Abstract
Identification of somatic mutations with high precision is one of the major challenges in the prediction of high-risk liver cancer patients. In the past, number of mutations calling techniques has been developed that include MuTect2, MuSE, Varscan2, and SomaticSniper. In this study, an attempt has been made to benchmark the potential of these techniques in predicting the prognostic biomarkers for liver cancer. Initially, we extracted somatic mutations in liver cancer patients using Variant Call Format (VCF) and Mutation Annotation Format (MAF) files from the cancer genome atlas. In terms of size, the MAF files are 42 times smaller than VCF files and containing only high-quality somatic mutations. Furthermore, machine learning-based models have been developed for predicting high-risk cancer patients using mutations obtained from different techniques. The performance of different techniques and data files has been compared based on their potential to discriminate high- and low-risk liver cancer patients. Based on correlation analysis, we selected 80 genes having significant negative correlation with the overall survival of liver cancer patients. The univariate survival analysis revealed the prognostic role of highly mutated genes. Single gene-based analysis showed that MuTect2 technique-based MAF file has achieved maximum hazard ratio (HRLAMC3) of 9.25 with P-value of 1.78E-06. Further, we developed various prediction models using risk-associated top-10 genes for each technique. Our results indicate that MuTect2 technique-based VCF files outperform all other methods with maximum Area Under the Receiver-Operating Characteristic curve of 0.765 and HR = 4.50 (P-value = 3.83E-15). Eventually, VCF file generated using MuTect2 technique performs better among other mutation calling techniques for the prediction of high-risk liver cancer patients. We hope that our findings will provide a useful and comprehensive comparison of various mutation-calling techniques for the prognostic analysis of cancer patients. In order to serve the scientific community, we have provided a Python-based pipeline to develop the prediction models using mutation profiles (VCF/MAF) of cancer patients. It is available on GitHub at https://github.com/raghavagps/mutation_bench.
Keywords: mutation calling techniques, prognosis, liver cancer, survival analysis, machine learning, regression
Key Points.
Albeit number of mutations calling techniques are available, it is hard to choose one to explore the role of mutations in cancer.
MuTect2, MuSE, Varscan2 and SomaticSniper based VCF and MAF files were used to traverse the mutations in liver cancer patients.
Univariate survival analysis was used to explore the prognostic role of mutations in liver cancer.
Various classification and regression models were developed to stratify patients with high- and low-risk of liver cancer.
MuTect2 based VCF file outperformed other mutation calling techniques.
Introduction
According to the World Health Organization, cancer is a life-threatening disease and the first leading cause of death worldwide. Global cancer statistics estimate that in the Year 2020, 19.3 million new cases and 10 million deaths have been occurred due to cancer [1]. Cancer is extremely heterogeneous; therefore, the same treatment strategy is not effective for individuals with similar types of cancers. Till now, there is no universal treatment available for all types of malignancies. However, several targeted therapies are available for cancer treatment, which majorly focus on the detection of mutations at the genetic level [2]. In the last few years, several therapies have been designed based on the mutated genes, for cancer treatment. For instance, B-Raf Proto-Oncogene, Serine/Threonine Kinase (BRAF) inhibitors (Sorafenib) are identified to treat melanoma patients with V600E mutation in the BRAF gene [3, 4]. Drugs like afatinib and erlotinib are used to target the mutation in the Epidermal Growth Factor Receptor (EGFR) in non-small cell lung cancer [5, 6]. Moreover, BRCA1/BRCA2 gene mutations in ovarian cancer patients have been treated by poly (ADP-ribose) polymerase inhibitor, i.e. olaparib [7]. Of note, research on the mutations associated with the genes in cancer patients is essential for identifying the correct mechanism of the disease. Due to the advancements in next-generation sequencing, such as whole-genome, whole-exome and mutation calling techniques, the detection of >98% of mutations associated with the disease using sequencing data is possible [8, 9]. The easy availability and low cost of next-generation sequencing techniques enable researchers to perform experiments on large cohorts of cancer patients [10].
The genetic variants are mainly categorized into single-nucleotide variant (SNV), insertion/deletion (indel) and structural variants (which incorporates copy number alterations, duplications and translocations). In the recent years, a huge number of somatic mutation calling algorithms (e.g. Mutect2, Varscan2, SomaticSniper, MuSE, Strelka2, etc.) have been developed to identify mutations at the genetic level using sequencing data [11–17]. Mutect2 calls somatic mutation such as single-nucleotide alterations and indels using the local assembly of haplotypes. SomaticSniper pipeline detects somatic SNVs using Bayesian algorithm to compare the genotype likelihoods in the tumour and normal samples. However, the Varscan2 mutation calling algorithm uses exomes, whole-genome sequencing data to capture germline variants, somatic mutations and copy number variants in tumour-normal data. Moreover, MuSE is a Markov Substitution model for Evolution and identify novel mutations in the large-scale tumour sequencing data.
Liver cancer is one of the deadliest diseases which is the seventh most common cancer among the 36 cancers reported by Global Cancer Statistics 2020 [1]. Ample treatment methods were developed in the past, but still the survival rate of liver cancer patients is very low, leading to a high-mortality rate [18]. Being the most comprehensive resource for cancer-related research, The Cancer Genome Atlas (TCGA) provides two types of file formats for mutation data such as Variant Call Format (VCF) and Mutation Annotation Format (MAF). VCF files are the raw mutation files that store and report the genomic sequence variations that directly came out of the various automated variant calling pipelines. On the other hand, MAF files are the processed version of the VCF files, which are curated by removing the false positives or by recovering the known calls that the automated pipelines may have missed. VCF files report mutations irrespective of their importance, but MAF files describe only the most affected ones by removing the low-quality mutations. In Genomic Data Commons (GDC) portal, both type of files available are generated using the four major mutation calling techniques named as MuTect2, MuSE, Varscan2 and SomaticSniper. Despite number of techniques available, it is difficult to understand which method and file is better to explore the role of mutations in cancer.
In this study, we have systematically evaluated the four mutation calling tools which are widely used in TCGA, to identify highly mutated genes associated with high-risk liver cancer patients. For this, we have collected VCF and MAF files of 418 liver cancer patients for all the mutation calling techniques. The gene-based annotations were identified using highly accurate and widely used methods ANNOtate VARiation (ANNOVAR) [19] and Maftools [20]. Correlation and survival analysis was performed to identify the mutated genes that can impact the survival of liver cancer patients. Finally, we have developed survival prediction and classification models using different machine learning algorithms on highly significant top-10 risk-associated genes, selected from four mutation calling techniques. Based on the inferences, we benchmarked different techniques which can provide a valuable reference and guidance to the researchers to choose a reliable somatic mutation algorithm to determine the mutation-associated genes having a significant impact on the survival of the cancer patients.
Material and methods
Overall study design
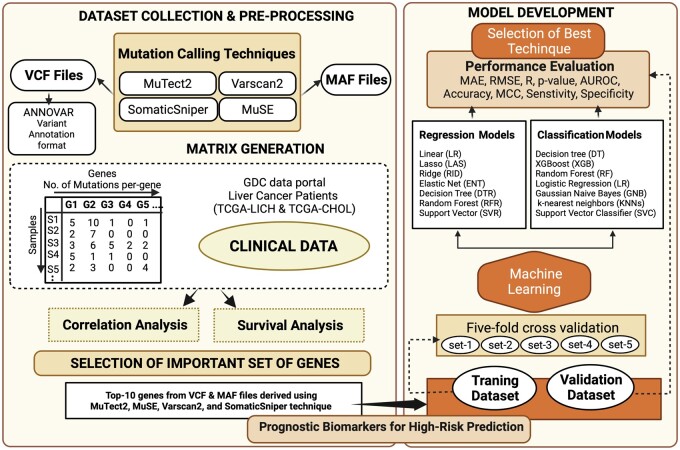
The complete pipeline of the study is shown in Fig. 1 with the step-by-step description.
