Abstract
Homemade eggplant salad, a traditional Greek appetizer, was inoculated with Escherichia coli O157:H7 NCTC 12900 supplemented with different concentrations of oregano essential oil (0.0, 0.7, 1.4, and 2.1% [vol/wt]) and stored at different temperatures (0, 5, 10, and 15°C). The product's pH was adjusted to 4.0, 4.5, or 5.0 with lemon juice. For each combination of the environmental factors, the bacterial counts were modeled, using the Baranyi model, as a function of time to estimate the kinetic parameters of the pathogen. A reduction of more than 1 log unit in E. coli O157:H7 counts was observed in all cases, and the death rate depended on the pH, the storage temperature, and the essential oil concentration. Separate quadratic models were developed with natural logarithms of the shoulder period and death rate as estimated by the growth model, as a function of temperature, pH, and oregano essential oil concentrations. These were further used to predict the population of E. coli O157:H7 NCTC 12900 from other inoculated eggplant salads at random conditions of temperature, pH, and oregano oil concentration. The predicted values were compared with viable-count measurements for validation.
Currently, there is a growing worldwide market in the sale of prepacked, chilled, ready-to-eat salads, with or without salad dressing, (e.g., mayonnaise or vinaigrette) (4). A food of similar manufacturing technology and composition is eggplant salad. The safety of these commodities is ensured mainly by low pHs (5, 28). Despite the intrinsic safety of these products, many pathogens, such as Escherichia coli O157:H7, a pathogenic bacterium which is responsible for hemorrhagic colitis and hemolytic-uremic syndrome, are able to survive (13, 28, 32, 34). The addition of essential oils (e.g., oregano essential oil) can provide safety for salad dressings (11, 18). Indeed, such natural supplements meet the current demands of consumers for minimizing chemical preservatives. Thus, it would be of interest to express quantitatively the effectiveness of such natural antimicrobial systems. This may be achieved by means of mathematical modeling of bacterial growth kinetics, providing a powerful tool for predicting the combined effect of environmental factors, including natural antimicrobials. Although growth kinetic models have been published (1, 25, 27), few studies involving modeling of microbial decline at suboptimal conditions are available. Recently, models of survival for Listeria monocytogenes, Yersinia enterocolitica, and Salmonella sp. under nonthermal inactivation conditions have been presented (8, 19, 21, 22, 23). Survival curves of bacterial populations have been proven to be sigmoidal or semisigmoidal with a “shoulder” and/or tailing region. Thus growth models, such as Gompertz and logistic models, have been shown to be applicable in cases of bacterial inactivation (19, 20, 21, 23). The aims of this study were (i) to monitor the survival-inactivation of E. coli O157:H7 in eggplant salad in the presence of oregano essential oil at different pHs and storage temperatures and (ii) to develop and validate a polynomial model which predicts the behavior of the pathogen in response to various combinations of pH, temperatures, and oregano essential oil concentration in eggplant salad.
MATERIALS AND METHODS
Bacterial strain and preparation of inoculum.
A nonpathogenic strain of E. coli O157:H7 NCTC 12900, provided by I. Ogden (Applied Food Microbiology Group, Department of Medical Microbiology of the University of Aberdeen, Aberdeen, Scotland), was used. The strain was phenotypically very similar to toxigenic strains of E. coli O157:H7, i.e., it originated from a verocytotoxigenic strain which lost its ability to produce toxin. The culture was maintained on nutrient agar (CM3; Oxoid Basingstoke, United Kingdom) slopes at 4°C. A loopful of culture was removed from a nutrient agar slope and transferred to 250 ml of brain heart infusion (BHI) broth (CM225; Oxoid) in a 1-liter flask. The strain was incubated overnight at 37°C. The final concentration of the microorganism was approximately 108 CFU ml−1.
Extraction of essential oil.
Five hundred grams of dried oregano (Origanum vulgare) was bought from the central market in Athens and placed in a 2-liter flask, and distilled water (1 liter) was added. A continuous steam distillation extraction head was attached to the flask. After steam distillation for approximately 3 h, the oil was collected and stored at 4°C.
Preparation of eggplant salad.
Two kilograms of eggplant salad was prepared with 12 eggplants, 6 slices of garlic (2 g), and 500 ml of Greek extra virgin olive oil. The eggplants were baked (at 180°C for 15 min) to soften, and then the inner parenchyma of each eggplant was removed and mixed with the other ingredients in a blender for 5 min at room temperature. The salad was then divided into portions (40 g), and amounts of lemon juice adequate to reduce the pH to 4.0, 4.5, and 5.0 were added. Different amounts of oregano essential oil were added to the above portions to give final concentrations of 0.0, 0.7, 1.4, and 2.1% (vol/wt). Finally, the samples were thoroughly mixed, inoculated with 2 ml of an overnight culture of E. coli O157:H7 NCTC 12900 at 37°C (target inoculum, 107 CFU ml−1), and stored at the appropriate experimental temperatures (0, 5, 10, and 15°C).
Enumeration of microorganisms.
For the enumeration of E. coli O157:H7, a 1-g portion of eggplant salad sample was suspended in 9 ml of Ringer's solution (strength, 1/4) and further serially diluted. A 0.1-ml volume of the appropriate dilution was spread on plates of xylose lysine decarboxylase (XLD) agar (catalog no. 1,05287; Merck, Darmstadt, Germany) and incubated at 37°C for 24 h. Additionally, plate count agar (PCA) plates were spread every two sampling times so as to compare the number of colonies on XLD agar with that on PCA agar as well as their morphologies.
Experimental design.
