Abstract
The diversity of microorganisms present in a sediment colonized by the phanerogam Zostera noltii has been analyzed. Microbial DNA was extracted and used for constructing two 16S rDNA clone libraries for Bacteria and Archaea. Bacterial diversity was very high in these samples, since 57 different sequences were found among the 60 clones analyzed. Eight major lineages of the Domain Bacteria were represented in the library. The most frequently retrieved bacterial group (36% of the clones) was δ-Proteobacteria related to sulfate-reducing bacteria. The second most abundant group (27%) was γ-Proteobacteria, including five clones closely related to S-oxidizing endosymbionts. The archaeal clone library included members of Crenarchaeota and Euryarchaeota, with nine different sequences among the 15 analyzed clones, indicating less diversity when compared to the Bacteria organisms. None of these sequences was closely related to cultured Archaea organisms.
Our objective in this study was to describe the diversity of the prokaryotic community inhabiting a marine sediment colonized by the marine phanerogam Zostera noltii located in the Bassin d'Arcachon, South-West France. This is a macrotidal coastal lagoon that represents the most extensive intertidal meadows of this rooted phanerogam in Western Europe (70 km2). This seagrass ecosystem is characterized by a high iron content, 111.5 (dry weight) μg/g (32) in the sediment, as well as strong tidal activity, which ensures regular mixing of the water body and exposes the sediment surface to the air for between 4 and 8 h each day. This environment is different from those that have previously been studied by molecular methods due to the presence of Z. noltii roots and rhizomes. A number of recent studies have shown that living seagrasses release dissolved organic carbon, which can significantly influence the composition and activity of the seagrass rhizosphere microflora (24, 36). For example, several studies have demonstrated high rates of heterotrophic nitrogen fixation in the rhizosphere of Z. noltii-colonized sediments which are coupled to the photosynthetic activity of the plants via the release of fixed carbon from the roots (20, 24, 36). Similarly, substantial stimulation of sulfate reduction rates in seagrass-colonized sediments in the light have been reported (4, 25). These data indicate a close interaction between the plants and the microbial community in the rhizosphere.
Samples.
Z. noltii-colonized sediment cores (5-cm diameter) were collected from Station A in the Bassin d'Arcachon in July 1996 and kept in the dark at −20°C until processed. Station A is located in the center of the Bay, in an open zone subjected to marine influences. Sediment cores that contained extensive rhizome material were sliced into sections (1-cm thick, from the surface), and horizon 2 (1 to 2 cm) was chosen for the microbial community analysis.
DNA extraction and purification.
A combination of the methods described by Zhou et al. (37) and Gray and Herwig (14) was used. Eight hundred milligrams of sediments were mixed with 2.16 ml of lysis buffer (100 mM Tris-HCl [pH 8.0], 100 mM Na EDTA [pH 8.0], 100 mM NaP [pH 8.0], 1.5 M NaCl, and 1% [wt/vol] cetyltrimethylammonium bromide) and 16 μl of proteinase K (14 mg/ml). Two-milliliter vials containing 2 g of 0.1-mm zirconia beads were filled with this mixture and shaken horizontally at 225 rpm for 30 min at 37°C. After shaking, 480 μl of 10% (wt/vol) sodium dodecyl sulfate were added. The content of the tubes were homogenized for 1 min on a Mini Bead Beater Cell Disrupter (Biospec Products, Bartlesville, Okla.) at maximum setting and then incubated in a 65°C water bath for 2 h with gentle end-over-end inversions every 10 to 15 min. Samples were centrifuged at 6,000 × g for 10 min, and the supernatants were collected. An equal volume of chloroform-isoamylalcohol (24:1) was added and then centrifuged at 16,000 × g for 5 min before 0.6 volumes of isopropanol were added to each tube, incubated at room temperature for 1 h, and centrifuged at 16,000 × g for 20 min. The supernatants were decanted, and the pellets were washed with 1 ml of 70% (vol/vol) ice-cold EtOH. The pellets were air dried and resuspended in 200 μl of sterile deionized water. One hundred microliters of crude DNA extract was purified with GENECLEAN Spin Kit (Bio 101, Inc., Vista, Calif.), resuspended into 40 μl of elution buffer, and electrophoresed on a 1% (wt/vol) agarose gel (Low EEO agarose; Pronadisa) in 1× Tris-acetate-EDTA at 4 V/cm. The gel was stained with 0.5 g of ethidium bromide and visualized with UV. The band larger than 23 kb was excised and purified twice with GENECLEAN columns. Finally, DNA was eluted into 40 μl of elution buffer.
