Abstract
Pseudoprogression (PsP) refers to treatment-related clinico-radiologic changes mimicking true progression (TP) that occurs in patients with glioblastoma (GBM), predominantly within the first 6 months after the completion of surgery and concurrent chemoradiation therapy (CCRT) with temozolomide. Accurate differentiation of TP from PsP is essential for making informed decisions on appropriate therapeutic intervention as well as for prognostication of these patients. Conventional neuroimaging findings are often equivocal in distinguishing between TP and PsP and present a considerable diagnostic dilemma to oncologists and radiologists. These challenges have emphasized the need for developing alternative imaging techniques that may aid in the accurate diagnosis of TP and PsP. In this review, we encapsulate the current state of knowledge in the clinical applications of commonly used metabolic and physiologic magnetic resonance (MR) imaging techniques such as diffusion and perfusion imaging and proton spectroscopy in distinguishing TP from PsP. We also showcase the potential of promising imaging techniques, such as amide proton transfer and amino acid-based positron emission tomography, in providing useful information about the treatment response. Additionally, we highlight the role of “radiomics”, which is an emerging field of radiology that has the potential to change the way in which advanced MR techniques are utilized in assessing treatment response in GBM patients. Finally, we present our institutional experiences and discuss future perspectives on the role of multiparametric MR imaging in identifying PsP in GBM patients treated with “standard-of-care” CCRT as well as novel/targeted therapies.
Keywords: amide proton transfer imaging, amino acid-based positron emission tomography, diffusion MR imaging, glioblastoma, perfusion MR imaging, proton MR spectroscopy, pseudoprogression, true progression
1 |. INTRODUCTION
Glioblastoma (GBM) is an aggressive brain tumor of astrocytic/neural stem cell origin and is the most common primary malignant brain neoplasm, representing 30% of all central nervous system tumors in adults.1 The current standard of care for newly diagnosed GBM comprises maximal safe tumor resection followed by adjuvant radiation therapy with concurrent temozolomide (TMZ) chemotherapy and six maintenance cycles of TMZ.2 Despite the multimodal first-line treatment, the prognosis of GBM patients is dismal, with a 5-year survival rate of about 5% and a median overall survival of only 14–16 months following diagnosis.3
In most cases (~80%), increased contrast enhancement and/or worsening surrounding T2 fluid-attenuated inversion recovery (T2-FLAIR) signal abnormality at the location of the original tumor or resection margins are observed within 3–6 months of completion of concurrent chemoradiation therapy (CCRT).4 While these neuroimaging changes may represent the true progression (TP) of GBM, it may also reflect predominant treatment effects/pseudoprogression (PsP) that is mediated by TMZ-induced enhanced vascular permeability and predominant inflammatory response.5,6 PsP, which appears in the absence of true tumor growth, is usually transient in nature and subsides or stabilizes naturally without requiring a change in therapy. The biology of PsP is not fully understood and several hypotheses have been proposed. The alkylating agent, TMZ, inhibits cell replication in the G2/M cell cycle phase and increases the number of breaks in DNA. It has been proposed that the combination of these effects causes an exaggerated cellular response within the tumor beds resulting in damage to the vascular endothelium, blood–brain barrier (BBB) disruption, and oligodendroglial cell injury that subsequently results in imaging changes consistent with PsP.7 The incidence of PsP ranges from 28% to 66% in GBM patients receiving CCRT8 and PsP is particularly frequent in patients harboring promoter methylation of O6-methylguanine-DNA methyltransferase (MGMT), a DNA repair enzyme that plays an important role in chemoresistance to alkylating agents such as TMZ.9
Classical radiation necrosis (RN) is a variant of treatment effects representing a local tissue reaction to radiation therapy in patients with GBM. It generally occurs 3–12 months after radiotherapy but can occur up to several years or decades later, differentiating it from PsP in terms of onset time and severity of disease.10 It has also been reported that PsP and RN share similar histopathological and molecular features.10
Intriguingly, PsP patients generally show a favorable response to TMZ treatment and tend to have improved clinical outcomes over TP patients.11 Therefore, patients with PsP are closely monitored with shorter interval follow-up magnetic resonance imaging (MRI) scans, usually every 4–6 weeks, and are symptomatically managed with a continuation of adjuvant TMZ. Conversely, patients with TP often require repeat biopsy/surgical resection, and/or switching to alternative therapies such as tumor-treating fields (TTFields) and immunotherapy.12 Thus, recognizing patients with PsP is also critical to avoid unnecessary repeat surgery and administration of expensive and potentially risky therapies. Exclusion of PsP is also of value to reduce the false-positive effects of a novel therapy, particularly in the context of clinical trials. For these reasons, it is imperative to differentiate TP from PsP in effectively managing the care of these patients. Response assessment in neuro-oncology (RANO) criteria13 based on conventional neuroimaging findings are often ambiguous in differentiating TP from PsP and hence present a significant diagnostic challenge (Figure 1).14,15 Therefore, there is a pressing need to develop quantitative imaging biomarkers for reliably assessing and redefining the treatment response in GBM patients.
FIGURE 1.
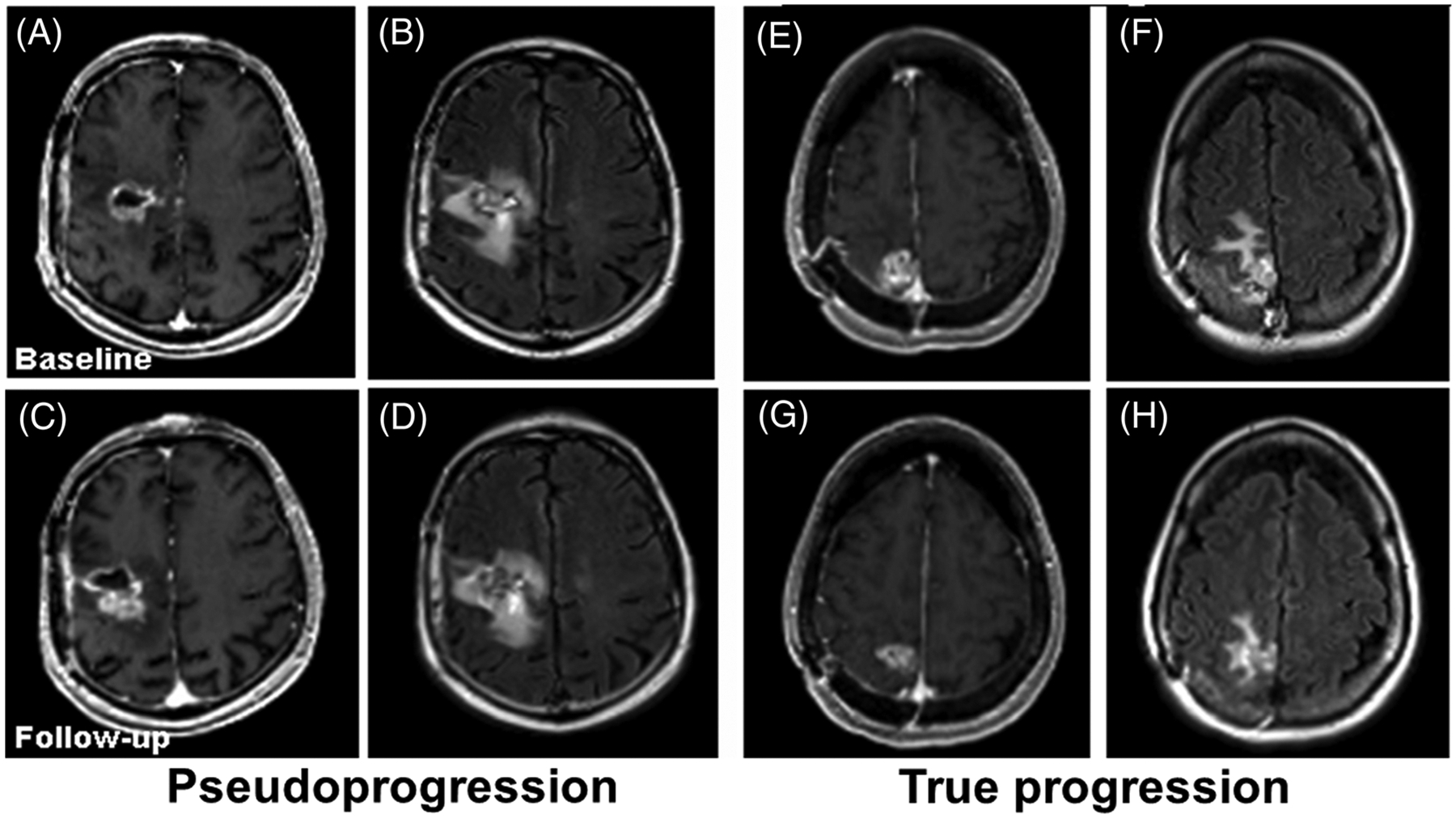
Representative contrast-enhanced T1-weighted (A, C, E and G) and fluid-attenuated inversion recovery (FLAIR) (B, D, F and H) images from two patients with glioblastomas (GBMs) at baseline (top row) and 1-month follow-up (bottom row). The images (A-D) are from a patient with pseudoprogression (PsP), showing increased tumor size at follow-up. The images (E-H) are from a patient with true progression (TP), showing decreased tumor size at follow-up, suggesting the limitation of conventional MR imaging in reliable distinction of TP from PsP in GBMs
The aim of the present review is to provide an overview of the more widely available as well as emerging novel metabolic and physiologic MRI techniques used in differentiation of TP and PsP in GBM patients. We also focus on the potential technical pitfalls of using these advanced MRI sequences in the clinical environment and present conceivable solutions regarding how to circumvent these challenges. In this era of artificial intelligence (AI), machine learning (ML)- and radiomics-based methods are continuously being investigated to extract more objective imaging features that may help us in evaluating treatment response with higher accuracy. Finally, we briefly discuss the emerging applications of AI in distinguishing TP from PsP.
2 |. MRI AND PET IMAGING APPROACHES FOR THE DIFFERENTIATION OF TP FROM PSP
2.1 |. Conventional MRI
Conventional MRI remains the most commonly used imaging modality in assessing treatment response to various therapeutic regimens in neuro-oncology. Patterns of contrast enhancement on postcontrast T1-weighted images and/or extent of hyperintense signal intensity on T2-FLAIR images are standard features that are used to monitor treatment response in GBM patients. However, these morphological features reflect only impairment in the BBB and are nonspecific in nature, hence can be seen in both TP and PsP, making these entities indistinguishable in most clinical scenarios. Revised criteria for assessment of treatment response in high-grade gliomas have been proposed by the RANO Working Group.13 These guidelines use two-dimensional biperpendicular measurements of enhancing lesions and/T2-hyperintense abnormality as the primary basis of response. However, these bidirectional measurements have been shown to overestimate tumor size.16 Additionally, high discordance rates between readers have been reported, presumably because of differences in head angulation and accurate identification of primary (longest) and secondary axes, biperpendicular diameters, especially when the enhancing lesion has an irregular shape and/or ill-defined boundaries.16 Moreover, the subjective interpretative nature and high rates of adjudication render the RANO criteria somewhat controversial in assessing treatment response in GBMs. In recent years, various attempts have been made to address these issues and to find more reliable distinguishing imaging features.
As such, a novel approach providing high-resolution treatment response assessment maps (TRAMs) was proposed to determine treatment outcomes in GBMs.17 This methodology is based on acquisition of two high-resolution three-dimensional (3D) T1-weighted images (at 3–5 and 60–75 min) after the injection of gadolinium-based contrast agents (GBCAs). Subsequently, these images are subtracted and color-coded to represent the spatial distribution of contrast accumulation and clearance. Active “viable” tumor regions/TP on TRAMs demonstrate effective clearance of contrast agent, whereas treatment effects/PsP with necrotic and occluded blood vessels tend to have contrast accumulation. TRAMs are relatively simple to acquire, readily interpretable, and are less prone to being confounded by susceptibility artifacts. This new technique has been used in determining treatment outcomes from high-grade glioma patients treated with CCRT17 (n = 4) and bevacizumab18 (n = 24). Although promising, this technique is associated with some limitations. An unavoidable limitation of TRAMs is the requirement to wait at least 1 hour after GBCA injection, which makes it challenging from a workflow perspective in busy academic medical centers. Moreover, the timings of postcontrast acquisitions are particularly important. Another significant disadvantage of TRAMs is their inability to depict nonenhancing tumor components. We believe that large, prospective, multicentric studies are warranted to confirm the clinical utility of TRAMs in the future.
2.2 |. Physiologic and metabolic MRI
In contrast to conventional anatomic MRI sequences, advanced MRI techniques are more sensitive to different biophysical processes in tissues and provide more comprehensive information about the tumor microenvironment, including cellular proliferation, tumor hemodynamics, vascular permeability, and tumor metabolism. The metabolic and physiologic MRI techniques include diffusion and perfusion MRI, proton MR spectroscopy (1H MRS), and amide proton transfer (APT). Readers are referred to excellent review articles for a detailed overview related to the clinical utilities of these MRI methods in the diagnosis, characterizing molecular profiles, describing infiltrative patterns, determining prognosis and assessing treatment response to established and novel therapies in high-grade gliomas.19–21
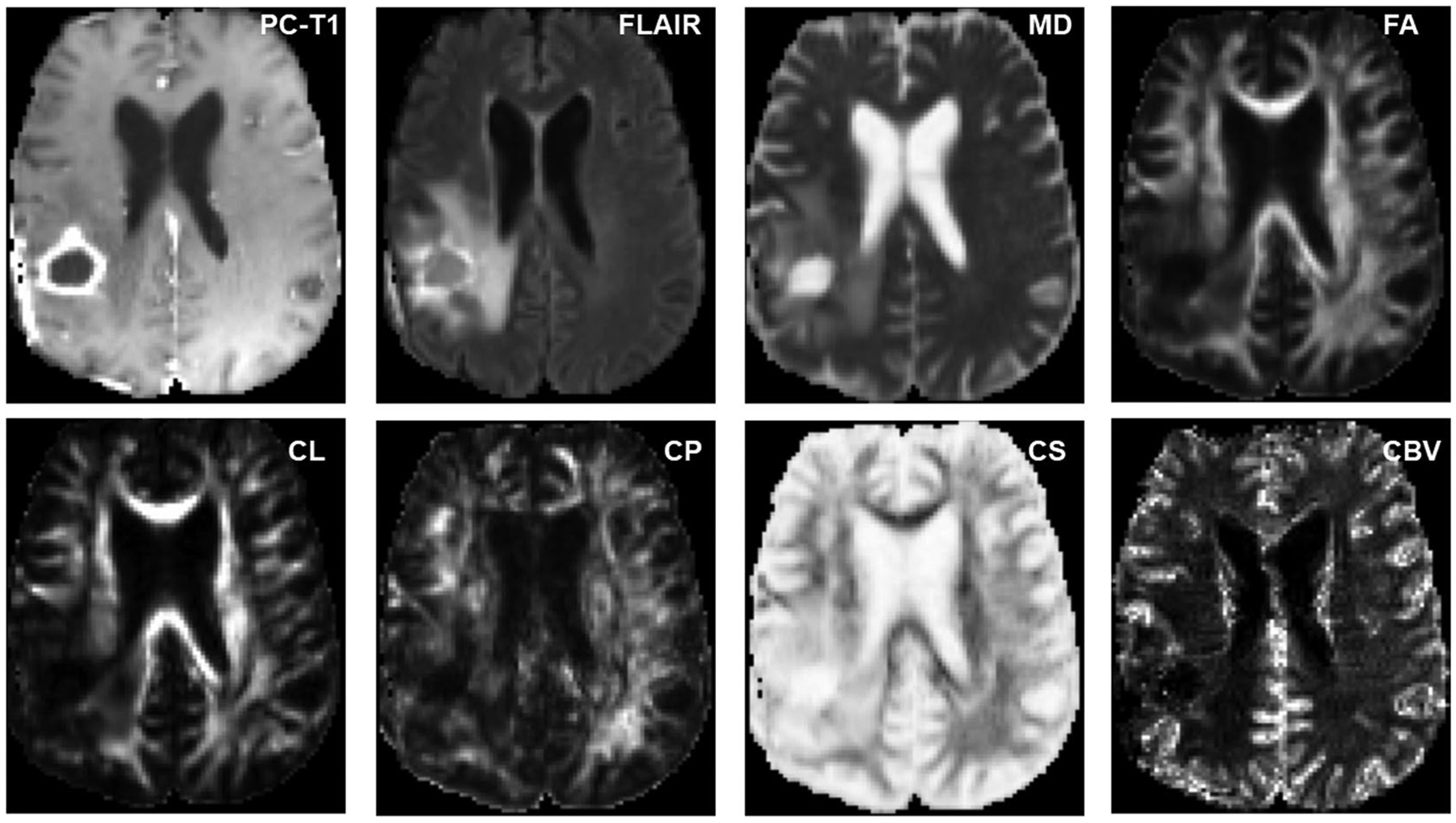
The general trends in structural, metabolic, and physiologic MRI-derived parameters in distinguishing TP from PsP in GBMs are shown as a block diagram in Figure 2. Currently, some of these are being routinely included in standard MRI acquisition protocols at many institutions. However, full implementation has been hindered by the lack of standardized data acquisition and postprocessing modules. There are considerable efforts currently in progress to implement these advanced imaging modalities into regular clinical workflows for improvised diagnostics.
