Abstract
Background
Listeriosis is a food-borne disease caused by the Gram-positive Bacillota (Firmicute) bacterium Listeria monocytogenes. Clinical L. monocytogenes isolates are often resistant to clinically used lincosamide clindamycin, thus excluding clindamycin as a viable treatment option.
Objectives
We have established newly developed lincosamide iboxamycin as a potential novel antilisterial agent.
Methods
We determined MICs of the lincosamides lincomycin, clindamycin and iboxamycin for L. monocytogenes, Enterococcus faecalis and Bacillus subtilis strains expressing synergetic antibiotic resistance determinants: ABCF ATPases that directly displace antibiotics from the ribosome and Cfr, a 23S rRNA methyltransferase that compromises antibiotic binding. For L. monocytogenes strains, either expressing VgaL/Lmo0919 or lacking the resistance factor, we performed time-kill kinetics and post-antibiotic effect assays.
Results
We show that the synthetic lincosamide iboxamycin is highly active against L. monocytogenes and can overcome the intrinsic lincosamide resistance mediated by VgaL/Lmo0919 ABCF ATPase. While iboxamycin is not bactericidal against L. monocytogenes, it displays a pronounced post-antibiotic effect, which is a valuable pharmacokinetic feature. We demonstrate that VmlR ABCF of B. subtilis grants significant (33-fold increase in MIC) protection from iboxamycin, while LsaA ABCF of E. faecalis grants an 8-fold protective effect. Furthermore, the VmlR-mediated iboxamycin resistance is cooperative with that mediated by the Cfr, resulting in up to a 512-fold increase in MIC.
Conclusions
While iboxamycin is a promising new antilisterial agent, our findings suggest that emergence and spread of ABCF ARE variants capable of defeating next-generation lincosamides in the clinic is possible and should be closely monitored.
Introduction
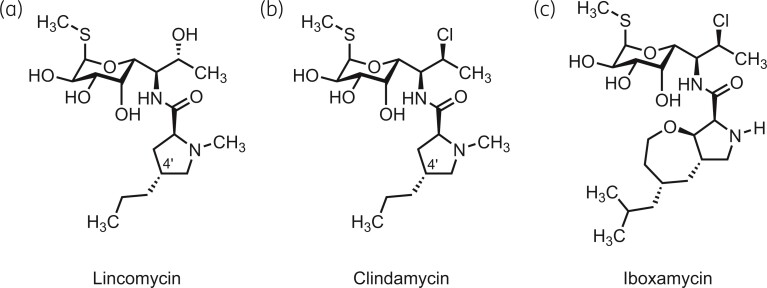
Lincosamides constitute an important class of antibiotics used both in veterinary and human medicine.1 These compounds inhibit protein synthesis by binding to and compromising the enzymatic activity of the peptidyl transferase centre (PTC) of the ribosome,2–5 resulting in bacteriostasis.6 Representatives of this antibiotic class share a common architecture and are typically comprised of a 4′-substituted l-proline residue connected via an amide bond to a unique S-glycosidic amino sugar moiety (Figure 1a and b). The first lincosamide to be discovered, lincomycin (Figure 1a), is a natural product produced by Streptomyces lincolnensis subsp. lincolnensis and is active against streptococcal, pneumococcal and staphylococcal infections.7 Its semi-synthetic derivative, clindamycin (Figure 1b), can be produced via a one-step stereoinvertive deoxychlorination of lincomycin.8 Clindamycin is more potent than lincomycin and is currently the lincosamide of choice for human medicine.9 Like lincomycin, clindamycin is mostly active against Gram-positive but not Gram-negative bacteria, which restricts the spectrum of its applications.10 A cis-4-ethyl-l-pipecolic acid amide of clindamycin, pirlimycin, has a similar spectrum of antibacterial activity11,12 and is approved for veterinary applications in the United States and European Union. Finally, a recently developed semisynthetic derivative of lincomycin (‘compound A’) was shown to be able to overcome clindamycin resistance in Staphylococcus aureus mediated by ribosomal RNA (rRNA) methylation by ErmA and ErmB antibiotic resistance determinants.13
Figure 1.
Chemical structures of lincosamide antibiotics lincomycin (a), clindamycin (b) and iboxamycin (c).
Iboxamycin (Figure 1c) is a newly developed lincosamide with an exceptionally broad spectrum of antibacterial activity.14 Featuring a fully synthetic, bicyclic oxepanoprolinamide aminoacyl fragment, iboxamycin improves upon previous lincosamides in its activity against both Gram-positive and Gram-negative pathogens.14 Iboxamycin was found to be more potent than clindamycin against Gram-positive pathogens and overcomes lincosamide resistance mediated by rRNA modification by Erm and Cfr 23S rRNA methyltransferases, both of which are highly clinically important and widespread antibiotic resistance determinants.15–18 While the presence of Cfr renders clinical isolates of S. aureus and Staphylococcus epidermidis virtually non-susceptible to clindamycin (MIC >128 mg/L), the resistance to iboxamycin conferred by Cfr (MIC of 2–8 mg/L compared with 0.06 mg/L for cfr−strains) is not sufficient to render the drug fully inactive in the context of infection treatment.14 Importantly, iboxamycin is also highly active against Enterococcus faecalis (MIC 0.06 mg/L as compared with 16 mg/L for clindamycin)—a species that is intrinsically resistant to ‘classical’ lincosamides as it encodes the LsaA antibiotic resistance (ARE) factor in its chromosomal genome,19 a member of the ABCF ATPase protein family that includes multiple resistance factors.20–22 LsaA provides resistance to pleuromutilin, lincosamide and streptogramin A (PLSA) antibiotics by displacing the drug from the ribosome,23 acting similarly to other ARE ABCFs.24–27 As evident from the 96- to 256-fold higher susceptibility to clindamycin and lincomycin in a ΔlsaA E. faecalis strain as compared with E. faecalis ectopically expressing LsaA,23 LsaA is a potent lincosamide resistance determinant. The high susceptibility of E. faecalis to iboxamycin suggests that this compound has the potential to overcome resistance mediated by other ARE ABCFs as well.