Figure 1:
Pipeline illustrating the overall overflow of the study.
Dataset collection
We obtained liver hepatocellular carcinoma (TCGA-LICH) and cholangiocarcinoma (TCGA-CHOL) mutation data from GDC data portal. Precisely, we collected the controlled access VCF files of liver cancer patients under the approval of dbGap (Project No. 17674) according to the GDC protocols [21]. In addition to that, we have also downloaded the MAF files of TCGA liver cancer patients. TCGA generate mutation profiles of cancer patients using four different mutation calling techniques, i.e. MuSE, Mutect2, Varscan2, and SomaticSniper. Currently, we have utilized VCF and MAF files of 418 liver cancer samples generated from four different mutation calling methods. Moreover, the clinical data like age, gender, tumour stage, overall survival (OS) time, and vital status were collected using TCGA assembler 2 [22].
Mutation annotations
We have used the ANNOVAR software (https://annovar.openbioinformatics.org/en/latest/) for the functional annotations of genetic variant mutations by utilizing the VCF files. First, we convert VCF files into ANNOVAR genetic variant files using the “convert2annovar.pl” script provided in the ANNOVAR package; the processed file contains five major columns: chromosome number, start position, end position, reference nucleotide, and altered nucleotides. It also provides three major type of annotations (i.e. gene-based, region-based, and filter-based annotations). Presently, we have utilized only gene-based annotations, in which we obtained mutations/gene/sample. We have developed an in-house Python script to count the number of mutations per-gene for each sample. Thus, we get per-gene mutations for each sample for the different mutation calling techniques. Similarly, in the case of MAF files, we counted the number of mutations/gene/sample. Finally, we generated matrices for each mutation calling technique from VCF and MAF files, in which number of mutations per gene per sample were reported.
Correlation analysis
To understand the impact of genetic mutations on survival of liver cancer patients, we have implemented correlation test. After computing the correlation coefficient and P-value corresponding to each gene, we filtered out the genes with the non-significant P-value, i.e. >0.05, and ranked the remaining genes on the bases of their correlation coefficients. We choose top-10 negatively correlated genes with significant P-value (i.e. <0.05) from each technique for VCF and MAF files for further analysis.
Survival analysis
In this study, we have performed survival analysis by the ‘survival’ package in R (version 3.5.1) using the cox proportional hazard (Cox PH) model. We performed univariate survival analysis to understand the overall impact of per-gene mutations on the survival of liver cancer patients. The log-rank test was used to estimate the significant survival distributions between high- and low-risk groups in terms of the P-value. In addition to that, we have computed hazard ratio (HR), 95% confidence interval and concordance index using the Cox PH model. Kaplan–Meier (KM) survival curves were used for the graphical representation of high-risk and low-risk groups [23].
Machine learning techniques
Classification models
In this study, we have implemented various machine learning techniques for the classification of high-risk and low-risk samples based on the number of mutations in the risk-associated genes. Classification algorithms included decision tree, support vector classifier, random forest, extreme gradient boosting, Gaussian naive Bayes, logistic regression, k-nearest neighbours and extra tree was implemented using scikit-learn library [24]. These classifiers belong to different families, such as rule-based, decision-tree, Bayesian, logistic regression, support vector machines, nearest-neighbours and boosting. Decision tree is a rule-based algorithm in which the outcome or the class assignment is based on a set of rules which are defined using the training dataset. This approach generates a model by building a decision tree, in which each node represents an attribute that further splits the data into two classes, and this process continues until all the instances belonging to a particular class get secluded [25]. Random forest and extra tree classifier belong to the ensemble family, in which numerous de-correlated decision trees are built on various subsets of samples to make an overall classification. These classifiers vary in terms of the construction of decision trees and the selection of thresholds to split the nodes [26, 27].
Moreover, logistic regression is a statistical approach that uses the logistic function to model the probabilities of the output variable using predictor variables [28]. Extreme gradient boosting algorithm belongs to the boosting class and tree-based approach; it implements an iterative process in which ensembles of decision trees are created where one tree is added at a time and fit to reduce the errors in the predictions resulted due to previous models [29]. Besides, k-nearest neighbour is a part of the nearest-neighbours family that works on the principle of proximity and assigns a class to the unknown variable based on its proximity to the data points in the training dataset [30]. Gaussian naïve Bayes is a stochastic approach based on the Bayes theorem and assumes that each feature is independent of the other with an equal contribution to the predictions [31]. In addition, support vector classifier belongs to the support vector machines family, which identifies the data points to create the hyperplanes that can separate the n-dimensional space into different classes [32].
Regression models
Furthermore, we implemented several regressors to develop regression models for the prediction of overall survival time of liver cancer patients. These models were developed by implementing various regressors including decision tree, random forest, linear, ridge, lasso, elastic net and support vector regressor from Python-library scikit-learn [24]. Decision tree regressor is a supervised-learning algorithm that uses a tree-like structure to predict the outcome. It utilizes the independent features to train a model in the design of a tree and make predictions for the unseen data [33]. Random forest regressor is an ensemble of multiple decision trees where each tree provides an output. The final output is derived by taking the average of all outcomes [34]. In this study, we have implemented ordinary least squares linear regression in which a linear model is developed with coefficients to minimize the residual sum of squares between the predicted and actual values [35]. Lasso or Least Absolute Shrinkage and Selection Operator regressor is an extension of linear regression with L1 regularization in which the loss function i.e. residual sum of squares is extended by the sum of the absolute values of model coefficients [36].
In addition, ridge regressor is also a modified version of linear regression with L2 regularization in which the loss function is altered to deal with the higher biasness of the model where the penalty of the sum of squares of the model coefficients is added to the loss function [37]. While, elastic net is the weighted combination of lasso and ridge regression, in which both L1 and L2 regularizations are considered [38]. Support vector regression supports both linear and non-linear regression, as it tries to fit the error between certain constraints. It is achieved by minimizing the coefficients to handle the error term in the constraints where the absolute error is less than or equal to the maximum error defined by a specified margin [39].
Performance evaluation
Cross-validation technique
We have implemented the 5-fold cross-validation to avoid overfitting, biasness and evaluate the performance of prediction models [40, 41]. In this method, the complete dataset was divided into 80:20 ratio, where 80% data called training dataset was used for internal validation and 20% data called validation dataset was used for external validation. The performance of the models on the training dataset was evaluated using 5-fold cross-validation technique. In this approach, the training dataset was divided into five equal non-overlapping sets where four sets were used for training the model and the remaining set was used for testing. This process was repeated five times so that each set tested once. We optimized the parameters of the model on the training dataset during internal validation to achieve the maximum performance. The overall performance or outcome was computed by taking the average of all the five sets. Finally, for external validation the tuned models were further evaluated on the 20% untouched validation dataset. The process of evaluation of models on the validation dataset is called external validation. The similar process was repeated for the cross validation of regression models, where the complete dataset was used for the 5-fold cross validation.