FIGURE 2.
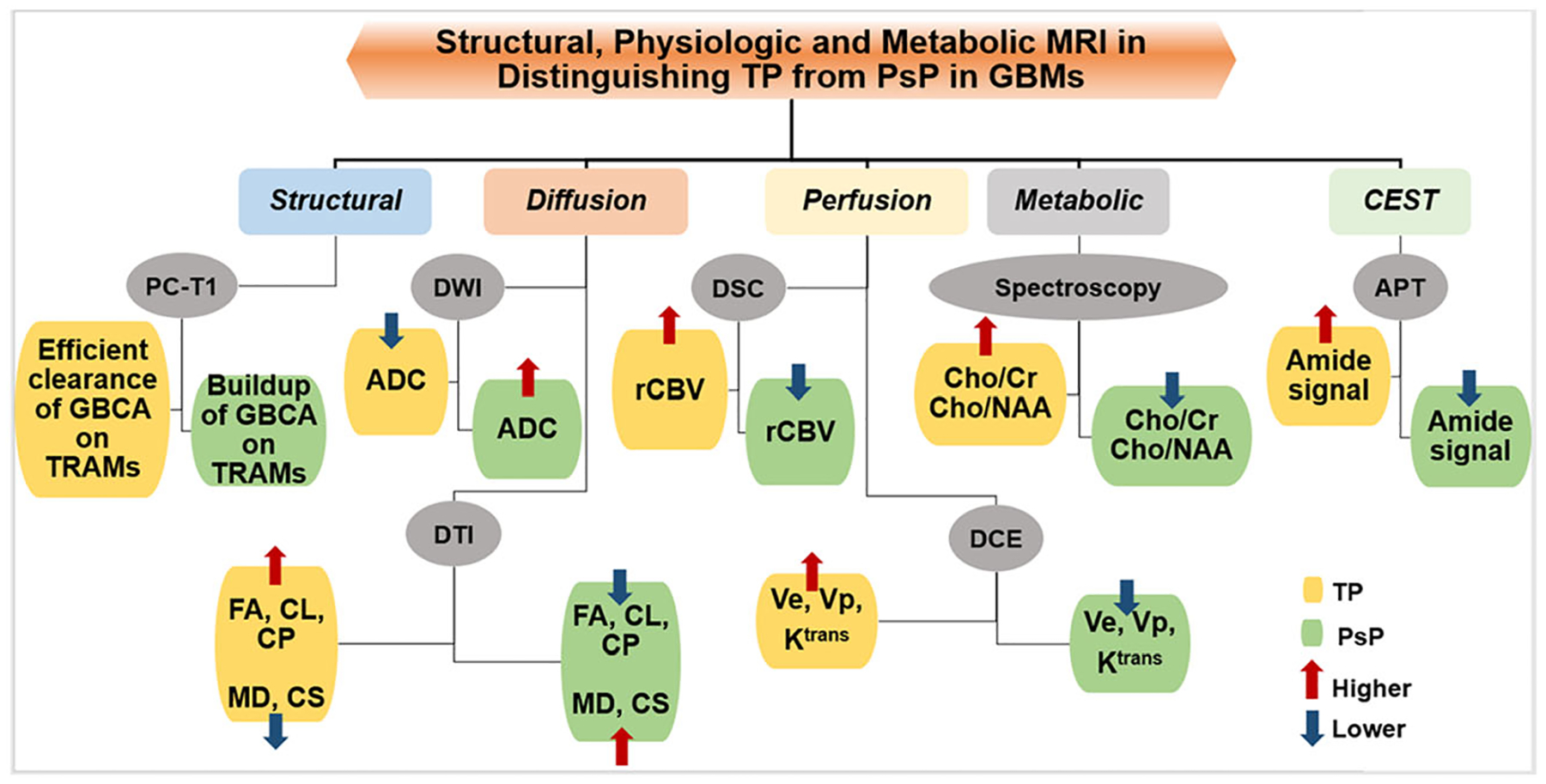
Block diagram showing the trends in structural, metabolic, and physiologic magnetic resonance imaging-derived parameters that are usually observed in distinguishing true progression (TP) from pseudoprogression (PsP) in glioblastomas. ADC, apparent diffusion coefficient; APT, amide proton transfer; CEST, chemical exchange saturation transfer; Cho, choline; CL, coefficient of linear anisotropy; CP, coefficient of planar anisotropy; Cr, creatine; CS, coefficient of spherical anisotropy; DCE, dynamic contrast-enhanced; DSC, dynamic susceptibility contrast; DTI, diffusion tensor imaging; DWI, diffusion-weighted imaging; FA, fractional anisotropy; GBCA, gadolinium-based contrast agent; Ktrans, volume transfer constant; MD, mean diffusivity; NAA, N-acetylaspartate; PC-T1, postcontrast T1-weighted images; rCBV, relative cerebral blood volume; TRAM, treatment response assessment map; Ve, volume fraction of extravascular-extracellular space in tissues; Vp, volume fraction of plasma space in tissues
2.3 |. Diffusion-weighted imaging
The biophysical mechanism of diffusion-weighted imaging (DWI) is based on random, microscopic, thermally induced translational motion of water molecules in biological tissues. Diffusion of water molecules follows the principles of Brownian motion that is caused by intermolecular collisions. The magnitude of this random movement is described by its apparent diffusion coefficient (ADC) measured in units of mm2/s. Several factors such as cellular packing, the presence of intracellular organelles, cell membranes, and macromolecules determine the ADC values. Moreover, alterations and redistributions of water molecules between intracellular and extracellular tissue compartments are also known to influence ADC values.22,23
In gliomas, cellular density and tumor grade are directly related to the degree of water restriction on DWI, resulting in an inverse relationship between ADC values and tumor grades. High-grade neoplasms harboring densely packed cancerous cells and diminished extracellular space demonstrate low ADC values.24 On DWI, changes in the diffusivity pattern of a tumor might be used to monitor tumor response to a therapeutic intervention. It is expected that successful CCRT, leading to necrosis or cellular lysis, would reduce tumor cellularity and therefore lead to increased ADC in GBMs representing predominantly treatment-related changes. On the other hand, the diffusivity of water molecules should be expected to decrease in TP with high cellularity and reduced extracellular space. Several studies25–27 have documented that PsP exhibits higher ADC from the enhancing regions of neoplasms than TP, partly because of lower cellular density and/or the presence of necrotic regions in PsP. In a study by Chu et al.,28 multiple parameters of ADC histograms, such as mean, minimum, skewness, and fifth percentile of cumulative ADC histograms, were used to distinguish TP from PsP. Among all these parameters, the fifth percentile of cumulative ADC histogram was found to be the best parameter for the differentiation of TP (n = 15) from PsP (n = 15).
Despite these promising results, some other groups28,29 have reported no significant differences in mean ADC values between the two groups of patients, probably because of the small number of patients (n = 30 and 17) in those studies. Additionally, a major limitation of those studies was the use of a region of interest (ROI)-based approach for data analysis. This subjective method does not reflect the heterogeneous nature of GBMs, with components that include varying degrees of cellular and nuclear pleomorphism, mitotic activity, vascular proliferation, necrosis, and microhemorrhages. This tissue heterogeneity tends to increase further in the post-treatment settings. Interobserver variations in ROI placement can also potentially alter quantitative findings. To overcome these limitations, novel voxel-wise approaches for image analysis such as parametric response maps (PRMs)30 and functional diffusion maps (FDMs)31 have been used to differentiate TP from PsP in proof-of-concept studies. However, these maps may not be appropriate for those GBMs that result in considerable changes in mass effect and brain shift between scans acquired at different time points. The diagnostic performances of DWI-derived parameters are summarized in Table 1.
TABLE 1.
Summary of the diagnostic performances of metabolic and physiologic MRI and PET imaging parameters in distinguishing TP from PsP in GBMs
| Imaging technique | Parameter | Study/reference | Cut-off value | Sensitivity | Specificity | Accuracy |
|---|---|---|---|---|---|---|
| DWI | ADCmean | Lee et al. 201225 TP (n = 10); PsP (n = 12) | 1.2 × 10−3 mm2/s | 80% | 83.3% | 81.2% |
| 5th percentile of ADC3000 | Chu et al. 201328 TP (n = 15); PsP (n = 15) | 0.645 × 10−3 mm2/s | 93.3% | 100% | 88.9% | |
| 5th percentile of ADC1000 | Chu et al. 201328 TP (n = 15); PsP (n = 15) | 0.929 × 10−3 mm2/s | 73.3% | 73.3% | 66.7% | |
| rADC | Reimer et al. 201730 TP (n = 28); PsP (n = 7) | 27.05% | 86% | 86% | 84.4% | |
| DTI | FA + CL + rCBVmax | Wang et al. 201632 TP (n = 21); mixed tumors (n = 12); PsP (n = 8) | 0.55 | 76% | 95% | 90.5% |
| DKI | rMK | Wu et al. 202133 TP (n = 24); PsP (n = 16) | 0.87 | 87.5% | 87.5% | 85.0% |
| DSC-PWI | CBV-PHP | Kim et al. 201034 TP (n = 14); mixed tumors (n = 10); PsP (n = 15) | 1.7 | 90.2% | 91.1% | 97.3% |
| CBV - MV | Kim et al. 201034 TP (n = 14); mixed tumors (n = 10); PsP (n = 15) | 2.6 | 96.5% | 93.1% | 98.7% | |
| ΔrCBV | Mangla et al. 201035 TP (n = 12); PsP (n = 7) | NA | 76.9% | 85.7% | 85% | |
| rCBV | Kong et al. 201136 TP (n = 33); PsP (n = 26) | 1.49 | 81.5% | 77.8% | NA | |
| rCBV lesion | Prager et al. 201537 TP (n = 58); PsP (n = 10) | 1.27 | 86.5% | 83.3% | 86.3% | |
| rCBV ROI | Prager et al. 201537 TP (n = 58); PsP (n = 10) | 1.74 | 91.9% | 66.7% | 79.7% | |
| ΔnCBV | Boxerman et al. 201738 TP (n = 9); PsP (n = 10) | NA | 100% | 85% | 97% | |
| nCBV – histograhic pattern | Baek et al. 201239 TP (n = 42); PsP (n = 37) | NA | 85.7% | 89.2% | 93.4% | |
| rCBV optimal threshold | Hu et al. 201240 TP (n = 16); PsP (n = 9) | 1.0 | 100% | 100% | 100% | |
| FTBhigh | Iv et al. 201941 TP (n = 30); PsP (n = 17) | >24.9% | 80% | 82% | 85% | |
| FTBlow | Iv et al. 201941 TP (n = 30); PsP (n = 17) | <28.5% | 50% | 94% | 77% | |
| rCBV | Iv et al. 201941 TP (n = 30); PsP (n = 17) | 1.53 | 70% | 88% | 81% | |
| DCE-MRI | Ktransmean | Thomas et al. 201542 TP (n = 24); PsP (n = 13) | 3.6 | 69% | 79% | 80.8% |
| vp mean | Thomas et al. 201542 TP (n = 24); PsP (n = 13) | 3.7 | 85% | 79% | NA | |
| vp 90% | Thomas et al. 201542 TP (n = 24); PsP (n = 13) | 3.9 | 85% | 92% | 90.4% | |
| 3D-EPSI | Cho/Cr + Cho/NAA | Verma et al. 201943 TP (n = 18); PsP (n = 9) | 0.40 | 94% | 87% | 93% |
| APT | APTmean | Ma et al. 201644 TP (n = 20); PsP (n = 12) | 2.42% | 85% | 100% | 98% |
| APT | APTmax | Ma et al. 201644 TP (n = 20); PsP (n = 12) | 2.54% | 95% | 91.7% | 97% |
| aF-FET-PET | TBRmax | Kebir et al. 201645 TP (n = 19); PsP (n = 7) | 1.9 | 84% | 86% | 85% |
| aF-FET-PET | TBRmean | Werner et al. 202146 TP (n = 12); PsP (n = 11) | 1.95 | 82% | 92% | 87% |
| aF-FET-PET | TBRmax | Werner et al. 202146 TP (n = 12); PsP (n = 11) | 2.85 | 64% | 92% | 78% |
| aF-FET-PET | TBRmean | Lohmann et al. 202147 TP (n = 18); PsP (n = 16) | 1.95 | 75% | 61% | 68% |
| aF-FET-PET | TBRmax | Lohmann P, et al. 202147 TP (n = 18); PsP (n = 16) | 2.25 | 81% | 67% | 74% |
Abbreviations: 3D-EPSI, three-dimensional echoplanar spectroscopic imaging; ADC, apparent diffusion coefficient; ADC3000, ADC using a ‘b’ value of 3000s/mm2; ADC1000, ADC using a ‘b’ value of 1000s/mm2; APT, amide proton transfer; Cho, choline; CL, coefficient of linear anisotropy; Cr, creatine; DCE-MRI, dynamic contrast-enhanced magnetic resonance imaging; DKI, diffusion kurtosis imaging; DSC-PWI, dynamic susceptibility contrast perfusion-weighted imaging; DTI, diffusion tensor imaging; DWI, diffusion-weighted imaging; FA, fractional anisotropy; FTB, fractional tumor burden; GBM, glioblastoma; Ktrans, volume transfer constant; MV, maximum value; NA, not available; NAA, N-acetylaspartate; nCBV, normalized cerebral blood volume; PET, positron emission tomography; PHP, peak height position; PsP, pseudoprogression; rADC, relative apparent diffusion coefficient; rCBV, relative cerebral blood volume; rMK, relative mean kurtosis; ROI, region of interest; TBRmax, maximum tumor/brain ratio; TBRmean, mean tumor/brain ratio; TP, true progression; Vp, volume fraction of extravascular-extracellular space in tissues; ΔnCBV, change in normalized CBV.
18F-FET-PET = O-(2-[18F]fluoroethyl)-L-tyrosine.
