Abstract
Butyrate is a preferred energy source for colonic epithelial cells and is thought to play an important role in maintaining colonic health in humans. In order to investigate the diversity and stability of butyrate-producing organisms of the colonic flora, anaerobic butyrate-producing bacteria were isolated from freshly voided human fecal samples from three healthy individuals: an infant, an adult omnivore, and an adult vegetarian. A second isolation was performed on the same three individuals 1 year later. Of a total of 313 bacterial isolates, 74 produced more than 2 mM butyrate in vitro. Butyrate-producing isolates were grouped by 16S ribosomal DNA (rDNA) PCR-restriction fragment length polymorphism analysis. The results indicate very little overlap between the predominant ribotypes of the three subjects; furthermore, the flora of each individual changed significantly between the two isolations. Complete sequences of 16S rDNAs were determined for 24 representative strains and subjected to phylogenetic analysis. Eighty percent of the butyrate-producing isolates fell within the XIVa cluster of gram-positive bacteria as defined by M. D. Collins et al. (Int. J. Syst. Bacteriol. 44:812–826, 1994) and A. Willems et al. (Int. J. Syst. Bacteriol. 46:195–199, 1996), with the most abundant group (10 of 24 or 42%) clustering with Eubacterium rectale, Eubacterium ramulus, and Roseburia cecicola. Fifty percent of the butyrate-producing isolates were net acetate consumers during growth, suggesting that they employ the butyryl coenzyme A-acetyl coenzyme A transferase pathway for butyrate production. In contrast, only 1% of the 239 non-butyrate-producing isolates consumed acetate.
The species diversity of the predominantly anaerobic bacterial communities from the human large bowel has been the subject of both conventional and molecular microbiological investigations (31, 32, 46, 49). These communities are believed to contribute to healthy gut function in a variety of ways, including protecting against pathogens and producing nutrients for the colonic mucosa (3, 11, 12, 15). We still know relatively little, however, about the contributions of individual anaerobic species to colonic fermentation and to the nutrition and health of the host.
Diet-derived substrates, particularly undigested fiber and starch reaching the large intestine, have major effects upon bacterial community structure and metabolism in the colon. Short-chain fatty acids (SCFA) formed by microbial fermentation have an important effect on colonic health (10, 44). Butyrate in particular has an important role in the metabolism and normal development of colonic epithelial cells and has been implicated in protection against cancer and ulcerative colitis (21). Butyrate is preferentially transported by gut epithelial cells (36), serves as a preferred energy source for colonocytes (37, 43), and has been shown to exert direct effects upon gene expression in mammalian cells through histone hyperacetylation and through interaction with butyrate response elements upstream of some genes (8, 45). Production of butyrate by mixed human fecal microflora in vitro is known to be strongly influenced by the growth substrate. Starch, for example, is strongly butyrogenic, whereas other polysaccharides such as pectin result in relatively less butyrate and more propionate and acetate (9). Thus, the relative production rates of these SCFA provide a potentially important link between diet and colonic health. Despite this, however, remarkably little is known about the physiologies, identities, and ecologies of the predominant species of butyrate-producing bacteria from the human large bowel. The most obvious explanation for the stimulation of butyrate synthesis by certain carbohydrates in vitro is direct selection for butyrate-producing components of the flora capable of utilizing the particular substrate. Greater knowledge of these bacteria should lead to a more mechanistic understanding of the effects of diet on butyrate production in the colon. The isolation and molecular characterization of butyrate-producing species from fecal samples of three human volunteers deliberately chosen to represent different ages and diets is described in this paper.
MATERIALS AND METHODS
Bacterial strains.
The following bacterial reference strains were used in restriction fragment length polymorphism (RFLP) analysis: Butyrivibrio fibrisolvens 1.230 (bovine rumen [41]), 2221 (bovine rumen, ATCC 19171 [6]), 16.4 (human feces [38]), Eubacterium multiforme (ATCC 25552), and E. limosum R5004 and Fusobacterium strains R5043, R7263, and R10012 (all from J. Brazier, University of Wales, Cardiff, United Kingdom).
Collection and processing of samples.
Freshly voided fecal samples were collected from three healthy individuals, an infant (initially sampled at 11 months of age), a lacto-ovo-vegetarian adult (adult B, aged 46), and an omnivorous adult (adult A, aged 32), on two occasions approximately 1 year apart. None of the individuals had received antibiotics or other drugs in the months prior to sampling.
Fecal samples were placed in sterile universal bottles and processed within 30 min of collection. A subsample of feces (1 g) was aseptically transferred into a further sterile preweighed bottle. To the samples, 9 ml of M2GSC diluent (modified Hobson [30]), containing (per 100 ml) 1 g of casitone, 0.25 g of yeast extract, 0.4 g of NaHCO3, 0.2 g of glucose, 0.2 g of cellobiose, 0.2 g of soluble starch, 30 ml of clarified rumen fluid, 0.1 g of cysteine, 0.045 g of K2HPO4, 0.045 g of KH2PO4, 0.09 g of (NH4)2SO4, 0.09 g of NaCl, 0.009 g of MgSO4 · 7H2O, 0.009 g of CaCl2, and 0.1 mg of resazurin, was added aseptically while the bottle was being flushed with CO2 according to the anaerobic Hungate method (25). This diluent corresponded to the first 10-fold dilution, which was then mixed by vortexing for 3 min to form an evenly distributed suspension. The first dilution was subsequently diluted by 10-fold serial dilutions through to a 10−9 dilution.
Anaerobic roll tubes (5) were prepared in 16- by 125-mm Hungate tubes (25) sealed with butyl septum stoppers (Bellco Glass Inc., Vineland, N.J.) using medium M2GSC containing 0.75% agar (Difco) (30) and inoculated with 0.5-ml aliquots of appropriate serial dilutions. Undiluted aliquots (1 ml) were dispensed into 1.5-ml Eppendorf tubes and centrifuged (13,000 × g, 10 min) to pellet the bacteria, which were stored at −70°C prior to DNA extraction. Roll tubes were incubated at 37°C for 48 h prior to the picking of 30 to 80 colonies from each fecal sample. Cultures picked and grown in broths of M2GSC were used for examination of gram-stained smears (23, 24), for determination of fermentation products by capillary gas chromatography (35), and for extraction of DNA (see below).
