in Chest Medicine . 2006;27(3):463–471. doi: 10.1016/j.ccm.2006.04.005. [DOI] [PubMed] [Google Scholar]
- 8.Smolonska J., Wijmenga C., Postma D. S., Boezen H. M. Meta-analyses on suspected chronic obstructive pulmonary disease genes. American Journal of Respiratory and Critical Care Medicine . 2009;180(7):618–631. doi: 10.1164/rccm.200905-0722OC. [DOI] [PubMed] [Google Scholar]
- 9.Castaldi P. J., Cho M. H., Cohn M., et al. The COPD genetic association compendium: a comprehensive online database of COPD genetic associations. Human Molecular Genetics . 2010;19(3):526–534. doi: 10.1093/hmg/ddp519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hardy J., Singleton A. Genomewide association studies and human disease. New England Journal of Medicine . 2009;360(17):1759–1768. doi: 10.1056/NEJMra0808700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lau J., Ioannidis J. P., Schmid C. H. Summing up evidence: one answer is not always enough. The Lancet . 1998;351(9096):123–127. doi: 10.1016/s0140-6736(97)08468-7. [DOI] [PubMed] [Google Scholar]
- 12.Ioannidis J. P., Boffetta P., Little J., et al. Assessment of cumulative evidence on genetic associations: interim guidelines. International Journal of Epidemiology . 2008;37(1):120–132. doi: 10.1093/ije/dym159. [DOI] [PubMed] [Google Scholar]
- 13.Langevin S. M., Ioannidis J. P. A., Vineis P., Taioli E. Assessment of cumulative evidence for the association between glutathione S-transferase polymorphisms and lung cancer: application of the Venice interim guidelines. Pharmacogenetics and Genomics . 2010;20(10):586–597. doi: 10.1097/FPC.0b013e32833c3892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tanni S. E., Pelegrino N. R., Angeleli A. Y., Correa C., Godoy I. Smoking status and tumor necrosis factor-alpha mediated systemic inflammation in COPD patients. Journal of Inflammation . 2010;7(1):p. 29. doi: 10.1186/1476-9255-7-29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhang B., Fadiel A. B., Long J., Zheng W. Genetic variants associated with breast-cancer risk: comprehensive research synopsis, meta-analysis, and epidemiological evidence. The Lancet Oncology . 2011;12(5):477–488. doi: 10.1016/s1470-2045(11)70076-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wacholder S., Chanock S., Closas M. G., El Ghormli L., Rothman N. Assessing the probability that a positive report is false: an approach for molecular epidemiology studies. Journal of the National Cancer Institute . 2004;96(6):434–442. doi: 10.1093/jnci/djh075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ma X., Zhang B., Zheng W. Genetic variants associated with colorectal cancer risk: comprehensive research synopsis, meta-analysis, and epidemiological evidence. Gut . 2014;63(2):326–336. doi: 10.1136/gutjnl-2012-304121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yoon H.-K., Hu H.-J., Rhee C.-K., et al. Polymorphisms inPDE4Dare associated with a risk of COPD in non-emphysematous Koreans. COPD: Journal of Chronic Obstructive Pulmonary Disease . 2014;11(6):652–658. doi: 10.3109/15412555.2014.898045. [DOI] [PubMed] [Google Scholar]