2.4 |. Advanced diffusion imaging techniques
To better understand the diffusion phenomenon in a complex biological tissue, a mathematical modeling of diffusion in a 3D space is usually employed. In diffusion tensor imaging (DTI), a tensor model of diffusion consists of a 3 × 3 matrix derived from diffusivity measurements in at least six noncollinear/nonplanar directions. However, the utilization of greater than six diffusion sensitizing directions increases the accuracy of tensor measurement for any arbitrary orientation. The diffusion tensor can be described by a diffusion ellipsoid whose main axis is parallel to the principal diffusion direction within a voxel. Because of the presence of natural barriers such as intracellular organelles, cellular membranes, and white matter fibers/tracts inside the brain, water molecules are unable to diffuse freely (anisotropic diffusion). Therefore, DTI benefits from this anisotropic movement of water molecules to estimate the axonal direction within a living system.48
The complete data modeling of DTI offers an estimation of several scalar parameters such as mean diffusivity (MD), fractional anisotropy (FA), axial diffusivity (AD), radial diffusivity (RD), coefficient of linear (CL), planar (CP), and spherical (CS) anisotropies describing the shape of a diffusion ellipsoid. However, the most commonly used DTI indices in neuro-oncology are MD and FA. The parameters MD and ADC are comparable and provide similar physiological information.49 FA signifies the degree of diffusion asymmetry present within a voxel and its value ranges from 0 (isotropic) to 1 (maximally anisotropic). Higher FA values indicate greater directionality of white matter fibers in the brain.50 The geometric indices (CL, CP, and CS) along with parameters (MD and FA) have been employed to distinguish GBMs from solitary metastases and brain lymphomas,51,52 necrotic GBMs from brain infections,53 and to assess response to immunotherapy in GBM patients,54 implying that directional organization of tissue microstructures may provide valuable information about tissue characterization.
Some studies32,55 have also used DTI to differentiate TP and PsP. In one such study,55 DTI metrics from four segmented regions of neoplasms (contrast enhancing, central core, immediate and peritumoral regions) were not helpful in distinguishing between TP (n = 17) and PsP (n = 7), probably because of the small sample size (n = 24). However, Wang et al.32 reported some encouraging findings from a study. The investigators classified all GBMs (n = 41) into three categories: TP (n = 21), mixed response (n = 12), and PsP (n = 8). The strength of this study was the availability of tumor specimens from all the cases following repeat surgery and, as such, histopathological features were used to determine the final diagnosis of TP (more than 75% neoplastic features), mixed tumors (25%–75% neoplastic features), or PsP (less than 25% neoplastic features). When the investigators analyzed the DTI data from contrast-enhancing regions (CERs), significant elevations in FA, CL, and CP, along with a decrease in CS, were observed in TP compared with PsP. Using ROC analyses in differentiating two groups of patients (TP [n = 21] and mixed tumors + PsP [n = 20]), the investigators observed an accuracy of 84% for CP and 78% for FA. To estimate the accuracy of their findings, the investigators also performed leave-one-out cross-validation tests. Collectively, these results indicate that DTI may be helpful in evaluating treatment response to “standard-of-care” CCRT in GBM patients. The diagnostic performances of DTI-derived parameters are summarized in Table 1.
The introduction of magnetic field gradients with high slew-rate and high-performance computing systems has transformed diffusion imaging to a new level that has greatly improved its diagnostic capability in the arena of neuro-oncology. The emergence of multiband imaging has enabled us to acquire diffusion sequences using multiple b-values, as well as permitting the implementation of relatively new techniques such as diffusion kurtosis imaging (DKI) within a reasonable acquisition time.56 DKI provides additional microstructural information by extending the DTI model to incorporate fourth-order gradient field terms in the diffusion signal. The kurtosis describes the degree of deviation from Gaussian distribution of spin displacement along an axis. When averaged over all directions, the mean kurtosis (MK) can be computed.57 GBMs are characterized by complex architectural integrity resulting from the presence of high cellular density, cell membranes, organelles, and vascular structures within the tumor beds, thus impeding the movement of water molecules. This in turn leads to higher non-Gaussianity and increased MK.58 Some studies have reported better diagnostic performance of DKI than DTI in classifying different grades of gliomas.59,60 Similarly, Wu et al.33 have shown higher accuracy for DKI-derived metrics than DTI metrics in differentiating TP (n = 24) from PsP (n = 16) in high-grade gliomas (Table 1). Another valuable technique known as intravoxel incoherent motion (IVIM) has also shown encouraging findings in the identification of PsP in GBM patients.61 IVIM allows simultaneous assessment of diffusion- and perfusion-related effects in tissues with a single diffusion-based imaging sequence.62
Although promising, we believe that the diagnostic performances of DKI and IVIM warrant further investigations before these techniques can be incorporated into the routine clinical workflow for neuro-oncological applications.
2.5 |. Perfusion MRI
Tumor neovascularization is characterized by the presence of a vast network of fragile and poorly organized blood vessels. Newly formed vessels are often large, abnormal, tortuous, permeable, and unequal in size, leading to increased tumor vascularity and perfusion.63 The most commonly used perfusion-weighted imaging (PWI) techniques in the settings of clinical neuro-oncology include dynamic susceptibility contrast (DSC)-PWI and dynamic contrast-enhanced (DCE)-MRI.
2.6 |. Dynamic susceptibility contrast-PWI
The DSC-PWI method involves the injection of a bolus of GBCA intravenously, followed by a series of rapidly scanned gradient-echo images over an organ of interest. DSC-PWI uses the T2* effect of paramagnetic contrast agent that causes a transient drop in signal intensity during the initial pass through vasculature by producing a local magnetic field distortion around the blood vessels. Several perfusion parameters such as blood volume, blood flow, and mean transit time can be obtained by measuring signal intensity as a function of time and fitting it to a mathematical model.64
The pathologic characteristics of tumor vasculature have been used to discriminate between TP and PsP. Multiple studies34–39 have shown the potential of DSC-PWI in distinguishing TP from PsP in GBM patients. In TP there is a marked increase in neoangiogenesis, which leads to an increase in lesion to white matter relative cerebral blood volume (rCBV) from contrast enhancing and peritumor regions of neoplasms. By contrast, PsP is characterized by an increased inflammatory response, local accumulation of edema, and abnormal vascular permeability leading to decreased rCBV.
In one study, Kong et al.36 achieved a significant difference in mean rCBV between patients classified as TP (n = 33) and PsP (n = 26) in a population of 59 patients treated with CCRT. With a threshold rCBV of 1.47, the investigators achieved a sensitivity of 81.5% and a specificity of 77.8% in distinguishing TP from PsP. In a related study (n = 68), Prager et al.37 found a sensitivity of 91.9%, and a specificity of 66.7% using a threshold rCBV of 1.74. In a prospective study,65 excellent correlations between high rCBV values and the presence of recurrent tumors (n = 24) were reported in which image-guided stereotactic biopsy specimens were obtained from patients who underwent repeat surgery after CCRT.
In a longitudinal study (n = 19), Mangla et al.35 compared rCBV at baseline (prior to treatment) and at 1-month postradiotherapy/TMZ treatment period. PsP patients (n = 7) exhibited a 41% decline in rCBV, while TP patients (n = 12) demonstrated a 12% elevation in rCBV post-treatment relative to baseline, leading to a sensitivity of 77% and a specificity of 86% in differentiating these two groups of patients. In another longitudinal study, Boxerman et al.38 found substantial overlap of mean rCBV at initial enhancement between TP (n = 9) and PsP (n = 10) lesions. However, changes in rCBV at first subsequent follow-up differed significantly between TP and PsP, suggesting that temporal variations in rCBV may be more helpful in distinguishing these two groups of patients.
Because GBMs are markedly heterogeneous in nature, ROI-based analysis of mean rCBV value may not be a robust measure for differentiating TP from PsP. To overcome these limitations, histogram and PRM methods have been used. In one study, Kim et al.34 used histogram analysis of CBV to calculate the volume fraction between high-grade recurrent tumors and post-treatment changes. Thirty-nine patients with recurrent tumors (tumor fraction ≥ 50%, n = 14), mixed tumors (tumor fraction ≥ 20% and < 50%, n = 10), and tumors showing predominant treatment-related changes (tumor fraction < 20%, n = 15) were evaluated in that study. The parameters that were analyzed from CER of neoplasms included histogram width, peak height position (PHP), and maximum value (MV). The parameters PHP and MV were significantly different between recurrent tumors and tumors, showing mixed response plus post-treatment changes. Moreover, PHP with an optimal threshold of 1.7 had a sensitivity of 90.2% and a specificity of 91.1% in differentiating two groups. Additionally, MV also had a high sensitivity of 96.5% and a specificity of 93.1% with an optimum threshold of 2.6 in distinguishing treatment change group from tumor recurrence and mixed groups. This study shows that histogram may be an appealing method of CBV analysis that can capture tumor heterogeneity more effectively. Using histogram-based analysis, serial DSC-PWI scans have also been performed to identify patients with PsP. In one such longitudinal study, Baek et al.39 analyzed DSC-PWI data within 4 weeks after the completion of CCRT and at a follow-up period (4–8 weeks of baseline study) from a cohort of patients with high-grade gliomas (n = 79). The investigators proposed that percentage changes in histogram-derived parameters, such as skewness and kurtosis, can be potentially useful imaging parameters to identify PsP (n = 37) or early TP (n = 42). In a novel thresholding method, Hu et al.40 subtracted precontrast voxel values from postcontrast to create a mask of enhancing voxels, which was then used to define the ROI for analyzing DSC-PWI data in distinguishing TP from PsP.
Alternatively, a PRM method has been proposed to address the issue of tumor heterogeneity. PRM analysis is a sensitive voxel-wise analytic method to compute regional changes in perfusion following therapy. PRMrCBV is derived for each voxel within the tumor, and regions of increasing or decreasing rCBV values are quantified individually. On the other hand, mean variations in rCBV values averaged all over the tumor (increasing and decreasing regions cancel out) will lack sufficient sensitivity in evaluating outcomes in GBM. Using this method, Tsien et al.66 concluded that PRMrCBV at week 3 during CCRT might represent an early imaging biomarker to distinguish TP from PsP in high-grade glioma patients. Another novel parameter known as fractional tumor burden (FTB) is defined as the volume fraction of tumor voxels exceeding a specified rCBV threshold. FTB providing per-voxel measurements rather than computing a single value to represent the entire contrast-enhancing volume of a tumor may be a robust method for overcoming the issue of tissue heterogeneity within the tumor. FTB has been shown to correlate with histologic tumor volume fraction in treated GBM better than rCBV.67 In a study by Iv et al. (n = 47),41 FTB was used to differentiate treatment effect (n = 17) from recurrent GBMs (n = 30) with high diagnostic performance. The investigators used rCBV threshold values of 1.0 and 1.75 to define three FTB classes: FTBlow (percentage of contrast-enhancing voxels with rCBV of ≤ 1.0); FTBmid (percentage of voxels with rCBV between 1.0 and 1.75); and FTBhigh (percentage of voxels with rCBV of ≥ 1.75). Additionally, mean rCBV values from the entire CERs of tumors were estimated in each case. Significant differences in FTBhigh, FTBlow, and mean rCBV values were observed between recurrent GBMs and treatment effect with moderate to high accuracies (FTBlow [70%], mean rCBV [81%], and FTBhigh [85%]). The findings from this study indicate that the diagnostic performance of FTB as a novel hemodynamic parameter is not significantly better than that of traditionally used mean rCBV values in discriminating TP from PsP. We believe that FTB maps using different threshold rCBV values should be investigated to improve their diagnostic performance. The diagnostic performances of DSC-PWI–derived parameters are summarized in Table 1. Lastly, ferumoxytol is a nanosized blood pool agent requiring no contrast agent leakage correction. rCBVmean using ferumoxytol has been found to be superior to that of GBCAs for evaluating treatment response in GBM patients.68
Collectively, these DSC-PWI studies have shown the clinical usefulness of rCBV in differentiating TP from PsP. However, the potential role of rCBV in evaluating treatment response in GBM has been limited as increased rCBV may not always necessarily be associated with TP; local inflammatory response may also result in elevated rCBV in the tumor beds, particularly in the setting of immunotherapy.69 Moreover, the wide variability in DSC-PWI acquisition and the use of different types of postprocessing software, as well as the techniques employed for contrast leakage and recirculation correction in different studies, are other potential causes of variable results reported in the literature. Moving forward, active efforts to standardize the acquisition and analysis of DSC-PWI data must be achieved to improve the consistency of interpreting rCBV findings across studies.
2.7 |. Dynamic contrast-enhanced MRI
DCE-MRI involves acquisition of T1-weighted images generally using a 3D-spoiled gradient recalled-echo (SPGR) or fast low-angle shot (FLASH) sequences before, during, and after the injection of GBCA. DCE-MRI measures T1 changes in tissues over time after bolus administration of GBCA. DCE-MRI is often performed for generating tissue perfusion parameters based on pharmacokinetic modeling that typically requires an additional precontrast T1-mapping protocol.70 In clinical settings, a pharmacokinetic model proposed by Tofts et al. is generally used to process DCE-MRI data.71 The most commonly used DCE-MRI–derived parameter is volume transfer constant (Ktrans), which determines the flux of contrast agent from intravascular to extravascular-extracellular space. Physiologically, Ktrans reflects the combined effects of vascular permeability and tumor perfusion. The other clinically important parameters include volume fraction of extravascular-extracellular space in tissues (ve) and volume fraction of plasma space in tissues (vp). The parameter ve has been shown to be positively associated with ADC because of reduced cellularity and greater extracellular space.72 The parameter vp may reflect angiogenic activity in tumors. Compared with DSC-PWI, DCE-MRI possesses greater spatial resolution, is less prone to susceptibility artifacts, and provides better estimation of vascular permeability.
In a retrospective DCE-MRI study on post-treatment GBM patients, Thomas et al.42 used Ktrans as well as vp to differentiate TP (n = 24) and PsP (n = 13). The investigators found a cut-off value of more than 3.6 for Ktrans yielding 69% sensitivity and 79% specificity for identifying TP and a cut-off value of less than 3.7 for vp yielding 85% sensitivity and 79% specificity for identifying PsP. In a related study by Yun et al.,73 the mean Ktrans and ve, but not the mean vp, were found to be significantly higher in TP (n = 17) than in PsP (n = 16). Although pharmacokinetic model-based quantitative parameters may be useful in measuring real physiological tissue properties, their clinical potential is hampered by a lack of consensus regarding the use of optimal pharmacokinetic model, water exchange effects, and an absence of clear criteria to compute arterial input functions.
Despite some clinical limitations and unsolved issues, the available evidence advocates for the potential of perfusion MRI in distinguishing TP from PsP in GBM patients. However, more well-controlled studies with standardized protocols are needed to validate these promising findings. The diagnostic performances of DCE-MRI–derived parameters are summarized in Table 1.
2.8 |. Combined use of diffusion and perfusion MRI
In GBMs, spatial and temporal intratumoral heterogeneity causes regional variations in metabolism, vasculature, oxygenation, and cytoarchitectural integrity that is reflected by a mismatch in the findings from different neuroimaging parameters. Therefore, the use of a single imaging technique or parameter may not always be reliable in characterizing GBMs and evaluating treatment response. Indeed, the multiparametric approach of data analysis is an upcoming method in which several quantitative MRI techniques are studied in combination to potentiate the individual value of each advanced MRI technique used in isolation in addressing the issue of inherent tumor heterogeneity.
Given that DTI- and PWI-derived parameters provide inherently different but complementary physiological information, it may be assumed that these parameters may interact synergistically in the combined data analysis, thus providing higher diagnostic power than what would be expected from individual parameters (Figures 3–5). In accordance with this hypothesis, several studies have reported the importance of merging the unique strengths of DTI and PWI together in discriminating necrotic GBMs from brain infections,53 histologic grades of nonenhancing gliomas,74 classification of brain neoplasms,51,52 discrimination of recurrent tumors from radiation necrosis,75 assessing tumor invasiveness,76 predicting survival,77 and evaluating response to immunotherapy in patients with GBM.54
FIGURE 3.
