SUMMARY
A total of 46 strains of Salmonella isolated from patients with sporadic diarrhoea or involved in foodborne outbreaks were analysed by PCR for genus identification and serotyping. Subtyping was performed using pulsed-field gel electrophoresis (PFGE) and multiple amplification of phage locus typing (MAPLT) for seven variable loci. Bacteria were identified as belonging to serotype Enteritidis (33 strains; 71·7%) or Typhimurium (13 strains; 28·3%). A high similarity coefficient (94·6%) was observed in the Salmonella Enteritidis group for which were found three related PFGE profiles and only one MAPLT; strains representing profile PA/P1/MI were prevalent (27; 81·8%). Two Salmonella Typhimurium isolates were untypeable by PFGE. The remaining 11 strains had eight PFGE and three MAPLT profiles. The discriminatory power of MAPLT was lower than that of PFGE. Salmonella Enteritidis of clonal nature is predominant in Paraná State, with the most prevalent profile PA/P1/M1 associated with sporadic diarrhoea and with seven of nine reported outbreaks. In conclusion, PFGE shows higher discriminatory power among Salmonella strains.
Key words: MAPLT, molecular epidemiology, PFGE, Salmonella
INTRODUCTION
Salmonella enterica subspecies enterica is an important cause of foodborne diseases and a major cause of outbreaks and sporadic cases of diarrhoea in humans, representing a public health problem worldwide [1, 2]. It is estimated that Salmonella causes 93·8 million cases of gastroenteritis worldwide each year, with 155 000 deaths [1]. The diagnosis of Salmonella infection is critical for detecting and controlling the illness. Although most infections caused by Salmonella are self-limiting and resolve without treatment, some patients develop a severe illness requiring pathogen-specific diagnosis, which is also important for a disease surveillance programme. Furthermore, Salmonella diagnosis is essential in investigations of outbreaks and of the source of associated infection, to prevent the spread of illness [1, 3, 4].
Classically, Salmonella infection diagnosis is based on phenotypical methods such as culture, biochemical profiling and serotyping [5, 6], which requires several days. In the last years, PCR-based methods for identification of most clinically relevant Salmonella serotypes were developed [7–10]. These molecular methods may offer an effective alternative to the traditional serotyping enabling faster diagnosis; this new approach may have a significant impact on the control of outbreaks of foodborne illnesses [6, 11]. Pulsed-field gel electrophoresis (PFGE), based on whole-genome cleavage by restriction enzymes generating large DNA fragments that are separated in agarose gels using alternating electric fields at different angles, is considered the gold standard for Salmonella subtyping and continues to be the most widely used method in outbreak investigations [3, 6, 11]. Nonetheless, alternative methods have been developed for subtyping Salmonella isolates [6, 11]. Among them, the multiple amplification of phage locus typing (MAPLT), a PCR-based assay for amplification of genomic sequences from temperate phages in Salmonella genome [12]. The assay was tested with different sets of primers and showed good discriminatory power for Salmonella serotypes Typhimurium and Enteritidis as well as the potential use as a tool for epidemiological studies [12, 13]. The aim of this work was to identify and characterise Salmonella strains previously isolated from patients with diarrhoea and to compare the discriminatory power of MAPLT and PFGE for subtyping the isolates.
METHODS
Bacterial isolates
A total of 46 Salmonella strains isolated from faecal cultures from patients with diarrhoea and identified by biochemical profile [5] were analysed. Samples were provided by Central State Laboratory (LACEN-PR), and were from patients with sporadic cases of diarrhoea or involved in foodborne outbreaks that occurred during the period of June 2010 to May 2013 in several localities of the Paraná state, southern Brazil [13; our unpublished data]. Thirty of these strains were serotyped and identified as serotype Enteritidis [14].
Growth conditions
Bacteria were grown in MacConkey agar (Oxoid; Basingstoke, UK) incubated overnight at 35°C or Tryptic Soy Broth (TSB, Difco; Sparks MD, USA), at 37°C for 14–16 h for PFGE.
DNA extraction
Overnight cultures were used for DNA extraction using the boiling method.