- 19.Chen W., Brehm J. M., Manichaikul A., et al. A genome-wide association study of chronic obstructive pulmonary disease in Hispanics. Annals of the American Thoracic Society . 2015;12(3):340–348. doi: 10.1513/AnnalsATS.201408-380OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Smolonska J., Koppelman G. H., Wijmenga C., et al. Common genes underlying asthma and COPD? genome-wide analysis on the Dutch hypothesis. European Respiratory Journal . 2014;44(4):860–872. doi: 10.1183/09031936.00001914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sakornsakolpat P., Prokopenko D., Lamontagne M., et al. Genetic landscape of chronic obstructive pulmonary disease identifies heterogeneous cell-type and phenotype associations. Nature Genetics . 2019;51(3):494–505. doi: 10.1038/s41588-018-0342-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lee J. H., Cho M. H., Hersh C. P., et al. Genetic susceptibility for chronic bronchitis in chronic obstructive pulmonary disease. Respiratory Research . 2014;15(1):p. 113. doi: 10.1186/s12931-014-0113-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hosgoodiii H. D., Díaz-Pea R., Blansky D., et al. PRDM15 is associated with risk of chronic obstructive pulmonary disease in a rural population in Chile. Respiration . 2020;99(4):307–315. doi: 10.1159/000506649. [DOI] [PubMed] [Google Scholar]
- 24.Díaz-Peña R., Boekstegers F., Silva R. S., et al. Amerindian ancestry influences genetic susceptibility to chronic obstructive pulmonary disease. Journal of Personalized Medicine . 2020;10(3):p. 93. doi: 10.3390/jpm10030093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Pillai S. G., Ge D., Zhu G., et al. A genome-wide association study in chronic obstructive pulmonary disease (COPD): identification of two major susceptibility loci. PLoS Genetics . 2009;5(3) doi: 10.1371/journal.pgen.1000421.1000421 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Cho M. H., Boutaoui N., Klanderman B. J., et al. Variants in FAM13A are associated with chronic obstructive pulmonary disease. Nature Genetics . 2010;42(3):200–202. doi: 10.1038/ng.535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Cho M. H., Castaldi P. J., Wan E. S., et al. A genome-wide association study of COPD identifies a susceptibility locus on chromosome 19q13. Human Molecular Genetics . 2012;21(4):947–957. doi: 10.1093/hmg/ddr524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Cho M. H., McDonald M.-L. N., Zhou X., et al. Risk loci for chronic obstructive pulmonary disease: a genome-wide association study and meta-analysis. The Lancet Respiratory Medicine . 2014;2(3):214–225. doi: 10.1016/s2213-2600(14)70002-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hobbs B. D., de Jong K., de Jong K., et al. Genetic loci associated with chronic obstructive pulmonary disease overlap with loci for lung function and pulmonary fibrosis. Nature Genetics . 2017;49(3):426–432. doi: 10.1038/ng.3752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hersh C. P., Dahl M., Ly N. P., Berkey C. S., Nordestgaard B. G., Silverman E. K. Chronic obstructive pulmonary disease in 1-antitrypsin PI MZ heterozygotes: a meta-analysis. Thorax . 2004;59(10):843–849. doi: 10.1136/thx.2004.022541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Dahl M., Hersh C. P., Ly N. P., Berkey C. S., Silverman E. K., Nordestgaard B. G. The protease inhibitor PI S allele and COPD: a meta-analysis. European Respiratory Journal . 2005;26(1):67–76. doi: 10.1183/09031936.05.00135704. [DOI] [PubMed] [Google Scholar]
- 32.Ding Z., Wang K., Li J., Tan Q., Tan W., Guo G. Association between glutathione S‐transferase gene M1 and T1 polymorphisms and chronic obstructive pulmonary disease risk: a meta‐analysis. Clinical Genetics . 2019;95(1):53–62. doi: 10.1111/cge.13373. [DOI] [PubMed] [Google Scholar]
- 33.Zhang L., Chang W.-W., Ding H., Su H., Wang H.-Y. Transforming growth factor-β1 polymorphisms and chronic obstructive pulmonary disease: a meta-analysis. International Journal of Tuberculosis & Lung Disease . 2011;15(10):1301–1307. doi: 10.5588/ijtld.10.0295. [DOI] [PubMed] [Google Scholar]
