SUMMARY
During May 2015, an increase in Salmonella Agona cases was reported from western Sydney, Australia. We examine the public health actions used to investigate and control this increase. A descriptive case-series investigation was conducted. Six outbreak cases were identified; all had consumed cooked tuna sushi rolls purchased within a western Sydney shopping complex. Onset of illness for outbreak cases occurred between 7 April and 24 May 2015. Salmonella was isolated from food samples collected from the implicated premise and a prohibition order issued. No further cases were identified following this action. Whole genome sequence (WGS) analysis was performed on isolates recovered during this investigation, with additional S. Agona isolates from sporadic-clinical cases and routine food sampling in New South Wales, January to July 2015. Clinical isolates of outbreak cases were indistinguishable from food isolates collected from the implicated sushi outlet. Five additional clinical isolates not originally considered to be linked to the outbreak were genomically similar to outbreak isolates, indicating the point-source contamination may have started before routine surveillance identified an increase. This investigation demonstrated the value of genomics-guided public health action, where near real-time WGS enhanced the resolution of the epidemiological investigation.
Key words: Outbreak, public health action, Salmonella Agona, sushi, whole genome sequencing
INTRODUCTION
Salmonellosis is a zoonotic disease which remains the second most commonly notified gastrointestinal disease in Australia, following campylobacteriosis [1]. Transmission to humans occurs via the consumption of raw or inadequately cooked foods of animal origin, or by food items and untreated water contaminated with infected faeces from a reservoir host [1–3]. Salmonella enterica subsp. enterica serovar Agona (Salmonella Agona) is recognised as a common global cause of salmonellosis in both animals and humans [4], and is a contaminant of farmed livestock and vegetables [5–7], and factory-prepared foods [8–11]. S. Agona has been ranked among the top 15 most frequent Salmonella serotypes in North America, seventh in Europe, and 15th in Australia during 2006 [4, 12]. Outbreaks of S. Agona have occurred in Ireland [8], UK [13, 14], USA [14–17], Israel [18], Finland [19], France [10], Germany [20] and Australia [21], with some sources of S. Agona identified as chickens [22, 23], cattle [24], Peruvian fishmeal [25], dry unsweetened cereals [16] and powered infant formula [10].
Whole genome sequencing (WGS) of foodborne bacteria of public health importance provides the ability to distinguish between isolates of interest within an individual outbreak [26, 27]. Enhanced management of outbreaks using WGS helps to confirm the contamination source, characterize transmission events using near real-time methodology and increased quality control of data [26, 27]. Along with the comprehensive genetic information provided by WGS, comes additional information regarding antibiotic resistance, virulence determinants and genome evolution [28]. As technology becomes less cost-prohibitive and turn-around times are reduced, WGS is expected to benefit communicable disease surveillance and outbreak investigations of foodborne disease [26, 29, 30]. It is essential, as the application of WGS advances quickly, that the public health community understands the benefits of the analytical methods used to explore their data [31].
Salmonellosis is notifiable within New South Wales (NSW) [32, 33], with records aggregated within the Notifiable Conditions Information Management System (NCIMS). Salmonella isolates from public and private pathology providers, and the NSW Department of Primary Industries (DPI) are routinely referred to the NSW Enteric Reference Laboratory, Centre for Infectious Diseases and Microbiology Laboratory Services for serotyping using the White-Kauffmann-Le Minor scheme (9th Ed 2007) [27]. During routine surveillance, the NSW Communicable Disease Branch identified nine cases of S. Agona in May 2015, all clustered geographically in western Sydney. This report examines the epidemiological and molecular investigation of an increase in S. Agona cases in NSW between January and July, 2015.
METHODS
Epidemiological investigation of the outbreak
A retrospective review of notifications in NCIMS was conducted, with de-identified data extracted for laboratory-confirmed S. Agona cases diagnosed within NSW between 1 January 2011 and 31 December 2014. Extracted variables included notification date, onset date and jurisdiction of residence. Average number of cases per year and 95% confidence intervals (95% CI) were calculated for the entire NSW population, with NSW population estimates obtained from NSW Health.
A descriptive case-series investigation was conducted on all notified cases of S. Agona, residing in the western Sydney area, and with a notification date of salmonellosis between March and June 2015. Telephone interviews of cases or surrogates (either a parent or guardian when the case was a child aged <16 years) were conducted using a standardised Salmonella questionnaire. Questions covered case demographics and illness, food consumption history, travel and environmental exposures 7 days prior to the onset of disease.