Molecular identification of Salmonella
The PCR system developed by Alvarez et al. [7] modified for identification only of the genus Salmonella and serotypes Enteritidis and Typhimurium was used. The triplex PCR was performed in a final volume of 25 µl, containing buffer Maxima Hot Start Taq DNA Polymerase® 1×, 1·5 mM MgCl2, 0·2 mM dNTPs, 1U of Maxima Hot Start Taq DNA Polymerase (Fermentas Life Sciences; Vilnius, LT), 1 µl of DNA and 0·4 µM of each primer (Table 1). The cycling program was as follows: 1 cycle at 95°C (2 min), 30 cycles at 95°C (1 min), 57°C (1 min), 72°C (2 min), and a final extension at 72°C (5 min). PCR was performed in a Veriti® Thermal Cycler (Applied Biosystems®) and detection of products was by electrophoresis in 2% agarose gel stained with ethidium bromide and visualised under UV light. Escherichia coli aEPEC 258FA [14], and Salmonella serotypes Typhimurium, Livingstone, Saint Paul, Newport, Muenster, London, Enteritidis, Javiana, Panama, Derby and Weltevreden – previously serotyped at the Brazilian National Reference Laboratory Fundação Oswaldo Cruz – served as controls.
Table 1.
Primer sequences and amplicon sizes
| Salmonella genus and serotype identification | |||
|---|---|---|---|
| Primer | Sequence (5′→3′) | Amplicon (bp) | Reference |
| OMPC (genus) | F-ATCGCTGACTTATGCAATCG R-CGGGTTGCGTTATAGGTCTG |
204 | [7] |
| ENT (Enteritidis) | F-TGTGTTTTATCTGATGCAAGAGG R-TGAACTACGTTCGTTCTTCTGG |
304 | [7] |
| TYPH (Typhimurium) | F-TTGTTCACTTTTTACCCCTGAA R-CCCTGACAGCCGTTAGATATT |
401 | [7] |
| MAPLT subtyping | |||
| c1ST64B | F-TGTGTCGTTTGAGTGACTGCG R-TTCTAGGCTGGCTGACTGC |
548 | [13] |
| SB21ST64B | F-CTGTAT GGTTATATCGATTATCTGG R-GATTTCCTTTGCCCAGATGACG |
478 | [13, 15] |
| SB26ST64B | F-GACACCATCAATGTATGGATCGC R-AGGTTATCTATAATTCCGACCTGG |
477 | [12] |
| SB28ST64B | F-TGCAGTCAAGAGGACGTCC R-TGCCGATATGCTGATCTGGC |
589 | [12] |
| SB46ST64B | F-CATTGATGGTATCGAAGTTCGCC R-ATCTTATCGGCAGCCATACC |
401 | [13] |
| gtrAp22 | F-AGACCTTTCCGAATCCGCTG R-TAATTGCCGAGAAAGTGATAAGGG |
336 | [13] |
| gtrBp22 | F-CTTTCTCGGCAATTAGCCTG R-TTAGCCAGCACCATATCCGC |
423 | [13] |
PFGE
This procedure was performed according to the PulseNet protocol [16, 17]. XbaI (Fermentas Life Sciences; Vilnius, LT) was used as primary restriction enzyme, while AvrII (Thermo Scientific; Waltham, USA) was used only for strains presenting the same XbaI PFGE profile. DNA fragments were resolved using the system CHEF-DR® III (Bio-Rad, Hercules CA, USA) under the following conditions: 6 V/cm for 18:30 h at 14°C, and pulse time ranging from 2·2 to 63·8 s. Salmonella serotype Braenderup (ATCC® BAA-664™) was used as a DNA size marker. DNA was stained with ethidium bromide and visualised under UV light. Gel images were analysed in software BioNumerics 7·5 (Applied Maths, Keistraat, Belgium), the banding pattern was compared by cluster analysis using the Dice similarity coefficient and UPGMA (unweighted pair group method with arithmetic mean) to generate a dendrogram with 1·5% optimisation and band position tolerance of 1·5%. Only DNA fragments ranging from 20·5 to 1135 kb were included. PFGE profiles were considered distinct when any difference in a DNA band pattern was observed [3].
