SUMMARY
The objectives were to present three approaches for calculating antimicrobial (AM) use in pigs that take into account the rearing period and rearing site, and to study the association between these measurements and phenotypical resistance and abundance of resistance genes in faeces samples from 10 finisher batches. The AM use was calculated relative to the rearing period of the batches as (i) ‘Finisher Unit Exposure’ at unit level, (ii) ‘Lifetime Exposure’ at batch level and (iii) ‘Herd Exposure’ at herd level. A significant effect on the occurrence of tetracycline resistance measured by cultivation was identified for Lifetime Exposure for the AM class: tetracycline. Furthermore, for Lifetime Exposure for the AM classes: macrolide, broad-spectrum penicillin, sulfonamide and tetracycline use as well as Herd Unit Exposure for the AM classes: aminoglycoside, lincosamide and tetracycline use, a significant effect was observed on the occurrence of genes coding for the AM resistance classes: aminoglycoside, lincosamide, macrolide, β-lactam, sulfonamide and tetracycline. No effect was observed for Finisher Unit Exposure. Overall, the study shows that Lifetime Exposure is an efficient measurement of AM use in finisher batches, and has a significant effect on the occurrence of resistance, measured either by cultivation or metagenomics.
Key words: Antimicrobial drugs, antimicrobial resistance in agricultural settings, cultivation, metagenomic, pigs
INTRODUCTION
The World Health Organization has declared that antimicrobial resistance (AMR) is one of the most worrying health threats to humans in the 21st century [1], as it adversely affects treatment options in human medicine [2]. Current AMR surveillance is based mainly on passive reporting of clinical diagnoses and phenotypical laboratory results for specific pathogens [3]. However, an approach that provides an insight into the phenotypical resistance may not be representative for the overall occurrence of resistance in the bacterial population it is derived from [3].
Swann et al. were the first to raise awareness of a potential link between veterinary use of antimicrobials (AMs) and bacterial AMR in humans [4]. This relationship has since been confirmed by several studies [5–8]. Different methods have been used to measure AMR in the livestock reservoir, including culture-based and molecular methods [3, 9]. Metagenomics sequencing and read mapping have recently been revealed as powerful methods for quantifying AMR in the normal flora of pigs [3]. Quantification of the presence of resistance genes is of relevance when attempting to quantify the contribution from pigs to human pathogens [7, 8, 10].
The association between veterinary use of AM and AMR in bacterial isolates from pigs has been demonstrated repeatedly [11–16] with variations in its occurrence being related to variations in AM use [12, 14–16]. Therefore, less frequent use of AMs in pig farms is a vital factor in reducing the occurrence of AMR in finishers [8, 17–19]. Thus, in order to develop effective tools for interventions and targets for AM reduction, a standardised method is required that closely reflects the dynamics of AMR. As with AMR, which may be measured in several different ways, AM usage may be quantified using different approaches [12, 14–16, 20].
AM use for finishers has traditionally been calculated based on data obtained either from farm records [16, 21–23] or from databases on prescribed medicines: at unit [14, 16], farm [12, 14, 16] or farm-owner level [20, 24]. An AM calculation restricted to a specific unit may lack essential information when compared with the exposure during the entire rearing period from a piglet's birth to the final fattening stage. This is often the case for the finisher unit, where AM use is particularly low [25]. In Denmark, farm-level (integrated) data will not be available for a vast proportion of farms delivering finishers for slaughter [26], as traditional integrated pig production has largely been replaced by multisite pig production where rearing (the farrowing unit → the weaning unit → the finisher unit) takes place on farms at different geographic sites that are owned either by the same farmer or by different farmers [26]. Therefore, the estimation of usage at farm level does not reflect the full exposure during the rearing period. The farm-owner level calculations may include AM use of no relevance if one or more farms are not part of the rearing system.
The periods set for the data extraction in the same studies were mostly 1 year prior to sampling [12, 14, 20]. However, changes in AMR amounts occur at much shorter time spans [13, 16, 27], and yearly calculations of AM use may therefore be insufficiently refined for the purpose of revealing associations at finisher batch level. Furthermore, regarding finisher production, each group of animals (batch) is moved from farrowing to weaning and then to the finisher unit primarily on an all-in/all-out basis. Thus, the variation in AM use between batches due to the occurrence of diseases will not be observed when the calculation of AM use covers a whole year.
By means of the Central Husbandry Register (CHR), the Danish Pig Movement Database (PMD) and the Danish Veterinary Medicines Statistic Program (VetStat), it is possible to calculate an approximation of the AM use per batch based on the rearing period and the rearing site/sites, thus capturing variation in AM use between batches. AMR results for finishers can therefore be obtained at time of slaughter, without consideration for rearing site(s). The objectives of the study were to: (i) develop three different approaches for calculating AM use at finisher batch level, taking into account the rearing period and rearing site, and (ii) compare the association with measured AMR in composite faeces samples from 10 batches of finishers.
METHODS
Data sources
Data on the farms and number of pigs were obtained from the national CHR, where all farms with production animals are recorded [28, 29]. The CHR stores information linked to a farm code (ID), which refers to a specific geographical location and includes, e.g. animal species, the number of animals per age group (sows, weaners, finishers), thus the number per unit on any given day. The pigs owned by a producer (the herd) can be kept at many geographical localities (farms). For the sake of simplicity, all animals owned by an individual producer are referred to as a herd throughout the study even though some herds are kept at many farms [24].
Data on movements of pigs between farms were obtained from the PMD, an integral part of the CHR. The PMD records the number of pigs, date, ID of origin farm and ID of destination farm for each movement [28, 29].
Data on AM use were obtained from VetStat, which contains data on all medicine prescribed by veterinarians for animals. Records are based on veterinarian prescriptions and contain information on active substances, amounts, target species, age groups, diagnosis groups and farm IDs [29]. The age groups in VetStat correspond to the age groups in the CHR, and the units, sows, weaners and finishers on a farm. In order to produce comparable data across records, active compounds were converted into a unit measuring how many kilograms of pig could be treated per day – Animal Defined Daily Doses per kilogram (ADDkg) [30].
