Abstract
MIR4435-2HG (LINC00978) is a long non-coding RNA (lncRNA) that acts as an oncogene in almost all cancers. This lncRNA participates in the molecular cascades involved in other disorders such as coronary artery diseases, osteonecrosis, osteoarthritis, osteoporosis, and periodontitis. MIR4435-2HG exerts its functions via the spectrum of different mechanisms, including inhibition of apoptosis, sponging microRNAs (miRNAs), promoting cell proliferation, increasing cell invasion and migration, and enhancing epithelial to mesenchymal transition (EMT). MIR4435-2HG can regulate several signaling pathways, including Wnt, TGF-β/SMAD, Nrf2/HO-1, PI3K/AKT, MAPK/ERK, and FAK/AKT/β‑catenin signaling pathways; therefore, it can lead to tumor progression. In the present review, we aimed to discuss the potential roles of lncRNA MIR4435-2HG in developing cancerous and non-cancerous conditions. Due to its pivotal role in different disorders, this lncRNA can serve as a potential biomarker in future investigations. Moreover, it may serve as a potential therapeutic target for the treatment of various diseases.
Keywords: MIR4435-2HG, lncRNA, Cancer, Biomarker
Introduction
Genetic alterations are one of the primary causes of cancer, leading to the deregulation of gene networks [1–3]. In recent years following developments in RNA sequencing technologies, this insight came into being that a large part of the genome transcribes into non-protein-coding RNAs [4]. Long non-coding RNAs (lncRNAs) are a subclass of functional RNAs which are longer than 200 nucleotides in sequence length without a protein-coding capacity [5–7]. In the beginning, lncRNA transcripts were regarded as ‘transcriptional noise’ or ‘junk’. Subsequent investigations revealed that lncRNAs are key players in human disorders, particularly in malignant conditions [8, 9]. Although lncRNAs do not translate into proteins, they play a meaningful function in regulating gene expression through different mechanisms such as remodeling of chromatin, modulating the activity of transcription factors, epigenetic regulation, post-transcriptional, and cell cycle regulation [10, 11]. MIR4435-2 Host Gene (MIR4435-2HG), also named LINC00978, AK001796, AWPPH, MIR4435-1HG, MORRBID, and AGD2, is an lncRNA that resides on chromosome 2q13 region and includes ten exons. MIR4435-2HG has 108 transcripts produced through alternative splicing (https://www.ensembl.org/Homo_sapiens/Gene/Summary?db=core;g=ENSG00000172965;r=2:111006015-111523376). Previous studies have reported that the MIR4435-2HG has an oncogenic role in the progression of different cancer types. In addition to the role of MIR4435-2HG in tumorigenesis, some studies suggest that it is involved in the pathogenesis of non-cancerous conditions such as coronary artery diseases [12], osteonecrosis [13], osteoarthritis [14], osteoporosis [15], and periodontitis [16]. Due to the important regulatory roles of MIR4435-2HG, in the present review, we provide comprehensive information about its function in cancer and other diseases.
MIR4435-2HG and cancer
Previous studies have shown that the expression level of MIR4435-2HG was upregulated in almost all cancers. MIR4435-2HG upregulation can promote tumor progression by increasing cell proliferation, invasion, migration, epithelial-mesenchymal transition (EMT), chemoresistance and suppression of apoptosis.
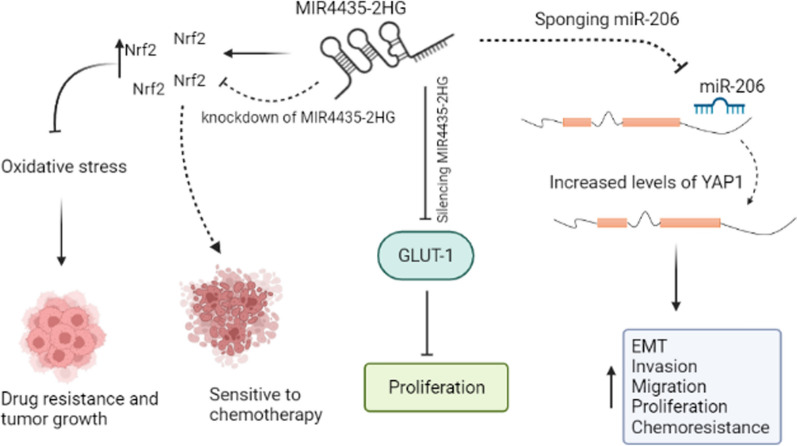
Colorectal cancer (CRC)
Overexpression of MIR4435-2HG has been reported in CRC tissues and cell lines in several studies [17–22]. Wen et al. have demonstrated that upregulation of MIR4435-2HG in CRC tissues was significantly correlated with the TNM stage [17]. Cancer-developing conditions such as chemoresistance, invasion, metastasis, migration, cancer stemness, and EMT can be regulated by Yes-related protein 1 (YAP1) transcription factor [23]. Dong et al. showed that MIR4435-2HG could regulate the expression of miR-206. On the other hand, they also indicated that YAP1 was a potential target for miR-206 (Fig. 1 and Table 1). MIR4435-2HG knockdown could block invasion, migration, and cell proliferation through the miR-206/YAP1 axis in the HCT116 and SW620 cell lines [18]. Previous studies reported that expression of nuclear factor erythroid 2-related factor 2 (Nrf2) and its regulator, heme oxygenase-1 (HO-1), increased after treating cancer cells with chemotherapeutic agents. These factors regulate the detoxification process and antioxidant enzymes, which results in the reduction of drug effects and an increase in drug resistance [24, 25]. In HCT116R cells (a cisplatin-resistant cell line), knockdown of MIR4435-2HG significantly induced cisplatin sensitivity, enhanced apoptosis, and inhibited cell proliferation via modulating Nrf2/HO-1 cascade (Fig. 1). Hence, it seems that MIR4435-2HG is involved in oxidative stress [19]. Another experiment has indicated that in patients with colon cancer, serum levels of glucose transporter 1 (GLUT‑1) and MIR4435-2HG were significantly higher than healthy controls. Moreover, silencing MIR4435-2HG inhibits cell proliferation through downregulation of GLUT-1 in the HT-29 cancerous cell line (Fig. 1) [20]. In our previous study, we showed a positive correlation between β-catenin and MIR4435-2HG expression that indicated mentioned lncRNA might regulate the Wnt signaling pathway via stabilization of β-catenin, which can lead to the progression of CRC [21]. Shen et al. showed that high expression of MIR4435-2HG was remarkably related to clinicopathological features, including stage, tumor size, tumor node and lymph node metastasis. Their results showed that the patients with higher levels of MIR4435-2HG had a worse prognosis than the patients with lower expression levels. In addition, MIR4435-2HG silencing remarkably reduced cell proliferation and enhanced cell apoptosis [22]. To conclude, MIR4435-2HG can promote CRC via different mechanisms.
