T-cell large granular lymphocyte leukemias (T-LGLL) is a heterogeneous disease with special histology, immune phenotype and genetic abnormalities. The 2017 World Health Organization (WHO) Classification recommends 2-20 x 10E + 9/L for LGL population in peripheral blood [1]. In practice, persons with leukopenia fails to meet this cutoff value, but they have other clinical features of T-LGLL, such as anemia, high proportion of clonal LGL population, and STAT3 mutations. We usually considered these oligolymphocytic large granular lymphocyte proliferations (LGLP) as T-LGLL.
Some recurrent mutations such as STAT3, STAT5B and TNFAIP3 are found in T-LGLL [2–4]. STAT3 mutations are reported in 30–75% of persons with T-LGLL resulting in STAT3 activation [2,5–8]. However, STAT3 activation is also reported in persons without a STAT3 mutation suggesting STAT3 is important in the pathogenesis of T-LGLL [9]. But STAT3 mutations are not included in the current diagnostic criteria of T-LGLL and T-LGLLs with and without STAT3 mutations are not distinguished in present classifications [1]. STAT5B activating mutations are reported previously in only 1 percent of persons with T-LGLL [3]. STAT5B mutations occurred mainly in CD4 positive T-LGLL [10]. Only one study detected TNFAIP3 mutations in 8 percent of persons with T-LGLL, and this finding needs to be confirmed by more studies [4].
Some studies report LGLP can be found in solid cancers and myeloid neoplasms (MN), especially in myelodysplastic syndromes (MDS) [11–14]. One hypothesis is that cancer-related neoantigens trigger an immune response activating cytotoxic T-cells. These activated polyclonal cells transform into monoclonal T-cell population under selection pressures [1,8]. Here, we compare the clinical and molecular features of T-LGLPs with or without MN. In addition, we also explored the distribution of LGLL related gene mutations.
1622 subjects including 983 with MDS, 195 with myeloproliferative neoplasm (MPN), 102 with MDS/MPN, 89 with acute myeloid leukemia (AML) and 253 cytopenias caused by other reasons were enrolled in the study From June 2018 to May 2020. Subjects provided informed consent in compliance with the Declaration of Helsinki. Flow cytometric immune phenotypic analyses were done on blood samples by usual multi-parameter flow cytometry. Antigens analyzed included CD2, CD3, CD4, CD5, CD7, CD8, CD16, CD56, CD57, TCR α/β and TCR γ/δ. T-cell receptor (TCR) rearrangement was assessed by multiplex polymerase chain reaction (PCR). Next generation sequencing (NGS) was done for coding sequences of 137 genes including STAT3, STAT5b and TNFAIP3 associated with hematologic neoplasms as described (Supplementary Table 1).
Diagnosis of T-LGLP requires these criteria: (1) expression of abnormal T-LGL immune phenotype such as CD3 +, CD8 +, CD5 dim+/− CD7 dim+/− and CD57 +; (2) clonal TCR gene rearrangement by PCR; (3) blood LGL concentration ≥ 0.5 × 10E + 9/L or <0.5 × 10E + 9/L if there was an unexplained cytopenia. When the 1st 2 criteria were met in someone with a WHO-defined myeloid neoplasm the assigned diagnosis was myeloid neoplasms with coexisting LGLP (MN-LGLP). Using these criteria 23 subjects were diagnosed as simple T-LGLP and 16 as MN-LGLP including 10 with MDS (Supplementary Table 2).
STAT3 mutations were detected in 19 subjects with T-LGLP 80 percent of which were in the SH2 domain. p.Y640F was the most common variant (Figure 1(A)). STAT3 mutations were not detected in MN-LGLP cohort but STAT5B mutations were detected in 3 higher-risk MDS subjects with CD8 positive T-LGLP including 2 with MDS with excess blasts (MSD-EB) and 1 with MDS with ring sideroblasts (MDS-RS). The subjects with MDS-RS had a TP53 mutation and a complex karyotype. An abnormal karyotype (-Y) was detected in only 1 subject in the T-LGLP cohort, but - Y is common in older males and may not really an abnormality. In contrast, seven of 12 subjects in the MN-LGLP cohort had an abnormal karyotype.