The study was carried out in two stages. In the first stage, a three-way analysis of variance experiment was designed. Four storage temperatures (0, 5, 10, and 15°C), three pH levels (4.0, 4.5, and 5.0) and four oregano essential-oil concentrations (0.0, 0.7, 1.4, and 2.1% [vol/wt]) were studied. Twelve random cases were duplicated (see Table 1). For each treatment, the time dependence of E. coli O157:H7 NCTC 12900 survival was monitored by the plate count spread method on XLD and PCA plates. In cases where colonies of E. coli O157:H7 were not evident on the agar plates from the 10-fold dilution, an enrichment technique was used for the resuscitation of possibly injured living cells, as follows. A 10-g portion of eggplant salad was suspended within 500 ml of Selenite Cystine enrichment broth (1,07709; Merck) and incubated at 35°C for 12 to 18 h. One milliliter of Selenite Cystine enrichment broth was serially diluted and spread on XLD plates. The objective of this stage was to measure the inactivation of E. coli O157:H7 for the development of the model.
TABLE 1.
Outputs of Baranyi modela for the survival of E. coli O157:H7 NCTC 12900 at various temperatures, pH values, and oregano essential-oil concentrations in eggplant salad
| Conditions
|
Output
|
|||||||
|---|---|---|---|---|---|---|---|---|
| Temp (°C) | pH | Oregano essential-oil concn (%, vol/wt) | Maximum DRb (h−1) | SPb (days) |
y0 (log10) CFU g−1
|
SE of fit | r2 | |
| Estimated | Observed | |||||||
| 0c | 4 | 0 | −0.315 | 4.620 | 7.607 | 7.823 | 0.462 | 0.925 |
| 0 | 4 | 0.7 | −1.060 | 4.385 | 7.280 | 7.562 | 0.300 | 0.984 |
| 0 | 4 | 1.4 | −2.781 | 0.000 | 7.753 | 7.477 | 0.618 | 0.954 |
| 0 | 4 | 2.1 | −2.654 | 0.000 | 6.073 | 6.778 | 0.616 | 0.917 |
| 0 | 4.5 | 0 | −0.377 | 13.699 | 7.812 | 8.013 | 0.456 | 0.877 |
| 0c | 4.5 | 0.7 | −0.590 | 10.184 | 7.369 | 7.739 | 0.544 | 0.940 |
| 0c | 4.5 | 1.4 | −0.937 | 7.259 | 7.145 | 7.631 | 0.512 | 0.950 |
| 0 | 4.5 | 2.1 | −2.781 | 0.000 | 7.753 | 7.477 | 0.618 | 0.954 |
| 0 | 5 | 0 | −0.097 | 0.000 | 8.047 | 7.806 | 0.513 | 0.645 |
| 0 | 5 | 0.7 | −0.589 | 13.510 | 7.653 | 8.009 | 0.727 | 0.867 |
| 0 | 5 | 1.4 | −1.075 | 11.730 | 7.535 | 7.839 | 0.315 | 0.976 |
| 0 | 5 | 2.1 | −0.829 | 8.835 | 6.462 | 7.265 | 0.488 | 0.930 |
| 5 | 4 | 0 | −0.523 | 11.277 | 7.445 | 8.079 | 0.353 | 0.970 |
| 5c | 4 | 0.7 | −0.564 | 4.587 | 7.353 | 7.562 | 0.499 | 0.949 |
| 5 | 4 | 1.4 | −1.622 | 0.000 | 7.617 | 7.45 | 0.404 | 0.976 |
| 5 | 4 | 2.1 | −2.586 | 0.000 | 5.597 | 6.778 | 1.036 | 0.772 |
| 5 | 4.5 | 0 | −0.404 | 15.086 | 7.607 | 7.826 | 0.295 | 0.932 |
| 5 | 4.5 | 0.7 | −0.557 | 11.742 | 7.569 | 7.875 | 0.377 | 0.967 |
| 5 | 4.5 | 1.4 | −0.616 | 2.955 | 7.342 | 7.559 | 0.588 | 0.913 |
| 5 | 4.5 | 2.1 | −1.613 | 0.000 | 7.247 | 7.477 | 0.326 | 0.984 |
| 5 | 5 | 0 | −0.188 | 10.914 | 7.877 | 8.217 | 0.287 | 0.882 |
| 5c | 5 | 0.7 | −1.092 | 17.954 | 7.613 | 8.017 | 0.290 | 0.970 |
| 5 | 5 | 1.4 | −0.691 | 10.925 | 7.579 | 7.792 | 0.414 | 0.963 |
| 5c | 5 | 2.1 | −1.060 | 8.881 | 6.590 | 7.322 | 0.434 | 0.941 |
| 10 | 4 | 0 | −0.835 | 12.546 | 7.357 | 7.798 | 0.601 | 0.903 |
| 10c | 4 | 0.7 | −0.548 | 4.944 | 7.454 | 7.562 | 0.484 | 0.942 |
| 10 | 4 | 1.4 | −1.607 | 0.000 | 7.205 | 7.450 | 0.355 | 0.978 |
| 10 | 4 | 2.1 | −2.559 | 0.000 | 5.740 | 6.778 | 0.936 | 0.805 |
| 10c | 4.5 | 0 | −0.174 | 9.972 | 7.642 | 8.064 | 0.438 | 0.770 |
| 10 | 4.5 | 0.7 | −0.666 | 14.380 | 7.545 | 7.881 | 0.464 | 0.938 |
| 10 | 4.5 | 1.4 | −0.323 | 0.000 | 7.736 | 7.531 | 1.024 | 0.621 |
| 10c | 4.5 | 2.1 | −2.573 | 0.000 | 7.075 | 7.477 | 0.475 | 0.968 |
| 10 | 5 | 0 | −0.049 | 0.000 | 7.846 | 8.230 | 0.222 | 0.689 |
| 10 | 5 | 0.7 | −0.069 | 0.000 | 7.897 | 7.845 | 0.267 | 0.735 |
| 10c | 5 | 1.4 | −1.018 | 11.844 | 7.333 | 7.814 | 0.493 | 0.928 |
| 10 | 5 | 2.1 | −2.060 | 11.595 | 6.250 | 7.265 | 0.595 | 0.870 |
| 15 | 4 | 0 | −0.508 | 4.374 | 7.653 | 7.851 | 0.586 | 0.913 |
| 15c | 4 | 0.7 | −1.301 | 5.033 | 7.327 | 7.562 | 0.280 | 0.983 |
| 15 | 4 | 1.4 | −1.041 | 0.000 | 6.751 | 7.450 | 0.959 | 0.856 |
| 15c | 4 | 2.1 | −4.839 | 0.000 | 6.300 | 6.778 | 0.765 | 0.882 |
| 15 | 4.5 | 0 | −0.636 | 6.325 | 7.813 | 7.903 | 0.633 | 0.931 |
| 15c | 4.5 | 0.7 | −0.903 | 13.231 | 7.581 | 8.152 | 0.351 | 0.963 |
| 15 | 4.5 | 1.4 | −0.399 | 0.000 | 7.787 | 7.604 | 0.503 | 0.936 |
| 15 | 4.5 | 2.1 | −0.989 | 0.000 | 6.593 | 7.477 | 0.601 | 0.931 |
| 15 | 5 | 0 | −0.192 | 8.521 | 7.796 | 8.000 | 0.450 | 0.826 |
| 15 | 5 | 0.7 | −0.439 | 10.205 | 7.576 | 7.816 | 0.454 | 0.944 |
| 15 | 5 | 1.4 | −0.714 | 9.404 | 7.235 | 7.681 | 0.567 | 0.915 |
| 15c | 5 | 2.1 | −0.506 | 0.000 | 6.976 | 7.212 | 0.870 | 0.768 |
See reference 3.