Study design
The study design used was first described by Munk et al. [3]. In brief, based on the average sizes for sow and finisher farms in 2009 [31, 32], herds with more than 500 sows and a production of at least 5000 finishers annually were selected using the CHR and PMD. In total, 376 herds met these criteria. From VetStat, the total amounts of AM and tetracycline used in these herds from June to November 2013 were calculated as ADDkg and adjusted according to the number of animals in the herds. To cover a wide span of AM use, the 376 herds were ranked, and the owners of the top and bottom 10% AM and tetracycline use quantiles were invited to participate consecutively until five herds within each quantile had accepted. Each of these 10 herds was located between one and seven farms. In total, 23 herds from the top quantile and 15 herds from the bottom quantile were invited.
In the finisher units of the 10 herds, composite pen-floor samples were collected consisting of single samples from 30 randomly chosen pens within the section(s) with finishers weighing between 80 and 100 kg (in Denmark, pigs are delivered for slaughter weighing 100–105 kg) [3,9]. For each herd, the group of pigs from which samples were collected represents a finisher batch. For each of the finisher batches, the 30 single faeces samples were pooled together, resulting in one pooled sample per batch. Sampling took place from March to June 2014.
Measurements of AM use
For each batch of finisher, three quantitative usage measurements of the AM classes: aminoglycoside, lincosamide, macrolide, broad-spectrum penicillin, sulfonamide and tetracycline were calculated. They were based on the day of sampling, as the start from which backward assessment of time periods in the rearing site(s) were established. The measurements varied in terms of exposure, as explained below.
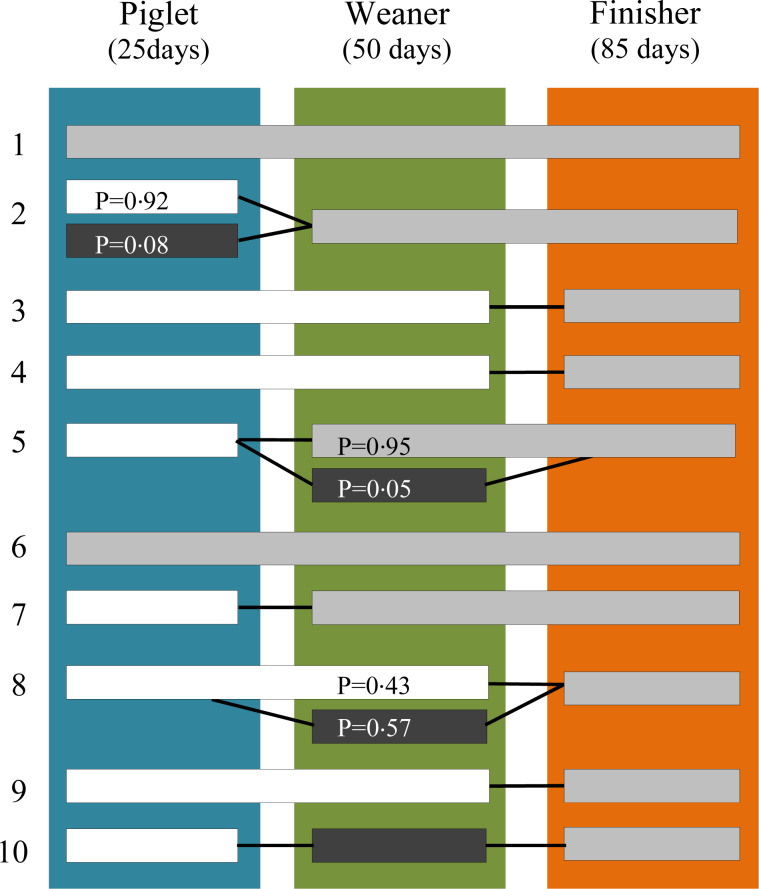
Danish national averages for pig production productivity for 2014 were applied for the rearing periods per unit (in days) [33–35], resulting in 25, 50 and 85 days in the farrowing (piglet), weaning (weaner) and finisher units (finisher), respectively. Starting with the finisher batches and the day of sampling, the PMD was used to trace the movements of the batches from sampling site back to birth site. Figure 1 shows the pathways of the 10 finisher batches.
Fig. 1.
The rearing pathway of each of the 10 finisher batches from birth site to finisher site compared with the day of sampling. The 10 horizontal bars depict the movements of the batches. A colour shift in a bar denotes that a farm has a different geographical location to the farm where sampling took place and P denotes the proportion of pigs being moved. Bars without P are equal to one. Numbers 1–5 denote the initially high users and 6–10 depict the initially low users. The three vertical coloured bars represent the assumed days of exposure to AM in the: sow-piglet (farrowing) unit, weaner unit and finisher unit.
Each farmer in the study had a health advisory contract with an individual veterinarian in accordance with Danish legislation. These contracts permit veterinarians to prescribe AMs for treatment of plausible diseases diagnosed at a farm visit for the subsequent 35 or 63 days (finishers only) at which point the next visit must take place. Therefore, taking into account the rearing period of 160 days, and the maximum period a prescription may cover (63 days), data from VetStat on prescribed AMs 9 months prior to sampling were extracted for the 22 farms comprising the finisher batches’ pathways from birth to sampling site (Fig. 1).
AM exposure was calculated for the classes: aminoglycoside, lincosamide, macrolide, broad-spectrum penicillin, tetracycline and sulfonamide, as these are most commonly used in Danish pig production [25]. By adopting the method used by Vigre et al. [24], the daily amounts of the six AM classes used were calculated for sows, weaners and finishers in each of the 22 farms based on two assumptions. First, as veterinarians are permitted to prescribe for potential diseases for 35 or 65 days, the interval between two prescriptions was used to calculate the daily use for each prescription of AM, assuming that an average amount of the prescribed AM was used on each day of the interval. The calculations were subsequently added together per day per AM class. Second, if an interval was shorter than 7 days, the following prescription date was used to set the number of days for the interval.