Figure 1.
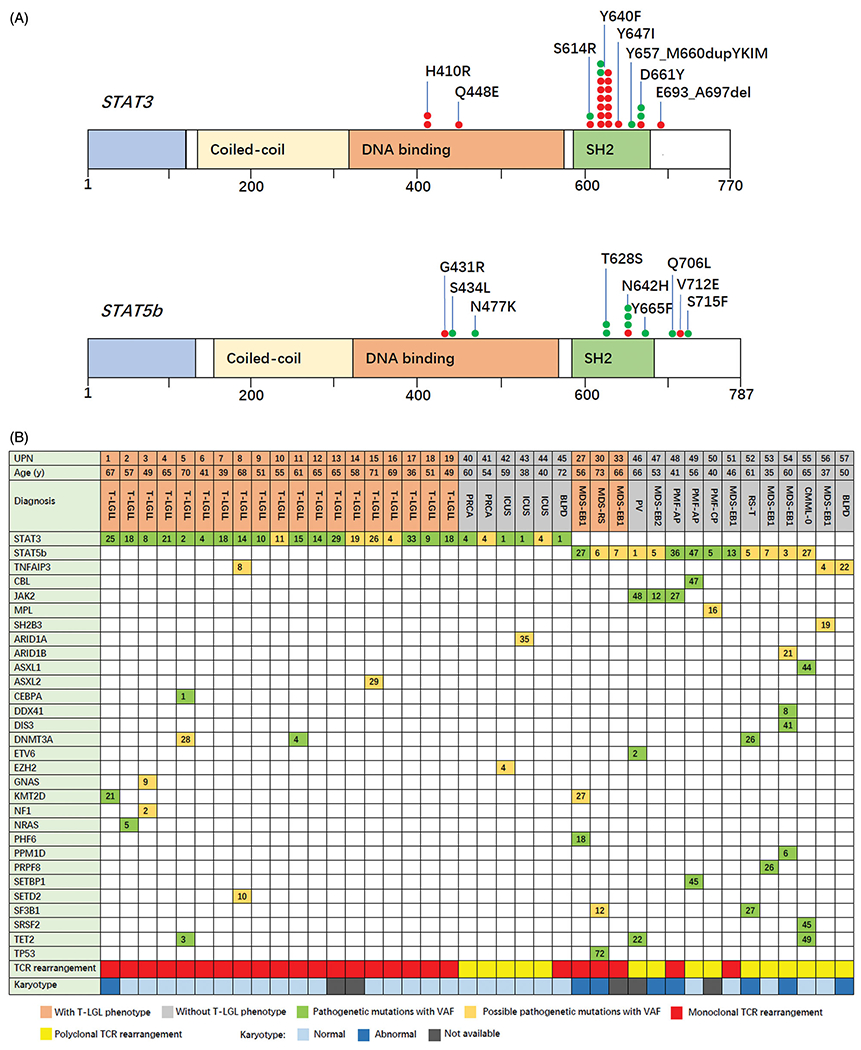
STAT3 and STAT5B gene variant sites and concomitant mutations. (A) The upper gene structure map represents the STAT3 variant site and the lower map, the STAT5B variant site. Red circles represent presence of large granular lymphocyte proliferation and the green circles, absence of large granular lymphocyte proliferation; (B) The Oncoplot represents the diagnosis and concomitant mutations of each subject with STAT3, STAT5B or TNFAIP3 variants. Number in the box represents the mutant gene variant allele fraction (%). AP: accelerated phase; BLPD: B-cell lympho-proliferative diseases; CMML: chronic myelomonocytic leukemia; CP: chronic phase; PMF: primary myelofibrosis; RS-T: ring sideroblasts and thrombocytosis; UPN: unique patient number.
