Abstract
Several actinomycetes isolated from nature were able to use both natural rubber (NR) and synthetic cis-1,4-polyisoprene rubber (IR) as a sole source of carbon. According to their degradation behavior, they were divided into two groups. Representatives of the first group grew only in direct contact to the rubber substrate and led to considerable disintegration of the material during cultivation. The second group consisted of weaker rubber decomposers that did not grow adhesively, as indicated by the formation of clear zones (translucent halos) around bacterial colonies after cultivation on NR dispersed in mineral agar. Taxonomic analysis of four selected strains based on 16S rRNA similarity examinations revealed two Gordonia sp. strains, VH2 and Kb2, and one Mycobacterium fortuitum strain, NF4, belonging to the first group as well as one Micromonospora aurantiaca strain, W2b, belonging to the second group. Schiff's reagent staining tests performed for each of the strains indicated colonization of the rubber surface, formation of a bacterial biofilm, and occurrence of compounds containing aldehyde groups during cultivation with NR latex gloves. Detailed analysis by means of scanning electron microscopy yielded further evidence for the two different microbial strategies and clarified the colonization efficiency. Thereby, strains VH2, Kb2, and NF4 directly adhered to and merged into the rubber material, while strain W2b produced mycelial corridors, especially on the surface of IR. Fourier transform infrared spectroscopy comprising the attenuated total reflectance technique was applied on NR latex gloves overgrown by cells of the Gordonia strains, which were the strongest rubber decomposers. Spectra demonstrated the decrease in number of cis-1,4 double bonds, the formation of carbonyl groups, and the change of the overall chemical environment, indicating that an oxidative attack at the double bond is the first metabolic step of the biodegradation process.
cis-1,4-polyisoprene, with an average molecular mass of about 106 Da, is the main constituent (>90% of dry weight) of natural rubber (NR) obtained from the latex of Hevea brasiliensis (rubber tree) for commercial purposes. Alternatively, cis-1,4-polyisoprene in the same mass range is synthesized chemically to obtain the so-called isoprene rubber (IR). These raw rubbers are usually converted into rubber products by the process of vulcanization that leads to cross-links between the polymer chains either by heating in the presence of elemental sulfur (e.g., during the manufacture of tires that also contain other kind of synthetic rubbers) or by irradiation and peroxidation, respectively, like in the case of NR latex gloves (21).
The microbial susceptibility of NR either in the raw or in the vulcanized state was sufficiently examined and reviewed (20, 28). Several microorganisms were isolated from such experiments, and pure cultures were tested for their rubber-degrading potential. Results showed that actinomycetes were almost the only organisms able to considerably decompose NR and to use the rubber hydrocarbon as a carbon source (6, 9, 12, 13, 17, 24). The first indication for the mechanism involved in the biodegradation of the cis-1,4-polyisoprene chain was obtained after analysis of the degradation products. Here, both NR latex gloves cultivated with a Nocardia strain (24) and raw NR latex treated with a crude extracellular extract of a Xanthomonas strain (25) led to the accumulation of oligomers with molecular masses between 103 and 104 Da. Infrared and nuclear magnetic resonance spectroscopy revealed the occurrence of carbonyl groups for each oligomer at both ends, suggesting cleavage of the polymeric chain by oxygenative attack at the double bonds. It is remarkable that analogous results were obtained with two taxonomically different microorganisms also exhibiting varying rubber-degrading properties (the Xanthomonas strain is a very weak decomposer of solid rubber in contrast to the Nocardia strain [23]).
Recently, several actinomycetes with chemotaxonomic characteristics of the genus Gordonia (formerly known as Gordona) could be isolated, showing enhanced solubilization, disintegration, and mineralization of NR, NR latex gloves, and IR (13). One of the strains was already taxonomically classified as the novel species Gordonia polyisoprenivorans Kd2T (DSM 44302T) (14).
In this report, special emphasis is given to the degradation mechanism of two still-unclassified Gordonia species with strong rubber-decomposing properties. In comparative studies with two other actinomycetes, different strategies towards the biodegradation of cis-1,4-polyisoprene are pointed out, thus providing a basis for the planning of future screening experiments.
MATERIALS AND METHODS
Rubbers.
NR latex concentrate (Neotex Latz) was obtained from Weber & Schaer (Hamburg, Germany) and IR (SK13) was from Continental AG (Hannover, Germany). NR latex gloves (rotiprotect) were purchased from Roth (Karlsruhe, Germany).
Microorganisms.