DNA extraction.
DNA was extracted from bacterial pellets by following the method of Ausubel et al. (1). Bacterial pellets were resuspended in 567 μl of 1× Tris–EDTA buffer, pH 8.0. Three microliters of 20-mg/ml proteinase K and 30 μl of 10% (wt/vol) sodium dodecyl sulfate were added, and the solution was mixed and incubated for 1 h at 37°C. One hundred microliters of 5 M NaCl was added, followed by 80 μl of cetyltrimethylammonium bromide-NaCl solution (10% [wt/vol] cetyltrimethylammonium bromide, 0.7 M NaCl). The solutions were mixed and incubated for 10 min at 65°C. An equal volume of chloroform-isoamyl alcohol (24:1) was added, and the solution was mixed thoroughly and centrifuged for 10 min at 14,000 × g. The supernate was extracted twice with phenol-chloroform-isoamyl alcohol (25:24:1) and isopropanol precipitated. The DNA pellets were resuspended in sterile distilled water and stored at 4°C for immediate use or aliquoted at −20°C for long-term storage.
PCR amplification of 16S rDNA.
Fifty nanograms of total genomic DNA was used as a target for amplification of approximately 1,500 bp of 16S ribosomal DNA (rDNA) using the eubacterial primers fD1, 5′ AGAGTTTGATCCTGGCTCAG 3′ (Escherichia coli positions 8 to 27), and rP2, 5′ ACGGCTACCTTGTTACGACTT 3′ (E. coli positions 1494 to 1513) (47). PCR amplification was carried out as described previously (26).
Selected strains were sequenced. Sequencing was carried out using an automated ABI 377 sequencer. For the sequence reactions various universal primers (Table 1) and a Big Dye Ready Reaction DyeDeoxy Terminator Cycle Sequencing kit (Perkin-Elmer) were employed. All selected strains were sequenced in full (approximately 1,500 bases of 16S rDNA).
TABLE 1.
Oligonucleotide primers used (positions are relative to those of E. coli) for 16S rDNA sequence analysis and ribotyping
| Primer | Sequence | Positions relative to those of E. coli |
|---|---|---|
| fD1a | 5′ AGAGTTTGATCCTGGCTCAG 3′ | 8–27 |
| rP2a | 5′ ACGGCTACCTTGTTACGACTT 3′ | 1513–1494 |
| EBA-1b | 5′ GCCACATTGGGACTGAGACA 3′ | 310–329 |
| But-3b | 5′ GGAATTCCTAGTGTAGCGGT 3′ | 673–692 |
| Uniseq1c | 5′ ATGTTGGGTTAAGTCCCGCAAC 3′ | 1082–1103 |
| P3-Modd | 5′ ATTAGATACCCTDGTAGTCC 3′ | 787–806 |
| PC5d | 5′ TACCTTGTTACGACTT 3′ | 1507–1487 |
Nucleotide sequence accession numbers.
The 16S rDNA sequences of isolates used in the phylogenetic analysis have been deposited in the EMBL data library under accession numbers AJ70469 to AJ270492. That of human B. fibrisolvens strain 16.4 was AJ250365 (38), and that of rumen B. fibrisolvens strain 1.230 was AJ270493 (46). The reference stains used in phylogenetic analysis were also from the EMBL database.
PCR-RFLP analysis.
PCR products were digested to completion with the enzymes HhaI, AluI, TaqI, and MspI and then analyzed by electrophoresis in a 2.0% (wt/vol) agarose gel (40).
Phylogenetic analysis.
The rDNA sequences corresponding to E. coli 16S rRNA bases 1 to 1500 (4) were compared directly with sequences in the EMBL and GenBank nonredundant nucleotide database using BLAST (19) and data of the Ribosomal Database Project (29). Database sequences with high similarity were then directly aligned over equalized lengths with the isolate sequences and used in phylogenetic analysis.
Sequences derived from previously cultured and described organisms of known phylogeny which corresponded to major subdivisions of the domain Bacteria were included in the phylogenetic analysis of the fecal isolates. The sequence data approximating to E. coli positions 1 to 1500 were aligned using CLUSTAL V (22), and a phylogenetic tree was generated using software from the PHYLIP package (17). The DNADIST program analyzed distances using the Kimura-Nei correction (27) trees generated from distance matrices that employed the neighbor-joining method (39). Sequence data for distance matrices and analysis were subjected to bootstrap resampling (data resampled 100 times) using the SEQBOOT program, and consensus trees were generated by the CONSENSE program (16).
RESULTS
Isolation of butyrate producers.
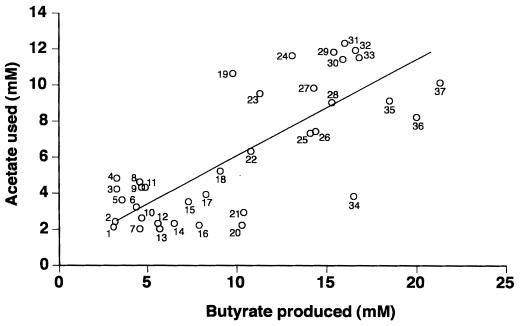
Bacteria were isolated from anaerobic roll tubes inoculated with freshly voided fecal samples from the three subjects. Each individual was sampled on two occasions, separated by approximately 1 year. A rumen fluid-based medium which has been shown to support growth of a wide range of anaerobic bacteria (14, 30) was used. A total of 313 colonies (20 to 30 from the first isolation and 80 from the second isolation) picked from the highest dilutions (10−8 or 10−9) were purified, grown in broth culture, and tested for the production of volatile fatty acids. Isolates producing detectable butyrate varied widely with respect to the quantity of butyrate produced in batch culture (Table 2). A net value of 2 mM butyrate was chosen as a cutoff, since this was considered the lowest value that could be distinguished unambiguously from butyrate concentrations present in uninoculated media. The proportion of isolates producing more than 2 mM butyrate ranged from 3 to 76% for the six samples analyzed. The largest proportion of high-concentration-butyrate-producing isolates was from the first infant sample, with around 50% of isolates producing >10 mM butyrate. The vegetarian adult fecal samples yielded the smallest proportion of butyrate producers and yielded no strains producing >5 mM butyrate. Net utilization of acetate (by >2 mM) present in the rumen fluid component of the medium was detected in 50% of butyrate-producing isolates. Comparison of the amounts of acetate utilized with butyrate produced gave a positive relationship, a correlation r2 value of 0.6, and a slope of 0.51 ± 0.071 (mean ± standard deviation) (Fig. 1). Thus, by averaging across all acetate-consuming strains, approximately 1 mol of acetate initially present in the medium was consumed for every 2 mol of butyrate produced. In marked contrast, only 2 out of 239 non-butyrate-producing strains showed net acetate consumption (Table 2).