- 34.Liao N., Zhao H., Chen M.-L., Xie Z.-F. Association between the TGF-β1 polymorphisms and chronic obstructive pulmonary disease: a meta-analysis. Bioscience Reports . 2017;37(4) doi: 10.1042/bsr20170747.BSR20170747 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ahmadi A., Ghaedi H., Salimian J., Jamalkandi S. A., Ghanei M. Association between chronic obstructive pulmonary disease and interleukins gene variants: a systematic review and meta-analysis. Cytokine . 2019;117:65–71. doi: 10.1016/j.cyto.2019.02.003. [DOI] [PubMed] [Google Scholar]
- 36.Liao N., Zhao H., Chen M.-L., Xie Z.-F. Association of the IL-13 polymorphisms rs1800925 and rs20541 with chronic obstructive pulmonary disease risk. Medicine . 2017;96(47) doi: 10.1097/md.0000000000008556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zhang Y.-F., Zhao A.-D. Common polymorphisms in IL-27 genes may contribute to risk of various human diseases in asian populations: a meta-analysis. Medical Science Monitor . 2016;22:766–775. doi: 10.12659/msm.895558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Shao Z. T., Zhang T. J., Su Q. M. Meta-analysis of association between-1562C/T polymorphism of the MMP-9 promoter and COPD risk. Acta Universitatis Medicinalis Nanjing . 2017;37(10):1357–1368. doi: 10.7655/NYDXBNS20171033. [DOI] [Google Scholar]
- 39.Yu X.-L., Zhang J., Zhao F., Pan X.-M. Retracted article: relationships of COX2 and MMP12 genetic polymorphisms with chronic obstructive pulmonary disease risk: a meta-analysis. Molecular Biology Reports . 2014;41(12):8149–8162. doi: 10.1007/s11033-014-3715-3. [DOI] [PubMed] [Google Scholar]
- 40.Cui K., Ge X., Ma H. Four SNPs in the CHRNA3/5 alpha-neuronal nicotinic acetylcholine receptor subunit locus are associated with COPD risk based on meta-analyses. PLoS One . 2014;9(7) doi: 10.1371/journal.pone.0102324.102324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zhang R., Li H., Zhao H., Chen W., Cheng D. Polymorphisms in a disintegrin and metalloprotease 33 gene and the risk of chronic obstructive pulmonary disease: a meta-analysis. Respirology . 2014;19(3):312–320. doi: 10.1111/resp.12235. [DOI] [PubMed] [Google Scholar]
- 42.Zhou D.-C., Zhou C.-F., Toloo S., Shen T., Tong S.-L., Zhu Q.-X. Association of a disintegrin and metalloprotease 33 (ADAM33) gene polymorphisms with the risk of COPD: an updated meta-analysis of 2,644 cases and 4,804 controls. Molecular Biology Reports . 2015;42(2):409–422. doi: 10.1007/s11033-014-3782-5. [DOI] [PubMed] [Google Scholar]
- 43.Wang C.-D., Chen N., Huang L., et al. Impact ofCYP1A1Polymorphisms on susceptibility to chronic obstructive pulmonary disease: a meta-analysis. BioMed Research International . 2015;2015:9. doi: 10.1155/2015/942958.942958 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ma T., Liu X., Liu Z. Functional polymorphisms in surfactant protein genes and chronic obstructive pulmonary disease risk: a meta-analysis. Genetic Testing and Molecular Biomarkers . 2013;17(12):910–917. doi: 10.1089/gtmb.2013.0308. [DOI] [PubMed] [Google Scholar]
- 45.Liao Y., Huang C., Wang J., Fan X. Association of surfactant-associated protein D gene polymorphisms with the risk of COPD: a meta-analysis. Clinics . 2019;74 doi: 10.6061/clinics/2019/e855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zeng X., Zhang L., Gu H., Gu Y. Association between TNF-α-308 G/A polymorphism and COPD susceptibility: a meta-analysis update. International Journal of Chronic Obstructive Pulmonary Disease . 2016;11:1367–1379. doi: 10.2147/copd.s105394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Cui K., Ge X. Y., Ma H. L. Association of-238G/A and-863C/A polymorphisms in the TNF-α gene with chronic obstructive pulmonary disease based on a meta-analysis. Genetics and Molecular Research . 2013;12(4):4981–4989. doi: 10.4238/2013.October.24.10. [DOI] [PubMed] [Google Scholar]