Performance measure parameters
To evaluate the performance of classification models, we have used standard parameters. We have calculated threshold dependent such as sensitivity, specificity, accuracy, F1-score, kappa and Matthews Correlation Coefficient (MCC), and independent parameters like Area Under the Receiver Operating Characteristic curve (AUROC). These parameters were calculated using the following Equations (1–5).
| (1) |
| (2) |
| (3) |
| (4) |
| (5) |
where Tp = True Positive; Tn = True Negative; Fp = False Positive; Fn = False Negative.
Similarly, to evaluate the regression models, we have used parameters such as mean absolute error (MAE), root-mean-square error (RMSE), correlation coefficient (R) and P-value; as previously used in different studies [42–44].
Results
In this study, we have used 418 TCGA liver cancer patients’ somatic mutation data (VCF files and MAF files) and survival data (OS time and vital status). The mutation data were taken from four different mutation calling techniques, i.e. MuSE, Mutect2, Varscan2 and SomaticSniper. ANNOVAR software and in-house scripts were used to extract the number of mutations/gene/sample from the VCF and MAF files. The total number of genes and mutations extracted from different techniques is shown in Table 1. We observed that in VCF files Mutect2 and SomaticSniper reported the highest number of genes and mutation counts, i.e. more than 25 000 genes and 5 million mutations. On the other hand, the reported number of genes and mutations in MAF files are comparatively less for each technique.
Table 1:
Total number of genes and mutations for each gene extracted from VCF and MAF files using different mutation calling techniques
| File type | Technique | Number of genes | Number of mutations |
|---|---|---|---|
| VCF | MuTect2 | 25 366 | 5 237 093 |
| MuSE | 19 425 | 379 368 | |
| Varscan2 | 19 422 | 576 231 | |
| SomaticSniper | 25 785 | 5 003 969 | |
| MAF | MuTect2 | 16 474 | 59 741 |
| MuSE | 15 712 | 51 184 | |
| Varscan2 | 15 950 | 54 877 | |
| SomaticSniper | 14 979 | 44 102 |
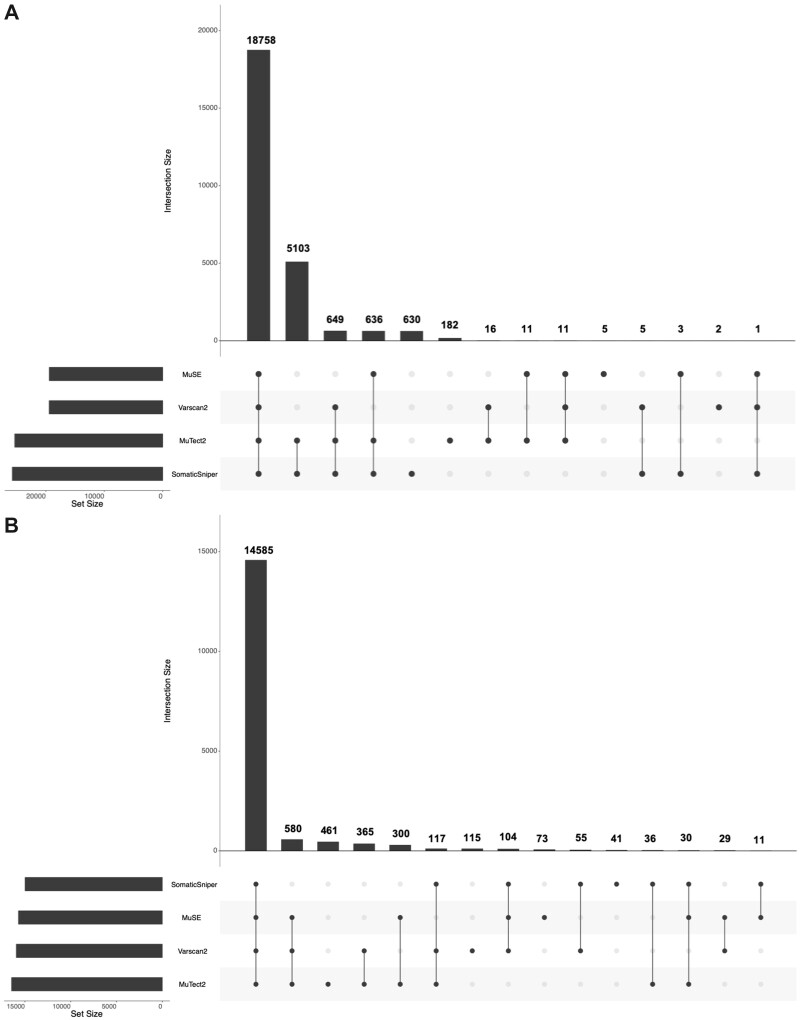
Further, in order to understand and visualize the distribution of genes corresponding to each technique, we developed an UpSet plot [45] as shown in Fig. 2. According to the plots, in VCF file, 18 758 genes were common in all the four techniques, whereas 182, 5, 2 and 630 genes are uniquely reported by MuTect2, MuSE, Varscan2 and SomaticSniper technique, respectively. Similarly, in the case of MAF files, 14 585 genes were shared by all the techniques, while 461 genes are unique in file by MuTect2 technique, 73 by MuSE, 115 by Varscan2 and 41 unique genes were reported by SomaticSniper technique.
Figure 2:
Upset-plot for distribution of genes in four techniques. (A) From VCF files and (B) from MAF files.
Comparison of MAF files
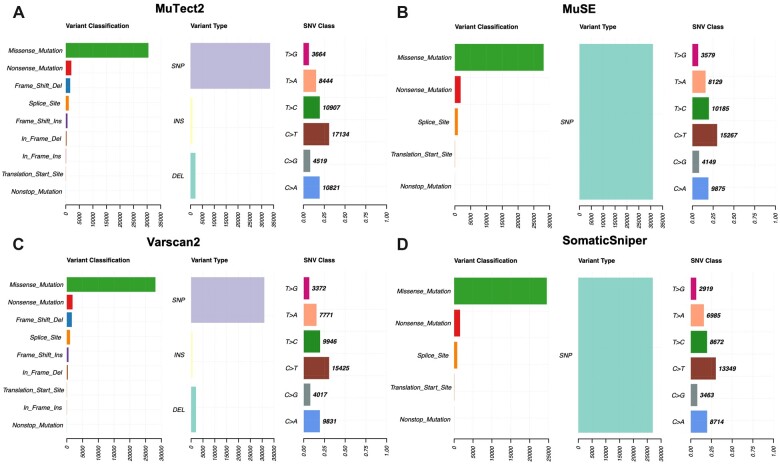
To compare different mutation calling techniques, we have taken processed and annotated MAF files from TCGA. We utilized the Maftools package to comprehensively analyse the somatic variants extracted from MuSE, Mutect2, Varscan2 and SomaticSniper mutation calling technique. From the analysis, we observed few changes in the mutation calling techniques for the same cohort of samples. For example, MuSE and SomaticSniper MAF files (Fig. 3A and B) only report SNPs on the other side Varscan2, and MuTect2 (Fig. 3C and D) represent SNPs, INS and DEL under the variant type.