Thus, the daily use of an AM was calculated as an average daily use per farm (F) per age group/unit (U) and adjusted in accordance with the number of pigs at risk per day per unit on each farm, as:
 |
where 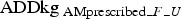 = the prescribed amount per unit per farm measured as ADDkg, daysprescription interval = the interval in days between the day of the initial prescription and the day of the subsequent prescription and pigsday_F_U = the number of sows, weaners or finishers on any given day. The number of sows substituted the number of piglets, as the latter is not registered in the CHR.
= the prescribed amount per unit per farm measured as ADDkg, daysprescription interval = the interval in days between the day of the initial prescription and the day of the subsequent prescription and pigsday_F_U = the number of sows, weaners or finishers on any given day. The number of sows substituted the number of piglets, as the latter is not registered in the CHR.
The number of 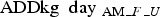 was then summarised per day for each AM class (AMc):
was then summarised per day for each AM class (AMc):
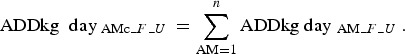 |
Based on the finisher batches’ rearing periods in days: days 1–85 in the finisher unit, days 86–135 in the weaning unit and days 136–160 in the sow unit, where day 1 corresponds to the day of sampling (Fig. 1), the number of 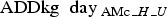 was summarised for each rearing period (R) per unit, and adjusted to suit the proportion (P) of animals being moved from a farm (Fig. 1):
was summarised for each rearing period (R) per unit, and adjusted to suit the proportion (P) of animals being moved from a farm (Fig. 1):
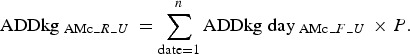 |
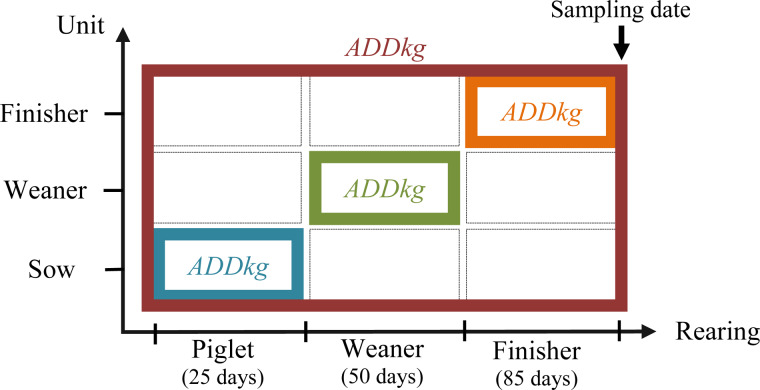
For each AM class, three measurements of exposure were calculated, given the rearing pathways, the rearing periods (Fig. 1) and the unit(s) in each farm (Fig. 2).
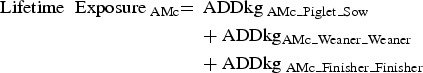 |
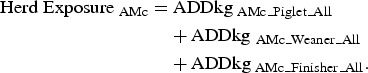 |
Fig. 2.
The exposure measurements of AM usage. The orange square applies to Finisher Unit Exposure and therefore comprises the AM usage in the finisher-rearing period in the finisher unit. Lifetime Exposure applies to the orange, green and blue squares, and therefore comprises the AM usage in the piglet-rearing period in the sow unit, the weaning-rearing period in the weaning unit and the finisher-rearing period in the finisher unit. The red square applies to Herd Exposure, and therefore comprises AM usage throughout the entire rearing period in all units.
For information on the units per farm included in the exposure measurements in the rearing period of the 10 finisher batches, see Figure S1 in the Supplementary data. For information on the obtained exposure variables, see Table S1 in the Supplementary data.
AMR measurements
In this study, the cultivation and metagenomic results of the pooled samples from 10 pig herds described by Munk et al. [3] were used as the AMR results.
Cultivation
In brief, for each pooled sample, faeces was suspended in isotonic saline prior to serial dilution. Aliquots of dilutions were plated onto selective and non-selective LB and MacConkey plates to quantify aerobic bacteria and Escherichia coli, respectively. Selective plates contained 8 mg/l tetracycline (T3383 tetracycline hydrochloride, former: Sigma-Aldrich, current: Thermo Fisher Scientific, Roskilde, Denmark) in LB and 16 mg/l ampicillin (A9393 Ampicillin, former: Sigma-Aldrich, current: Thermo Fisher Scientific, Roskilde, Denmark) or tetracycline in MacConkey. All assays were performed in triplicate. For each triplicate set, a weighted average of resistance proportion was calculated based on cfu counts per dilution [3]. For information on the obtained outcome variables, see Table S1 in the Supplementary data.
Metagenomics
In brief, AMR genes for the classes: aminoglycoside, lincosamide, macrolide, β-lactam, sulfonamide and tetracycline were obtained using whole community sequencing (WCS), and measured as reads per kilobase reference per million (RPKM). For information on the genes within each AMR class, see Table S2 in the Supplementary data.
In order to compare the 10 faeces samples, the raw read counts were normalised to the size of the dataset for each AM class with the following formula:
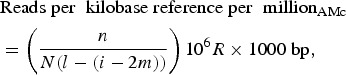 |
where n = number of mapped reads, N = total number of reads, l = gene length, i = insert size, m = minimum mapping length, R = reads and bp = base pair.
The normalisation takes into account the fact that the pooling and sequencing of several indexed samples produces varying DNA library sizes, resulting in comparable RPKM values and independence of sequencing depth. For information on the obtained outcome variables, see Table S1 in the Supplementary data.
Data analyses
The quantitative effect of broad-spectrum penicillin and tetracycline use, measured as Finisher Unit Exposure, Lifetime Exposure and Herd Exposure, on ampicillin and tetracycline resistance obtained by cultivation was calculated using simple linear regression, and as a measure of model fit, the coefficient of determination (R2) was applied. The quantitative effect of exposure for the AM classes: aminoglycoside, lincosamide, macrolide, broad-spectrum penicillin, sulfonamide and tetracycline, measured as Finisher Unit Exposure, Lifetime Exposure and Herd Exposure, on similar AMR gene abundance was calculated using simple linear regression, and as a measure of model fit, the coefficient of determination (R2) was applied. The assessment of homoscedasticity was performed by visual inspection of the plots, including measured values and regression lines.