Fig. 1.
Schematic representation of MIR4435-2HG functions in CRC. Increased level of MIR4435-2HG blocks miR-206 which leads to elevation of YAP1 and enhanced cell invasion, migration, proliferation and chemoresistance. In HCT116R cells, the activation of Nrf2 pathway leads to drug resistance; however, knockdown of MIR4435-2HG sensetives cancer cell to chemotherapy through inhibition of Nrf2. Furthermore, MIR4435-2HG can increase CRC progression via promoted GLUT-1
Table 1.
MIR4435-2HG participates in the pathogenesis of different cancers via the regulation of different miRNAs (∆: knock-down, EMT: Epithelial-Mesenchymal Transition, TNBC: Triple‐negative breast cancer, NSCLC: non‑small cell lung cancer, HNSCC: head and neck squamous cell carcinoma)
| Cancer type | MIR4435-2HG | miRNA | Target gene | Function | References |
|---|---|---|---|---|---|
| Colorectal cancer | Up-regulated | ↓miR-206 | ↑YAP1 | ∆ MIR4435-2HG: ↓Invasion, ↓Migration, ↓Cell proliferation, ↓EMT, ↓CRC growth, ↓Liver metastasis | [18] |
| Gastric cancer | Up-regulated | ↓miR-497 | ↑ NTRK3 | ∆ MIR4435-2HG: ↓Cell proliferation, ↓Metastasis, ↑Apoptosis | [28] |
| Gastric cancer | Up-regulated | ↓miR-138-5p | ↑SOX4 | ∆ MIR4435-2HG: ↓Invasion, ↓Migration, ↓Cell proliferation, ↓EMT, ↑Apoptosis, ↓Tumor growth | [29] |
| Hepatocellular carcinoma | Up-regulated | ↑miRNA-487a | – | ↑MIR4435-2HG: ↑miRNA-487a, ↑Cell proliferation, | [35] |
| Hepatocellular carcinoma | Up-regulated | ↓miR‑136‑5p | ↑B3GNT5 | ∆ MIR4435-2HG: ↓Invasion, ↓Migration, ↓Cell proliferation | [38] |
| NSCLC | Up-regulated | ↓miR-6754-5p | – | ∆ MIR4435-2HG: ↓Invasion, ↓Migration, ↓Cell proliferation, ↑Apoptosis | [45] |
| Breast cancer | Up-regulated | ↓miR-22-3p | ↑TMEM9B | ∆ MIR4435-2HG: ↓viability, ↓Invasion, ↓Migration, ↓Cell proliferation, ↓EMT | [48] |
| Ovarian cancer | Up-regulated | ↓miR-128-3p | ↑CDK14 | ∆ MIR4435-2HG: ↓Cell proliferation, ↓Invasion, ↓Migration, ↓Tumor growth, ↑Apoptosis | [51] |
| Glioma cancer | Up-regulated | ↓miR-1224-5p | ↑TGFBR2 | ∆ MIR4435-2HG: ↓Cell proliferation, ↓Invasion, ↓Tumor growth | [58] |
| Glioma cancer | Up-regulated | ↓miR- 125a- 5p | ↑TAZ | ∆ MIR4435-2HG: ↓Migration, ↓Cell proliferation, ↑Apoptosis, ↓Wnt pathway, ↓Tumor volume | [61] |
| Cervical cancer | Up-regulated | ↓miR-128-3p | ↑MSI2 | ∆ MIR4435-2HG: ↓Cell proliferation, ↓Invasion, ↓Migration | [66] |
| Bladder cancer | Up-regulated | ↓miR-4288 | – | ∆ MIR4435-2HG: ↓Cell proliferation, ↓Invasion, ↓Migration | [71] |
| HNSCC | Up-regulated | ↓miR‑383‑5p | ↑RBM3 | ∆ MIR4435-2HG: ↓Cell proliferation, ↓Invasion, ↓Migration, ↓EMT, ↓Tumor growth | [70] |
| Melanoma | Up-regulated | ↓miR-802 | ↑FLOT2 | ↑MIR4435-2HG: ↑Cell proliferation, ↑Invasion, ↑Migration | [72] |
| TNBC | Up-regulated | ↑miRNA‐21 | - | ↑MIR4435-2HG: ↑cell viability, ↑cell proliferation, ↑chemoresistance | [49] |
| NSCLC | Up-regulated | ↓miRNA-204 | ↑CDK6 | ∆ MIR4435-2HG: ↓cell proliferation, ↓invasion, ↓migration | [46] |
Gastric cancer (GC)
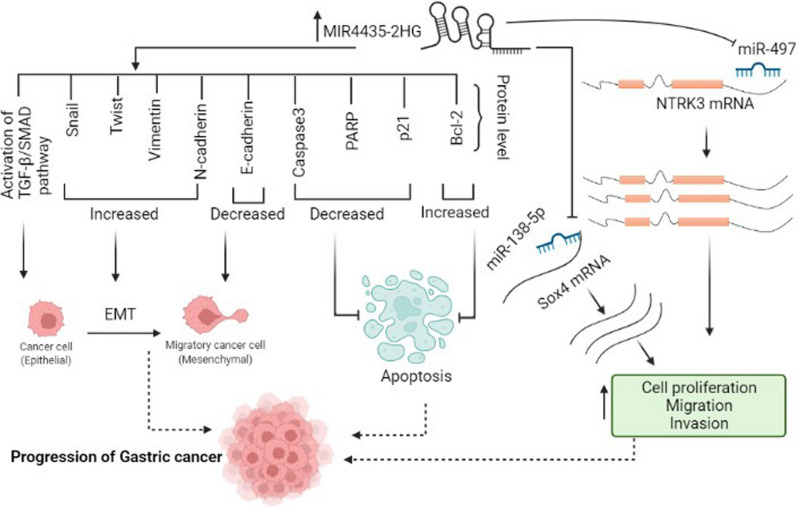