Listeriosis is a dangerous food-borne bacterial disease caused by the Gram-positive Bacillota (formerly Firmicute) bacterium Listeria monocytogenes, which infects people through contaminated meat, fish and dairy products.28,29 While it is a relatively rare infection that mainly affects people with weakened immune systems, or who are pregnant,30 the majority of listeriosis cases require hospitalization and mortality rates can be as high as 20%–30% even with antibiotic treatment.31,32 Antibiotic treatment options for L. monocytogenes infections include cell wall synthesis disruptors ampicillin and vancomycin, folic acid synthesis inhibitors sulfamethoxazole and trimethoprim, and protein synthesis inhibitors, such as gentamicin and azithromycin.33L. monocytogenes strains reported in recent years are often resistant to clindamycin, with the resistant fraction ranging from 29% to 76%, depending on the collection,34–37 thus excluding clindamycin as a viable option for treatment of L. monocytogenes infections. Importantly, just as E. faecalis encodes the ABCF ATPase LsaA, L. monocytogenes encodes the ARE ABCF PLSA resistance factor VgaL/Lmo0919 in its core genome.38 As with LsaA, VgaL operates on the ribosome,23 and loss of VgaL results in increased susceptibility to lincosamides, with the Δlmo0919 L. monocytogenes strain being 8- to 16-fold more susceptible to lincomycin as compared with the isogenic wild type (WT).23 Finally, a model Bacillota, Bacillus subtilis, also encodes an ARE ABCF PLSA resistance factor—VmlR.27,39
In this report, using lincomycin and clindamycin as reference compounds, we (i) characterized the efficacy of iboxamycin against L. monocytogenes; (ii) probed its ability to specifically counter resistance mediated by ARE ABCF Lmo0919 in L. monocytogenes, LsaA in E. faecalis and VmlR in B. subtilis; (iii) characterized its bactericidal/bacteriostatic mechanism of action; and, finally, (iv) assessed the strength of its post-antibiotic effect (PAE).
Materials and methods
Synthesis of iboxamycin
Iboxamycin was prepared according to the method reported by Mason and colleagues.40
Strains and media
WT L. monocytogenes 10403S was provided by Daniel A. Portnoy, WT L. monocytogenes EGD-e was provided by Jörgen Johansson, construction of L. monocytogenes EDG-e Δlmo0919 was described earlier,23E. faecalis ΔlsaA (lsa::Kan) strain TX533219 was provided by Barbara E. Murray, E. faecalis ΔlsaA pCIEspec and E. faecalis ΔlsaA pCIEspec LsaA were described earlier.23 WT 168 trpC B. subtilis (laboratory stock) was used. B. subtilis strains trpC ΔvmlR (VHB5) and ΔvmlR thrC::Phy-spank-vmlR (VHB44) were described earlier.27 To construct B. subtilis thrC::Phy-spank-cfr (VHB138) and ΔvmlR thrC::Phy-spank-cfr (VHB139), a PCR product encoding Staphylococcus sciuri cfr gene optimized to Escherichia coli codon usage41 was PCR-amplified from the pBRCfr plasmid using primers VHT25 (5′-CGGATAACAATTAAGCTTAGTCGACTTAAGGAGGTGTGTCTCATGAACTTTAACAACAAAACCAAATAC-3′) and VHT26 (5′-GTTTCCACCGAATTAGCTTGCATGCTCACTGGGAGTTCTGATAGTTACCATACA-3′). The second PCR fragment encoding a kanamycin-resistance marker, a polylinker downstream of the Phy-spank promoter and the lac repressor ORF—all inserted in the middle of the thrC gene—was PCR-amplified from pHT009 plasmid using primers pHT002_F (5′-GTCGACTAAGCTTAATTGTTATCCGCTCACAATTACACACATTATGCC-3′) and pHT002_R (5′-GCATGCAAGCTAATTCGGTGGAAACGAGGTCATC-3′). The two fragments were ligated using the NEBuilder HiFi DNA Assembly master mix (New England BioLabs, Ipswich, MA, USA) yielding the pHT009-cfr plasmid (VHp439), which was used to transform either WT 168 trpC2 or ΔvmlR (VHB5) strain. Selection for kanamycin resistance yielded the desired VHB138 and VHB139 strain.
Growth assays, MIC, cidality and PAE assays with L. monocytogenes were performed in MH-F broth, E. faecalis MIC assays were performed in BHI broth and B. subtilis MIC assays were performed in LB broth. The media were prepared as per EUCAST guidelines (https://www.eucast.org/fileadmin/src/media/PDFs/EUCAST_files/Disk_test_documents/2020_manuals/Media_preparation_v_6.0_EUCAST_AST.pdf) and contained 95% Mueller–Hinton broth (MHB) media (Sigma, Lot# BCCB5572), 5% lysed horse blood (defibrinated 50% stock, Hatunalab AB cat. no. 139) and 20 mg/mL β-NAD (Sigma, Lot# SLCD5502). Prior to use the 50% horse blood stock was freeze thawed five times and clarified via centrifugation twice for 30 min at 18 000 rpm at 4°C and then filtrated using 0.2 μm membrane filter, aliquotted and stored at −20°C. Solid agar plates were prepared from BHI broth media (VMR, Lot# G0113W) supplemented with 1% (final concentration) agar.