TABLE 2.
Occurrence of butyrate-producing and acetate-consuming strains in six human fecal samples
| Individualc | Sample | Butyrate production (mM) among human fecal isolates numbering:
|
No. (%)a of strains showing net consumption of acetateb among strains producing:
|
|||||
|---|---|---|---|---|---|---|---|---|
| 0–2.0 | 2.1–5.0 | 5.1–10.0 | 10.1–15.0 | >15.0 | >2 mM butyrate | <2 mM butyrate | ||
| Infant | 1 | 5 | 0 | 5 | 4 | 7 | 15/16 | 0/5 |
| 2 | 63 | 9 | 4 | 4 | 0 | 7/17 | 2/63 | |
| Adult A | 1 | 15 | 11 | 1 | 0 | 0 | 5/12 | 0/15 |
| 2 | 55 | 8 | 8 | 5 | 3 | 10/24 | 0/55 | |
| Adult B | 1 | 25 | 2 | 0 | 0 | 0 | 0/2 | 0/25 |
| 2 | 76 | 3 | 0 | 0 | 0 | 0/3 | 0/76 | |
| Total | 239 | 33 | 18 | 13 | 10 | 37/74 (50) | 2/239 (1) | |
Number of strains relative to total numbers. N.B., most (90%) strains not showing acetate consumption produced more than 2 mM acetate.
Defined as >2 mM acetate, net.
Adult A is omnivorous; adult B is vegetarian.
FIG. 1.
Summary of butyrate synthesis and acetate utilization in a selection of human colonic butyrate-producing isolates. Key to strains: 1, A1-86; 2, A1-189; 3, A1-816; 4, L2-6; 5, L2-21; 6, A2-204; 7, A2-207; 8, L2-15; 9, A1-87; 10, A1-815; 11, L2-10; 12, A2-225; 13, L1-81; 14, L1-83; 15, A2-194; 16, L2-7; 17, L1-92; 18, A2-215; 19, L1-93; 20, L2-39; 21, L2-61; 22, L1-911; 23, A2-223; 24, L1-872; 25, A2-173; 26, A2-165; 27, L1-910; 28, A2-227; 29, L1-94; 30, L1-9171; 31, L1-8151; 32, L1-97; 33, A2-181; 34, L1-91; 35, L1-82; 36, A2-183; and 37, L1-952.
Ribotypes of the butyrate producers based on PCR-RFLP analysis.
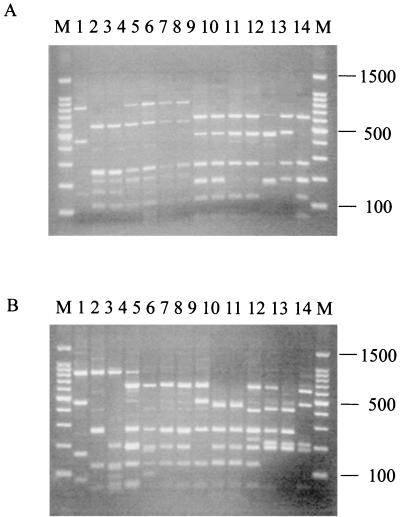
In order to group similar isolates, all 74 butyrate-producing isolates (30 from the first set of samples and 44 from the second set of fecal isolations, taken approximately 1 year later), were examined by 16S rDNA ribosomal profiling and compared with reference strains available within the laboratory (see Materials and Methods). The 16S rDNAs were amplified by PCR using universal primers (47), and the amplified material was cleaved initially with the restriction enzymes HhaI, AluI, TaqI, and MspI (33). The RFLP profiles obtained with the four restriction enzymes indicated that the enzyme AluI allowed the best discrimination between the isolates, distinguishing 18 different ribotypes. The collected results from ribotyping for all six fecal isolations are shown in Table 3. It is apparent that samples from different individuals and from different sampling times show very little overlap with respect to the most abundant ribotypes present. Although none of the 18 ribotypes observed had a profile identical to that of any of the type strains, some of the isolates exhibited bands identical in size with those of type strains. One of the main identifiable bands is at 610 bp, which is characteristic of Eubacterium and Butyrivibrio species (Fig. 2).
TABLE 3.
Distribution of the 74 butyrate-producing strains isolated in this study in each of the 18 PCR-RFLP ribotypes obtained with the restriction enzyme AluI
| Individual | Sample | No. of butyrate-producing strains from the indicated isolation in RFLP group:
|
Total | |||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 | 11 | 12 | 13 | 14 | 15 | 16 | 17 | 18 | |||
| Infant | 1 | 14 | 1 | 1 | 16 | |||||||||||||||
| 2 | 9 | 4 | 2 | 1 | 1 | 17 | ||||||||||||||
| Adult A | 1 | 12 | 12 | |||||||||||||||||
| 2 | 3 | 5 | 4 | 2 | 3 | 4 | 2 | 1 | 24 | |||||||||||
| Adult B | 1 | 1 | 1 | 2 | ||||||||||||||||
| 2 | 1 | 1 | 1 | 3 | ||||||||||||||||
FIG. 2.