Multiple amplification of phage loci typing
This assay is based on the presence or absence of particular phage loci. A set of seven MAPLT primer pairs (Table 1) targeting prophage loci within the Salmonella genome were selected for subtyping of the isolates. These primers generated different MAPLT profiles with Salmonella Enteritidis or Salmonella Typhimurium and were capable of distinguishing the strains [12, 13, 15, 18]. The reagents and equipment used for PCR were the same described above. Primer pairs SB46ST64B and SB28ST64B were tested in a duplex PCR using the following cycling program: 1 cycle at 95°C (2 min), 30 cycles at 95°C (30 s), 58°C (1 min), 72°C (2 min), and a final extension at 72°C (5 min). Primer pairs gtrBp22, c1ST64B, SB21ST64B, and SB26ST64B were used in separated reactions under the same cycling conditions; for gtrAp22 the annealing temperature was 61°C (1 min).
Discriminatory index
The discriminatory ability (D) of PFGE was calculated according to the method of Hunter and Gaston [19].
RESULTS
Molecular identification of Salmonella
Results of PCR for identification of Salmonella indicated that all strains, including the isolates from patients with diarrhoea and the positive controls belonged to this genus, as expected. No amplification was observed for E. coli (not shown). On the other hand, among the Salmonella strains used as controls, PCR for molecular serotyping yielded positive results only for serotypes Enteritidis and Salmonella Typhimurium confirming the specificity of the primers. Among the 46 strains isolated from patients, 33 (71·7%) were characterised at the molecular level as belonging to serotype Enteritidis, including those previously identified using antisera [14]. The remaining 13 (28·3%) strains were identified as serotype Typhimurium (not shown).
PFGE profiling
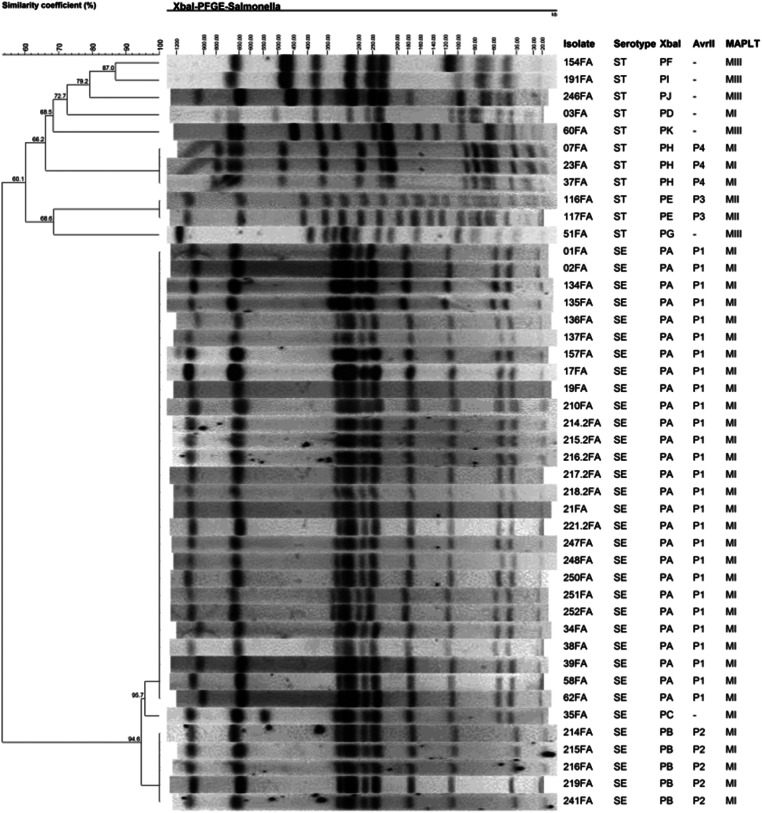
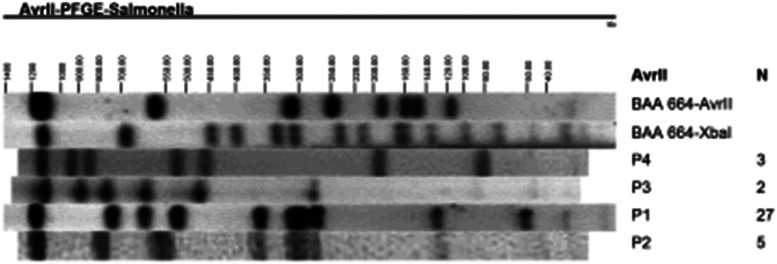
A total of 11 distinct PFGE profiles containing from 10 to 19 DNA fragments were observed and named PA to PK (Fig. 1). Strains of serotype Enteritidis had three related profiles (PA, PB and PC). PA was predominant, shared by 27 (81·8%) strains of this serotype; profile B, distinguished from PA by the absence of a fragment of approximately 54 kb, was detected in five (15·2%) strains, and only one strain (3·0%) had profile C which differed from PA by the presence of a band of approximately 550 kb (Fig. 1). The enzyme AvrII, used only for strains belonging to the same XbaI PFGE profile, separated Salmonella Enteritidis into two groups (P1 and P2; Fig. 2), confirming the results obtained with XbaI. This finding resulted in PFGE groups: PA/P1, PB/P2 and PC/–. Amongst the 13 strains of Salmonella Typhimurium two were untypeable, and the remaining 11 strains produced eight PFGE profiles (PD to PK). Profiles PH and PE contained three and two strains, respectively. The other strains showed unique PFGE profiles (Fig. 1). AvrII profiling indicated that all strains of PE belonged to profile P3 (PE/P3), and strains PH to profile P4 (PH/P4). The coefficient of genetic similarity was 94·6% for Salmonella Enteritidis and 60·1% for Salmonella Typhimurium (Fig. 1). A low discriminatory index (D = 0·32) of PFGE was found for Salmonella Enteritidis.
