Abstract
The genome of an organism is regulated in concert with the organized action of various genetic regulators at different hierarchical levels. Small non-coding RNAs are one of these regulators, among which microRNAs (miRNAs), a distinguished sRNA group with decisive functions in the development, growth and stress-responsive activities of both plants as well as animals, are keenly explored over a good number of years. Recent studies in plants revealed that apart from the silencing activity exhibited by miRNAs on their targets, miRNAs of specific size and structural features can direct the phasing pattern of their target loci to form phased secondary small interfering RNAs (phasiRNAs). These trigger-miRNAs were identified to target both coding and long non-coding RNAs that act as potent phasiRNA precursors or PHAS loci. The phasiRNAs produced thereby exhibit a role in enhancing further downstream regulation either on their own precursors or on those transcripts that are distinct from their genetic source of origin. Hence, these tiny regulators can stimulate an elaborative cascade of interacting RNA networks via cis and trans-regulatory mechanisms. Our review focuses on the comprehensive understanding of phasiRNAs and their trigger miRNAs, by giving much emphasis on their role in the regulation of plant defense responses, together with a summary of the computational tools available for the prediction of the same.
Keywords: Gene regulation, PhasiRNAs, Plant resistance, R genes, Computational tools
Introduction
Plants are persistently exposed to an extensive range of environmental stresses. Being sessile in nature, they had developed different mechanisms to recognize and transform various stress stimuli into adaptive responses. The conjoined functioning of regulatory networks in the genome of organisms is at the helm of controlling their adaptation to different types of biotic and abiotic stresses (Zheng et al. 2015). Plant genomes are a resort for a huge number of small RNAs (sRNAs), which exist as key regulators of several important biological processes, including plant development, stress resistance, and epigenetic modifications. The sRNAs are classified and distinguished mainly based on their biogenesis and loci of origin as microRNAs (miRNAs), small interfering RNAs (siRNAs), phased secondary small interfering RNAs (phasiRNAs), and heterochromatic small interfering RNAs (hc-siRNAs) (Fei et al. 2013). In plants, these endogenous small RNAs are 18 to 24 nucleotide (nt) sized transcripts, where most of them are derived from either hairpin precursors or double-stranded RNA (dsRNA) precursors with the help of dicer-like (DCL) proteins and RNA-dependent RNA polymerases (RDRs). After processing, these sRNA duplexes associate with argonaute (AGO) protein, which is a principal component of RNA-induced silencing complex (RISC) that directs the sRNA to its respective coding or non-coding target-transcripts according to sequence complementarity (Komiya 2017; Tabara et al. 2018).
miRNAs represent a major class of sRNAs that regulate different stages of plant growth and stress responses by post-transcriptional gene regulation mediated through transcript cleavage or translational repression (Yu et al. 2019b). The targeted gene silencing mediated by these tiny regulators is well exploited for the production of disease-resistant varieties as well (Nair and Alagu 2020). Both the accessibility to multiple targets and co-functional activities exhibited by miRNAs divulge their potentiality in complementing adaptability to diverse stress conditions, especially against certain dreadful pathogens (Nair et al. 2020). Even though miRNAs are similar to siRNAs when considering their size, they differ to a great extent in their biogenesis, structure of precursors and in their way of action (Kamthan et al. 2015). Most of these sRNA populations in the plant genome are descended from either repetitive sequences or transposable elements, termed heterochromatic siRNAs or repeat-associated siRNAs (Lindbo 2012). There also exists a different group of sRNAs termed secondary siRNAs, whose biosynthesis is induced by other sRNA-mediated cleavages on their precursors (de Felippes 2019). Among them, trans-acting small interfering RNAs (tasiRNAs) are typical secondary siRNAs derived from miRNA-targeted transcripts, that can target the transcripts other than their original source, hence named trans-acting (Chen 2012). Such secondary siRNAs that were first renowned as tasiRNAs now come under a much broader family called phasiRNAs, which exhibit a remarkable role in different aspects of plant growth, development and stress responses (Table 1). They represent endogenous eukaryotic small RNAs that exist at an interlude of 21–26 nucleotides and whose production is triggered by miRNAs along with the help of other components for siRNA production (Fei et al. 2013; Komiya 2017).
Table 1.
Role of phasiRNAs in plants
| S. No. | Precursor | Initiator miRNA | Functional involvement | Host plant | References |
|---|---|---|---|---|---|
| 1 | TAS1 | miR173 | Thermotolerance | Arabidopsis thaliana | Li et al. 2014 |
| 2 | TAS1a | miR173 | Response to cold stress | Arabidopsis thaliana | Calixto et al. 2019 |
| 3 | TAS3 | miR390 | Regulate lateral root growth and symbiotic nodulation | Medicago truncatula | Hobecker et al. 2017 |
| 4 | TAS3 | miR390 | Aluminium stress response | Linum usitatissimum | Dmitriev et al. 2017 |
| 5 | TAS3 | miR390 | Seed growth and development | Glycine max | Xu et al. 2014 |
| 6 | TAS3 | miR156, miR390 | Regulate developmental timing | Physcomitrella patens | Cho et al. 2012 |
| 7 | TAS3 | miR390 | Regulate lateral root growth | Arabidopsis thaliana | Marin et al. 2010 |
| 8 | TAS3 | miR2118, miR2275 | Development of inflorescence | Oryza sativa | Johnson et al. 2009 |
| 9 | TAS4 | miR828 | Leaf trichome development | Arabidopsis thaliana | Guan et al. 2014 |
| Fibre development | Gossypium hirsutum | ||||
| 10 | TAS4 | miR828 | Anthocyanin biosynthesis | Arabidopsis thaliana | Tiwari et al. 2021 |
| 11 | TAS6 | miR156, miR529 | Slice Zinc-finger domain transcript | Physcomitrella patens | Arif et al. 2012 |
| 12 | MIST1 | miRNA825-5p | Defense response | Arabidopsis thaliana | López-Márquez et al. 2020 |
| 13 | NBS-LRR | miR482 | Defense response | Gossypium raimondii | Zhu et al. 2013 |
| 14 | NAC transcription factor | miR1514 | Response to stress |
Phaseolus vulgaris, Medicago truncatula |
Sosa-Valencia et al. 2017 |
| 15 | PMS1T | miR2118 | Photosensitive male sterility | Oryza sativa | Fan et al. 2016 |
| 16 | MLA1 | miR9863 | Defense response |
Hordeum vulgare, Nicotiana benthamiana |
Liu et al. 2014 |
| 17 | NBS-LRR | miR1507, miR2109, miR2118 | Regulate symbiotic association | Medicago truncatula, Glycine max | Zhai et al. 2011 |
| 18 | WSGAR | miR9678 | Regulate seed germination | Triticum aestivum | Guo et al. 2018 |
| 19 | PPR | miR161 | Defense response | Arabidopsis thaliana | Hou et al. 2019 |
A substantial increase in genome sequencing allied with downstream small RNA sequence analysis enhanced the characterization of a large number of sRNAs, notably phasiRNAs as well as their targets. With each new transcriptome sequenced, apart from the prediction of novel miRNAs and their targets, there arises a new opportunity to explore the extreme potential and least explored function of miRNAs acting as the initiator sRNA for phasiRNA production. So far, a few protein-coding transcripts and long non-coding RNAs (lncRNAs) are reported as pronounced sources that act as the loci for phasiRNA generation (Fei et al. 2013). The initial discovery and characterization of miRNA-triggered trans-acting secondary siRNAs were made in Arabidopsis thaliana, where it was found that sRNAs induce the production of siRNAs that silence the expression of transcripts other than their precursors (Vazquez et al. 2004). As yet, four families of tasiRNA producing loci (TAS genes) are uncovered in A. thaliana, among which miR173 targets most of TAS1 and TAS2 family members whereas, miR390 and miR828 have a significant role in TAS3 and TAS4 induction, respectively. Among these, TAS3-derived tasiRNAs are found to be profoundly conserved among a broad range extending from bryophytes to angiosperms (Deng et al. 2018). Later on, phasiRNAs which not only promote the cleavage of their targets in trans fashion, but also act in a cis manner were discovered that ultimately designated tasiRNAs as a subclass coming under the so-called phasiRNA group. Several additional TAS gene families have been discovered in species other than A. thaliana, which paved the way for an augmented chance for exposing many undiscovered phasiRNA loci in plants by exploiting the advanced genome sequencing facilities (de Felippes 2019).
An elevation in the plant disease resistance beyond a specific limit most probably causes a threat to proper growth and development of the same. Several studies manifested that the miRNA-phasiRNA pathway in plants may act as a regulatory hub in controlling both plant growth and disease resistance in harmony, by taking control over defense-related genes and by mediating cross-talk between the attacking pathogen (Liu et al. 2020). Studies conducted in legumes and members of Solanaceae reported PHAS loci on mRNAs, which code for various nucleotide-binding site leucine-rich repeat (NBS-LRR) proteins associated with plant disease resistance (Zhai et al. 2011; Park and Shin 2015; Zhao et al. 2015). Apart from NBS-LRR disease resistance proteins, some of the other coding transcripts from which the phasiRNA production is well evaluated are, MYB transcription factors (Xia et al. 2012; Rock 2013; Guan et al. 2014; Zheng et al. 2015), auxin response factors (ARFs) (Williams et al. 2005; Hunter et al. 2006; Arikit et al. 2014), calcium ATPase transporters (Wang et al. 2011), F-box genes (Xia et al. 2015b; Chen et al. 2018), pentatricopeptide repeat (PPR) genes (Xia et al. 2013; Arikit et al. 2014; Zheng et al. 2015) etc. Aside from coding transcripts, there are studies that expose the existence of PHAS loci on lncRNAs. A study on A. thaliana disclosed about eight miRNA-triggered lncRNA-phasiRNA pathways, out of which three of them appeared to be conserved in species like Glycine max, Oryza sativa and Gossypium hirsutum (Yu et al. 2019a). Works on Litchi chinensis also divulged PHAS loci on lncRNA genes that are triggered by miRNA and accompany both alternative splicing as well as polyadenylation for gene silencing (Ma et al. 2018). Up to this date, though several studies have been conducted to reveal the defense response regulation of plants by lncRNAs, there still exists a huge gap in the area explaining how these non-coding transcripts are regulated. Hence, the increasing evidence of lncRNAs acting as potent PHAS loci may imply the role of miRNA-phasiRNA pathways in controlling these long transcripts during pathogen invasion.