- 48.Li H., Fu W.-P., Hong Z.-H. Microsomal epoxide hydrolase gene polymorphisms and risk of chronic obstructive pulmonary disease: a comprehensive meta-analysis. Oncology Letters . 2013;5(3):1022–1030. doi: 10.3892/ol.2012.1099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Xie X., Zhang Y., Ke R., et al. Vitamin D-binding protein gene polymorphisms and chronic obstructive pulmonary disease susceptibility: a meta-analysis. Biomedical Reports . 2015;3(2):183–188. doi: 10.3892/br.2014.392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wang Y. L., Kong H., Xie W. P., Wang H. Association of vitamin D-binding protein variants with chronic obstructive pulmonary disease: a meta-analysis. Genetics and Molecular Research . 2015;14(3):10774–10785. doi: 10.4238/2015.September.9.16. [DOI] [PubMed] [Google Scholar]
- 51.Feng H.-h., Mao L., Pan K., Zhang L., Rui D.-s. Association between F+1 polymorphism in a disintegrin and metalloprotease 33 (ADAM33) gene and chronic obstructive pulmonary disease susceptibility: an evidence-based meta-analysis. Gene . 2019;719 doi: 10.1016/j.gene.2019.144009.144009 [DOI] [PubMed] [Google Scholar]
- 52.Du Y., Xue Y., Xiao W. Association of IREB2 gene rs2568494 polymorphism with risk of chronic obstructive pulmonary disease: a meta-analysis. Medical Science Monitor . 2016;22:177–182. doi: 10.12659/msm.894524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Zhou H., Ying X., Liu Y., Ye S., Yan J., Li Y. Genetic polymorphism of heme oxygenase 1 promoter in the occurrence and severity of chronic obstructive pulmonary disease: a meta-analysis. Journal of Cellular and Molecular Medicine . 2017;21(5):894–903. doi: 10.1111/jcmm.13028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Ma Y., Tong X., Liu Y., Liu S., Xiong H., Fan H. ACE gene polymorphism is associated with COPD and COPD with pulmonary hypertension: a meta-analysis. International Journal of Chronic Obstructive Pulmonary Disease . 2018;13:2435–2446. doi: 10.2147/copd.s168772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Fu M., Xie W. J., Ming W. C. A Meta-analysis on investigating the association of ADRB2 genetic polymorphisms with chronic obstructive pulmonary disease. Journal of Clinical and Pathological Research . 2018;38(10):2151–2158. doi: 10.3978/j.issn.2095-6959.2018.10.016. [DOI] [Google Scholar]
- 56.Feng X. L., He J., Li W. C., Li X. Y., Ren S. Q. Association between fam13a gene polymorphism and the risk of chronic obstructive pulmonary disease:a meta-analysis. Medical Journal of Qilu . 2017;32(03):315–320. doi: 10.13362/j.qlyx.201703020. [DOI] [Google Scholar]
- 57.Wu T. Y., Chen H. B., Cheng D. Meta-analysis on relationship between HHIP rs13118928 single nucleotide polymorphism and susceptibility to chronic obstructive pulmonary disease. Chinese Journal of Lung Diseases (Electronic Edition) . 2019;12(4):420–425. doi: 10.3877/cma.j.issn. [DOI] [Google Scholar]
- 58.Liang H., He J., Li X. Y. Tumor necrosis factor-α+ 489 polymorphism and risk of chronic obstructive pulmonary disease: a meta-analysis. Chinese Journal of Lung Diseases (Electronic Edition) . 2016;09(03):286–291. doi: 10.3877/cma.j.issn.1674-6902.2016.03.012. [DOI] [Google Scholar]
- 59.Wang Y. S., Liu L., Xu X. Y., et al. Association of interleukin-1β-511C/T promoter polymorphism with COPD risk: a meta-analysis. Genetics and Molecular Research . 2015;14(2):4477–4484. doi: 10.4238/2015.May.4.5. [DOI] [PubMed] [Google Scholar]