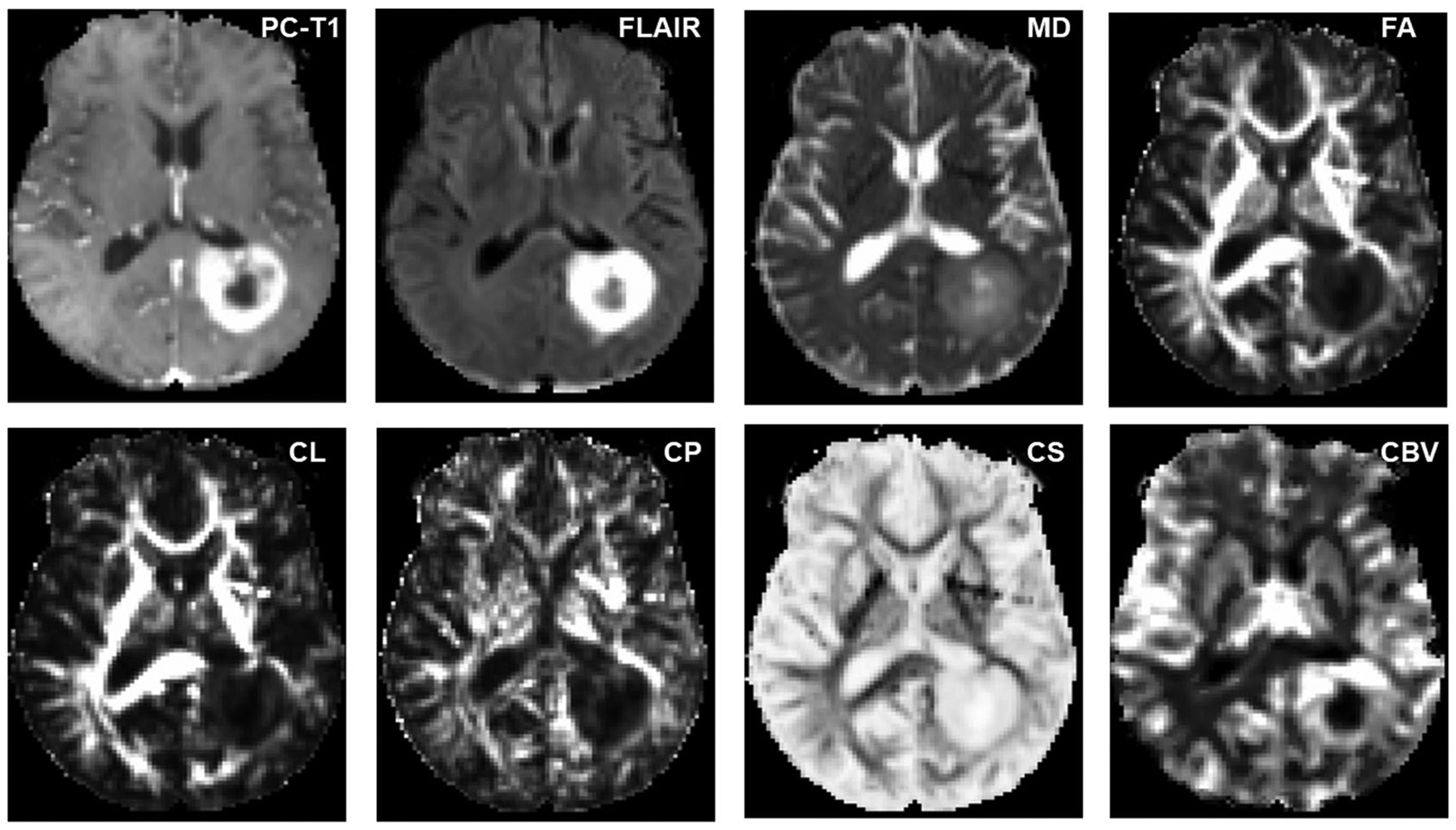
Axial MR images from a glioblastoma patient with true progression. The postcontrast T1 (PC-T1)-weighted image shows a ring-enhancing lesion in the left parietal region. Coregistered diffusion tensor imaging-derived parametric maps and cerebral blood volume (CBV) maps are shown. Increased mean diffusivity (MD) and coefficient of spherical anisotropy (CS), and decreased fractional anisotropy (FA), coefficient of linear anisotropy (CL), and coefficient of planar anisotropy (CP), are observed from the enhancing part compared with the contralateral normal white matter regions. The enhancing part of the lesion is also demonstrating high blood volume on CBV maps. FLAIR, fluid-attenuated inversion recovery
FIGURE 5.
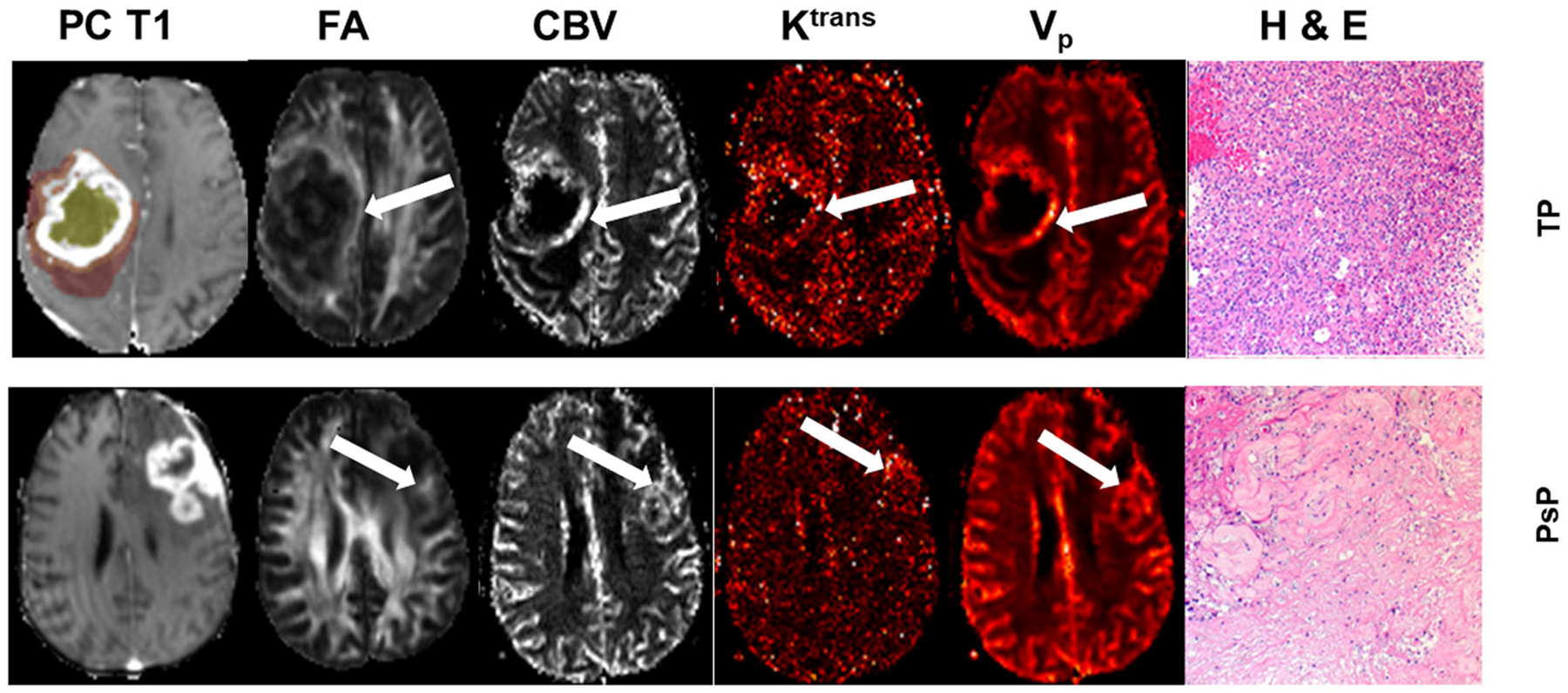
Axial postcontrast T1 (PC-T1)-weighted image from a patient with true progression (TP) (top row), demonstrating a neoplasm in the right parietal region infiltrating into the lateral ventricles. The regions of interest are overlaid on the image, with the colors indicating the following defined regions: white, contrast-enhancing region; yellow, central core; orange, immediate peritumoral region; and brown, distal peritumoral region. Fractional anisotropy (FA), cerebral blood volume (CBV), volume transfer constant (Ktrans), and volume fraction of plasma space in tissues (Vp) values from the enhancing regions (arrows) in TP are higher than those of pseudoprogression (PsP) (bottom row). A photomicrograph of hematoxylin–eosin (H & E) stain from the TP case demonstrates areas of high tumor cellularity, pseudopalisading necrosis, endothelial proliferation, and increased mitotic activity, whereas PsP shows predominant treatment-related changes, including extensive geographic necrosis and vascular fibrinoid necrosis
In an earlier study, Fink et al.78 observed significantly higher rCBVmax from lesions characterized as tumor recurrence (n = 30) compared with those with predominant treatment effects (n = 10). However, the investigators did not find any significant difference in ADC ratio (minimum value of ADC from lesions normalized to contralateral white matter regions) between these two groups. In another study (n = 68), Prager et al.37 observed significantly higher rCBV and lower ADC in TP compared with PsP. Notably, the diagnostic performance of rCBV was better than that of ADC in distinguishing these two groups of tumors. Together, the findings from these two studies support the notion that intratumoral heterogeneity causes mismatch and incongruity in the phenotypic information obtained from different physiologic MRI-derived parameters, thus emphasizing the importance of a multiparametric analytical approach for achieving better diagnostic accuracy. Indeed, when rCBV and ADC parameters were included in the combined model, Prager et al.37 obtained a slightly better sensitivity and specificity than rCBV or ADC alone in distinguishing TP from PsP. Unfortunately, only 60% of all patients had the availability of both DWI and DSC-PWI data in that study.
In a similar study from our group (n = 41),32 follow-up GBM patients presenting with new enhancing lesions within 6 months of completion of CCRT were classified into TP (n = 21), mixed tumors (n = 12), and PsP (n = 8) groups based on histopathological findings from tumor specimens obtained from repeat surgery. Significantly elevated values of FA, CP, CL, and rCBVmax were noticed in TP compared with those with PsP plus mixed tumors from CER of neoplasms with variable sensitivities (62%–71%) and specificities (75%–90%) of individual parameters. However, the best logistic regression model to differentiate TP from PsP comprised FA, CL, and rCBVmax, resulting in an accuracy of 90%, a sensitivity of 76%, and a specificity of 95%. Moving forward, we used a combination of DTI and PWI parameters in evaluating treatment response to EGFRvIII targeted chimeric antigen receptor T-cell therapy in patients with recurrent GBM (n = 10) in a more recent study.54 When percent changes in individual imaging parameters were assessed from enhancing lesions at different follow-up periods relative to baseline, no definite trends were observed, indicating that imaging parameters when used in isolation may have a limited role in assessing the heterogeneity of treatment response. However, when we used probabilities of tumor progression derived from a combination of FA, CL, and rCBVmax, we were able to objectively characterize each lesion as either TP or PsP at each individual time point. Collectively, these findings suggest that a multiparametric approach may provide a more accurate assessment of treatment response than an individual parameter or technique in GBM patients.
2.9 |. Proton MR spectroscopy
A number of studies,79,80 including from our group,81–84 have described the usefulness of 1H MRS in characterizing brain tumors. Several data analytical methods have been proposed to report 1H MRS data for studying brain tumor metabolism. To report 1H MRS data, it has been a common practice to use metabolite ratios for quantification purposes, as metabolite ratios are more easily measured than metabolite concentrations. These metabolite ratios correct for several unknown and hard to obtain or uncontrollable experimental conditions such as static (B0) and radiofrequency (B1) field inhomogeneities, regional susceptibility variations, instrumental gain drifts, localization method differences, variations in voxel size and partial volume effects causing contamination with metabolite free cerebral-spinal fluid components. Generally, there are two approaches for reporting metabolite ratios, and these include normalized (with reference to contralateral normal brain regions) and non-normalized methods (with reference to ipsilateral regions).
While it has been reported that the inclusion of a metabolite from a voxel encompassing contralateral normal brain region in the denominator can potentially introduce some variability such as noise propagation and localization,85 we believe that reporting spectroscopy data as a normalized metabolite ratio is a better option. This is because normalization of the area under the peak or concentration of metabolites from tumor regions with respect to similar metabolite measures from contralateral normal brain regions may account for intersubject variability and thus facilitate unbiased group comparisons. Our supposition is corroborated by an earlier study, in which normalized metabolite ratios were found to be better than non-normalized metabolite ratios in characterizing gliomas.86 In another study, normalized metabolite ratios provided improved discrimination of recurrent tumors from RN over non-normalized metabolite ratios.87
Using standard multivoxel 1H MRS methods covering both solid/contrast enhancing, and peritumoral regions, several studies87–89 have shown the clinical potential of 1H MRS in differentiating recurrent tumors from RN in GBM patients. Collectively, these studies have shown that metabolite ratios of choline (Cho; a marker of increased cellular proliferation that is generally elevated in brain tumors90) relative to N-acetylaspartate (NAA; reflects neuronal integrity and viability, and is decreased in tumors90) and/or creatine (Cr; a marker of energy metabolism90), either from the ipsilateral voxel from tumors or from the voxel encompassing contralateral normal brain regions, were significantly higher in recurrent tumors than in RN. In one study,91 Chotumor/NAAtumor was found to be the best parameter in distinguishing recurrent tumors (n = 20) from RN (n = 13) with a sensitivity of 85%, a specificity of 69.2%, and an accuracy of 92%.
Despite the success of reporting 1H MRS data as metabolite ratios, relative quantification methods may introduce some potential errors and can lead to misinterpretation of 1H MRS data.85 On the other hand, quantifying the absolute concentration of metabolites allows unambiguous interpretation of data. There are several methods for estimating the absolute concentration of metabolites that include an external standard reference, tissue water signal as an internal reference, phantom replacement method and principle of reciprocity. Interested readers are referred to excellent articles available in the literature for a detailed overview on computing absolute concentrations of metabolites using 1H MRS.92–94 Tissue water signal as an internal reference has been the most commonly used method for estimating metabolite concentrations for characterizing brain tumors in previous studies.95,96
Because the pathogeneses of RN and PsP are comparable,10,97 it may be hypothesized that these two entities would present similar metabolic patterns such that widely available reports of abnormal metabolite profiles from RN may be extrapolated to understand differences between TP and PsP (Figure 6). Indeed, the investigators of a longitudinal study98 observed elevated levels of lipids and decreased Cho/NAA in PsP compared with TP patients. 3D echo planar spectroscopic imaging (3D-EPSI) allows acquisition of high-resolution whole brain metabolite maps.99,100 The utility of 3D-EPSI has been reported in several applications in neuro-oncology.54,101–105 Using the 3D-EPSI sequence, we recently reported promising findings in distinguishing TP (n = 18) from PsP (n = 9) in GBM patients.43 While comparing metabolite ratios from CER, immediate peritumor region (IPR) and distal peritumor region (DPR) of neoplasms, significantly elevated Cho/NAA was observed from all of these regions in TP patients compared with those with PsP (Table 1). Additionally, significantly elevated Cho/Cr levels from CER were observed in TP compared with PsP. Higher Cho/NAA from peritumoral regions (IPR and DPR) in TP in comparison with PsP suggests that TP lesions were associated with a greater degree of neoplastic infiltration and/or greater damage to neuronal integrity in regions beyond the CER. These findings imply that the mapping of metabolite ratios from peritumoral areas should also be considered when differentiating TP from PsP. We believe that assessment of the real extent of neoplastic spread will enable the formulation of more aggressive treatment strategies for the management of TP patients.
FIGURE 6.