Food and environmental investigation of the outbreak
Based on the responses of interviewed cases, the NSW DPI conducted investigations at two separate sushi premises (Sushi-A and Sushi-B), both operating within one large shopping complex in western Sydney. The NSW DPI inspected both Sushi-A and Sushi-B on 17 June 2015, with follow-up inspections conducted at Sushi-B. Food and environmental samples were collected during each inspection and sent to the NSW Enteric Reference Laboratory for Salmonella identification and further molecular typing of isolates. Management and staff members at both premises were questioned by the NSW DPI during these inspections, regarding food preparation techniques, cleaning processes and staff illnesses.
Retrospective WGS analysis
A retrospective WGS analysis was performed on clinical and food isolates recovered during this descriptive cases-series investigation, along with additional S. Agona isolates from sporadic-clinical cases and routine food samples (collected by the NSW DPI) in NSW, during the period January to July 2015. The genomic deoxyribonucleic acid (DNA) was prepared using an automated workstation Chemagic Prepito-D (PerkinElmer). The DNA sequencing libraries were prepared using the Nextera XT DNA preparation kit (Illumina) and the sequencing was performed on the NextSeq 500 (Illumina) with 2 × 150 base-pairs (bp) paired-end chemistry. Sequencing data quality was checked by FastQC v1.0.0 (BaseSpace Labs, Illumina). Single nucleotide polymorphisms (SNP) were identified using CLC Genomics Workbench v8·0 (Qiagen) by mapping reads to the reference genome S. Agona 460004 2–1 (GenBank: NZ_CP011259). The significant thresholds for SNP calling were set for a minimum coverage at 20 and a minimum variant frequency at 80%.
The generated SNP list from each of the isolates were exported as Excel files and a binary type SNP matrix representing presence and absence of SNP in each isolates was generated. The clustering analysis was performed using a categorical coefficient with a UPGMA (unweighted pair group method with arithmetic mean) in BioNumerics (Applied Maths). To validate the clustering analysis, de novo assembly was performed in the CLC Genomics Workbench with a minimum contig-size set at 200 bp. The assembly was then mapped against the reference genome for SNP calling and phylogeny inferred using a web-based server (https://cge.cbs.dtu.dk/services/CSIPhylogeny/). Maximum likelihood trees at 100 bootstraps were generated from SNP fasta file using MEGA-6 [34].
RESULTS
Progression of the outbreak
Historically, an annual average of 28·3 cases of S. Agona (N = 113; 95% CI 17·2–39·3) were notified in NSW between 1 January 2011 and 31 December 2014. Within the epidemiological investigation period (March–June 2015), 19 cases residing in the greater western Sydney area were notified, and were included in the descriptive case-series investigation. Sixteen of these 19 clinical cases (84%) were contactable for an interview, of which, six had consumed sushi that had been purchased from one of two different outlets within a large shopping complex in western Sydney. Onset of illness of these six outbreak cases occurred between 7 April and 24 May 2015. These six outbreak cases had a median age of 8 years (age range 3–45 years), with five reporting symptoms of diarrhoea (median duration of 5·5 days: range 2–8 days), and four with bloody stools. Two cases, both aged <14 years, presented to the emergency department with one case hospitalised for three days due to the severity of symptoms.
Of the remaining 10 clinical cases of the descriptive case-series investigation, no common exposures were identified during interviews. Salmonellosis experienced by a mother and her new-born baby were possibly hospital related; two unrelated cases had recently returned home from international travel (Vanuatu and Thailand); two regularly attended child-care; and one was a resident of an aged care facility. No high-risk exposures were identified for three cases. Repeated attempts to contact the remaining three cases for an interview were unsuccessful.
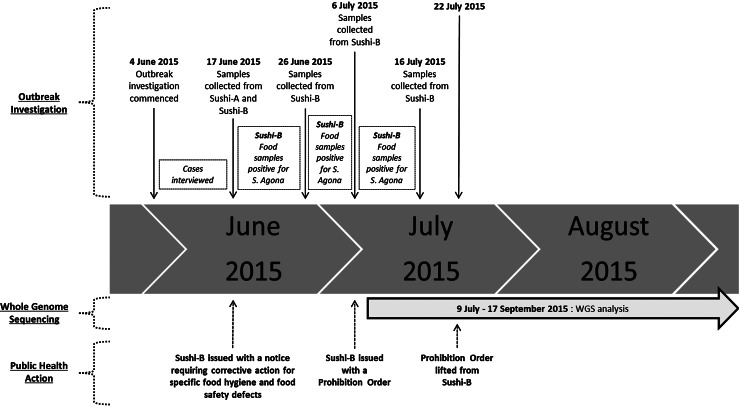
On 17 June 2015, food and environmental samples were collected from two different sushi venues (Sushi-A and Sushi-B) operating within a large shopping complex in western Sydney, as both were implicated by the cases. Samples from both Sushi-A and Sushi-B were representative of the complete sushi handling process, including raw ingredients and their storage containers, prepared cooked ingredients and their storage containers, and the final sushi product for consumption. Salmonella was not detected from any of the food or environmental samples collected from Sushi A. Cooked tuna sushi rolls from Sushi-B were found positive for S. Agona culture, while the individual ingredients of the sushi rolls and environmental samples were each negative (Table 1). Due to specific food hygiene and food safety defects identified, Sushi-B was issued with a notice requiring corrective action. On 6 July 2015, a Prohibition Order was issued (immediately following the availability of laboratory results) to Sushi-B, prohibiting the handling or production of cooked tuna sushi rolls intended for sale on the premises (Fig. 1).