WPS Workbench, Version: 3.1.1.0.0, and Microsoft Excel 2010 were applied in data processing, and data analyses were performed using R, version 3.
RESULTS
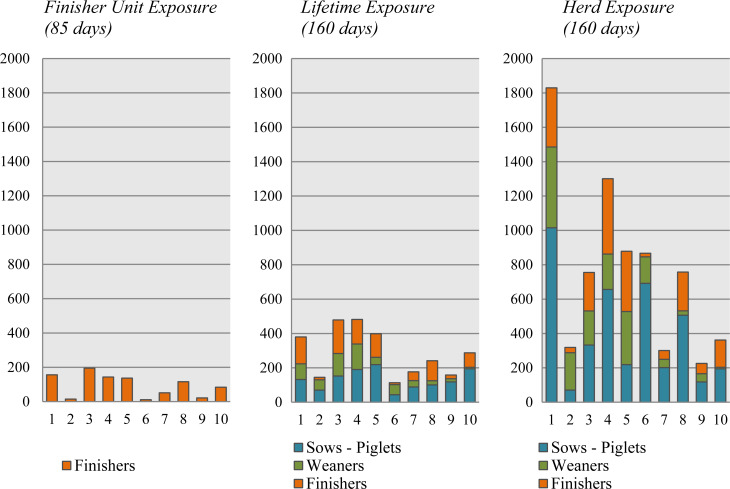
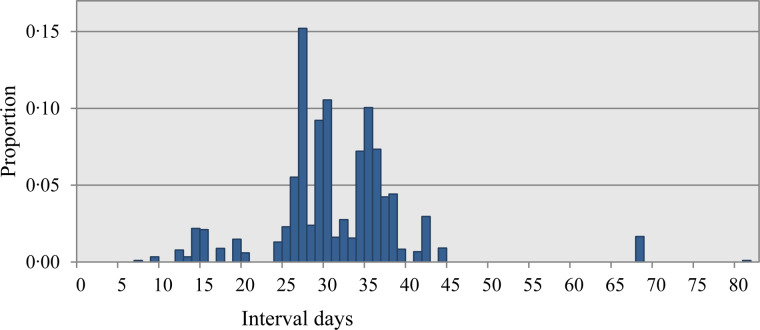
The three different quantitative measurements of total AM use presented in Figure 3 show use within the applied rearing periods. The Lifetime Exposure is lower than Herd Exposure, as the former consists of a part of the latter. The difference is caused by the level at which data were obtained; the Lifetime Exposure was obtained at rearing batch level, while the Herd Exposure was obtained at rearing herd level. A similar pattern applies to the Finisher Unit Exposure compared with the Lifetime Exposure, since the latter was obtained at batch level, whereas the former was obtained at unit level. Figure 4 shows the distribution of the intervals in days between prescriptions. Most of the intervals are between 24 and 45 days, with fewer between 7 and 21 days. The short prescription intervals were observed mainly in the initially high-user farms. Of these intervals, two-thirds were due to a subsequent prescription of the same AM, with the remaining third caused by a different prescribed AM.
Fig. 3.
Finisher Unit Exposure, Lifetime Exposure and Herd Exposure. The total AM use and the distribution between sow piglets, weaners and finishers within the 10 finisher batches. Numbers 1–5 denote the initially high users and 6–10 depict the initially low users.
Fig. 4.
The distribution of prescription interval days – days between two prescriptions.
Association between AM use and phenotypical measured resistance
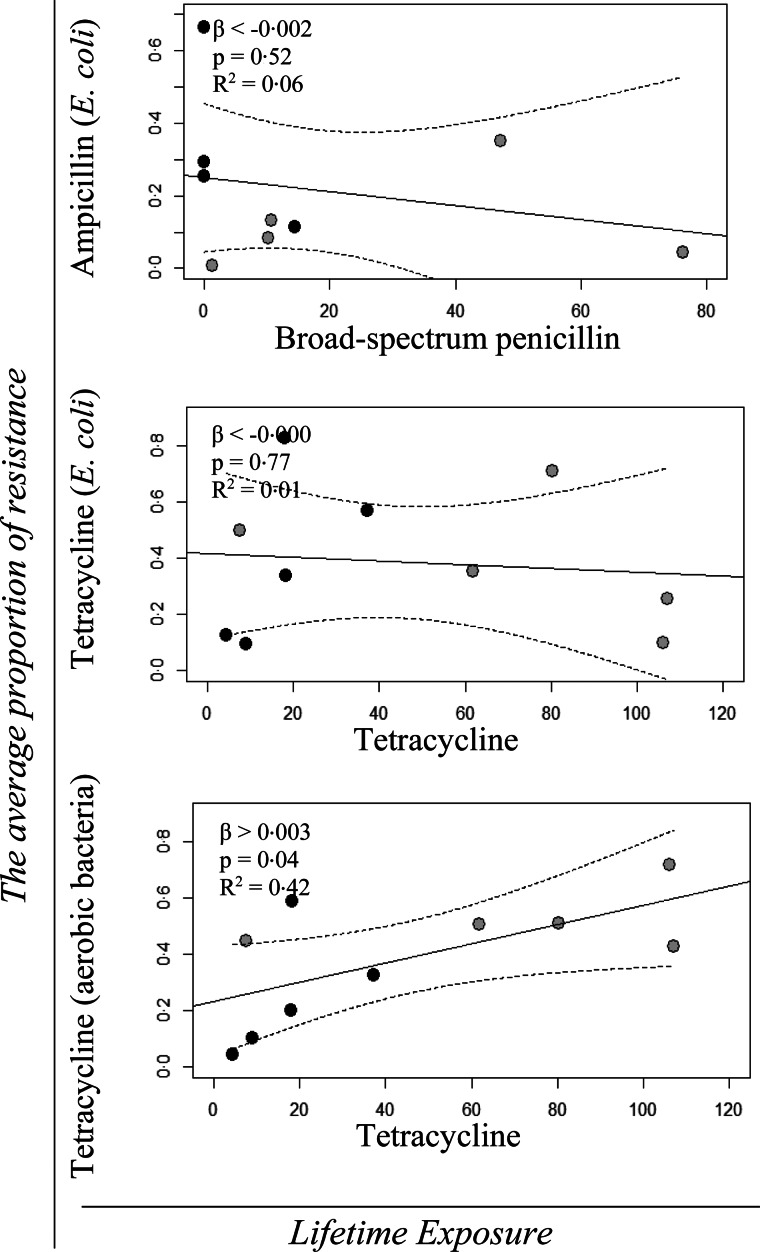
Figure 5 shows the regression models with 95% confidence intervals of the quantitative effect of Lifetime Exposure for broad-spectrum penicillin and tetracycline use on the average proportion of ampicillin and tetracycline-resistant E. coli (MacConkey), respectively, and of Lifetime Exposure for tetracycline use on the average proportion of tetracycline-resistant aerobic bacteria (LB). The β-values, P-values and R2 values for the regression analyses are shown in the same figure. The regression analyses showed only a significant effect of Lifetime Exposure for tetracycline use on the average proportion of tetracycline-resistant aerobic bacteria (Fig. 5).