Liquid growth assays
L. monocytogenes was pre-grown on BHI agar plates at 37°C for 48 h. Individual fresh colonies were used to inoculate 2 mL of MH-F broth in 15 mL round bottom tubes, which were then incubated overnight at 37°C with shaking at 180 rpm. The overnight cultures were diluted then with MH-F broth to final OD600 of 0.005 and incubated for 8 h in a water bath shaker (Eppendorf™ Inova™ 3100 High-Temperature) at 37°C with shaking at 160 rpm. bacterial growth was monitored by OD600 measurements every 30 min.
Antibiotic susceptibility testing
The MIC antibiotic susceptibility testing was performed according to EUCAST guidelines (http://www.eucast.org/ast_of_bacteria/mic_determination), as described earlier.23
L. monocytogenes strains were grown in MH-F broth inoculated with 5 × 105 cfu/mL (OD600 of approximately 0.0015) with increasing concentrations of antibiotics. After 24–48 h of incubation at 37°C without shaking, the presence or absence of bacterial growth was scored by eye.
E. faecalis strains were grown in BHI media supplemented with 2 mg/mL kanamycin (to prevent lsa revertants), 0.1 mg/mL spectinomycin (to maintain the pCIEspec plasmid), 100 ng/mL of cCF10 peptide (to induce expression of LsaA protein) as well as increasing concentrations of antibiotics, was inoculated with 5 × 105 cfu/mL (OD600 of approximately 0.0005) of E. faecalis ΔlsaA (lsa::Kan) strain TX5332 transformed either with empty pCIEspec plasmid, or with pCIEspec encoding LsaA. After 16–20 h at 37°C without shaking, the presence or absence of bacterial growth was scored by eye.
B. subtilis strains were grown in LB medium supplemented with increasing concentrations of antibiotics. The cultures were inoculated with 5 × 105 cfu/mL (OD600 of approximately 0.0005), and after 16–20 h at 37°C without shaking the presence or absence of bacterial growth was scored by eye.
Time-kill kinetics assay
The protocol was based on CLSI42 and Svetlov et al.43 Exponential L. monocytogenes cultures in MH-F broth (OD600 ≈ 0.3) were diluted to 105 cfu/mL (OD600 = 0.001) in 10 mL of MH-F broth either supplemented with appropriate antibiotic at 4-fold MIC concentration or without antibiotics (positive growth control), and the resultant cultures were incubated at 37°C without shaking. Aliquots (1 mL) were taken at incremental incubation times (0, 2, 4, 6, 8 and 10 h), spun down at 4000 rpm for 5 min at room temperature and cell pellets were gently washed twice with 900 μL of 1 × PBS. Cell pellets were resuspended in 100 μL of 1 × PBS, 10-fold serial dilutions were prepared in 96-well plates (10−1–10−8), and 10 μL resultant 10-fold seral dilutions were spotted on BHI agar plates. Colony forming units were scored after 24–48 h incubation at 37°C.
PAE assay
Exponential cultures of L. monocytogenes strains in MH-F blood broth media (OD600 ≈ 0.3) were diluted to 105 cfu/mL (≈ OD600 of 0.001) in 5 mL of MH-F media either supplemented with appropriate antibiotic at 4-fold MIC concentration or without antibiotics (positive growth control) and incubated at 37°C without shaking for 2 h. After the 2 h pretreatment, antibiotics were removed by 1:100 dilution of 100 μL into 10 mL of fresh prewarmed MH-F blood broth media. At incremental timepoints (0, 2, 4, 6, 8 and 10 h), 1 mL of the 100 × diluted cell culture was harvested, centrifuged for 5 min at 4000 rpm, 900 μL of the medium was removed, and the pellets were resuspended in the remaining 100 μL. The volume was adjusted to 1 mL with 1 × PBS. Control cultures without antibiotics were handled similarly. Cell solutions were then serially diluted 10-fold to 10−8, and 10 μL was spotted on BHI agar plates. Plates for individual timepoints were incubated at room temperature until the last set of plates were spotted (10 h timepoint), and then incubated at 37°C. The plates were scored after 24 and 48 h incubation at 37°C and imaged using ImageQuant LAS 4000 (GE Healthcare). The last timepoint (24 h) was processed separately analogously to 0–10 h timepoints (see above).
Results
L. monocytogenes is highly susceptible to iboxamycin despite VgaL/Lmo0919 ABCF resistance factor
To test the lincosamide susceptibility of L. monocytogenes we used two widely used WT strains, both belonging to serovar 1/2a: EGD-e44 and 10403S, a streptomycin-resistant variant of 10403.45 The two WTs are genomically distinct, e.g. the virulence master-regulator PrfA is overexpressed in EGD-e and the prophage content differs between the two strains.46 In addition to the two WTs, we also tested an L. monocytogenes EDG-e derivative that was genomically modified to abrogate the expression of VgaL/Lmo0919 PLSA resistance factor (EDG-e Δlmo0919).23
Both WT L. monocytogenes strains are much more susceptible to iboxamycin (MIC of 0.125–0.5 mg/L) as compared with clindamycin (MIC of 1 mg/L) and lincomycin (MIC of 2–8 mg/L) (Table 1). In agreement with the higher susceptibility of Δlmo0919 EDG-e to lincomycin,23 this strain is 2–8-fold more susceptible to iboxamycin than the corresponding WT. This indicates that while VgaL does confer some protection from iboxamycin, the high potency of the synthetic antibiotic would likely allow the drug to overcome resistance in clinical settings. A likely explanation is that increased affinity of the synthetic drug for the ribosome renders antibiotic displacement by ABCF ATPases inefficient.