This review mainly focuses on the biogenesis of phasiRNAs and scrutinizes their regulatory aspects over a wide spectrum of biological processes, mainly defense response, accompanied by other genetic components like miRNAs, coding as well as non-coding transcripts in plants. The review also covers the so far developed bioinformatics tools used for the prediction of PHAS loci, their initiator miRNAs, candidate phasiRNAs produced and their respective targets. Understanding the role of phasiRNAs in the diverse genetic regulatory cascades can contribute novel and significant insights to sRNA-mediated regulatory network by determining new genetic elements and their interactions that expose the underlying genetic basis of plant development and defense responses in-depth.
Biogenesis of phasiRNAs
TasiRNAs were first identified in A. thaliana, where they represented a class of endogenous short interfering RNAs that have a role in regulating the expression of mRNAs with little similarity to their precursor transcript. The study revealed that a precursor non-coding RNA, TAS1A (At2g27400), which is first transformed to dsRNA, is later destined to form 21-nt entities of secondary siRNAs. Also, these sRNAs were found to be different from already characterized sRNAs like miRNAs and siRNAs in certain aspects. When compared to miRNAs that are derived from short double-stranded RNA precursors, tasiRNAs are derived from long double-stranded RNAs. Compared to siRNAs, tasiRNAs differ mainly in the components needed for their biogenesis, functional pathway and most importantly, post-transcriptional silencing of target transcripts other than their source of origin (Vazquez et al. 2004). In general, phasiRNAs are produced by miRNA-triggers that cleave their targets resulting in the generation of phased small RNAs that can regulate gene expression both in cis and trans positions (Vargas-Asencio and Perry 2019). These siRNAs are created in a phased pattern in a definite top to toe fashion from a particular nucleotide; hence they are termed phased siRNAs (Fei et al. 2013; Deng et al. 2018).
Regulatory elements involved in phasiRNA production
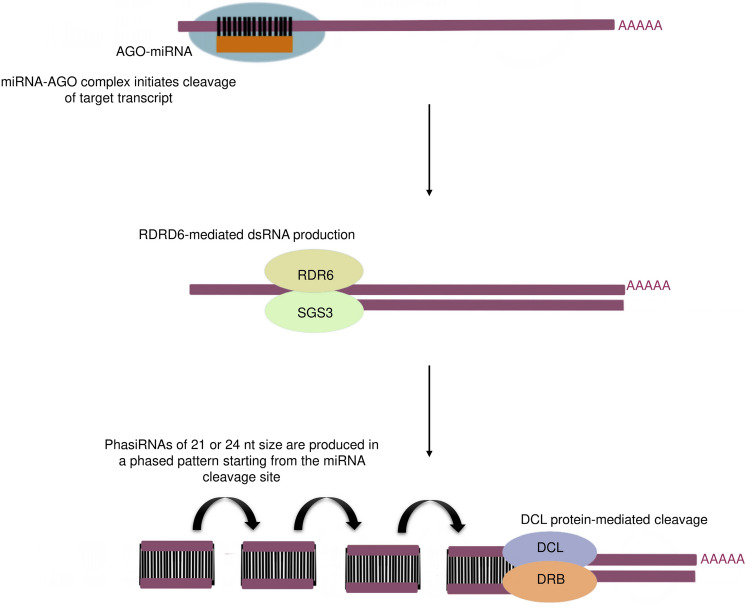
Major proteins involved in phasiRNA biogenesis are AGO proteins, RNA-dependent RNA polymerase 6 (RDR6), Suppressor of Gene Silencing 3 (SGS3) proteins, DCL proteins and Double-stranded RNA binding (DRB) proteins (Fig. 1). Among them, AGO proteins form complexes with miRNAs to regulate gene expression at both transcriptional and post-transcriptional levels by mediating target RNA cleavage or translational inhibition. In phasiRNA biogenesis, their corresponding loci are cleaved by specific miRNA-AGO complexes (Cervera-Seco et al. 2019). In A. thaliana, a single cleavage spawned by the combination of 22-nt miRNAs and AGO1 causes tasiRNA production from TAS1, TAS2 and TAS4 loci. Whereas, the processing of TAS3 appeared to be a dual-targeting process performed by 21-nt long miRNAs and AGO7 (de Felippes et al. 2017). Subsequently produced phasiRNAs are then carried by the AGO1 complex to execute either cleavage of their targets or cause the production of other secondary siRNAs from their corresponding targets. Whereas DCL1-mediated 21-nt siRNAs, which later implement DNA methylation of their own loci, are generally integrated with the AGO4 family (Deng et al. 2018). RDR6, another important protein involved in the phasiRNA biogenesis pathway, recruits AGO-miRNA cleaved PHAS loci to synthesize dsRNAs (Deng et al. 2018). Though the function of SGS3 still remains perplexing, there are studies showing a considerable decrease in phasiRNA production in mutants of this specific protein (Deng et al. 2018). Also, in vitro experiments in A. thaliana exposed the role of SGS3 in protecting the miRNA-cleaved TAS loci from degradation and thereby permitting RDR6 activity (Yoshikawa et al. 2013). DCL proteins have a role in transforming dsRNAs created by RDR6 into 21-nt siRNAs. Though DCL4 remains the commonly found DCL protein in this pathway, there are studies in A. thaliana that expose the role of other proteins like DCL2 and DCL3 in tasiRNA biogenesis when DCL4 is absent (Gasciolli et al. 2005). Besides these, DCL1 proteins contribute to 21-nt tasiRNA production and are renowned for tasiRNA-mediated DNA methylation at TAS3 loci (Wu et al. 2012). Also, a set of 24-nt phasiRNAs was brought into light at the reproductive stage of O. sativa, which was processed by another DCL3b protein homologous to DCL3 in A. thaliana (Song et al. 2012). DRB proteins are established to interact with DCL proteins and thus, have a hand in both miRNA and tasiRNA biogenesis (Pegler et al. 2019). DRB1 and DRB4, which are two of the best characterized plant DRB proteins, aid DCL1 and DCL4 to produce miRNA and tasiRNA respectively, from their dsRNA precursors (Eamens et al. 2012). In addition, immunoprecipitation experiment in A. thaliana demonstrated the co-immunoprecipitation of DCL4 with DRD4, which highlights the possible interactions between the same (Nakazawa et al. 2007).
Fig. 1.
Biogenesis of phasiRNAs is initiated by the combined cleavage activity of the miRNA-AGO complex, which further triggers the RDR6/SGS3-mediated dsRNA synthesis. Consequently, these dsRNAs are transformed to 21 nt or 24 nt phasiRNAs that are produced in a phased pattern by the DCL-mediated cleavage aided by the DRB proteins
One-hit and two-hit models of phasiRNA production
Preliminary studies to unravel the pathway of tasiRNA synthesis were performed in A. thaliana, which endowed the significance of miR173 and miR390 in directing cleavage at TAS1, TAS2 and TAS3 loci, respectively to produce tasiRNAs (Allen et al. 2005). Further attempts to discern the mechanism involved in phasiRNA biogenesis led to the identification of a ‘two-hit’ model in Physcomitrella patens. In P. patens, tasiRNAs were found to ensue from flanking sequences of two miR390-complementary sites at TAS3 loci (Axtell et al. 2006). The existence of two cleavable sites flanking the tasiARFs (TAS3 transcripts that regulate auxin response factor) in Pinus taeda propounds that a two-hit model of tasiRNA biogenesis may also appear in angiosperms. For A. thaliana, the situation is contrary where among the two complementary sites, only 3' region is proficient for AGO-mediated cleavage, whereas 5' is not. Furthermore, the mutations at any of these sites affected the biogenesis process substantially (Axtell et al. 2006). Thus, the two-hit model of tasiRNA biogenesis involving dual miRNA binding sites appears to be a direct selective trigger of transcripts for tasiRNA production.