Estimated by the model. No model was developed for the parameter yend (see Results).
Cases replicated.
In the second stage, a similar independent experiment was conducted to validate the model. In total, two storage temperatures (7 and 10°C), five pH levels (4, 4.2, 4.3, 4.5, and 4.7), and seven concentrations of oregano essential oil (0.05, 0.5, 0.7, 1.0, 1.4, 1.7, and 2.1 [vol/wt]) were tested against the pathogen in a new batch of eggplant salad, prepared identically. The population of E. coli O157:H7 was assessed immediately after the inoculation and at various times during the storage period, which lasted 25 days.
Model development and validation.
The data from plate counts were transformed to log10 values. At each combination of oregano essential oil, storage temperature, and pH, 10 to 15 bacterial-count points were plotted against time. The Baranyi model (2, 3) was fitted to the logarithm of the viable-cell concentration. For curve fitting, the in-house program DMFit (Institute of Food Research, Reading, United Kingdom), which was kindly provided by J. Baranyi, was used.
The Baranyi model (2, 3) is based on four parameters: a parameter expressing the lag phase, which in the case of inactivation curves will be regarded as the shoulder period, or survival period (SP); DR, the death rate (log10 CFU per gram per day); y0, representing the upper asymptote, which corresponds to the initial bacterial counts (log10 CFU per gram; and yend, representing the lower asymptote, which corresponds to final bacterial counts (log10 CFU per gram). Using response surface methodology (6, 9, 17, 24), these parameters can be further expressed as a quadratic function of temperature, pH, and essential oil concentration using the following equation:
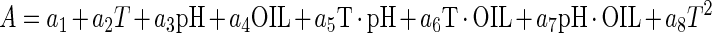 |
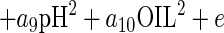 |
where A is any of the Baranyi model parameters or a logarithmic transformation (ln) of them in order to stabilize their variance (34); ai (where “i” represents any number from 1 to 10) are the coefficients to be estimated; T is temperature; OIL is the essential oil concentration (percent); e is a random error; and pH has its usual meaning.
The fitted parameters can be used to estimate the population of E. coli O157:H7, under given conditions and at given times, by interpolation. This is performed by obtaining predictions for DR and SP at specific conditions (temperature, pH, oregano essential oil concentration) and further simulating a survival curve in accordance with the Baranyi model. The indices employed for the evaluation of the performance of the developed predictive model were the bias and accuracy factors (B and A, respectively) (30), as well as their modified forms (equations 1 and 2 respectively), proposed by J. Baranyi (personal communication):
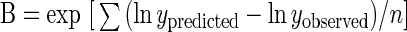 |
1 |
 |
2 |
where y is the response variable and n is the number of observations. The bias factor is a multiplicative factor by which a model over- or underpredicts, and the accuracy factor is a measure of the average difference between observed and predicted values (30). Perfect agreement between predictions and observations leads to bias and accuracy factors equal to 1, while the reverse is true only for the accuracy factor, due to its definition (see equation 2). Values higher than 1 for the bias factor indicate that predicted values are larger than observed ones, while values below 1 indicate the opposite. Although to date, interpretation of bias and accuracy factors has been used for comparison of time-based measurements, such as generation time (1) and time to 1,000-fold increase (10), in the present study, the same concept is applied to viable-count data. The latter is the result of time-based measurements, i.e., shoulder period and death rate, given by the developed secondary models.
The two complementary indices are termed the percent discrepancy (%D) (equation 3) and the percent bias (%B) (equation 4), proposed by Baranyi (personal communication) and calculated as follows:
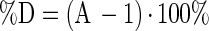 |
3 |
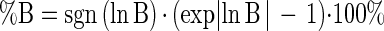 |
4 |
sgn (ln B) is equal to +1, 0, or −1 when ln B is positive, zero, or negative, respectively. By the definition of the percent bias, it is evident that the sign of the factor is determined by the value of sgn (ln B). If %B is positive, then, on average, the model predicts higher values than the real observations; the opposite is true for negative %B values.
RESULTS
Effect of essential oil alone and combined with pH and storage temperature.