PCR amplification and cloning of PCR products.
Universal primers for PCR amplification of bacterial and archaeal 16S rRNA genes were used in this study (forward primer 27f for Bacteria [16], forward primer 21F for Archaea [7], and universal reverse primer 1492r [16] for both Bacteria and Archaea). PCR was carried out as described previously (1). Two 16S rDNA clone libraries were constructed, one for 16S rDNA amplified with primers specific for Bacteria organisms (clone library B2M) and one for Archaea organisms (clone library A2M). The PCR products obtained from different PCRs were cloned by using the original TA Cloning Kit (Invitrogen) and following the manufacturer's recommendations. Inserts were PCR reamplified with specific primers for the vector and purified with the QIA-quick PCR Purification Kit (QIAgen) according to the manufacturer's protocol. The DNA was recovered in 30 μl of water and diluted to a concentration between 0.3 and 0.5 μg/μl.
Sequencing of 16S rDNAs and data analysis.
Nucleotide sequences of PCR products were determined by using the ABI PRISM Dye Terminator Cycle Sequencing Ready Reaction Kit (Perkin-Elmer) according to the manufacturer's indications. Sixty clones were selected from B2M, and 15 clones were selected from A2M. For these clones, the 3′ end was partially sequenced with primers Bact1055 and Arc915 (3) as the sequencing primers for Bacteria and Archaea, respectively. The 16S rRNA genes of six clones (B2M-54, B2M-23, B2M-58, B2M-60, B2M-61, and B2M-68) were totally sequenced by using primers 27f (16), Bact335 (3), Bact785 (3), and 1492r (16). The phylogenetic affiliation of the obtained sequences was carried out by BLAST at the National Center for Biotechnology Information (NCBI) web site (www.ncbi.nlm.nhi.gov) (2). Similarity percentages were calculated after manual alignment of the clone sequences with that of the closest relative provided by NCBI. Sequences were submitted to the CHECK_CHIMERA program at the Ribosomal Database Project (RDP) (18). For tree calculations, sequences were aligned by using the Clustal W program (Genetics Computer Group [GCG] Package), and their similarity matrix was calculated by the Jukes-Cantor method with the MEGA (Molecular Evolutionary Genetic Analysis) 1.01 program obtained from the Institute of Molecular Evolutionary Genetics, the Pennsylvania State University, University Park.
Bacterial 16S rDNA clone library.
Out of the 60 bacterial clones analyzed, five were of chloroplast origin, which is not surprising considering that the analyzed sediment was densely colonized by Z. noltii roots, which have attached diatoms (11). The remaining 55 (Table 1) were of bacterial origin and fell into eight major lineages of the Domain Bacteria: α-, β-, δ-, ɛ-, and γ-Proteobacteria, Cytophaga, Spirochaeta, and gram-positive organisms with a high G+C content. Among these 55 bacterial sequences analyzed, there were 52 unique sequences, indicating that, firstly, the bacterial diversity in these sediments was very high and, secondly, that we are far from establishing the total bacterial diversity of the analyzed samples. None of the clones was identical to any known 16S rRNA sequence from cultured organisms or environmental clones.
TABLE 1.