Table 1.
Food and environmental samples collected by the NSW Department of Primary Industries (DPI) from Sushi-B between 17 June and 16 July 2015
| Date | Samples collected | Sample type | Test result (S = satisfactory, U = unsatisfactory) |
|---|---|---|---|
| 17·06·15 | Total of 15 environmental samples | Environmental | S |
| Total of 12 food samples (including raw whole shell eggs) | Food | S | |
| 2 × cooked tuna sushi rolls (cucumber + avocado) | Food | U (S. Agona detected) | |
| Raw chicken pieces | Food | U (S. Kiambu detected) | |
| 26·06·15 | Total of 16 environmental samples | Environmental | S |
| Total of 11 food samples | Food | S | |
| Cooked tuna and avocado sushi roll | Food | U (S. Agona detected) | |
| Cooked tuna and cucumber sushi roll | Food | U (S. Agona detected) | |
| Cooked tuna mix (tuna + mayonnaise + sugar) | Food | U (S. Agona detected) | |
| 06·07·15 | Total of 5 environmental samples | Environmental | S |
| Total of 11 food samples (including opened mayonnaise) | Food | S | |
| Cooked tuna and avocado sushi roll | Food | U (S. Agona detected) | |
| Cooked tuna mix (tuna + mayonnaise + sugar) – large container | Food | U (S. Agona detected) | |
| Cooked tuna mix (tuna + mayonnaise + sugar) – small container | Food | U (S. Agona detected) | |
| 16·07·15 | Multiple environmental samples | Environmental | S |
| Total of 22 food samples (including all ingredients for new trial batch of cooked tuna sushi rolls) | Food | S |
Fig. 1.
Time line of the outbreak investigation, retrospective WGS analysis and public health action.
It was revealed that a staff-member at Sushi-B had been ill during early May 2015, with symptoms including vomiting and diarrhoea whilst at home on a rostered day-off; no stool sample was collected for testing. This staff member, when questioned by the NSW DPI, confirmed that they had consumed sushi rolls while at work between 28 and 30 April 2015, as staff members at Sushi-B are entitled to one sushi roll per shift. This staff member could not recall which varieties of sushi were eaten on those days but indicated that it could have been cooked tuna, salmon or chicken. Daily work duties for this staff member included preparing sushi rolls at the designated preparation bench and serving customers.
Whole genome sequencing
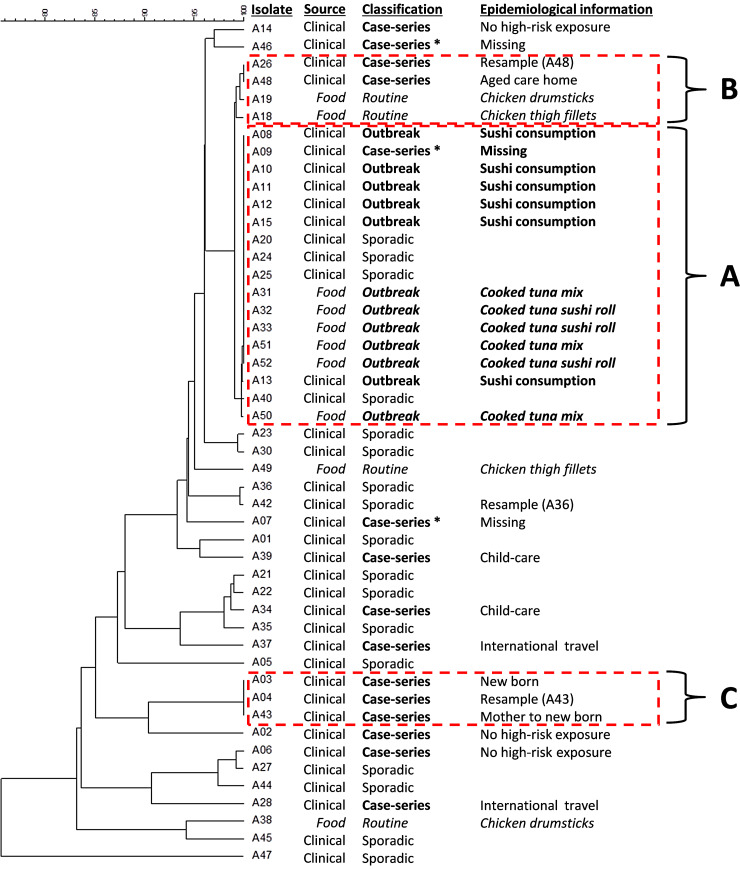
Forty-eight isolates serotyped as S. Agona and referred to the NSW Enteric Reference Laboratory between January and July 2015 were subjected to WGS. These included 38 clinical isolates (from 35 notified cases) and 10 food isolates. The 38 clinical isolates encompassed 21 clinical isolates collected during the descriptive case-series investigation (from the 19 S. Agona notifications of residents in the western Sydney area, March–June 2015, and included two repeat samples) and 17 sporadic-clinical isolates (from S. Agona notifications in the wider region of NSW, January–July 2015, and included one repeat sample). Based on the epidemiological results of this investigation, six of the 21 clinical isolates collected during the descriptive case-series investigation were classified as outbreak isolates (recovered from people who had consumed sushi purchased from within a large shopping complex in western Sydney). Of the 10 food isolates, six were recovered from samples collected from Sushi-B, March–July 2015, and four were collected as part of routine survey work by the NSW DPI, January–March 2015 (Fig. 2).