Fig. 5.
Univariable linear regression plots (solid line) and 95% confidence interval (dotted lines) of the average resistance proportion of ampicillin and tetracycline resistance from cultivation of Escherichia coli on MacConkey agar with and without ampicillin and tetracycline, as a function of Lifetime Exposure for the AM classes; broad-spectrum penicillin and tetracycline, respectively, and the average resistance proportion of tetracycline resistance from cultivation of aerobic bacteria on LB agar with and without tetracycline, as a function of Lifetime Exposure for the AMc; tetracycline. The grey points denote the initially high users and the black points depict the initially low users. The effect (β), the P-value (P) and the R2 value are shown in the top left corner of each model.
No significant effect was observed for Finisher Unit Exposure or Herd Exposure (result not shown).
Association between AM use and metagenomic measured resistance
No significant effect was observed of Finisher Unit Exposure for any of the AM classes on their respective AMR gene classes (result not shown).
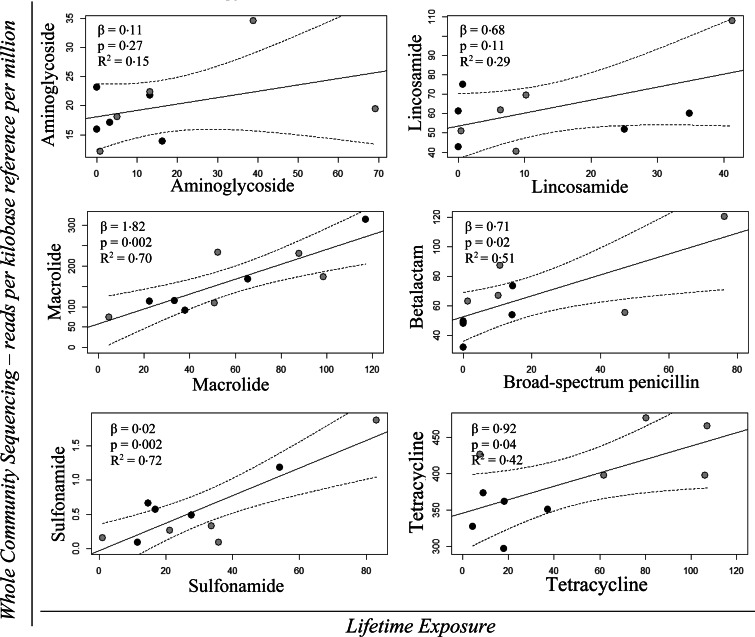
Figure 6 shows the regression models with 95% confidence intervals for the quantitative effect of Lifetime Exposure for the AM classes: aminoglycoside, lincosamide, macrolide, broad-spectrum penicillin, sulfonamide and tetracycline on the AMR gene classes: aminoglycoside, lincosamide, macrolide, β-lactam, sulfonamide and tetracycline. The β-values, P-values and R2 values for the regression analyses are shown in the same figure. The regression analyses revealed a significant effect of Lifetime Exposure for the AM classes: macrolide, broad-spectrum penicillin, sulfonamide and tetracycline on AMR genes for macrolide, β-lactam, sulfonamide and tetracycline, respectively (Fig. 6).
Fig. 6.
Univariable linear regression plots (solid line) with 95% confidence interval (dotted lines) of WCS – RPKM of the AMR genes for: aminoglycoside, lincosamide, macrolide, β-lactam, sulfonamide and tetracycline as a function of Lifetime Exposure for the AM classes: aminoglycoside, lincosamide, macrolide, broad-spectrum penicillin, sulfonamide and tetracycline, respectively. The grey points denote the initially high users and the black points depict the initially low users. The effect (β), the P-value (P) and the R2 value are shown in the top left corner of each model.
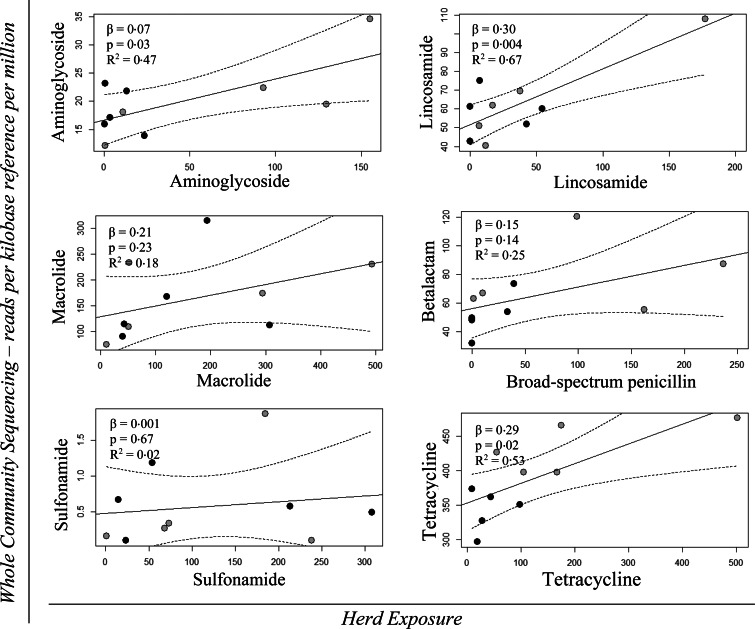
Figure 7 shows the regression models with 95% confidence intervals of the quantitative effect for Herd Exposure for the AM classes: aminoglycoside, lincosamide, macrolide, broad-spectrum penicillin, sulfonamide and tetracycline on the AMR gene classes: aminoglycoside, lincosamide, macrolide, β-lactam, sulfonamide and tetracycline. The β-values, P-values and R2 values for the regression analyses are shown in the same figure. The regression analyses revealed a significant effect of Herd Exposure for the AM classes: aminoglycoside, lincosamide and tetracycline on the AMR genes for aminoglycoside, lincosamide and tetracycline, respectively (Fig. 7).
