Abstract
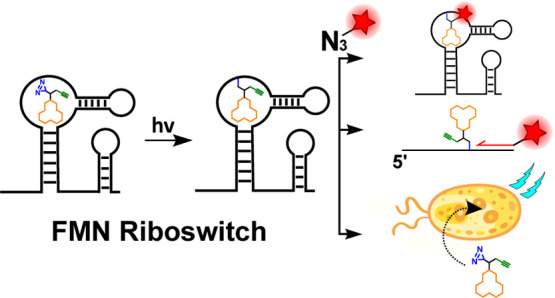
Riboswitches are structural RNA elements that control gene expression. These naturally occurring RNA sensors are of continued interest as antibiotic targets, molecular sensors, and functional elements of synthetic circuits. Here, we describe affinity-based profiling of the flavin mononucleotide (FMN) riboswitch to characterize ligand binding and structural folding. We designed and synthesized photoreactive ligands and used them for photoaffinity labeling. We showed selective labeling of the FMN riboswitch and used this covalent interaction to quantitatively measure ligand binding, which we demonstrate with the naturally occurring antibiotic roseoflavin. We measured conditional riboswitch folding as a function of temperature and cation concentration. Furthermore, combining photoaffinity labeling with reverse transcription revealed ligand binding sites within the aptamer domain with single-nucleotide resolution. The photoaffinity probe was applied to cellular extracts of Bacillus subtilis to demonstrate conditional folding of the endogenous low-abundant ribD FMN riboswitch in biologically derived samples using quantitative PCR. Lastly, binding of the riboswitch-targeting antibiotic roseoflavin to the FMN riboswitch was measured in live bacteria using the photoaffinity probe.
Introduction
RNA is a multifaceted biomolecule that exhibits many crucial cellular functions ranging from architectural to catalytic.1−3 These functions go far beyond the initially proposed role of RNA as information carrier. Recent discoveries have revealed the role of RNA in chromatin regulation,4 post-transcriptional regulation,5 gene silencing6 and enhancing,7 and transcription and translational control through riboswitches,8−14 among other functions. The newly appreciated importance of RNA in pathological processes has sparked interest in the development of selective small-molecule drugs that can modulate cellular RNA activity.15−18 To study the diverse functions, structure, and druggability of RNA, there is a need for new chemical tools.3,19−23
Early pioneering studies demonstrated the broad potential of (photo)chemical methods to investigate nucleic acid structure and function.24−28 An emerging approach to study RNA is applying small chemical probes that bind RNA with high affinity and that form covalent bonds between the probe and RNA of interest. The covalently bound small-molecule probe can be subsequently modified for analysis. Recent reports have employed this concept to investigate small-molecule binding to pre-mRNAs29 and microRNAs30,31 (CHEM-Clip) and artificial synthetic aptamers (PEARL-seq).32 Here, we explore if we can apply similar principles to characterize ligand binding and structural folding of bacterial riboswitches (Figure 1A).
Figure 1.
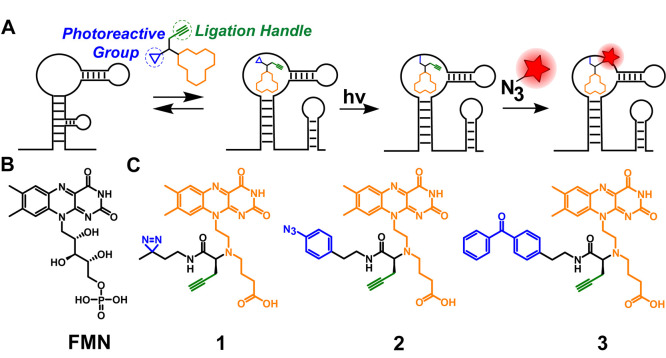
Schematic overview of affinity-based profiling of riboswitches. (A) Schematic illustration of the photoaffinity labeling workflow. Cross-linking is achieved through light activation of the photoreactive group, and the alkyne ligation handle enables downstream analysis. (B) Molecular structure of the natural ligand flavin mononucleotide (FMN). (C) Structures of the three designed and synthesized bifunctional photoaffinity probes 1, 2, and 3. The orange parts represent the RNA-binding moiety, the blue parts the photoactivatable groups, and the green parts the attached alkyne ligation handle.
Riboswitches are cis-regulatory structural RNA elements that are found in the 5′ untranslated region of mRNAs. They control gene expression in several organisms and are broadly distributed across bacteria.10,11,33,34 They consist of an aptamer domain and an expression platform. Binding of the cognate ligand to the aptamer induces a conformational change in the expression platform, thereby altering the expression of the downstream gene.10,35 The aptamer is highly conserved and serves as a sensor for its target metabolite. These sensors must be sufficiently dynamic to respond and adapt precisely to specific signals, to rapidly produce the intended effect for cell survival.
One widely distributed class of RNA regulatory elements in bacteria is the flavin mononucleotide (FMN) riboswitch with FMN (Figure 1B) as primary regulatory ligand.8,36 This riboswitch controls expression of genes required for biosynthesis and transport of riboflavin, an important vitamin for both bacteria and humans.37 FMN regulates the expression of these downstream genes upon binding to the aptamer.8
Mechanistic data of riboswitches are often obtained with synthetic RNA, using a combination of physical methods, such as nuclear magnetic resonance (NMR) studies,38−40 X-ray diffraction crystallography,36,41 fluorescence spectroscopy,42,43in vitro profiling methods,8,41,44,45 and genetic approaches.46,47 Though biologically vital, the structural features of riboswitches can prove challenging to study and remain of continued interest.11,33
Chemical methods that covalently capture the interaction between a riboswitch and its ligand could further assist in elucidating riboswitch properties and druggability.
Here, we set out to develop chemical photoaffinity probes to study the interactions between the FMN riboswitch and its ligands on purified RNA, as well as in bacterial extracts and live bacteria. Inspired by activity-based protein profiling48,49 and recent reports of photoaffinity labeling of RNA,29,32,50−52 we designed three photoreactive FMN ligands (Figure 1C). Labeling of the FMN riboswitch was observed for two of three synthesized probes. Competition experiments with its cognate ligand FMN and the naturally occurring antibiotic roseoflavin demonstrate the potential use of photoaffinity probes to screen for riboswitch inhibitors. Conditional riboswitch folding, controlled by temperature and cation concentration, was analyzed with the probe. Furthermore, photo-cross-linking sites could be identified with single-nucleotide resolution, potentially helping to pinpoint the binding site within the riboswitch aptamer domain. Finally, quantification of probe binding in bacterial extracts and live bacteria was achieved using bead enrichment and RT-qPCR, demonstrating the potential use of photoaffinity labeling to measure riboswitch inhibitor binding in vivo.