Phylogenetic identification of the partial 16S rDNA of Bacteria clones
| Clone | % Similarity to known organisma | Length (bp)b |
|---|---|---|
| δ-Proteobacteria | ||
| B2M26 | 96.3 Desulfosarcina variabilis | 348 |
| B2M16 | 95.5 Desulfonema magnum | 333 |
| B2M56 | 95.1 Desulfonema magnum | 368 |
| B2M62 | 93.8 Desulfosarcina variabilis | 274 |
| B2M36 | 93.3 Desulfonema magnum | 404 |
| B2M1 | 93.2 Desulfobulbus sp. | 351 |
| B2M37 | 92.9 Syntrophobacter sp. | 353 |
| B2M53 | 92.8 Desulfobulbus sp. | 319 |
| B2M44 | 92.4 Desulfonema magnum | 397 |
| B2M9 | 92.1 Desulfobulbus rhabdoformis | 407 |
| B2M23 | 92.0 Geobacter arculus | 319 |
| B2M24 | 91.9 Desulfurosoma succinoxidans | 322 |
| B2M33 | 91.1 Desulfosarcina variabilis | 369 |
| B2M47 | 91.1 Desulforomonas thiophila | 346 |
| B2M57 | 90.7 Desulfosarcina variabilis | 365 |
| B2M65 | 90.6 Geobacter sp. | 331 |
| B2M17 | 88.9 Nitrospina gracilis | 341 |
| B2M12 | 88.6 Desulforhabdus amnigenus | 414 |
| B2M21 | 88.0 Desulforhabdus amnigenus | 367 |
| B2M20 | 87.0 Desulforhabdus acetothermus | 345 |
| B2M6 | 86.6 Desulforhabdus acetothermus | 358 |
| B2M11 | 86.6 Desulfobulbus rhabdoformis | 405 |
| B2M4 | 84.7 Desulfonatronovibrio hydrogenovorans | 393 |
| B2M55 | 79.8 Desulforhabdus acetothermus | 367 |
| γ-Proteobacteria | ||
| B2M15 | 94.6 Methylobacter sp. | 325 |
| B2M18 | 94.1 Pseudomonas sp. | 408 |
| B2M48 | 93.4 Aeromonas salmonicida | 349 |
| B2M32 | 93.2 Pseudomonas fluorescens | 413 |
| B2M60 | 91.9 Codakia orbicularis symbiont | 1,390 |
| B2M52 | 91.9 Beggiatoa sp. | 347 |
| B2M71 | 91.9 Azotobacter paspali | 344 |
| B2M61 | 91.3 Lucina nassula symbiont | 1,368 |
| B2M23 | 91.3 Solemya velum symbiont | 1,455 |
| B2M54 | 91.2 Anodontia phillipiana gill symbiont | 1,415 |
| B2M67 | 90.9 Methylophaga thalassica | 358 |
| B2M28 | 90.8 Codakia orbicularis symbiont | 1,334 |
| B2M31 | 90.8 Photobacterium profundum | 306 |
| B2M66 | 83.9 Dichelobacter nodosus | 353 |
| B2M70 | 82.7 Alcanivorax borkumensis | 301 |
| Cytophaga/Flexibacter | ||
| B2M45 | 97.1 Bacteroides fragilis | 313 |
| B2M41 | 94.1 Capnocytophaga gingivalis | 340 |
| B2M39 | 91.7 Microscilla arenaria | 390 |
| α-Proteobacteria | ||
| B2M25 | 95.8 Sphingomonas terrae | 356 |
| B2M72 | 95.0 Sulfitobacter pontiacus | 339 |
| B2M58 | 93.3 Rhodobium marinum | 315 |
| B2M68 | 81.8 Rhizobium fredii | 1,098 |
| B2M51 | 78.3 Rhodoplanes roseus | 295 |
| ɛ-Proteobacteria | ||
| B2M5 | 93.6 Arcobacter skirrowi | 344 |
| B2M7 | 93.6 Arcobacter skirrowi | 344 |
| B2M13 | 93.6 Arcobacter skirrowi | 344 |
| B2M2 | 88.9 Arcobacter skirrowi | 298 |
| Spirochaeta | ||
| B2M19 | 92.7 Spirochaeta africana | 382 |
| B2M38 | 92.7 Spirochaeta africana | 382 |
| β-Proteobacteria | ||
| B2M14 | 92.2 Methylophilus methylotrophus | 336 |
| Gram positive, high G+C content | ||
| B2M43 | 86.2 Streptomyces griseocarneum | 298 |
% Similarity was determined with the best match in the databases.
The length is that of the analyzed sequence.