Figure 3:
Visualization of mutation summary (variants classification, type and SNVs) for (A) MuTect2, (B) MuSE, (C) Varscan2 and (D) SomaticSniper MAF files.
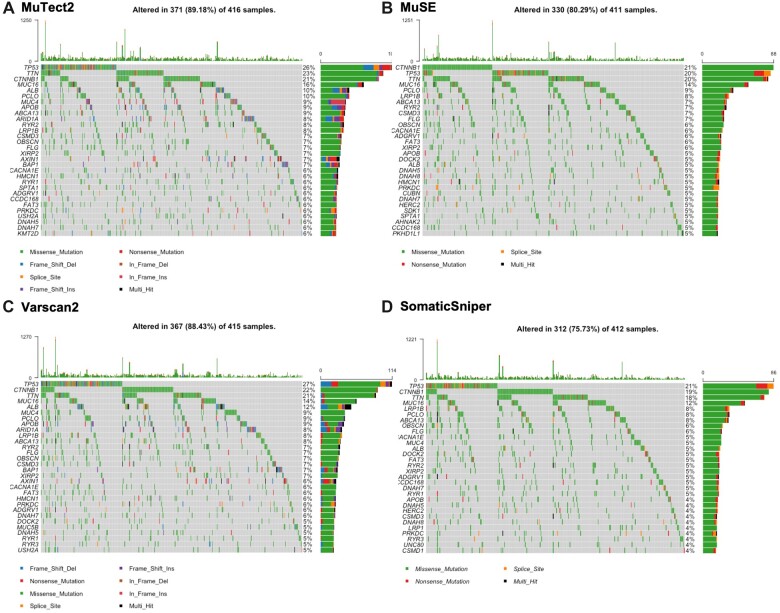
In Varscan2 and MuTect2, the variant classification distribution represents nine types of mutations such as Missense_Mutation, Nonsense_Mutation, Splice_Site, Translational_Start_Site, Frame_Shift_Ins, Frame_Shift_Del, In_Frame_Ins, In_Frame_Del and Nonstop_Mutations, while MuSE and SomaticSniper MAF files consist of Missense_Mutation, Nonsense_Mutation, Splice_Site, Translational_Start_Site and Nonstop_Mutations. The SNV class visualizes the single-nucleotide variants in the TCGA cohort, we observed that all the methods present diverse distribution of SNV as shown in (Fig. 3). Oncoplots generated by the Maftools visualization module illustrating the somatic landscape of the cancer patients for Varscan2, MuTect2, MuSE and SomaticSniper MAF files. In Fig. 4, we display the topmost mutated genes with their mutation percentage (≥5%) in total number of samples. From the results, we observed that TP53 is a highly mutated gene and have almost 20% or >20% mutations among different techniques.
Figure 4:
Oncoplot visualization of mutation frequency of top-most mutated genes. The rows represented the genes with percent mutations, and columns display the samples. (A) Illustrates the oncoplot of MuTect2 technique and indicates that 89.18% of samples having mutated genes. (B) Illustrates the oncoplot of MuSE technique and shows that 80.29% of samples having mutated genes. (C) Presents the oncoplot of Varscan2 approach and shows that 88.43% of samples having mutated genes. (D) Illustrates the oncoplot of SomaticSniper technique and indicates that 75.73% of samples having alerted/mutated genes.
Correlation analysis
By implementing the correlation test we ranked the genes and choose top-10 genes having significant negative correlation with the overall survival time. The procedure was repeated for all the four techniques using MAF and VCF files of liver cancer patients, which lead to 80 genes in total. The complete correlation analysis is provided in Supplementary Table S1.
Prognostic biomarkers for high-risk prediction
Single gene
Univariate survival analysis was performed using the Cox PH model. We have measured the HR and P-value for the negatively correlated genes obtained from each mutation calling technique. In the case of VCF files, single gene-based analysis revealed that the genes extracted from SomaticSniper technique has achieved the maximum HR and P-value followed by Varscan2, MuTect2 and MuSE corresponding to genes CLDN20, FAM160A2, SNHG10 and CLMP, respectively (see Table 2). Similar analysis was done for MAF files for each technique where HR, P-values was calculated. In the case of MAF files, Mutect2 technique achieved the maximum performance followed by Varscan2, MuSE and SomaticSniper for genes LAMC3, SYDE1, ITGB8 and CAD, respectively (see Table 2). Supplementary Table S2 contains the comprehensive results for all the risk-associated genes derived from each technique for VCF and MAF files.
Table 2:
HR for risk-associated top-10 genes from VCF and MAF files derived using MuTect2, MuSE, Varscan2 and SomaticSniper technique
| MuTect2 |
MuSE |
Varscan2 |
SomaticSniper |
||||
|---|---|---|---|---|---|---|---|
| Gene | HR | Gene | HR | Gene | HR | Gene | HR |
| (P-value) | (P-value) | (P-value) | (P-value) | ||||
| VCF files | |||||||
| SNHG10 | 5.49 (3.94E-06) | CLMP | 3.01 (1.67E-05) | FAM160A2 | 6.81 (4.01E-05) | CLDN20 | 7.06 (6.62E-07) |
| WIZ | 2.69 (9.71E-07) | BIRC6 | 2.80 (4.46E-04) | LOC100420587 | 5.45 (1.31E-07) | NR2C2AP | 5.17 (3.16E-05) |
| MGAT4EP | 2.49 (4.46E-04) | LINC02210-CRHR1 | 2.03 (6.42E-03) | SPDYA | 3.08 (7.70E-04) | ATG9B | 3.34 (2.59E-04) |
| LINC00304 | 2.39 (7.40E-05) | DHX8 | 2.00 (2.90E-02) | BRSK2 | 2.55 (1.01E-03) | HAUS5 | 2.79 (2.22E-05) |
| CACNG7 | 1.93 (5.72E-04) | LINC00972 | 1.91 (9.31E-03) | ADGRF4 | 2.21 (1.23E-02) | LOC100287329 | 2.58 (8.23E-04) |
| OR52B6 | 1.83 (1.12E-03) | PAX7 | 1.90 (8.29E-04) | LINC00972 | 2.11 (2.18E-03) | P4HTM | 2.18 (2.43E-02) |
| TYK2 | 1.80 (2.21E-03) | TAS1R2 | 1.61 (2.63E-02) | TM4SF18 | 2.07 (1.40E-02) | OR6C76 | 2.12 (1.18E-03) |
| PIGO | 1.79 (1.66E-02) | SNTG1 | 1.53 (3.37E-02) | OR5AS1 | 1.86 (1.43E-02) | CLK2 | 1.94 (3.58E-02) |
| S100A12 | 1.71 (1.10E-02) | CNTN5 | 1.34 (2.25E-01) | PDE11A | 1.72 (2.74E-03) | FAM187B | 1.64 (1.51E-02) |
| DNAJC9-AS1 | 1.08 (6.51E-01) | ZNF521 | 1.26 (2.63E-01) | LOC101929073 | 1.29 (2.98E-01) | NOMO3 | 1.34 (1.45E-01) |
| MAF files | |||||||
| LAMC3 | 9.25 (1.78E-06) | ITGB8 | 8.37 (5.69E-07) | SYDE1 | 8.46 (3.71E-05) | CAD | 5.56 (8.10E-04) |
| EVC2 | 4.30 (8.66E-05) | TBX3 | 8.10 (6.06E-05) | ALPP | 4.33 (1.44E-03) | TOP2A | 4.63 (2.73E-03) |
| NYNRIN | 3.94 (1.22E-03) | SIPA1L3 | 4.90 (5.54E-05) | KIAA2026 | 3.85 (1.49E-03) | KIAA2026 | 4.01 (2.62E-03) |
| KIAA2026 | 3.85 (1.49E-03) | CAD | 4.45 (3.58E-03) | CAD | 3.32 (1.91E-02) | EVC2 | 4.00 (1.04E-03) |