Table 1.
Broth microdilution MIC testing of lincosamide antibiotics against L. monocytogenes, E. faecalis and B. subtilis strains
| Species/strain | Antibiotic MIC, mg/L | ||
|---|---|---|---|
| lincomycin | clindamycin | iboxamycin | |
| L. monocytogenes 10403S | 4–8 | 2 | 0.125–0.25 |
| L. monocytogenes EDG-e | 8 | 1–2 | 0.125–0.5 |
| L. monocytogenes EDG-e Δlmo0919 | 0.25–1 | 0.125–0.5 | 0.0625 |
| E. faecalis ΔlsaA pCIEspec | 0.125 | 0.125 | 0.0625 |
| E. faecalis ΔlsaA pCIEspec LsaA | 16–32 | 16 | 0.5 |
| B. subtilis WT 168 | 80 | 4 | 2 |
| B. subtilis ΔvmlR | 2.5 | 0.125 | 0.06 |
| B. subtilis ΔvmlR thrC::Phy-spank-vmlR (IPTG: 1 mM) | 160 | 8 | 4 |
| B. subtilis thrC::Phy-spank-cfr (IPTG: 1 mM) | >640 | 640 | 16–32 |
| B. subtilis ΔvmlR thrC::Phy-spank-cfr (IPTG: 1 mM) | >640 | 320 | 2 |
In the case of L. monocytogenes strains, MIC testing was carried out in MH-F broth and growth inhibition was scored after 48 h incubation at 37°C. E. faecalis MIC testing was carried out in BHI broth supplemented with 2 mg/mL kanamycin (to prevent lsa revertants), 0.1 mg/mL spectinomycin (to maintain the pCIEspec plasmid) and 100 ng/mL of cCF10 peptide (to induce expression of LsaA protein). B. subtilis MIC testing was carried out in either LB medium or LB supplemented with 1 mM IPTG to induce expression of either VmlR or Cfr protein, and growth inhibition was scored after 16–20 h at 37°C. The MIC experiments were performed as three (L. monocytogenes strains) or two (B. subtilis and E. faecalis strains) biological replicates.
Importantly, expression of Lmo0919 is not constitutive: it is elicited by antibiotic-induced ribosomal stalling on the regulatory short open reading frame upstream of the lmo0919 gene.38 Therefore, the difference in iboxamycin susceptibility between WT and Δlmo0919 EDG-e strains reflects both the ability of Lmo0919 to protect the ribosome from the antibiotic as well as the efficiency of iboxamycin-mediated induction of Lmo0919. To deconvolute these two effects, in the following experiments we used engineered E. faecalis and B. subtilis strains that allow for ectopic inducible expression of ARE ABCFs: E. faecalis LsaA and B. subtilis VmlR, respectively.
E. faecalis ABCF LsaA grants a moderate protective effect against iboxamycin
To test the ability of other ABCF PLSA resistance factors to confer resistance to iboxamycin, we compared a pair of E. faecalis strains: one lacking the chromosomally encoded LsaA (ΔlsaA pCIEspec) and the other allowing cCF10-peptide-inducible expression of LsaA (ΔlsaA pCIEspec LsaA).23 Using this experimental set up, we could specifically assess the ability of LsaA to protect the strain from lincosamides. While expression of LsaA dramatically increases resistance to clindamycin and lincomycin (96- to 256-fold, respectively), it results in a mere 8-fold protective effect against iboxamycin (MIC of 0.0625 and 0.5 mg/L, respectively) (Table 1), demonstrating that iboxamycin can also largely overcome LsaA-mediated resistance. Our current results are in agreement with our earlier MIC measurements for WT E. faecalis lsaA+ OG1RF strain (lincomycin 32 mg/L, clindamycin 16–32 mg/L)23 and WT E. faecalis ATCC 29212 lsaA+ strain (clindamycin 16 mg/L and iboxamycin 0.6 mg/L).14
B. subtilis ABCF VmlR acts cooperatively with rRNA methyltransferase Cfr to grant significant protection against iboxamycin
Next we tested a set of B. subtilis strains: WT 168 B. subtilis, ΔvmlR (VHB5) as well as a ΔvmlR strain in which VmlR is expressed under the control of IPTG-inducible Phy-spank promoter (VHB44)47 (Table 1). Disruption of vmlR results in a 33-fold increase in iboxamycin susceptibility (MIC of 2 and 0.06 mg/L, respectively), and resistance is restored upon ectopic expression of VmlR (MIC of 4 mg/L, 2-fold increase over the WT levels). The iboxamycin susceptibility of Δlmo0919 L. monocytogenes EDG-e and ΔvmlR B. subtilis is near-identical, indicating that the 16-/4-fold difference in iboxamycin susceptibility between WT L. monocytogenes and B. subtilis is due to the different efficiency of resistance granted by Lmo0919 and VmlR, respectively.
Importantly, VmlR loss results in the same relative increase in susceptibility to all lincosamides tested—iboxamycin, clindamycin and lincomycin; 32–33-fold—regardless of the potency of the lincosamide (Table 1). This suggests that if the affinity of iboxamycin to the target were to be decreased by, for instance, rRNA modification, direct target protection by the ABCF could cooperatively lead to high levels of resistance. To probe this hypothesis, we have characterized the lincosamide susceptibility of B. subtilis strains that express Cfr 23S rRNA methyltransferase under the control of IPTG-inducible Phy-spank promotor, either in the presence or absence of the chromosomally encoded VmlR. Ectopic expression of Cfr in vmlR+ B. subtilis effected a cooperative resistance to iboxamycin, resulting in MICs of 16–32 mg/L as opposed to 2 mg/L when either of these resistance determinants are expressed individually (Table 1). As expected, Cfr also granted high levels of lincomycin and clindamycin resistance when ectopically expressed in both WT and ΔvmlR strains (MIC ranging from 320 to excess of 640 mg/L).