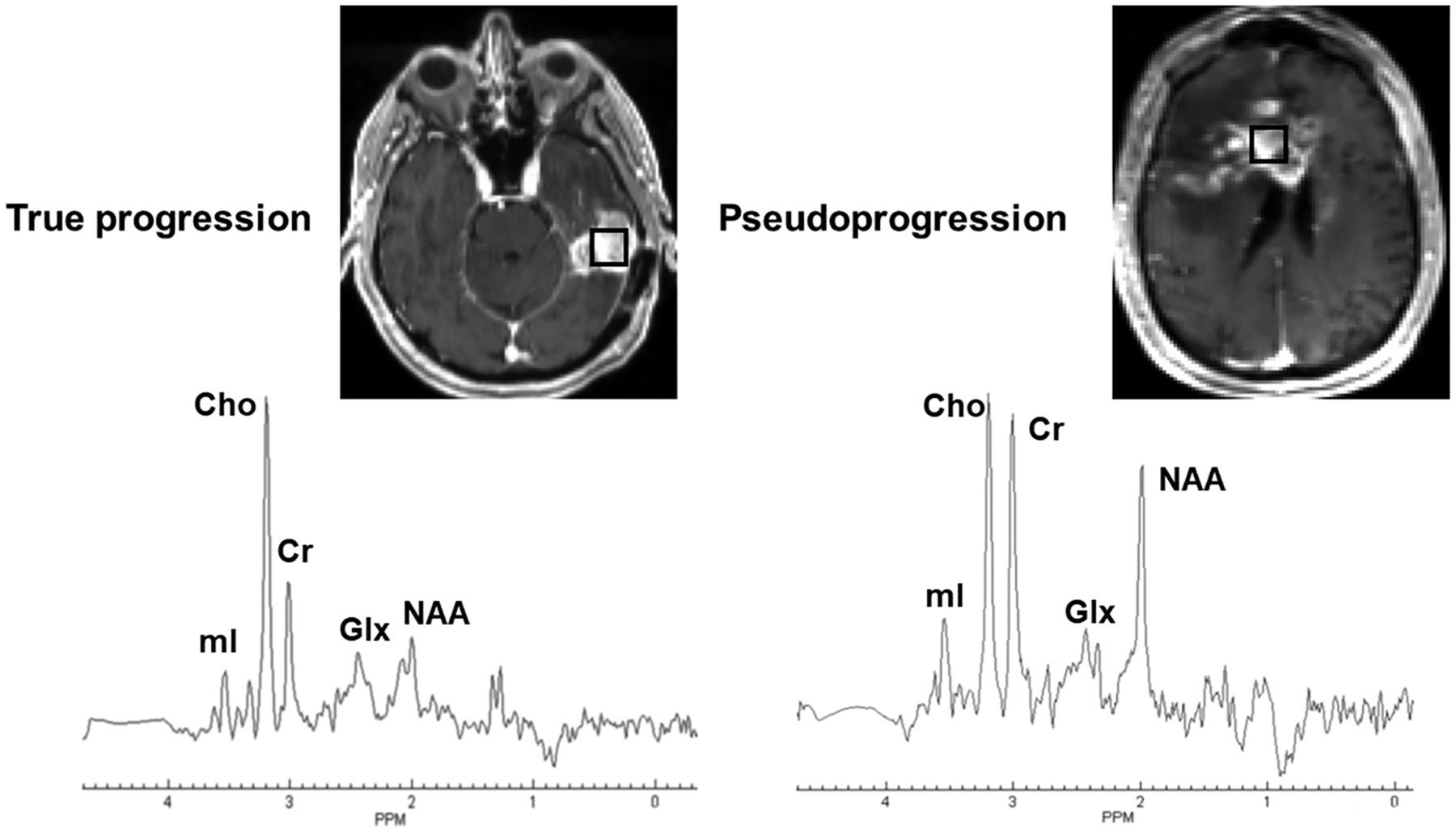
Left panel (true progression [TP]): the axial postcontrast T1-weighted image demonstrates a nodular contrast-enhancing lesion in the left temporal lobe. The square box overlaid on the enhancing lesion is a combination of 16 voxels (nominal voxel size = 4.3 × 4.3 × 5.6 mm3). Corresponding summed 1H MRS spectrum from these 16 voxels exhibiting various metabolites. Right panel (pseudoprogression [PsP]): the axial postcontrast T1-weighted image shows a heterogeneously enhancing lesion in the right more than left frontal lobes involving the genu and body of corpus callosum. The square box overlaid on the enhancing lesion is a combination of 16 voxels (nominal voxel size = 4.3 × 4.3 × 5.6 mm3). Corresponding summed 1H MRS spectrum from these 16 voxels exhibiting various metabolites. Please note the presence of higher Cho/NAA and Cho/Cr ratios in TP than in PsP. Both these patients were scanned on a 3-T MR system after the completion of maximal resection of tumor followed by standard-of-care chemoradiation therapy and were exhibiting new contrast-enhancing lesion in the resection bed within 6 months of standard treatment. The whole-brain 1H MRS was acquired using a three-dimensional echoplanar spectroscopic imaging sequence (a modified spin-echo sequence with GRAPPA). The typical sequence parameters were: repetition time/echo time = 1700/17.6 ms; scanning time = ~17 min that included interleaved acquisition of metabolite and water reference scans. Cho, choline; Cr, creatine; Glx, glutamate + glutamine; mI, myo-inositol; NAA, N-acetylaspartate
2.10 |. Amide proton transfer imaging
Chemical exchange saturation transfer (CEST) is a somewhat novel metabolic imaging technique that allows detection of both exogenous and endogenous molecules possessing labile proton groups such as hydroxyls (-OH), amides (-NH), and amines (-NH2) that are present in very low concentrations (μM to mM range). These exchangeable protons are selectively saturated using a continuous radiofrequency irradiation. Subsequently, when these saturated protons exchange with bulk water protons, a decline in the bulk water signal is observed over a period of time. The degree of CEST effect depends on the concentration of molecules, number of labile proton groups per molecule, and the exchange rate of these labile protons. Fluctuations in environmental conditions such as temperature and pH can also vary the CEST effect by altering the exchange rate.
APT-weighted imaging is a CEST method widely used to map the endogenous mobile proteins and peptides in tissues by exploiting the labile amide protons.106 These amide protons resonate at about 8.3 ppm on the MR spectrum, and hence have a chemical shift 3.5 ppm downfield from bulk water resonance (4.8 ppm). Because of the slow exchange rate (10–30 s−1), it is feasible to obtain almost complete saturation of amide protons utilizing a low-power and long-duration saturation pulse. The slow exchange rate of amide protons also permits acquisition of good quality APT-weighted images, even at a clinical field strength of 3 T.
It is widely believed that malignant cells express higher amounts of mobile peptide components that contribute to higher APT signals from tumor tissues.107 APT contrast has been shown in demarcating the neoplastic lesions from normal brain tissues in both preclinical and clinical studies.108 A few studies109,110 have also demonstrated the strong clinical potential of APT-weighted imaging in delineating malignant neoplastic infiltration from peritumoral vasogenic edema, in distinguishing histopathological grades, and in discriminating high-grade gliomas from CNS lymphomas. Some studies44 have also documented significantly higher APT-weighted signal intensities in TP (n = 20) than in PsP (n = 12) cases, mainly due to the presence of higher mobile proteins/peptides concentrations in TP (Table 1).
Collectively, these studies suggest that APT is rapidly evolving as a novel molecular MRI technique in neuro-oncology. However, several technical factors impact the tissue contrast generated by APT-weighted imaging.111 Water longitudinal relaxation time (T1) has been shown to influence the APT signal. Depending on the levels of direct water saturation, the T1 effect on APT signal is either linear or complex. This relationship further depends on the field strength, irradiation power and steady state or nonsteady state acquisition schemes. Furthermore, magnetization transfer (MT) and nuclear Overhauser enhancement (NOE) saturation transfer effects are known to impact the APT signal intensity.112 Because of these reasons, we believe that more research is needed to establish whether APT can be used routinely for post-treatment imaging in GBM patients.
2.11 |. Amino acid-based positron emission tomography imaging
Over the last several years, [18F]-2-Fluoro-2-deoxy-D-glucose ([18F]-FDG) has been the most commonly used positron emission tomography (PET) tracer with which to study brain tumors.113,114 Elevated [18F]-FDG uptake in neoplastic cells reflects increased expression of glucose transporters and/or enzymatic activity of hexokinase.115 However, normal brain parenchyma shows a natural higher uptake of [18F]-FDG, which undermines the diagnostic accuracy of FDG tracer for accurate delineation of brain tumor margins, especially from adjacent gray matter regions. On the other hand, several amino acid-based PET imaging tracers have emerged as alternative candidates for metabolic imaging of brain tumors.116 These tracers are characterized by high tumor to brain contrast based on their relatively high specificity for neoplastic cells and low accumulation in normal brain tissues. Frequently used amino acid tracers include O-(2-[18F]fluoroethyl)-L-tyrosine ([18F]-FET), [11C]methyl-L-methionine ([11C]-MET), and 3,4-dihydroxy-6-[18F]fluoro-L-phenylalanine ([18F]-FDOPA) targeting energy-independent amino acid transporters of L-type (LAT) that are known to be upregulated in brain tumors.117 In addition to amino acid-based tracers, 18F-fluorothymidine ([18F]-FLT), a pyrimidine analogue, has been used as a surrogate marker for cellular proliferation in neuro-oncology because of its preferential uptake by rapidly dividing neoplastic cells where it indicates the activity of thymidine kinase-1 (a key enzyme involved in DNA synthesis).118
In a prospective study using FLT-PET imaging, Brahm et al.119 did not observe any differences between TP (n = 7) and PsP (n = 7) in GBM patients who were treated with CCRT, mainly because of the fact that FLT uptake in GBMs reflects not only the trapping of FLT in proliferating neoplastic cells, but also the disruption of BBB integrity.120 The limited diagnostic utility of FLT-PET in distinguishing TP from PsP might be attributed to the fact that BBB leakage is known to occur in both TP and PsP. On the other hand, amino acid-based PET imaging tracers can cross the intact BBB, allowing the depiction of tumor regions beyond the contrast enhancement that is seen on MR imaging.121 Using [18F]-FET as a PET imaging tracer, Kebir and his colleagues45 were successful in distinguishing TP (n = 19) from PsP (n = 7) in GBM patients treated with CCRT. Some other studies46,47 have also shown the potential of [18F]-FET-PET imaging in distinguishing TP from PsP in GBMs (Table 1). The plausible explanation might be that active tumor cells express higher concentrations of mobile protein and peptide components,122 providing a higher contrast in TP than in PsP. The other potential significance of using [18F]-FET-PET has been that FET tracer exhibits a high uptake by neoplastic cells and less uptake by inflammatory cells.123
2.12 |. Combined analysis of physiologic MRI and molecular biomarkers
The recent emergence of molecular biomarkers in neuro-oncology has had a considerable bearing on the clinical management of GBM patients. Numerous genetic and molecular alterations in GBMs have been associated with the development of PsP, including an isocitrate dehydrogenase (IDH) mutation, a p53 mutation, and an MGMT promoter methylation.124
The methylation (deactivated) status of MGMT gene promoter is an important biomarker of tumor response to TMZ chemotherapy.125 Patients harboring methylated MGMT promoters are associated with more favorable prognosis and survival outcomes than those with unmethylated genotype.126,127 It is essential to develop imaging biomarkers that can help us in identifying the MGMT status of GBMs noninvasively. While some studies128,129 have documented lower ADC in tumors with unmethylated MGMT promoters, some other studies130,131 have found no significant relationships between ADC and MGMT promoter methylation status. In a multiparametric analysis,132 a combination of tumor location, extent of necrosis, ADC, and cerebral blood flow had the greatest accuracy in identifying MGMT promoter methylation status.
While evaluating the potential of diffusion and perfusion imaging parameters in predicting PsP within GBM patients based on stratification via MGMT methylation status, Yoon et al.133 noticed a trend towards higher accuracy in the MGMT promoter methylation group than in the unmethylation group. Additionally, the pairing of MGMT methylation status with imaging parameters was found to have a better accuracy for predicting PsP compared with MGMT methylation status alone. Furthermore, the probability of PsP was highest (95.7%) when the 90th percentile of normalized CBV (nCBV90) was below 4.02 in the MGMT promoter methylation group. In another study,36 the unmethylated MGMT promoter group had a significant difference in mean rCBV between TP and PsP, although the methylated MGMT promoter group had no significant difference. Collectively, these studies suggest that diffusion and perfusion imaging-derived parameters may reveal variable investigative values in predicting PsP in GBM patients stratified by MGMT promoter methylation status.
2.13 |. Radiomics and ML
Radiomics is an emerging translational field that automatically produces mineable high dimensionality data from clinical images.134 Readily interpretable and quantitative features are extracted from a predefined ROI encompassing both solid and peritumoral regions of neoplasms in a typical fashion. Then ML is used to train classifier models using various radiomic features. ML is a group of computational procedures that are based upon predictive models, which identify imaging patterns in a set of training data that are statistically associated with a clinical outcome. The training cohort is used to instruct the computer algorithm to detect patterns of features that are subsequently examined in a validation cohort to evaluate the performance of the algorithm in correctly predicting the presence or absence of a feature and its association with outcome. A feature selection module diminishes the number of highly associated features and/or noise, thus increasing the prediction accuracy.
Several studies135–137 have shown the potential of radiomic signatures, derived from conventional MRI features, in distinguishing TP from PsP. In a recent study, Kim et al.137 incorporated DWI and DSC-PWI parameters in a radiomic model and showed improved diagnostic performance in distinguishing TP (n = 35) from PsP (n = 26). Moreover, their predictive model had good generalizability and showed robustness in a multicenter setting. In a recent histopathology-validated ML study involving quantitative multiparametric (structural and physiologic parameters) analysis from independent discovery (n = 40; TP = 23, PsP = 6, mixed tumors = 11) and replication (n = 23; TP = 12, PsP = 4, mixed tumors = 7) cohorts of GBM patients who underwent second resection due to progressive radiographic changes suspicious for recurrence, Akbari et al.138 proposed a method to define multiparametric features of the resected tissues after deformable registration of postoperative to preoperative images using publicly available software139 known as Deformable Registration Attribute Matching and Mutual-Saliency weighting (DRAMMS). The areas in preoperative images that corresponded to the resected tissues in postsurgery images were defined in ROIs in an effort to correlate imaging and histopathological features from the same regions within neoplasms. Based on the histopathological assessment (degree of pseudopalisading necrosis, microvascular proliferation, dystrophic calcification, vascular hyalinization, quantitative assessment of Ki-67, and number of mitotic figures), the whole resected specimens were scored from 1 to 6. Score 1 represented less than 10% neoplastic features, score 2 10%–25% neoplastic features, score 3 25%–50% neoplastic features, score 4 50%–75% neoplastic features, score 5 75%–90% neoplastic features, and a score of 6 more than 90% neoplastic features. The scores of 1–2 were defined as PsP, 3–4 as mixed tumors (a mixture of TP and PsP), and 5–6 as TP. The investigators of this study employed deep learning and conventional feature extraction methods to extract quantitative imaging characteristics from multiparametric MRI. They found good correlations between the imaging features and histopathologic characteristics of the resected tissue specimens for TP, PsP, and mixed tumors (Figure 7). The accuracy of the proposed imaging signatures was 87% for predicting PsP and 84% for predicting TP in leave-one-out cross-validation tests, whereas in the discovery/validation cohort, the accuracy was 87% for predicting PsP and 78% for TP. Moreover, seven of 10 TP and eight of 10 PsP patients were correctly diagnosed by the predictive model, resulting in an accuracy of 75% when interinstitutional validation was performed from 20 GBM patients.
FIGURE 7.