Subjects in the T-LGLP cohort had higher percent blood LGLs and a higher blood concentration, more severe anemia and lower percent reticulocytes compared with MN-LGLP cohort (Table 1). Eight subjects had pure red cell aplasia (PRCA). Average number of mutations was 1.48 and the average variant allele fraction (VAF) < 10 percent in the T-LGLP cohort except for STAT3 mutation. Subjects in the MN-LGLP cohort had more mutations and higher VAFs (Table 1, Figure 1(B)).
Table 1.
Comparison of clinical features between T-LGLP and MN-LGLP.
| Co-variate | T-LGLP (n = 23) | MNa-LGLP (n = 1 6) | p-value |
|---|---|---|---|
| Male | 15 | 11 | .818 |
| Age, years (median, range) | 58 (33–71) | 62 (33–75) | .558 |
| WBC × 10E + 9/L (median, range) | 3.96 (0.65–27.7) | 2.7 (1.11–28) | .475 |
| ANC × 10E + 9/L (median, range) | 1.38 (0.09–21.86) | 1.05 (0.43–18) | .711 |
| ALC × 10E + 9/L (median, range) | 1.85 (0.53–5.45) | 1.29 (0.47–4.58) | .061 |
| LGL % | 36 (6–66) | 17.3 (4–24.5) | .001 |
| ALGLC × 10E + 9/L (median, range) | 1.12 (0.3–4.15) | 0.49 (0.15–1.78) | .005 |
| ALGLC ≥ 0.5 × 10E + 9/L | 21 | 8 | .011 |
| Hemoglobin g/L (median, range) | 63 (41–139) | 103 (54–140) | .001 |
| MCV, fL (median, range) | 99.3 (78.7–118) | 105.9 (78.3–124.8) | .166 |
| Reticulocytes % (median, range) | 0.7 (0.14–4.4) | 2.25 (0.81–6.09) | .000 |
| PLT, × 109/L (median, range) | 232 (4–287) | 74.5 (22–935) | .07 |
| PRCA phenotype | 8 | 0 | .004 |
| Abnormal karyotype | 1/21 | 7/12 | .002 |
| STAT3 mutation | 19 | 0 | .000 |
| STAT5B mutation | 0 | 3 | .061 |
| TNFAIP3 mutation | 1 | 0 | .335 |
| N mutations (mean ± SD) | 1.48 ± 1.04 | 3 ± 1.79 | .002 |
| VAF other than STAT3/5B, % (mean ± SD) | 9.79 ± 9.96 | 24.85 ± 16.99 | .000 |
ALC: Absolute lymphocyte concentration; ALGLC: Absolute large granular lymphocyte concentration; ANC: Absolute neutrophil concentration; MCV: mean corpuscular volume; WBC: white blood cell concentration.
Myeloid neoplasms include MDS with single lineage dysplasia (n = 1), MDS with multi-lineage dysplasia (n = 3), MDS with excess blasts (n = 5), MDS with ring sideroblasts (n = 1), primary myelofibrosis (n = 3), essential thrombocythemia (n = 1), atypical chronic myeloid leukemia (n = 1), and systemic mastocytosis (n = 1).