Taken together, our B. subtilis MIC results demonstrate that despite the cooperative action of the two resistance determinants, iboxamycin is a much more potent antibiotic against cfr+ abcf+ strains as compared with lincomycin and clindamycin.
Iboxamycin is bacteriostatic against L. monocytogenes and displays a strong PAE
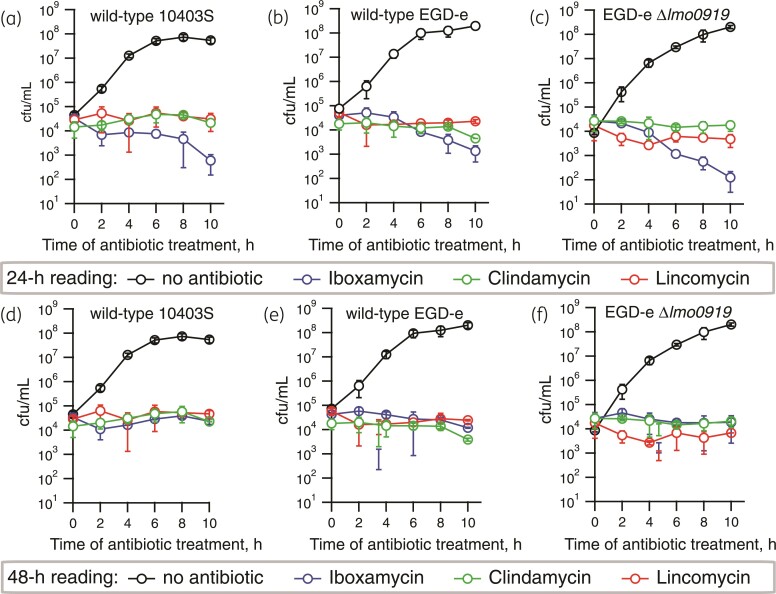
Macrolide antibiotics that tightly bind the ribosome and dissociate slowly are bactericidal, while macrolides that dissociate rapidly are bacteriostatic.43 As with lincomycin and clindamycin, iboxamycin was shown to be bacteriostatic against a panel of bacterial species.14 However, since effects on L. monocytogenes were not assessed in the original report—and the species is highly susceptible to iboxamycin—we tested for potential bactericidal effects of iboxamycin against this pathogen. The three L. monocytogenes strains that we used for the MIC measurements—WT 10403 and EGD-e as well as ABCF-deficient EDG-e Δlmo0919—were treated with 4 × MIC concentration of either iboxamycin, clindamycin and lincomycin for increasing periods of time (from 2 to 24 h), washed, and then plated on BHI agar plates that contained no antibiotic. The bacterial growth expressed in cfu was scored after either 24 or 48 h incubation of plates at 37°C. When the colony counting was performed after 24 h, we observed potentially bactericidal behaviour of iboxamycin, with almost a two log10 drop in cfu after the 10 h treatment with the antibiotic (Figure 2a–c). Importantly, no similar cfu decrease was observed for either clindamycin or lincomycin (Figure 2a–c). However, this apparent cfu drop effect of iboxamycin disappeared after 48 h of incubation (Figure 2d–f), suggesting slow regrowth rather than cidality, indicative of the so-called PAE.48,49
Figure 2.
Iboxamycin is bacteriostatic against L. monocytogenes. Exponentially growing L. monocytogenes type strains; 10403S (a and d), EDG-e (b and e) or VgaA-deficient EDG-e Δlmo0919 (c and e) were treated with 4 × MIC of either iboxamycin, clindamycin or lincomycin or no antibiotic as control. Cells were harvested at given timepoints and washed before plating. After 24 (a–c) or 48 h (d–f) of incubation, colonies were counted to determine cfu/mL. All experiments were carried out in MH-F broth at 37°C without shaking, data points are from three biological replicates and standard deviation is indicated with error bars.
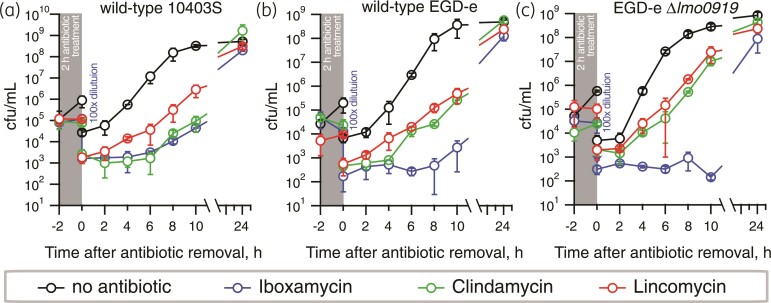
PAE is characterized by the time after antibiotic removal where no growth of the treated bacteria is observed. This prolonged action of iboxamycin has been previously noted for S. aureus and Enterococcus faecium.14 Therefore, we next performed PAE experiments in L. monocytogenes, demonstrating that, indeed, iboxamycin displays pronounced PAE, suppressing the growth of the WT 10403S and WT EGD-e for 6 and 8 h, respectively (Figure 3b and c). Clindamycin demonstrates a weaker PAE against EGD-e (2 h) and similar PAE against 10403S. No clear PAE is detectible for lincomycin. Compared with the isogenic WT, EDG-e Δlmo0919 displays similar PAE in the case of clindamycin, and, possibly, somewhat more pronounced PAE in the case of iboxamycin.