PCR-RFLP profiles of a selection of human butyrate-producing isolates and standard strains digested with the restriction endonuclease AluI. (A) Lanes 1 to 14 contain human colonic isolates A2-165 (ribotype 7), A2-168 (ribotype 11), A2-166 (ribotype 11), A2-228 (ribotype 14), A2-175 (ribotype 14), A2-178 (ribotype 14), T2-132 (ribotype 14), A1-89 (ribotype 4), A2-171 (ribotype 4), A2-181 (ribotype 4), A2-183 (ribotype 4), T2-87 (ribotype 17), T2-145 (ribotype 18), and T1-815 (ribotype 5), respectively. (B) Lanes 1 to 12 contain human colonic isolates L2-6 (ribotype 7), L2-12 (ribotype 10), L2-10 (ribotype 9), L2-21 (ribotype 3b), L1-92 (ribotype 3a), L2-9 (ribotype 3b), L2-16 (ribotype 3b), L1-81 (ribotype 1), L2-7 (ribotype 8), L2-65 (ribotype 8), L1-83 (ribotype 2), and B. fibrisolvens 16.4, respectively. Lanes 13 and 14 contain rumen isolates B. fibrisolvens 1.230 and B. fibrisolvens 2221 (ATCC 19971), respectively. Size markers (1-kb ladder; Promega) are shown in lanes marked “M.” The 610-bp band in panel A, lanes 4 to 7, and in panel B, lanes 4, 7, and 8 and the 400-bp fragment in panel B, lane 11, represent partial products. Ribotype groups 6, 12, 13, 15, and 16 are not represented in this figure. Their AluI banding profiles (in base pairs) are 610, 475, 210, 190, and 50 (ribotype 6); 610, 240, and 220 (ribotype 12); 475, 265, 220, and 190 (ribotype 13); 400, 265, 220, 190, and 127 (ribotype 15); and 797, 610, 265, and 190 (ribotype 16). The band pattern for ribotype 6 is similar to that in panel A, lanes 8 to 11, while that for ribotype 12 is similar to that in panel A, lanes 4 to 7. The band pattern for ribotype 13 is similar to that in panel A, lane 12, while that for ribotype 15 is similar to that in panel B, lane 13. The band pattern for ribotype 16 is similar to that in panel A, lanes 4 to 7.
The 25 strains of ribotypes 1, 2, and 3 were found uniquely in the flora from the infant, and ribotype 1 accounted for 14 of the 16 strains isolated from the first (preweaning) infant sample. Only two ribotypes (ribotypes 7 and 14) were recovered from more than one individual, and only four ribotypes (ribotypes 3, 4, 7, and 14) were recovered from more than one sample.
Phylogenetic comparison.
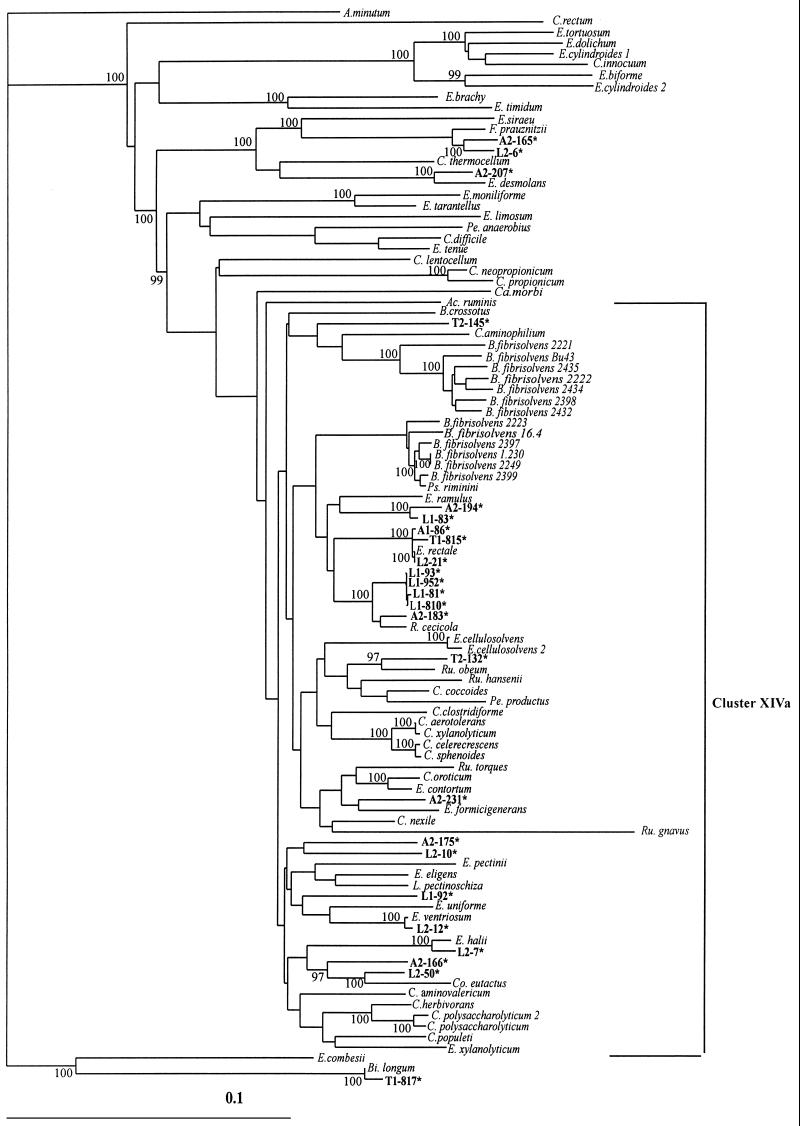
Complete 16S rDNA sequences were determined for 24 isolates representative of the most common ribotypes identified by RFLP analysis. These included strains producing large (>10 mM), medium (>5 and <10 mM), and small (<5 mM) amounts of butyrate in vitro (Table 4). These sequences were compared with available database sequences by a BLAST search (Table 4), and a phylogenetic tree was constructed (Fig. 3). Twenty of the sequences were found to belong within cluster XIVa of gram-positive bacteria. A few isolates were shown to be closely related (>97% sequence identity) to Eubacterium rectale (A1-86, L2-21, and T1-815), Eubacterium ventriosum (L2-12), or Fusobacterium prauznitzii (A2-165 and L2-6). The remaining 18 of the 24 sequenced isolates however shared 95% or less 16S rDNA sequence identity with their nearest relative.