Fig. 2.
Maximum likelihood tree of S. Agona isolates based on SNP. * representing clinical cases who were originally included in the descriptive case-series investigation and non-contactable for an interview during this investigation.
The maximum likelihood phylogenetic tree of S. Agona, based on the SNP analysis of these 48 isolates, grouped 17 isolates together with a 0–1 SNP variation across clustered genomes (Fig. 2). These included the six outbreak isolates, and six food isolates collected from Sushi-B during the outbreak (Cluster-A, Fig. 2). Also within Cluster-A were four sporadic-clinical isolates collected in NSW between January and March 2015, and a single clinical isolate recovered from a non-contactable individual who was originally included in the descriptive case-series investigation. All clinical isolates within Cluster-A had similar genetic relatedness to food isolates recovered from Sushi-B during this outbreak (Fig. 2).
Two isolates of S. Agona obtained from raw retail chicken samples (collected between February and March 2015) and re-sampled isolates collected from an elderly individual at an aged care home (date of onset 3 April 2015) (Cluster-B, Fig. 2) shared similar genomes with the outbreak isolates (Cluster-A, Fig. 2). The SNP variation between isolates of Cluster-A and Cluster-B did not exceed 4–8 bp across the entire genome. A mother (date of onset 25 March 2015) and her new-born (date of onset 22 March 2015), who were not related to the outbreak, had indistinguishable S. Agona genomes by SNP analysis (Cluster-C, Fig. 2). S. Agona recovered from cases with overseas travel histories were genetically unrelated to the outbreak isolates of Cluster-A (Fig. 2).
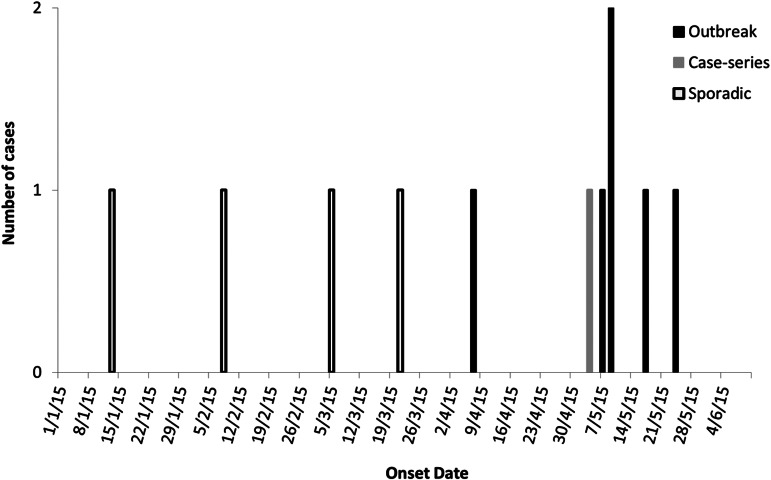
In total, 11 clinical isolates were grouped within Cluster-A by genetic analysis. This included (a) six outbreak isolates; (b) a single clinical isolate recovered from a non-contactable individual who was part of the descriptive case-series investigation; and (c) four sporadic-clinical isolates from cases that were notified outside the epidemiological investigation period and/or area. Common exposure to contaminated foods was confirmed between 7 April and 24 May 2015 during the epidemiological investigation of the outbreak. However, as indicated by the grouping of sporadic-clinical isolates with similar genetic patterns to the cases of the outbreak (Cluster-A), the duration of this point-source contamination issue at Sushi-B cannot be confirmed without supporting epidemiological information (Fig. 3).