- 60.Tang B., Kang L., Chen M. Association between-589C/T polymorphism in IL-4 gene and the risk of COPD: a meta-analysis. Chinese Journal of Evidence-Based Medicine . 2016;16(6):663–667. doi: 10.7507/1672-2531.20160103. [DOI] [Google Scholar]
- 61.Dou L., Yu B., Han K., Xiao M., Liu Y., Lv F. Association of IL-6 polymorphisms with chronic obstructive pulmonary disease risk: meta-analysis of genetic association, gene expression and expression quantitative trait locus analysis. Personalized Medicine . 2016;13(4):395–403. doi: 10.2217/pme-2016-0003. [DOI] [PubMed] [Google Scholar]
- 62.Zhou H., Wu Y., Jin Y., et al. Genetic polymorphism of matrix metalloproteinase family and chronic obstructive pulmonary disease susceptibility: a meta-analysis. Scientific Reports . 2013;3(1) doi: 10.1038/srep02818.2818 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Jiang S., Yang Z. H., Chen Y. Y., et al. MMP-9 genetic polymorphism may confer susceptibility to COPD. Genetics and Molecular Research . 2016;15(2) doi: 10.4238/gmr.15026272. [DOI] [PubMed] [Google Scholar]
- 64.Wang Z. C., Hua L. Meta-analysis of SOD gene polymorphism and susceptibility to COPD. Medical Information . 2018;31(17):57–62. doi: 10.3969/j.issn.1006-1959.2018.17.018. [DOI] [Google Scholar]
- 65.Wang W., Li P., Chen Y., Yang J. Association between β2-adrenergic receptor-16Arg/gly gene polymorphism and chronic obstructive pulmonary disease risk:systematic review and meta-analysis. Iranian journal of public health . 2014;43(7):877–888. [PMC free article] [PubMed] [Google Scholar]
- 66.Nielsen A. O., Jensen C. S., Arredouani M. S., Dahl R., Dahl M. Variants of the ADRB2 gene in COPD: systematic review and meta-analyses of disease risk and treatment response. COPD: Journal of Chronic Obstructive Pulmonary Disease . 2017;14(4):451–460. doi: 10.1080/15412555.2017.1320370. [DOI] [PubMed] [Google Scholar]
- 67.Tian D. B., Cai S. X., Pan G. C., Zhang Y. F., Xiao J. H. Association between the rs3795879 G/A polymorphism of the SERPINE2 gene and chronic obstructive pulmonary disease: a meta-analysis. Genetics and Molecular Research . 2015;14(3):7920–7928. doi: 10.4238/2015.July.14.18. [DOI] [PubMed] [Google Scholar]
- 68.Saccone N. L., Culverhouse R. C., Schwantes-An T.-H., et al. Multiple independent loci at chromosome 15q25.1 affect smoking quantity: a meta-analysis and comparison with lung cancer and COPD. PLoS Genetics . 2010;6(8) doi: 10.1371/journal.pgen.1001053.1001053 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Khanna R., Nandy D., Senapati S. Systematic review and meta-analysis to establish the association of common genetic variations in vitamin D binding protein with chronic obstructive pulmonary disease. Frontiers in Genetics . 2019;10:p. 413. doi: 10.3389/fgene.2019.00413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Gong Y., Fan L., Wan H., et al. Lack of association between the TGF-β1 gene and development of COPD in Asians: a case-control study and meta-analysis. Lung . 2011;189(3):213–223. doi: 10.1007/s00408-011-9294-3. [DOI] [PubMed] [Google Scholar]
- 71.Hu G. P., Peng G. Y., Hu J. X., Ran P. X. Association of tumor necrosis factor alpha 308 G/A gene promoter polymorphism with the presence of chronic obstructive pulmonary disease: a meta-analysis. Chinese Journal of Tuberculosis and Respiratory Diseases . 2007;30(8):588–594. doi: 10.3760/j.issn:1001-0939.2007.08.007. [DOI] [PubMed] [Google Scholar]
- 72.Zhan P., Wang J., Wei S.-Z., et al. TNF-308 gene polymorphism is associated with COPD risk among Asians: meta-analysis of data for 6,118 subjects. Molecular Biology Reports . 2011;38(1):219–227. doi: 10.1007/s11033-010-0098-y. [DOI] [PubMed] [Google Scholar]