We screened the sequencing data of another 1583 subjects without LGLP detecting STAT3 mutations (n = 6), STAT5B mutations (n = 10) and TNFAIP3 mutations (n = 2) in 18 subjects (Supplementary Table 3). Clonal TCR gene rearrangement was detected in 3 subjects. Diagnoses of the 6 subjects with STAT3 mutations included idiopathic PRCA (n = 2), idiopathic cytopenia of undetermined significance (ICUS; n = 3) and CD5-/CD10- small B-cell lymphoma, unclassifiable (n = 1; Figure 1(B)). Average VAF of STAT3 mutations was 2.53% (range, 1.17–3.89%) which was significantly lower than 15.66% (range, 6.9–24.42%) of VAFs of the T-LGLL cohort (p = .001). Diagnoses of the 10 subjects with STAT5B mutations included MDS-EB (n = 4), primary myelofibrosis, accelerated phase (PMF; n = 2), PMF, chronic phase (n = 1), polycythemia vera (PV; n = 1), chronic myelomonocytic leukemia-0 (CMML; n = 1) and MDS/MPN with ring sideroblasts and thrombocytosis (RS-T, n = 1). The most common variants were in the SH2 domain with p.N642H (n = 3) and p.T628S (n = 2; Figure 1(A)). 5 subjects with STAT5B mutations had co-mutations of other JAK-STAT signal pathway genes including JAK2V617F (n = 3), CBLC404Y (n = 1) and MPLY591N (n = 1; Figure 1(B)). The 2 subjects with a TNFAIP3 mutation had MDS-EB1 and splenic marginal zone lymphoma (SMZL). A SH2B3 mutation was found in 1 subject. These results clearly indicate that STAT3, STAT5B and TNFAIP3 mutations are not specific for LGLL.
Our results have several interesting implications. 1st, there are different clinical and molecular features between MN-LGLP and simple T-LGLP. In particular, STAT3 mutations were detected in most subjects with T-LGLP but not in subjects with MN-LGLP. These data indicate that the pathogenesis of LGLP in two cohorts is different. Although the definition of LGL leukemia subsets according to associated diseases does not improve the diagnosis and treatment of this rare disease, the presence of a driver gene contributes to distinguish whether LGLP is a real leukemia or a transient response to neoantigens stimulation.
2nd, the median number of LGL in T-LGLP cohort was 1.12 x 10E + 9/L with only 4 subjects greater than 2 x 10E + 9/L, the frequency of STAT3 mutations was as high as 83%. It suggests that these oligolymphocytic T-LGLP have the same pathogenesis as other typical T-LGLL with STAT3 mutation. We suggest that T-LGLL with STAT3 mutations should be classified into one subtype of T-LGLLs, instead of being defined according to the arbitrarily LGL threshold. It is reasonable to stratify this heterogeneous disease according to molecular profiles, which can be relevant for targeted treatment options.
3nd, subjects with a low STAT3 mutation VAF typically present with unexplained cytopenia(s). Although no abnormal T-LGL phenotype was detected it is impossible to exclude a STAT3 mutation as the cause of the cytopenia(s). These persons may eventually develop T-LGLP. Consequently, we recommend that persons with an unexplained cytopenia(s) should be screened with NGS or sensitive techniques to sequence STAT3.
Lastly, eight of 13 subjects with STAT5B mutations had an advanced myeloid neoplasm including MDS-EB (n = 6) and PMF, accelerated phase (n = 2). A previous study has shown that STAT5BN642H mutation is associated with myeloid neoplasms with eosinophilia [15]. To our knowledge, this is the first report that STAT5B mutations are associated with advanced myeloid neoplasms. STAT5B is often co-mutated with other JAK-STAT signaling pathway genes. We speculate continuous STAT5B activation may induce mutation. Due to the small number of subjects, these findings need to be confirmed by more studies.
Our research has several limitations. 1st, as a national tertiary blood disease hospital our subjects may reflect selection biases. 2nd, most subjects are lost to long-term follow-up because they return to the referral hospital.
In summary, our data indicate STAT3 mutations occur predominately in persons with simple T-LGLP and unexplained cytopenia(s) whereas STAT5B mutations occur predominately in persons with advanced myeloid neoplasms and who often have co-mutations in other JAK-STAT signaling pathway genes.
Supplementary Material
Funding
Supported in part by National Natural Science Funds (No.81870104, No.81530008, No.81770129), Tianjin Natural Science Funds (No.18JCZDJC34900, No. 19JCQNJC09400), CAMS Initiative Fund for Medical Sciences (No. 2016-I2M-1-001). RPG acknowledges support from the National Institute of Health Research NIHR) Biomedical Research Centre funding scheme.