Several studies reported that the expression level of MIR4435-2HG was significantly increased in GC tissues, plasma samples, and different cell lines compared to the normal controls [26–29]. TGF-β/SMAD is one of the pathways involved in the progression of metastasis in gastric cancer [30]. Min et al. showed that MIR4435-2HG expression was significantly correlated with TNM stage, tumor size, and lymphatic metastasis. Knockdown of MIR4435-2HG elevated the expression of E-cadherin protein while the expression levels of vimentin, slug, N-cadherin, and twist proteins were inhibited. On the other hand, MIR4435-2HG knockdown leads to the inhibition of transforming growth factor beta (TGF-β) and phosphorylated SMAD2 (p-SMAD2) in gastric cancer cell lines. This observation suggests that knockdown of MIR4435-2HG can elevate EMT, and apoptosis and inhibit cell cycle progression, invasion, and migration via the regulation of TGF-β/SMAD signaling pathway (Fig. 2) [27, 31, 32]. Yuan et al. reported that MIR4435-2HG could target miR-497. Interestingly, tropomyosin receptor kinase C (NTRK3) plays a critical role in cancer progression and is a direct target of miR-497. MIR4435-2HG acts as a molecular sponge of miR-497, which leads to an increase in NTRK3. It can be concluded that the elevation of MIR4435-2HG could enhance tumorigenesis via miR-497/NTRK3 axis (Fig. 2 and Table 1) [28]. Gao et al. demonstrated that high expression of MIR4435-2HG was associated with poor survival rate in GC patients. They also reported the enhancement of apoptosis and suppression of cell proliferation, migration, invasion and EMT after MIR4435-2HG knockout in gastric carcinoma cells. It was suggested that overexpression of MIR4435-2HG affects the expression of SRY-box transcription factor 4(SOX4) via sponging miR-138-5p. Therefore, MIR4435-2HG plays an oncogenic role in GC by targeting the miR-138-5p/SOX4 axis (Fig. 2 and Table 1) [29].
Fig. 2.
MIR4435-2HG exerts its oncogenic role via different mechanisms in the progression of GC. MIR4435-2HG acts as a molecular sponge for miR-497 and miR-138-5p; therefore, it enhances the expression of NTRK3 and SOX4 (respectively), which results in cell proliferation, migration, and invasion. Also, an elevated level of MIR4435-2HG activates the TGF-β/SMAD signaling pathway, promotes EMT and suppresses apoptosis
Hepatocellular carcinoma (HCC)
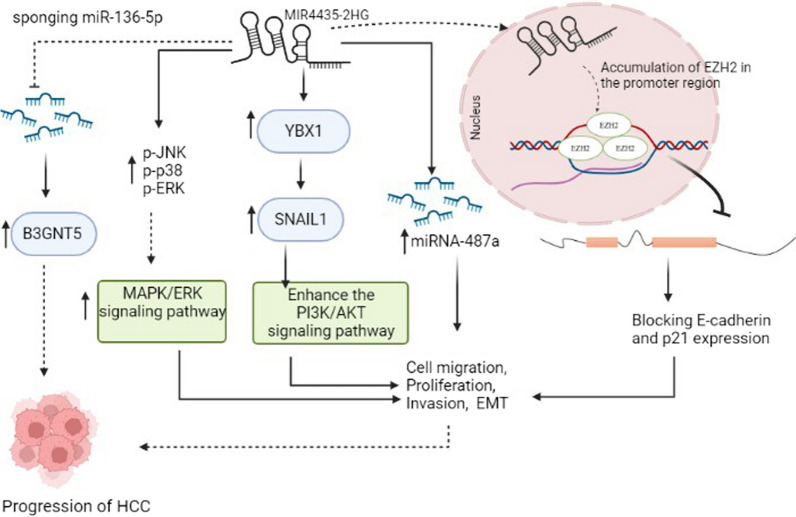
MIR4435-2HG upregulation has also been detected in hepatocellular carcinoma tissues and cell lines. [33–38]. In vitro and in vivo studies performed by Zhao et al. showed that upregulation of MIR4435-2HG increased migration, cell proliferation, metastasis, and tumor growth in hepatocellular carcinoma cells via regulating the interaction of Y-box binding protein 1 (YBX1) with snail family transcriptional repressor 1 (SNAIL1) and phosphatidylinositol-4,5-bisphosphate 3-kinase catalytic subunit alpha)PIK3CA(. Previous studies reported that YBX1 could induce EMT, SNAIL1 mRNA translation, and promote metastasis. YBX1 can stimulate PIK3CA transcription and enhance the PI3K/AKT signaling pathway by binding its promoter in cancer cells (Fig. 3) [34, 39, 40]. Another study showed that miRNA-487a and MIR4435-2HG were elevated in HCC tumor samples compared to adjacent tissues, and a positive correlation was detected between the genes expression. The overexpression of MIR4435-2HG in the HCC SNU-398 and SNU-182 cell lines promoted cell proliferation through upregulation of miRNA-487a (Fig. 3 and Table 1) [35]. Polycomb repressive complex 2 (PRC2) consists of multiple subunits including, Enhancer of Zeste Homolog 2 (EZH2) that displays methyltransferase activity. Previous studies showed that EZH2 was remarkably upregulated in many cancers, including HCC. Using chromatin immunoprecipitation (ChIP), Xueying et al. showed that E-cadherin and p21 are molecular targets of