Though the two-hit model represents an acceptable pathway for tasiRNA production from TAS3 loci, their biogenesis from other TAS loci is not explained by the same. When considering TAS1, TAS2 and TAS4 loci, each of them appears to have only a single miRNA target site, which is thus referred to as the ‘one-hit model’ of secondary siRNA production (Felippes and Weigel 2009). While miR173 seems to trigger TAS1 and TAS2, miR828 contributes to the initiation of TAS4-derived tasiRNA generation (Montgomery et al. 2008b; Felippes and Weigel 2009). Besides, the tasiRNA forming region in TAS3 arises from the region upstream to miRNA cleavage, and that of TAS1, TAS2 and TAS4 are produced downstream or 3' regions of the respective cleavage motifs (Montgomery et al. 2008b). A substitute model was later discovered, which states that a single miRNA targeting is ample for the secondary siRNA production to be equitable. The same model appears to be adequate for miR390-mediated siRNA production from TAS3 locus in A. thaliana, which previously claimed the requirement of two miRNA complementary sites for siRNA biogenesis. The study also uncovered that miRNA-mediated cleavage is redundant and mere interaction between miRNA and the respective target is enough for RDR6 recruitment followed by downstream pathway proceedings (de Felippes et al. 2017). Meanwhile, the two-hit model is assumed to evolve for reinforcing the efficiency and accuracy of the secondary siRNA biogenesis pathway (de Felippes et al. 2017). Though the study exposes a basic mechanism that justifies the perplexity of heterogeneous plant phasiRNA production, there are discrepancies in certain pathway components like AGO7, 21-nt miRNA complex specifically interacting with TAS3 locus whereas, the AGO1, 22-nt miRNA complex interacting with the other TAS loci. This in turn demands additional in-depth studies to be performed to refine out a transparent basic model, including each of the varying components and their descending functional activities in the phasiRNA pathway.
The size of miRNA is considered as a pivotal factor that enhances secondary siRNA generation (Chen et al. 2010; Cuperus et al. 2010). In plants, the canonical 21-nt miRNAs are predominantly produced compared to other classes of miRNAs, particularly with those that constitute 22-nt (Chen et al. 2010). Beyond the bounds, exploring genome-scale small RNAs through several studies disclosed the significant role of 22-nt miRNAs in secondary siRNA biogenesis (de Felippes 2019). A study on Nicotiana benthamiana treated with Agrobacterium together with a modified 22-nt miR173 and miR828 construct revealed that, unlike their 21-nt form, these modified miRNAs could commence phasiRNA production. The addition of an extra nucleotide at the 3' region of miR319, which is generally processed as 21-nt, bestows the modified 22-nt miR319 with the siRNA producing capacity (Chen et al. 2010). In most instances, 22-nt miRNAs are produced as a consequence of an asymmetric bulge present in the miRNA/miRNA* precursor pair, which results in the formation of a duplex with 22/21-nt conformation (de Felippes 2019). Experiments in A. thaliana proclaimed that three 22-nt miRNAs, namely miR173, miR393 and miR472, are generated from asymmetrically paired miRNA duplex precursors. However, there are some exceptions where miRNA precursor duplexes with symmetric mispair also contribute to the generation of 22-nt miRNAs, like that of miR828 in A. thaliana (Cuperus et al. 2010). Despite the so far mentioned factors like miRNA size and miRNA duplex structure, another important character that may contribute to the stability as well as precision in the biogenesis pathway is the interaction between 5' region of miRNA and AGO protein. As already quoted, the ‘two-hit’ model of secondary siRNA biogenesis from TAS3 exposed that, unlike other TAS loci, TAS3 loci cognates with 21-nt miRNA, predominantly miR390, instead of 22-nt miRNAs and only forms the effector complex with AGO7. This strong selectivity between AGO7 and miR390 is imparted by a 5' Adenosine and the same element excludes other AGO proteins from interacting with miR390 (Montgomery et al. 2008a). Furthermore, preference for a 5' Uracil instead of Adenosine intercepts AGO1 from interacting with TAS3-specific miR390 (Montgomery et al. 2008a).
Regulatory role of phasiRNAs in plant defense responses
The static nature of plants always demands quick feedback on pathogen attacks. Therefore, the multifarious roles of sRNAs in regulating gene expression subsist as a prime factor for rapid responses against invading pathogens. Apart from the brisk as well as systematic gene regulation performed by sRNAs, what makes them exceptional is that some sRNAs, precisely miRNAs, can produce phasiRNAs that in turn transform many numbers of transcripts right away. This property of modulating several transcripts by phasiRNAs in plant immune responses remains as an extensive but still less explored area to date.
miRNA-resistance gene (R gene)–phasiRNA pathways in plant diseases
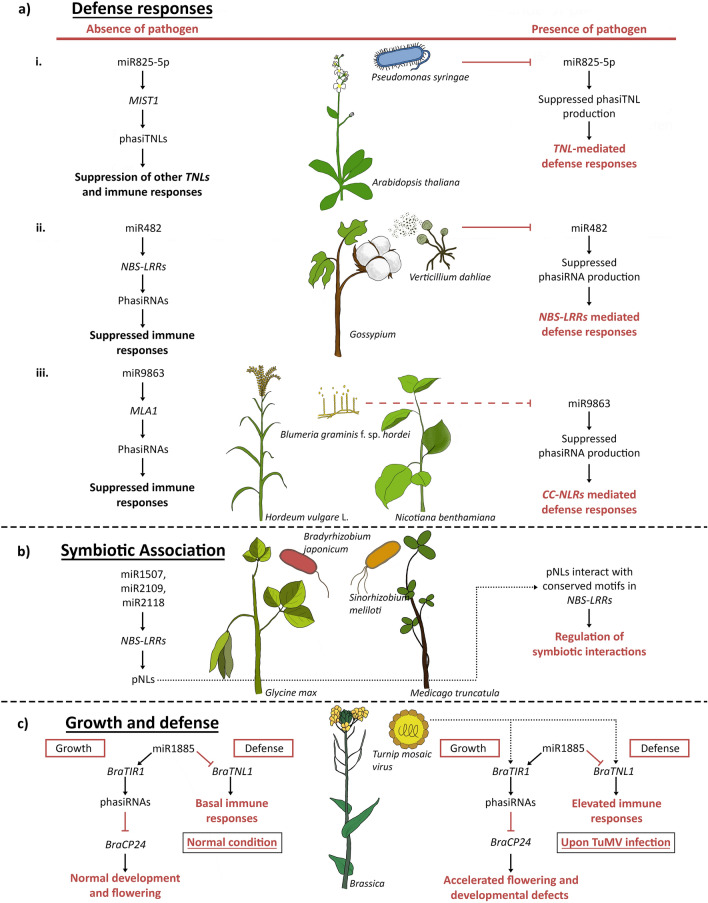
Plant resistance (R) genes or NBS-LRRs are a well-portrayed gene family constituting intracellular receptors that can discern effector molecules produced by pathogens and thereby initiate plant immunity (Fei et al. 2013). A dual-line regulation of NBS-LRRs by miRNAs along with phasiRNAs was disclosed in A. thaliana, where several Toll/interleukin-1-NBS-LRRs (TNLs) were found to be targeted by the miRNA-derived phasiRNAs (Fig. 2a). The study exposed the targeted silencing of a conserved motif called TIR2 in MIST1 (microRNA-silenced TNL1) gene of A. thaliana by a 22-nt miRNA, miR825-5p, which thereby suppresses undesired activation of these defense-related genes when there is no invading pathogen. Examining the resistance of A. thaliana with a varied expression of miR825-5p against a bacterial pathogen, namely Pseudomonas syringae, also defended the fact that tuning of MIST1 by the production of miRNA-derived phasiTNLs is a key element in allotting immunity against the harmful outsiders (López-Márquez et al. 2020). Similar production of phasiRNAs is obeyed in cotton plants infected by a fungal pathogen, Verticillium dahliae, where the NBS-LRR defense genes are silenced by miR482 via the production of phasiRNAs from the same, in the absence of pathogen (Fig. 2a). On the other hand, infection by V. dahliae repressed the expression of miR482, that in turn reduced the generation of phasiRNAs, thereby enhancing the expression as well as activity of NBS-LRR genes against the respective fungal pathogen (Zhu et al. 2013). A study on another fungal pathogen, Blumeria graminis f. sp. hordei that cause powdery mildew disease in plants, also exposed the post-transcriptional regulation mediated by phasiRNA production from Mla1 (Mildew resistance locus a) transcripts, which generally code for coiled-coil NBS-LRR receptors (CC-NLRs) (Fig. 2a). The study revealed the role of miR9863 in silencing the MLA1 gene in both Hordeum vulgare L. and N. benthamiana by cleaving the Mla1 transcript as well as by initiating phasiRNA production from the same, which thereby leads to their attenuation. The most interesting factor here is, examining the amino acid sequence alignment manifested a conservation of miR9863 target sites on Mla alleles and R genes in wheat along with their associated species (Liu et al. 2014). This selective expression of the miR9863 family unfolds a novel phase for exploring the miRNA-phasiRNA-defense gene regulatory network in Triticeae species (Liu et al. 2014). An independent study ensued later represented an elevated amino acid sequence identity between phasiRNA producing NLR loci (HORVU3Hr1G105020) in barley and wheat CNL9 gene that codes for a CC-NLR gene called SGS3, that was previously reported to impart resistance to stem rust disease in wheat caused by Ug99 lineage of stem rust fungus (Hunt et al. 2019). This sequence conservation among different species unfolds the likelihood of their functional similarity and thereby enhances the prediction of more phasiRNA producing loci that are yet to be annotated in plants, which might be crucial for plant defense responses. Deciphering the role of sRNAs in regulating the defense response in potatoes under the attack of the dreadful Potato virus Y (PVY) also highlighted the appearance of various differentially expressing phasiRNAs produced from NBS-LRR loci (Križnik et al. 2017).
Fig. 2.