Strains VH2 (DSM 44266), Kb2 (DSM 44215) and NF4 (DSM 44216) were isolated as reported previously (13). Strain W2b (DSM 44438) was isolated from the same fouling water inside of a deteriorated old tire on a farmer's field in Münster, Germany, like strain G. polyisoprenivorans Kd2T (DSM 44302) (14).
Cultures.
Liquid cultures were carried out in Erlenmeyer flasks containing mineral salts medium (MSM), as described previously (18). NR latex gloves were cut into pieces with masses of 0.25 g and added either untreated or after extraction with acetone (1 to 2 days) to 50 ml of MSM in 500-ml flasks and subsequently autoclaved. IR was treated as follows: 3 g was extracted with 100 ml of acetone (1 to 2 days) and dissolved in 100 ml of chloroform to yield a 3% IR solution. Rectangular thin aluminum pieces with a surface area of about 1 cm2 were dipped into the solution several times and dried in a stream of air. The procedure was repeated until both sides of the aluminum pieces were coated completely with IR material. The coated pieces were sterilized with 96% ethanol and added to 30 ml of already autoclaved MSM solution in 300-ml flasks. Cells were precultivated for 3 to 6 days at 30°C in Luria-Bertani complex medium, washed twice in saline solution, and inoculated in small amounts into the rubber-containing cultures. Inoculated flasks were shaken at 180 rpm and 30°C. Solid cultures were prepared in petri dishes containing MSM agar. The mineral agar was overlayed by a thin layer of NR latex concentrate being dispersed into MSM agar at a concentration of 0.02% (wt/vol). Alternatively, the latex concentrate was spread as a thin film directly on the mineral agar. Incubation of inoculated latex plates took place at 37°C.
Analysis of 16S rDNA.
Extraction of genomic DNA of strain W2b was carried out as described previously (2). An additional step at the beginning of the procedure comprised the treatment of the cell pellet with 1% (wt/vol) lysozyme in 567 μl of Tris-EDTA buffer for 12 h at 30°C before adding 30 μl of 10% (wt/vol) sodium dodecyl sulfate. Extraction of genomic DNA of the other strains as well as amplification of the 16S rRNA of all strains were performed as described previously (16). In the case of strain NF4, purified PCR products were sequenced by using the Taq DyeDeoxy Terminator Cycle Sequencing kit (Applied Biosystems) according to the manufacturer's protocol. Sequence reactions were electrophoresed by using the Applied Biosystems 373A DNA sequencer. In the case of strain W2b, the nucleotide sequences were determined with a 4000L DNA sequencer (LI-COR Inc., Biotechnology Division, London, Nebr.) and a Thermo Sequenase fluorescence-labelled primer cycle-sequencing kit (Amersham Life Science, Little Chalfont, United Kingdom) as specified by the manufacturers. The 16S rDNA sequences were aligned manually with published sequences from representatives of actinomycetes obtained from EMBL.
Staining with Schiff's reagent.
Staining of NR latex gloves with Schiff's reagent was recently shown (26). The analogous procedure applied here was as follows. In a tightly stoppered bottle, 10 ml of the fuchsin reagent was added to a sample, and the purple color was developed for 10 to 30 min at room temperature. An amount of excess reagent was then discarded, and 10 ml of the sulfite solution was added in order to suppress the nonspecific color reaction of the blank sample. The composition of the fuchsin reagent (4) was the following: 2 g of fuchsin dissolved in 50 ml of glacial acetic acid plus 10 g of Na2S2O5 plus 100 ml of 0.1 N HCl plus 50 ml of H2O. The composition of the sulfite solution was 5 g of Na2S2O5 plus 5 ml of concentrated HCl (37 to 38%) in a 100-ml aqueous solution.
Scanning electron microscopy.
NR latex gloves and IR-coated thin aluminum pieces were taken from liquid cultures at varying cultivation periods and fixed with 2.5% glutaraldehyde in 0.1 M phosphate-buffered saline (PBS; pH 7.3) according to Sørensen (1). After washing with PBS, the cultures were postfixed in 1% osmium tetroxide in 0.1 M PBS (pH 7.3) and dehydrated in graded ethanol (30, 50, 70, 90, and 96% and absolute ethanol). The dehydrated samples were subjected to critical point drying with liquid CO2 according to the standard procedure. Subsequently, the samples were mounted on aluminum specimen stubs by using electrically conducting carbon (PLANO, Wetzlar, Germany) and sputter-coated with a gold layer having a thickness of approximately 15 nm by using argon gas as the ionizing plasma. Imaging was performed with an S-450 scanning electron microscope (SEM; Hitachi Ltd., Tokyo, Japan) with secondary electrons at a 20-kV acceleration voltage and at room temperature. Micrographs were recorded from a high-resolution cathode-ray tube using negative film (Agfapan, APX 100; Agfa-Geraert AG, Leverkusen, Germany).