MIR4435-2HG. As shown in Fig. 3, MIR4435-2HG enhances the promoter methylation of E-cadherin and p21 genes via mediating the accumulation of EZH2 in the promoter region. It can be concluded that MIR4435-2HG increases HCC progression via blocking E-cadherin and p21 expression through EZH2-mediated epigenetic silencing (Fig. 3) [36, 41, 42]. Zhang et al. reported that high expression of MIR4435-2HG correlates with poor HCC prognosis. They also indicated that MIR4435-2HG knockdown strongly induced apoptosis, cell cycle arrest and significantly decreased HCC cell proliferation capacity. Inhibition of MIR4435-2HG led to a decrease of phosphorylated JNK (p-JNK), phospho-p38 (p-p38), and phospho-ERK (p-ERK). It seems that MIR4435-2HG induces the progression of HCC by activating the MAPK/ERK signaling pathway (Fig. 3) [37]. Zhu et al. identified the target genes of MIR4435-2HG. They also confirmed interactions between MIR4435‑2HG, miR‑136‑5p, and B3GNT5, one of the downstream targets of miR‑136‑5p, using luciferase reporter assays. MiR‑136‑5p acts as a tumor suppressor in various cancers such as liver cancer. MIR4435‑2HG could sponge miR‑136‑5p while the expression of UDP-GlcNAc:betaGal beta-1,3-N-acetylglucosaminyltransferase 5 (B3GNT5) was upregulated in liver cancer tissues. It can be concluded that MIR4435‑2HG, by sponging miR‑136‑5p, can directly reverse its inhibitory effects on target genes such as B3GNT5, thereby facilitates the progression of liver cancer via the MIR4435‑2HG/miR‑136‑5p/ B3GNT5 axis (Fig. 3 and Table 1) [38].
Fig. 3.
The impact of MIR4435-2HG in the progression of HCC through different mechanisms. MIR4435-2HG increases promoter methylation of E-cadherin and p21 via mediating the accumulation of EZH2 in the promoter region. MIR4435-2HG increases HCC progression via blocking E-cadherin and p21 expression. In addition, MIR4435-2HG promotes cell proliferation, migration, invasion, and EMT via activation of MAPK/ERK and PI3K/AKT signaling pathways. The effect of MIR4435-2HG is exerted via sponging miR-136-5p and interaction with miR-487a
Lung cancer (LC)
Qiaoyuan et al. showed that the MIR4435-2HG expression was downregulated after treating LC cells with resveratrol. They showed that cell cycle arrest occurred in G0/G1 phase following MIR4435-2HG knockdown. They also indicated that inhibition of MIR4435-2HG in lung cancerous cell lines enhanced the anticancer effects of resveratrol [43]. Another experiment revealed EMT suppression following MIR4435-2HG knockdown. Notably, MIR4435-2HG prevents the destruction of β-catenin by the proteasome system, however, MIR4435-2HG knockdown resulted in the decreased β-catenin transactivation and subsequent inhibition of the Wnt/β-catenin signaling pathway [44]. MIR4435-2HG can potentially sponge miR-6754-5p in non‑small cell lung cancer (NSCLC). In NSCLC samples, the miR-6754-5p expression was downregulated and negatively correlated with MIR4435-2HG expression. It can be concluded that the MIR4435-2HG plays an oncogenic role in NSCLC via blocking the miR-6754-5p function (Table 1) [45]. Recently, Wu et al. introduced miR-204 as a target for MIR4435-2HG in NSCLC. MIR4435-2HG leads to the progression of NSCLC through sponging miR-204. Silencing of MIR4435-2HG promoted the expression of miR-204 and therefore decreased the expression of cyclin dependent kinase 6 (CDK6), resulting in the enhancement of cell proliferation, invasion and migration in the A549 cell line [46].