Coordinated role of phasiRNAs and their initiator miRNAs during plant–microbe interactions: a Timely activation and inactivation of R genes by miRNA triggered phasiRNAs during different plant-pathogen interactions. The production of phasiRNAs from NBS-LRR genes in the absence of pathogens helps in preventing unnecessary activation of defense genes and thereby blocks autoimmunity in plants. On the other hand, upon pathogen invasion, the trigger-miRNAs get inactivated, which in turn suppressess phasiRNA production and results in the active NBS-LRR gene responses. b Interaction of phasiRNAs with NBS-LRR gene motifs enhances symbiotic associations exhibited by plants-microbes. c miRNA-phasiRNA mediated controlled regulation of plant growth and defense activities by interacting with, tasiRNA producing genes (BraTIR1), photosynthesis-related genes (BraCP24) and R genes (BraTNL1)
Regulatory elements that oversee plant–microbe symbiotic interactions and how this multidirectional beneficial association is maintained by concealing the detrimental pathogenic impacts are still obscure. A study on symbiosis prevailed in two leguminous plants, namely Medicago truncatula and Glycine max, revealed the production of phasi-NBS-LRRs (pNLs) triggered by miRNAs that target several conserved motifs in the NBS-LRR gene family, which have a pivotal role in regulating symbiotic interactions (Fig. 2b). The study mentioned above exposed three 22-nt miRNA families, including miR1507, miR2109 and miR2118, as the significant contributors to pNL trigger. Interestingly, miR2109 in M. truncatula was found to be a 22 nt miRNA, whereas, in G. max, it turned out to be a 21 nt miRNA (Zhai et al. 2011). This variation in the length of phasiRNA triggers in different leguminous species might represent the variability in their potential to initiate phasiRNA production and this may even contribute to the regulation of subsisting nodulation specificity exhibited by individual plants for selective narrowing of their host range. Also, the miR2118 family, which produces phasiRNAs from NBS-LRRs in leguminous plants, is previously reported to enhance phasiRNA production in grasses but from intergenic non-coding transcripts. These differences in the target PHAS loci of the same miRNA-trigger propose that gene regulation by phasiRNAs perhaps has been an evolutionarily selected regulatory scheme in plants that developed functional diverseness upon passage of time (Xia et al. 2015a).
Growth and timely immune activities in plants demand the execution of appropriate resources that systematically regulate the same. The well-timed activation and repression of the R genes are in part under the control of phasiRNAs and their miRNA triggers. As already mentioned, R genes that are generally inactive in the absence of pathogen infection by the compatible generation of phasiRNAs, get activated once a foreign threat invades the host. Hence, exploring other verges of a regulatory cascade, where the strategy used to suppress the biogenesis of initiator-miRNA followed by phasiRNAs when the plant is under pathogen attack is of prime importance. Examining an A. thaliana double mutant cpr1 aba1, which exhibits a defective miRNA and phasiRNA biogenesis, unveiled the negative regulatory role of R protein named SNC1 along with a transcriptional co-repressor termed TPR1, on the mentioned sRNA biogenesis (Cai et al. 2018). The comprehensive regulation of R genes following the presence or absence of pathogens is an amalgam of multifarious processes and has been a prominent topic contemplated by plant defense-related studies. Thus, further exploration of the miRNA-phasiRNA-R gene cascade is very relevant for, interestingly a small number of miRNAs that targets some of the R genes can give rise to a profusion of phasiRNAs, which in turn regulate the expression of a vast number of other R genes and defense-related regulatory elements. Breeding approaches always necessitate the development of crop varieties with a stabilized degree of both yield and disease resistance, as a rise in plant immune responses imparts drastic effects on plant growth. miR1885 in Brassica were found to have control over both defense and growth-related genes, where the BraTNL1 defense gene expression is directly suppressed by miR1885 upon infection with Turnip mosaic virus, whereas the BraCP24 gene which is related to plant growth was found to be regulated by tasiRNAs produced from BraTIR1 PHAS locus, again triggered by miR1885 (Fig. 2c). Generally, miR1885 shows a marginal expression level at normal conditions, which is then hiked according to either pathogen invasion or plant developmental processes (Cui et al. 2020). On this account, addressing the global regulation of the R gene by miRNAs and the respective phasiRNAs produced will earn a great significance in the field of plant breeding since there is an extensive area to be still explored, which holds a repository of complex genetic elements that interacts with each other for the synchronized maintenance of plant growth and defense responses.
Other least explored but significant miRNA-plant gene-phasiRNA pathways in plant diseases
PPR is a large gene family that codes for RNA-binding proteins, which constitute an amino acid sequence motif of approximately 30–40 in size (Manna 2015). Apart from the common resistance gene analogs such as NBS-LRRs, RLKs (receptor-like kinases) and RLPs (receptor-like proteins), several present-day studies have revealed a similar function of PPR genes in synchronizing plant-pathogen interactions (Sekhwal et al. 2015). A study on the interaction between dreadful oomycete pathogen, namely Phytophthora capsici and model plant A. thaliana, exposed the secondary siRNA-triggering activity of miR161 from distinct PPR genes. These secondary siRNAs were later carried in extracellular vesicles directed towards pathogen genes to suppress their growth and pathogenicity. Also, mutant A. thaliana deficient in these secondary siRNAs seems hypersensitive to the P. capsici infection. Strikingly, a counter activity displayed by P. capsici by producing an effector molecule termed PSR2 appears to hinder the secondary siRNA biogenesis and thereby enhance the infection. Here, when the PPR-secondary siRNA pathway discloses a plant immune response that might be an anciently subsisted one as it prevails in almost all eudicots, the activity of PSR2 highlights the evolution of pathogens in bringing out effective molecules to obstruct the host defense along time (Hou et al. 2019). Independent infection studies on A. thaliana with P. syringae, a bacterial pathogen, as well as Botrytis cinerea, a fungal pathogen, stated that miR400-triggered cleavage of PPR genes manifest the susceptibility of the plant towards the mentioned bacterial and fungal pathogens to more extend. Both experiments, including a transgenic A. thaliana, which overexpresses miR400 and A. thaliana with mutated PPR genes, exhibited drastic disease traits compared to their wild varieties (Park et al. 2014). The explicit or implicit action of miR400, similar to other PPR-phasiRNA inducing miRNAs like miR161 and miR173, unveils the possibilities of substantial research exploring the role of the large plant PPR gene family along with their miRNA triggers as well as downstream phasiRNAs and targets during plant-pathogen interactions (Xia et al. 2013; Park et al. 2014). The abrupt evolutionary extension of PPR gene families exposes the need for advancement in several regulatory mechanisms, including the post-transcriptional regulation mediated by phasiRNA generation from specific transcripts that may encounter moderately the expansive nature of this large gene family (Howell et al. 2007).
An integrated study including various plant species pertaining to predominant phylogenetic groups like algae, mosses, gymnosperms, monocots, dicots, etc. exposed the presence of diverse PHAS loci in their coding and non-coding transcripts (Zheng et al. 2015). Unlike the disease resistance R gene PHAS loci, which are extensively prevalent in almost all plant species, the study revealed certain PHAS loci specific to particular species. One such example is a phasiRNA generating gene, namely squamosal promoter-binding protein gene, which set out to be a PHAS locus only in grapevine, though it exists as a conserved target transcript for miR156 in several plant species. This arbitrary appearance of different phasiRNAs among plant species proposes the chance of the recent evolutionary emergence of functional PHAS loci as key riboregulators that control various unambiguous molecular activities contributing to systematic plant growth and development. The aforementioned study also compared the phasiRNA expression in several viral infected plants, which revealed that certain phasiRNAs get activated or suppressed upon pathogen invasion (Zheng et al. 2015). ARFs, a group of DNA-binding proteins that enhance the activity of the phytohormone auxin, which regulates practically every facet of plant development spanning from embryogenesis to senescence and recently proven to have a role in plant disease resistance, are a least explored PHAS loci (Ghanashyam and Jain 2009; Zheng et al. 2015). The plant-viral infection model considered in the above-mentioned study unraveled the inhibition and activation of several auxin-signaling PHAS loci in generating phasiRNAs upon infection with Papaya ringspot virus. Among them, ARF3 and certain AFB (AUXIN SIGNALING F-BOX) genes exhibited decreased phasiRNA production, whereas a SAUR-like ARF showed a drastic increase in phasiRNA generation upon viral infection (Zheng et al. 2015). Here, the increase in phasiRNA expression observed during viral infection reflects the possibility of their role in synchronizing various defense activities like those which contribute to restrained viral replication, whereas a decline in phasiRNA production upon viral invasion supports that the precursors of these phasiRNAs might be defense-related genes, which should be active during pathogen attack.
A substantial disclosure of the regulatory role exhibited by extensive non-coding transcripts in the genome of both flora and fauna demands a comprehensive elucidation of the functional aspects of the same. Several studies reinforce lncRNAs, which have a characteristic length that exceeds 200 bp and with a structure similar to mRNAs though they are deficient in appreciable coding potential, to have a considerable role in plant developmental as well as stress response activities (Budak et al. 2020). An interesting experiment conducted in rice revealed the potential of large intergenic non-coding RNAs (lincRNAs) in producing phasiRNAs that specifically interact with an AGO protein, namely MEIOSIS ARRESTED AT LEPTOTENE1 (MEL1) (Komiya et al. 2014). Besides the study mentioned above, which considers the interaction between germline-specific MEL1 protein and phasiRNA generated from lincRNA precursors in rice, another fascinating study conducted in rice uncovered a lncRNA called PMS1T, related to photosensitive male sterility acting as a phasiRNA precursor upon interaction with an initiator miR2118 (Fan et al. 2016). While both the studies stated above are associated with the reproductive facets of rice, recent works are being administered focusing on the stress-responsive regulation of the lncRNA-phasiRNA pathway in plants. One such notable work determining lncRNAs acting as PHAS loci was performed in A. thaliana, where the target prediction of these phasiRNAs unveiled their interactions with transcripts involved in plant developmental and stress-responsive activities (Yu et al. 2019a). While the implication of lncRNAs in the multifaceted defense system in plants is widely explored these days, experiments on how they are regulated still lack a proper interpretation (Zaynab et al. 2018). Recent findings that manifest the generation of phasiRNAs from lncRNAs, which are destined for their regulation during several plant development events, unlock the chance for a new leeway to inspect the potential role of phasiRNAs in regulating lncRNAs involved in plant defense responses and thereby disclose the detailed persona of how plants modulate their anti-pathogenic activities via non-coding RNA mediated surveillance.