Fig. 3.
Epidemiological curve of S. Agona cases (Cluster-A) identified from NSW, Australia with an onset date between January and June 2015.
DISCUSSION
Rapid epidemiological investigation identified contaminated cooked tuna sushi rolls consumed from an individual sushi outlet in western Sydney as the likely vehicle for this outbreak. Once the food outlet had been identified, our multidisciplinary team promptly detected the contamination issue and instigated action to eliminate the ongoing risk to the community. We highlight the public health importance of WGS, as well as the additional value that this modern technology provides. The WGS analysis identified five additional clinical isolates that may have been linked to the outbreak (one clinical isolate recovered from an individual who was originally included in the descriptive case-series investigation and was not contactable for an interview; and four sporadic-clinical isolates). It was speculated that the original contamination-source for this outbreak could have been raw retail chicken meat, based on the limited SNP variation between the isolates of the outbreak (Cluster-A) and the routinely collected chicken meat samples in Cluster-B (Fig. 2), with the timing of this cross-contamination event remaining uncertain.
The WGS offers increased discriminative power and enhances traditional descriptive epidemiology with detailed phylogenetic relationships between isolates [35]. During this particular outbreak investigation, the SNP analysis, which included clinical isolates that were not originally considered to be part of the outbreak, may have increased the total outbreak size by 183%. However, it is worth noting that the ‘threshold’ for the maximum SNP differences of Salmonella isolates related to an outbreak cluster remains not well defined [27]. Previous WGS reports of Salmonella outbreaks have had SNP differences of up to 12–30-bp to help define a cluster [27, 29, 35]. Using a similar threshold, our outbreak would have included an elderly individual from the aged care home (Cluster-B, Fig. 2), who lacked supporting epidemiological information to link them to this outbreak at Sushi-B. WGS data should not be used in isolation to define an outbreak, and should be used to compliment traditional epidemiological data [29].
The SNP variation of S. Agona isolates obtained from raw retail chicken meat routinely sampled by the NSW DPI during 2015 (Cluster-B, Fig. 2) and our outbreak (Cluster-A, Fig. 2) was between 4-bp and 8-bp over an entire genome of over 4 million bp [17]. The low number of SNP variation between our outbreak isolates and strains recovered from the raw chicken meat suggests that these different isolates may share a common source of origin, such as a single producer or processor of chicken meat. In light of the specific food hygiene and food safety defects identified at Sushi-B by the NSW DPI, we hypothesise that there could have been a breakdown of proper hygiene protocol at the implicated sushi outlet while processing contaminated raw chicken meat. This breakdown of proper hygiene resulted in the cross-contamination of the cooked tuna mix and/or containers used exclusively for storage of this product. S. Agona is commonly associated with poultry and poultry products [5], and has a considerable ability to form biofilms on plastic materials [11]. The resistant and persistent nature of Salmonella within biofilms [36] may provide some insight for the extended duration of this localised contamination issue at the sushi outlet. Another possible source for cross-contamination could have been an ill staff-member at Sushi-B. However, without supporting clinical or microbiological evidence from this ill staff-member, it remains uncertain whether this staff member was the source of contamination at Sushi-B, or an additional undiagnosed case linked to this outbreak.
Based on epidemiological results, the duration of this point-source contamination event at Sushi-B was reported to be between 7 April and 24 May 2015. However, as indicated by the clustering of sporadic-clinical isolates within Cluster-A (Fig. 2), this point-source contamination issue may have started earlier than first suspected (Fig. 3). This hypothesis was unable to be investigated further during the investigation, as these additional cases linked to the outbreak by WGS were notified with S. Agona between 6 and 8 months prior to the WGS-based analysis. Any epidemiological information obtained from these additional cases at the time of this outbreak investigation would have been unreliable due to poor recall of food consumption more than half a year ago. If these additional cases had been interviewed at the time of their illness (as all notified cases of Salmonella are not routinely interviewed by NSW Health), it may have been possible to link them to the outbreak at Sushi-B, or obtain additional information to help identify the original source that led to the cross-contamination event at Sushi-B.