Results and Discussion
Design and Synthesis of FMN Riboswitch Photoaffinity Probes
To measure the interactions between the FMN riboswitch and its ligand, we designed and synthesized photoreactive derivatives of FMN that can covalently label RNA when bound in the aptamer region of the riboswitch and exposed to UV light (Figure 1A and C). Seminal work by Vicens, Batey, and co-workers53 provided a set of principles to ensure specific and productive binding of FMN analogues to the FMN riboswitch that we followed.53 Importantly, they showed that the ribityl-phosphate chain attached to position 10 of the isoalloxazine ring of FMN (Figure 1B) can be modified without perturbing the interactions between the FMN riboswitch and its ligand. Additionally, it was found that the hydroxyl groups of the ribityl chain could be removed without losing affinity for the riboswitch.53 Lastly, it was shown that the terminal phosphate group can be replaced with a carboxylic anion moiety.53 On the basis of these structural constraints, we designed probes 1, 2, and 3, which all contain the isoalloxazine core that is crucial for binding (Figure 1C).53 The ribityl chain was replaced with a linker containing a terminal carboxylate and tertiary amine to which a photoreactive group and alkyne are appended. The tertiary amine was also found to be beneficial for enhanced affinity for the FMN riboswitch and bacterial uptake.53 Three photoaffinity probes were designed and synthesized, containing the frequently used diazirine, aryl azide, and benzophenone photoreactive groups (Figure 1C).54 When the probes bind in the FMN riboswitch aptamer domain, activation of the photoreactive groups can induce the formation of a covalent bond with the FMN RNA. For convenient downstream analysis of these probe–RNA complexes, an alkyne ligation handle was included in the designs of probes 1, 2, and 3 (Figure 1C).
Performance of Photoaffinity Probes
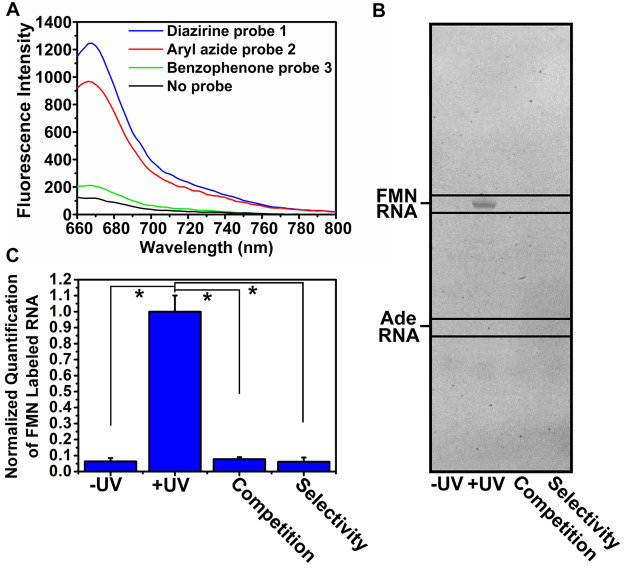
The performance of photoaffinity probes 1, 2, and 3 was examined by measuring probe binding to an in vitro transcribed FMN riboswitch (see the SI for details) upon UV irradiation. After ligating a Cy5 fluorophore to the probe–RNA complexes using copper(I)-catalyzed azide–alkyne cycloaddition, the fluorescence intensity was measured to examine probe binding at concentrations ranging 2–100 μM (Figures 2A and S1). At a concentration of 10 μM, diazirine probe 1 efficiently labeled the riboswitch, as was apparent from a significant fluorescent signal (Figures 2A and S1).
Figure 2.
Efficiency and selectivity of FMN probes 1, 2, and 3. (A) Fluorescence intensity measurements after labeling FMN riboswitch aptamer with 10 μM 1, 2, or 3 and subsequent “click” reaction with Cy5-azide. (B) Denaturing RNA gel showing selective binding of probe 1 to the FMN RNA aptamer. (C). Quantification of band intensities of the PAGE experiment in B in triplicate. “Competition” is labeling in the presence of 10 μM FMN natural ligand. “Selectivity” is labeling on the adenine riboswitch. Error bars represent standard deviations based on three technical replicates. Statistical significance was calculated using an unpaired two-tailed Student’s t test (*p < 0.05).
Aryl azide probe 2 displayed efficient labeling as well, whereas benzophenone probe 3 showed only minor fluorescence. A potential explanation is that the size of the photo-cross-linking group is crucial for optimal probe binding, with the bulky benzophenone group being too sterically hindered to effectuate labeling.54 Moreover, it is possible that the “click” reaction works less efficiently for ligated probe 3, which would result in lower fluorescence. Because of the superior performance of probe 1 compared to probe 3 and slightly better performance than probe 2, we decided to use probe 1 for all further experiments.
To test the selectivity of probe 1 for the FMN riboswitch, labeling was analyzed with denaturing polyacrylamide gel electrophoresis (PAGE) (Figures 2B,C and S2). Both UV irradiation and fluorophore ligation were required to observe labeling of the FMN riboswitch by probe 1. To show that probe 1 selectively bound to the FMN riboswitch, 10 μM FMN natural ligand was incubated as a competitor for the probe to bind to the riboswitch aptamer. The observed labeling signal disappeared, indicating that the probe was competed out of the aptamer domain (Figures 2B,C and S2). This might imply a selective interaction between probe 1 and the FMN riboswitch. To further demonstrate selectivity, probe 1 was incubated with the adenine riboswitch, instead of the FMN riboswitch. No labeling of the adenine riboswitch was detected, underlining the selectivity of probe 1 for the FMN riboswitch aptamer (Figures 2B,C and S2).
Structure Probing of the Riboswitch–Probe Interaction
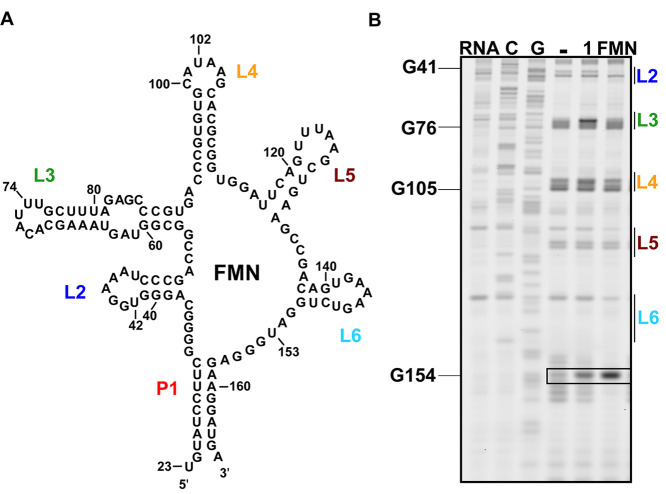
To study the binding interaction between probe 1 and the FMN riboswitch, SHAPE analysis3,55,56 was performed (see the SI for details). For SHAPE, FMN and probe 1 were separately incubated at 100 μM with the riboswitch at 37 °C for 30 min, after which 50 mM 2-methylnicotinic acid imidazolide (NAI)57 was added to each sample. The samples were incubated at 37 °C for 10 min, and RNA was isolated using precipitation. Reverse transcription (RT) was performed and analyzed using PAGE (Figures 3 and S3). Increased reactivity at U153 was observed for both FMN and probe 1. This is believed to be typical for ligand binding to the aptamer and caused by an altered conformation in which U153 is bulged out and exposed to solvent;44 it might indicate that probe 1 displays similarity in binding to the aptamer as compared to FMN. Slight differences in SHAPE profile were observed as well at nucleotides U42, U74, and U102, which are located in L2, L3, and L4, respectively, and may be caused by differences in structure of the side-chain. Increased reactivity in these loop regions was observed previously for FMN structural analogues on which probe 1 was based.53 This was further analyzed by molecular docking of probe 1 to the FMN aptamer (Figure S4), which showed similar binding of the isoalloxazine core to FMN and no noticeable differences at nucleotide U42.
Figure 3.