| SUPT20H | 3.41 (7.53E-03) | EVC2 | 4.16 (2.97E-04) | BRINP2 | 2.83 (2.43E-02) | KTN1 | 2.56 (1.09E-01) |
| BRINP2 | 2.83 (2.43E-02) | ARHGEF11 | 3.17 (2.37E-02) | TP53 | 1.60 (9.85E-03) | EPHA3 | 2.25 (1.67E-01) |
| LRP1B | 1.93 (7.81E-03) | BRINP2 | 2.80 (2.56E-02) | PCDH15 | 1.48 (2.81E-01) | KIF26B | 2.03 (1.66E-01) |
| TP53 | 1.48 (3.60E-02) | PCDH15 | 1.72 (1.20E-01) | TG | 1.46 (4.53E-01) | PCDH15 | 1.76 (1.78E-01) |
| TG | 1.46 (4.53E-01) | TG | 1.46 (4.55E-01) | PLCB1 | 1.25 (7.00E-01) | TP53 | 1.63 (1.20E-02) |
| PCDH15 | 1.43 (3.30E-01) | CSMD3 | 1.24 (4.54E-01) | XIRP2 | 1.11 (7.55E-01) | TG | 1.18 (8.17E-01) |
Multiple gene
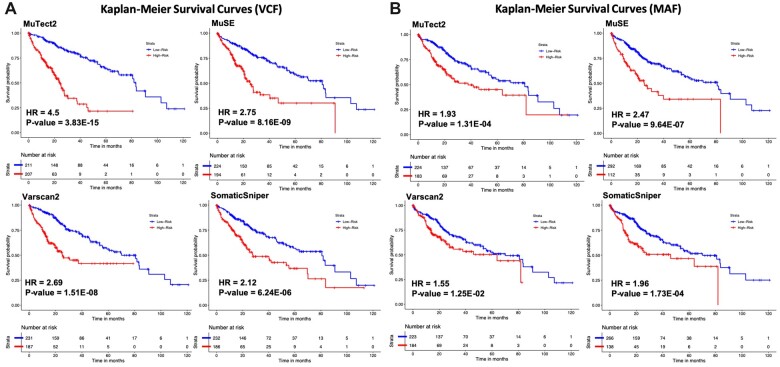
In order to explore the effect of mutations in the selected genes altogether, we have predicted the survival time to estimate the high-risk group in liver cancer patients. Using the predicted OS time, HR and P-value was computed with Cox PH model for each technique that corresponds to each file type. We achieved the highest HR = 4.50 with highly significant P-value of 3.83E-15 for the VCF files generated using the MuTect2 technique (see Fig. 5A). However, in the case of MAF files, MuSE technique performed best among the other techniques with HR = 2.47 and P-value = 9.64E-07 (see Fig. 5B). Additionally, KM survival plots clearly represents the segregation of high- and low-risk groups; the comparison of different mutation calling techniques based on two file formats is shown in Fig. 5.
Figure 5:
KM survival curves for the risk estimation of liver cancer patients based on the combined effect of mutation. (A) Survival plots for the VCF files and (B) survival plots for the MAF files.
Prediction of overall survival of patients
To predict the overall survival for liver cancer patients, we have used number of mutations in the top-10 genes as the input feature and developed regression models for VCF and MAF files for each technique, using seven different regressors such as, random forest, ridge, lasso, decision tree, elastic net, linear and support vector regressor. Table 3 exhibits the performance of best performing regressor in each file type. Performance of all the regressors for each file type and technique is reported in Supplementary Table S3. In the case of MuTect2 technique, the predicted survival time using VCF files have achieved minimum error in terms of MAE of 12.52 months and significant correlation of 0.57 between the true and predicted OS, whereas in MAF file, the MAE is 16.47 months with a correlation of 0.37. In addition, MuSE technique also achieved the minimum MAE of 13.88 months for VCF files (See Table 3). Although, we observed that in the case of other mutation calling techniques such as Varscan2 and SomaticSniper, the error rate is comparatively high. In addition, as shown in Table 3 for VCF as well as MAF files, MuTect2 technique outperformed the other techniques in terms of MAE, RMSE and R-value.
Table 3:
Performance of best regressors on top-10 genes from VCF and MAF files extracted using all techniques
| Technique | File type | MAE | RMSE | R | P-value |
|---|---|---|---|---|---|
| MuTect2 | VCF | 12.52 | 19.58 | 0.57 | 7.00E-37 |
| MAF | 16.47 | 22.16 | 0.37 | 1.31E-14 | |
| MuSE | VCF | 13.88 | 20.38 | 0.51 | 1.38E-29 |
| MAF | 16.89 | 22.48 | 0.34 | 1.68E-12 | |
| Varscan2 | VCF | 14.57 | 20.78 | 0.48 | 4.77E-26 |
| MAF | 16.53 | 22.26 | 0.36 | 9.11E-14 | |
| SomaticSniper | VCF | 15.76 | 21.82 | 0.40 | 3.31E-17 |
| MAF | 16.72 | 22.26 | 0.33 | 8.46E-12 |
Discrimination of low- and high-risk patients
Initially, the dataset was divided into two groups, i.e. high-risk and low-risk group based on the median OS. Samples with survival time less than the median OS were designated to the high-risk group, whereas the remaining was assigned to the low-risk group. To assess the ability of the number of mutations/gene/sample and to classify the patients into the high- and low-risk groups, various classification models were developed on top 10 genes for each technique and file type. Number of mutations corresponding to each gene reported through different technique was used to develop models for the stratification of high- and low-risk group. In order to compare the two file types derived from four different mutations calling techniques, we have reported the performance of models based on the best classifier, i.e. logistic regression as shown in Table 4. While the performance of all the other classifiers generated on each technique for both the files were reported in Supplementary Table S4. As shown in table below, in the case of VCF as well as MAF file, Mutect2 outperformed the other techniques by achieving highest AUROC of 0.765 and 0.659 on the validation dataset, respectively. In terms of average, VCF file-based models have higher performance in comparison to the models developed on MAF files with an AUROC of 0.699 ± 0.061 on validation dataset. In conclusion, for VCF and MAF files, MuTect2 technique performed best among other techniques in terms AUROC, F1, Kappa and MCC values (see Table 4).