Figure 3.
Iboxamycin displays strong PAE against L. monocytogenes. To determine the time taken for antibiotic treated L. monocytogenes strains to resume growth after a 2 h antibiotic treatment, exponentially growing type strains; 10403S (a), EDG-e (b) or VgaA-deficient EDG-e Δlmo0919 (c) were treated with 4x MIC of either iboxamycin, clindamycin, lincomycin, or no antibiotic as control, for 2 h. Cells were then diluted by 100-fold to remove the antibiotic, and samples taken every 2 h subsequently for viability counting. All experiments were carried out in MH-F broth at 37°C with shaking at 180 rpm, data points are from three biological replicates and standard deviation is indicated with error bars.
Discussion
In this report we have evaluated the efficiency of the oxepanoprolinamide iboxamycin against L. monocytogenes. The antibiotic can largely overcome the intrinsic PLSA resistance of this species that is mediated by the ribosome-associated ATPase VgaL/Lmo0919, and can similarly counteract the intrinsic resistance mediated by ARE ABCF LsaA in E. faecalis. ARE ABCF PLSA resistance factors are broadly distributed among bacterial pathogens,20,22,50,51 and therefore the ability of iboxamycin to largely counteract the ABCF-mediated resistance is a valuable feature of the new antibiotic. However, given that B. subtilis VmlR does confer significant levels of iboxamycin resistance (33-fold increase in MIC) and is cooperative with the Cfr rRNA methyltransferase resistance determinant, emergence and spread of ABCF ARE variants capable of defeating next-generation lincosamides in the clinic is possible and should be closely monitored.
Furthermore, we demonstrate that iboxamycin displays a strong PAE against L. monocytogenes, compromising bacterial regrowth for many hours post-antibiotic removal. In clinical settings the longer PAE would allow the design of dosing regiments with larger dosing intervals, resulting in fewer daily administrations of the drug.52,53 The PAE is considerably stronger than that of clindamycin while lincomycin displays no PAE. It is possible that the strength of the PAE reflects how tightly the antibiotic binds to the target, the ribosome—and how slowly it dissociates from it. The pronounced PAE suggests that development of even more tight-binding lincosamides could produce effectively bactericidal drugs in the context of infection. Further biochemical studies are necessary to substantiate this hypothesis. Experiments in L. monocytogenes infection models are necessary to further assess the potential of iboxamycin as a novel drug for the treatment of listeriosis.
Acknowledgements
We are grateful to Daniel A. Portnoy for sharing WT L. monocytogenes 10403S, Jörgen Johansson for sharing WT L. monocytogenes EGD-e, Barbara E. Murray for sharing E. faecalis ΔlsaA (lsa::Kan) strain TX533219 and Birte Vester for sharing the S. sciuri cfr-encoding plasmid.41
Contributor Information
Tetiana Brodiazhenko, University of Tartu, Institute of Technology, 50411 Tartu, Estonia.
Kathryn Jane Turnbull, Department of Clinical Microbiology, Rigshospitalet, 2200 Copenhagen, Denmark.
Kelvin J Y Wu, Department of Chemistry and Chemical Biology, Harvard University, Cambridge, MA, USA.
Hiraku Takada, Department of Experimental Medicine, University of Lund, 221 84 Lund, Sweden; Faculty of Life Sciences, Kyoto Sangyo University, Kamigamo, Motoyama, Kita-ku, Kyoto 603-8555, Japan.
Ben I C Tresco, Department of Chemistry and Chemical Biology, Harvard University, Cambridge, MA, USA.
Tanel Tenson, University of Tartu, Institute of Technology, 50411 Tartu, Estonia.
Andrew G Myers, Department of Chemistry and Chemical Biology, Harvard University, Cambridge, MA, USA.
Vasili Hauryliuk, University of Tartu, Institute of Technology, 50411 Tartu, Estonia; Department of Experimental Medicine, University of Lund, 221 84 Lund, Sweden.
Funding
This work was supported by the funds from the European Regional Development Fund through the Centre of Excellence for Molecular Cell Technology (V.H., T.T.); the Estonian Research Council (grant PRG335 to V.H., T.T.); Swedish Research council (project grants 2017-03783 and 2021-01146, grant 2018-00956 within the RIBOTARGET consortium under the framework of JPIAMR); and the Ragnar Söderberg foundation (V.H.). K.J.Y.W. was supported by a National Science Scholarship (PhD) by the Agency for Science, Technology and Research, Singapore.
Transparency declarations
A.G.M. is an inventor in a provisional patent application submitted by the President and Fellows of Harvard College covering oxepanoprolinamide antibiotics described in this work. A.G.M. has filed the following international patent applications: WO/2019/032936 ‘Lincosamide Antibiotics and Uses Thereof’ and WO/2019/032956 ‘Lincosamide Antibiotics and Uses Thereof’. All other authors: none to declare.