SHAPE analysis of interaction of probe 1 with the FMN riboswitch. (A) Secondary structure of the FMN riboswitch from Bacillus subtilis, with loop regions annotated in color. (B) PAGE of the SHAPE experiment with probe 1 and the FMN ligand. The boxed area shows the area of the gel where characteristic changes in the SHAPE profile are observed for ligand binding. For triplicate experiments and quantification of band intensities see Figure S3.
Together, these results imply that the isoalloxazine core of photoreactive probe 1 likely interacts with the FMN riboswitch in a similar manner to the natural ligand and that the diazirine group in the side-chain might cause slight changes in structure.
Competitive Photoaffinity Labeling of the FMN Riboswitch
To show that the photoaffinity labeling approach can be used to study the interactions of small molecules with a riboswitch aptamer, a competition experiment was performed. To this end, a series of concentrations of the natural ligand FMN and the FMN riboswitch targeting bacterial antibiotic roseoflavin58 were preincubated with the FMN riboswitch. Subsequently, probe 1 (10 μM) was added, cross-linked, modified with fluorescein-azide, and finally analyzed using PAGE, and band intensities were quantified to determine the dose-dependent inhibition of labeling. IC50 values were calculated using a sigmoidal fit (see the SI for details). Both FMN and roseoflavin were capable of fully outcompeting probe 1, with an apparent IC50 of 0.4 ± 0.01 and 7.0 ± 0.18 μM, respectively (Figures 4A, S5, and S6). This observed ∼20-fold difference is in accordance with earlier observed dissociation constants (KD) of FMN and roseoflavin for the ribD riboswitch from Bacillus subtilis (B. subtilis).58
Figure 4.
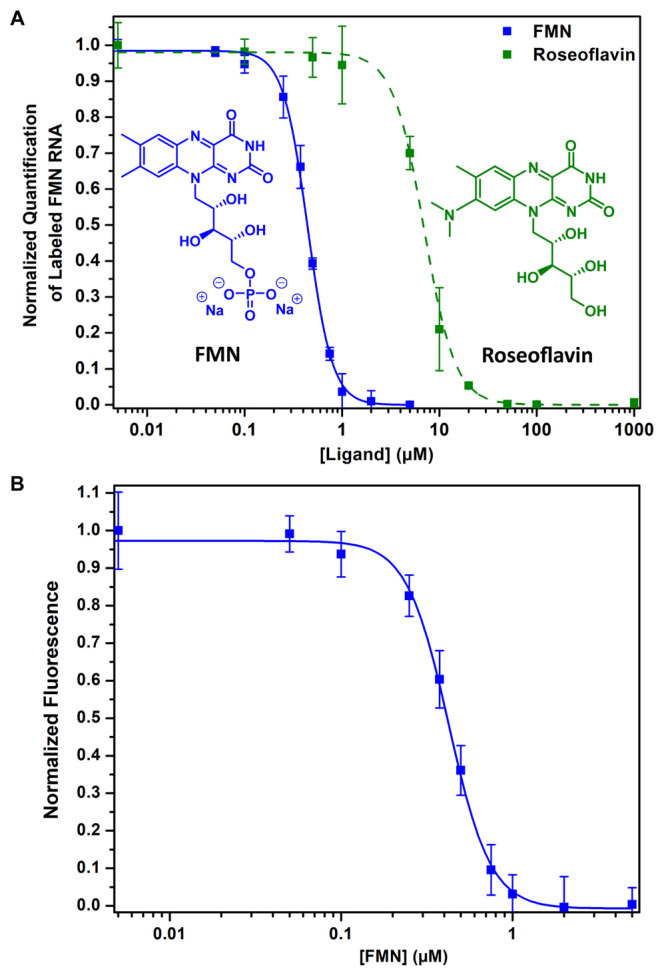
Competitive photoaffinity labeling. (A) Dose-dependent inhibition of labeling with the FMN ligand and roseoflavin. After photolabeling, the probe is modified with a fluorescein fluorophore and binding is determined by PAGE and subsequently quantified by measuring band intensity in triplicates (see Figures S5 and S6). (B) Competitive photoaffinity labeling with the FMN ligand quantified using a microplate reader. Error bars represent standard deviations based on three technical replicates.
To further improve the utility of this approach, a fluorescently labeled FMN riboswitch was quantified on a microplate reader, yielding similar results with an apparent IC50 of 0.4 ± 0.01 μM for FMN and avoiding the need for PAGE (Figure 4B). We expect that this feature will be found useful for screening for new small-molecule riboswitch binders.59,60
Conditional Aptamer Folding
Riboswitch conformations are conditional and depend on temperature and ion concentrations.34 When the riboswitch unfolds at increased temperature or low cation concentration, the interaction between RNA and ligand is lost.61 Bacteria are suggested to exploit these phenomena to fine-tune their gene regulation.14
Since probe 1 exclusively reports on the riboswitch bound state, we aimed to exploit this feature to study folding of the riboswitch aptamer. To examine if probe 1 can measure changes in riboswitch conformations, we applied the probe to the FMN riboswitch under varied temperature and cation conditions. Probe 1 was incubated with the FMN riboswitch at temperatures in the range 4–72 °C and exposed to UV light to initiate photo-cross-linking. The amount of captured riboswitch was quantified using a click reaction with fluorescein azide and subsequent PAGE analysis (Figure S7 and see the SI for details). At increased temperature the amount of captured riboswitch decreased, which might be attributed to unfolding of the aptamer domain, with the corresponding midpoint at 46 ± 0.4 °C, using a sigmoidal fit (see the SI for details) (Figure 5A). The interaction between RNA and probe decreased above 37 °C (Figure 5A), and no significant interaction between the probe and riboswitch was observed above 62 °C, which might imply that the riboswitch was fully unfolded at this temperature (Figure 5A).61 These observations are in line with previously reported data.44 These experiments are conducted well below the reported activation temperature of diazirine photoreactive groups,62 yet slight differences in photochemistry cannot be excluded and should be taken into consideration.
Figure 5.
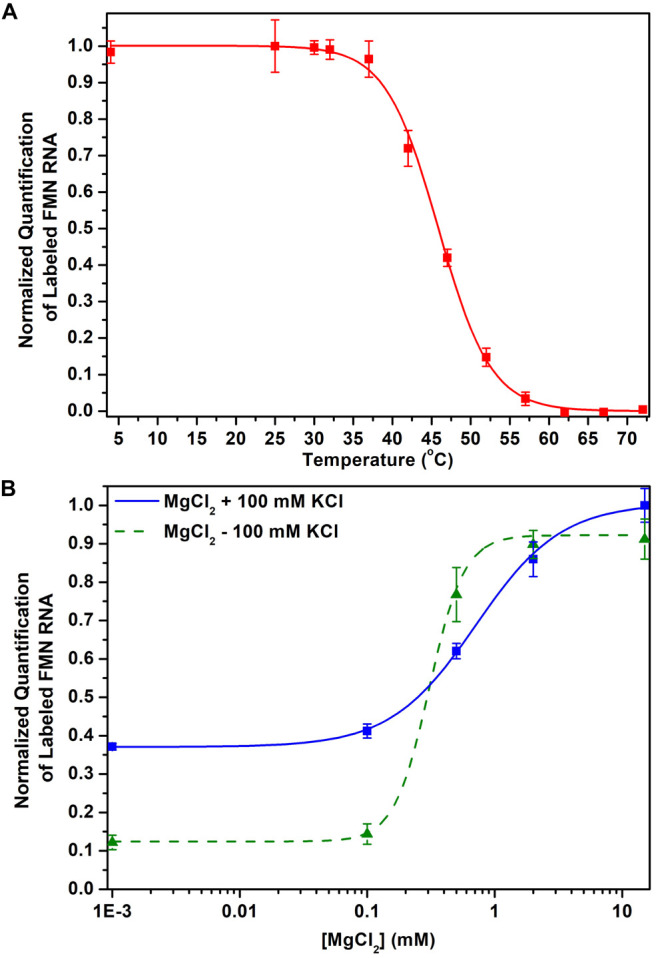
Measurements of FMN riboswitch folding with probe 1. (A) A decrease in FMN riboswitch labeling is observed at increased temperatures. (B) FMN riboswitch labeling is highly dependent on cation concentration, which is attributed to potential (un)folding of the aptamer domain. Error bars represent standard deviations based on three technical replicates.