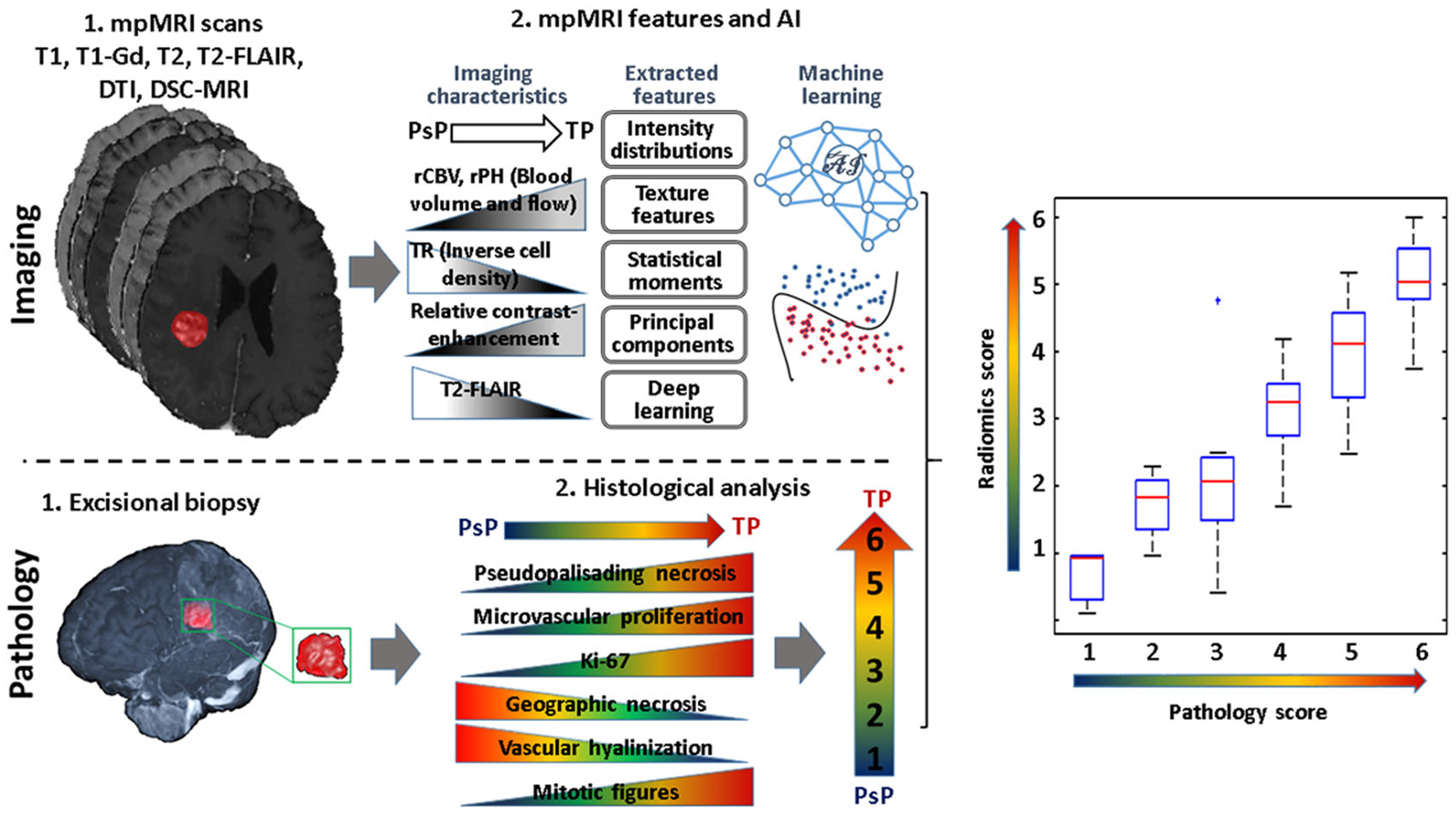
Processing pipeline of the artificial intelligence radiographic biomarkers and histopathologic analyses. Top row (imaging): (1) multiparametric (mp) MRI scans: T1-weighted precontrast and postcontrast, T2-weighted, T2 fluid-attenuated inversion recovery (T2-FLAIR), diffusion tensor imaging (DTI), dynamic susceptibility contrast (DSC)-MRI and defining the resected tissues; (2) defining the resected enhancing tissues after registration of the preoperative with postoperative images using Deformable Registration Attribute Matching and Mutual-Saliency weighting (DRAMMS) software. Features are extracted from each region, quantifying intensity, shape, principal component analysis, statistics, and texture. Machine learning analysis is performed. Bottom row (pathology): (1) excisional biopsy; (2) histological characteristics and defined pathology scores from resected tumor specimen. Right panel: a plot showing a correlation between pathology scores and imaging scores via artificial intelligence (radiomics scores)
As radiomics is a relatively young and evolving field, it is associated with some technical challenges and limitations that hamper its implementation in routine clinical settings. Owing to variations in data acquisition, data processing schemes and lack of external validations, the findings from ML methods may not always be generalized or transferred from one patient cohort to another patient cohort. The issue of reproducibility of radiomic features has become even more pertinent in retrospective studies involving publicly available repositories such as the cancer genome atlas-GBM (TCGA-GBM) and Ivy GBM atlas project (Ivy-GAP), where imaging data are collected and archived from different MR systems and sites. Currently, several efforts are underway to standardize the image acquisition protocols, data processing pipelines, and radiomic feature extraction modules across multiple sites. Additionally, the availability of open-source platforms such as the Cancer Imaging Phenomics Toolkit, which was specifically developed for neuro-oncology applications,140 and the accessibility of more widespread packages such as Pyradiomics,141 can provide the research community with a standardized workflow for radiomic feature extraction and analysis.
As GBMs are extremely heterogenous in nature, the critical aspect of the radiomic pipeline is the segmentation of such tumors into entire tumor volumes or subvolumes known as “tumor habitats”. A variety of problems that complicates image segmentation of a GBM includes normal anatomic variations, postsurgical anatomic variations, neoplastic infiltration and irregular tumor boundaries, inadequate contrast, as well as the presence of tissue and technical artifacts within an image. Manual tumor segmentation is not only a labor-intensive process but is also impacted by interobserver variability and subjectivity.142 While some radiomic studies have used automatic and semiautomatic methods for segmentation,143 the existing segmentation algorithms are not consistent among different research groups and can have a substantial impact on the radiomic analyses as well as on developing reliable, and reproducible, diagnostic, predictive, and prognostic models.
Another potential problem in the field of radiomics concerns the small data size, which increases the risk of overfitting the data. Relatively small datasets can lead to significant variability or bias; thus, an ML algorithm trained based on a small dataset may suffer from lower performance and reproducibility when applied to a large dataset. To solve this issue, retraining the algorithm is generally required. Another possible solution is utilizing incremental learning and adjusting the computerized systems in an automatic way. Random sampling, choosing simple models, and removing outliers from data are some other potential strategies that can be used with small datasets to improve the accuracy of a classification model.144
We believe that with continuous progress in ML algorithms and the availability of large datasets, the upcoming field of radiomics and ML will have a vital role in evaluating treatment response in GBM patients with high accuracy.
3 |. CONCLUSIONS
While distinguishing TP from PsP in GBMs, large variations in the diagnostic performances of metabolic and physiologic MRI parameters have been observed from several previous studies. These variations may be attributed to several factors, including inherent spatial and temporal heterogeneity of GBMs, small sample size, variability and subjectivity in the tissue segmentation methods, and inconsistencies in drawing the ROIs from different tissue compartments of tumors. Notwithstanding some of the above-described shortcomings and unresolved issues, metabolic and physiological MRI techniques provide quantifiable, unbiased, and physiologically relevant information in post-treatment characterization of GBMs. When used in combination (i.e., a multiparametric approach), these advanced MRI techniques offer more comprehensive information with enhanced accuracy in differentiating TP from PsP. However, data acquisition protocols for MRI and PET imaging are variable across different imaging sites, which adversely impact the generalizability of imaging findings. To facilitate widespread clinical acceptance, standardization, and harmonization of methodology, guidelines have been provided for data acquisition and analysis, quality assessment and data interpretation for MR diffusion,145 perfusion,146 spectroscopy,147,148 and PET imaging149 techniques. Additional improvement in this field requires data sharing, and large multi-institutional and validation studies.
FIGURE 4.
Axial MR images from a glioblastoma patient with pseudoprogression (PsP). The postcontrast T1 (PC-T1)-weighted image shows a ring-enhancing lesion in the right parietal lobe. Coregistered diffusion tensor imaging-derived parametric maps and cerebral blood volume (CBV) maps are shown. Decreased mean diffusivity (MD) and coefficient of spherical anisotropy (CS), and increased fractional anisotropy (FA), coefficient of linear anisotropy (CL), and coefficient of planar anisotropy (CP), are noticed from the enhancing part compared with normal white matter. Please also note the presence of lower CBV from contrast-enhancing regions compared with that from a true progression (TP) patient as shown above suggesting a lower degree of perfusion and neovascularization in PsP compared with TP. FLAIR, fluid-attenuated inversion recovery
ACKNOWLEDGMENTS
The authors are deeply grateful to Dr. Beverly Collins, Department of Radiology, Perelman School of Medicine at the University of Pennsylvania, Philadelphia, PA, USA, for graciously reviewing and proofreading the final version of the manuscript. This work was supported by funding obtained from the Penn Center for Precision Medicine (PCPM), Perelman School of Medicine at the University of Pennsylvania, Philadelphia, USA (PI: Sanjeev Chawla, PhD, DABMP).
Abbreviations used:
- 1H MRS
proton MR spectroscopy
- 3D
three-dimensional
- AD
axial diffusivity
- ADC
apparent diffusion coefficient
- APT
amide proton transfer
- CCRT
concurrent chemoradiation therapy
- CER
contrast-enhancing region
- CEST
chemical exchange saturation transfer
- Cho
choline
- CL
coefficient of linear anisotropy
- CP
coefficient of planar anisotropy
- Cr
creatine
- CS
coefficient of spherical anisotropy
- DCE
dynamic contrast enhanced
- DKI
diffusion kurtosis imaging
- DPR
distal peritumoral region
- DRAMMS
Deformable Registration Attribute Matching and Mutual-Saliency weighting
- DSC
dynamic susceptibility contrast
- DTI
diffusion tensor imaging
- DWI
diffusion-weighted imaging
- EPSI
echo planar spectroscopic imaging
- FA
fractional anisotropy
- FDM
functional diffusion map
- FET
fluoroethyl-L-tyrosine
- FLAIR
fluid-attenuated inversion recovery
- FLT
fluorothymidine
- GBCA
gadolinium-based contrast agent
- GBM
glioblastoma
- IDH
isocitrate dehydrogenase
- IPR
immediate peritumoral region
- IVIM
intravoxel incoherent motion
- Ktrans
volume transfer constant
- MD
mean diffusivity
- MET
methyl-L-methionine
- MGMT
O6-methylguanine-DNA methyltransferase
- MIDAS
metabolite imaging and data analysis system
- MK
mean kurtosis
- MPRAGE
magnetization-prepared rapid acquisition of gradient echo
- MRI
magnetic resonance imaging
- MT
magnetization transfer
- MV
maximum value
- NAA
N-acetylaspartate
- nCBV90
90th percentile of normalized cerebral blood volume
- NOE
nuclear Overhauser enhancement
- PET
positron emission tomography
- PHP
peak height position
- PRM
parametric response map
- PsP
pseudoprogression
- PWI
perfusion-weighted imaging
- RANO
response assessment in neuro-oncology
- rCBV
relative cerebral blood volume
- RD
radial diffusivity
- RN
radiation necrosis
- ROI
region of interest
- TMZ
temozolomide
- TP
true progression
- TRAM
treatment response assessment map
- TTFields
tumor-treating fields
- Ve
volume fraction of extravascular-extracellular space in tissues
- Vp
volume fraction of plasma space in tissues
REFERENCES
- 1.Ostrom QT, Gittleman H, Farah P, et al. CBTRUS statistical report: Primary brain and central nervous system tumors diagnosed in the United States in 2006–2010. Neuro Oncol. 2013;15(Suppl 2):1–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Rossignol J, Srinageshwar B, Dunbar GL. Current therapeutic strategies for glioblastoma. Brain Sci. 2019;10(1):15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Li R, Li H, Yan W, et al. Genetic and clinical characteristics of primary and secondary glioblastoma is associated with differential molecular subtype distribution. Oncotarget. 2015;6:7318–7324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Topkan E, Topuk S, Oymak E, et al. Pseudoprogression in patients with glioblastoma multiforme after concurrent radiotherapy and temozolomide. Am J Clin Oncol. 2012;35:284–289. [DOI] [PubMed] [Google Scholar]
- 5.Chaskis C, Neyns B, Michotte A, et al. Pseudoprogression after radiotherapy with concurrent temozolomide for high-grade glioma: clinical observations and working recommendations. Surg Neurol. 2009;72:423–428. [DOI] [PubMed] [Google Scholar]
- 6.Thust SC, van den Bent MJ, Smits M. Pseudoprogression of brain tumors. J Magn Reson Imaging. 2018;48(3):571–589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Franceschi E, Tosoni A, Pozzati E, Brandes AA. Association between response to primary treatments and MGMT status in glioblastoma. Expert Rev Anticancer Ther. 2008;8:1781–1786. doi: 10.1586/14737140.8.11.1781 [DOI] [PubMed] [Google Scholar]
- 8.Fink J, Born D, Chamberlain MC. Pseudoprogression: relevance with respect to treatment of high-grade gliomas. Curr Treat Options Oncol. 2011;12: 240–252. doi: 10.1007/s11864-011-0157-1 [DOI] [PubMed] [Google Scholar]
- 9.Motegi H, Kamoshima Y, Terasaka S, et al. IDH1 mutation as a potential novel biomarker for distinguishing pseudoprogression from true progression in patients with glioblastoma treated with temozolomide and radiotherapy. Brain Tumor Pathol. 2013;30:67–72. [DOI] [PubMed] [Google Scholar]
- 10.Brandsma D, van den Bent MJ. Pseudoprogression and pseudoresponse in the treatment of gliomas. Curr Opin Neurol. 2009;22:633–638. [DOI] [PubMed] [Google Scholar]
- 11.Roldan GB, Scott JN, McIntyre JB, et al. Population-based study of pseudoprogression after chemoradiotherapy in GBM. Can J Neurol Sci. 2009;36: 617–622. doi: 10.1017/S0317167100008131 [DOI] [PubMed] [Google Scholar]
- 12.Gerstner ER, McNamara MB, Norden AD, et al. Effect of adding temozolomide to radiation therapy on the incidence of pseudo-progression. J Neurooncol. 2009;94:97–101. [DOI] [PubMed] [Google Scholar]
- 13.Wen PY, Macdonald DR, Reardon DA, et al. Updated response assessment criteria for high-grade gliomas: response assessment in neuro-oncology working group. J Clin Oncol. 2010;28:1963–1972. [DOI] [PubMed] [Google Scholar]
- 14.Hygino da Cruz LC Jr, Rodriguez I, Domingues RC, et al. Pseudoprogression and pseudoresponse: imaging challenges in the assessment of posttreatment glioma. Am J Neuroradiol. 2011;32:1978–1985. doi: 10.3174/ajnr.A2397 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kazda T, Hardie JG, Pafundi DH, et al. Evaluation of RANO response criteria compared to clinician evaluation in WHO grade III anaplastic astrocytoma: implications for clinical trial reporting and patterns of failure. J Neurooncol. 2015;122:197–203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dempsey MF, Condon BR, Hadley DM. Measurement of tumor “size” in recurrent malignant glioma: 1D, 2D, or 3D? Am J Neuroradiol. 2005;26: 770–776. [PMC free article] [PubMed] [Google Scholar]
- 17.Zach L, Guez D, Last D, et al. Delayed contrast extravasation MRI: a new paradigm in neuro-oncology. Neuro Oncol. 2015;17:457–465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Daniels D, Guez D, Last D, et al. Early biomarkers from conventional and delayed-contrast MRI to predict the response to Bevacizumab in recurrent high-grade gliomas. Am J Neuroradiol. 2016;37:2003–2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bonm AV, Ritterbusch R, Throckmorton P, et al. Clinical imaging for diagnostic challenges in the management of gliomas: a review. J Neuroimaging. 2020;30:139–145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Li C, Gan Y, Chen H, et al. Advanced multimodal imaging in differentiating glioma recurrence from post-radiotherapy changes. Int Rev Neurobiol. 2020;151:281–297. [DOI] [PubMed] [Google Scholar]
- 21.Goncalves FG, Chawla S, Mohan S. Emerging MRI techniques to redefine treatment response in patients with glioblastoma. J Magn Reson Imaging. 2020;52:978–997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chawla S, Kim S, Wang S, et al. Diffusion-weighted imaging in head and neck cancers. Future Oncol. 2009;5:959–975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kim M, Kim HS. Emerging techniques in brain tumor imaging: what radiologists need to know. Korean J Radiol. 2016;17:598–619. doi: 10.3348/kjr.2016.17.5.598 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sugahara T, Korogi Y, Kochi M, et al. Usefulness of diffusion-weighted MRI with echo-planar technique in the evaluation of cellularity in gliomas. J Magn Reson Imaging. 1999;9:53–60. doi: [DOI] [PubMed] [Google Scholar]
- 25.Lee WJ, Choi SH, Park CK, et al. Diffusion-weighted MR imaging for the differentiation of true progression from pseudoprogression following concomitant radiotherapy with temozolomide in patients with newly diagnosed high-grade gliomas. Acad Radiol. 2012;19:1353–1361. [DOI] [PubMed] [Google Scholar]
- 26.Hein PA, Eskey CJ, Dunn JF, et al. Diffusion-weighted imaging in the follow-up of treated high-grade gliomas: tumor recurrence versus radiation injury. Am J Neuroradiol. 2004;25:201–209. [PMC free article] [PubMed] [Google Scholar]