The most abundant group (24 clones, 44% of the total) was δ-Proteobacteria, and organisms of this group comprised sequences (20 clones, 36% of the total) related to sulfur- and sulfate-reducing bacteria (SRB). This was also the predominant group in the 16S rDNA clone libraries constructed by Gray and Herwig (14) and Ravenslach et al. (27) when analyzing marine sediments from creosote-contaminated Eagle Harbor (Puget Sound, Washington) and permanently cold sediments collected off the coast of Spitsbergen (Arctic Ocean), respectively. The predominance of SRB in marine sediments has been previously demonstrated by direct quantification using rRNA slot blot and fluorescent in situ hybridization (FISH) analysis (17, 31). The importance of SRB in the oxidation of organic carbon in marine sediments is well established, since sulfate is one of the main electron acceptors present in these environments. In addition, SRB are able to utilize different electron donors available in marine sediments, such as carbon compounds released by plant roots (in this case, Z. noltii) or fermentation products of other bacteria, such as acetate, lactate, butanol, and formate (33). Furthermore, several studies (6, 19, 21) have shown that some SRB are oxygen tolerant, which enables them to colonize microaerophilic habitats, such as plant roots.
Molecular approaches have been used previously to describe the diversity of SRB in marine environments since sulfate reduction is a major route of organic matter mineralization in these environments (15, 34). A number of studies have indicated that marine sediments are inhabited by a great diversity of SRB, and new sequences and groups are presently being described (8, 9, 28).
From the same sediment analyzed in this work several SRB have been isolated: Desulfospira joergensenii (12), Desulfocapsa sulfoexigens (13), as well as SRB related to the genera Desulfovibrio, Desulfobacter, Desulfobacula, and Desulfobacterium (A. Cifuentes, J. Antón, R. de Wit, and F. Rodriguez-Valera, unpublished observation). On the other hand, Desulfovibrio zosterae (23) was isolated from surface-sterilized roots of the benthic macrophyte Zostera marina. Interestingly, none of the SRB 16S rDNA sequences directly retrieved from the environment match those of the isolates mentioned above. Indeed, we have not recovered sequences related to the family Desulfovibrionaceae. These results contrast with those of Sahm et al. (30), who recovered as the most abundant 16S rDNA sequences the ones that corresponded to SRB frequently isolated from the same environment (an Arctic marine sediment) affiliated with the Desulfotalea cluster.
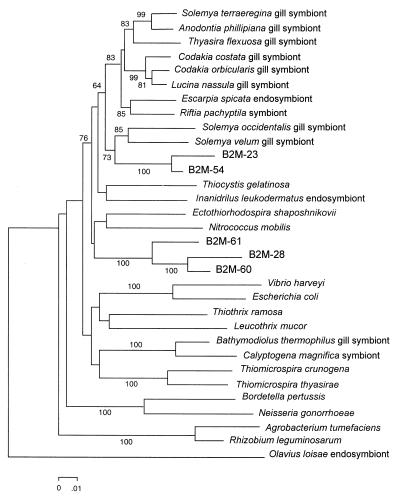
The second most abundant group (around 27% of the clones analyzed) in our clone library was formed by sequences related to γ-Proteobacteria organisms. This group included sequences related to bacteria that are readily isolated from marine environments, like Pseudomonas spp., Aeromonas spp., and Photobacterium spp. Remarkably, six of the γ-Proteobacteria-related sequences (clones B2M-23, B2M-28, B2M-54, B2M-60, B2M-61, and B2M-68) were related to sulfur-oxidizing bacterial endosymbionts. These clones were fully sequenced, and their phylogenetic relationship was established as shown in Fig. 1. Clone B2M-68, which presented a 77.8% similarity to the partial sequence (positions 1081 to 1399) of an Escarpia spicata endosymbiont, was found to be related (81.8% similarity) to the α-proteobacterium Rhizobium fredii when the complete 16S rDNA was analyzed. When sequencing the complete gene, we found that these sequences were only distantly related to the endosymbionts (similarities from 90.8 to 91.9%) but still clearly associated with them, as shown in Fig. 1. These sequences have been also found in other marine sediments (27), but whether they correspond to free-living bacteria remains unknown. However, they account for an important part of the bacterial diversity recovered in this study. It is not unreasonable to speculate that these bacteria, due to their phylogenetic relationships, are indeed sulfur oxidizers that are known to be associated with SRB and may play an important role in the detoxification of sulfide, which is toxic to most living organisms.
FIG. 1.