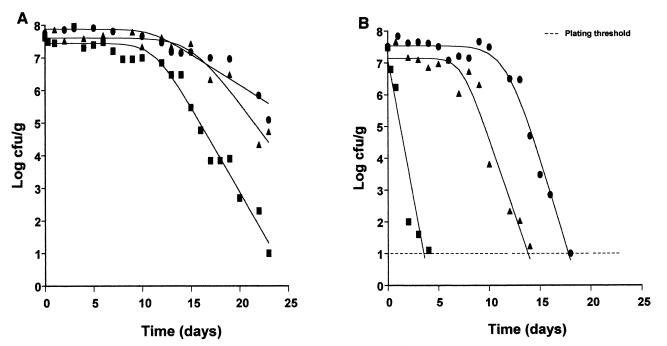
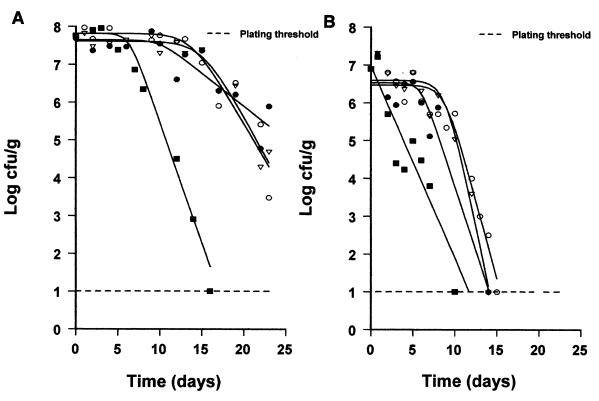
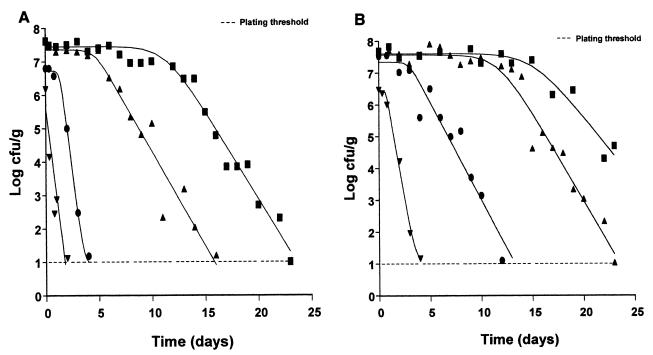
A total of 48 survival curves corresponding to different combinations of pH, temperature, and oregano oil concentrations were generated with the Baranyi model. In Table 1, the outputs of kinetic characteristics of the model are shown. The temperature of storage, the amount of oregano essential oil, and the pH of the eggplant salad influenced the kinetic characteristics of E. coli O157:H7 (Table 2). Indeed, the three-way analysis of variance showed that the death rate and survival period were affected by all the above-mentioned factors (Table 2). In general, the addition of oregano essential oil in eggplant salad resulted in an increase in the death rate of E. coli (Table 2; Fig. 1 through 3) and a reduction in the survival period of E. coli. The Baranyi model parameter y0, corresponding to the initial bacterial population, was not significantly affected by the tested factors. Within 25 days (sampling period), the inactivation curves did not show any tailing region. Thus, no model was developed for the parameter yend. The above-mentioned effects on the death rate and survival period of E. coli O157:H7 were fitted using the quadratic model and demonstrated through response surfaces in Fig. 4. In most cases, the behavior of E. coli O157:H7 followed similar patterns, meaning a survival period (shoulder of curve) and then a decline (Fig. 1 and 2). This profile was identical throughout the storage period, regardless of the enumeration medium, i.e., XLD or PCA. Indeed, the counts did not differ more than 0.5 log unit (results not shown).
TABLE 2.
F Values for the single factors and their interactions, which affect the survival kinetics of E. coli O157:H7 NCTC 12900 in eggplant salad
| Factor(s)a | Response variable
|
|||
|---|---|---|---|---|
| Death rate (h−1)
|
Survival period (days)
|
|||
| F | P | F | P | |
| A | 2.694 | 0.056 | 5.392 | 0.003 |
| B | 144.561 | 0.000 | 42.908 | 0.000 |
| C | 220.309 | 0.000 | 48.044 | 0.000 |
| A · B | 5.431 | 0.000 | 1.560 | 0.179 |
| A · C | 5.687 | 0.000 | 1.928 | 0.070 |
| B · C | 37.892 | 0.000 | 12.396 | 0.000 |
| A · B · C | 7.596 | 0.000 | 2.368 | 0.009 |
A, temperature; B, pH; C, oregano essential-oil concentration (percentage, vol/wt).
FIG. 1.
Survival curves of E. coli O157:H7 NCTC 12900 in eggplant salad stored at 5°C, at pHs 4.0 (■), 4.5 (▴), and 5.0 (●), without (A) and with (B) 1.4% (vol/wt) oregano essential oil, fitted with the Baranyi model.
FIG. 3.
Survival curves of E. coli O157:H7 NCTC 12900 in eggplant salad stored at 0°C (○), 5°C (▿), 10°C (●), and 15°C (■) at pH 4.5 without oregano essential oil (A) and at pH 5.0 with 1.4% (vol/wt) oregano essential oil (B), fitted with the Baranyi model.
FIG. 4.
Quadratic response surfaces, predicting the survival period (A) and death rate (B) of E. coli O157:H7 NCTC 12900 in eggplant salad, as a function of temperature (T)–oregano essential oil (OIL%) and pH–oregano essential oil, respectively.
FIG. 2.
Survival curves of E. coli O157:H7 NCTC 12900 in eggplant salad stored at 5°C without oregano essential oil (■) and with 0.7% (▴), 1.4% (●) and 2.1% (▾) (vol/wt) oregano essential oil at pH 4.0 (A) and at pH 4.5 (B), fitted with the Baranyi model.
Development and validation of the model.