Fig. 1.
Dendrogram depicting genetic similarity of Salmonella isolates based on PFGE profiles of chromosomal DNA digested with XbaI. Results of PFGE of AvrII-digested DNA and MAPLT are also indicated. Strains belonging to MAPLT group MI were positive for all loci tested: SB46ST64B+, SB28ST64B+, gtrAp22+, gtrBp22+, c1ST64B+, SB21ST64B+, SB26ST64B+; MII: SB46ST64B+, c1ST64B+, SB21ST64B+; MIII: gtrAp22+, gtrBp22+. SE and ST denote respectively Salmonella Enteritidis and Salmonella Typhimurium.
Fig. 2.
PFGE profiles of AvrII-digested Salmonella isolates chromosomal DNA. PFGE profiles obtained with AvrII are indicated. N refers to number of strains exhibiting the profile.
Molecular profiling by MAPLT
The MAPLT results classified the strains into three groups named MI to MIII distinguished by the number and type of loci present (Fig. 1). All Salmonella Enteritidis strains belonged to profile MI, resulting in groups PA/P1/MI, PB/P2/MI and PC/–/MI. All 13 Salmonella Typhimurium strains were typeable by MAPLT; four strains had profile MI, two profile MII and seven had profile MIII.
The molecular profiling revealed two clusters of Salmonella Enteritidis, the largest of them containing 27 strains with the profile PA/P1/MI (Fig. 1). Among these, six were isolated from patients with sporadic diarrhoea and 20 from patients involved in six outbreaks of foodborne diseases that occurred between July 2011 and February 2013 (respectively, strains 134–137FA; 214·2–218·2 and 221·2FA; 248, 250–252FA; 01–02FA; 38–39FA; 17 and 21FA). Of note, the other strain (34FA) with profile PA/P1/MI was isolated from a foodborne outbreak that occurred in January 2011. From patients of this outbreak, strain 35FA was also isolated which exhibited profile PC/-/MI (Fig. 1). The PFGE profiles PA/P1/MI and PC/-/MI are related, differing by only one DNA band, while MAPLT was identical for these two strains. The second cluster of Salmonella Enteritidis contained strains 214-216FA, 219FA and 241FA (PB/P2/MI), isolated from patients from an outbreak that occurred in November 2010. Only one outbreak, in June 2010, was associated with Salmonella Typhimurium strains 116-117FA (PE/P3/MII; Fig. 1). All other Salmonella Typhimurium were isolated from sporadic cases of diarrhoea.
DISCUSSION
Salmonella is one of the most frequent bacterial causes of acute gastroenteritis, a major cause of morbidity and mortality worldwide, and an important public health concern [1, 2, 20, 21]. The development of rapid and sensitive methods for the detection and characterisation of Salmonella may have a significant impact on the disease burden caused by this pathogen [6].