FTIR-ATR spectroscopy.
NR latex glove material overgrown by the Gordonia strains was first subjected to cleaning with distilled water in order to remove the microbial biofilm from the sample surface. For this purpose, the sample was mounted and fixed on a carrier, and the biofilm was scraped off very carefully with a soft cotton bud during rinsing with water. Spectra were recorded by an IFS 88 Fourier transform infrared (FTIR) spectrometer (Bruker Optics GmbH, Karlsruhe, Germany) with the attenuated total reflectance (ATR) technique, as previously reported (19). The angle of incidence was set at 45° by using a ZnSe crystal with 20 active internal reflections. Sixty scans were coadded with a resolution set at 4 cm−1. For advanced interpretation, the second derivatives of the absorbance spectra were calculated to exhibit frequency shifts and band feature alterations. In second-derivative spectra, the bands of interest appear negative. For comparative analysis, spectra were standardized by applying a vector normalization. No further spectral processing was used to ensure band frequency and band shape quality. For spectral control, measurements in the transmission mode had been performed by using ZnSe disks as sample holders.
Nucleotide sequence accession number.
The 16S rRNA gene sequence data of strain W2b have been submitted to the EMBL nucleotide sequence database and are under accession no. AJ245712.
RESULTS
Screening for rubber-degrading bacteria.
Several environmental samples were tested for microbial growth on NR being available either as a latex overlay or as a latex film on mineral agar plates. Thereby, two different microbial strategies could be recognized. Bacterial colonies appearing on the latex overlay plates produced clearing zones (translucent halos) through the opaque mineral agar. These colonies were not able to considerably grow on mineral agar plates containing a latex film on their surface. On the other hand, bacteria appearing adhesively on the latex film were neither able to grow on latex overlay plates nor able to produce clearing zones in any way. Enrichment procedures described previously (13) led to the isolation of pure cultures of several potent rubber-degrading actinomycetes. These isolates also showed adhesive growth on latex film plates and no growth on latex overlay plates, and strains VH2, Kb2, and NF4 were selected for further analysis. From the other group of bacteria producing clearing zones on latex overlay plates, one with a remarkable zone area, strain W2b, was selected.
Taxonomic classification of selected strains.
Chemotaxonomic studies on strains VH2 and Kb2 comprising analysis of cell wall components, fatty acids, mycolic acids, and 16S rRNA genes revealed novel species within the genus Gordonia. The data were similar to those obtained for G. polyisoprenivorans Kd2T (14) and will be published separately for the species characterization.
The chemotaxonomic markers of strain NF4 were consistent with those of species of the genus Mycobacterium, i.e., menaquinone MK-9(H2) and mycolic acids of a chain length with about 90 carbon atoms. The cleavage product of the mycolic acid esters by trimethylsulfonium hydroxide combined with the fatty acid pattern (15) revealed a typical pattern found in members of the Mycobacterium fortuitum group. For a definite species identification, the 16S rDNA was sequenced and compared to all mycobacterium sequences available. The obtained partial sequence (500 nucleotides) revealed a 100% sequence similarity to M. fortuitum subsp. fortuitum DSM 46621, and the strain was deposited at the DSMZ (Deutsche Sammlung von Mikroorganismen und Zellkulturen, Braunschweig, Germany) as M. fortuitum NF4 (DSM 44216).
The first analysis of strain W2b by light microscopy showed morphological features, like well-developed, branched, septate mycelium that were about 0.5 μm, and nonmotile single spores, which were consistent to the genus Micromonospora according to Kawamoto (10). Subsequent analysis of 16S rDNA led to the description of almost the complete sequence, consisting of 1,477 nucleotides. According to results of the EMBL database search, the sequence revealed a 99.8% similarity to Micromonospora fulvopurpureus, 99.7% similarity to Micromonospora aurantiaca, and 99.6% similarity to Micromonospora globosa. According to Koch et al. (11), M. aurantiaca is a valid type strain and both M. fulvopurpureus and M. globosa are invalidly described species. Due to the high similarity value of >99.5%, a classification to M. aurantiaca was carried out. The next highest similarity, 99.1%, was to Micromonospora chalcea. The W2b strain was deposited in the DSMZ as M. aurantiaca W2b (DSM 44438).
Staining of NR latex gloves with Schiff's reagent.