Phylogenetic tree from complete 16S rDNA gene of clones related to sulfur-oxidizing bacterial endosymbionts and reference strains. The tree was built with the neighbor-joining method by using the Jukes-Cantor distance estimation. Bootstrap values higher than 50% are indicated at the main nodes. Nucleotide sequence accession numbers are as follows: A. phillipiana gill symbiont, L25711; A. tumefaciens, AH007805; B. pertussis, AF142327; B. thermophilus gill symbiont, M99445; C. costata gill symbiont, L25712; C. magnifica symbiont, L25718; C. orbicularis gill symbiont, X84979; E. coli, U00096; E. shaposhnikovii, M59151; E. spicata endosymbiont, U77482; L. mucor, X87277; L. nassula gill symbiont, X95229; N. gonorrhoeae, AJ239304; N. mobilis, g530889; O. loisae endosymbiont, AF104475; R. leguminosarum, U89831; R. pachyptila symbiont, U77478; S. occidentalis gill symbiont, U41049; S. terraeregina gill symbiont, U62131; S. velum gill symbiont, M90415; T. crunogena, AF064545; T. flexuosa gill symbiont, L01575; T. gelatinosa, Y11317; T. ramosa, g987512; T. thyasirae, AF016046; V. harveyi, X56578; and Y. leukodermatus endosymbiont, U24110.
The rest of the groups were less abundant: Cytophaga (three clones), α-Proteobacteria (five clones), Spirochaeta (two clones), β-Proteobacteria (one clone), and gram-positive and high G+C content bacteria (one clone).
We found two repeated sequences (clones B2M-5, B2M-7, and B2M-13, 93.6% similarity with Arcobacter skirrowi; clones B2M-19 and B2M-38, 92.7% similarity to Spirochaeta africana). Llobet-Brossa et al. (17), when analyzing by FISH the microbial composition of Wadden Sea sediments, reported a considerable proportion (1.3%) of bacteria detectable with a FISH probe specifically designed for Arcobacter spp. This bacterium had not previously been reported to be significant in marine sediments. Arcobacter was only detected in the upper layer of the sediments which, according to these authors, was not surprising, considering their ability to respire nitrate. Marine sediments are typical habitats of spirochetes (5). However, sequences related to Spirochaeta spp. have not been found previously in sediment 16S rDNA libraries (14, 27) but, on the other hand, Rosselló-Mora et al. (29) found sequences affiliated to the spirochete phylum when analyzing by denaturing gradient gel electrophoresis anoxic sediments from the Black Sea.
Archaeal 16S rDNA clone library.
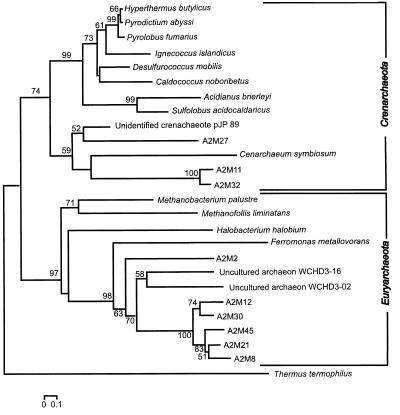
Fifteen archaeal clones were partially sequenced, five (clones A2M-27 [identical to clone A2M-28], A2M-11 [identical to clone A2M-17], and A2M-32) of which were phylogenetically associated with Crenarchaeota sequences and ten (clones A2M-2 [identical to clones A2M-3 and A2M-4], A2M-8, A2M-12, A2M-21, A2M-30, A2M-36, A2M-45, and A2M-57) of which were phylogenetically associated with Euryarchaeota (see Fig. 2).
FIG. 2.
Relationships between partial 16S archaeal rDNA gene clones and reference strains. The tree was constructed as that in Fig. 1. T. thermophilus was used as the outgroup. Nucleotide sequence accession numbers are as follows: A. brierleyi, X90477; C. noboribetus, D85038; C. symbiosum, U51469; crenarchaeotal sp. clone pJP 89, L25305; D. mobilis, M36474; F. metallovorans, AJ224936; H. salinarium, AJ002947; H. butylicus, X99553; I. islandicus, X99562; M. palustre, AF093061; M. liminatans, Y16428; P. abyssi, X99559; P. fumarius, X99555; S. acidocaldarius, D14053; T. waimanguensis, AF098975; T. thermophilus, X07998; uncultured archaeon WCHD3-16, AF050618; and uncultured archaeon WCHD3-02, AF050616.