TABLE 4.
| Strain | Ribotype group | Species identified | % Homologyb | SCFA production (mM)
|
H2 production (μ mol ml−1) | ||
|---|---|---|---|---|---|---|---|
| Butyrate | Acetate | Lactate | |||||
| L1-81 | 1 | Unidentified | NA | 5.7 | −2.0 | 1.4 | 5.0 |
| L1-810 | 1 | Unidentified | NA | 5.5 | 41.0 | 25.5 | 0.1 |
| L1-952 | 1 | Unidentified | NA | 21.3 | −10.0 | 7.1 | 6.5 |
| L1-93 | 1 | Unidentified | NA | 9.8 | −10.6 | 5.7 | NDc |
| L1-83 | 2 | Unidentified | NA | 6.5 | −2.3 | 2.1 | 2.7 |
| L1-92 | 3a | Unidentified | NA | 8.3 | −3.9 | 0.1 | 12.0 |
| L2-21 | 3a | Eubacterium rectale | 98 | 3.6 | −3.6 | ND | ND |
| L2-50 | 3b | Unidentified | NA | 12.7 | 11.1 | 0.97 | 4.3 |
| A1-86 | 4 | Eubacterium rectale | 98 | 4.7 | −2.1 | 26.5 | 3.2 |
| A2-183 | 4 | Roseburia cecicola | 95 | 20.0 | −8.2 | 9.7 | 14.8 |
| T1-815 | 5 | Eubacterium rectale | 98 | 3.3 | 0.7 | 18.7 | 2.6 |
| T1-817 | 6 | Bifidobacterium infantis or longum | 98 | 2.1 | 7.2 | 9.2 | 2.1 |
| L2-6 | 7 | Fusobacterium prauznitzii | 98 | 3.3 | −4.8 | ND | ND |
| A2-165 | 7 | Fusobacterium prauznitzii | 98 | 14.4 | −7.4 | 2.9 | 0 |
| L2-7 | 8 | Eubacterium spp. | 97 | 7.9 | −2.2 | ND | ND |
| L2-10 | 9 | Unidentified | NA | 4.9 | −4.3 | ND | ND |
| L2-12 | 10 | Eubacterium ventriosum | 98 | 4.6 | −1.7 | ND | ND |
| A2-166 | 11 | Unidentified | NA | 9.7 | 2.4 | ND | ND |
| A2-207 | 12 | Eubacterium spp. | 96 | 4.6 | −2.0 | 0 | 1.7 |
| A2-231 | 13 | Unidentified | NA | 5.6 | 5.2 | ND | ND |
| A2-175 | 14 | Unidentified | NA | 5.0 | −1.8 | ND | ND |
| T2-132 | 14 | Unidentified | NA | 2.2 | 47.6 | 6.7 | 0.1 |
| A2-194 | 15 | Unidentified | NA | 7.3 | −3.5 | 7.1 | 0 |
| T2-145 | 18 | Unidentified | NA | 2.7 | 1.0 | ND | ND |
Data for SCFA (butyrate and acetate) and for production of lactate and H2 were from two separate experiments. None of the strains produced propionate, with the possible exception of L1-83 (3 mM).
Percent homology indicates the maximum base sequence identity obtained in database searches. NA, not applicable, indicates no value above 95% identity.
ND, not determined.
FIG. 3.
Phylogenetic tree showing the relationships of 16S rDNA sequences from human butyrate-producing isolates falling within cluster XIVa of the Clostridium subphylum of low-G+C-content gram-positive bacteria (7, 48). The scale bar represents genetic distance (10 substitutions per 100 nucleotides). The tree was constructed using the neighbor-joining analysis of a distance matrix obtained from a multiple-sequence alignment. Bootstrap values (expressed as percentages of the value for 100 replications) are shown at branch points; values of 97% or more were considered significant. Sequences derived from the database are shown in italics (e.g., B. fibrisolvens). Atopobium minutum is used as the outgroup sequence.
The largest group of 10 newly isolated strains clusters with Eubacterium ramulus, Eubacterium rectale, and Roseburia cecicola. This group includes all sequenced strains belonging to ribotypes 1 and 4, together with one ribotype 3 strain and strains of ribotypes 5 and 15. Eight additional strains showed relationships to other Eubacterium species, including Eubacterium halii, Eubacterium uniforme, Eubacterium formicigenerans, and Eubacterium ventriosum, or to other members of the XIVa cluster, including two strains (L2-50 and A2-166) that are related to Coprococcus eutactus. The strains that lie outside the XIVa cluster were still related to gram-positive bacteria. These included A2-207 and the two strains (L2-6 and A2-165) belonging to ribotype 7 that are close to F. prauznitzii, as noted above. The remaining strain, T1-817, was related to Bifidobacterium longum, which is not expected to produce butyrate, but the amounts of butyrate produced by this strain were extremely small.
DISCUSSION
To our knowledge, this is the first study directed specifically at establishing the identities of the butyrate-producing bacteria in the human gut. In one respect the diversity of the butyrate-producing strains as revealed by phylogenetic analysis of 16S rDNA sequences was less than might have been anticipated. All isolates were related to gram-positive bacteria, 80% of which belonged to cluster XIVa, and none were related to gram-negative bacteria, since the few F. prauznitzii-like isolates reported here are in fact also related to gram-positive bacteria at the genotypic level. On the other hand, the RFLP analyses revealed extraordinary diversity and variability of the major strain types recovered between the different individuals and sampling times. Much more extensive work would be required to establish whether this diversity reflects differences in diet, age, or genotype between the individuals tested or simply large fluctuations in the predominant types within individuals between different times of sampling. While it is likely that the bacteria isolated include many of the predominant butyrate producers from the human colon, further work will be required to establish the occurrence of related strains in a larger number of individuals. This could be approached by using specific molecular probes to avoid possible cultural bias.