Table 4:
Performance of logistic regression based models on top-10 genes from VCF and MAF files extracted using all techniques on validation dataset
| Technique | File type | AUROC | F1 | Kappa | MCC |
|---|---|---|---|---|---|
| MuTect2 | VCF | 0.765 | 0.767 | 0.421 | 0.442 |
| MAF | 0.659 | 0.661 | 0.259 | 0.335 | |
| MuSE | VCF | 0.735 | 0.737 | 0.400 | 0.421 |
| MAF | 0.621 | 0.667 | 0.225 | 0.277 | |
| Varscan2 | VCF | 0.656 | 0.661 | 0.250 | 0.348 |
| MAF | 0.653 | 0.661 | 0.308 | 0.309 | |
| SomaticSniper | VCF | 0.638 | 0.672 | 0.276 | 0.277 |
| MAF | 0.617 | 0.667 | 0.225 | 0.243 | |
| Average | VCF | 0.699 ± 0.061 | 0.709 ± 0.051 | 0.337 ± 0.086 | 0.372 ± 0.075 |
| MAF | 0.638 ± 0.022 | 0.664 ± 0.003 | 0.254 ± 0.039 | 0.291 ± 0.040 |
Discussion
Liver cancer is a global problem and occurs after severe liver diseases [46]. Chronic liver diseases are associated with cancer development and prompt progressive mutations at the genomic level [47, 48]. Previous studies report that liver cancer is associated with poor prognosis and a high mortality rate amongst the most frequent cancer types [49, 50]. Nowadays, several mutation calling techniques are available to identify the mutation landscape in tumour/normal patients. Hitherto, there is not an appropriate comparison of mutation detection methods for the predictive and prognostic analysis. In this study, we examine the performance of four widely used mutation calling techniques such as MuTect2, MuSE, Varscan2 and SomaticSniper using TCGA liver cancer cohort. We have performed correlation and survival analysis for the identification of prognostic biomarkers (i.e. risk-associated genes) in liver cancer patients. In addition, we have applied various machine learning techniques in order to compare all the methods for predicting high-risk liver cancer patients. First, we have used VCF and MAF files generated by the different mutation calling methods. We have used the most popular software (ANNOVAR and Maftools) to identify the gene-associated mutations in liver cancer samples. From the analysis, we observed that the VCF files of Mutect2 and SomaticSniper report highest number of mutated genes and cover over 5 million mutations. Whereas, MAF files reports comparatively less mutated genes for each technique as shown in Table 1.
Then, we performed correlation analysis in order to understand the impact of mutations on the survival of liver cancer patients. The univariate survival analysis revealed that risk-associated genes such as LncRNA SNGH10, CLMP, FAM160A2 and CLDN20 achieved the highest HR value in MuTect2, MuSE, Varscan2 and SomaticSniper technique, respectively. A study by Lan et al. also strengthen our findings and revealed that oncogenic lncRNA SNGH10 is associated with the poor survival in the liver cancer patients [51]. In addition, the down-regulation of SNGH10 is also associated with the poor survival in non-small cell lung cancer patients with HR = 2.09 and P = 0.02 [52]. Our study also corresponds with the previous studies and exhibits that the mutations in SNGH10 gene is associated with poor outcome in liver cancer patients with HR 5.49 and P-value 3.94E-06. Whereas, the differential expression of CLMP gene is associated with the progression of breast cancer [53]. Yang et al. also reported the significance of CLDN20 gene in the survival of breast cancer patients with HR 1.38 and P-value 0.047 [54]. Our analysis also revealed the role of CLMP and CLDN20 gene in the survival of liver cancer patients. Further, in the case of MAF files, the univariate survival analysis reveals that SYDE1, LAMC3, ITGB8, CAD, EVC2, NYNRIN, BRSK2 and TP53 genes significantly reduces the overall survival. As shown by the recent study, the overexpressed SYDE1 oncogene acts as an important diagnostic and prognostic biomarker in glioma patients [55]. Moreover, the down-regulation of LAMC3 is correlated with the poor prognosis and metastasis in the ovarian cancer patients [56]. A study also reveals that mutations associated with LAMC3 genes may cause paroxysmal nocturnal haemoglobinuria, a rare disorder of clonal stem cell in foetus, which may lead to high mortality rate infection and premature birth [57, 58]. We also observed that mutations associated with LAMC3 significantly reduce the survival of patients with HR = 9.25 and P-value of 1.78E-06. In addition, ITGB8 is shown to be highly upregulated in high-grade ovarian cancer patients, which leads to shorter OS with significant HR = 1.42 [59]. Paul etal. also reveals that EVC2 gene is highly mutated in breast cancer patients and dysregulates pathways like mTOR, CDK/RB, cAMP/PKA, WNT, etc. [60]. Our study showed that mutations associated with EVC2 genes reduce the overall survival of the patients with HR = 4.3 and P-value of 8.66E-05.
Researchers have shown that the overexpression of BRSK2 gene is correlated with the patients survival and prognosis in pancreatic cancer [61]. Of note, a number of studies report that TP53 is the highly mutated gene among most of the human cancers and affect the survival of cancer patients [62–66]. In our study, we also found that the number of mutations associated with TP53 gene is very high among the liver cancer patients and covers almost 20% mutations. Correlation and survival analysis showed that the mutation associated with TP53 significantly reduces the overall survival with HR = 1.63 and P = 1.20E-02. While considering the combined effect of the selected genes in each file, MuTect2 technique outperformed all the other techniques in VCF file with HR = 4.50 (P = 3.83E-15), whereas MuSE technique outperformed other mutation calling methods with HR = 2.47 (P = 9.64E-07) in the case of MAF files (Fig. 5). Furthermore, to compare the different mutation calling techniques, we develop various survival prediction and classification models using the top-10 risk-associated genes. Logistic regression-based model developed on 10 selected genes from VCF file of MuTect2 technique performed best among the other techniques in stratification of patients in high- and low-risk group with AUROC of 0.765 on validation dataset. In addition, MuSE also perform quite well with an AUROC of 0.735 on validation dataset, whereas Varscan2 and SomaticSniper-based models does not perform well on both VCF and MAF files. We examined the models developed using different machine learning techniques, and the results indicate that the error is not due to machine learning techniques as the performance measure AUROC was similar on training and validation dataset which signifies that these models are reliable, and no overfitting has been observed. Similarly, Mutect2 technique-based VCF reported the minimum error of 12.52 months using decision tree regressor, while predicting the OS time using different methods of regression (see Supplementary Table S3). Our results revealed that the VCF file generated using MuTect2 mutation calling technique provides the comprehensive information which can be used for the risk estimation of liver cancer cohort. Furthermore, this needs to be confirmed on the other cancer cohorts to explore the prognostic potential of mutations in different type of cancers. In order to aid the scientific community working in this era, we have developed a complete Python-based end-to-end pipeline (https://github.com/raghavagps/mutation_bench), where users need to provide only VCF/MAF files and can compare the performances of various prediction models developed on different mutation calling techniques.
Important findings
We examined the results to understand the limitations and propose some possible suggestions. We found that the classification and regression models developed using VCF/MAF file obtained from the MuTect2 technique performed better than the models developed using other mutation calling techniques. Of note, we can conclude that MuTect2 is a better mutation calling technique than the other techniques compared in this study. Additionally, our findings also indicate that the models based on VCF files perform better than models developed on MAF files for most of the mutation calling techniques except Varscan2. Since VCF file comprises information in the raw form, it is bigger in size in comparison to the MAF file which is a processed version. Hence, the number of mutations reduced drastically when we convert the VCF to MAF, but at the same time, performances declined too, i.e. during the conversion of VCF to MAF format, valuable and efficient variants may get dropped. Therefore, there is a need to develop an efficient method that converts VCF to MAF format without dropping useful information. Moreover, we identify that gene-based prognostic biomarkers are different for different techniques as well as for VCF and MAF format. Ideally, these variant calling techniques should display the same mutations in a given gene as well as the same biomarkers. It exhibits that the set of mutations in a given gene varies with the mutation calling techniques. Thus, there is a need to develop better variant calling methods or to identify the consensus mutations. A recent study [67] also revealed the importance of consensus mutations over hybrid models.