- 27.Brandes AA, Franceschi E, Tosoni A, et al. MGMT promoter methylation status can predict the incidence and outcome of pseudoprogression after concomitant radiochemotherapy in newly diagnosed glioblastoma patients. J Clin Oncol. 2008;26:2192–2197. [DOI] [PubMed] [Google Scholar]
- 28.Chu HH, Choi SH, Ryoo I, et al. Differentiation of true progression from pseudoprogression in glioblastoma treated with radiation therapy and concomitant temozolomide: comparison study of standard and high-b-value diffusion-weighted imaging. Radiology. 2013;269:831–840. [DOI] [PubMed] [Google Scholar]
- 29.Asao C, Korogi Y, Kitajima M, Hirai T, et al. Diffusion-weighted imaging of radiation-induced brain injury for differentiation from tumor recurrence. Am J Neuroradiol. 2005;26:1455–1460. [PMC free article] [PubMed] [Google Scholar]
- 30.Reimer C, Deike K, Graf M, et al. Differentiation of pseudoprogression and real progression in glioblastoma using ADC parametric response maps. PLoS One. 2017;12:e0174620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hamstra DA, Galban CJ, Meyer CR, et al. Functional diffusion map as an early imaging biomarker for high-grade glioma: correlation with conventional radiologic response and overall survival. J Clin Oncol. 2008;26:3387–3394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wang S, Martinez-Lage M, Sakai Y, et al. Differentiating tumor progression from pseudoprogression in patients with glioblastomas using diffusion tensor imaging and dynamic susceptibility contrast MRI. Am J Neuroradiol. 2016;37:28–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wu XF, Liang X, Wang XC, et al. Differentiating high-grade glioma recurrence from pseudoprogression: Comparing diffusion kurtosis imaging and diffusion tensor imaging. Eur J Radiol. 2021;135:109445. [DOI] [PubMed] [Google Scholar]
- 34.Kim HS, Kim JH, Kim SH, et al. Posttreatment high-grade glioma: usefulness of peak height position with semiquantitative MR perfusion histogram analysis in an entire contrast-enhanced lesion for predicting volume fraction of recurrence. Radiology. 2010;256:906–915. [DOI] [PubMed] [Google Scholar]
- 35.Mangla R, Singh G, Ziegelitz D, et al. Changes in relative cerebral blood volume 1 month after radiation-temozolomide therapy can help predict overall survival in patients with glioblastoma. Radiology. 2010;256:575–584. [DOI] [PubMed] [Google Scholar]
- 36.Kong DS, Kim ST, Kim EH, et al. Diagnostic dilemma of pseudoprogression in the treatment of newly diagnosed glioblastomas: the role of assessing relative cerebral blood flow volume and oxygen-6-methylguanine-DNA methyltransferase promoter methylation status. Am J Neuroradiol. 2011;32: 382–387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Prager AJ, Martinez N, Beal K, et al. Diffusion and perfusion MRI to differentiate treatment-related changes including pseudoprogression from recurrent tumors in high-grade gliomas with histopathologic evidence. Am J Neuroradiol. 2015;36:877–885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Boxerman JL, Ellingson BM, Jeyapalan S, et al. Longitudinal DSC-MRI for distinguishing tumor recurrence from pseudoprogression in patients with a high-grade glioma. Am J Clin Oncol. 2017;40:228–234. doi: 10.1097/COC.0000000000000156 [DOI] [PubMed] [Google Scholar]
- 39.Baek HJ, Kim HS, Kim N, et al. Percent change of perfusion skewness and kurtosis: a potential imaging biomarker for early treatment response in patients with newly diagnosed glioblastomas. Radiology. 2012;264:834–843. [DOI] [PubMed] [Google Scholar]
- 40.Hu LS, Eschbacher JM, Heiserman JE, et al. Reevaluating the imaging definition of tumor progression: perfusion MRI quantifies recurrent glioblastoma tumor fraction, pseudoprogression, and radiation necrosis to predict survival. Neuro Oncol. 2012;14:919–930. doi: 10.1093/neuonc/nos112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Iv M, Liu X, Lavezo J, et al. Perfusion MRI-based fractional tumor burden differentiates between tumor and treatment effect in recurrent glioblastomas and informs clinical decision-making. Am J Neuroradiol. 2019;40:1649–1657. doi: 10.3174/ajnr.A6211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Thomas AA, Arevalo-Perez J, Kaley T, et al. Dynamic contrast enhanced T1 MRI perfusion differentiates pseudoprogression from recurrent glioblastoma. J Neurooncol. 2015;125:183–190. doi: 10.1007/s11060-015-1893-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Verma G, Chawla S, Mohan S, et al. Three-dimensional echo planar spectroscopic imaging for differentiation of true progression from pseudoprogression in patients with glioblastoma. NMR Biomed. 2019;32:e4042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ma B, Blakeley JO, Hong X, et al. Applying amide proton transfer-weighted MRI to distinguish pseudoprogression from true progression in malignant gliomas. J Magn Reson Imaging. 2016;44:456–462. doi: 10.1002/jmri.25159 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kebir S, Fimmers R, Galldiks N, et al. Late pseudoprogression in glioblastoma: diagnostic value of dynamic O-(2-[18F]fluoroethyl)-L-tyrosine PET. Clin Cancer Res. 2016;22:2190–2196. doi: 10.1158/1078-0432.CCR-15-1334 [DOI] [PubMed] [Google Scholar]
- 46.Werner JM, Weller J, Ceccon G, et al. Diagnosis of pseudoprogression following lomustine-temozolomide chemoradiation in newly diagnosed glioblastoma patients using FET-PET. Clin Cancer Res. 2021;27:3704–3713. doi: 10.1158/1078-0432.CCR-21-0471 [DOI] [PubMed] [Google Scholar]
- 47.Lohmann P, Elahmadawy MA, Gutsche R, et al. FET PET radiomics for differentiating pseudoprogression from early tumor progression in glioma patients post-chemoradiation. Cancers. 2020;12:3835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Gerstner ER, Sorensen AG. Diffusion and diffusion tensor imaging in brain cancer. Semin Radiat Oncol. 2011;21:141–146. doi: 10.1016/j.semradonc.2010.10.005 [DOI] [PubMed] [Google Scholar]
- 49.Abdullah KG, Lubelski D, Nucifora PG, et al. Use of diffusion tensor imaging in glioma resection. Neurosurg Focus. 2013;34:E1. [DOI] [PubMed] [Google Scholar]
- 50.Lazar M Mapping brain anatomical connectivity using white matter tractography. NMR Biomed. 2010;23:821–835. doi: 10.1002/nbm.1579 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wang S, Kim S, Chawla S, et al. Differentiation between glioblastomas and solitary brain metastases using diffusion tensor imaging. Neuroimage. 2009;44:653–660. doi: 10.1016/j.neuroimage.2008.09.027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wang S, Kim S, Chawla S, et al. Differentiation between glioblastomas, solitary brain metastases, and primary cerebral lymphomas using diffusion tensor and dynamic susceptibility. Am J Neuroradiol. 2011;32:507–514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Chawla S, Wang S, Mohan S, et al. Differentiation of brain infection from necrotic glioblastoma using combined analysis of diffusion and perfusion MRI. J Magn Reson Imaging. 2019;49:184–194. [DOI] [PubMed] [Google Scholar]
- 54.Wang S, O’Rourke DM, Chawla S, et al. Multiparametric magnetic resonance imaging in the assessment of anti-EGFRvIII chimeric antigen receptor T cell therapy in patients with recurrent glioblastoma. Br J Cancer. 2019;120:54–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Agarwal A, Kumar S, Narang J, et al. Morphologic MRI features, diffusion tensor imaging and radiation dosimetric analysis to differentiate pseudo-progression from early tumor progression. J Neurooncol. 2013;112:413–420. doi: 10.1007/s11060-013-1070-1 [DOI] [PubMed] [Google Scholar]
- 56.Fieremans E, Jensen JH, Helpern JA. White matter characterization with diffusional kurtosis imaging. Neuroimage. 2011;58:177–188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Jensen JH, Helpern JA, Ramani A, et al. Diffusional kurtosis imaging: the quantification of non-gaussian water diffusion by means of magnetic resonance imaging. Magn Reson Med. 2005;53:1432–1440. [DOI] [PubMed] [Google Scholar]
- 58.Steven AJ, Zhuo J, Melhem ER. Diffusion kurtosis imaging: an emerging technique for evaluating the microstructural environment of the brain. Am J Roentgenol. 2014;202:W26–W33. [DOI] [PubMed] [Google Scholar]
- 59.Tan Y, Zhang H, Zhao RF, et al. Comparison of the values of MRI diffusion kurtosis imaging and diffusion tensor imaging in cerebral astrocytoma grading and their association with aquaporin-4. Neurol India. 2016;64:265–272. [DOI] [PubMed] [Google Scholar]
- 60.Pogosbekian EL, Pronin IN, Zakharova NE, et al. Feasibility of generalised diffusion kurtosis imaging approach for brain glioma grading. Neuroradiology. 2021;63(8):1241–1251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Liu ZC, Yan LF, Hu YC, et al. Combination of IVIM-DWI and 3D-ASL for differentiating true progression from pseudoprogression of glioblastoma multiforme after concurrent chemoradiotherapy: study protocol of a prospective diagnostic trial. BMC Med Imaging. 2017;17:10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Paschoal AM, Leoni RF, Dos Santos AC, et al. Intravoxel incoherent motion MRI in neurological and cerebrovascular diseases. NeuroImage Clin. 2018; 20:705–714. doi: 10.1016/j.nicl.2018.08.030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Wang N, Jain RK, Batchelor TT. New directions in anti-angiogenic therapy for glioblastoma. Neurotherapeutics. 2017;14:321–332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Jahng GH, Li KL, Ostergaard L, Calamante F. Perfusion magnetic resonance imaging: a comprehensive update on principles and techniques. Korean J Radiol. 2014;15:554–577. doi: 10.3348/kjr.2014.15.5.554 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Hu LS, Baxter LC, Smith KA, et al. Relative cerebral blood volume values to differentiate high-grade glioma recurrence from posttreatment radiation effect: direct correlation between image-guided tissue histopathology and localized dynamic susceptibility-weighted contrast-enhanced perfusion MR imaging measurements. Am J Neuroradiol. 2009;30:552–558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Tsien C, Galban CJ, Chenevert TL, et al. Parametric response map as an imaging biomarker to distinguish progression from pseudoprogression in high-grade glioma. J Clin Oncol. 2010;28:2293–2299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Prah MA, Al-Gizawiy MM, Mueller WM, et al. Spatial discrimination of glioblastoma and treatment effect with histologically-validated perfusion and diffusion magnetic resonance imaging metrics. J Neurooncol. 2018;136:13–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Gahramanov S, Raslan AM, Muldoon LL, et al. Potential for differentiation of pseudoprogression from true tumor progression with dynamic susceptibility-weighted contrast-enhanced magnetic resonance imaging using ferumoxytol vs. gadoteridol: a pilot study. Int J Radiat Oncol Biol Phys. 2011;79:514–523. doi: 10.1016/j.ijrobp.2009.10.072 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Skolnik AD, Wang S, Gopal PP, Mohan S. Commentary: pitfalls in the neuroimaging of glioblastoma in the era of antiangiogenic and immuno/targeted therapy. Front Neurol. 2018;9:51. doi: 10.3389/fneur.2018.00051 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Sourbron SP, Buckley DL. Classic models for dynamic contrast-enhanced MRI. NMR Biomed. 2013;26:1004–1027. [DOI] [PubMed] [Google Scholar]
- 71.Tofts PS, Brix G, Buckley DL, et al. Estimating kinetic parameters from dynamic contrast-enhanced T(1)-weighted MRI of a diffusable tracer: standardized quantities and symbols. J Magn Reson. 1999;10:223–232. doi: [DOI] [PubMed] [Google Scholar]
- 72.Meyer HJ, Leifels L, Schob S, et al. Histogram analysis parameters identify multiple associations between DWI and DCE MRI in head and neck squamous cell carcinoma. Magn Reson Imaging. 2018;45:72–77. [DOI] [PubMed] [Google Scholar]
- 73.Yun TJ, Park CK, Kim TM, et al. Glioblastoma treated with concurrent radiation therapy and temozolomide chemotherapy: differentiation of true progression from pseudoprogression with quantitative dynamic contrast-enhanced MR imaging. Radiology. 2015;274:830–840. [DOI] [PubMed] [Google Scholar]
- 74.Liu X, Tian W, Kolar B, et al. MR diffusion tensor and perfusion-weighted imaging in preoperative grading of supratentorial nonenhancing gliomas. Neuro Oncol. 2011;13:447–455. doi: 10.1093/neuonc/noq197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Masch WR, Wang PI, Chenevert TL, et al. Comparison of diffusion tensor imaging and magnetic resonance perfusion imaging in differentiating recurrent brain neoplasm from radiation necrosis. Acad Radiol. 2016;23:569–576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Li C, Wang S, Yan JL, et al. Characterizing tumor invasiveness of glioblastoma using multiparametric magnetic resonance imaging. J Neurosurg. 2019; 132:1465–1472. [DOI] [PubMed] [Google Scholar]
- 77.Stringfield O, Arrington JA, Johnston SK, et al. Multiparameter MRI predictors of long-term survival in glioblastoma multiforme. Tomography. 2019;5: 135–144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Fink JR, Carr RB, Matsusue E, et al. Comparison of 3 Tesla proton MR spectroscopy, MR perfusion and MR diffusion for distinguishing glioma recurrence from posttreatment effects. J Magn Reson Imaging. 2012;35:56–63. [DOI] [PubMed] [Google Scholar]
- 79.Bulik M, Jancalek R, Vanicek J, et al. Potential of MR spectroscopy for assessment of glioma grading. Clin Neurol Neurosurg. 2013;115:146–153. [DOI] [PubMed] [Google Scholar]
- 80.Ohba S, Murayama K, Abe M, et al. Magnetic resonance imaging and proton magnetic resonance spectroscopy for differentiating between enhanced gliomas and malignant lymphomas. World Neurosurg. 2019;127:e779–e787. [DOI] [PubMed] [Google Scholar]
- 81.Chawla S, Wang S, Wolf RL, et al. Arterial spin-labeling and MR spectroscopy in the differentiation of gliomas. Am J Neuroradiol. 2007;28: 1683–1689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Chawla S, Oleaga L, Wang S, et al. Role of proton magnetic resonance spectroscopy in differentiating oligodendrogliomas from astrocytomas. J Neuroimaging. 2010;20:3–8. doi: 10.1111/j.1552-6569.2008.00307.x [DOI] [PubMed] [Google Scholar]
- 83.Chawla S, Zhang Y, Wang S, et al. Proton magnetic resonance spectroscopy in differentiating glioblastomas from primary cerebral lymphomas and brain metastases. J Comput Assist Tomogr. 2010;34:836–841. [DOI] [PubMed] [Google Scholar]
- 84.Chawla S, Krejza J, Vossough A, et al. Differentiation between oligodendroglioma genotypes using dynamic susceptibility contrast perfusion-weighted imaging and proton MR spectroscopy. Am J Neuroradiol. 2013;34:1542–1549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Li BS, Wang H, Gonen O. Metabolite ratios to assumed stable creatine level may confound the quantification of proton brain MR spectroscopy. Magn Reson Imaging. 2003;21:923–928. [DOI] [PubMed] [Google Scholar]