The actively growing colonies of each of the selected strains were visualized on the surfaces of NR latex gloves after cultivation in liquid culture. The purple color produced by the reagent around the colonies was evidence that isoprene oligomers containing aldehyde groups were produced and accumulated during the microbial degradation, as was recently pointed out (26). After a cultivation period of 4 to 6 weeks with the strains Gordonia sp. strain VH2, Gordonia sp. strain Kb2, and M. fortuitum NF4, the entire glove surface was colored (noninoculated controls remained completely unstained), indicating the formation of a bacterial biofilm on the glove surface. However, colonization efficiency of M. aurantiaca W2b was still very low after this period and could be enhanced when acetone-extracted glove material was used as a carbon source. Tests performed after an incubation period of 6 weeks additionally revealed staining of very small pieces of glove material in the Gordonia cultures, especially in that of strain VH2, as a result of the beginning rubber disintegration process.
Analysis by SEM.
Degradation behavior of each of the selected strains was examined by SEM with respect to colonization, disintegration of rubber, and biofilm formation. Thereby, the adhesively growing strains Gordonia sp. strain VH2, Gordonia sp. strain Kb2, and M. fortuitum NF4 differed significantly from strain M. aurantiaca W2b.
Growth on IR is illustrated in the micrographs shown in Fig. 1. A section from the surface of a noninoculated IR control is shown in Fig. 1A. Figure 1B demonstrates colonization of this material by cells of Gordonia sp. strain VH2 after 4 days, proceeding by producing specific colony craters on the surface and by penetrating into the material. An analogous behavior was also observed for the cells of Gordonia sp. strain Kb2 after 1 week (not shown). However, colonization by Gordonia sp. strain VH2, which was the strongest rubber decomposer, hereby proceeded very fast, so that after 1 week the rubber surface was already completely coated by a biofilm (Fig. 1C), and after 4 weeks, >50% of the material was degraded. Figure 1D illustrates growth of M. fortuitum NF4 after 1 week. Beside analogous colony crater formation (not visible in this section), cells were directly embedded and merged into the rubber matrix. After 4 weeks, a complete NF4 biofilm was also formed. On the other hand, M. aurantiaca W2b cells tended to produce mycelial corridors on the material's surface after 1 week and to penetrate the rubber with hyphae (Fig. 1E). Destruction of the material increased during the cultivation period of 4 weeks (Fig. 1F), but a classical biofilm, as in case of the adhesively growing strains, was definitely not formed.
FIG. 1.
Secondary electron micrographs of rubber-degrading strains on synthetic cis-1,4-polyisoprene (IR). Shown are the noninoculated control (A), growth of Gordonia sp. strain VH2 after 4 days (B) and after 1 week (C), growth of M. fortuitum NF4 after 1 week (D), and the growth of M. aurantiaca W2b after 1 (E) and 4 (F) weeks. Bars, 5 μm (A, B, D to F) and 50 μm (C).
Figure 2 illustrates colonization and disintegration of untreated NR latex gloves by the rubber-degrading strains. Compared to the noninoculated control (Fig. 2A), growth of Gordonia sp. strain VH2 led to a considerable material disintegration after 2 weeks (Fig. 2B), as indicated on the micrograph by the tearing apart of threads of rubber material. Obviously, a similar behavior was observed with Gordonia sp. strain Kb2 (not shown). On the other hand, M. fortuitum NF4 produced some kind of elevations on the rubber surface after the same cultivation period so that borders between cells and rubber material could not be distinguished any more (Fig. 2C). Around these elevations, dispersed cells were recognizable, but the rubber surface was not affected in any way. Growth of strain M. aurantiaca W2b on NR latex gloves was rather poor (Fig. 2D). However, an increase in the roughness of the rubber surface in comparison to that of the control (Fig. 2A) could be recognized. Biofilm formation after 6 weeks on NR latex gloves is demonstrated for the strains Gordonia sp. strain VH2 (Fig. 2E) and M. fortuitum NF4 (Fig. 2F). Strain M. aurantiaca W2b did not produce an overall biofilm like the other strains. It was generally recognized that biofilms on IR exhibited a softer appearance, whereas those on NR latex gloves exhibited a harder appearance.
FIG. 2.
Secondary electron micrographs of rubber-degrading strains on NR latex glove material. Shown are the noninoculated control (A); growth after 2 weeks for Gordonia sp. strain VH2 (B), M. fortuitum NF4 (C), and M. aurantiaca W2b (D); and biofilms formed after 6 weeks for Gordonia sp. strain VH2 (E) and M. fortuitum NF4 (F). Bars, 5 μm (A to D) and 50 μm (E, F).