Supplementary data
Supplementary data is available at Biology Methods and Protocols online.
Supplementary Material
Acknowledgements
Authors are thankful to the Department of Computational Biology, IIIT-Delhi for infrastructure, Department of Biotechnology (DBT) and Department of Science and Technology (DST-INSPIRE) for financial support and fellowships.
Author contributions
S.P., A.D. and G.P.S.R. collected and processed the datasets. S.P., A.D. and G.P.S.R. implemented the algorithms. S.P., A.D. and G.P.S.R. developed the prediction models. S.P., A.D. and G.P.S.R. analysed the results. S.P., A.D. and G.P.S.R. penned the manuscript. G.P.S.R. conceived and coordinated the project and provided overall supervision to the project. All authors have read and approved the final manuscript.
Funding
The current work has not received any specific grant from any funding agencies.
Conflict of interest statement
The authors declare no competing financial and non-financial interests.
Contributor Information
Sumeet Patiyal, Department of Computational Biology, Indraprastha Institute of Information Technology, New Delhi 110020, India.
Anjali Dhall, Department of Computational Biology, Indraprastha Institute of Information Technology, New Delhi 110020, India.
Gajendra P S Raghava, Department of Computational Biology, Indraprastha Institute of Information Technology, New Delhi 110020, India.
References
- 1. Sung H, Ferlay J, Siegel RL. et al. Global cancer statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA A Cancer J Clin 2021;71:209–49 [DOI] [PubMed] [Google Scholar]
- 2. Gerlinger M, Rowan AJ, Horswell S. et al. Intratumor heterogeneity and branched evolution revealed by multiregion sequencing. N Engl J Med 2012;366:883–92 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Taylor SS. Protein kinases: a diverse family of related proteins. Bioessays 1987;7:24–9. [DOI] [PubMed] [Google Scholar]
- 4. Flaherty KT, Puzanov I, Kim KB. et al. Inhibition of mutated, activated BRAF in metastatic melanoma. N Engl J Med 2010;363:809–19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Lynch TJ, Bell DW, Sordella R. et al. Activating mutations in the epidermal growth factor receptor underlying responsiveness of non-small-cell lung cancer to gefitinib. N Engl J Med 2004;350:2129–39 [DOI] [PubMed] [Google Scholar]
- 6. Hirsch FR, Scagliotti GV, Mulshine JL. et al. Lung cancer: current therapies and new targeted treatments. Lancet 2017;389:299–311 [DOI] [PubMed] [Google Scholar]
- 7. Audeh MW, Carmichael J, Penson RT. et al. Oral poly(ADP-ribose) polymerase inhibitor olaparib in patients with BRCA1 or BRCA2 mutations and recurrent ovarian cancer: a proof-of-concept trial. Lancet 2010;376:245–51 [DOI] [PubMed] [Google Scholar]
- 8. LaDuca H, Farwell KD, Vuong H. et al. Exome sequencing covers >98% of mutations identified on targeted next generation sequencing panels. PLoS One 2017;12:e0170843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Lelieveld SH, Spielmann M, Mundlos S. et al. Comparison of exome and genome sequencing technologies for the complete capture of protein-coding regions. Hum Mutat 2015;36:815–22 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Hartley T, Wagner JD, Warman-Chardon J, et al. ; Care4Rare Canada Consortium. Whole-exome sequencing is a valuable diagnostic tool for inherited peripheral neuropathies: outcomes from a cohort of 50 families. Clin Genet 2018;93:301–9 [DOI] [PubMed] [Google Scholar]
- 11. Koboldt DC, Zhang Q, Larson DE. et al. VarScan 2: somatic mutation and copy number alteration discovery in cancer by exome sequencing. Genome Res 2012;22:568–76 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Kim S, Scheffler K, Halpern AL. et al. Strelka2: fast and accurate calling of germline and somatic variants. Nat Methods 2018;15:591–4 [DOI] [PubMed] [Google Scholar]
- 13. Alioto TS, Buchhalter I, Derdak S. et al. A comprehensive assessment of somatic mutation detection in cancer using whole-genome sequencing. Nat Commun 2015;6:10001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. do Valle IF, Giampieri E, Simonetti G. et al. Optimized pipeline of MuTect and GATK tools to improve the detection of somatic single nucleotide polymorphisms in whole-exome sequencing data. BMC Bioinformatics 2016;17:341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Cibulskis K, Lawrence MS, Carter SL. et al. Sensitive detection of somatic point mutations in impure and heterogeneous cancer samples. Nat Biotechnol 2013;31:213–9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Fan Y, Xi L, Hughes DS. et al. MuSE: accounting for tumor heterogeneity using a sample-specific error model improves sensitivity and specificity in mutation calling from sequencing data. Genome Biol 2016;17:178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Larson DE, Harris CC, Chen K. et al. SomaticSniper: identification of somatic point mutations in whole genome sequencing data. Bioinformatics 2012;28:311–7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Revathidevi S, Munirajan AK.. Akt in cancer: mediator and more. Semin Cancer Biol 2019;59:80–91 [DOI] [PubMed] [Google Scholar]
- 19. Wang K, Li M, Hakonarson H.. ANNOVAR: functional annotation of genetic variants from high-throughput sequencing data. Nucleic Acids Res 2010;38:e164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Mayakonda A, Lin DC, Assenov Y. et al. Maftools: efficient and comprehensive analysis of somatic variants in cancer. Genome Res 2018;28:1747–56 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Grossman RL, Heath AP, Ferretti V. et al. Toward a shared vision for cancer genomic data. N Engl J Med 2016;375:1109–12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Wei L, Jin Z, Yang S. et al. TCGA-assembler 2: software pipeline for retrieval and processing of TCGA/CPTAC data. Bioinformatics 2018;34:1615–7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Goel MK, Khanna P, Kishore J.. Understanding survival analysis: Kaplan-Meier estimate. Int J Ayurveda Res 2010;1:274–8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Pedregosa F, Varoquaux G, Gramfort A. et al. Scikit-learn: machine learning in python. J Mach Learn Res 2012;12:2825–30 [Google Scholar]
- 25. Kamiński B, Jakubczyk M, Szufel P.. A framework for sensitivity analysis of decision trees. Cent Eur J Oper Res 2018;26:135–59 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Denisko D, Hoffman MM.. Classification and interaction in random forests. Proc Natl Acad Sci USA 2018;115:1690–2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Geurts P, Ernst D, Wehenkel L.. Extremely randomized trees. Mach Learn 2006;63:3–42 [Google Scholar]
- 28. van der Ploeg T, Austin PC, Steyerberg EW.. Modern modelling techniques are data hungry: a simulation study for predicting dichotomous endpoints. BMC Med Res Methodol 2014;14:137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Chen TaG C. XGBoost: A Scalable Tree Boosting System. In: Proceedings of the 22nd ACM SIGKDD International Conference on Knowledge Discovery and Data Mining, 2016, 785–94
- 30. Nigsch F, Bender A, van Buuren B. et al. Melting point prediction employing k-nearest neighbor algorithms and genetic parameter optimization. J Chem Inf Model 2006;46:2412–22 [DOI] [PubMed] [Google Scholar]
- 31. Jahromi AH, Taheri M.. A non-parametric mixture of Gaussian naive Bayes classifiers based on local independent features. Artificial Intelligence and Signal Processing Conference (AISP). 2017, 209–12 [Google Scholar]