- 73.Zhang S., Wang C., Xi B., Li X. Association between the tumour necrosis factor-α−308G/A polymorphism and chronic obstructive pulmonary disease: an update. Respirology . 2011;16(1):107–115. doi: 10.1111/j.1440-1843.2010.01879.x. [DOI] [PubMed] [Google Scholar]
- 74.Xie Z. J., Xin T. Q., Xie W. J. Meta analysis of association between polymorphism of TNF-α-308 and susceptibility to COPD. Chinese Journal of Ethnomedicine and Ethnopharmacy . 2018;27(15):59–62. [Google Scholar]
- 75.Li Y. L., Chen J. M., Xu J., Xu X. N. Meta analysis on tumor necrosis factor-α308 G/A polymorphism and cervical cancer susceptibility. Jilin Medical Journal . 2016;37(3):547–550. doi: 10.3969/j.issn.1004-0412.2016.03.013. [DOI] [Google Scholar]
- 76.Cui K., Ge X. Y., Ma H. L. Association of the TNF-α+489 G/A polymorphism with chronic obstructive pulmonary disease risk in Asians: meta-analysis. Genetics and Molecular Research . 2015;14(2):5210–5220. doi: 10.4238/2015.May.18.12. [DOI] [PubMed] [Google Scholar]
- 77.Li X. L., Xu J., Wang J. L., Zhang D. D. Meta analysis of association between TNF-α + 489 G/A polymorphism and chronic obstructive pulmonary disease susceptibility in China. Heilongjiang Medical Journal . 2017;41(12):1163–1165. doi: 10.3969/j.issn.1004-5775.2017.12.006. [DOI] [Google Scholar]
- 78.Hu G., Yao W., Zhou Y., et al. Meta- and pooled analyses of the effect of glutathione S-transferase M1 and T1 deficiency on chronic obstructive pulmonary disease. International Journal of Tuberculosis & Lung Disease: The Official Journal of the International Union Against Tuberculosis and Lung Disease . 2008;12(12):1474–1481. [PubMed] [Google Scholar]
- 79.Xue H., Su J., Sun K., Xie W., Wang H. Glutathione S-transferase M1 and T1 gene polymorphism and COPD risk in smokers: an updated analysis. Molecular Biology Reports . 2012;39(4):5033–5042. doi: 10.1007/s11033-011-1300-6. [DOI] [PubMed] [Google Scholar]
- 80.Mei J. J., Liang Y., Shen N., He B. [Association between interleukin-1B polymorphisms and chronic obstructive pulmonary disease: a meta-analysis] Zhonghua Yixue Zazhi . 2013;93(12):910–915. [PubMed] [Google Scholar]
- 81.Xiao M., Guo L., Wang T., et al. Systematic review/meta-analysis interleukin-1B-31T/C promoter polymorphism and chronic obstructive pulmonary disease risk: a meta-analysis. Archives of Medical Science . 2014;3(3):434–438. doi: 10.5114/aoms.2014.43737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Xie Z.-K., Huang Q.-P., Huang J., Xie Z.-F. Association between the IL1B, IL1RN polymorphisms and COPD risk: a meta-analysis. Scientific Reports . 2014;4(1) doi: 10.1038/srep06202.6202 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Li D. D., Guo S. J., Jia L. Q., Wen F. Q. Interleukin-1β gene polymorphisms and the susceptivity to chronic obstructive pulmonary disease: a systematic review. Chinese Journal of Lung Diseases . 2013;6(2):15–18. doi: 10.3877/cma.j.issn.1674-6902.2013.02.006. [DOI] [Google Scholar]
- 84.Chen L., Wang T., Liu L., Shen Y., Wan C., Wen F. Matrix metalloproteinase-9-1562C/T promoter polymorphism confers risk for COPD: a meta-analysis. PLoS One . 2013;8(3) doi: 10.1371/journal.pone.0060523.60523 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Hu G., Shi Z., Hu J., Zou G., Peng G., Ran P. Association between polymorphisms of microsomal epoxide hydrolase and COPD: results from meta-analyses. Respirology . 2008;13(6):837–850. doi: 10.1111/j.1440-1843.2008.01356.x. [DOI] [PubMed] [Google Scholar]