Breast cancer (BC)
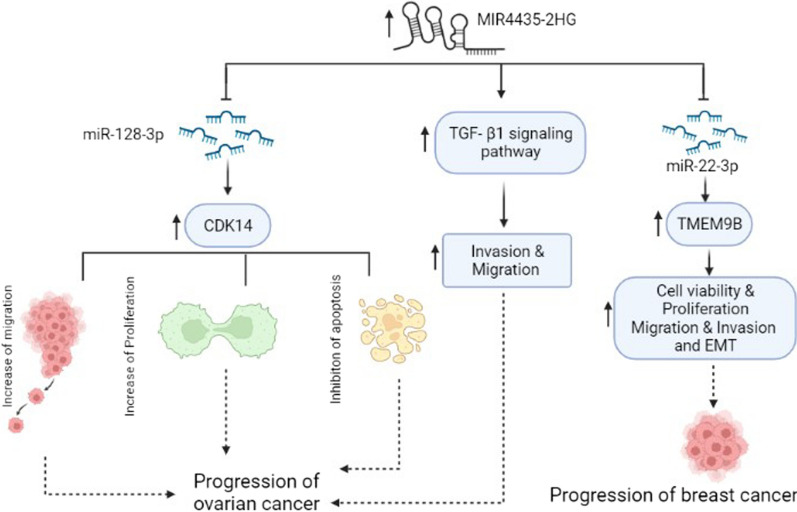
One of the pioneer investigations for the assessment of MIR4435-2HG has been conducted in the BC tissues and cell lines by Lin et al. They indicated that MIR4435-2HG was over-expressed in breast cancer tissues and cell lines compared with corresponding controls and therefore may act as an oncogene. They reported that hormone receptor status and MIR4435-2HG expression were negatively correlated [47]. Consistent with the above study, Jing et al. showed that MIR4435-2HG was upregulated in breast cancer tissues and cell lines. They indicated that MIR4435-2HG could enhance many cellular parameters such as proliferation, EMT, migration, and invasion via regulating the miR-22-3p/TMEM9B axis (Fig. 4 and Table 1) [48]. Liu et al. demonstrated that the plasma level of MIR4435-2HG was remarkably higher in patients with Triple-negative breast cancer (TNBC) than healthy controls, and its expression level was positively correlated to miR-21. It was concluded that overexpression of MIR4435-2HG increased cell viability, proliferation and induced chemoresistance via interaction with miR-21 in MDA-MB-231 and BT-20 cell lines [49].
Fig. 4.
The role of MIR4435-2HG in the pathogenesis of ovarian and breast cancers. In ovarian cancer, MIR4435-2HG regulates migration, invasion and apoptosis through modulating the expression of CDK14. The effect of MIR4435-2HG is exerted via sponging miR-128-3p. In addition, MIR4435-2HG enhances the progression of ovarian cancer via activation of the TGF-β signaling pathway. In breast cancer, MIR4435-2HG sponges miR-22-3p and results in a subsequent increase in TMEM9B expression and tumorigenesis
Ovarian cancer (OC)
It has been shown that MIR4435-2HG was upregulated in OC tissues and cell lines [50, 51]. It is suggested that MIR4435-2HG can distinguish stage I and II OC patients from healthy controls. Gong et al. reported that the expression level of TGF-β1 was upregulated in OC tissues and positively correlated with MIR4435-2HG expression. Using in vitro studies, they indicated the overexpression of MIR4435-2HG in UWB1.289 and UWB1.289 + BRCA1 cells led to upregulation of TGF- β1. Taken together, MIR4435-2HG could increase OC progression through overexpression of TGF-β1 (Fig. 4) [50]. Lijuan et al. indicated that the MIR4435-2HG and cyclin dependent kinase 14 (CDK14) were upregulated while miR-128-3p was down-regulated in cell lines and OC tissue samples. On the other hand, the expression of MIR4435-2HG was negatively associated with miR-128-3p in OC tissue. They showed that MIR4435-2HG could target miR-128-3p therefore, it might be concluded that MIR4435-2HG acts as the miR-128-3p sponge. CDK14 is a downstream target of miR-128-3p. In vitro studies confirmed that miR-128-3p targeted CDK14 and suppressed its expression. Knockdown of MIR4435-2HG promoted the expression of miR-128-3p, which led to decreased CDK14 expression (Fig. 4 and Table 1) [51].
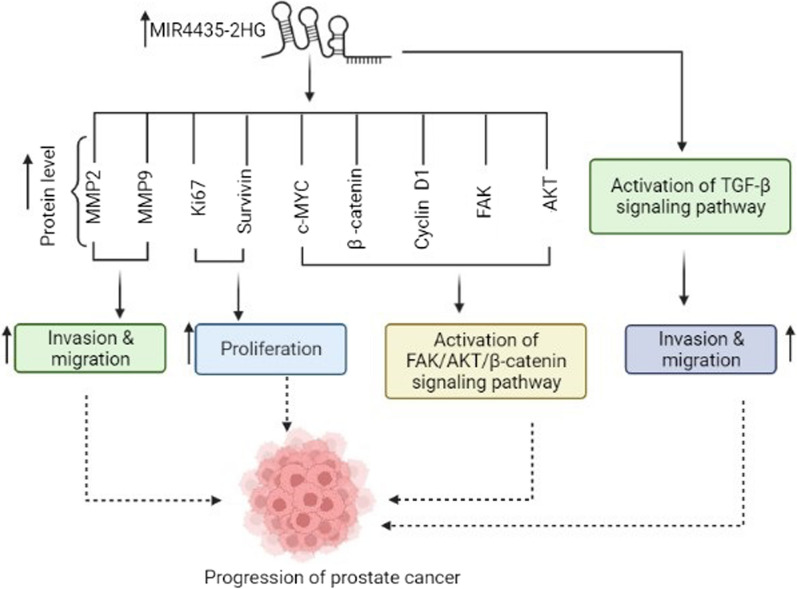
Prostate cancer (PC)
The expression level of MIR4435-2HG is reported to be enhanced in prostate cancer. It is suggested that this lncRNA causes cancer progression through various mechanisms such as FAK/AKT/β‑catenin and TGF-β1 pathways [52, 53]. Moreover, overexpression of ST8 alpha-N-acetyl-neuraminide alpha-2,8-sialyltransferase 1 (ST8SIA1) can increase the tumor cell proliferation, migration, and invasion in prostate cancer, colorectal cancer, and breast cancer via the promotion of the FAK‑AKT‑mTOR signaling pathway [52, 54, 55]. Knockdown of MIR4435-2HG suppressed cell proliferation, invasion, and migration by blocking the activation of the FAK/AKT/β‑catenin pathway in PC cell lines. Xing et al. indicated that knockdown of ST8SIA1 suppressed the effects of MIR4435-2HG in tumor progression. It seems that MIR4435-2HG contributes to tumorigenesis via the MIR4435-2HG/ST8SIA1 axis [52, 56]. Hui et al. demonstrated that the plasma level of TGF- β1 was remarkably higher in patients with PC than healthy controls, and TGF- β1 expression level was positively correlated to MIR4435-2HG. They reported that the effects of MIR4435-2HG on cell migration and invasion decreased following the inhibition of TGF-β1 (Fig. 5) [53].