Fig. 7.
Univariable linear regression plots (solid plot) with 95% confidence interval (dotted lines) of WCS – RPKM of the AMR genes for: aminoglycoside, lincosamide, macrolide, β-lactam, sulfonamide and tetracycline as a function of Herd Exposure for the AM classes: aminoglycoside, lincosamide, macrolide, broad-spectrum penicillin, sulfonamide and tetracycline, respectively. The grey points denote the initially high users and the black points depict the initially low users. The effect (β), the P-value (P) and the R2 value are shown in the top left corner of each model.
The regression models were re-analysed, excluding the most extreme data point of each set, to assess the robustness of the significant results. The significant effect of Lifetime Exposure for the AM classes: macrolide, sulfonamide and tetracycline on the occurrence of macrolide, sulfonamide and tetracycline resistance genes, respectively, remained under the 5% significance level. This was not the case for the significant effect of broad-spectrum penicillin usage on the occurrence of β-lactam resistance genes (result not shown). Only the effect of Herd Exposure for tetracycline on tetracycline-resistant genes remained under the 5% significance level (result not shown). The coefficient of determination (R2) analyses of the models with significant results revealed that the variation of AMR that could be explained by the Lifetime Exposure measurements varied from 0·42 (tetracycline) to 0·72 (sulfonamide) for the variables (Figs 5 and 6), and from 0·47 (aminoglycoside) to 0·67 (lincosamide) for the Herd Exposure measurements (Fig. 7).
DISCUSSION
Independent variables
The exposure measurements were derived from different data sources, thereby integrating any errors of the sources in the daily ADDkg.
The length of the rearing period influences the exposure measurements; however, this study did not determine whether the rearing periods should be longer or overlapped in order to assess the effect on the occurrence of AMR genes. Nevertheless, shorter rearing periods reduce the AM amounts, resulting in less variation in the exposure measurements between finisher batches. Longer rearing periods have the opposite effect.
The intervals were calculated based on two assumptions. First, as the intervals were not drug-specific, a prescription of tetracycline followed by a prescription of macrolide counted as an interval because it was assumed that any drug shifts were due to a different disease or a change, as the veterinarian could not observe a sufficient effect caused by the first drug. This is supported by veterinarians’ permission to prescribe AMs. Consequently, a new situation is likely to have occurred on the farm if AMs were prescribed with intervals shorter than 35 days. Short intervals result in high daily ADDkg, causing a substantial increase in the measured AM use, as the prescribed amount of AM is divided by fewer days. Second, all prescription intervals of <7 days were assumed to be due to technical issues at the pharmacies, e.g. shortages of a drug or a shift in batches at the pharmacy, or caused by the veterinarian issuing two identical prescriptions (the sub-diagnosis differs) that were handed in on different dates.
Dependent variables
Cultivation using aerobic bacteria and E. coli, respectively, are traditional phenotypical methods for assessing the occurrence of AMR in populations of animals [9]. However, the approaches only provide insight into a fraction of the intestinal microbiota, and thus potentially underestimate the actual reservoir of AMR in a population [3, 7, 9].
WCS measures the presence of known resistance genes throughout the microbial community, and therefore does not determine the genetic location of the AMR genes and thereby whether they are likely to transfer from the animal to the human reservoir [3]. A distinction between the two is important since ubiquitous AMR genes may not pose a risk to humans [19]. By contrast, any AMR genes in excessive abundance in habitats with high AM use, such as pig farms, are more likely to be relevant for AMR development [7, 19]. Furthermore, as the ResFinder database contains only AMR genes found in culturable bacteria, a considerable number of ubiquitous functional AMR genes may have been missed [3]. Therefore, although not all AMR genes necessarily constitute a risk to humans, different amounts in faeces from pigs represent an available gene pool from which zoonotic bacteria may obtain resistance genes [19].
Result discussion
The 10 finisher batches varied in rearing pathways (Fig. 1) and the herds in which they were produced differed concerning the number of farms per herd. Thus, on that basis, no pattern or common feature could be observed. Given that the study included only 10 finisher batches, no attempt was made to conduct an analytical adjustment of a potential confounding effect from herd or farm management.
It is notable that the results of the regression models for four out of six combinations of Lifetime Exposure vs AMR obtained by WCS (Fig. 6) gave significant results, as did three out of six combinations of Herd Exposure vs AMR obtained by WCS (Fig. 7). In order to crudely evaluate the reliability of the results, the individual observations in each of the significant plots that appeared to have the strongest influence on the result were removed and the model was re-analysed. The Lifetime Exposure measurement still provided significant results for all but broad-spectrum penicillin, while only Herd Exposure remained significant for tetracycline, indicating that the former method of measuring AM use is more sensitive. Furthermore, the measurement of Herd Exposure is highly influenced by differences in the units present in each farm, rather than differences in AM use. By comparing the pathways of the finisher batches, large differences can be observed in the number of units per farm, which would greatly affect the measurement (Fig. 1 and Supplementary Fig. S1). The fact that overall the R2 models of Lifetime Exposure explain more of the variation in AMR compared with Herd Exposure indicates that Lifetime Exposure has the strongest effect on AMR in finisher batches. No effect of Finisher Unit Exposure on AMR could be demonstrated; however, use of AM in this unit is limited in terms of both AM classes and amounts [25].