- 86.Lee J., Nordestgaard B. G., Dahl M. EPHX1 polymorphisms, COPD and asthma in 47,000 individuals and in meta-analysis. European Respiratory Journal . 2011;37(1):18–25. doi: 10.1183/09031936.00012110. [DOI] [PubMed] [Google Scholar]
- 87.An L., Xia H., Zhou P., Hua L. Exploration of association between EPHX1 and chronic obstructive pulmonary disease on the basis of combined data mining. Genetics and Molecular Research . 2016;15(2) doi: 10.4238/gmr.15028639. [DOI] [PubMed] [Google Scholar]
- 88.Chen L., Shen Y., Liu L., Li X., Wang T., Wen F. Interleukin-13-1112 C/T promoter polymorphism confers risk for COPD: a meta-analysis. PLoS One . 2013;8(7) doi: 10.1371/journal.pone.0068222.68222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Aierken H., Wang J., Wushouer Q., et al. Polymorphisms of the ADAM33 gene and chronic obstructive pulmonary disease risk: a meta-analysis. The Clinical Respiratory Journal . 2014;8(1):108–115. doi: 10.1111/crj.12046. [DOI] [PubMed] [Google Scholar]
- 90.Li D. D., Guo S. J., Jia L. Q., Wen F. Q. Association of a disintegrin and metalloproteinase 33 (ADAM33) gene polymorphisms with chronic obstructive pulmonary disease in the Chinese population: a meta-analysis. Genetics and Molecular Research . 2014;13(3):6391–6397. doi: 10.4238/2014.August.25.2. [DOI] [PubMed] [Google Scholar]
- 91.Luo X. M., Yan S. M., Ge W. H. Meta-analysis on ADAM33 gene polymorphism and chronic obstructive pulmonary disease. Pharmaceutical and Clinical Research . 2013;21(5):575–579. [Google Scholar]
- 92.Nandy D., Sharma N., Senapati S. Systematic review and meta-analysis confirms significant contribution of surfactant protein D in chronic obstructive pulmonary disease. Frontiers in Genetics . 2019;10:p. 339. doi: 10.3389/fgene.2019.00339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Horita N., Miyazawa N., Tomaru K., Inoue M., Ishigatsubo Y., Kaneko T. Vitamin D binding protein genotype variants and risk of chronic obstructive pulmonary disease: a meta-analysis. Respirology . 2015;20(2):219–225. doi: 10.1111/resp.12448. [DOI] [PubMed] [Google Scholar]
- 94.Xiao M., Wang T., Zhu T., Wen F. Dual role of vitamin D-binding protein 1F allele in chronic obstructive pulmonary disease susceptibility: a meta-analysis. Genetics and Molecular Research . 2015;14(2):3534–3540. doi: 10.4238/2015.April.17.1. [DOI] [PubMed] [Google Scholar]
- 95.Salimi Asl M., Ahmadi A., Salimian J., Shohani S., Jamalkandi S. A., Ghanei M. TNF-α−308 G/A variant and susceptibility to chronic obstructive pulmonary disease: a systematic review and meta-analysis. Cytokine . 2019;123 doi: 10.1016/j.cyto.2019.154763. [DOI] [PubMed] [Google Scholar]
- 96.Shi C., Zhao H. Association between tumor necrosis factor-308 G/A polymorphism and chronic obstructive pulmonary disease risk in Chinese population: evidence from a meta-analysis. Clinical Laboratory . 2019;65(10) doi: 10.7754/Clin.Lab.2019.190313. [DOI] [PubMed] [Google Scholar]
- 97.Van Durme Y. M. T. A., Eijgelsheim M., Joos G. F., et al. Hedgehog-interacting protein is a COPD susceptibility gene: the Rotterdam study. European Respiratory Journal . 2010;36(1):89–95. doi: 10.1183/09031936.00129509. [DOI] [PubMed] [Google Scholar]
- 98.Jakoby W. B. The GlutathioneS-transferases: a group of multifunctional detoxification proteins. Advances in Enzymology and Related Areas of Molecular Biology . 1978;46:383–414. doi: 10.1002/9780470122914.ch6. [DOI] [PubMed] [Google Scholar]