The estimated kinetic parameters of the Baranyi model (DR and SP), for each individual survival response (48 in total) were further expressed as functions of the controlling factors. Indeed, by using response surface methodology, it was possible to generate equations predicting the survival of E. coli O157:H7 as a function of the environmental factors temperature, pH, and oregano oil concentration (OIL) (Fig. 4). For each of the above-mentioned response variables (DR and SP), regression analysis of variance versus the average as obtained from the replicates was performed to select the proper transformation for the dependent variables of models (35). Among the tested combinations (data not shown), only [variance(y)/average(y)]2 versus average(y) indicated no significant correlation (t = 0.123), and thus log transformations (35) are suitable. Consequently, the significant (P < 0.05) coefficients and the correlation coefficients (r2) of the equations expressing the dependence of the bacterial survival kinetic parameters on the studied factors are as follows:
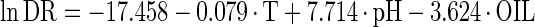 |
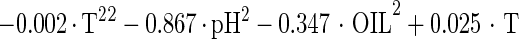 |
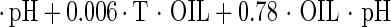 |
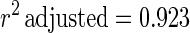 |
Standard error of fit=0.567
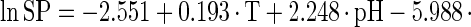 |
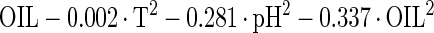 |
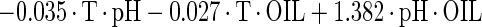 |
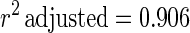 |
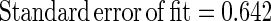 |
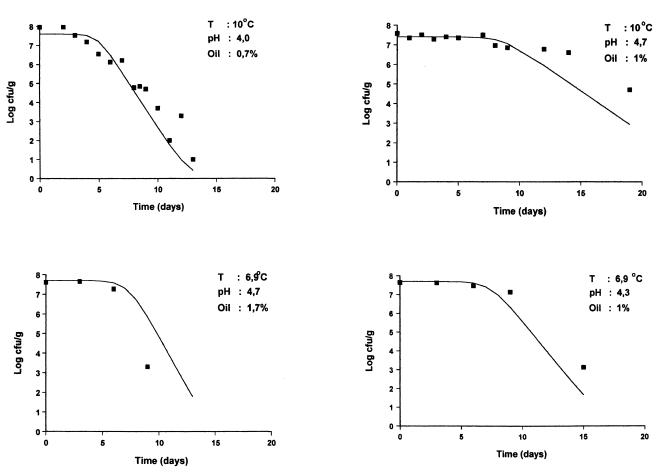
To validate the model, the observed populations of E. coli obtained from the second stage of the study were compared to the predicted populations, which were estimated by using the Baranyi model (2, 3) with kinetic parameters calculated at some random combinations of pH, oil concentration, and temperature at different time intervals. The observed and predicted counts of E. coli are shown in Table 3. Moreover, some typical comparisons are visualized (Fig. 5) by plotting the observed data points and the predicted inactivation curves of the bacterium on the same graph.
TABLE 3.
Predicted and observed populations of E. coli O157:H7 NCTC 12900 in eggplant salad under random environmental conditions at various time intervals
| Conditions
|
Time (days) | Population (log10 CFU g−1)
|
|||
|---|---|---|---|---|---|
| Temp (°C) | pH | Essential oil concn (%) | Predicted | Observed | |
| 6.9 | 4.5 | 0.5 | 0 | 7.70 | 7.62 |
| 3 | 7.70 | 7.61 | |||
| 9 | 7.66 | 7.38 | |||
| 6.9 | 4.7 | 1.0 | 0 | 7.70 | 7.60 |
| 6 | 7.60 | 7.55 | |||
| 9 | 7.20 | 7.34 | |||
| 10 | 4.7 | 1.7 | 0 | 7.70 | 7.60 |
| 3 | 7.65 | 7.65 | |||
| 6 | 5.41 | 7.26 | |||
| 9 | 3.11 | 3.30 | |||
| 10 | 4.0 | 0.5 | 0 | 7.50 | 7.64 |
| 6 | 7.38 | 7.09 | |||
| 9 | 6.52 | 5.00 | |||
| 10 | 4.5 | 1.4 | 0 | 7.60 | 7.57 |
| 0.6 | 7.60 | 6.54 | |||
| 0.8 | 7.60 | 4.08 | |||
| 2 | 7.60 | 3.95 | |||
| 10 | 4.0 | 0.7 | 0 | 7.60 | 7.96 |
| 3 | 7.59 | 7.53 | |||
| 7 | 5.56 | 6.20 | |||
| 8 | 4.60 | 4.78 | |||
| 9 | 3.65 | 4.69 | |||
| 10 | 2.69 | 3.69 | |||
| 12 | 0.98 | 3.30 | |||
| 13 | 0.43 | 1.00 | |||
| 10 | 4.5 | 0.7 | 0 | 7.60 | 6.91 |
| 2 | 7.60 | 7.78 | |||
| 3 | 7.60 | 7.88 | |||
| 6 | 7.47 | 7.15 | |||
| 8 | 6.89 | 6.28 | |||
| 9 | 6.50 | 5.79 | |||
| 10 | 6.11 | 6.20 | |||
| 11 | 5.71 | 5.20 | |||
| 12 | 5.32 | 4.69 | |||
| 14 | 4.53 | 1.80 | |||
| 16 | 3.74 | 1.00 | |||
FIG. 5.
Survival of E. coli O157:H7 NCTC 12900 (data points) in home-made eggplant salad and predicted survival curves by Baranyi model.
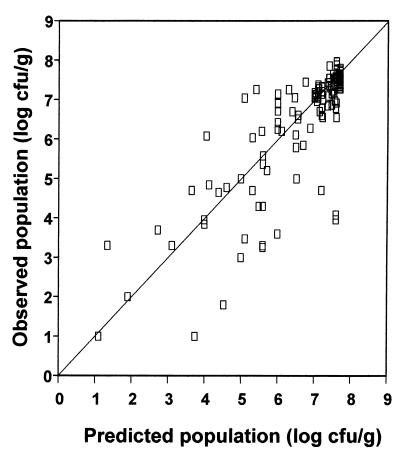
Simulation curves were generated by the predicted survival kinetics of E. coli O157:H7 based on the developed models. The entire validation is represented in Fig. 6, where predicted values are plotted against the actual measured population. The bias and accuracy factors (30; J. Baranyi, personal communication) are 1.033 and 1.233, respectively. The %B is 3.3%, i.e., positive and very low, indicating that the population, on average, was slightly overestimated by the models (Fig. 6). Accordingly, the %D between the predicted and the observed population was 23.3%. The above numbers indicate that the models produce “fail-safe” predictions (30).