Footnotes
Supplemental data for this article can be accessed here.
Disclosure statement
No potential conflict of interest was reported by the author(s).
References
- [1].Lamy T, Moignet A, Loughran TP Jr.. LGL leukemia: from pathogenesis to treatment. Blood. 2017;129(9):1082–1094. [DOI] [PubMed] [Google Scholar]
- [2].Koskela HL, Eldfors S, Ellonen P, et al. Somatic STAT3 mutations in large granular lymphocytic leukemia. N Engl J Med. 2012;366(20):1905–1913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Rajala HL, Eldfors S, Kuusanmäki H, et al. Discovery of somatic STAT5b mutations in large granular lymphocytic leukemia. Blood. 2013;121(22):4541–4550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Johansson P, Bergmann A, Rahmann S, et al. Recurrent alterations of TNFAIP3 (A20) in T-cell large granular lymphocytic leukemia. Int J Cancer. 2016;138(1):121–124. [DOI] [PubMed] [Google Scholar]
- [5].Fasan A, Kern W, Grossmann V, et al. STAT3 mutations are highly specific for large granular lymphocytic leukemia. Leukemia. 2013;27(7):1598–1600. [DOI] [PubMed] [Google Scholar]
- [6].Jerez A, Clemente MJ, Makishima H, et al. STAT3 mutations indicate the presence of subclinical T-cell clones in a subset of aplastic anemia and myelodysplastic syndrome patients. Blood. 2013;122(14):2453–2459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Rajala HL, Olson T, Clemente MJ, et al. The analysis of clonal diversity and therapy responses using STAT3 mutations as a molecular marker in large granular lymphocytic leukemia. Haematologica. 2015;100(1):91–99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Durrani J, Awada H, Kishtagari A, et al. Large granular lymphocytic leukemia coexists with myeloid clones and myelodysplastic syndrome. Leukemia. 2020;34(3):957–962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Teramo A, Gattazzo C, Passeri F, et al. Intrinsic and extrinsic mechanisms contribute to maintain the JAK/STAT pathway aberrantly activated in T-type large granular lymphocyte leukemia. Blood. 2013;121(19):3843–3854. [DOI] [PubMed] [Google Scholar]
- [10].Andersson EI, Tanahashi T, Sekiguchi N, et al. High incidence of activating STAT5B mutations in CD4-positive T-cell large granular lymphocyte leukemia. Blood. 2016;128(20):2465–2468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Saunthararajah Y, Molldrem JL, Rivera M, et al. Coincident myelodysplastic syndrome and T-cell large granular lymphocytic disease: clinical and pathophysiological features. Br J Haematol. 2001;112(1):195–200. [DOI] [PubMed] [Google Scholar]
- [12].Huh YO, Medeiros LJ, Ravandi F, et al. T-cell large granular lymphocyte leukemia associated with myelodysplastic syndrome: a clinicopathologic study of nine cases. Am J Clin Pathol. 2009;131(3):347–356. [DOI] [PubMed] [Google Scholar]
- [13].Viny AD, Maciejewski JP. High rate of both hematopoietic and solid tumors associated with large granular lymphocyte leukemia. Leuk Lymphoma. 2015;56(2):503–504. [DOI] [PubMed] [Google Scholar]
- [14].Komrokji RS, Ali NA, Sallman D, et al. Characterization of myelodysplastic syndromes (MDS) with T-cell large granular lymphocyte proliferations (LGL) [published online ahead of print. Leukemia. 2020;34(11):3097–3099. [DOI] [PubMed] [Google Scholar]
- [15].Cross NCP, Hoade Y, Tapper WJ, et al. Recurrent activating STAT5B N642H mutation in myeloid neoplasms with eosinophilia. Leukemia. 2019;33(2):415–425. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