Traditional serotyping is based on the identification of the somatic (O) and flagellar (H) antigens through bacterial agglutination with specific antisera [5, 6, 11]. Nonetheless, serotyping has several drawbacks, such as false-positive reactions resulting from weak, non-specific agglutination; untypeable isolates due to autoagglutination and loss of antigen expression in rough and immotile strains and the use of expensive antisera [6, 11]. Thus, alternative systems for serotyping Salmonella have been described. Alvarez et al. [7] developed a fast and reliable PCR system for detection of specific sequences of genus Salmonella, and from Salmonella serotypes 4,5,12:i:-, Enteritidis, Typhimurium, Typhimurium DT104 and U302, and Salmonella C2 serogroup. Considering that S. enterica are classified in more than 2500 serotypes, PCR serotyping methods are still limited in terms of the number of serotypes that can be identified. On the other hand, these assays allow for identification of the most common serotypes and several multiplex assays are available [6–9]. Furthermore, PCR-based identification of genera and the most frequent serotypes in each geographic region may enable fast and cost-effective diagnosis of Salmonella besides being a valuable support for epidemiological surveillance. This notion is especially important in developing countries and remote regions where the serotyping is not performed on a routine basis. In this study, three of the primer pairs proposed by Alvarez et al. [7] were selected, which allowed for detection of S. enterica and the most common serotypes. For all the 46 isolates, two bands of amplified DNA were observed, enabling their classification as Salmonella serotype Enteritidis (33, 71·7%) or Typhimurium (13, 28·3%; Fig. 1). The serotyping PCR identified these serotypes with high specificity and may be useful for low-resource laboratories. Salmonella Enteritidis or Typhimurium are the most frequently isolated serotypes in Brazil [14, 22–24] and in several other regions worldwide [4, 20, 21, 25–29]. Moreover, eight of nine outbreaks analysed were associated to Salmonella Enteritidis.
The predominance of this serotype in food-borne illness outbreaks was also observed in Brazil by other authors [14, 23, 30] and was also described in other countries [20, 31]. Salmonella serotyping is useful in a surveillance programme as a starting point for identification and source tracking of outbreaks, and enables worldwide comparisons [4, 6, 11, 32]. Nonetheless, it does not provide the level of discrimination required for outbreak investigations because it does not have enough capacity to fingerprint strains [6, 11]. Typing methods must be able not only to identify outbreak-associated strains but also to distinguish those from closely related but genetically distinct strains of the same serotype, requiring a high-resolution typing system to accomplish this task [18]. Several DNA-based fingerprinting methods have been used for subtyping bacteria. Due to the high discriminating capacity, robustness and reproducibility, PFGE is considered the gold standard for subtyping of foodborne bacterial pathogens [6, 16]. Nevertheless, PFGE is time-consuming and labour-intensive and does not show the same sensitivity for all serotypes [6]. MAPLT is a PCR-based methodology which detects prophage loci within Salmonella genome [12]. It is fast and easy to perform and does not require specialised equipment; this feature is an advantage over PFGE. Additionally, the use of a PCR-based methodology may be useful in outbreak investigations where the source must be readily identified [18].