FIG. 6.
Comparison of the observed population of E. coli O157:H7 NCTC 12900 in eggplant salad under various pHs, storage temperatures, and oregano essential oil concentrations with the population predicted by the quadratic model based on Baranyi estimates.
It seems that the accuracy of our models was higher for large populations (Fig. 6), which correspond to early stages of storage, indicating that good predictions are obtained in all cases during the shoulder of survival curve. In contrast, the variance of correlated values increased as the measured population decreased. The latter observations are confirmed by segregating the total validation data and calculating %B and %D separately for the shoulder and inactivation periods. Indeed, 78 data points in the shoulder region give %B equal to 2.67% and %D equal to 11.05%, while the %B and %D estimated from 83 points within the inactivation phase are 5.83 and 33.01%, respectively. Thus, it is evident that the total %B and %D are highly affected by the %B and %D of the shoulder and inactivation phases.
DISCUSSION
The survival of many pathogens, including E. coli O157:H7, in acidic products such as mayonnaise-based salad dressing or vinaigrette depends on a variety of extrinsic factors (temperature and oxygen limitation) as well as intrinsic factors (e.g., the acidity in the aqueous phase, the organic acid content of the acidulant used [e.g., lemon juice or vinegar], the actual pH, the amount and type of oil used, etc.) (13, 16, 28, 29).
Thus, the use of essential oils can be considered an additional intrinsic determinant (hurdle) for their safety (18). The use of essential oils in foods as preservatives is limited (11, 18, 31); possible reasons for this limitation may be the strong smell of these substances when used at effective doses and the decrease in their effectiveness when they are added to complicated food ecosystems (11) compared with microbiological media. In salads and dressings, spices, which are the main source of essential oils, are part of the product formulation as flavoring agents, and thus the problem is moderated. In the present study, oregano essential oil is examined as an alternative natural additive and found to contribute to the intrinsic safety of eggplant salad, acting synergistically with low pHs and storage temperatures. Additionally, concentrations of essential oil as low as 0.7% appeared to be effective and organoleptically acceptable as well. Indeed, a higher degree of inactivation of E. coli O157:H7, in both the presence and the absence of essential oil, was evident at low pH (Fig. 1 and 2; Table 2). This can be attributed (i) to the fact that the essential oil becomes more hydrophobic at low pH and thus can be dissolved better in the lipid phase of the bacterial membrane (16) and (ii) to the synergistic effect of lemon juice (citric acid) added, due to the higher undissociated form of the latter at such a low pH. On the other hand, the extended survival of E. coli in most cases in this study is consistent with previous results in acidified environments. In particular, E. coli has been proven to survive at pHs of <4.0, in synthetic gastric fluid, apple juice, and Trypticase soy broth (TSB), in the presence of HCl or/and organic acids (32). Similar results were reported in TSB when pH was adjusted by addition of HCl (7).
As far as the effect of temperature on the death rate of salmonellae and E. coli is concerned, similar results have been reported by other researchers (12, 13, 18, 28). In particular, during storage of such products at relatively high temperatures (15 to 22°C), a marked decrease in the bacterial population was observed, while lower temperatures protected Salmonella sp. and E. coli. These findings are consistent with the results of the present study, where survival of E. coli was greater as the temperature decreased (Fig. 3). It has been suggested that the protective effect of low temperatures on survival of acidification arises from alteration of the kinetics of protein denaturation (5).
Predictive microbiology has been used to describe the effect of environmental factors and interactive effects on the growth, survival, and inactivation of food-borne bacteria (1, 6, 7, 19, 20, 23, 24). In such cases, the deviation from the observed data could be mainly attributed to (i) the indication that many bacterial inactivation curves were not linear (19, 20, 21, 22, 23), (ii) the fact that these models have been constructed with data derived from broths only and not from foods, and (iii) the fact that validation was based mainly on literature data. Indeed, laboratory medium-produced growth models overestimate, on average, the responses assessed in foods (6, 10). Moreover, the deviation of laboratory medium results from those obtained with foods is also reflected in the inhibitory effect of some natural antimicrobials, e.g., the inhibitory effect of essential oils, which is significantly reduced in foods compared to studies with broths (11). Inactivation curves with an atypical linear form were also evident in our study (Fig. 1 through 3). In the majority of cases, an initial shoulder was evident, followed by an exponential reduction phase. Several mathematical models, such as the Gompertz, Baranyi, and logistic models, previously used for describing bacterial growth have also been used in studies with bacterial decline (18, 19, 20, 21, 23). Although the better overall “performance” of the Baranyi model over the Gompertz model with microbial growth curves has been established (3, 24, 26), the use of one model or the other in the case of inactivation curves should be guided by specific needs (14). Moreover, the former model would theoretically be capable of fitting all commonly shaped survival curves, such as linear, sigmoidal, and semisigmoidal (linear with a tailing region) curves. It has also been suggested that the Baranyi model may be useful as an alternative model for describing the inactivation of bacteria in suboptimal environments, such as those where natural antimicrobials are present (18).