To further explore structural changes with probe 1, we focused on the influence of cation concentrations, which is known to affect riboswitch conformations.61 First, the MgCl2 concentration was reduced from 15 mM to 0 mM, resulting in an apparent decrease of fluorescently labeled riboswitch as observed by PAGE (Figures 5B and S8). A sharp decrease in labeled riboswitch was observed below 2 mM of MgCl2, with a [Mg2+]1/2 of 0.7 ± 0.06 mM calculated using a sigmoidal fit, which is in accordance with the literature value of 0.8 ± 0.1 mM.44 The presence of monovalent cations such as potassium can partially compensate for a lack of magnesium.63 To study this, we repeated the prior experiment in the presence of 100 mM KCl. We found increased riboswitch labeling compared to low magnesium concentrations with a [Mg2+]1/2 of 0.3 ± 0.02 mM, which could signal that potassium can partially assist riboswitch folding in the absence of magnesium (Figures 5B and S8). These experiments demonstrate the potential utility of directly measuring conditional riboswitch folding using photoaffinity probes.44
Primer Extension of Labeled Transcripts
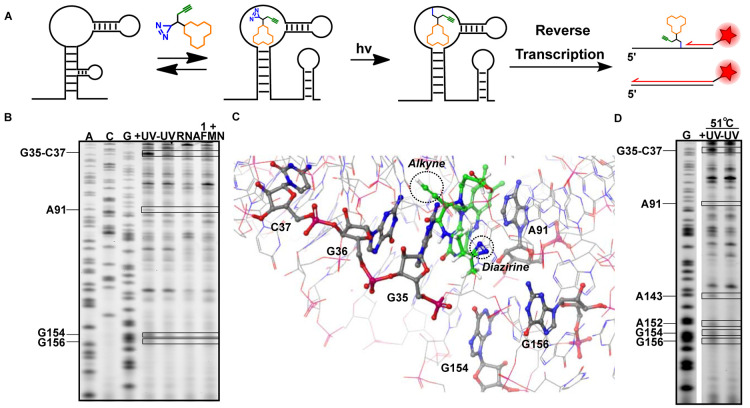
We postulated that transforming transient binding interactions into covalent interactions could enable the identification of the binding site of the probe within the riboswitch with single-nucleotide resolution. After photo-cross-linking, primer extension can be performed with reverse transcriptase and a fluorescently labeled primer, which stops at the covalent bond of the probe to the riboswitch (Figure 6A).55 The exact nucleotide to which the probe is photo-cross-linked can then be deduced by performing sequence analysis using electrophoresis. To this end, 10 μM of probe 1 was incubated with 2 μM FMN riboswitch RNA in folding buffer at 37 °C for 30 min, after which UV irradiation was performed to photo-cross-link the probe to RNA. The RNA was purified and reverse transcription was performed (see the SI for further details).
Figure 6.
Structural insights in binding of probe 1 to the FMN riboswitch aptamer domain. (A). Schematic illustration of the reverse transcription (RT) termination assay. (B) PAGE gel after performing the RT termination assay. Boxed bands show termination sites only observed with probe treatment. (C) Docking of probe 1 in the FMN riboswitch aptamer of Fusobacterium nucleatum (PDB: 3F2Q)36 with observed labeled nucleotides annotated according to B. subtilis numbering. (D) PAGE analysis of the RT termination assay at increased temperature. Sequencing lanes were obtained by incorporating dideoxynucleotides. The “+UV” lane shows results with photo-cross-linking and “–UV” shows the control without photo-cross-linking. “RNA” is the control with untreated RNA. The “1 + FMN” lane shows results of the termination assay in the presence of the competing FMN ligand. For triplicate experiments and quantification of band intensities see Figure S9.
New stops were observed at G35-C37, A91, G154, and G156 after photo-cross-linking of the probe to the FMN riboswitch (Figures 6B,C and S9). These stops were only observed after UV irradiation and are located in the aptamer domain of the FMN riboswitch.36 When the experiment was repeated in the presence of competing concentrations of natural ligand FMN, the RT stops disappeared, indicating that the observed interactions are specific (Figure 6B).
Nucleotides G35 and G36 are known to interact with the ribityl chain of FMN36 and are therefore expected to be in close proximity to the photoreactive diazirine group, explaining stops observed at G35-C37. Nucleotides G154 and G156 are positioned around the isoalloxazine ring of FMN in the ligand-bound state.36 The flexibility of the diazirine linker could potentially explain the appearance of stops at these positions. Interestingly, nucleotide A91 is reported to base-stack with A115, and the isoalloxazine ring of FMN intercalates between these two nucleotides.36 The stop observed at A91 is possibly a result of this interaction. Molecular docking of probe 1 into the FMN riboswitch showed that the probe is centered within a cluster of nucleotides at which the RT stops are observed (Figures 6C and S10).
To elucidate temperature-dependent changes in conformation, we repeated the experiment at 51 °C. Two additional stops were observed at A143 and A152 (Figures 6D and S9C). The uracil-like edge of the isoalloxazine ring system of the ligand forms specific Watson–Crick hydrogen bonds with the highly conserved nucleotide A152.36 No stop was observed at this position at 37 °C, which could mean that the elevated temperature is weakening this base-pair interaction. Interestingly, slight differences in SHAPE reactivity at this nucleotide have been previously observed at elevated temperature, but not below 65 °C.44 The additional stops observed at positions A143 and A152 could be the result of differences in flexibility of the ligand and FMN as well, positioning probe 1 and L6 (Figure 3A) of the riboswitch in closer proximity at elevated temperatures.
Photoaffinity Labeling in Bacterial Extracts and Live Bacteria
To show that we can determine ligand binding to the FMN riboswitch in endogenous samples, we first performed photoaffinity labeling on cell extracts from B. subtilis (see the SI for details). Extracts were incubated at 37 °C with 100 μM probe 1 and exposed to 365 nm light to initiate covalent labeling. Samples were incubated with biotin azide and a click mixture. Biotinylated RNA was isolated with Streptavidin magnetic beads and reverse transcribed. Using PCR with gene-specific primers for the ribD gene that is under control of the FMN riboswitch,64 successful labeling was demonstrated (Figure 7A). The amount of captured RNA was quantified using qPCR. Employing probe 1, we found a 10-fold enrichment compared to a DMSO control, showing that we can selectively capture the ribD FMN riboswitch in bacterial extracts. When using control linker 4 (Figure 7B and C) instead of probe 1, significantly lower amounts of ribD were enriched, indicating a high degree of selectivity provided by the flavin scaffold of probe 1. To further demonstrate the selectivity and show the potential to measure binding of RNA modulators in cell extracts, we repeated the experiment in the presence of 100 μM competing FMN ligand. As expected, a significant decrease in enrichment compared to probe 1 alone was observed (Figure 7C). To investigate if we can potentially measure conditional folding of the FMN riboswitch in endogenous samples, we repeated the experiment at 46 and 72 °C. A temperature-dependent decrease in enriched ribD RNA when using probe 1 was observed compared to a negative control of DMSO only, which might potentially be explained by partial unfolding of the FMN riboswitch at higher temperatures (Figure 7D).