Phylogenetic analysis shows that the butyrate-producing strains isolated that belong to cluster XIVa are widely distributed within this cluster (Fig. 2). The majority of isolates appear to be broadly related to known species, but only 8 out of the 24 sequenced strains had greater than 95% 16S rDNA identity with type strains. This and the lack of recognized species type strains in some branches of the tree indicate that new uncharacterized phylogenetic groups are represented among our isolates. Of the three clusters, the Eubacterium rectale-R. cecicola group comprises the highest proportion of sequenced isolates (10 out of 24, 42%) while overall ribotypes 1 and 4, whose sequenced representatives fall within this group, account for 27 out of the 74 (36%) isolates examined. Within the main grouping the Eubacterium rectale subgroup appears to contain isolates from all of the individuals sampled whereas the R. cecicola subgroup was found mainly in infant samples. It would be of particular interest to establish in future work whether R. cecicola-related strains belonging to ribotype 1 are characteristic of preweaned infants.
A number of molecular approaches are now available for rapid bacterial strain typing. RFLP analyses based on hybridization with 16S rDNA probes have been widely used and depend on detection of variations in chromosomal sequences flanking multiple copies of rRNA target genes (2, 28). The approach used here is quite different in that it depends on PCR amplification of internal 16S rDNA fragments and cleavage with restriction enzymes (50). This approach proved very valuable here in revealing the diversity of butyrate-producing strains, and two ribotypes, 1 and 7, conformed well to branches within the phylogenetic tree obtained subsequently from sequence analysis. In general, however, it must be recognized that PCR-RFLP analysis based on a single enzyme will often group together genetically distant strains since extensive sequence changes can occur between strains without affecting sites for a particular restriction enzyme. The RFLP classes must therefore be seen as a useful comparative method for examining diversity rather than as a definitive phylogenetic typing approach, unless results from many enzymes are combined (33).
Fifty percent of butyrate-producing isolates examined here showed some net utilization of acetate in M2GSC medium. Such utilization is consistent with the operation of the butyryl coenzyme A-acetyl coenzyme A transferase route for butyrate synthesis (20). On average across the strains studied in vitro here, approximately 1 mol of acetate initially present in the medium was utilized for every 2 mol of butyrate produced. We were not able to estimate how much additional acetate may have been produced and utilized by each strain for butyrate synthesis. Some acetate may, of course, be used for biosynthetic reactions rather than butyrate synthesis. The high proportion of acetate-utilizing strains seen here suggests that a significant fraction of colonic acetate may be rerouted into butyrate in the human colon. Preliminary studies involving 13C-labeled acetate suggest that appreciable butyrate may be derived from exogenous acetate in mixed fecal fermentations (S. H. Duncan et al., unpublished data). Since for some of the strains isolated here growth was absolutely dependent on inclusion of acetate in the medium (S. H. Duncan et al., unpublished data), these strains would not have been recovered through isolations with media lacking acetate. The alternative butyrate kinase pathway is known to operate in many Clostridium spp. and in certain ruminal B. fibrisolvens strains, including the type strain 2221, and strains depending on this pathway would not be expected to utilize acetate (13, 20). It is also worth noting that only 1% of the non-butyrate-producing isolates examined showed net acetate utilization and that nearly all (95%) of the acetate utilizers recovered were butyrate producers.
Recent work by Diez-Gonzalez et al. (13) suggested that rumen butyrate-producing Butyrivibrio species could be divided into two distinct groups, based upon pathways that they utilized to produce butyrate and that these could be distinguished by production or consumption of acetate. Rumen Butyrivibrio strains have also been classified as lactate producing or non-lactate producing (42). None of the human isolates obtained in this study fell within either group of Butyrivibrio strains. However, the present work does confirm that one isolate, B. fibrisolvens 16.4, obtained previously from fermentor studies with the human fecal flora (38), is related to the group of ruminal B. fibrisolvens strains that includes B. fibrisolvens 1.230 and 2223.
We found no simple correlation between the phylogenetic position of the butyrate-producing isolates examined here and their metabolic behavior. Thus, for example, ribotype 1 strains included one acetate producer (L1-810) in addition to the predominant acetate-utilizing strains. In addition, we were able to detect significant production of lactate both in acetate-producing strains (e.g., L1-810) and in acetate-utilizing strains (e.g., L1-952) from human feces. On the other hand it is worth noting that ribotypes 1 and 7 contained particularly high proportions of strains that produced >10 mM butyrate in vitro (11 out of 14 [78%] and 6 out of 9 [67%], respectively, compared to 24 out of 74 [32%] butyrate producers over all ribotypes).
Recent investigations by fluorescent in situ hybridization with group-specific probes (18) showed that the highest proportion of human fecal organisms detected fell within the Clostridium coccoides-Eubacterium rectale group (7.2 × 1010 cells/g [dry weight] of feces), which forms part of clostridial cluster XIVa. We have shown here that most of the butyrate-producing isolates in this study fall into cluster XIVa and that they are related to the Clostridium coccoides-Eubacterium rectale group. Further understanding of the phylogeny and physiology of this group of organisms will be crucial in understanding the role of the anaerobic microflora in colonic metabolism and gut health.
ACKNOWLEDGMENTS
We thank Tony Richardson and Peter Dewey for their contributions to the SCFA analyses and Moira Johnston for DNA sequencing.
This work was supported by SERAD (Scottish Executive Rural Affairs Department), by a BORC (Boyd Orr Research Consortium) Ph.D. studentship to A.B., and by a SERAD flexible fund grant.