In the present study, model development and validation were performed with data from challenge tests in food (eggplant salad). It is important to validate models with data independent of those used for developing the model. Usually, studies involving the development and/or validation of predictive models in broth and foods are based on literature data and data from challenge tests as well (6, 10, 15, 18, 20, 23, 33, 35). The use of a quadratic model (based on estimates from the Baranyi growth model) could provide relatively reasonable predictions of E. coli O157:H7 responses to pH, temperature, and oregano essential oil concentration, in eggplant salad, at least when applied to a self-consistent system. The above independent variables were regarded as some of the main intrinsic and extrinsic factors which determined the survival of the pathogen in this product. However, the contrasting indications between %B and %D may be associated with (i) the fact that the majority of data points (Fig. 6) are distributed in large populations (7 to 9 log units), i.e., in the shoulder region, and this may heavily influence the accuracy measures of the model; (ii) possible differences in batches of eggplant salad for the validation of the model due to the variability of factors related to product formulation (e.g., olive oil concentration, type of oregano oil, proportion of solid garlic, etc.). The models presented were developed considering the pH as a stable vector (it was not altered during storage), which is directly dependent on the amount of lemon juice (citric acid) added. The effect of pH was also considered in other growth modeling studies with Y. enterocolitica and E. coli (1, 27). In contrast, the death rate of E. coli was modeled in the present study.
The bias and accuracy factors were first introduced as indices for the performance of kinetic models, and as such, they are suitable for comparing time-based measures of microbial responses (e.g., maximum specific growth rates, generation times, time for a 1,000-fold increase in cell numbers, etc.) (10, 30). In the present study, an effort was made to expand the application of bias and accuracy factors by comparing bacterial counts obtained at specific combinations of the environmental factors tested. These data derived indirectly from time-based kinetic parameters (death rate and survival period) of the bacterial population, as predicted by two separate polynomial models. The calculated bias and accuracy factors, as well as the proposed percent bias and discrepancy, seem to be consistent with the graphical comparison of predicted and observed bacterial counts (Fig. 6), indicating that the specific approach of performance indices is also suitable for the evaluation of such predictive models.
With respect to the aim of this study, i.e., modeling the effects of temperature, pH, and especially oregano essential-oil on the survival of E. coli O157:H7, the development of polynomial models, based on Baranyi model estimates of survival kinetics, appeared to be a promising means of predicting the responses of this bacterium in eggplant salad.
ACKNOWLEDGMENTS
Part of this research was funded by DGXII FAIR CT96-1066.
We thank J. Baranyi for valuable comments related to the mathematical processing of our results.
REFERENCES
- 1.Adams M R, Little C L, Easter M C. Modelling the effect of pH, acidulant and temperature on the growth rate of Yersinia enterocolitica. J Appl Bacteriol. 1991;71:65–71. [PubMed] [Google Scholar]
- 2.Baranyi J, Roberts T A. A dynamic approach to predicting bacterial growth in food. Int J Food Microbiol. 1994;23:277–294. doi: 10.1016/0168-1605(94)90157-0. [DOI] [PubMed] [Google Scholar]
- 3.Baranyi J, Roberts T A, McClure P. A non-autonomous differential equation to model bacterial growth. Food Microbiol. 1993;10:43–59. [Google Scholar]
- 4.Brockelhurst T F. Delicatessen salads and chilled prepared fruit and vegetable products. In: Man C M D, Jones A A, editors. Shelf life evaluation of foods. London, United Kingdom: Blackie Academic & Professional; 1995. pp. 87–126. [Google Scholar]
- 5.Brown M H, Booth I R. Acidulants and low pH. In: Russell N J, Gould G W, editors. Food preservatives. London, United Kingdom: Blackie Academic & Professional; 1991. pp. 22–43. [Google Scholar]
- 6.Buchanan R L, Bagi L K, Goins R V, Philips J G. Response surface models for the growth kinetics of Escherichia coli O157:H7. Food Microbiol. 1992;10:303–315. [Google Scholar]
- 7.Buchanan R L, Edelson S G. Culturing enterohemorrhagic Escherichia coli in the presence and absence of glucose. Appl Environ Microbiol. 1996;62:4009–4013. doi: 10.1128/aem.62.11.4009-4013.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Buchanan R L, Golden M H. Model for the non-thermal inactivation of Listeria monocytogenes in a reduced oxygen environment. Food Microbiol. 1995;12:203–212. [Google Scholar]
- 9.Buchanan R L, Klawitter L A. The effect of temperature, initial pH, and sodium chloride on the growth kinetics of Escherichia coli O157:H7. Food Microbiol. 1992;9:185–196. [Google Scholar]
- 10.Dalgaard P, Jorgensen L V. Predicted and observed growth of Listeria monocytogenes in seafood challenge tests and in naturally contaminated cold-smoked salmon. Int J Food Microbiol. 1998;40:106–115. doi: 10.1016/s0168-1605(98)00019-1. [DOI] [PubMed] [Google Scholar]
- 11.Davidson P M. Chemical preservatives and natural antimicrobial compounds. In: Doyle M P, Beuchat L R, Montville T J, editors. Food microbiology fundamentals and frontiers. Washington, D.C.: ASM Press; 1997. pp. 520–556. [Google Scholar]
- 12.Erickson J P, Jenkins P. Comparative Salmonella spp. and Listeria monoytogenes inactivation rates in four commercial mayonnaise products. J Food Prot. 1991;54:913–916. doi: 10.4315/0362-028X-54.12.913. [DOI] [PubMed] [Google Scholar]