Figure 7.
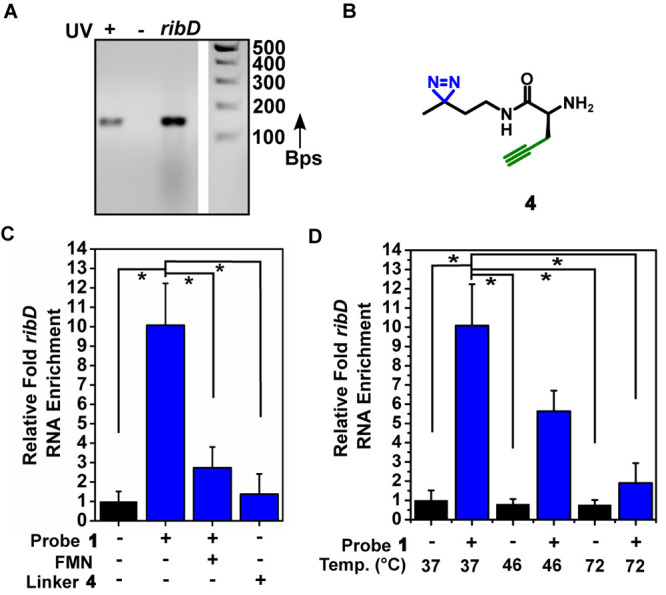
Photoaffinity labeling of ribD FMN riboswitch in B. subtilis extracts. (A) Agarose gel analysis of RT-PCR of enriched ribD mRNA using probe 1. Amplification is observed only when probe 1 is activated by UV (+) and not without UV (−). (B) Molecular structure of control diazirine linker 4. (C) Relative enrichment of ribD mRNA compared to the control (DMSO), in the presence of competing FMN, and with control diazirine linker 4. (D) Enrichment of ribD mRNA at increasing temperatures. Error bars represent standard errors based on three biological replicates, each consisting of two technical replicates. Statistical significance was calculated using an unpaired two-tailed Student’s t test (*p < 0.05).
To assess the potential utility of photoaffinity labeling for measuring riboswitch binding in vivo, probe 1 was incubated with live Escherichia coli (E. coli) CS1562 containing a plasmid carrying the E. coli ribB riboswitch (pTXTL-sroGp2eGFP, see Figure S11 and the SI for details). This strain is efflux impaired to secure a sufficient intracellular probe concentration (Figure S12).46
In an initial experiment, 25 μM probe 1 was incubated for 30 min with bacteria and then irradiated to initiate cross-linking. Bacteria were lysed, and extracted RNA was exposed to biotin azide and a click mixture. After Streptavidin pulldown, enriched ribB riboswitch RNA was quantified by qPCR. A ∼5-fold enrichment compared to a DMSO control was observed (Figure 8A). To further analyze the selectivity of labeling and potential of measuring inhibitor binding in vivo with photoaffinity labeling, competition experiments with roseoflavin were performed. Bacteria were preincubated with increasing concentrations of roseoflavin, after which photoaffinity labeling was performed with probe 1. A dose-dependent effect on enrichment was observed with an apparent IC50 of 5 ± 0.64 μM (Figure 8B). No decrease in enriched housekeeping gene (cysG) was observed at high concentrations of roseoflavin (Table. S1), supporting that the decrease in enrichment might be attributed to competitive binding to the riboswitch aptamer.
Figure 8.
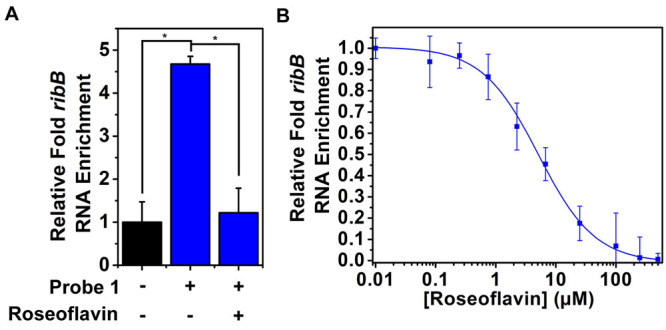
Photoaffinity labeling of ribB riboswitch in live E. coli CS1562 pTXTL-sroGp2eGFP. (A) Relative enrichment of ribB riboswitch RNA with probe 1 compared to the control (DMSO) and in the presence of competing roseoflavin (100 μM). (B) Dose-dependent inhibition of enrichment with roseoflavin in live E. coli CS1562 pTXTL-sroGp2eGFP. Error bars represent standard deviations based on three biological replicates, each consisting of two technical replicates. Statistical significance was calculated using an unpaired two-tailed Student’s t test (*p < 0.05).
Together, these experiments demonstrate the possibility to measure riboswitch binding in biologically derived samples and could further assist in elucidating riboswitch properties and druggability.
Conclusion
In summary, we have prepared photoaffinity probes to study the interaction between small-molecule ligands and riboswitches. We have applied our photoaffinity probe 1 to the FMN riboswitch and were able to selectively label the aptamer region of the riboswitch. The utility of the photoaffinity probe was demonstrated by quantitatively measuring the interaction between the riboswitch and small-molecule ligands, which we think might find applicability in drug discovery, as riboswitches make attractive antibiotic targets.46,59,60 By measuring riboswitch–ligand interactions at varied temperatures and cation concentrations, we showed the potential use of photoaffinity labeling for determining conditional riboswitch folding. Photoaffinity labeling yields a new addition to the riboswitch profiling toolbox, as it directly measures the interaction between ligand and RNA, unlike other profiling methods that indirectly observe altered RNA reactivity.3,33,65 Using reverse transcription and gel electrophoresis, we were able to determine the RNA binding site with single-nucleotide resolution. Interestingly, by conducting the experiment under elevated temperature conditions we observed altered ligand binding interactions that, to the best of our knowledge, have not been reported before using traditional RNA profiling methods. Lastly, by applying the photoaffinity probe to bacterial extracts and live bacteria, we were able to competitively measure ligand binding using bead enrichment and qPCR. We believe these experiments demonstrate the applicability of photoaffinity labeling to investigate riboswitch properties in vitro and in complex biological samples.66
Acknowledgments
We would like to thank Ing. Frank Nelissen for help with biochemical experimentation and the Biophysical Department at Radboud University and Dr. Hans Heus for generously granting us access to their facilities. We are grateful to Prof. Wiktor Szymanski and Prof. Arnold Driessen from the University of Groningen for gifting us E. coli CS1562. We thank Mart Bartelds for help with plasmid design.
Supporting Information Available
The Supporting Information is available free of charge at https://pubs.acs.org/doi/10.1021/jacs.2c02685.