- 86.Di Costanzo A, Scarabino T, Trojsi F, et al. Proton MR spectroscopy of cerebral gliomas at 3 T: spatial heterogeneity, and tumour grade and extent. Eur Radiol. 2008;18:1727–1735. [DOI] [PubMed] [Google Scholar]
- 87.Rabinov JD, Lee PL, Barker FG, et al. In vivo 3-T MR spectroscopy in the distinction of recurrent glioma versus radiation effects: initial experience. Radiology. 2002;225:871–879. [DOI] [PubMed] [Google Scholar]
- 88.Schlemmer HP, Bachert P, Henze M, et al. Differentiation of radiation necrosis from tumor progression using proton magnetic resonance spectroscopy. Neuroradiology. 2002;44:216–222. [DOI] [PubMed] [Google Scholar]
- 89.Chernov MF, Ono Y, Abe K, et al. Differentiation of tumor progression and radiation-induced effects after intracranial radiosurgery. Acta Neurochir Suppl. 2013;116:193–210. [DOI] [PubMed] [Google Scholar]
- 90.McKnight TR. Proton magnetic resonance spectroscopic evaluation of brain tumor metabolism. Semin Oncol. 2004;31:605–617. [DOI] [PubMed] [Google Scholar]
- 91.Smith EA, Carlos RC, Junck LR, Tsien CI, Elias A, Sundgren PC. Developing a clinical decision model: MR spectroscopy to differentiate between recurrent tumor and radiation change in patients with new contrast-enhancing lesions. Am J Roentgenol. 2009;192:W45–W52. doi: 10.2214/AJR.07.3934 [DOI] [PubMed] [Google Scholar]
- 92.Wiebenga OT, Klauser AM, Nagtegaal GJ, et al. Longitudinal absolute metabolite quantification of white and gray matter regions in healthy controls using proton MR spectroscopic imaging. NMR Biomed. 2014;27:304–311. doi: 10.1002/nbm.3063 [DOI] [PubMed] [Google Scholar]
- 93.Alger JR. Quantitative proton magnetic resonance spectroscopy and spectroscopic imaging of the brain: a didactic review. Top Magn Reson Imaging. 2010;21:115–128. doi: 10.1097/RMR.0b013e31821e568f [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Jansen JF, Backes WH, Nicolay K, et al. 1H MR spectroscopy of the brain: absolute quantification of metabolites. Radiology. 2006;240:318–332. [DOI] [PubMed] [Google Scholar]
- 95.Durmo F, Rydelius A, Cuellar Baena S, et al. Multivoxel (1)H-MR spectroscopy biometrics for preoprerative differentiation between brain tumors. Tomography. 2018;4:172–181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Panigrahy A, Krieger MD, Gonzalez-Gomez I, et al. Quantitative short echo time 1H-MR spectroscopy of untreated pediatric brain tumors: preoperative diagnosis and characterization. Am J Neuroradiol. 2006;27:560–572. [PMC free article] [PubMed] [Google Scholar]
- 97.Brandsma D, Stalpers L, Taal W, et al. Clinical features, mechanisms, and management of pseudoprogression in malignant gliomas. Lancet Oncol. 2008;9:453–461. [DOI] [PubMed] [Google Scholar]
- 98.Sawlani V, Taylor R, Rowley K, et al. Magnetic resonance spectroscopy for differentiating pseudo-progression from true progression in GBM on concurrent chemoradiotherapy. Neuroradiol J. 2012;25:575–586. [DOI] [PubMed] [Google Scholar]
- 99.Ebel A, Soher BJ, Maudsley AA. Assessment of 3D proton MR echo-planar spectroscopic imaging using automated spectral analysis. Magn Reson Med. 2001;46:1072–1078. doi: 10.1002/mrm.1301 [DOI] [PubMed] [Google Scholar]
- 100.Maudsley AA, Darkazanli A, Alger JR, et al. Comprehensive processing, display and analysis for in vivo MR spectroscopic imaging. NMR Biomed. 2006;19:492–503. doi: 10.1002/nbm.1025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Roy B, Gupta RK, Maudsley AA, et al. Utility of multiparametric 3-T MRI for glioma characterization. Neuroradiology. 2013;55:603–613. doi: 10.1007/s00234-013-1145-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Parra NA, Maudsley AA, Gupta RK, et al. Volumetric spectroscopic imaging of glioblastoma multiforme radiation treatment volumes. Int J Radiat Oncol Biol Phys. 2014;90:376–384. doi: 10.1016/j.ijrobp.2014.03.049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Chawla S, Wang S, Kim S, et al. Radiation injury to the normal brain measured by 3D-echo-planar spectroscopic imaging and diffusion tensor imaging: initial experience. J Neuroimaging. 2015;25:97–104. [DOI] [PubMed] [Google Scholar]
- 104.Mohan S, Chawla S, Wang S, et al. Assessment of early response to tumor-treating fields in newly diagnosed glioblastoma using physiologic and metabolic MRI: initial experience. CNS Oncol. 2016;5:137–144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Shim H, Holder CA, Olson JJ. Magnetic resonance spectroscopic imaging in the era of pseudoprogression and pseudoresponse in glioblastoma patient management. CNS Oncol. 2013;2:393–396. doi: 10.2217/cns.13.39 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Zhou J, Lal B, Wilson DA, Laterra J, van Zijl PCM. Amide proton transfer (APT) contrast for imaging of brain tumors. Magn Reson Med. 2003;50: 1120–1126. doi: 10.1002/mrm.10651 [DOI] [PubMed] [Google Scholar]
- 107.Yan K, Fu Z, Yang C, et al. Assessing amide proton transfer (APT) MRI contrast origins in 9 L gliosarcoma in the rat brain using proteomic analysis. Mol Imaging Biol. 2015;17:479–487. doi: 10.1007/s11307-015-0828-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Zhou J, Heo HY, Knutsson L, et al. APT-weighted MRI: techniques, current neuro applications, and challenging issues. J Magn Reson Imaging. 2019; 50:347–364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Jiang S, Yu H, Wang X, et al. Molecular MRI differentiation between primary central nervous system lymphomas and high-grade gliomas using endogenous protein-based amide proton transfer MR imaging at 3 Tesla. Eur Radiol. 2016;26:64–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Kamimura K, Nakajo M, Yoneyama T, et al. Amide proton transfer imaging of tumors: theory, clinical applications, pitfalls, and future directions. Jap J Radiol. 2019;37:109–116. [DOI] [PubMed] [Google Scholar]
- 111.van Zijl PC, Yadav NN. Chemical exchange saturation transfer (CEST): what is in a name and what isn’t? Magn Reson Med. 2011;65:927–948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Zhang XY, Wang F, Li H, et al. Accuracy in the quantification of chemical exchange saturation transfer (CEST) and relayed nuclear Overhauser enhancement (rNOE) saturation transfer effects. NMR Biomed. 2017;30(7). doi: 10.1002/nbm.3716 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Werner JM, Lohmann P, Fink GR, et al. Current landscape and emerging fields of PET imaging in patients with brain tumors. Molecules. 2020;25(6): 1471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Goldberg MF, Chawla S, Alavi A, et al. PET and MR imaging of brain tumors. PET Clin. 2008;3:293–315. [DOI] [PubMed] [Google Scholar]
- 115.Som P, Atkins HL, Bandoypadhyay D, et al. A fluorinated glucose analog, 2-fluoro-2-deoxy-D-glucose (F-18): nontoxic tracer for rapid tumor detection. J Nucl Med. 1980;21:670–675. doi: 10.1097/00004728-198012000-00045 [DOI] [PubMed] [Google Scholar]
- 116.Drake LR, Hillmer AT, Cai Z. Approaches to PET imaging of glioblastoma. Molecules. 2020;25(3):568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Wiriyasermkul P, Nagamori S, Tominaga H, et al. Transport of 3-fluoro-L-alpha-methyl-tyrosine by tumor-upregulated L-type amino acid transporter 1: a cause of the tumor uptake in PET. J Nucl Med. 2012;53:1253–1261. [DOI] [PubMed] [Google Scholar]
- 118.Shields AF, Grierson JR, Dohmen BM, et al. Imaging proliferation in vivo with [F-18]FLT and positron emission tomography. Nat Med. 1998;4: 1334–1336. [DOI] [PubMed] [Google Scholar]
- 119.Brahm CG, den Hollander MW, Enting RH, et al. Serial FLT PET imaging to discriminate between true progression and pseudoprogression in patients with newly diagnosed glioblastoma: a long-term follow-up study. Eur J Nucl Med Mol Imaging. 2018;45:2404–2412. doi: 10.1007/s00259-018-4090-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Nowosielski M, DiFranco MD, Putzer D, et al. An intra-individual comparison of MRI, [18F]-FET and [18F]-FLT PET in patients with high-grade gliomas. PLoS One. 2014;9:e95830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Galldiks N, Lohmann P, Albert NL, et al. Current status of PET imaging in neuro-oncology. Neurooncol Adv. 2019;1:vdz010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Raucher D Tumor targeting peptides: novel therapeutic strategies in glioblastoma. Curr Opin Pharmacol. 2019;47:14–19. doi: 10.1016/j.coph.2019.01.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Hutterer M, Nowosielski M, Putzer D, et al. [18F]-fluoro-ethyl-L-tyrosine PET: a valuable diagnostic tool in neuro-oncology, but not all that glitters is glioma. Neuro Oncol. 2013;15:341–351. doi: 10.1093/neuonc/nos300 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Yang Z, Ling F, Ruan S, et al. Clinical and prognostic implications of 1p/19q, IDH, BRAF, MGMT promoter, and TERT promoter alterations, and expression of Ki-67 and p53 in human gliomas. Cancer Manag Res. 2021;13:8755–8765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Hegi ME, Diserens AC, Gorlia T, et al. MGMT gene silencing and benefit from temozolomide in glioblastoma. N Engl J Med. 2005;352:997–1003. [DOI] [PubMed] [Google Scholar]
- 126.Brandes AA, Tosoni A, Franceschi E, et al. Recurrence pattern after temozolomide concomitant with and adjuvant to radiotherapy in newly diagnosed patients with glioblastoma: correlation with MGMT promoter methylation status. J Clin Oncol. 2009;27:1275–1279. [DOI] [PubMed] [Google Scholar]
- 127.Balana C, Capellades J, Pineda E, et al. Pseudoprogression as an adverse event of glioblastoma therapy. Cancer Med. 2017;6:2858–2866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Moon WJ, Choi JW, Roh HG, et al. Imaging parameters of high grade gliomas in relation to the MGMT promoter methylation status: the CT, diffusion tensor imaging, and perfusion MR imaging. Neuroradiology. 2012;54:555–563. [DOI] [PubMed] [Google Scholar]
- 129.Romano A, Calabria LF, Tavanti F, et al. Apparent diffusion coefficient obtained by magnetic resonance imaging as a prognostic marker in glioblastomas: correlation with MGMT promoter methylation status. Eur Radiol. 2013;23:513–520. doi: 10.1007/s00330-012-2601-4 [DOI] [PubMed] [Google Scholar]
- 130.Choi YS, Ahn SS, Kim DW, et al. Incremental prognostic value of ADC histogram analysis over MGMT promoter methylation status in patients with glioblastoma. Radiology. 2016;281:175–184. [DOI] [PubMed] [Google Scholar]
- 131.Gupta A, Prager A, Young RJ, et al. Diffusion-weighted MR imaging and MGMT methylation status in glioblastoma: a reappraisal of the role of preoperative quantitative ADC measurements. Am J Neuroradiol. 2013;34:E10–E11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Han Y, Yan LF, Wang XB, et al. Structural and advanced imaging in predicting MGMT promoter methylation of primary glioblastoma: a region of interest based analysis. BMC Cancer. 2018;18:215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Yoon RG, Kim HS, Paik W, Shim WH, Kim SJ, Kim JH. Different diagnostic values of imaging parameters to predict pseudoprogression in glioblastoma subgroups stratified by MGMT promoter methylation. Eur Radiol. 2017;27:255–266. doi: 10.1007/s00330-016-4346-y [DOI] [PubMed] [Google Scholar]
- 134.Aerts HJ, Velazquez ER, Leijenaar RT, et al. Decoding tumour phenotype by noninvasive imaging using a quantitative radiomics approach. Nat Commun. 2014;5:4006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Qian X, Tan H, Zhang J, et al. Identification of biomarkers for pseudo and true progression of GBM based on radiogenomics study. Oncotarget. 2016; 7:55377–55394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Jang BS, Jeon SH, Kim IH, et al. Prediction of pseudoprogression versus progression using machine learning algorithm in glioblastoma. Sci Rep. 2018; 8:12516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Kim JY, Park JE, Jo Y, et al. Incorporating diffusion- and perfusion-weighted MRI into a radiomics model improves diagnostic performance for pseudoprogression in glioblastoma patients. Neuro Oncol. 2019;21:404–414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Akbari H, Rathore S, Bakas S, et al. Histopathology-validated machine learning radiographic biomarker for noninvasive discrimination between true progression and pseudo-progression in glioblastoma. Cancer. 2020;126:2625–2636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Ou Y, Akbari H, Bilello M, da X, Davatzikos C. Comparative evaluation of registration algorithms in different brain databases with varying difficulty: results and insights. IEEE Trans Med Imaging. 2014;33:2039–2065. doi: 10.1109/TMI.2014.2330355 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Rathore S, Mohan S, Bakas S, et al. Multi-institutional noninvasive in vivo characterization of IDH, 1p/19q, and EGFRvIII in glioma using neuro-Cancer Imaging Phenomics Toolkit (neuro-CaPTk). Neurooncol Adv. 2020;2:iv22–iv34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.van Griethuysen JJM, Fedorov A, Parmar C, et al. Computational radiomics system to decode the radiographic phenotype. Cancer Res. 2017;77: e104–e107. doi: 10.1158/0008-5472.CAN-17-0339 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Gillies RJ, Kinahan PE, Hricak H. Radiomics: images are more than pictures, they are data. Radiology. 2016;278:563–577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Kumar V, Gu Y, Basu S, et al. Radiomics: the process and the challenges. Magn Reson Imaging. 2012;30:1234–1248. doi: 10.1016/j.mri.2012.06.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Chaddad A, Kucharczyk MJ, Daniel P, et al. Radiomics in glioblastoma: current status and challenges facing clinical implementation. Front Oncol. 2019;9:374. doi: 10.3389/fonc.2019.00374 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Ellingson BM, Bendszus M, Boxerman J, et al. Consensus recommendations for a standardized brain tumor imaging protocol in clinical trials. Neuro Oncol. 2015;17:1188–1198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Welker K, Boxerman J, Kalnin A, et al. ASFNR recommendations for clinical performance of MR dynamic susceptibility contrast perfusion imaging of the brain. Am J Neuroradiol. 2015;36:E41–E51. doi: 10.3174/ajnr.A4341 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Lin A, Andronesi O, Bogner W, et al. Minimum Reporting Standards for in vivo Magnetic Resonance Spectroscopy (MRSinMRS): Experts’ consensus recommendations. NMR Biomed. 2021;34:e4484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Wilson M, Andronesi O, Barker PB, et al. Methodological consensus on clinical proton MRS of the brain: Review and recommendations. Magn Reson Med. 2019;82:527–550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Law I, Albert NL, Arbizu J, et al. Joint EANM/EANO/RANO practice guidelines/SNMMI procedure standards for imaging of gliomas using PET with radiolabelled amino acids and [(18)F]FDG: version 1.0. Eur J Nucl Med Mol Imaging. 2019;46:540–557. [DOI] [PMC free article] [PubMed] [Google Scholar]