In this study, a significant quantitative association could be demonstrated for Lifetime Exposure of tetracycline on AMR across methods of obtaining resistance (Figs 5 and 6), which is in alignment with Munk et al., who demonstrated a correlation in AMR between cultivation and metagenomics [3].
The measurements were calculated as total amounts throughout the rearing period for the six specified AM classes, rather than a measurement for each rearing period. Consequently, the measurements do not distinguish between different AM use patterns within the three rearing periods. Batches with the same total AM class usage might relate to use in different units; thus, the rearing period, and the occurrence of AMR for these finisher batches, may not be identical due to rapid change in the occurrence of AMR [13, 36].
In this study, three different approaches for calculating use of AM at finisher batch level were developed. Using an optimal and standardised calculation method is important for setting drug use targets and for direct possible interventions with an expected effect on AMR. Lifetime Exposure has the advantage of being independent of production type/rearing site. Furthermore, it follows finisher batches through rearing site(s) in the actual rearing periods, hereby capturing variations in AM use between batches. In contrast to this, the analyses did not consider other factors e.g. duration of treatment or dispensing type, which are known to have an impact on the development and spread of AMR [16]. Overall, this study showed that the entire rearing period should be taken into account when studying the association between AMR and AM use, and revealed that the method developed for calculating Lifetime Exposure is an efficient measurement of the effect of AM use on AMR found in finisher batches.
By using metagenomics, we measured the relative abundance of specific genes in the faeces. Even if more DNA fragments were sequenced per sample, there is a likelihood that genes with a very low relatively abundance would not be detected. In the estimation of the association between AM and AMR, the abundance of resistance genes was aggregated to phenotypical level. The estimated associations should only be interpreted at this level and not to gene level. Due to the limited size of this study, an estimation of the quantitative association between the AM usage and abundance of the specific genes was not carried out.
Overall, the study has generated quantitative knowledge of how the usage of AM through the entire rearing period affects the occurrence of AMR in animals at the end of the production. This knowledge will be valuable when assessing effects of alternative AM usage on the occurrence of AMR in the pig production. The validity of these assessments will improve the robustness and precision of the decisions about interventions targeted against reducing the occurrence of AMR in pigs.
ACKNOWLEDGMENTS
The authors thank all the farmers for participating in this study.
FUNDING
This work was supported by the Danish Ministry of Food, Agriculture and Fisheries.
Supplementary material
For supplementary material accompanying this paper visit http://dx.doi.org/10.1017/S0950268817001285.
click here to view supplementary material
CONFLICTS OF INTEREST
None.
REFERENCES
- 1.WHO. The evolving threat of antimicrobial resistance – options for action. World Health Organization, 2012. http://www.who.int/patientsafety/implementation/amr/publication/en/index.html (accessed 11 January 2017).
- 2.ECDC/EMEA. Joint Technical Report: the bacterial challenge: time to react, 2009. http://ecdc.europa.eu/en/publications/_layouts/forms/Publication_DispForm.aspx?ID=199&List=4f55ad51-4aed-4d32-b960-af70113dbb90 (accessed 11 January 2017).
- 3.Munk P, et al. A sampling and metagenomic sequencing-based methodology for monitoring antimicrobial resistance in swine herds. Journal of Antimicrobial Chemotherapy 2017; 72(2): 385–392. doi: 10.1093/jac/dkw415. [DOI] [PubMed] [Google Scholar]
- 4.Swann MM, et al. Joint Committee on the use of Antibiotics in Animal Husbandry and Veterinary Medicine (Swann Report). London: H.M.S.O, 1969. [Google Scholar]
- 5.Martel J-L, et al. New trends in regulatory rules and surveillance of antimicrobial resistance in bacteria of animal origin. Veterinary Research 2001; 32: 381–392. doi: 10.1051/vetres:2001131. [DOI] [PubMed] [Google Scholar]
- 6.Catry B, et al. Antimicrobial resistance in livestock. Journal of Veterinary Pharmacology and Therapeutics 2003; 26: 81–93. doi: 10.1046/j.1365-2885.2003.00463.x. [DOI] [PubMed] [Google Scholar]
- 7.Marshall BM, Levy SB. Food animals and antimicrobials: impacts on human health. Clinical Microbiology Reviews 2011; 24: 718–733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hu Y, et al. The bacterial mobile resistome transfer network connecting the animal and human microbiomes. Applied and Environmental Microbiology 2016; 82: 6672–6681. doi: 10.1128/AEM.01802-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Schmidt GV, et al. Sampling and pooling methods for capturing herd level antibiotic resistance in swine feces using qPCR and CFU approaches. PLoS ONE 2015; 10: e0131672. doi: 10.1371/journal.pone.0131672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hammerum AM, et al. Characterization of extended-spectrum beta-lactamase (ESBL)-producing Escherichia coli obtained from Danish pigs, pig farmers and their families from farms with high or no consumption of third- or fourth-generation cephalosporins. Journal of Antimicrobial Chemotherapy 2014; 69: 2650–2657. doi: 10.1093/jac/dku180 [doi]. [DOI] [PubMed] [Google Scholar]
- 11.Aarestrup FM. Veterinary drug usage and antimicrobial resistance in bacteria of animal origin. Basic and Clinical Pharmacology and Toxicology 2005; 96: 271–281. [DOI] [PubMed] [Google Scholar]
- 12.Jensen VF, et al. Correlation between apramycin and gentamicin use in pigs and an increasing reservoir of gentamicin-resistant Escherichia coli. Journal of Antimicrobial Chemotherapy 2006; 58: 101–107. [DOI] [PubMed] [Google Scholar]