When tested with phenotypically closely related but epidemiologically distinct Salmonella isolates, MAPLT has shown to provide good resolution with greater discriminative power in terms of the number of profiles in comparison with the profiles generated by PFGE [12, 13], or at least equal [18]. In the present study, we have selected seven phage loci amongst the most variable for Salmonella Enteritidis or Salmonella Typhimurium [12, 13, 15]. Although three PFGE profiles were observed for Salmonella Enteritidis, only one was obtained with MAPLT (Fig. 1) indicating that the assay was not able to distinguish these strains. For Salmonella Typhimurium, eight PFGE profiles were found compared with three by means of MAPLT (Fig. 1), also indicating that the latter assay has weaker discriminatory power than PFGE does. Often particular loci within an assay do not provide sufficient allelic diversity for maximum separation of isolates [18]; therefore, it is possible that a large number and/or a different set of MAPLT primers may enable better discrimination. By contrast, other PCR-based methods for Salmonella typing such as RAPD (randomly amplified polymorphic DNA) or VNTRs (variable number of tandem repeats) have a potential for use in low-resource settings and may be more discriminatory, thus deserving future research. Additionally, PFGE showed low discriminatory power (D = 0·32) for Salmonella Enteritidis with only three groups among the 33 strains suggesting high genetic homogeneity, with a similarity coefficient of 94·6% (Fig. 1). These results indicate that these strains are strongly related genetically. High similarity among Salmonella Enteritidis isolates was also observed in other studies [24, 33, 34], indicating the clonal nature of these bacteria. Strains with the PFGE profile PA (Fig. 1) were associated with seven of eight outbreaks caused by Salmonella Enteritidis. There is still few studies on molecular typing of Salmonella isolates in Brazil [35–38] but apparently the same PFGE profile was also the most common among Salmonella Enteritidis strains associated with foodborne outbreaks occurred in Rio Grande do Sul State, southern Brazil [24], suggesting the propagation of one clone of the bacteria in the south of the country. Interestingly, most Salmonella Enteritidis isolated from sporadic cases in Taiwan presented the PFGE profile apparently identical to PA found in this study [39]. The same pattern was also the most common pulsetype (SENTXB.0001) found in an international multi-centre study including nine European countries [40], and was also observed among Salmonella Enteritidis presenting PFGE pattern 9 (strains 8, 14 and 16) isolated in Turkey [27]. In another study of human and animal isolates performed in Pennsylvania, USA, PA corresponded to pattern JEGX01·0002 one of the 20 most common human Salmonella PFGE patterns and found only amongst human isolates. Additionally, Salmonella Typhimurium pattern PD apparently corresponds to JPXX01·0001 of the same study which was found among humans and animal isolates [41].
These data suggest that Salmonella Enteritidis strains that have the PA PFGE profile are widely disseminated around the world. Salmonella serotypes differed regarding the PFGE profiles and the level of genetic similarity (94·6% for Salmonella Enteritidis and 60·1% for Salmonella Typhimurium). Although subtyping by MAPLT was less discriminatory than PFGE, the technique is simple with a potential for rapid subtyping of Salmonella, especially in countries where funds are in short supply. The strains with profile PA/P1/MI were predominant, representing 81·8% of Salmonella Enteritidis, thus supporting the common occurrence worldwide of this subtype in both sporadic and outbreak diarrhoea.
ACKNOWLEDGEMENTS
This work was supported by Fundação Araucária and the Brazilian Program of National Institutes of Science and Technology – INCT/Brazilian Research Council – CNPq/MCT.
DECLARATION OF INTEREST
None.
REFERENCES
- 1.Majowicz SE, et al. The global burden of nontyphoidal Salmonella gastroenteritis. Clinical Infectious Diseases 2010; 50: 882–889. [DOI] [PubMed] [Google Scholar]
- 2.Scallan E, et al. Foodborne Illness acquired in the United States—Major Pathogens. Emerging Infectious Diseases 2011; 17: 7–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Barrett TJ, Gerner-Smidt P, Swaminathan B. Interpretation of pulsed-field gel electrophoresis patterns in foodborne disease investigations and surveillance. Foodborne Pathogens and Disease 2006; 3: 20–31. [DOI] [PubMed] [Google Scholar]