REFERENCES
- 1.Ausubel F M, Brent R, Kingston R E, Moore D M, Seidman J G, Smith J A, Struhl K. Current protocols in molecular biology. I. New York, N.Y: J. Wiley and Sons, Inc.; 1994. p. 2.4. [Google Scholar]
- 2.Avgustin G, Wright F, Flint H J. Genetic diversity and phylogenetic relationships among strains of Prevotella (Bacteroides) ruminicola from the rumen. Int J Syst Bacteriol. 1994;47:284–288. doi: 10.1099/00207713-44-2-246. [DOI] [PubMed] [Google Scholar]
- 3.Bengmark S. Econutrition and health maintenance—a new concept to prevent GI inflammation, ulceration and sepsis. Clin Nutr. 1996;15:1–10. doi: 10.1016/s0261-5614(96)80253-6. [DOI] [PubMed] [Google Scholar]
- 4.Brosius J, Palmer M L, Kennedy P J, Noller H F. Complete nucleotide sequence of a 16S ribosomal gene from Escherichia coli. Proc Natl Acad Sci USA. 1978;75:4801–4805. doi: 10.1073/pnas.75.10.4801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bryant M P. Commentary on the Hungate technique for cultivation of anaerobic bacteria. Am J Clin Nutr. 1972;25:1324–1328. doi: 10.1093/ajcn/25.12.1324. [DOI] [PubMed] [Google Scholar]
- 6.Bryant M P, Small N. The anaerobic monotrichous butyric acid-producing curved rod-shaped bacteria of the rumen. J Bacteriol. 1956;72:16–21. doi: 10.1128/jb.72.1.16-21.1956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Collins M D, Lawson P A, Willems A, Cordoba J J, Fernandez-Garayzabal J, Garcia P, Cai J, Hippe H, Farrow J A E. The phylogeny of the genus Clostridium: proposal of five new genera and eleven new species combinations. Int J Syst Bacteriol. 1994;44:812–826. doi: 10.1099/00207713-44-4-812. [DOI] [PubMed] [Google Scholar]
- 8.Csordas, A. 1995. Toxicology of butyrate and short-chain fatty acids, p. 105–125. In M. J. (ed.), Role of gut bacteria in human toxicology and pharmacology. Taylor and Francis, London, United Kingdom.
- 9.Cummings J H, Pomare E W, Branch W J, Naylor C P E, MacFarlane G T. Short-chain fatty acids in human large intestine, portal, hepatic and venous blood. Gut. 1987;28:1221–1227. doi: 10.1136/gut.28.10.1221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cummings J H, MacFarlane G T. The colonic flora, fermentation and large bowel digestive function. In: Philips S F, Pemberton J H, Shorter R G, editors. The large intestine: physiology, pathophysiology, and disease. New York, N.Y: Raven Press; 1991. pp. 51–91. [Google Scholar]
- 11.Cummings J H, MacFarlane G T. Colonic microflora: nutrition and health. Nutrition. 1997;13:476–478. doi: 10.1016/s0899-9007(97)00114-7. [DOI] [PubMed] [Google Scholar]
- 12.Cummings J H, MacFarlane G T. Role of intestinal bacteria in nutrient metabolism. Clin Nutr. 1997;16:3–11. doi: 10.1177/0148607197021006357. [DOI] [PubMed] [Google Scholar]
- 13.Diez-Gonzalez F, Bond D R, Jennings E, Russell J B. Alternative schemes of butyrate production in Butyrivibrio fibrisolvens and their relationship to acetate utilization, lactate production and phylogeny. Arch Microbiol. 1999;171:324–330. doi: 10.1007/s002030050717. [DOI] [PubMed] [Google Scholar]
- 14.Duncan S H, Flint H J, Stewart C S. Inhibitory activity of gut bacteria against Escherichia coli O157 mediated by dietary plant metabolites. FEMS Microbiol Lett. 1998;164:283–288. doi: 10.1111/j.1574-6968.1998.tb13099.x. [DOI] [PubMed] [Google Scholar]
- 15.Engelhardt W, Bartels J, Kirschberger S, Meyer zu Duttingdof H D, Busche R. Role of short-chain fatty acids in the hind gut. Vet Q. 1998;20:S52–S59. [PubMed] [Google Scholar]
- 16.Felsenstein J. Confidence limits on phylogenies: an approach using the bootstrap. Evolution. 1985;39:783–791. doi: 10.1111/j.1558-5646.1985.tb00420.x. [DOI] [PubMed] [Google Scholar]
- 17.Felsenstein J. PHYLIP—Phylogeny Inference Package (version 3.2) Cladistics. 1989;5:164–166. [Google Scholar]
- 18.Franks A H, Harmsen H J M, Raangs G C, Jansen G J, Welling G W. Variations of bacterial populations in human feces quantified by fluorescent in situ hybridization with group-specific 16S rRNA-targeted oligonucleotide probes. Appl Environ Microbiol. 1998;64:3336–3345. doi: 10.1128/aem.64.9.3336-3345.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gish W, States D J. Identification of protein coding regions by database similarity search. Nat Genet. 1993;3:266–272. doi: 10.1038/ng0393-266. [DOI] [PubMed] [Google Scholar]
- 20.Gottschalk G. Bacterial fermentations. In: Starr M P, editor. Bacterial metabolism. New York, N.Y: Springer-Verlag; 1979. pp. 167–224. [Google Scholar]
- 21.Hague A, Singh B, Paraskeva C. Butyrate acts as a survival factor for colonic epithelial cells: further fuel for the in vivo versus in vitro debate. Gastroenterology. 1997;112:1036–1040. doi: 10.1053/gast.1997.v112.agast971036. [DOI] [PubMed] [Google Scholar]
- 22.Higgins D G, Bleasby A J, Fuchs R. CLUSTALV: improved software for multiple sequence alignment. Comput Appl Biosci. 1992;8:189–191. doi: 10.1093/bioinformatics/8.2.189. [DOI] [PubMed] [Google Scholar]
- 23.Hobson P N. Rumen bacteria. Methods Microbiol. 1969;3B:133–149. [Google Scholar]
- 24.Holdeman L V, Cato E P, Moore W E C. Anaerobe laboratory manual. 4th ed. Blacksburg: Virginia Polytechnic Institute and State University; 1977. [Google Scholar]
- 25.Hungate R E. The rumen and its microbes. New York, N.Y: Academic Press; 1966. [Google Scholar]
- 26.Innis M A, Gelfand D H, Sninsky J J, White T J. PCR products: a guide to methods and applications. London, United Kingdom: Academic Press, Inc.; 1990. [Google Scholar]
- 27.Kimura M. A simple method for estimating evolutionary rates of base substitutions through comparative studies of nucleotide sequences. J Mol Evol. 1980;16:111–120. doi: 10.1007/BF01731581. [DOI] [PubMed] [Google Scholar]