- 13.Cavaco LM, et al. Selection and persistence of CTX-M-producing Escherichia coli in the intestinal flora of pigs treated with amoxicillin, ceftiofur, or cefquinome. Antimicrobial Agents and Chemotherapy 2008; 52: 3612–3616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Vieira AR, et al. Association between tetracycline consumption and tetracycline resistance in Escherichia coli from healthy Danish slaughter pigs. Foodborne Pathogens and Disease 2009; 6: 99–109. [DOI] [PubMed] [Google Scholar]
- 15.Chantziaras I, et al. Correlation between veterinary antimicrobial use and antimicrobial resistance in food-producing animals: a report on seven countries. Journal of Antimicrobial Chemotherapy 2014; 69: 827–834. [DOI] [PubMed] [Google Scholar]
- 16.Colloneau L, et al. Guidance on the selection of appropriate indicators for quantification of antimicrobial usage in humans and animals. Zoonoses and Public Health 2017; 64: 165–174. [DOI] [PubMed] [Google Scholar]
- 17.Aarestrup FM, et al. Effect of abolishment of the use of antimicrobial agents for growth promotion on occurrence of antimicrobial resistance in fecal enterococci from food animals in Denmark. Antimicrobial Agents and Chemotherapy 2001; 45: 2054–2059. doi: 10.1128/AAC.45.7.2054-2059.2001 [doi]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Agersø Y, Aarestrup FM. Voluntary ban on cephalosporin use in Danish pig production has effectively reduced extended-spectrum cephalosporinase-producing Escherichia coli in slaughter pigs. Journal of Antimicrobial Chemotherapy 2013; 68: 20569–20572. [DOI] [PubMed] [Google Scholar]
- 19.Martinez JL, Coque TM, Baquero F. What is a resistance gene? Ranking risk in resistomes. Nature Reviews Microbiology 2015; 13: 116–123. doi: 10.1038/nrmicro3399. [DOI] [PubMed] [Google Scholar]
- 20.Emborg HD, et al. Tetracycline consumption and occurrence of tetracycline resistance in Salmonella typhimurium phage types from Danish pigs. Microbial Drug Resistance 2007; 13: 289–294. doi: 10.1089/mdr.2007.746. [DOI] [PubMed] [Google Scholar]
- 21.Rosengren LB, et al. Associations between feed and water antimicrobial use in farrow-to-finish swine herds and antimicrobial resistance of fecal Escherichia coli from grow-finish pigs. Microbial Drug Resistance 2007; 13: 261–270. [DOI] [PubMed] [Google Scholar]
- 22.Timmerman T, et al. Quantification and evaluation of antimicrobial drug use in group treatments for fattening pigs in Belgium. Preventive Veterinary Medicine 2006; 74: 251–263. doi: 10.1016/j.prevetmed.2005.10.003. [DOI] [PubMed] [Google Scholar]
- 23.Sjölund M, et al. Quantitative and qualitative antimicrobial usage patterns in farrow-to-finish pig herds in Belgium, France, Germany and Sweden. Preventive Veterinary Medicine 2016; 130: 41–50. [DOI] [PubMed] [Google Scholar]
- 24.Vigre H, et al. Use of register data to assess the association between use of antimicrobials and outbreak of postweaning multisystemic wasting syndrome (PMWS) in Danish pig herds. Preventive Veterinary Medicine 2010; 93: 98–109. [DOI] [PubMed] [Google Scholar]
- 25.DANMAP. Use of antimicrobial agents and occurrence of antimicrobial resistance in bacteria from food animals, foods and humans in Denmark. 2015. http://www.danmap.org/Downloads/Reports.aspx (accessed 11 January 2017).
- 26.SEGES. ANNUAL REPORT 2015. 2016. http://eng.vsp.lf.dk/~/media/Files/PDF - Aarsberetning VSP English/VSPårsberetningUK2015.pdf (accessed 11 January 2017).
- 27.Holman DB, Chénier MR. Impact of subtherapeutic administration of tylosin and chlortetracycline on antimicrobial resistance in farrow-to-finish swine. FEMS Microbiology Ecology 2013; 85: 1–13. doi: 10.1111/1574-6941.12093. [DOI] [PubMed] [Google Scholar]
- 28.Houe H, Gardner IA, Nielsen LR. Use of information on disease diagnoses from databases for animal health economic, welfare and food safety purposes: strengths and limitations of recordings. Acta Veterinaria Scandinavica 2011; 53: S7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Stege H, et al. VETSTAT – the Danish system for surveillance of the veterinary use of drugs for production animals. Preventive Veterinary Medicine 2003; 57: 105–115. [DOI] [PubMed] [Google Scholar]
- 30.Jensen VF, Jacobsen E, Bager F. Veterinary antimicrobial-usage statistics based on standardized measures of dosage. Preventive Veterinary Medicine 2004; 64: 201–215. doi: 10.1016/j.prevetmed.2004.04.001. [DOI] [PubMed] [Google Scholar]
- 31.Christiansen MG. No 1429: The Structural Development in Danish Pig Production 2014 [in Danish only]. SEGES Pig Research Centre. 2014; 11. http://vsp.lf.dk/Publikationer/Kilder/Notater/2014/1429.aspx (accessed 11 January 2017).
- 32.Jensen T, et al. Assessment of future production systems for pigs [in Danish only. Summary available in English]. Videncenter for Svineproduktion 2011. http://vsp.lf.dk/Publikationer/Kilder/lu_Rapporter/38.aspx (accessed 11 January 2017).
- 33.SEGES. ANNUAL REPORT 2014. 2015. http://eng.vsp.lf.dk/~/media/Files/PDF - Aarsberetning VSP English/Årsberetning 2014_UK.pdf (accessed 11 January 2017).
- 34.Maribo H. The feeding of suckling piglets [in Danish only]. SEGES Pig Research Centre. 2010; 2. http://vsp.lf.dk/Viden/Foder/Tildeling af foder/Fodring af/Fodring af pattegrise.aspx?full=1 (accessed 11 January 2017).
- 35.Jessen O. No 1523: National average pig productivity 2014 [in Danish only]. 2015; 1–16. http://vsp.lf.dk/~/media/Files/PDF - Publikationer/Notater 2015/Notat_1523.pdf (accessed 11 January 2017).
- 36.Abatih EN, et al. Impact of antimicrobial usage on the transmission dynamics of antimicrobial resistant bacteria among pigs. Journal of Theoretical Biology 2009; 256: 561–573. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
For supplementary material accompanying this paper visit http://dx.doi.org/10.1017/S0950268817001285.
click here to view supplementary material