The extensive research in the field of miRNA and phasiRNA studies laid the foundations for the emergence of two potent artificial sRNA-mediated silencing tools, artificial miRNAs (amiRNAs) and synthetic tasiRNAs (syn-tasiRNAs). AmiRNAs are constructed by modifying the duplex miRNA-precursor sequences to produce amiRNA/amiRNA* duplexes (Samad et al. 2017). Whereas syn-tasiRNAs are generated by replacing specific sequences in tasiRNA precursors with the sequence of interest. Both these artificial sRNA constructs follow their biogenesis pathway and initiate an explicit sRNA-mediated silencing of their targets (Sanan-Mishra et al. 2021). The exceptional advantages of amiRNAs and syn-tasiRNAs, for instance, minimal off-target silencing, availability of tools for automated designing of desired sRNA construct, proficiency in multiplexing several artificial sRNAs in a single sRNA construct and multiple targeting, allocate these two sRNA silencing approaches preferable than conventional dsRNA silencing approach as well as are well-recognized even in the era of CRISPR/CAS9-mediated gene silencing approach (Carbonell 2019). Currently, several amiRNA and syn-tasiRNA mediated resistance has been successfully implemented against plant pathogens, namely Rice black streaked dwarf virus in O. sativa (Sun et al. 2016), Blumeria graminis in H. vulgare (Liu et al. 2014), Turnip mosaic virus (TuMV) and Cucumber mosaic virus (CMV) in A. thaliana, Tomato spotted wilt virus (TSWV) in N. benthamiana and Solanum lycopersicum, Potato spindle tuber viroid (PSTVd) in N. benthamiana (Chen et al. 2016; Carbonell and Daròs 2017; Carbonell et al. 2019). The recent recognizable impact of the artificial sRNA-mediated gene silencing approach on plant resistance against dreadful pathogens frames amiRNAs and syn-tasiRNAs as one of the most efficient and significant tools in plant genetic engineering. Nevertheless, the shortfall in the detailed exploration of biogenesis pathway of these sRNAs, specifically tasiRNAs/phasiRNAs, hinders the accuracy and competence of silencing mediated by the same. This demands a detailed scrutinization of molecular mechanisms underlying the biogenesis and regulatory aspects of miRNAs and phasiRNAs during plant-pathogen interactions, along with developing a robust sRNA delivery system to introduce the desired artificial sRNAs into plants.
Computational tools for the identification of phasiRNAs and their associated genetic elements in plants
Apart from features like the length of the sequences and their conservation amid species, structural attributes of the precursors are considered as an essential factor by computational tools to delineate miRNAs from other sRNAs (Li et al. 2010). At the same time, as the precursors of phasiRNAs are in a deficit of a specific secondary structure, their detection demands a discrete computational approach for their identification. Phasing patterns of phasiRNAs contemplate as a robust feature for detecting the same (Morgado and Johannes 2017). The introductory study on developing a computational algorithm for tasiRNA loci detection from RNA sequence data was performed on A. thaliana by Chen et al. (2007). TasiRNA production is mostly influenced by miRNA-initiated cleavage and phasing patterns. Nevertheless, miRNA-targeting is not considered as a significant rationale for tasiRNA prediction, as all miRNA target loci do not ensue in small RNA clusters. Whereas the phasing pattern of sRNAs in 21-nt increments was considered for algorithm development in the respective study on A. thaliana (Chen et al. 2007). Subsequently, a web-based server named pssRNAMiner was developed, which detects not only the phased small RNA clusters but also unfolds the possible phase-initiators that can initiate cleavage at TAS loci (Dai and Zhao 2008). SoMART is another web server that is associated with several Solanaceae databases, which comprises a set of tools, namely a ‘Slicer detector’ to predict the sRNA that targets specific genes, a ‘dRNA mapper’, that identifies degradome RNA products in the given input data and thereby enhances the verification of sRNA-triggered cleavage sites in the same, a ‘PreMIR detector’ that exposes miRNA precursors or tasiRNA precursors and finally an ‘sRNA mapper’, which map the sRNAs against the given input data (Li et al. 2012). Similarly, another sRNA software package constituting several sRNA analyses tools, the UEA sRNA workbench, generates novel workflows from sRNA NGS data (Stocks et al. 2012). The preliminary UEA sRNA workbench had several tools for performing functions like, prediction of the precursors as well as mature miRNAs, comparing sRNA expression levels, prediction of phased tasiRNAs based on the prevailing algorithm by Chen et al. (2007) and ultimately, visualization of predicted sRNA loci, phased as well as unphased 21-nt sRNAs in each input samples separately using VisSR tool (Stocks et al. 2012). One of the remarkable modifications found in the latest version of UEA sRNA workbench is the PAREsnip tool, which executes the identification of sRNA-mRNA interactions with the help of a degradation profile derived from a PARE (Parallel Analysis of RNA Ends) experiment and thereby supplements that target prediction (Mohorianu et al. 2017). ShortStack, a stand-alone tool, can productively annotate sRNAs from heterogeneous data sets and can perform a significant role in revealing de novo small RNA clusters, miRNAs and hairpin-associated loci, check phasing or repeated orientation of aligned sRNAs, identify loci with small-sized sequence constitution, repetitiveness, etc. The prime factor that complements ShortStack from other mentioned tools is its ability to operate downstream for sRNA annotation by a single command (Axtell 2013). Subsequently, simple shell command has been preferred as an essential feature in most of the newly developed software, as it enables researchers with little knowledge about bioinformatics applications to get accustomed to using the same. One such ready-to-use software is Unitas, which performs complete annotation of sRNA datasets, reinforcing non-coding RNA sequences of numerous species accessible in the Ensembl databases (Gebert et al. 2017). As the sensitivity of Unitas depends barely on the number of background reads, this software is considered as an ideal tool for identifying less abundant phasiRNA loci.
Elevation in the disclosure of phasiRNAs as a significant regulator in plants imposed the need for a robust tool that can explicitly predict phasiRNAs. PhaseTank is such an integrated tool, which efficiently delineates phasiRNA loci as well as their regulatory networks. The quantification of identified PHAS loci, extensive annotation of phasiRNAs and their downstream functional aspects by one command analysis accentuate the significance of PhaseTank as a potential phasiRNA prediction tool. Moreover, PhaseTank not only predicts the targets of identified PHAS loci, but also trigger miRNAs, which altogether makes its features exceptional as it could perform both de novo predictions of phasiRNAs as well as unfold their regulatory aspects. Though agreement between the performance of PhaseTank and Unitas is acceptable while predicting the commonly occurring 21-nt phasiRNAs, when it comes to the case of less abundant 22-nt and 24-nt phasiRNAs, Unitas exhibited more sensitivity than PhaseTank along with fewer false positives (Guo et al. 2015). PHASIS, a recently developed computational suite, relies on a modular approach rather than a single ‘one-command’ approach as it enhances the inclusion of different connected analyses together with extended flexibility for discrete data or study conditions. This suite provides a consolidated remedy for parallel analysis of hundreds of samples on a single run, which thereby conserves time to a great extent. The major tools incorporated in PHASIS are; 1) “phasdetect”- which executes the de novo prediction of PHAS loci as well as their precursor RNAs with the aid of sRNA libraries and a corresponding reference genome/transcriptome provided by the user, 2) “phasmerge” – which summarizes the output data of “phasdetect” by comparing the list of PHAS loci in each library as well as ancillary data of the same and this tool also links PHAS loci with its respective annotations, 3) “phastrigs” – is the tool that potentially identifies the sRNA triggers of detected PHAS loci and their precursors by interpreting the “phasmerge” output along with a list of miRNAs provided by the user. This tool thereby excludes the requirement of additional PARE experimental data for trigger detection, at the same time, if PARE data is available, “phasetrigs” can expose more reliable experimentally supported triggers that hold up more credible downstream analysis (Kakrana et al. 2017).
Though the advancement of several novel computational tools transpired in line with a large amount of experimentally elucidated genomic data, a need for an alignment-free approach is of prime importance as it enhances the annotation of complex sRNA data comprising unaligned sequences. One such approach was developed by Patel et al. (2018), using machine learning methods for the prediction of reproductive phasiRNAs in the Poaceae family, mainly rice and maize. The study created a Random Forest classifier that can predict and discriminate reproductive phasiRNAs from other sRNAs, on the basis of sequence-based as well as positional characteristics extracted from a given sequence data. This machine-learning approach, with an accuracy of > 80%, also possesses the ability to determine the specifics about AGO-phasiRNA interaction by taking 5' nucleotide of the small RNA sequences into consideration (Patel et al. 2018). A more recently developed web server, PhasiRNAnalyzer, appears to have more advanced features compared to other conventional phasiRNA detectors. This tool can even predict the differentially expressed phasiRNAs along with other regulatory components associated with the identified phasiRNAs and their targets, exposing the complete functional significance of specific phasiRNA pathways (Fei et al. 2021). The list of so far developed phasiRNA prediction tools is depicted in Fig. 3.
Fig. 3.