Three of the Crenarchaeota-related clones (A2M-11 and A2M-17, which are identical, and A2M-32) were distantly related to Cenarchaeum symbiosum, a marine archaeon that inhabits the tissues of a temperate water sponge (26). Two of the remaining Crenarchaeota-related clones (A2M-27 and A2M-28, which are identical) were associated to an environmental clone retrieved from a hot spring in Yellowstone National Park.
Euryarchaeota-related clones were associated with environmental clones retrieved from an aquifer contaminated with hydrocarbons and chlorinated solvents undergoing intrinsic bioremediation (10). Our clones were related to sequences WCHD3-02 and WCHD3-16 retrieved from the methanogenic redox sampling zone.
The presence of Euryarchaeota in sediments has been widely reported since methanogens are known to inhabit this kind of environment. Although the molecular approaches to the study of archaeal diversity in marine sediments are very limited, some authors have also retrieved methanogen-related 16S rDNA sequences from these environments. Munson et al. (22) recovered a wide range of Euryarchaeota sequences when studying a salt marsh sediment sample that showed active methanogenesis and sulfate reduction. These authors were unable to find any sequence related to those of Crenarchaeota organisms. However, in this study, several Crenarchaeota-related clones were identified. The presence of Crenarchaeota-related sequences in marine sediments has been described previously (35).
Since we are working with partial sequences that present very low similarity with cultured Archaea organisms, it is not possible from these limited data to comment on their ecological role in these sediments. However, it is evident that Archaea organisms other than methanogens may be an important part of the prokaryotic community in marine sediments.
Nucleotide sequence accession numbers.
The sequences from this study are available through GenBank under accession no. AF218422 to AF218430 and AF223253 to AF223307.
Acknowledgments
This work was supported by European Commission Grant ENV4-CT96-0218 and Spanish Government CICYT AMB96-2484-CE. European Land-Ocean Interaction Studies (ELOISE) contribution number 128. A.C. was a Generalitat Valenciana fellowship holder.
REFERENCES
- 1.Acinas S G, Antón J, Rodríguez-Valera F. Diversity of free-living and attached bacteria in offshore Western Mediterranean waters as depicted by analysis of genes encoding 16S rRNA. Appl Environ Microbiol. 1999;65:514–522. doi: 10.1128/aem.65.2.514-522.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Altschul S F, Madden T L, Schäffer A A, Zhang J, Zhang Z, Miller W, Lipman D J. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Amann R I, Ludwig W, Schleifer K-H. Phylogenetic identification and in situ detection of individual microbial cells without cultivation. Microbiol Rev. 1995;59:143–169. doi: 10.1128/mr.59.1.143-169.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Blaabjerg V, Mouritsen K N, Finster K. Diel cycles of sulfate reduction in sediments of a Zostera marina bed (Denmark) Aquat Microb Ecol. 1998;15:97–102. [Google Scholar]
- 5.Canale-Parola E. Free-living saccharolytic spirochetes: the genus Spirochaeta. In: Balows A, Trüper H G, Dworkin M, Harder W, Schleifer K-H, editors. The prokaryotes. 2nd ed. New York, N.Y: Springer-Verlag; 1992. pp. 3524–3536. [Google Scholar]
- 6.Canfield D E, DesMarais D J. Aerobic sulfate reduction in microbial mats. Science. 1991;251:1471–1473. doi: 10.1126/science.11538266. [DOI] [PubMed] [Google Scholar]
- 7.DeLong E F. Archaea in coastal marine environments. Proc Natl Acad Sci USA. 1992;89:5685–5689. doi: 10.1073/pnas.89.12.5685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Devereux R, Kane M D, Winfrey J, Stahl D A. Genus- and group-specific hybridization probes for determinative and environmental studies of sulfate-reducing bacteria. Syst Appl Microbiol. 1992;15:601–609. [Google Scholar]