Further experimental details, including PCR, in vitro transcription, affinity-based profiling, SHAPE assay, RT termination assay, bacterial growth, RNA extraction, qPCR, bacterial transformations, synthetic procedures, and spectral data.(PDF)
Author Contributions
All authors have given approval to the final version of the manuscript.
This work was supported by startup funds from the Institute of Molecules and Materials, Radboud University, and an NWO-XS grant from the Dutch Research Council to W.A.V.
The authors declare no competing financial interest.
Supplementary Material
References
- Cech T. R.; Steitz J. A. The Noncoding RNA Revolution - Trashing Old Rules to Forge New Ones. Cell 2014, 157 (1), 77–94. 10.1016/j.cell.2014.03.008. [DOI] [PubMed] [Google Scholar]
- Morris K. V.; Mattick J. S. The Rise of Regulatory RNA. Nat. Rev. Genet. 2014, 15, 423–437. 10.1038/nrg3722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Velema W. A.; Kool E. T. The Chemistry and Applications of RNA 2′-OH Acylation. Nat. Rev. Chem. 2020, 4 (1), 22–37. 10.1038/s41570-019-0147-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Isoda T.; Moore A. J.; He Z.; Chandra V.; Aida M.; Denholtz M.; Piet van Hamburg J.; Fisch K. M.; Chang A. N.; Fahl S. P.; Wiest D. L.; Murre C. Non-Coding Transcription Instructs Chromatin Folding and Compartmentalization to Dictate Enhancer-Promoter Communication and T Cell Fate. Cell 2017, 171 (1), 103–119. 10.1016/j.cell.2017.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin Q.-F.; Yang L.; Zhang Y.; Xiang J.-F.; Wu Y.-W.; Carmichael G. G.; Chen L.-L. Long Noncoding RNAs with SnoRNA Ends. Mol. Cell 2012, 48 (2), 219–230. 10.1016/j.molcel.2012.07.033. [DOI] [PubMed] [Google Scholar]
- Wutz A. Gene Silencing in X-Chromosome Inactivation: Advances in Understanding Facultative Heterochromatin Formation. Nat. Rev. Genet. 2011, 12 (8), 542–553. 10.1038/nrg3035. [DOI] [PubMed] [Google Scholar]
- Andersson R.; Gebhard C.; Miguel-Escalada I.; Hoof I.; Bornholdt J.; Boyd M.; Chen Y.; Zhao X.; Schmidl C.; Suzuki T.; Ntini E.; Arner E.; Valen E.; Li K.; Schwarzfischer L.; Glatz D.; Raithel J.; Lilje B.; Rapin N.; Bagger F. O.; Jørgensen M.; Andersen P. R.; Bertin N.; Rackham O.; Burroughs A. M.; Baillie J. K.; Ishizu Y.; Shimizu Y.; Furuhata E.; Maeda S.; Negishi Y.; Mungall C. J.; Meehan T. F.; Lassmann T.; Itoh M.; Kawaji H.; Kondo N.; Kawai J.; Lennartsson A.; Daub C. O.; Heutink P.; Hume D. A.; Jensen T. H.; Suzuki H.; Hayashizaki Y.; Müller F.; Forrest A. R. R.; Carninci P.; Rehli M.; Sandelin A.; Consortium T. F. An Atlas of Active Enhancers across Human Cell Types and Tissues. Nature 2014, 507 (7493), 455–461. 10.1038/nature12787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winkler W. C.; Cohen-Chalamish S.; Breaker R. R. An MRNA Structure That Controls Gene Expression by Binding FMN. Proc. Natl. Acad. Sci. U. S. A. 2002, 99 (25), 15908–15913. 10.1073/pnas.212628899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nahvi A.; Sudarsan N.; Ebert M. S.; Zou X.; Brown K. L.; Breaker R. R. Genetic Control by a Metabolite Binding MRNA. Chem. Biol. 2002, 9 (9), 1043–1049. 10.1016/S1074-5521(02)00224-7. [DOI] [PubMed] [Google Scholar]
- Serganov A.; Nudler E. A Decade of Riboswitches. Cell 2013, 152 (1), 17–24. 10.1016/j.cell.2012.12.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Batey R. T. Riboswitches: Still a Lot of Undiscovered Country. RNA 2015, 21 (4), 560–563. 10.1261/rna.050765.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren A.; Rajashankar K. R.; Patel D. J. Fluoride Ion Encapsulation by Mg2+ Ions and Phosphates in a Fluoride Riboswitch. Nature 2012, 486 (7401), 85–89. 10.1038/nature11152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perez-Gonzalez C.; Lafontaine D. A.; Penedo J. C. Fluorescence-Based Strategies to Investigate the Structure and Dynamics of Aptamer-Ligand Complexes. Front. Chem. 2016, 4, 33. 10.3389/fchem.2016.00033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reining A.; Nozinovic S.; Schlepckow K.; Buhr F.; Fürtig B.; Schwalbe H. Three-State Mechanism Couples Ligand and Temperature Sensing in Riboswitches. Nature 2013, 499 (7458), 355–359. 10.1038/nature12378. [DOI] [PubMed] [Google Scholar]
- Warner K. D.; Hajdin C. E.; Weeks K. M. Principles for Targeting RNA with Drug-like Small Molecules. Nat. Rev. Drug Discovery 2018, 17, 547–558. 10.1038/nrd.2018.93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Falese J. P.; Donlic A.; Hargrove A. E. Targeting RNA with Small Molecules: From Fundamental Principles towards the Clinic. Chem. Soc. Rev. 2021, 50 (4), 2224–2243. 10.1039/D0CS01261K. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer S. M.; Williams C. C.; Akahori Y.; Tanaka T.; Aikawa H.; Tong Y.; Childs-Disney J. L.; Disney M. D. Small Molecule Recognition of Disease-Relevant RNA Structures. Chem. Soc. Rev. 2020, 49 (19), 7167–7199. 10.1039/D0CS00560F. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas J. R.; Hergenrother P. J. Targeting RNA with Small Molecules. Chem. Rev. 2008, 108 (4), 1171–1224. 10.1021/cr0681546. [DOI] [PubMed] [Google Scholar]
- Kubota M.; Tran C.; Spitale R. C. Progress and Challenges for Chemical Probing of RNA Structure inside Living Cells. Nat. Chem. Biol. 2015, 11 (12), 933–941. 10.1038/nchembio.1958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taemaitree L.; Shivalingam A.; El-Sagheer A. H.; Brown T. An Artificial Triazole Backbone Linkage Provides a Split-and-Click Strategy to Bioactive Chemically Modified CRISPR SgRNA. Nat. Commun. 2019, 10 (1), 1610. 10.1038/s41467-019-09600-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ankenbruck N.; Courtney T.; Naro Y.; Deiters A. Optochemical Control of Biological Processes in Cells and Animals. Angewandte Chemie - International Edition. 2018, 57, 2768–2798. 10.1002/anie.201700171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Resendiz M. J. E.; Schön A.; Freire E.; Greenberg M. M. Photochemical Control of RNA Structure by Disrupting π-Stacking. J. Am. Chem. Soc. 2012, 134 (30), 12478–12481. 