The probability of a Salmonella infection to the consumer from contaminated foods is related to the infectious dose ingested by the person. It is estimated that a person is required to ingest 100–1000 bacteria to initiate an infection, with a 10–20% chance of infection related to 100 organisms (model-based prediction) and a 60–80% chance related to 10 000-fold increase of organisms [3]; furthermore, PPIs (Proton Pump Inhibitors) appear to increase the susceptibility of an individual to Salmonella [37]. During this outbreak, the low number of cases identified may have been a result of a highly variable organism counts within the contaminated cooked tuna mix. It is possible that following the original cross-contamination event, the infrequent number of notified cases could have been related to mixing and/or diluting of cooked tuna mix between batches; from inadequate cleaning and sanitising of storage containers used exclusively for the cooked tuna mix; or from mild illness where the patient did not seek medical attention.
Pulse-field gel electrophoresis (PFGE) and multilocus variable-number tandem-repeat analysis (MLVA) have been commonly used for analysis of Salmonella outbreaks [29]. PFGE has been the ‘gold standard’, with high discriminatory power and established guidelines for interpreting banding patterns; however, remains relatively slow, labour intensive and produces results that are subjective to interpretation [26, 29]. When available for individual serotypes, MLVA is fast, high-throughput, highly discriminatory and provides results that can be harmonised between laboratories [26, 27, 29]. While MLVA has become the standard method for strain typing of Salmonella Typhimurium in Australia, there is no established MLVA scheme for less common serovars such as S. Agona. In contrast, WGS provides ultimate discriminatory power and an additional ability to infer the timeline of an outbreak, with the potential to replace routine strain typing methods in the future.
During this outbreak investigation, we demonstrated the significant potential for WGS to assist with routine epidemiological investigations, particularly with regards to the prevention of further cases through timely identification of the outbreak source (assuming WGS was routine) and important insights into the contamination timeline. This will fundamentally change the epidemiological understanding of transmission dynamics, and open up new, more targeted strategies for disease control and prevention [26]. Even for this investigation where the final results for the WGS analytical component were not available until 8 weeks following the lifting of the prohibition order at Sushi-B (Fig. 1), the information has been valuable in better understanding the epidemiology of the outbreak. Use of this technology will become more applicable to real-time investigations in the future as speed of sequencing of bacterial genomes improves from weeks and days to perhaps hours [29, 38].
There are significant challenges associated with application of WGS to public health surveillance. Standardisation of testing and reporting, data sharing and governance, and the bioinformatics infrastructure capable of handling large volumes of data are among the most urgent ones [26]. Costs are also an issue when considering the inclusion of WGS as part of outbreak investigations (especially when WGS costs are additional to the costs of conventional laboratory methods), and are not necessarily affordable for routine use. In time, WGS has the potential to allow epidemiological hypotheses to be tested in near real-time, supporting enhanced management and control of communicable diseases and effective public health action [26]. With declining costs, improved speed of WGS and minimising the number of laboratory tests conducted, this modern technology will have a more permanent role within routine surveillance of notifiable diseases, with high-quality data influencing decision-making and action during an outbreak investigation.
Limitations of this outbreak investigation should be noted. They included poor recall of some cases and surrogates during interviews, a low sample size for the epidemiological investigation, and the descriptive nature of the epidemiological study. This descriptive study measured the occurrences of outcomes rather than incorporating an analytical design that could investigate associations between exposures and outcomes. WGS analysis was completed after the outbreak was concluded, and it was not ideal to follow-up the additional cases within Cluster-A due to the time that had lapsed. Also, due to the passive nature of the notification surveillance system in NSW, this investigation was biased towards people with more severe illness, who were more likely to be tested and had a positive culture. In spite of these limitations, our findings present strong evidence of a traditional epidemiological investigation supported by high-throughput sequencing technology, which implicated a single sushi outlet as the source of this outbreak, and assisted with the hypothesis that this outbreak was caused by cross contamination from raw chicken.
CONCLUSION
This multidisciplinary outbreak investigation was responsible for correcting various hygiene and food safety control defects identified at a sushi outlet. This outlet was responsible for a point-source foodborne outbreak of S. Agona in western Sydney, Australia between April and June 2015. The high-throughput genome sequencing was also utilised retrospectively and identified additional cases that may have been linked to the outbreak, extending the timeframe of the outbreak and providing additional insight as to the possible source for the outbreak. The genomics-enhanced public health action added value to the descriptive epidemiological investigation and will most certainly play an increasing role in public health surveillance of foodborne diseases, especially as costs of this technology becomes more acceptable and turn-around times of testing are reduced.