References
- 1. Schwarz S, Shen J, Kadlec Ket al. Lincosamides, streptogramins, phenicols, and pleuromutilins: mode of action and mechanisms of resistance. Cold Spring Harb Perspect Med 2016; 6: a027037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Matzov D, Eyal Z, Benhamou RIet al. Structural insights of lincosamides targeting the ribosome of Staphylococcus aureus. Nucleic Acids Res 2017; 45: 10284–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Tu D, Blaha G, Moore PBet al. Structures of MLSBK antibiotics bound to mutated large ribosomal subunits provide a structural explanation for resistance. Cell 2005; 121: 257–70. [DOI] [PubMed] [Google Scholar]
- 4. Dunkle JA, Xiong L, Mankin ASet al. Structures of the Escherichia coli ribosome with antibiotics bound near the peptidyl transferase center explain spectra of drug action. Proc Natl Acad Sci U S A 2010; 107: 17152–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Schlunzen F, Zarivach R, Harms Jet al. Structural basis for the interaction of antibiotics with the peptidyl transferase centre in eubacteria. Nature 2001; 413: 814–21. [DOI] [PubMed] [Google Scholar]
- 6. Spížek J, Řezanka T. Lincosamides: Chemical structure, biosynthesis, mechanism of action, resistance, and applications. Biochem Pharmacol 2017; 133: 20–8. [DOI] [PubMed] [Google Scholar]
- 7. Macleod AJ, Ross HB, Ozere RLet al. Lincomycin: a new antibiotic active against staphylococci and other Gram-positive cocci: clinical and laboratory studies. Can Med Assoc J 1964; 91: 1056–60. [PMC free article] [PubMed] [Google Scholar]
- 8. Birkenmeyer RD, Lincomycin KF. XI. Synthesis and structure of clindamycin. A potent antibacterial agent. J Med Chem 1970; 13: 616–9. [DOI] [PubMed] [Google Scholar]
- 9. Phillips I. Past and current use of clindamycin and lincomycin. J Antimicrob Chemother 1981; 7Suppl A: 11–8. [DOI] [PubMed] [Google Scholar]
- 10. Smieja M. Current indications for the use of clindamycin: a critical review. Can J Infect Dis 1998; 9: 22–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Ahonkhai VI, Cherubin CE, Shulman MAet al. In vitro activity of U-57930E, a new clindamycin analog, against aerobic Gram-positive bacteria. Antimicrob Agents Chemother 1982; 21: 902–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Birkenmeyer RD, Kroll SJ, Lewis Cet al. Synthesis and antimicrobial activity of clindamycin analogues: pirlimycin, a potent antibacterial agent. J Med Chem 1984; 27: 216–23. [DOI] [PubMed] [Google Scholar]
- 13. Hirai Y, Maebashi K, Yamada Ket al. Characterization of compound A, a novel lincomycin derivative active against methicillin-resistant Staphylococcus aureus. J Antibiot (Tokyo) 2021; 74: 124–32. [DOI] [PubMed] [Google Scholar]
- 14. Mitcheltree MJ, Pisipati A, Syroegin EAet al. A synthetic antibiotic class overcoming bacterial multidrug resistance. Nature 2021; 599: 507–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Long KS, Poehlsgaard J, Kehrenberg Cet al. The Cfr rRNA methyltransferase confers resistance to phenicols, lincosamides, oxazolidinones, pleuromutilins, and streptogramin A antibiotics. Antimicrob Agents Chemother 2006; 50: 2500–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Schwarz S, Werckenthin C, Kehrenberg C. Identification of a plasmid-borne chloramphenicol-florfenicol resistance gene in Staphylococcus sciuri. Antimicrob Agents Chemother 2000; 44: 2530–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Uchiyama H, Weisblum B. N-Methyl transferase of Streptomyces erythraeus that confers resistance to the macrolide-lincosamide-streptogramin B antibiotics: amino acid sequence and its homology to cognate R-factor enzymes from pathogenic bacilli and cocci. Gene 1985; 38: 103–10. [DOI] [PubMed] [Google Scholar]
- 18. Maravic G. Macrolide resistance based on the Erm-mediated rRNA methylation. Curr Drug Targets Infect Disord 2004; 4: 193–202. [DOI] [PubMed] [Google Scholar]
- 19. Singh KV, Weinstock GM, Murray BE. An Enterococcus faecalis ABC homologue (Lsa) is required for the resistance of this species to clindamycin and quinupristin-dalfopristin. Antimicrob Agents Chemother 2002; 46: 1845–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Wilson DN, Hauryliuk V, Atkinson GCet al. Target protection as a key antibiotic resistance mechanism. Nat Rev Microbiol 2020; 18: 637–48. [DOI] [PubMed] [Google Scholar]
- 21. Murina V, Kasari M, Takada Het al. ABCF ATPases involved in protein synthesis, ribosome assembly and antibiotic resistance: structural and functional diversification across the tree of life. J Mol Biol 2019; 431: 3568–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Ero R, Kumar V, Su Wet al. Ribosome protection by ABC-F proteins-molecular mechanism and potential drug design. Protein Sci 2019; 28: 684–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Crowe-McAuliffe C, Murina V, Turnbull KJet al. Structural basis of ABCF-mediated resistance to pleuromutilin, lincosamide, and streptogramin A antibiotics in Gram-positive pathogens. Nat Commun 2021; 12: 3577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Murina V, Kasari M, Hauryliuk Vet al. Antibiotic resistance ABCF proteins reset the peptidyl transferase centre of the ribosome to counter translational arrest. Nucleic Acids Res 2018; 46: 3753–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Sharkey LK, Edwards TA, O’Neill AJ. ABC-F proteins mediate antibiotic resistance through ribosomal protection. mBio 2016; 7: e01975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Su W, Kumar V, Ding Yet al. Ribosome protection by antibiotic resistance ATP-binding cassette protein. Proc Natl Acad Sci U S A 2018; 115: 5157–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Crowe-McAuliffe C, Graf M, Huter Pet al. Structural basis for antibiotic resistance mediated by the Bacillus subtilis ABCF ATPase VmlR. Proc Natl Acad Sci U S A 2018; 115: 8978–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Radoshevich L, Cossart P. Listeria monocytogenes: towards a complete picture of its physiology and pathogenesis. Nat Rev Microbiol 2018; 16: 32–46. [DOI] [PubMed] [Google Scholar]