- 28.Lawson P A, Gharbia S E, Shah H N, Clark D R. Intrageneric relationships of members of the genus Fusobacterium as determined by reverse transcriptase sequencing of small-subunit rRNA. Int J Syst Bacteriol. 1991;41:347–354. doi: 10.1099/00207713-41-3-347. [DOI] [PubMed] [Google Scholar]
- 29.Maidak B L, Larsen N, McCaughey M J, Overbeek R, Olsen G J, Forgel K, Blandy J, Woese C R. The Ribosomal Database Project. Nucleic Acids Res. 1994;22:3485–3487. doi: 10.1093/nar/22.17.3485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Miyazaki K, Martin J C, Marinsek-Logar R, Flint H J. Degradation and utilization of xylans by the rumen anaerobe Prevotella bryantii (formerly P. ruminicola subsp. brevis) B14. Anaerobe. 1997;3:373–381. doi: 10.1006/anae.1997.0125. [DOI] [PubMed] [Google Scholar]
- 31.Moore W E C, Holdeman L V. Human fecal flora: the normal flora of 20 Japanese-Hawaiians. Appl Environ Microbiol. 1974;27:961–979. doi: 10.1128/am.27.5.961-979.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Moore W E C, Moore L H. Intestinal floras of populations that have a high risk of colon cancer. Appl Environ Microbiol. 1995;61:3202–3207. doi: 10.1128/aem.61.9.3202-3207.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Moyer C L, Dobbs F C, Carl D M. Estimation of diversity and community structure through restriction fragment length polymorphism distribution analysis of bacterial 16S rDNA genes from a microbial mat at an active, hydrothermal vent system, Loihi Seamount, Hawaii. Appl Environ Microbiol. 1996;60:871–879. doi: 10.1128/aem.60.3.871-879.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Pryde S E, Richardson A J, Stewart C S, Flint H J. Molecular analysis of the microbial diversity present in the colonic wall, colonic lumen, and cecal lumen of a pig. Appl Environ Microbiol. 1999;65:5372–5377. doi: 10.1128/aem.65.12.5372-5377.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Richardson A J, Calder A G, Stewart C S, Smith A. Simultaneous determination of volatile and non-volatile acidic fermentation products of anaerobes by capillary gas chromatography. Lett Appl Microbiol. 1989;9:5–8. [Google Scholar]
- 36.Ritzhaupt A, Ellis A, Hosie K B, Shirazi-Beechey S P. The characterization of butyrate transport across pig and human colonic luminal membrane. J Physiol. 1998;507:819–830. doi: 10.1111/j.1469-7793.1998.819bs.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Roediger W E W. The colonic epithelium in ulcerative colitis: an energy deficient disease? Lancet. 1980;ii:712–715. doi: 10.1016/s0140-6736(80)91934-0. [DOI] [PubMed] [Google Scholar]
- 38.Rumney C, Duncan S H, Henderson C, Stewart C S. Isolation and characteristics of a wheatbran degrading butyrivibrio from human faeces. Lett Appl Microbiol. 1995;20:232–236. doi: 10.1111/j.1472-765x.1995.tb00435.x. [DOI] [PubMed] [Google Scholar]
- 39.Saitou N, Nei M. The neighbor joining method: a new method for constructing phylogenetic trees. Mol Biol Evol. 1987;4:406–425. doi: 10.1093/oxfordjournals.molbev.a040454. [DOI] [PubMed] [Google Scholar]
- 40.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. I. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. pp. 5.30–5.32. [Google Scholar]
- 41.Scott K P, Barbosa T M, Forbes K J, Flint H J. High-frequency transfer of naturally occurring chromosomal tetracycline resistance element in the ruminal anaerobe Butyrivibrio fibrisolvens. Appl Environ Microbiol. 1997;6:3405–3411. doi: 10.1128/aem.63.9.3405-3411.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Shane B S, Gouws L, Kistner A. Cellulolytic bacteria occurring in the rumen of sheep conditioned to low-protein teff hay. J Gen Microbiol. 1969;55:445–457. doi: 10.1099/00221287-55-3-445. [DOI] [PubMed] [Google Scholar]
- 43.Sheppach W, Bartram H P, Richter F. Role of short-chain fatty acids in the prevention of colorectal cancer. Eur J Cancer. 1998;31A:107–1080. doi: 10.1016/0959-8049(95)00165-f. [DOI] [PubMed] [Google Scholar]
- 44.Szylit O, Andrieux C. Physiological and pathophysiological effects of carbohydrate fermentation. World Rev Nutr Diet. 1993;74:88–122. doi: 10.1159/000422603. [DOI] [PubMed] [Google Scholar]
- 45.Tran C P, Familari M, Parker L M, Whitehead R H, Giraud A S. Short-chain fatty acids inhibit intestinal trefoil factor gene expression in colon cancer cells. Am J Physiol-Gastrointest Liver Physiol. 1998;38:G85–G93. doi: 10.1152/ajpgi.1998.275.1.G85. [DOI] [PubMed] [Google Scholar]
- 46.Wang R F, Cao W W, Cernaglia C E. PCR detection and quantitation of predominant and anaerobic bacteria in human and animal fecal samples. Appl Environ Microbiol. 1996;62:1242–1247. doi: 10.1128/aem.62.4.1242-1247.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Weisburg W G, Barns S M, Pelletier D A, Lane D J. 16S ribosomal DNA for phylogenetic study. J Bacteriol. 1991;173:697–703. doi: 10.1128/jb.173.2.697-703.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Willems A, Amat-Marco M, Collins M D. Phylogenetic analysis of Butyrivibrio strains reveals three distinct groups of species within the Clostridium subphylum of the gram-positive bacteria. Int J Syst Bacteriol. 1996;46:195–199. doi: 10.1099/00207713-46-1-195. [DOI] [PubMed] [Google Scholar]
- 49.Wilson K H, Blitchington R B. Human colonic biota studied by ribosomal DNA sequence analysis. Appl Environ Microbiol. 1996;62:2273–2278. doi: 10.1128/aem.62.7.2273-2278.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wood J, Scott K P, Avgustin G, Newbold C J, Flint H J. Estimation of the relative abundance of different Prevotella ribotypes in gut samples by restriction enzyme profiling of PCR-amplified 16S rRNA gene sequences. Appl Environ Microbiol. 1998;64:3683–3689. doi: 10.1128/aem.64.10.3683-3689.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]