Fig. 5.
MIR4435-2HG contributes to the pathogenesis of prostate cancer via different mechanisms. Elevation of MIR4435-2HG induces cell proliferation, invasion, migration and activation of TGF-β and FAK/AKT/ β-catenin signaling pathways via modulating the expression of MMP2, MMP9, Ki67, SURVIVIN, c-MYC, β-catenin, Cycline D1, FAK and AKT
Glioma cancer
One of the members of the TGF-β/Smad signaling pathway is transforming growth factor-beta receptor type II (TGFBR2) that acts as a cancer suppressor [57]. Xu et al. reported that the expression level of MIR4435-2HG was upregulated in patients with glioblastoma (GBM). In contrast, the expression level of miR-1224-5p was suppressed in GBM cancer cell lines. They used bioinformatics predictions and in vitro methods to show that this lncRNA acts as a sponge for miR-1224-5p. On the other hand, one of the direct targets of miR-1224-5p is TGFBR2 gene, and the mRNA level of its gene was enhanced in GBM cancer cell lines. Taken together, it can be argued that MIR4435-2HG can promote cancer progression by targeting the miR-1224-5p/TGFBR2 axis (Table 1) [58]. TGF-β signaling pathway is an essential factor for EMT. In patients with glioma cancer, a positive correlation was detected between the plasma levels of TGF-β and MIR4435-2HG. Therefore, it may be concluded that MIR4435-2HG is involved in the progression of glioma occur through TGF-β signaling pathway [59]. As a transcription coactivator, tafazzin, phospholipid-lysophospholipid transacylase (TAZ) is one of the most important downstream effectors of the Hippo signaling pathway that regulates cell proliferation, migration, and apoptosis [60]. The expression level of TAZ was upregulated in the brain tissue of glioma patients. Shen et al. indicated that the expression of MIR4435- 2HG was positively correlated with TAZ expression, while miR- 125a- 5p expression was negatively correlated with TAZ expression in the brain tissue (Table 1) [61].
Leukemia cancers
Rho‑associated protein kinase 2 (ROCK2) is an important therapeutic target, and its upregulation was confirmed in many cancers, including T‑cell acute lymphoblastic leukemia (T‑ALL) [62, 63]. The expression of MIR4435-2HG and ROCK2 was positively correlated in patients with T‑ALL. The overexpression of MIR4435-2HG remarkably increased ROCK2 expression at both protein and mRNA levels; also, the overexpression of ROCK2 significantly upregulated MIR4435-2HG expression in T‑ALL cells. It seems that MIR4435-2HG inhibits apoptosis and increases cell proliferation in T‑ALL cells through interactions with ROCK2 [64]. Zhigang et al. reported that MIR4435-2HG was overexpressed in human acute myeloid leukemia (AML), which was correlated with a poor survival rate. They showed that MIR4435-2HG acts as a transcriptional repressor of BIM pro-apoptotic gene and, via this mechanism, regulates the lifespan of myeloid cells. In vivo study demonstrated that the loss of MIR4435-2HG in genetic mice models promoted the expression levels of BIM that increased cell death in mature and immature myeloid cells [65].
Other cancers
The overexpression of MIR4435-2HG has also been reported in other cancers, including cervical cancer (CC) [66], clear cell renal cell carcinoma (ccRCC) [67, 68], esophageal squamous cell (ESCC) [69], head and neck squamous cell carcinoma (HNSCC) [70], bladder cancer (BCa) [71], melanoma [72] and nasopharyngeal carcinoma (NPC) [73].
MIR4435-2HG could target miR-128-3p and negatively modulated its expression in cervical cancer (CC). As shown in Fig. 5d, miR-128-3p negatively regulates the expression level of musashi RNA binding protein 2 (MSI2) gene. Therefore, it can be concluded that knockdown of MIR4435-2HG suppresses migration, invasion, and proliferation of CC cells via regulating the miR-128-3p/MSI2 axis (Table 1) [66]. Jianquan et al. reported that MIR4435-2HG knockdown not only increased apoptosis and cell cycle arrest in G0/G1 phase but also decreased invasion and migration in clear cell renal cell carcinoma [67]. Zhu et al. suggested that MIR4435-2HG could directly interact with miR-513a-5p and repressed its expression in ccRCC. Knockdown of MIR4435-2HG inhibited proliferation, metastasis and tumour progression by downregulating Kruppel like factor 6 (KLF6) as the direct target of miR-513a-5p [68].
Knockdown of lncRNA MIR4435-2HG regulates cell cycle, cell proliferation, and cells growth via modulating MDM2/p53 signaling pathway in patients with ESCC [69]. Wang et al. demonstrated that the expression level of MIR4435-2HG was upregulated in BCa tissues and cell lines compared to the control samples. They indicated that MIR4435-2HG served as a competing endogenous RNA (ceRNA) and sponged miR-4288. Using in vitro methods, they showed that knockdown of MIR4435-2HG significantly inhibited tumor growth by sponging miR-4288 [71]. Shu et al. reported that the high expression level of MIR4435-2HG was significantly associated with advanced tumor metastasis node in patients with HNSCC. In vivo and in vitro investigations indicated that knockdown of MIR4435-2HG decreased invasion, cell proliferation, and EMT in HNSCC cancer cell lines. As shown in Table 1, MIR4435-2HG executes these functions via modulating miR‑383‑5p. On the other hand, one target of miR‑383‑5p is RNA binding motif protein 3 (RBM3). It can be concluded that HNSCC progression can be regulated by MIR4435-2HG/miR‑383‑5p/RBM3 axis [70]. It has been established that flotillin-2 (FLOT2) has a critical role in the progression of human cancers through different mechanisms [74, 75]. According to bioinformatics analysis, miR-802 targets FLOT2 gene. Han et al. showed that MIR4435-2HG sponged miR-802 which leads to increased expression of FLOT2 and tumor progression (Table 1) [72]. The experimental studies showed that MIR4435-2HG inhibited apoptosis while facilitating migration, and cell proliferation in NPC cells. The mentioned lncRNA exerts this function by inhibiting of phosphatase and tensin homolog (PTEN) as a tumor suppressor gene [73].