10.1021/ja306304w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rovira A. R.; Fin A.; Tor Y. Chemical Mutagenesis of an Emissive RNA Alphabet. J. Am. Chem. Soc. 2015, 137 (46), 14602–14605. 10.1021/jacs.5b10420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han H.; Dervan P. B. Visualization of RNA Tertiary Structure by RNA-EDTA.Fe(II) Autocleavage: Analysis of TRNA(Phe) with Uridine-EDTA.Fe(II) at Position 47. Proc. Natl. Acad. Sci. U. S. A. 1994, 91 (11), 4955–4959. 10.1073/pnas.91.11.4955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han H.; Dervan P. B. Sequence-Specific Recognition of Double Helical RNA and RNA.DNA by Triple Helix Formation. Proc. Natl. Acad. Sci. U. S. A. 1993, 90 (9), 3806–3810. 10.1073/pnas.90.9.3806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mrksich M.; Dervan P. B. Design of a Covalent Peptide Heterodimer for Sequence-Specific Recognition in the Minor Groove of Double-Helical DNA. J. Am. Chem. Soc. 1994, 116 (8), 3663–3664. 10.1021/ja00087a088. [DOI] [Google Scholar]
- Bispink L.; Matthaei H. Photoaffinity Labeling of 23 S RRNA InEscherichia Coli Ribosomes with Poly(U)-Coded Ethyl 2-Diazomalonyl-Phe-TRNA. FEBS Lett. 1973, 37 (2), 291–294. 10.1016/0014-5793(73)80480-6. [DOI] [PubMed] [Google Scholar]
- Schwartz I.; Gordon E.; Ofengand J. Photoaffinity Labeling of TRNA Binding Sites in Macromolecules. Photoaffinity Labeling of the Ribosomal A Site with S-(p-Azidophenacyl)Valyl-TRNA. Biochemistry 1975, 14 (13), 2907–2914. 10.1021/bi00684a018. [DOI] [PubMed] [Google Scholar]
- Chen J. L.; Zhang P.; Abe M.; Aikawa H.; Zhang L.; Frank A. J.; Zembryski T.; Hubbs C.; Park H.; Withka J.; Steppan C.; Rogers L.; Cabral S.; Pettersson M.; Wager T. T.; Fountain M. A.; Rumbaugh G.; Childs-Disney J. L.; Disney M. D. Design, Optimization, and Study of Small Molecules That Target Tau Pre-MRNA and Affect Splicing. J. Am. Chem. Soc. 2020, 142 (19), 8706–8727. 10.1021/jacs.0c00768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costales M. G.; Hoch D. G.; Abegg D.; Childs-Disney J. L.; Velagapudi S. P.; Adibekian A.; Disney M. D. A Designed Small Molecule Inhibitor of a Non-Coding RNA Sensitizes HER2 Negative Cancers to Herceptin. J. Am. Chem. Soc. 2019, 141 (7), 2960–2974. 10.1021/jacs.8b10558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Velagapudi S. P.; Costales M. G.; Vummidi B. R.; Nakai Y.; Angelbello A. J.; Tran T.; Haniff H. S.; Matsumoto Y.; Wang Z. F.; Chatterjee A. K.; Childs-Disney J. L.; Disney M. D. Approved Anti-Cancer Drugs Target Oncogenic Non-Coding RNAs. Cell Chem. Biol. 2018, 25 (9), 1086–1094. 10.1016/j.chembiol.2018.05.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mukherjee H.; Blain J. C.; Vandivier L. E.; Chin D. N.; Friedman J. E.; Liu F.; Maillet A.; Fang C.; Kaplan J. B.; Li J.; Chenoweth D. M.; Christensen A. B.; Petersen L. K.; Hansen N. J. V.; Barrera L.; Kubica N.; Kumaravel G.; Petter J. C. PEARL-Seq: A Photoaffinity Platform for the Analysis of Small Molecule-RNA Interactions. ACS Chem. Biol. 2020, 15 (9), 2374–2381. 10.1021/acschembio.0c00357. [DOI] [PubMed] [Google Scholar]
- Ames T. D.; Breaker R. R. Bacterial Riboswitch Discovery and Analysis. Chem. Biol. Nucleic Acids. 2010, 433–454. 10.1002/9780470664001.ch20. [DOI] [Google Scholar]
- Sherwood A. V.; Henkin T. M. Riboswitch-Mediated Gene Regulation: Novel RNA Architectures Dictate Gene Expression Responses. Annu. Rev. Microbiol. 2016, 70 (1), 361–374. 10.1146/annurev-micro-091014-104306. [DOI] [PubMed] [Google Scholar]
- Breaker R. R. Prospects for Riboswitch Discovery and Analysis. Mol. Cell 2011, 43 (6), 867–879. 10.1016/j.molcel.2011.08.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Serganov A.; Huang L.; Patel D. J. Coenzyme Recognition and Gene Regulation by a Flavin Mononucleotide Riboswitch. Nature 2009, 458 (7235), 233–237. 10.1038/nature07642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LeBlanc J. G.; Milani C.; de Giori G. S.; Sesma F.; van Sinderen D.; Ventura M. Bacteria as Vitamin Suppliers to Their Host: A Gut Microbiota Perspective. Curr. Opin. Biotechnol. 2013, 24 (2), 160–168. 10.1016/j.copbio.2012.08.005. [DOI] [PubMed] [Google Scholar]
- Warhaut S.; Mertinkus K. R.; Höllthaler P.; Fürtig B.; Heilemann M.; Hengesbach M.; Schwalbe H. Ligand-Modulated Folding of the Full-Length Adenine Riboswitch Probed by NMR and Single-Molecule FRET Spectroscopy. Nucleic Acids Res. 2017, 45 (9), 5512–5522. 10.1093/nar/gkx110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee M.-K.; Gal M.; Frydman L.; Varani G. Real-Time Multidimensional NMR Follows RNA Folding with Second Resolution. Proc. Natl. Acad. Sci. U. S. A. 2010, 107 (20), 9192–9197. 10.1073/pnas.1001195107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helmling C.; Klötzner D.-P.; Sochor F.; Mooney R. A.; Wacker A.; Landick R.; Fürtig B.; Heckel A.; Schwalbe H. Life Times of Metastable States Guide Regulatory Signaling in Transcriptional Riboswitches. Nat. Commun. 2018, 9 (1), 944. 10.1038/s41467-018-03375-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liberman J. A.; Suddala K. C.; Aytenfisu A.; Chan D.; Belashov I. A.; Salim M.; Mathews D. H.; Spitale R. C.; Walter N. G.; Wedekind J. E. Structural Analysis of a Class III PreQ1 Riboswitch Reveals an Aptamer Distant from a Ribosome-Binding Site Regulated by Fast Dynamics. Proc. Natl. Acad. Sci. U. S. A. 2015, 112 (27), E3485–E3494. 10.1073/pnas.1503955112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarkar B.; Ishii K.; Tahara T. Microsecond Folding of PreQ1 Riboswitch and Its Biological Significance Revealed by Two-Dimensional Fluorescence Lifetime Correlation Spectroscopy. J. Am. Chem. Soc. 2021, 143 (21), 7968–7978. 10.1021/jacs.1c01077. [DOI] [PubMed] [Google Scholar]