- 99.Wu W., Peden D., Sanchez D. D. Role of GSTM1 in resistance to lung inflammation. Free Radical Biology and Medicine . 2012;53(4):721–729. doi: 10.1016/j.freeradbiomed.2012.05.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Gwilt C., Donnelly L., Rogers D. The non-neuronal cholinergic system in the airways: an unappreciated regulatory role in pulmonary inflammation? Pharmacology & Therapeutics . 2007;115(2):208–222. doi: 10.1016/j.pharmthera.2007.05.007. [DOI] [PubMed] [Google Scholar]
- 101.Bierut L. J. Nicotine dependence and genetic variation in the nicotinic receptors. Drug and Alcohol Dependence . 2009;104 doi: 10.1016/j.drugalcdep.2009.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Wang J. C., Cruchaga C., Saccone N. L., et al. Risk for nicotine dependence and lung cancer is conferred by mRNA expression levels and amino acid change in CHRNA5. Human Molecular Genetics . 2009;18(16):3125–3135. doi: 10.1093/hmg/ddp231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Gosman M. M. E., Marike Boezen H., van Diemen C. C., et al. A disintegrin and metalloprotease 33 and chronic obstructive pulmonary disease pathophysiology. Thorax . 2007;62(3):242–247. doi: 10.1136/thx.2006.060988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Ju C.-R., Liu W., Chen R.-C. Serum surfactant protein D: biomarker of chronic obstructive pulmonary disease. Disease Markers . 2012;32(5):281–287. doi: 10.1155/2012/509063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Churg A., Dai J., Tai H., Xie C., Wright J. L. Tumor necrosis factor-α is central to acute cigarette smoke-induced inflammation and connective tissue breakdown. American Journal of Respiratory and Critical Care Medicine . 2002;166(6):849–854. doi: 10.1164/rccm.200202-097OC. [DOI] [PubMed] [Google Scholar]
- 106.Wilson A. G., Symons J. A., McDowell T. L., McDevitt H. O., Duff G. W. Effects of a polymorphism in the human tumor necrosis factor α promoter on transcriptional activation. Proceedings of the National Academy of Sciences . 1997;94(7):3195–3199. doi: 10.1073/pnas.94.7.3195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Wu M.-L., Layne M. D., Yet S.-F. Heme oxygenase-1 in environmental toxin-induced lung disease. Toxicology Mechanisms and Methods . 2012;22(5):323–329. doi: 10.3109/15376516.2012.666685. [DOI] [PubMed] [Google Scholar]
- 108.Shinohara T., Kaneko T., Nagashima Y., Ueda A., Tagawa A., shigatsubo Y. Adenovirus-mediated transfer and overexpression of heme oxygenase 1 cDNA in lungs attenuates elastase-induced pulmonary emphysema in mice. Human Gene Therapy . 2005;16(3):318–327. doi: 10.1089/hum.2005.16.318. [DOI] [PubMed] [Google Scholar]
- 109.Pepicelli C. V., Lewis P. M., McMahon A. P. Sonic hedgehog regulates branching morphogenesis in the mammalian lung. Current Biology . 1998;8(19):1083–1086. doi: 10.1016/s0960-9822(98)70446-4. [DOI] [PubMed] [Google Scholar]
- 110.Zhou X., Baron R. M., Hardin M., et al. Identification of a chronic obstructive pulmonary disease genetic determinant that regulates HHIP. Human Molecular Genetics . 2012;21(6):1325–1335. doi: 10.1093/hmg/ddr569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Daiger S. P., Schanfield M. S., Sforza L. L. C. Group-specific component (Gc) proteins bind vitamin D and 25-hydroxyvitamin D. Proceedings of the National Academy of Sciences . 1975;72(6):2076–2080. doi: 10.1073/pnas.72.6.2076. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Table 1: statistically significant variants from the other two GWASs. Supplementary Table 2: statistically nonsignificant gene-variant COPD overall associations. Supplementary Table 3: conflicting gene-variant COPD overall associations.
Data Availability Statement
All data generated or analyzed during this study are included in this published article and its supplementary information files.