MIR4435-2HG and non-cancerous diseases
Accumulating evidence reveals that MIR4435-2HG not only is involved in cancer progression but also plays a critical role in the development of other diseases. In this section, we investigated the role of MIR4435-2HG in non-cancerous disorders.
Coronary artery diseases (CAD)
The serum level of MIR4435-2HG remarkably increased in CAD patients compared to healthy controls. Clinical studies revealed that treatment with statins drugs, atorvastatin, and rosuvastatin, reduced MIR4435-2HG level significantly in CAD patients. This reduction was mainly observed in patients treated with rosuvastatin [12].
Osteoarthritis
The MIR4435-2HG transcript level was lower in plasma samples of patients with osteoarthritis than in healthy controls. Knockdown of MIR4435-2HG decreased proliferation and promoted cell apoptosis in chondrocytes, while overexpression of MIR4435-2HG enhanced proliferation of chondrocytes and suppressed apoptosis. After treatment with anti-inflammatory drugs (such as naproxen), reducing the joint burden and exercise, the expression level of MIR4435-2HG was increased [14].
Osteoporosis
Guang et al. reported that MIR4435-2HG was downregulated in plasma of patients suffering osteoporosis compared to healthy controls. They also showed a positive correlation between MIR4435-2HG and bone turnover markers, procollagen-1 N-terminal peptide (P1NP) and tartrate-resistant acid phosphatase 5b (TRACP-5b). The phenotype of osteoblasts can be regulated by type I collagen α1/α2 ratio. Knockdown of MIR4435-2HG suppressed α1 expression but upregulated α2. In contrast, upregulation of MIR4435-2HG elevated α1 but decreased α2. It can be concluded that MIR4435-2HG can affect the phenotype of osteoblasts via alteration in type I collagen α1/α2 ratio [15].
Osteonecrosis of the femoral head (ONFH)
Runt‑related transcription factor 2 (RUNX2) has been identified as a marker of osteoblastic differentiation [76, 77]. Decreased expression of RUNX2 led to the development of non‑traumatic ONFH [78]. The investigation showed that the expression level of MIR4435-2HG in both serum and mesenchymal stem cells (MSCs) samples was significantly downregulated. Silencing and overexpression of MIR4435-2HG in hMSC‑BM cells could lead to inhibition and promotion of RUNX2 expression, respectively. To conclude, MIR4435-2HG participates in the progression of non‑traumatic ONFH through elevated RUNX2 [13].
Periodontitis
Xiaofang et al. demonstrated that the expression level of MIR4435-2HG was elevated in plasma samples of patients with periodontitis compared to healthy controls. They showed that the expression level of MIR4435-2HG was remarkably downregulated after treatment (administration of both oral and topical antibiotics, root planning and scaling). However, after two years of follow-up, the expression of MIR4435-2HG was significantly elevated in patients with recurrence of periodontitis [16].
Human immunodeficiency virus (HIV) infection
Expression of MIR4435-2HG is also involved in immune responses against HIV-1 infection. Hartana et al. investigated the expression level of this lncRNA in myeloid dendritic cells (mDCs) obtained from HIV-1 elite controllers (ECs), in whom the virus replication is under control in the absence of antiretroviral treatment, compared to HIV-1-negative healthy controls and those who were treated using antiretroviral therapy. They found that regulatory associated protein of MTOR complex 1 (RPTOR) gene, a major component of the mammalian target of rapamycin (mTOR) signaling pathway, via induction of an epigenetic alteration. Taken together, upregulation of MIR4435-2HG in mDCs from ECs influences immunometabolic activities through different mechanisms, including altered glycolysis, oxidative phosphorylation, epigenetic modifications [79].
Diagnostic value of MIR4435-2HG
Several studies have shown that evaluating lncRNAs expression in serum, plasma, and other body fluids may serve as diagnostic or prognostic biomarkers in different disorders that are non-invasive and convenient compared to biopsy and imaging methods. For example, Fu et al. showed the elevated levels of MIR4435-2HG both in tumor tissues and serum samples of gastric cancer patients. They suggested that this lncRNA may be a potential biomarker in gastric cancer [27]. Receiver Operating Characteristic (ROC) Curve Analysis plays a central role in evaluating the diagnostic ability of tests to discriminate the true state of subjects. The diagnostic value of MIR4435-2HG has been evaluated in some tumors and other diseases. MIR4435-2HG can differentiate tumor samples from corresponding controls and distinguish disease status in other non-cancerous conditions. As shown in Table 2, MIR4435-2HG has the best diagnostic power in osteoarthritis subjects.
Table 2.