- Uhm H.; Kang W.; Ha K. S.; Kang C.; Hohng S. Single-Molecule FRET Studies on the Cotranscriptional Folding of a Thiamine Pyrophosphate Riboswitch. Proc. Natl. Acad. Sci. U. S. A. 2018, 115 (2), 331–336. 10.1073/pnas.1712983115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vicens Q.; Mondragón E.; Batey R. T. Molecular Sensing by the Aptamer Domain of the FMN Riboswitch: A General Model for Ligand Binding by Conformational Selection. Nucleic Acids Res. 2011, 39 (19), 8586–8598. 10.1093/nar/gkr565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sherlock M. E.; Sudarsan N.; Breaker R. R. Riboswitches for the Alarmone PpGpp Expand the Collection of RNA-Based Signaling Systems. Proc. Natl. Acad. Sci. U. S. A. 2018, 115 (23), 6052–6057. 10.1073/pnas.1720406115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howe J. A.; Wang H.; Fischmann T. O.; Balibar C. J.; Xiao L.; Galgoci A. M.; Malinverni J. C.; Mayhood T.; Villafania A.; Nahvi A.; Murgolo N.; Barbieri C. M.; Mann P. A.; Carr D.; Xia E.; Zuck P.; Riley D.; Painter R. E.; Walker S. S.; Sherborne B.; de Jesus R.; Pan W.; Plotkin M. A.; Wu J.; Rindgen D.; Cummings J.; Garlisi C. G.; Zhang R.; Sheth P. R.; Gill C. J.; Tang H.; Roemer T. Selective Small-Molecule Inhibition of an RNA Structural Element. Nature 2015, 526 (7575), 672–677. 10.1038/nature15542. [DOI] [PubMed] [Google Scholar]
- Lünse C. E.; Mayer G.. Reporter Gene-Based Screening for TPP Riboswitch Activators BT - Antibiotics: Methods and Protocols; Springer: New York, NY, 2017; pp 227–235. [DOI] [PubMed] [Google Scholar]
- Li N.; Overkleeft H. S.; Florea B. I. Activity-Based Protein Profiling: An Enabling Technology in Chemical Biology Research. Curr. Opin. Chem. Biol. 2012, 16 (1), 227–233. 10.1016/j.cbpa.2012.01.008. [DOI] [PubMed] [Google Scholar]
- Heal W. P.; Dang T. H. T.; Tate E. W. Activity-Based Probes: Discovering New Biology and New Drug Targets. Chem. Soc. Rev. 2011, 40, 246–257. 10.1039/C0CS00004C. [DOI] [PubMed] [Google Scholar]
- Ayele T. M.; Loya T.; Valdez-Sinon A. N.; Bassell G. J.; Heemstra J. M.. Imaging and Tracking MRNA in Live Mammalian Cells via Fluorogenic Photoaffinity Labeling. bioRxiv 2020, 10.1101/2020.02.10.942482 (accessed: 2022–05–20). [DOI] [Google Scholar]
- Arguello A. E.; DeLiberto A. N.; Kleiner R. E. RNA Chemical Proteomics Reveals the N6-Methyladenosine (M6A)-Regulated Protein-RNA Interactome. J. Am. Chem. Soc. 2017, 139 (48), 17249–17252. 10.1021/jacs.7b09213. [DOI] [PubMed] [Google Scholar]
- Balaratnam S.; Rhodes C.; Bume D. D.; Connelly C.; Lai C. C.; Kelley J. A.; Yazdani K.; Homan P. J.; Incarnato D.; Numata T.; Schneekloth Jr J. S. A Chemical Probe Based on the PreQ1 Metabolite Enables Transcriptome-Wide Mapping of Binding Sites. Nat. Commun. 2021, 12 (1), 5856. 10.1038/s41467-021-25973-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vicens Q.; Mondragón E.; Reyes F. E.; Coish P.; Aristoff P.; Berman J.; Kaur H.; Kells K. W.; Wickens P.; Wilson J.; Gadwood R. C.; Schostarez H. J.; Suto R. K.; Blount K. F.; Batey R. T. Structure-Activity Relationship of Flavin Analogues That Target the Flavin Mononucleotide Riboswitch. ACS Chem. Biol. 2018, 13 (10), 2908–2919. 10.1021/acschembio.8b00533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith E.; Collins I. Photoaffinity Labeling in Target- and Binding-Site Identification. Future Med. Chem. 2015, 7 (2), 159–183. 10.4155/fmc.14.152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merino E. J.; Wilkinson K. A.; Coughlan J. L.; Weeks K. M. RNA Structure Analysis at Single Nucleotide Resolution by Selective 2′-Hydroxyl Acylation and Primer Extension (SHAPE). J. Am. Chem. Soc. 2005, 127 (12), 4223–4231. 10.1021/ja043822v. [DOI] [PubMed] [Google Scholar]
- Habibian M.; Velema W. A.; Kietrys A. M.; Onishi Y.; Kool E. T. Polyacetate and Polycarbonate RNA: Acylating Reagents and Properties. Org. Lett. 2019, 21 (14), 5413–5416. 10.1021/acs.orglett.9b01526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spitale R. C.; Crisalli P.; Flynn R. A.; Torre E. A.; Kool E. T.; Chang H. Y. RNA SHAPE Analysis in Living Cells. Nat. Chem. Biol. 2013, 9, 18–20. 10.1038/nchembio.1131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee E. R.; Blount K. F.; Breaker R. R. Roseoflavin Is a Natural Antibacterial Compound That Binds to FMN Riboswitches and Regulates Gene Expression. RNA Biol. 2009, 6 (2), 187–194. 10.4161/rna.6.2.7727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blount K. F.; Breaker R. R. Riboswitches as Antibacterial Drug Targets. Nat. Biotechnol. 2006, 24 (12), 1558–1564. 10.1038/nbt1268. [DOI] [PubMed] [Google Scholar]
- Panchal V.; Brenk R. Riboswitches as Drug Targets for Antibiotics. Antibiotics. 2021, 10 (1), 45. 10.3390/antibiotics10010045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garst A. D.; Edwards A. L.; Batey R. T. Riboswitches: Structures and Mechanisms. Cold Spring Harb. Perspect. Biol. 2011, 3 (6), a003533. 10.1101/cshperspect.a003533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Musolino S. F.; Pei Z.; Bi L.; DiLabio G. A.; Wulff J. E. Structure-Function Relationships in Aryl Diazirines Reveal Optimal Design Features to Maximize C-H Insertion. Chem. Sci. 2021, 12 (36), 12138–12148. 10.1039/D1SC03631A. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen H. T.; Hori N.; Thirumalai D. Theory and Simulations for RNA Folding in Mixtures of Monovalent and Divalent Cations. Proc. Natl. Acad. Sci. U. S. A. 2019, 116 (42), 21022–21030. 10.1073/pnas.1911632116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wickiser J. K.; Winkler W. C.; Breaker R. R.; Crothers D. M. The Speed of RNA Transcription and Metabolite Binding Kinetics Operate an FMN Riboswitch. Mol. Cell 2005, 18 (1), 49–60. 10.1016/j.molcel.2005.02.032. [DOI] [PubMed] [Google Scholar]
- Tijerina P.; Mohr S.; Russell R. DMS Footprinting of Structured RNAs and RNA-Protein Complexes. Nat. Protoc. 2007, 2, 2608–2623. 10.1038/nprot.2007.380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Broft P.; Dzatko S.; Krafcikova M.; Wacker A.; Hänsel-Hertsch R.; Dötsch V.; Trantirek L.; Schwalbe H. In-Cell NMR Spectroscopy of Functional Riboswitch Aptamers in Eukaryotic Cells. Angew. Chemie Int. Ed. 2021, 60 (2), 865–872. 10.1002/anie.202007184. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.