Progressive list of bioinformatics tools developed for phasiRNA prediction and annotation
The lack of unique precursor structure and partially unveiled biogenesis of phasiRNAs still impart quite a few computational challenges for precisely predicting valid PHAS loci and their corresponding phasiRNAs. Though there are a number of computational tools available for the prediction of phasiRNAs at present, each one operates on the basis of discrete features, which thus mandate the requirement of integrating essential components of these individual tools to generate a combined framework with better performance.
Conclusion
Growing evidences complement the role of sRNAs, namely miRNAs and phasiRNAs, in reprogramming the expression of plant genes associated with growth, reproduction and stress responses via transcriptional and post-transcriptional gene silencing. Significant studies on these midget RNAs exposed their impact in providing both abiotic and biotic stress tolerances like drought tolerance, salt tolerance, heavy-metal tolerance, tolerance to intense temperature and resistance to pathogens, respectively. The characterization and functional annotation of these miRNA-phasiRNA duos, which can act as mobile regulatory elements, have prime importance in developing stress-tolerant plant varieties. The recent development of artificial miRNAs (amiRNAs) and synthetic-tasiRNAs (syn-tasiRNAs) are two pinnacle discoveries for the selective silencing of both endogenous RNAs as well as the invading pathogen RNAs in plants. Hence, in-depth mining of phasiRNAs and their trigger miRNAs associated with different biological activities is in the front line to unravel the complex regulatory networks initiated by them and also for their targeted manipulation. Furthermore, there exists an insistent need for the development of an explicit integrated computational tool for the prediction, characterization and functional annotation of phasiRNAs. The extensive interpretation of dynamic phasiRNA biogenesis, their targets and functional relevance implements a novel approach to regulate the expression of transcripts that are crucial for the development of superior plants with proper growth, better yield and with exceptional adaptation to environmental stresses.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- Allen E, Xie Z, Gustafson AM, Carrington JC. MicroRNA-directed phasing during trans-acting siRNA biogenesis in plants. Cell. 2005;121:207–221. doi: 10.1016/j.cell.2005.04.004. [DOI] [PubMed] [Google Scholar]
- Arif MA, Fattash I, Ma Z, et al. DICER-LIKE3 activity in Physcomitrella patens DICER-LIKE4 mutants causes severe developmental dysfunction and sterility. Mol Plant. 2012;5:1281–1294. doi: 10.1093/mp/sss036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arikit S, Xia R, Kakrana A, Huang K, Zhai J, Yan Z, Valdés-López O, Prince S, Musket TA, Nguyen HT, Stacey G, Meyers BC. An atlas of soybean small RNAs identifies phased siRNAs from hundreds of coding genes. Plant Cell. 2014;26:4584–4601. doi: 10.1105/tpc.114.131847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Axtell MJ. ShortStack: comprehensive annotation and quantification of small RNA genes. RNA. 2013;19:740–751. doi: 10.1261/rna.035279.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Axtell MJ, Jan C, Rajagopalan R, Bartel DP. A two-hit trigger for siRNA biogenesis in plants. Cell. 2006;127:565–577. doi: 10.1016/j.cell.2006.09.032. [DOI] [PubMed] [Google Scholar]
- Budak H, Kaya SB, Cagirici HB. Long non-coding RNA in plants in the era of reference sequences. Front Plant Sci. 2020;11:276. doi: 10.3389/fpls.2020.00276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai Q, Liang C, Wang S, et al. The disease resistance protein SNC1 represses the biogenesis of microRNAs and phased siRNAs. Nat Commun. 2018;9:5080. doi: 10.1038/s41467-018-07516-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calixto CPG, Tzioutziou NA, James AB, et al. Cold-dependent expression and alternative splicing of arabidopsis long non-coding RNAs. Front Plant Sci. 2019;10:235. doi: 10.3389/fpls.2019.00235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carbonell A. Secondary Small interfering RNA-based silencing tools in plants: an update. Front Plant Sci. 2019;10:687. doi: 10.3389/fpls.2019.00687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carbonell A, Daròs J-A (2017) Artificial microRNAs and synthetic trans-acting small interfering RNAs interfere with viroid infection. Mol Plant Pathol 18:746–753. [DOI] [PMC free article] [PubMed]
- Cervera-Seco L, Marques MAC, Sanz-Carbonell A, Marquez-Molins J, Carbonell A, Darï SJA, Gomez G (2019) Identification and characterization of stress-responsive TAS3-derived TasiRNAs in Melon. Plant Cell Physiol 60:2382–2393. 10.1093/pcp/pcz131 [DOI] [PubMed]
- Chen C, Zeng Z, Liu Z, Xia R. Small RNAs, emerging regulators critical for the development of horticultural traits. Hort Res. 2018;5:63. doi: 10.1038/s41438-018-0072-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen HM, Chen LT, Patel K, Li YH, Baulcombe DC, Wu SH (2010) 22-Nucleotide RNAs trigger secondary siRNA biogenesis in plants. Proc Natl Acad Sci USA 107:15269–15274. Doi: 10.1073/pnas.1001738107 [DOI] [PMC free article] [PubMed]
- Chen HM, Li YH, Wu SH. Bioinformatic prediction and experimental validation of a microRNA-directed tandem trans-acting siRNA cascade in Arabidopsis. Proc Natl Acad Sci USA. 2007;104:3318–3323. doi: 10.1073/pnas.0611119104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen L, Cheng X, Cai J, Zhan L, Wu X, Liu Q, Wu X (2016) Multiple virus resistance using artificial trans-acting siRNAs. J Virol Methods 228:16–20. Doi: 10.1016/j.jviromet.2015.11.004 [DOI] [PubMed]
- Chen X. Small RNAs in development—insights from plants. Curr Opin Genet Dev. 2012;22:361–367. doi: 10.1016/j.gde.2012.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho SH, Coruh C, Axtell MJ (2012) miR156 and miR390 Regulate tasiRNA Accumulation and Developmental Timing in Physcomitrella patens. Plant Cell 24:4837–4849. Doi: 10.1105/tpc.112.103176 [DOI] [PMC free article] [PubMed]
- Cui C, Wang JJ, Zhao JH, Fang YY, He XF, Guo HS, Duan CG (2020) A brassica miRNA regulates plant growth and immunity through distinct modes of action. Mol Plant 13:231–245. Doi: 10.1016/j.molp.2019.11.010 [DOI] [PubMed]
- Cuperus JT, Carbonell A, Fahlgren N, Garcia-Ruiz H, Burke RT, Takeda A, Sullivan CM, Gilbert SD, Montgomery TA, Carrington JC. Unique functionality of 22-nt miRNAs in triggering RDR6-dependent siRNA biogenesis from target transcripts in Arabidopsis. Nat Struct Mol Biol. 2010;17:997–1003. doi: 10.1038/nsmb.1866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dai X, Zhao PX. pssRNAMiner: a plant short small RNA regulatory cascade analysis server. Nucleic Acids Res. 2008;36:W114–W118. doi: 10.1093/nar/gkn297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Felippes FF. Gene regulation mediated by microRNA-triggered secondary small RNAs in plants. Plants. 2019;8:112. doi: 10.3390/plants8050112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Felippes FF, Marchais A, Sarazin A, Oberlin S, Voinnet O. A single miR390 targeting event is sufficient for triggering TAS3-tasiRNA biogenesis in arabidopsis. Nucleic Acids Res. 2017;45:5539–5554. doi: 10.1093/nar/gkx119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng P, Muhammad S, Cao M, Wu L. Biogenesis and regulatory hierarchy of phased small interfering RNAs in plants. Plant Biotechnol J. 2018;16:965–975. doi: 10.1111/pbi.12882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dmitriev AA, Kudryavtseva A v, Bolsheva NL, et al (2017) miR319, miR390, and miR393 Are Involved in Aluminum Response in Flax (Linum usitatissimum L.). Biomed Res Int 2017:4975146. Doi: 10.1155/2017/4975146 [DOI] [PMC free article] [PubMed]
- Eamens AL, Wook Kim K, Waterhouse PM. DRB2, DRB3 and DRB5 function in a non-canonical microRNA pathway in Arabidopsis thaliana. Plant Signal Behav. 2012;7:1224–1229. doi: 10.4161/psb.21518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan Y, Yang J, Mathioni SM, Yu J, Shen J, Yang X, Wang L, Zhang Q, Cai Z, Xu C, Li X, Xiao J, Meyers BC, Zhang Q. PMS1T, producing phased small-interfering RNAs, regulates photoperiod-sensitive male sterility in rice. Proc Natl Acad Sci USA. 2016;113:15144–15149. doi: 10.1073/pnas.1619159114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fei Q, Xia R, Meyers BC. Phased, secondary, small interfering RNAs in posttranscriptional regulatory networks. Plant Cell. 2013;25:2400–2415. doi: 10.1105/tpc.113.114652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fei Y, Feng J, Wang R, Zhang B, Zhang H, Huang J. PhasiRNAnalyzer: an integrated analyser for plant phased siRNAs. RNA Biol. 2021;18:1–8. doi: 10.1080/15476286.2021.1879543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Felippes FF, Weigel D. Triggering the formation of tasiRNAs in Arabidopsis thaliana: the role of microRNA miR173. EMBO Rep. 2009;10:264–270. doi: 10.1038/embor.2008.247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gasciolli V, Mallory AC, Bartel DP, Vaucheret H. Partially redundant functions of arabidopsis DICER-like enzymes and a role for DCL4 in producing trans-acting siRNAs. Curr Biol. 2005;15:1494–1500. doi: 10.1016/j.cub.2005.07.024. [DOI] [PubMed] [Google Scholar]