Analysis by FTIR-ATR spectroscopy.
NR latex glove material was significantly disintegrated by the two Gordonia strains VH2 and Kb2 after a cultivation period of 8 weeks. Rubber material from such cultures was used for further analysis by means of FTIR-ATR spectroscopy. As it was shown recently (19), this method allows a nondestructive in situ analysis of surfaces coated by microbial biofilms. Here, analysis of NR latex gloves yielded additional evidence for the formation of a biofilm on the material's surface. This was demonstrated by the absorbance spectra, where specific marker bands for bacteria could be clearly attributed compared to the noninoculated control (spectra not shown). These bands included the protein region with amide I and amide II bands at 1,652 and 1,545 cm−1, respectively, the fatty acid region from 2,800 to 3,000 cm−1, and the polysaccharide bands in the region from 900 to 1,200 cm−1 to refer to the most prominent bands (19). These bands were subsequently used as marker bands for proving removal of biofilm by the procedure described above. Absorbance spectra of a noninoculated NR latex glove recorded in the transmission and ATR mode corresponded well with the data from the literature for natural rubber (Hevea rubber SMR-5) (8).
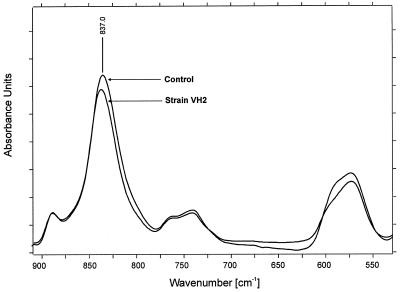
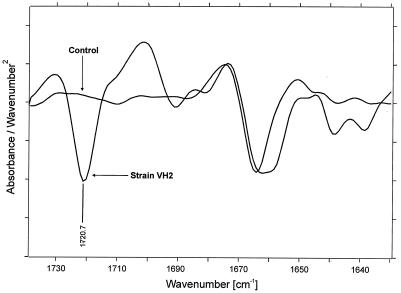
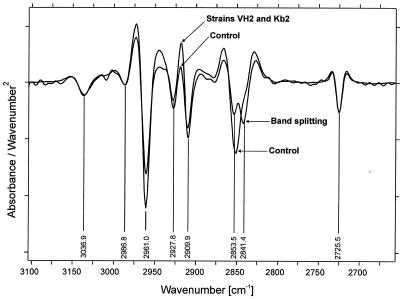
When the absorbance spectrum of the control was compared to those obtained from the samples after biofilm removal, several spectral differences became obvious. In the absorption area corresponding to the cis-1,4 double bonds in the polyisoprene chain, a relative decrease of the δ(=CH2) band at 835 cm−1 was observed for the sample compared to that of the control (Fig. 3) after normalization of the most prominent bands at 1,446, 1,373, 1,130, and 1,085 cm−1 (not shown). In the region of 1,650 to 1,750 cm−1 comprising the absorption of the carbonyl groups (Fig. 4), the appearance of a ketone band at 1,720 cm−1 (according to the literature) with a weak shoulder at 1,710 cm−1 also became obvious for the sample, as well as a broadening of the band at 1,660 cm−1, which indicates formation of aldehyde groups in the lower frequency region. Moreover, a change of the overall chemical environment became evident in the sample spectra after detection of further changes in the regions around the ν(CHx) stretching vibrations (2,800 to 3,000 cm−1) and the δ(CHx) deformation vibrations (1,400 to 1,500 cm−1). In these areas, several shifts in respect to band positions and band ratios could be determined. Among them, a splitting of the νs(CH2) band at 2,853 cm−1 into two bands at 2,855 and 2,841 cm−1 was characteristic (Fig. 5), indicating a formation of two different bonding environments. In all cases, no significant further differences between the spectra of Gordonia sp. strain VH2 and Gordonia sp. strain Kb2 occurred. Spectra in Fig. 4 and 5 are presented as second derivatives of absorbance spectra for a better interpretation and clarity of the results.
FIG. 3.
FTIR absorbance spectrum of NR latex gloves in the region of 550 to 900 cm−1 comprising the δ(=CH2) cis-1,4 double bond. The comparison is of the noninoculated control and the sample treated with Gordonia sp. strain VH2.
FIG. 4.
FTIR second-derivative spectrum of NR latex gloves in the region of 1,600 to 1,750 cm−1 comprising the carbonyl functional group (−C=O). The comparison is of the noninoculated control and the sample treated with Gordonia sp. strain VH2.
FIG. 5.