- 4.Herikstad H, Motarjemi Y, Tauxe RV. Salmonella surveillance: a global survey of public health serotyping. Epidemiology and Infection 2002; 129: 1–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bopp CA, et al. Escherichia, Salmonella and Shigella identification. In: Murray PR, et al. , eds. Manual of Clinical Microbiology. Washington: ASM Press, 2003, pp. 654–671. [Google Scholar]
- 6.Wattiau P, Boland C, Bertrand S. Methodologies for Salmonella enterica subsp. enterica subtyping: gold standards and alternatives. Applied and Environmental Microbiology 2011; 77: 7877–7885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Alvarez J, et al. Development of a multiplex PCR technique for detection and epidemiological typing of Salmonella in human clinical samples. Journal of Clinical Microbiology 2004; 42: 1734–1738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kim S, et al. Multiplex PCR-based method for identification of common clinical serotypes of Salmonella enterica subsp. enterica. Journal of Clinical Microbiology 2006; 44: 3608–3615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hong Y, et al. Rapid screening of Salmonella enterica serovars Enteritidis, Hadar, Heidelberg and Typhimurium using a serologically-correlative allelotyping PCR targeting the O and H antigen alleles. BMC Microbiology 2008; 8: 178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Leader BT, et al. High-throughput molecular determination of Salmonella enterica Serovars by use of multiplex PCR and capillary electrophoresis analysis. Journal of Clinical Microbiology 2009; 47: 1290–1299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Barco L, et al. Salmonella source attribution based on microbial subtyping. International Journal of Food Microbiology 2013; 16: 193–203. [DOI] [PubMed] [Google Scholar]
- 12.Ross IL, Heuzenroeder MW. Discrimination within phenotypically closely related definitive types of Salmonella enterica serovar Typhimurium by the multiple amplification of phage locus typing technique. Journal of Clinical Microbiology 2005; 43: 1604–1611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ross IL, Heuzenroeder WM. A comparison of two PCR-based typing methods with pulsed-field gel electrophoresis in Salmonella enterica serovar Enteritidis. International Journal of Medical Microbiology 2009; 299: 410–420. [DOI] [PubMed] [Google Scholar]
- 14.Assis FEA, et al. Impact of Aeromonas and diarrheagenic Escherichia coli screening in patients with diarrhea in Paraná, southern Brazil. The Journal of Infection in Developing Countries 2014; 8: 1609–1614. [DOI] [PubMed] [Google Scholar]
- 15.Ross IL, Parkinson IH, Heuzenroeder MW. The use of MAPLT and MLVA analyses of phenotypically closely related isolates of Salmonella enterica serovar Typhimurium. International Journal of Medical Microbiology 2009; 299: 37–41. [DOI] [PubMed] [Google Scholar]
- 16.Ribot EM, et al. Standardization of pulsed-field gel electrophoresis protocols for the subtyping of Escherichia coli O157:H7, Salmonella, and Shigella for PulseNet. Foodborne Pathogens and Disease 2006; 3: 59–67. [DOI] [PubMed] [Google Scholar]
- 17.PNL05. Standard operating procedure for PulseNet PFGE of Escherichia coli O157:H7, Escherichia coli non-O157 (STEC), Salmonella serotypes, Shigella sonnei and Shigella flexneri. In: PNL05 last updated April 2013: Centers for Disease Control and Prevention (CDC), 2013.
- 18.Ross IL, Young CC, Heuzenroeder MW. New options for rapid typing of Salmonella enterica Serovars for outbreak investigation. In: Kumar Y, ed. Salmonella – A Diversified Superbug. InTech, 2012, pp. 523–541. [Google Scholar]
- 19.Hunter PR, Gaston MA. Numerical index of the discriminatory ability of typing systems: an application of Simpson's index of diversity. Journal of Clinical Microbiology 1988; 26: 2465–2466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Centers for Disease Control and Prevention (CDC). Surveillance for Foodborne Disease Outbreaks United States, 2013: Annual Report. Atlanta, Georgia: US Department of Health and Human Services. Centers for Disease Control and Prevention (CDC), 2015. [Google Scholar]
- 21.EFSA, ECDC (European Food Safety Authority and European Centre for Disease Prevention and Control). The European Union summary report on trends and sources of zoonoses, zoonotic agents and food-borne outbreaks in 2013. European Food Safety Authority Journal 2015; 13: 3991, 165 pp. doi: 10.2903/j.efsa.2015.3991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Fernandes SA, et al. Salmonella serovars isolated from humans in Sao Paulo state, Brazil, 1996–2003. Revista do Instituto de Medicina Tropical de São Paulo 2006; 48: 179–184. [DOI] [PubMed] [Google Scholar]