- Gebert D, Hewel C, Rosenkranz D. Unitas: the universal tool for annotation of small RNAs. BMC Genom. 2017;18:644. doi: 10.1186/s12864-017-4031-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghanashyam C, Jain M. Role of auxin-responsive genes in biotic stress responses. Plant Signal Behav. 2009;4:846–848. doi: 10.4161/psb.4.9.9376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guan X, Pang M, Nah G, Shi X, Ye W, Stelly DM, Chen ZJ. miR828 and miR858 regulate homoeologous MYB2 gene functions in Arabidopsis trichome and cotton fibre development. Nat Commun. 2014;5:3050. doi: 10.1038/ncomms4050. [DOI] [PubMed] [Google Scholar]
- Guo Q, Qu X, Jin W. PhaseTank: genome-wide computational identification of phasiRNAs and their regulatory cascades. Bioinformatics. 2015;31:284–286. doi: 10.1093/bioinformatics/btu628. [DOI] [PubMed] [Google Scholar]
- Guo G, Liu X, Sun F, et al. Wheat miR9678 affects seed germination by generating phased siRNAs and modulating abscisic acid/gibberellin signaling. Plant Cell. 2018;30:796–814. doi: 10.1105/tpc.17.00842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hobecker KV, Reynoso MA, Bustos-Sanmamed P, et al. The MicroRNA390/tas3 pathway mediates symbiotic nodulation and lateral root growth. Plant Physiol. 2017;174:2469–2486. doi: 10.1104/pp.17.00464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hou Y, Zhai Y, Feng L, Karimi HZ, Rutter BD, Zeng L, Choi DS, Zhang B, Gu W, Chen X, Ye W, Innes RW, Zhai J, Ma W. A Phytophthora effector suppresses trans-kingdom RNAi to promote disease susceptibility. Cell Host Microbe. 2019;25:153–165.e5. doi: 10.1016/j.chom.2018.11.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howell MD, Fahlgren N, Chapman EJ, Cumbie JS, Sullivan CM, Givan SA, Kasschau KD, Carrington JC. Genome-wide analysis of the RNA-DEPENDENT RNA POLYMERASE6/DICER-LIKE4 pathway in arabidopsis reveals dependency on miRNA and tasiRNA-directed targeting. Plant Cell. 2007;19:926–942. doi: 10.1105/tpc.107.050062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hunt M, Banerjee S, Surana P, Liu M, Fuerst G, Mathioni S, Meyers BC, Nettleton D, Wise RP. Small RNA discovery in the interaction between barley and the powdery mildew pathogen. BMC Genom. 2019;20:610. doi: 10.1186/s12864-019-5947-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hunter C, Willmann MR, Wu G, Yoshikawa M, de la Luz G-N, Poethig SR. Trans-acting siRNA-mediated repression of ETTIN and ARF4 regulates heteroblasty in arabidopsis. Development. 2006;133:2973–2981. doi: 10.1242/dev.02491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson C, Kasprzewska A, Tennessen K, et al (2009) Clusters and superclusters of phased small RNAs in the developing inflorescence of rice. Genome Res 19:1429–1440. Doi: 10.1101/gr.089854.108 [DOI] [PMC free article] [PubMed]
- Kakrana A, Li P, Patel P, et al (2017) PHASIS: A computational suite for de novo discovery and characterization of phased, siRNA-generating loci and their miRNA triggers. bioRxiv. Doi: 10.1101/158832
- Kamthan A, Chaudhuri A, Kamthan M, Datta A. Small RNAs in plants: recent development and application for crop improvement. Front Plant Sci. 2015;6:208. doi: 10.3389/fpls.2015.00208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Komiya R. Biogenesis of diverse plant phasiRNAs involves an miRNA-trigger and Dicer-processing. J Plant Res. 2017;130:17–23. doi: 10.1007/s10265-016-0878-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Komiya R, Ohyanagi H, Niihama M, Watanabe T, Nakano M, Kurata N, Nonomura KI. Rice germline-specific Argonaute MEL1 protein binds to phasiRNAs generated from more than 700 lincRNAs. Plant J. 2014;78:385–397. doi: 10.1111/tpj.12483. [DOI] [PubMed] [Google Scholar]
- Križnik M, Petek M, Dobnik D, Ramšak Ž, Baebler Š, Pollmann S, Kreuze J, Žel J, Gruden K (2017) Tolerance to PVY infection in potato is conditioned by perturbation of small RNA-gibberellin regulatory network. bioRxiv 130757. Doi: 10.1101/130757 [DOI] [PMC free article] [PubMed]
- Li L, Xu J, Yang D, Tan X, Wang H. Computational approaches for microRNA studies: a review. Mamm Genome. 2010;21:1–12. doi: 10.1007/s00335-009-9241-2. [DOI] [PubMed] [Google Scholar]
- Li F, Orban R, Baker B. SoMART: a web server for plant miRNA, tasiRNA and target gene analysis. Plant J. 2012;70:891–901. doi: 10.1111/j.1365-313X.2012.04922.x. [DOI] [PubMed] [Google Scholar]
- Li S, Liu J, Liu Z, et al (2014) HEAT-INDUCED TAS1 TARGET1 mediates thermotolerance via HEAT STRESS TRANSCRIPTION FACTOR A1a-directed pathways in arabidopsis. Plant Cell 26:1764–1780. Doi: 10.1105/tpc.114.124883 [DOI] [PMC free article] [PubMed]
- Lindbo JA (2012) A historical overview of RNAi in plants. Methods Mol Biol 894:1–16. Doi: 10.1007/978-1-61779-882-5_1 [DOI] [PubMed]
- Liu J, Cheng X, Liu D, Xu W, Wise R, Shen QH. The miR9863 family regulates distinct Mla alleles in barley to attenuate NLR receptor-triggered disease resistance and cell-death signaling. PLoS Genet. 2014;10:e1004755. doi: 10.1371/journal.pgen.1004755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y, Teng C, Xia R, Meyers BC. PhasiRNAs in plants: their biogenesis, genic sources, and roles in stress responses, development, and reproduction. Plant Cell. 2020;32:3059–3080. doi: 10.1105/tpc.20.00335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- López-Márquez D, Del-Espino Á, López-Pagán N, Rodríguez-Negrete EA, Rubio-Somoza I, Ruiz-Albert J, Bejarano ER, Beuzón CR (2020) MiRNA and phasiRNAs-mediated regulation of TIR-NBS-LRR defense genes in Arabidopsis thaliana. bioRxiv 2020.03.02.972620. Doi: 10.1101/2020.03.02.972620
- Nair MM, Alagu M (2020) MicroRNAs as fine-tuners of gene regulation in plant-microbe interactions. Curr Sci 119:1282–1290. Doi: 10.18520/cs/v119/i8/1282-1290
- Nair MM, TS K, Alagu M (2020) Bioinformatics insights into microRNA mediated gene regulation in Triticum estivum during multiple fungal diseases. Plant Gene 21:100219. Doi: 10.1016/j.plgene.2019.100219
- Ma W, Chen C, Liu Y, Zeng M, Meyers BC, Li J, Xia R. Coupling of microRNA-directed phased small interfering RNA generation from long noncoding genes with alternative splicing and alternative polyadenylation in small RNA-mediated gene silencing. New Phytol. 2018;217:1535–1550. doi: 10.1111/nph.14934. [DOI] [PubMed] [Google Scholar]
- Manna S. An overview of pentatricopeptide repeat proteins and their applications. Biochimie. 2015;113:93–99. doi: 10.1016/j.biochi.2015.04.004. [DOI] [PubMed] [Google Scholar]
- Marin E, Jouannet V, Herz A, et al. miR390, Arabidopsis TAS3 tasiRNAs, and their AUXIN RESPONSE FACTOR targets define an autoregulatory network quantitatively regulating lateral root growth. Plant Cell. 2010;22:1104–1117. doi: 10.1105/tpc.109.072553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohorianu, Stocks MB, Applegate C, et al. The UEA small RNA workbench: a suite of computational tools for small RNA analysis. Methods Mol biol. 2017;1580:193–224. doi: 10.1007/978-1-4939-6866-4_14. [DOI] [PubMed] [Google Scholar]
- Montgomery TA, Howell MD, Cuperus JT, Li D, Hansen JE, Alexander AL, Chapman EJ, Fahlgren N, Allen E, Carrington JC. Specificity of ARGONAUTE7-miR390 interaction and dual functionality in TAS3 trans-acting siRNA formation. Cell. 2008;133:128–141. doi: 10.1016/j.cell.2008.02.033. [DOI] [PubMed] [Google Scholar]
- Montgomery TA, Yoo SJ, Fahlgren N, Gilbert SD, Howell MD, Sullivan CM, Alexander A, Nguyen G, Allen E, Ahn JH, Carrington JC. AGO1-miR173 complex initiates phased siRNA formation in plants. Proc Natl Acad Sci USA. 2008;105:20055–20062. doi: 10.1073/pnas.0810241105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morgado L, Johannes F. Computational tools for plant small RNA detection and categorization. Brief Bioinform. 2017;20:1181–1192. doi: 10.1093/bib/bbx136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakazawa Y, Hiraguri A, Moriyama H, Fukuhara T. The dsRNA-binding protein DRB4 interacts with the Dicer-like protein DCL4 in vivo and functions in the trans-acting siRNA pathway. Plant Mol Biol. 2007;63:777–785. doi: 10.1007/s11103-006-9125-8. [DOI] [PubMed] [Google Scholar]