FTIR second-derivative spectrum of NR latex gloves in the region of 2,800 to 3,000 cm−1 comprising the ν(CHx) stretching vibrations. The comparison is of the noninoculated control and the samples treated with Gordonia sp. strain VH2 and Gordonia sp. strain Kb2.
DISCUSSION
Screening procedures for the isolation of rubber-degrading bacteria led to pure cultures of various actinomycetes employing two different strategies towards utilization of this solid, hydrophobic carbon source. The first group of bacteria expressed adhesive growth on rubber materials. Thereby, the strains grew in direct contact to the rubber and formed a biofilm during cultivation. Growth on latex films that were spread on mineral agar plates as well as staining with Schiff's reagent and investigation by SEM showed this phenomenon. Colonization of the rubber started by direct merging of the cells into the substrate and indicated a high hydrophobicity of the cell surface. Adhesive growth of coryneform bacteria is probably related to the presence and the chain length of mycolic acids, as was recently reported (3). Taxonomic classification of three selected strains with adhesive growth behavior, strains VH2, Kb2, and NF4, based on 16S rRNA similarity examinations, revealed two Gordonia and one Mycobacterium species. Both genera exhibit a coryneform morphology and are known to contain long mycolic acids. Although all three isolates were similar in their ability to form colony craters at the rubber surface and to finally produce a biofilm after a similar cultivation period, only the two Gordonia strains, especially strain VH2, showed a visible disintegration of vulcanized rubber material (Fig. 2B), even after a prolonged cultivation period of more than 6 weeks. On the other hand, cells of M. fortuitum NF4 were, from the beginning, exceptionally tightly attached to the rubber material (Fig. 1D and 2C), suggesting immobilization of the cells in the rubber matrix. It is well known that biofilms are additionally embedded in a polymer matrix containing polysaccharides and proteins, which is synthesized by the cells itself, and that the adhesion to surfaces is a general microbial strategy for survival as well as for utilization of solid substrates, especially in low-nutrient environments (5). Considering this and that mycobacteria are generally known as slow growers, even on nutrient-rich complex media, the growth strategy of forming biofilms can be very advantageous with respect to competition with other faster-growing microorganisms. In the case of the Gordonia strains, a strong rubber-degrading mechanism was additionally obvious. The behavior of these strains resembles that of Tsuchii's Nocardia sp. strain 835A which was studied in detail (24, 26, 27). According to the author's comments (23), the strain did not express extracellular degradation activity and was tightly bound to rubber pieces in the initial stage of growth, leading to a strong decomposition of solid rubber during cultivation. These findings correspond well to the results obtained for the Gordonia strains. It is therefore suggested that the rubber-degrading activity of all these strains is bound to the cell surface due to the expected inability of cells to transport solid rubber directly into the cell before cleavage into smaller molecules.
Secondly, one other bacterium employed a different strategy and formed clear zones on latex overlay plates. This suggests that the rubber-degrading activity did not occur at the surface of the cells. In the case of the selected and classified M. aurantiaca W2b, analysis by SEM yielded a strong indication thereof (Fig. 1E and F). The colonization behavior was clearly different from that of the other three strains. Growth on IR did not proceed by embedding into the rubber matrix, but by producing mycelial corridors on the surface of the rubber. This indicates an excretion of rubber-decomposing factors. However, in contrast to IR, the surface of NR latex gloves was not significantly affected by this microorganism (Fig. 2D), even after a prolonged cultivation period, suggesting either difficulties in breaking up cross-links in the vulcanizate or inhibition by microbicidal substances added during the manufacture of the gloves. The latter possibility is favored by the enhancement of colonization efficiency when acetone-extracted glove material was used as substrate. During the past decades, several bacteria could be isolated exhibiting the same property in forming clearing zones on latex overlay plates, like M. aurantiaca W2b. However, if the clear zone technique alone is applied as the screening method for the isolation of rubber-degrading bacteria as previously done (9, 12, 17), adhesively growing strains will not be included.