- 32. Rosasco L, De Vito E, Caponnetto A. et al. Are loss functions all the same? Neural Comput 2004;16:1063–76 [DOI] [PubMed] [Google Scholar]
- 33. Strobl C, Malley J, Tutz G.. An introduction to recursive partitioning: rationale, application, and characteristics of classification and regression trees, bagging, and random forests. Psychol Methods 2009;14:323–48 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Smith PF, Ganesh S, Liu P.. A comparison of random forest regression and multiple linear regression for prediction in neuroscience. J Neurosci Methods 2013;220:85–91 [DOI] [PubMed] [Google Scholar]
- 35. Hidalgo B, Goodman M.. Multivariate or multivariable regression? Am J Public Health 2013;103:39–40 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Reid S, Tibshirani R.. Sparse regression and marginal testing using cluster prototypes. Biostatistics 2016;17:364–76 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. de Vlaming R, Groenen PJ.. The current and future use of ridge regression for prediction in quantitative genetics. Biomed Res Int 2015;2015:1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Liu M, Vemuri BC.. A robust and efficient doubly regularized metric learning approach. Comput Vis ECCV 2012;7575:646–59 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Dey S, Eslamy M, Yoshida T. et al. A support vector regression approach for continuous prediction of ankle angle and moment during walking: an implication for developing a control strategy for active ankle prostheses. IEEE Int Conf Rehabil Robot 2019;2019:727–33 [DOI] [PubMed] [Google Scholar]
- 40. Patiyal S, Agrawal P, Kumar V. et al. NAGbinder: an approach for identifying N-acetylglucosamine interacting residues of a protein from its primary sequence. Protein Sci 2020;29:201–10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Kaur H, Dhall A, Kumar R. et al. Identification of platform-independent diagnostic biomarker panel for hepatocellular carcinoma using large-scale transcriptomics data. Front Genet 2019;10:1306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Dhall A, Patiyal S, Kaur H. et al. Computing skin cutaneous melanoma outcome from the HLA-alleles and clinical characteristics. Front Genet 2020;11:221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Bhalla S, Kaur H, Dhall A. et al. Prediction and analysis of skin cancer progression using genomics profiles of patients. Sci Rep 2019;9:15790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Schemper M. The relative importance of prognostic factors in studies of survival. Stat Med 1993;12:2377–82 [DOI] [PubMed] [Google Scholar]
- 45. Lex A, Gehlenborg N, Strobelt H. et al. UpSet: visualization of Intersecting Sets. IEEE Trans Vis Comput Graph 2014;20:1983–92 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Davis GL, Dempster J, Meler JD. et al. Hepatocellular carcinoma: management of an increasingly common problem. Proc (Bayl Univ Med Cent) 2008;21:266–80 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Muller M, Bird TG, Nault JC.. The landscape of gene mutations in cirrhosis and hepatocellular carcinoma. J Hepatol 2020;72:990–1002 [DOI] [PubMed] [Google Scholar]
- 48. Farazi PA, DePinho RA.. Hepatocellular carcinoma pathogenesis: from genes to environment. Nat Rev Cancer 2006;6:674–87 [DOI] [PubMed] [Google Scholar]
- 49. Lin L, Yan L, Liu Y. et al. The burden and trends of primary liver cancer caused by specific etiologies from 1990 to 2017 at the global, regional, national, age, and sex level results from the global burden of disease study 2017. Liver Cancer 2020;9:563–82 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Balogh J, Victor D, Asham EH. et al. Hepatocellular carcinoma: a review. J Hepatocell Carcinoma 2016;3:41–53 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Lan T, Yuan K, Yan X. et al. LncRNA SNHG10 facilitates hepatocarcinogenesis and metastasis by modulating its homolog SCARNA13 via a positive feedback loop. Cancer Res 2019;79:3220–34 [DOI] [PubMed] [Google Scholar]
- 52. Liang M, Wang L, Cao C. et al. LncRNA SNHG10 is downregulated in non-small cell lung cancer and predicts poor survival. BMC Pulm Med 2020;20:273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Nilchian A, Johansson J, Ghalali A. et al. CXADR-mediated formation of an AKT inhibitory signalosome at tight junctions controls epithelial-mesenchymal plasticity in breast cancer. Cancer Res 2019;79:47–60 [DOI] [PubMed] [Google Scholar]
- 54. Yang G, Jian L, Chen Q.. Comprehensive analysis of expression and prognostic value of the claudin family in human breast cancer. Aging (Albany NY) 2021;13:8777–96 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Han Z, Zhuang X, Yang B. et al. SYDE1 acts as an oncogene in glioma and has diagnostic and prognostic values. Front Mol Biosci 2021;8:714203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Lei SM, Liu X, Xia LP. et al. Relationships between decreased LAMC3 and poor prognosis in ovarian cancer. Zhonghua Fu Chan Ke Za Zhi 2021;56:489–97 [DOI] [PubMed] [Google Scholar]
- 57. De Angelis C, Byrne AB, Morrow R. et al. Compound heterozygous variants in LAMC3 in association with posterior periventricular nodular heterotopia. BMC Med Genomics 2021;14:64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Qian X, Liu X, Zhu Z. et al. Variants in LAM. C3 causes occipital cortical malformation. Front Genet 2021;12:616761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. He J, Liu Y, Zhang L. et al. Integrin subunit beta 8 (ITGB8) upregulation is an independent predictor of unfavorable survival of high-grade serous ovarian carcinoma patients. Med Sci Monit 2018;24:8933–40 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Paul MR, Pan TC, Pant DK. et al. Genomic landscape of metastatic breast cancer identifies preferentially dysregulated pathways and targets. J Clin Invest 2020;130:4252–65 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Lou Dr W, Niu G.. BRSK2 expression as a prognosis marker in pancreatic cancer patients. J Clin Oncol 2009;27:e15603 [Google Scholar]
- 62. Olivier M, Hollstein M, Hainaut P.. TP53 mutations in human cancers: origins, consequences, and clinical use. Cold Spring Harb Perspect Biol 2010;2:a001008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Petitjean A, Achatz MIW, Borresen-Dale AL. et al. TP53 mutations in human cancers: functional selection and impact on cancer prognosis and outcomes. Oncogene 2007;26:2157–65 [DOI] [PubMed] [Google Scholar]
- 64. Monti P, Menichini P, Speciale A. et al. Heterogeneity of T. P53 mutations and P53 protein residual function in cancer: does it matter? Front Oncol 2020;10:593383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Ungerleider NA, Rao SG, Shahbandi A. et al. Breast cancer survival predicted by TP53 mutation status differs markedly depending on treatment. Breast Cancer Res 2018;20:115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Rosenberg S, Okamura R, Kato S. et al. Survival implications of the relationship between tissue versus circulating tumor DNA TP53 mutations-A perspective from a real-world precision medicine cohort. Mol Cancer Ther 2020;19:2612–20 [DOI] [PubMed] [Google Scholar]
- 67. Wang M, Luo W, Jones K. et al. SomaticCombiner: improving the performance of somatic variant calling based on evaluation tests and a consensus approach. Sci Rep 2020;10:12898. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.