- Park JH, Shin C. The role of plant small RNAs in NB-LRR regulation. Brief Funct Genom. 2015;14:268–274. doi: 10.1093/bfgp/elv006. [DOI] [PubMed] [Google Scholar]
- Park YJ, Lee HJ, Kwak KJ, Lee K, Hong SW, Kang H. MicroRNA400-guided cleavage of Pentatricopeptide repeat protein mRNAs Renders Arabidopsis thaliana more susceptible to pathogenic bacteria and fungi. Plant Cell Physiol. 2014;55:1660–1668. doi: 10.1093/pcp/pcu096. [DOI] [PubMed] [Google Scholar]
- Patel P, Mathioni S, Kakrana A, Shatkay H, Meyers BC. Reproductive phasiRNAs in grasses are compositionally distinct from other classes of small RNAs. New Phytol. 2018;220:851–864. doi: 10.1111/nph.15349. [DOI] [PubMed] [Google Scholar]
- Pegler JL, Oultram JMJ, Curtin SJ, Grof CPL, Eamens AL. Further disruption of the tas3 pathway via the addition of the AGO7 mutation to the DRB1, DRB2 or DRB4 mutations severely impairs the reproductive competence of arabidopsis thaliana. Agronomy. 2019;9:680. doi: 10.3390/agronomy9110680. [DOI] [Google Scholar]
- Rock CD. Trans-acting small interfering RNA4: key to nutraceutical synthesis in grape development? Trends Plant Sci. 2013;18:601–610. doi: 10.1016/j.tplants.2013.07.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samad AFA, Sajad M, Nazaruddin N, et al. MicroRNA and transcription factor: key players in plant regulatory network. Front Plant Sci. 2017;8:565. doi: 10.3389/fpls.2017.00565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanan-Mishra N, Abdul Kader Jailani A, Mandal B, Mukherjee SK. Secondary siRNAs in plants: biosynthesis, various functions, and applications in virology. Front Plant Sci. 2021;12:610283. doi: 10.3389/fpls.2021.610283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sekhwal MK, Li P, Lam I, Wang X, Cloutier S, You FM. Disease resistance gene analogs (RGAs) in plants. Int J Mol Sci. 2015;16:19248–19290. doi: 10.3390/ijms160819248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song X, Li P, Zhai J, Zhou M, Ma L, Liu B, Jeong DH, Nakano M, Cao S, Liu C, Chu C, Wang XJ, Green PJ, Meyers BC, Cao X. Roles of DCL4 and DCL3b in rice phased small RNA biogenesis. Plant J. 2012;69:462–474. doi: 10.1111/j.1365-313X.2011.04805.x. [DOI] [PubMed] [Google Scholar]
- Sosa-Valencia G, Romero-Pérez PS, Palomar VM, et al. Insights into the function of the phasiRNA-triggering miR1514 in response to stress in legumes. Plant Signal Behav. 2017;12:e1284724–e1284724. doi: 10.1080/15592324.2017.1284724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stocks MB, Moxon S, Mapleson D, Woolfenden HC, Mohorianu I, Folkes L, Schwach F, Dalmay T, Moulton V. The UEA sRNA workbench: a suite of tools for analysing and visualizing next generation sequencing microRNA and small RNA datasets. Bioinformatics. 2012;28:2059–2061. doi: 10.1093/bioinformatics/bts311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun L, Lin C, Du J, et al. Dimeric artificial microRNAs mediate high resistance to RSV and RBSDV in transgenic rice plants. Plant Cell. Plant Cell, Tissue Organ Cult. 2016;126:127–139. doi: 10.1007/s11240-016-0983-8. [DOI] [Google Scholar]
- Tabara M, Ohtani M, Kanekatsu M, Moriyama H, Fukuhara T. Size Distribution of small interfering RNAs in various organs at different developmental stages is primarily determined by the dicing activity of dicer-like proteins in plants. Plant Cell Physiol. 2018;59:2228–2238. doi: 10.1093/pcp/pcy144. [DOI] [PubMed] [Google Scholar]
- Tiwari B, Habermann K, Arif MA, et al. Identification of small RNAs during high light acclimation in arabidopsis thaliana. Front Plant Sci. 2021;12:656657. doi: 10.3389/fpls.2021.656657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vargas-Asencio JA, Perry KL. A small RNA-mediated regulatory network in arabidopsis thaliana demonstrates connectivity between phasiRNA regulatory modules and extensive co-regulation of transcription by miRNAs and phasiRNAs. Front Plant Sci. 2019;10:1710. doi: 10.3389/fpls.2019.01710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vazquez F, Vaucheret H, Rajagopalan R, Lepers C, Gasciolli V, Mallory AC, Hilbert JL, Bartel DP, Crété P (2004) Endogenous trans-Acting siRNAs regulate the accumulation of arabidopsis mRNAs. Mol Cell 16:69–79. Doi: 10.1016/j.molcel.2004.09.028 [DOI] [PubMed]
- Wang Y, Itaya A, Zhong X, Wu Y, Zhang J, van der Knaap E, Olmstead R, Qi Y, Ding B. Function and evolution of a MicroRNA that regulates a Ca2+-ATPase and triggers the formation of phased small interfering RNAs in tomato reproductive growth. Plant Cell. 2011;23:3185–3203. doi: 10.1105/tpc.111.088013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams L, Carles CC, Osmont KS, Fletcher JC. A database analysis method identifies an endogenous trans-acting short-interfering RNA that targets the Arabidopsis ARF2, ARF3, and ARF4 genes. Proc Natl Acad Sci USA. 2005;102:9703–9708. doi: 10.1073/pnas.0504029102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu L, Mao L, Qi Y. Roles of dicer-like and argonaute proteins in TAS-derived small interfering RNA-triggered DNA methylation. Plant Physiol. 2012;160:990–999. doi: 10.1104/pp.112.200279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xia R, Meyers BC, Liu Z, Beers EP, Ye S, Liu Z. MicroRNA superfamilies descended from miR390 and their roles in secondary small interfering RNA biogenesis in eudicots. Plant Cell. 2013;25:1555–1572. doi: 10.1105/tpc.113.110957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xia R, Xu J, Arikit S, Meyers BC. Extensive families of miRNAs and PHAS loci in norway spruce demonstrate the origins of complex phasiRNA networks in seed plants. Mol Biol Evol. 2015;32:2905–2918. doi: 10.1093/molbev/msv164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xia R, Ye S, Liu Z, Meyers BC, Liu Z. Novel and recently evolved MicroRNA clusters regulate expansive F-BOX gene networks through phased small interfering RNAs in wild diploid strawberry. Plant Physiol. 2015;169:594–610. doi: 10.1104/pp.15.00253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xia R, Zhu H, An Y, Beers EP, Liu Z. Apple miRNAs and tasiRNAs with novel regulatory networks. Genome Biol. 2012;13:R47. doi: 10.1186/gb-2012-13-6-r47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu M, Li Y, Zhang Q, et al. Novel miRNA and phasiRNA biogenesis networks in soybean roots from two sister lines that are resistant and susceptible to SCN race 4. PLoS One. 2014;9:e110051–e110051. doi: 10.1371/journal.pone.0110051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshikawa M, Iki T, Tsutsui Y, Miyashita K, Poethig RS, Habu Y, Ishikawa M. 3’ fragment of miR173-programmed RISC-cleaved RNA is protected from degradation in a complex with RISC and SGS3. Proc Natl Acad Sci USA. 2013;110:4117–4122. doi: 10.1073/pnas.1217050110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu L, Guo R, Jiang Y, Ye X, Yang Z, Meng Y, Shao C (2019a) Identification of novel phasiRNAs loci on long non-coding RNAs in Arabidopsis thaliana. Genomics 111:1668–1675. Doi: 10.1016/j.ygeno.2018.11.017 [DOI] [PubMed]
- Yu Y, Zhang Y, Chen X, Chen Y. Plant noncoding RNAs: hidden players in development and stress responses. Annu Rev Cell Dev Biol. 2019;35:407–431. doi: 10.1146/annurev-cellbio-100818-125218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaynab M, Fatima M, Abbas S, Umair M, Sharif Y, Raza MA. Long non-coding RNAs as molecular players in plant defense against pathogens. Microb Pathog. 2018;121:277–282. doi: 10.1016/j.micpath.2018.05.050. [DOI] [PubMed] [Google Scholar]
- Zhai J, Jeong DH, De Paoli E, Park S, Rosen BD, Li Y, González AJ, Yan Z, Kitto SL, Grusak MA, Jackson SA, Stacey G, Cook DR, Green PJ, Sherrier DJ, Meyers BC. MicroRNAs as master regulators of the plant NB-LRR defense gene family via the production of phased, trans-acting siRNAs. Genes Dev. 2011;25:2540–2553. doi: 10.1101/gad.177527.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao M, Cai C, Zhai J, Lin F, Li L, Shreve J, Thimmapuram J, Hughes TJ, Meyers BC, Ma J (2015) Coordination of MicroRNAs, PhasiRNAs, and NB-LRR genes in response to a plant pathogen: insights from analyses of a set of soybean Rps gene near-isogenic lines. Plant Genome 8:eplantgenome2014.09.0044. Doi: 10.3835/plantgenome2014.09.0044 [DOI] [PubMed]
- Zheng Y, Wang Y, Wu J, Ding B, Fei Z. A dynamic evolutionary and functional landscape of plant phased small interfering RNAs. BMC Biol. 2015;13:32. doi: 10.1186/s12915-015-0142-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu QH, Fan L, Liu Y, Xu H, Llewellyn D, Wilson I. miR482 regulation of NBS-LRR defense genes during fungal pathogen infection in cotton. PLoS One. 2013;8:e84390. doi: 10.1371/journal.pone.0084390. [DOI] [PMC free article] [PubMed] [Google Scholar]