- 9.Devereux R, Mundfrom G W. A phylogenetic tree of 16S rRNA sequences from sulfate-reducing bacteria in a sandy marine sediment. Appl Environ Microbiol. 1994;60:3437–3439. doi: 10.1128/aem.60.9.3437-3439.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dojka M A, Hugenholtz P, Haack S K, Pace N R. Microbial diversity in a hydrocarbon- and chlorinated-solvent-contaminated aquifer undergoing intrinsic bioremediation. Appl Environ Microbiol. 1998;64:3869–3877. doi: 10.1128/aem.64.10.3869-3877.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Donnelly A P, Herbert R A. Microbial interactions in the rhizosphere of seagrass communities in shallow coastal lagoons. J Appl Microbiol. 1999;85:151–160. doi: 10.1111/j.1365-2672.1998.tb05294.x. [DOI] [PubMed] [Google Scholar]
- 12.Finster K, Liesack W, Tindall B J. Desulfospira joergensenii, gen. nov., sp. nov., a new sulfate-reducing bacterium isolated from marine surface sediment. Syst Appl Microbiol. 1997;20:201–208. [Google Scholar]
- 13.Finster K, Liesack W, Thamdrup B. Elemental sulfur and thiosulfate disproportionation by Desulfocapsa sulfoexigens sp. nov., a new anaerobic bacterium isolated from marine surface sediment. Appl Environ Microbiol. 1998;64:119–125. doi: 10.1128/aem.64.1.119-125.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gray J P, Herwig R P. Phylogenetic analysis of the bacterial communities in marine sediments. Appl Environ Microbiol. 1996;62:4049–4059. doi: 10.1128/aem.62.11.4049-4059.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jørgensen B B, Revsbech N P. Oxygen uptake, bacterial distribution and carbon-nitrogen-sulfur cycling in sediments from the Baltic Sea-North Sea transition. Ophelia. 1989;31:29–49. [Google Scholar]
- 16.Lane D J. 16S/23S rRNA sequencing. In: Stackebrand E, Goodfellow M, editors. Nucleic acid techniques in bacterial systematics. Chichester, United Kingdom: John Wiley and Sons; 1991. [Google Scholar]
- 17.Llobet-Brossa E, Roselló-Mora R, Amann R I. Microbial community composition of Wadden Sea sediments as revealed by fluorescence in situ hybridization. Appl Environ Microbiol. 1998;64:2691–2696. doi: 10.1128/aem.64.7.2691-2696.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Maidak B L, Olsen G J, Overbeek R, McCaughey M J, Woese R C. The RDP (Ribosomal Database Project) Nucleic Acids Res. 1997;25:109–111. doi: 10.1093/nar/25.1.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Manz W, Eisenbrecher M, Neu T R, Szewzyk U. Abundance and spatial organization of Gram-negative sulfate-reducing bacteria in activated sludge investigated by in situ probing with specific 16S rRNA targeted oligonucleotides. FEMS Microbiol Ecol. 1998;25:43–61. [Google Scholar]
- 20.McGlatherw K J, Risgaard-Petersen N, Christian P B. Temporal and spatial variation in nitrogen fixation activity in the eelgrass Zostera marina rhizosphere. Mar Ecol Prog Ser. 1998;168:245–258. [Google Scholar]
- 21.Minz D, Fishbain S, Green S J, Muyzer G, Cohen Y, Rittmann B E, Stahl D A. Unexpected population distribution in a microbial mat community: sulfate-reducing bacteria localized to the highly oxic chemocline in contrast to a eukaryote preference for anoxia. Appl Environ Microbiol. 1999;65:4659–4665. doi: 10.1128/aem.65.10.4659-4665.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Munson M A, Nedwell D B, Embley T M. Phylogenetic diversity of Archaea in sediment samples from a coastal salt marsh. Appl Environ Microbiol. 1997;63:4729–4733. doi: 10.1128/aem.63.12.4729-4733.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Nielsen J T, Liesack W, Finster K. Desulfovibrio zosterae sp. nov., a new sulfate reducer isolated from surface-sterilized roots of the seagrass Zostera marina. Int J Syst Bacteriol. 1999;49:859–865. doi: 10.1099/00207713-49-2-859. [DOI] [PubMed] [Google Scholar]