ACKNOWLEDGEMENTS
The authors thank Alan Edwards (NSW DPI) and John Shields (NSW DPI) who also assisted in the outbreak investigation. This work was completed while C.K.T. was a Master of Applied Epidemiology (MAE) scholar at the National Centre for Immunisation Research and Surveillance of Vaccine Preventable Diseases, at the The Children's Hospital at Westmead, and the National Centre for Epidemiology and Population Health, at The Australian National University. The National Centre for Immunisation Research and Surveillance of Vaccine Preventable Diseases is supported by the Australian Government Department of Health, the NSW Ministry of Health and The Children's Hospital at Westmead. The opinions expressed in this paper are those of the authors, and do not necessarily represent the views of these agencies.
DECLARATION OF INTEREST
None.
REFERENCES
- 1.Fearnley E, et al. Salmonella in chicken meat, eggs and humans; Adelaide, South Australia, 2008. International Journal of Food Microbiology 2011; 146: 219–227. [DOI] [PubMed] [Google Scholar]
- 2.Ashbolt R, Kirk M. Salmonella Mississippi infections in Tasmania: the role of native Australian animals and untreated drinking water. Epidemiology and Infection 2006; 134: 1257–1265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Heyman D. Control of Communicable Diseases Manual, 20th edn. Washington, DC: American Public Health Association Press, 2015. [Google Scholar]
- 4.Hendriksen RS, et al. Global monitoring of Salmonella serovar distribution from the World Health Organization global foodborne infections network country data bank: results of quality assured laboratories from 2001 to 2007. Foodborne Pathogens and Disease 2011; 8: 887–900. [DOI] [PubMed] [Google Scholar]
- 5.Dione MM, Geerts S, Antonio M. Characterisation of novel strains of multiply antibiotic-resistant Salmonella recovered from poultry in Southern Senegal. The Journal of Infection in Developing Countries 2011; 6: 436–442. [DOI] [PubMed] [Google Scholar]
- 6.Haley C, et al. Salmonella prevalence and antimicrobial susceptibility from the national animal health monitoring system swine 2000 and 2006 studies. Journal of Food Protection 2012; 75: 428–436. [DOI] [PubMed] [Google Scholar]
- 7.Quiroz-Santiago C, et al. Prevalence of Salmonella in vegetables from Mexico. Journal of Food Protection 2009; 72: 1279–1282. [DOI] [PubMed] [Google Scholar]
- 8.Nicolay N, et al. Salmonella enterica serovar Agona European outbreak associated with a food company. Epidemiology and Infection 2011; 139: 1272–1280. [DOI] [PubMed] [Google Scholar]
- 9.Anderson P, et al. Molecular analysis of Salmonella serotypes at different stages of commercial turkey processing. Poultry Science 2010; 89: 2030–2037. [DOI] [PubMed] [Google Scholar]
- 10.Brouard C, et al. Two consecutive large outbreaks of Salmonella enterica serotype Agona infections in infants linked to the consumption of powdered infant formula. The Pediatric Infectious Disease Journal 2007; 26: 148–152. [DOI] [PubMed] [Google Scholar]
- 11.Diez-Garcia M, Capita R, Alonso-Calleja C. Influence of serotype on the growth kinetics and the ability to form biofilms of Salmonella isolates from poultry. Food Microbiology 2012; 31: 173–180. [DOI] [PubMed] [Google Scholar]
- 12.The National Enteric Pathogens Surveillance System (NEPSS). Communicable diseases surveillance – Additional reports. Communicable Diseases Intelligence Quarterly Report 2006; 30: 486–494. [Google Scholar]
- 13.Threlfall EJ, et al. Application of pulsed-field gel electrophoresis to an international outbreak of Salmonella Agona. Emerging Infectious Diseases 1996; 2: 130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Killalea D, et al. International epidemiological and microbiological study of outbreak of Salmonella Agona infection from a ready to eat savoury snack—I: England and Wales and the United States. British Medical Journal 1996; 313: 1105–1107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Centers for Disease Control Prevention. Multistate outbreak of Salmonella serotype Agona infections linked to toasted oats cereal-United States, April-May, 1998. MMWR Morbidity and Mortality Weekly Report 1998; 47: 462. [PubMed] [Google Scholar]
- 16.Russo ET, et al. A recurrent, multistate outbreak of Salmonella serotype Agona infections associated with dry, unsweetened cereal consumption, United States, 20083. Journal of Food Protection 2013; 76: 227–230. [DOI] [PubMed] [Google Scholar]