- 29. Schlech WF 3rd, Lavigne PM, Bortolussi RAet al. Epidemic listeriosis–evidence for transmission by food. N Engl J Med 1983; 308: 203–6. [DOI] [PubMed] [Google Scholar]
- 30. Southwick FS, Purich DL. Intracellular pathogenesis of listeriosis. N Engl J Med 1996; 334: 770–6. [DOI] [PubMed] [Google Scholar]
- 31. de Noordhout CM, Devleesschauwer B, Angulo FJet al. The global burden of listeriosis: a systematic review and meta-analysis. Lancet Infect Dis 2014; 14: 1073–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Mylonakis E, Hohmann EL, Calderwood SB. Central nervous system infection with Listeria monocytogenes. 33 years’ experience at a general hospital and review of 776 episodes from the literature. Medicine (Baltimore) 1998; 77: 313–36. [DOI] [PubMed] [Google Scholar]
- 33. Temple ME, Nahata MC. Treatment of listeriosis. Ann Pharmacother 2000; 34: 656–61. [DOI] [PubMed] [Google Scholar]
- 34. Caruso M, Fraccalvieri R, Pasquali Fet al. Antimicrobial susceptibility and multilocus sequence typing of Listeria monocytogenes isolated over 11 years from food, humans, and the environment in Italy. Foodborne Pathog Dis 2020; 17: 284–94. [DOI] [PubMed] [Google Scholar]
- 35. Andriyanov PA, Zhurilov PA, Liskova EAet al. Antimicrobial resistance of Listeria monocytogenes strains isolated from humans, animals, and food products in Russia in 1950-1980, 2000-2005, and 2018-2021. Antibiotics (Basel) 2021; 10: 1206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Rugna G, Carra E, Bergamini Fet al. Distribution, virulence, genotypic characteristics and antibiotic resistance of Listeria monocytogenes isolated over one-year monitoring from two pig slaughterhouses and processing plants and their fresh hams. Int J Food Microbiol 2021; 336: 108912. [DOI] [PubMed] [Google Scholar]
- 37. Tirziu E, Herman V, Nichita Iet al. Diversity and antibiotic resistance profiles of Listeria monocytogenes serogroups in different food products from the Transylvania Region of Central Romania. J Food Prot 2022; 85: 54–59. [DOI] [PubMed] [Google Scholar]
- 38. Dar D, Shamir M, Mellin JRet al. Term-seq reveals abundant ribo-regulation of antibiotics resistance in bacteria. Science 2016; 352: aad9822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Ohki R, Tateno K, Takizawa Tet al. Transcriptional termination control of a novel ABC transporter gene involved in antibiotic resistance in Bacillus subtilis. J Bacteriol 2005; 187: 5946–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Mason JD, Terwilliger DW, Pote ARet al. Practical Gram-scale synthesis of iboxamycin, a potent antibiotic candidate. J Am Chem Soc 2021; 143: 11019–25. [DOI] [PubMed] [Google Scholar]
- 41. Ntokou E, Hansen LH, Kongsted Jet al. Biochemical and computational analysis of the substrate specificities of Cfr and RlmN methyltransferases. PLoS One 2015; 10: e0145655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. CLSI . Performance Standards for Antimicrobial Susceptibility Testing—Twenty-Sixth Edition: M100. 2016.
- 43. Svetlov MS, Vazquez-Laslop N, Mankin AS. Kinetics of drug-ribosome interactions defines the cidality of macrolide antibiotics. Proc Natl Acad Sci U S A 2017; 114: 13673–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Glaser P, Frangeul L, Buchrieser Cet al. Comparative genomics of Listeria species. Science 2001; 294: 849–52. [DOI] [PubMed] [Google Scholar]
- 45. Edman DC, Pollock MB, Hall ER. Listeria monocytogenes L forms. I. Induction maintenance, and biological characteristics. J Bacteriol 1968; 96: 352–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Bécavin C, Bouchier C, Lechat Pet al. Comparison of widely used Listeria monocytogenes strains EGD, 10403S, and EGD-e highlights genomic variations underlying differences in pathogenicity. mBio 2014; 5: e00969-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Britton RA, Eichenberger P, Gonzalez-Pastor JEet al. Genome-wide analysis of the stationary-phase sigma factor (sigma-H) regulon of Bacillus subtilis. J Bacteriol 2002; 184: 4881–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Walkup GK, You Z, Ross PLet al. Translating slow-binding inhibition kinetics into cellular and in vivo effects. Nat Chem Biol 2015; 11: 416–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Bundtzen RW, Gerber AU, Cohn DLet al. Postantibiotic suppression of bacterial growth. Rev Infect Dis 1981; 3: 28–37. [DOI] [PubMed] [Google Scholar]
- 50. Mohamad M, Nicholson D, Saha CKet al. Sal-type ABC-F proteins: intrinsic and common mediators of pleuromutilin resistance by target protection in staphylococci. Nucleic Acids Res 2022; 50: 2128–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Sharkey LKR, O’Neill AJ. Antibiotic resistance ABC-F proteins: bringing target protection into the limelight. ACS Infect Dis 2018; 4: 239–46. [DOI] [PubMed] [Google Scholar]
- 52. ter Braak EW, de Vries PJ, Bouter KPet al. Once-daily dosing regimen for aminoglycoside plus β-lactam combination therapy of serious bacterial infections: comparative trial with netilmicin plus ceftriaxone. Am J Med 1990; 89: 58–66. [DOI] [PubMed] [Google Scholar]
- 53. Gilbert DN. Once-daily aminoglycoside therapy. Antimicrob Agents Chemother 1991; 35: 399–405. [DOI] [PMC free article] [PubMed] [Google Scholar]