- 23.Mürmann L, et al. Quantification and molecular characterization of Salmonella isolated from food samples involved in salmonellosis outbreaks in Rio Grande do Sul, Brazil. Brazilian Journal of Microbiology 2008; 39: 529–534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Oliveira FA, et al. Characterization of Salmonella Enteritidis isolated from human samples. Food Research International 2012; 45: 1000–1003. [Google Scholar]
- 25.Jackson BR, et al. Outbreak-associated Salmonella enterica serotypes and food commodities, United States, 1998–2008. Emerging Infectious Diseases 2013; 19: 1239–1244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hendriksen RS, et al. Global monitoring of Salmonella serovar distribution from the world health organization global foodborne infections network country data Bank: results of quality assured laboratories from 2001 to 2007. Foodborne Pathogens and Disease 2011; 8: 887–900. [DOI] [PubMed] [Google Scholar]
- 27.Ozdemir K, Acar S. Plasmid profile and pulsed-field gel electrophoresis analysis of Salmonella enterica isolates from humans in Turkey. PLoS ONE 2014; 9: e95976. doi: 10.1371/journal.pone.0095976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Centers for Disease Control and Prevention (CDC). National Salmonella Surveillance Annual Report, 2012. Atlanta, Georgia: US Department of Health and Human Services. Centers for Disease Control and Prevention, CDC, 2014. [Google Scholar]
- 29.Ke B, et al. Serovar distribution, antimicrobial resistance profiles, and PFGE typing of Salmonella enterica strains isolated from 2007–2012 in Guangdong, China. BMC Infectious Diseases 2014; 14: 338. doi: 10.1186/1471-2334-14-338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Souza RB, et al. Ciprofloxacin susceptibility reduction of Salmonella strains isolated from outbreaks. Brazilian Journal of Microbiology 2010; 41: 497–500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Greig JD, Ravel A. Analysis of foodborne outbreak data reported internationally for source attribution. International Journal of Food Microbiology 2009; 130: 77–87. [DOI] [PubMed] [Google Scholar]
- 32.Ranieri ML, et al. Comparison of typing methods with a new procedure based on sequence characterization for Salmonella serovar prediction. Journal of Clinical Microbiology 2013; 51: 1786–1797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Thong KL, et al. Molecular analysis of Salmonella enteritidis by pulsed-field gel electrophoresis and ribotyping. Journal of Clinical Microbiology 1995; 33: 1070–1074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ngoi ST, Thong KL. Molecular characterization showed limited genetic diversity among Salmonella Enteritidis isolated from humans and animals in Malaysia. Diagnostic Microbiology and Infectious Disease 2013; 77: 304–311. [DOI] [PubMed] [Google Scholar]
- 35.Ghilardi AC, Tavechio AT, Fernandes SA. Antimicrobial susceptibility, phage types, and pulse types of Salmonella Typhimurium, in Sao Paulo, Brazil. Memórias do Instituto Oswaldo Cruz 2006; 101: 281–286. [DOI] [PubMed] [Google Scholar]
- 36.Oliveira FA, et al. Clonal relationship among Salmonella enterica serovar Enteritidis involved in foodborne outbreaks in Southern Brazil. Food Control 2009; 20: 606–610. [Google Scholar]
- 37.Kottwitz LBM, et al. Molecular characterization and resistance profile of Salmonella Enteritidis PT4 and PT9 strains isolated in Brazil. Journal of Medical Microbiology 2011; 60: 1026–1031. [DOI] [PubMed] [Google Scholar]
- 38.Campioni F, Moratto Bergamini AM, Falcao JP. Genetic diversity, virulence genes and antimicrobial resistance of Salmonella Enteritidis isolated from food and humans over a 24-year period in Brazil. Food Microbiology 2012; 32: 254–264. [DOI] [PubMed] [Google Scholar]
- 39.Pang JC, et al. Pulsed-field gel electrophoresis, plasmid profiles and phage types for the human isolates of Salmonella enterica serovar Enteritidis obtained over 13° years in Taiwan. Journal of Applied Microbiology 2005; 99: 1472–1483. [DOI] [PubMed] [Google Scholar]
- 40.Gatto A, et al. Distribution of molecular subtypes within Salmonella enterica serotype Enteritidis phage type 4 and S. Typhimurium definitive phage type 104 in nine European countries, 2000–2004: results of an international multi-centre study. Epidemiology and Infection 2006; 134: 729–736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sandt CH, et al. A comparison of non-typhoidal Salmonella from humans and food animals using pulsed-field gel electrophoresis and antimicrobial susceptibility patterns. PLoS ONE 2013; 8: e77836. doi: 10.1371/journal.pone.0077836. [DOI] [PMC free article] [PubMed] [Google Scholar]