- 17.Hoffmann M, et al. Complete genome sequence of Salmonella enterica subsp. enterica serovar Agona 460004 2–1, associated with a multistate outbreak in the United States. Genome Announcements 2015; 3: e00690–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Shohat T, et al. International epidemiological and microbiological study of outbreak of Salmonella Agona infection from a ready to eat savoury snack—II: Israel. British Medical Journal 1996; 313: 1107–1109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lindqvist N, Siitonen A, Pelkonen S. Molecular follow-up of Salmonella enterica subsp. enterica serovar Agona infection in cattle and humans. Journal of Clinical Microbiology 2002; 40: 3648–3653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Rabsch W, et al. Molecular epidemiology of Salmonella enterica serovar Agona: characterization of a diffuse outbreak caused by aniseed-fennel-caraway infusion. Epidemiology and Infection 2005; 133: 837–844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Merritt TD, Herlihy C. Salmonella outbreak associated with chicks and ducklings at childcare centres. The Medical Journal of Australia 2003; 179: 63–64. [DOI] [PubMed] [Google Scholar]
- 22.Carli KT, Eyigor A, Caner V. Prevalence of Salmonella serovars in chickens in Turkey. Journal of Food Protection 2001; 64: 1832–1835. [DOI] [PubMed] [Google Scholar]
- 23.Cardinale E, et al. Risk factors for Salmonella enterica subsp. enterica infection in Senegalese broiler-chicken flocks. Preventive Veterinary Medicine 2004; 63: 151–161. [DOI] [PubMed] [Google Scholar]
- 24.Sorensen O, et al. Salmonella spp. shedding by Alberta beef cattle and the detection of Salmonella spp. in ground beef. Journal of Food Protection 2002; 65: 484–491. [DOI] [PubMed] [Google Scholar]
- 25.Clark GM, Gangarosa AKJ, Thompson M. Epidemiology of an international outbreak of Salmonella Agona. The Lancet 1973; 302: 490–493. [DOI] [PubMed] [Google Scholar]
- 26.Sintchenko V, Holmes EC. The role of pathogen genomics in assessing disease transmission. BMJ 2015; 350: h1314. [DOI] [PubMed] [Google Scholar]
- 27.Octavia S, et al. Delineating community outbreaks of Salmonella enterica serovar Typhimurium by use of whole-genome sequencing: insights into genomic variability within an outbreak. Journal of Clinical Microbiology 2015; 53: 1063–1071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Roetzer A, et al. Whole genome sequencing versus traditional genotyping for investigation of a Mycobacterium tuberculosis outbreak: a longitudinal molecular epidemiological study. PLoS Medicine 2013; 10: e1001387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Leekitcharoenphon P, et al. Evaluation of whole genome sequencing for outbreak detection of Salmonella enterica. PLoS ONE 2014; 9: e87991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Moran-Gilad J, et al. Proficiency testing for bacterial whole genome sequencing: an end-user survey of current capabilities, requirements and priorities. BMC Infectious Diseases 2015; 15: 1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hatherell H-A, et al. Interpreting whole genome sequencing for investigating tuberculosis transmission: a systematic review. BMC Medicine 2016; 14: 1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Communicable Diseases Network Australia (CDNA)- Australian National Notifiable Diseases and Case Definitions (http://www.health.gov.au/internet/main/publishing.nsf/Content/cdna-casedefinitions.htm). Accessed October 2015.
- 33.Merritt T, Unicomb L. A review of Salmonella surveillance in New South Wales, 1998–2000. New South Wales Public Health Bulletin 2004; 15: 178–181. [DOI] [PubMed] [Google Scholar]
- 34.Tamura K, et al. MEGA6: molecular evolutionary genetics analysis version 6.0. Molecular Biology and Evolution 2013; 30: 2725–2729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Inns T, et al. A multi-country Salmonella Enteritidis phage type 14b outbreak associated with eggs from a German producer: ‘near real-time’ application of whole genome sequencing and food chain investigations, United Kingdom, May to September 2014. Euro Surveillance 2015; 20: 21098. [DOI] [PubMed] [Google Scholar]
- 36.Steenackers H, et al. Salmonella biofilms: an overview on occurrence, structure, regulation and eradication. Food Research International 2012; 45: 502–531. [Google Scholar]
- 37.Bavishi C, Dupont H. Systematic review: the use of proton pump inhibitors and increased susceptibility to enteric infection. Alimentary Pharmacology and Therapeutics 2011; 34: 1269–1281. [DOI] [PubMed] [Google Scholar]
- 38.Köser CU, et al. Routine use of microbial whole genome sequencing in diagnostic and public health microbiology. PLoS Pathogens 2012; 8: e1002824. [DOI] [PMC free article] [PubMed] [Google Scholar]