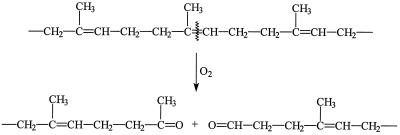
Application of the FTIR-ATR spectroscopy on NR latex gloves being used as a carbon source during cultivation of the Gordonia strains revealed insights into the biodegradation mechanism of these strong rubber decomposers. Spectra demonstrated a decrease in the number of cis-1,4 double bonds in the polyisoprene chain (Fig. 3), the appearance of ketone and aldehyde groups in the samples (Fig. 4), and the formation of two different bonding environments (Fig. 5). All these observations can be interpreted as a consequence from an oxidative reduction of the polymer chain length, thus leading to a change of the overall chemical environment. Accordingly, the biodegradation mechanism of the Gordonia strains can be described as follows: scission of the polymer chain at the cis-1,4 double bond by oxygen attack to produce carbonyl groups with an aldehyde on the one side and a ketone on the other side of each molecule (Fig. 6). Previously, an analogous mechanism was proposed for the NR latex glove degrading Nocardia sp. strain 835A according to findings obtained after analysis of extracted degradation products (24). Considering further literature data, in which an analogous oxidative cleavage is known to occur after degradation of NR latex, either extracellularly by a Xanthomonas sp. (25) or by inorganic activation of molecular oxygen (22), as well as the positive detection of aldehyde groups by the Schiff's reagent test on the surface of rubber being cultivated with each of the employed strains, it is assumed that the first metabolic step is common among all cis-1,4-polyisoprene-degrading bacteria, irrespective of their colonization strategy, implying scission of the cis-1,4 double bond by activated oxygen. However, enzymes catalyzing this reaction were hitherto neither isolated nor characterized or genetically assigned.
FIG. 6.
Proposed scheme for the biodegradation of cis-1,4-polyisoprene by oxygen attack at the double bond. (Reprinted from reference 24 with permission from the publisher.)
Physiological studies on several aspects of the biodegradation of cis-1,4-polyisoprene are nowadays still at the beginning. Therefore, a closer taxonomic classification of the selected strains was performed in order to establish a basis for future biochemical and genetic examinations in this field. Another aspect is that such bacteria could, in the future, contribute to biotechnological solutions of rubber waste treatment. As pointed out previously (7), an interesting approach would be to combine partial microbial degradation with physicochemical methods in order to obtain rubber material from waste that is suitable for recycling.
ACKNOWLEDGMENTS
We thank Cathrin Spröer from the DSMZ-Deutsche Sammlung von Mikroorganismen und Zellkulturen (Braunschweig, Germany) for carrying out the sequencing reaction for strain NF4 and Gudrun Kiefermann from the Institut für Medizinische Physik und Biophysik (Münster, Germany) for her expert photographic work. Provision of the description of the method for the staining with Schiff's reagent by Akio Tsuchii from the National Institute of Bioscience and Human-Technology, Higashi, Tsukuba, Ibaraki, Japan, is gratefully acknowledged.
REFERENCES
- 1.Arnold M. Histochemie. Einführung in die Grundlagen und Prinzipien der Methoden. Berlin, Germany: Springer; 1968. [Google Scholar]
- 2.Ausubel F M, Brent R, Kingston R E, Moore D D, Seidman J G, Smith J A, Struhl K. Current protocols in molecular biology. New York, N.Y: Wiley; 1995. [Google Scholar]
- 3.Bendinger B, Rijnaarts H H M, Altendorf K, Zehnder A J B. Physicochemical cell surface and adhesive properties of coryneform bacteria related to the presence and chain length of mycolic acids. Appl Environ Microbiol. 1993;59:3973–3977. doi: 10.1128/aem.59.11.3973-3977.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ehrlich G, Taylor H E, Waelsch H. The effect of surface-active substances on the fuchsin reaction of higher fatty aldehydes. J Biol Chem. 1948;173:547–551. [PubMed] [Google Scholar]
- 5.Flemming H-C. Relevance of biofilms for the biodeterioration of surfaces of polymeric materials. Polymer Degrad Stabil. 1998;59:309–315. [Google Scholar]