- 24.O'Donohue M J D, Moriarty D J W, McRae J C. Nitrogen fixation in the sediments and rhizosphere of the seagrass Zostera capricornii. Microb Ecol. 1991;22:53–64. doi: 10.1007/BF02540212. [DOI] [PubMed] [Google Scholar]
- 25.Pollard P C, Moriarty D J W. Organic carbon decomposition, primary and bacterial productivity and sulphate reduction in tropical seagrass beds of the Gulf of Carpenteria, Australia. Mar Ecol Prog Ser. 1991;69:149–159. [Google Scholar]
- 26.Preston C, Wu K Y, Molinski T F, DeLong E F. A psychrophilic crenarchaeon inhabits a marine sponge: Cenarchaeum symbiosum gen. nov., sp. nov. Proc Natl Acad Sci USA. 1996;93:6241–6246. doi: 10.1073/pnas.93.13.6241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ravenschlag K, Sahm K, Pernthaler J, Amann R. High bacterial diversity in permanently cold, marine sediments. Appl Environ Microbiol. 1999;65:3982–3989. doi: 10.1128/aem.65.9.3982-3989.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rooney-Varga J N, Genthner B R, Devereux R, Willis S G, Friedman S D, Hines M E. Phylogenetic and physiological diversity of sulfate-reducing bacteria isolated from a salt marsh sediment. Syst Appl Microbiol. 1998;21:557–568. doi: 10.1016/s0723-2020(98)80068-4. [DOI] [PubMed] [Google Scholar]
- 29.Rosselló-Mora R, Thamdrup B, Schäfer H, Weller R, Amann R. The response of the microbial community of marine sediments to organic carbon input under anaerobic conditions. Syst Appl Microbiol. 1999;22:237–248. doi: 10.1016/S0723-2020(99)80071-X. [DOI] [PubMed] [Google Scholar]
- 30.Sahm K, Knoblauch C, Amann R. Phylogenetic affiliation and quantification of psychrophilic sulfate-reducing isolates in marine arctic sediments. Appl Environ Microbiol. 1999;65:3976–3981. doi: 10.1128/aem.65.9.3976-3981.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sahm K, MacGregor B J, Jørgensen B B, Stahl D A. Sulfate reduction and vertical distribution of sulfate-reducing bacteria quantified by rRNA slot-blot hybridization in a coastal marine sediment. Environ Microbiol. 1999;1:65–74. doi: 10.1046/j.1462-2920.1999.00007.x. [DOI] [PubMed] [Google Scholar]
- 32.Schaub B E M, Stal L J. Phosphate buffering in the sediments of a coastal lagoon (Bassin d'Arcachon, France). Annual report: the ROle of BUffering capacities in STabilising coastal lagoon ecosistems (ROBUST). Arcachon, France: R. de Wit; 1998. [Google Scholar]
- 33.Smith W. Ecological actions of sulfate-reducing bacteria. In: Odom J M, Singleton R Jr, editors. The sulfate-reducing bacteria: contemporary perspectives. New York, N.Y: Springer-Verlag; 1993. pp. 161–188. [Google Scholar]
- 34.Upton A C, Nedwell D B, Parkes R J, Harvey S M. Seasonal benthic microbial activity in the Southern North Sea: oxygen uptake and sulfate reduction. Mar Ecol Prog Ser. 1993;101:273–281. [Google Scholar]
- 35.Vetriani C, Reysenbach A L, Dore J. Recovery and phylogenetic analysis of archaeal rRNA sequences from continental shelf sediments. FEMS Microbiol Lett. 1998;161:83–88. doi: 10.1111/j.1574-6968.1998.tb12932.x. [DOI] [PubMed] [Google Scholar]
- 36.Welsh D T, Bourguès S, de Wit R, Herbert R A. Seasonal variations in nitrogen fixation (acetylene reduction) and sulfate reduction rates in the rhizosphere of Zostera noltii: nitrogen fixation by sulfate reducing bacteria. Mar Biol. 1996;125:619–628. [Google Scholar]
- 37.Zhou J, Bruns M A, Tiedje J M. DNA recovery from soils of diverse composition. Appl Environ Microbiol. 1996;62:316–322. doi: 10.1128/aem.62.2.316-322.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]