- 13.Erickson J P, Stamer J W, Hayes M, Mckenna D N, Van Alstine L A. An assessment of Escherichia coli O157:H7 contamination risks in commercial mayonnaise from pasteurized eggs and environmental sources, and behaviour in low-pH dressings. J Food Prot. 1995;58:1059–1064. doi: 10.4315/0362-028X-58.10.1059. [DOI] [PubMed] [Google Scholar]
- 14.Geeraerd A, Herremans C, Van Impe J. Proceedings Science and Technology (International Institute of Refrigeration). Predictive microbiology applied to chilled food preservation. European Commission C2. 1997. Structural model requirements to describe microbial inactivation; pp. 280–287. Quimper, France. [Google Scholar]
- 15.Hudson J A. Comparison of response surface models for Listeria monocytogenes strains under aerobic conditions. Food Res Int. 1994;27:53–59. [Google Scholar]
- 16.Juven B J, Kanner J, Schved F, Weisslowicz H. Factors that interact with the antibacterial action of thyme essential oil and its active constituents. J Appl Bacteriol. 1994;76:626–631. doi: 10.1111/j.1365-2672.1994.tb01661.x. [DOI] [PubMed] [Google Scholar]
- 17.Khuri A I, Cornell G A. Response surfaces. In: Khuri A I, Cornell J A, editors. Determining optimum conditions. New York, N.Y: Marcel & Dekker; 1987. pp. 149–205. [Google Scholar]
- 18.Koutsoumanis K, Lampropoulou K, Taoukis P, Nychas G-J E. Modelling the effect of oregano (Origanum vulgare) essential oil on the death-survival of Salmonella enteritidis in homemade tarama salad. In: Tijskens L M N, Hertog M L A T M, editors. Applications of modelling as an innovative technology in agri-food chain model. International Symposium Wageningen; 1998. pp. 171–178. [Google Scholar]
- 19.Linton R H, Carter W H, Pierson M D, Hackney C R. Use of a modified Gompertz equation to model nonlinear survival curves for Listeria monocytogenes Scott A. J Food Prot. 1995;58:946–954. doi: 10.4315/0362-028X-58.9.946. [DOI] [PubMed] [Google Scholar]
- 20.Linton R H, Carter W H, Pierson M D, Hackney C R, Eifert J D. Use of a modified Gompertz equation to predict the effects of temperature, pH, and NaCl on the inactivation of Listeria monocytogenes Scott A heated in infant formula. J Food Prot. 1995;59:16–23. doi: 10.4315/0362-028X-59.1.16. [DOI] [PubMed] [Google Scholar]
- 21.Little C L, Adams M R, Anderson W A, Cole M B. Application of a log-logistic model to describe the survival of Yersinia enterocolitica at sub-optimal pH and temperature. Int J Food Microbiol. 1994;22:63–71. doi: 10.1016/0168-1605(94)90008-6. [DOI] [PubMed] [Google Scholar]
- 22.Little C L, Adams M R, Easter M C. The effect of pH, acidulant and temperature on the survival of Yersinia enterocolitica. Lett Appl Microbiol. 1991;14:148–152. [PubMed] [Google Scholar]
- 23.Little C L, Knøhel S. Growth and survival of Yersinia enterocolitica, Salmonella and Bacillus cereus in brie stored at 4, 8 and 20°C. Int J Food Microbiol. 1994;24:137–145. doi: 10.1016/0168-1605(94)90113-9. [DOI] [PubMed] [Google Scholar]
- 24.McClure P J, Beaumont A L, Sutherland J P, Roberts T A. Predictive modelling of growth of Listeria monocytogenes. The effects on growth of NaCl, pH, storage temperature and NaNO2. Int J Food Microbiol. 1997;34:221–232. doi: 10.1016/s0168-1605(96)01193-2. [DOI] [PubMed] [Google Scholar]
- 25.McMeekin T A, Olley J, Ross T, Ratkowsky D A. Predictive microbiology: theory and application. Somerset, United Kingdom: Research Studies Press Ltd; 1993. [Google Scholar]
- 26.Membré J-M, Ross T, McMeekin T A. Behavior of Listeria monocytogenes under combined chilling processes. Lett Appl Microbiol. 1999;28:216–220. doi: 10.1046/j.1365-2672.1999.00499.x. [DOI] [PubMed] [Google Scholar]
- 27.Presser K A, Ratkowsky D A, Ross T. Modelling the growth rate of Escherichia coli as a function of pH and lactic acid concentration. Appl Environ Microbiol. 1997;63:2355–2360. doi: 10.1128/aem.63.6.2355-2360.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Radford S A, Board R G. Review: fate of pathogens in home-made mayonnaise and related products. Food Microbiol. 1993;10:269–278. [Google Scholar]
- 29.Radford S A, Tassou C C, Nychas G J E, Board R G. The influence of different oils on the death rate of Salmonella enteriditis in homemade mayonnaise. Lett Appl Microbiol. 1991;12:125–128. [Google Scholar]
- 30.Ross T. Indices for performance evaluation of predictive models in food microbiology. J Appl Bacteriol. 1996;81:501–508. doi: 10.1111/j.1365-2672.1996.tb03539.x. [DOI] [PubMed] [Google Scholar]
- 31.Tassou C C, Drosinos E H, Nychas G J E. Effects of essential oil from mint (Mentha piperita) on Salmonella enteritidis and Listeria monocytogenes in model food systems at 4 and 10°C. J Appl Bacteriol. 1995;78:593–600. doi: 10.1111/j.1365-2672.1995.tb03104.x. [DOI] [PubMed] [Google Scholar]
- 32.Uljas H E, Ingham S C. Survival of Escherichia coli O157:H7 in synthetic gastric fluid after cold and acid habituation in apple juice or Trypticase soy broth acidified with hydrochloric acid or organic acids. J Food Prot. 1998;61:939–947. doi: 10.4315/0362-028x-61.8.939. [DOI] [PubMed] [Google Scholar]
- 33.Walls I, Scott V N, Bernard D T. Validation of predictive mathematical models describing growth of Staphylococcus aureus. J Food Prot. 1995;59:11–15. doi: 10.4315/0362-028X-59.1.11. [DOI] [PubMed] [Google Scholar]
- 34.Weagant S D, Bryant J L, Bark D H. Survival of Escherichia coli O157:H7 in mayonnaise and mayonnaise-based sauces at room and refrigerated temperatures. J Food Prot. 1994;57:629–631. doi: 10.4315/0362-028X-57.7.629. [DOI] [PubMed] [Google Scholar]
- 35.Zwietering M H, Cuppers H G A M, de Wit J C, van't Riet K. Evaluation of data transformations and validation of a model for the effect of temperature on bacterial growth. Appl Environ Microbiol. 1994;60:195–203. doi: 10.1128/aem.60.1.195-203.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]