- 6.Heisey R M, Papadatos S. Isolation of microorganisms able to metabolize purified natural rubber. Appl Environ Microbiol. 1995;61:3092–3097. doi: 10.1128/aem.61.8.3092-3097.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Holst O, Stenberg B, Christiansson M. Biotechnological possibilities for waste tyre-rubber treatment. Biodegradation. 1998;9:301–310. doi: 10.1023/a:1008337708006. [DOI] [PubMed] [Google Scholar]
- 8.Hummel D O, Scholl F. Atlas of polymers and plastic analysis. 2nd ed. Vol. 2. Weinheim, Germany: Verlag Chemie; 1984. [Google Scholar]
- 9.Jendrossek D, Tomasi G, Kroppenstedt R M. Bacterial degradation of natural rubber: a privilege of actinomycetes? FEMS Microbiol Lett. 1997;150:179–188. doi: 10.1016/s0378-1097(97)00072-4. [DOI] [PubMed] [Google Scholar]
- 10.Kawamoto I. Genus Micromonospora Ørskov 1923. In: Williams S T, Sharpe M E, Holt J G, editors. Bergey's manual of systematic bacteriology. Vol. 4. Baltimore, Md: The Williams & Wilkins Co.; 1989. pp. 2442–2450. [Google Scholar]
- 11.Koch C, Kroppenstedt R M, Stackebrandt E. Intrageneric relationships of the actinomycete genus Micromonospora. Int J Syst Bacteriol. 1996;46:383–387. [Google Scholar]
- 12.Leeflang K W H. Microbiologic degradation of rubber. J Am Water Works Assoc. 1963;53:1523–1535. [Google Scholar]
- 13.Linos A, Steinbüchel A. Microbial degradation of natural and synthetic rubbers by novel bacteria belonging to the genus Gordona. Kautsch Gummi Kunstst. 1998;51:496–499. [Google Scholar]
- 14.Linos A, Steinbüchel A, Spröer C, Kroppenstedt R M. Gordonia polyisoprenivorans sp. nov., a rubber degrading actinomycete isolated from automobile tire. Int J Syst Bacteriol. 1999;49:1785–1791. doi: 10.1099/00207713-49-4-1785. [DOI] [PubMed] [Google Scholar]
- 15.Müller K-D, Schmid E N, Kroppenstedt R M. Improved identification of mycobacteria by using the Microbial Identification System in combination with additional trimethylsulfonium hydroxide pyrolysis. J Clin Microbiol. 1998;36:2477–2480. doi: 10.1128/jcm.36.9.2477-2480.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Rainey F A, Ward-Rainey N, Kroppenstedt R M, Stackebrandt E. The genus Nocardiopsis represents a phylogenetically coherent taxon and a distinct actinomycete lineage: proposal of Nocardiopsaceae fam. nov. Int J Syst Bacteriol. 1996;46:1088–1092. doi: 10.1099/00207713-46-4-1088. [DOI] [PubMed] [Google Scholar]
- 17.Rook J J. Microbiological deterioration of vulcanized rubber. Appl Microbiol. 1955;3:302–309. doi: 10.1128/am.3.5.302-309.1955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Schlegel H G, Kaltwasser H, Gottschalk G. Ein Submersverfahren zur Kultur wasserstoffoxidierender Bakterien: Wachstumsphysiologische Untersuchungen. Arch Mikrobiol. 1961;38:209–222. [PubMed] [Google Scholar]
- 19.Schmitt J, Flemming H-C. FTIR-spectroscopy in microbial and material analysis. Int Biodeterior Biodegrad. 1998;41:1–11. [Google Scholar]
- 20.Seal K J. The biodeterioration and biodegradation of naturally occurring and synthetic plastic polymers. Biodeterior Abstr. 1988;2:295–317. [Google Scholar]
- 21.Subramaniam A. The chemistry of natural rubber latex. Immunol Allergy Clin N Am. 1995;15:1–20. [Google Scholar]
- 22.Tangpakdee J, Mizokoshi M, Endo A, Tanaka Y. Novel method for preparation of low molecular weight natural rubber latex. Rubber Chem Technol. 1998;71:795–802. [Google Scholar]
- 23.Tsuchii A. Microbial degradation of natural rubber. In: Steinbüchel A, editor. Biochemical principles and mechanisms of biosynthesis and biodegradation of polymers. Weinheim, Germany: Wiley-VCH; 1999. pp. 258–264. [Google Scholar]
- 24.Tsuchii A, Suzuki T, Takeda K. Microbial degradation of natural rubber vulcanizates. Appl Environ Microbiol. 1985;50:965–970. doi: 10.1128/aem.50.4.965-970.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tsuchii A, Takeda K. Rubber-degrading enzyme from a bacterial culture. Appl Environ Microbiol. 1990;56:269–274. doi: 10.1128/aem.56.1.269-274.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tsuchii A, Takeda K, Tokiwa Y. Colonization and degradation of rubber pieces by Nocardia sp. Biodegradation. 1996;7:41–48. doi: 10.1007/BF00056424. [DOI] [PubMed] [Google Scholar]
- 27.Tsuchii A, Takeda K, Tokiwa Y. Degradation of the rubber in truck tires by a strain of Nocardia. Biodegradation. 1997;7:405–413. doi: 10.1007/BF00056424. [DOI] [PubMed] [Google Scholar]
- 28.Zyska B J. Rubber. In: Rose A H, editor. Microbial biodeterioration, economic microbiology. Vol. 6. London, England: Academic Press; 1981. pp. 323–385. [Google Scholar]