Diagnostic value of MIR4435-2HG in cancers and non-cancerous conditions [ALL: Acute lymphoblastic leukemia, AUC: Area under the Curve]
| Disease | Expression | Number of samples | Sensitivity | Specificity | AUC | Sample | References |
|---|---|---|---|---|---|---|---|
| Gastric cancer | Up | 51 cancer patients and 53 healthy controls | 90.2 | 74.5 | 88.2 | Plasma | [26] |
| Gastric cancer | Up | 72 cancer patients and adjacent non-cancerous tissues | 80 | 70 | 83.1 | Serum | [27] |
| Hepatocellular cancer | Up | 58 cancer patients and 45 healthy controls | 75.9 | 95.9 | 91 | Serum | [36] |
| Colorectal cancer | Up | 70 cancer patients and adjacent non-cancerous tissues | 72 | 80 | 81 | Tissue | [21] |
| Colon cancer | Up | 46 cancer patients and 42 healthy controls | – | – | 84.8 | Serum | [20] |
| Ovarian carcinoma | Up | 66 cancer patients and 54 healthy controls | – | – | 88.2 | Plasma | [50] |
| ALL | Up | 32 cancer patients and 32 healthy controls | – | – | 89.5 | Bone marrow | [64] |
| Renal cell carcinoma | Up | 118 cancer patients and adjacent non-cancerous tissues | – | – | 94.6 | Tissue | [67] |
| Osteoarthritis | Down | 78 osteoarthritis and 58 healthy controls | – | – | 96 | joint fluid | [14] |
| Osteoporosis | Down | 88 osteoporosis patients and 57 healthy control | – | – | 92 | plasma | [15] |
| Non-traumatic ONFH | Down | 36 ONFH patients and 30 healthy controls | – | – | 81.8 | Serum | [13] |
Conclusion
MIR4435-2HG participates in the progression of different human disorders. MIR4435-2HG exerts its functions via the spectrum of different mechanisms, including inhibition of apoptosis, sponging miRNAs, promotion of cell proliferation, increasing cell invasion and migration, and enhancement of EMT. As mentioned above, different miRNAs such as miR-6754-5p, miR-1224-5p, miR-802, and miR-128-3p can be sponged by MIR4435-2HG. On the other hand, MIR4435-2HG can lead to tumor progression by affecting Wnt, TGF-β/SMAD, Nrf2/HO-1, PI3K/AKT, MAPK/ERK, and FAK/AKT/β‑catenin signaling pathways. Several studies have shown that MIR4435-2HG acts as an oncogene in different types of cancer.
The overexpression of MIR4435-2HG in all cancer types that have been studied so far indicates the key role of this lncRNA in cancer progression as an oncogene. Cell proliferation, EMT, invasion, migration, and suppressed apoptosis are key hallmarks of cancer that can be affected by MIR4435-2HG expression. Besides, several studies confirmed the effectiveness of MIR4435-2HG silencing in inhibiting tumor growth in colorectal cancer, esophageal squamous cell carcinoma, gastric cancer, hepatocellular carcinoma, lung cancer, neuroglioma, and prostate cancer.
In contrast, in non-cancerous conditions such as periodontitis, osteoporosis, osteoarthritis, and osteonecrosis of the femoral head, the expression level of MIR4435-2HG has been downregulated. However, in coronary artery diseases, the expression level of MIR4435-2HG was elevated.
The expression level of MIR4435-2HG alters in response to many drugs, including statins (atorvastatin and rosuvastatin), oral and topical antibiotics, anti-inflammatory drugs (such as naproxen) and chemopreventive agent resveratrol. This subject indicates that MIR4435-2HG has a pivotal function in molecular mechanisms involved in disease development. Therefore, it can be concluded that MIR4435-2HG may serve as a potential therapeutic target for the treatment of various diseases.
Despite fundamental improvement in cancer diagnosis methods, recurrence and metastasis occur in many patients suffering from cancer, therefore, the discovery of new diagnostic biomarkers could be helpful in this regard [80, 81]. Moreover, according to the literature, the diagnostic value of MIR4435-2HG was acceptable in both cancerous and non-cancerous conditions. Detection and measurement of MIR4435-2HG in body fluids such as serum, plasma, and joint fluid suggest that this lncRNA could be used as a non-invasive marker.
Although previous studies have emphasized the role of MIR4435-2HG in the progression of different diseases, few studies has been conducted to describe the possible mechanisms involved in its regulation. Therefore, understanding the mechanisms involved in MIR4435-2HG regulation may shed light on the diagnosis and treatment of several related diseases.
In conclusion, MIR4435-2HG has a pivotal role in cancer progression and critical function in non-neoplastic conditions. Future studies may explain the role of this lncRNA as a potential biomarker and therapeutic target in human disorders, especially in tumors.
Acknowledgements
We thank the members of the departments of Clinical Biochemistry and Medical Genetics for helpful discussions.
Abbreviations
- lncRNA
Long non-coding RNA
- miRNAs
MicroRNAs
- EMT
Epithelial-mesenchymal transition
- MIR4435-2HG
MIR4435-2 Host Gene
- YAP1
Yes-related protein 1
- Nrf2
Nuclear factor erythroid 2-related factor 2
- HO-1
Hemeoxygenase-1
- GLUT‑1
Glucose transporter 1
- p-SMAD2
Phosphorylated SMAD2
- NTRK3
Tropomyosin rseceptor kinase C
- SOX4
SRY-box transcription factor 4
- YBX1
Y-box binding protein 1
- SNAIL1
Snail family transcriptional repressor 1
- PIK3CA
Phosphatidylinositol-4,5-bisphosphate 3-kinase catalytic subunit alpha
- PRC2
Polycomb repressive complex
- EZH2
Enhancer of zeste homolog
- ChIP
Chromatin immunoprecipitation
- p-JNK
Phosphorylated JNK
- p-ERK
Phospho-ERK
- MAPK
Mitogen-activated protein kinase 1
- B3GNT5
UDP-GlcNAc:betaGal beta-1,3-N-acetylglucosaminyltransferase 5
- CDK6
Cyclin dependent kinase 6
- TGF- β1
Transforming growth factor-β1
- CDK14
Cyclin dependent kinase 14
- FAK
Focal adhesion kinase
- ST8SIA1
ST8 alpha-N-acetyl-neuraminide alpha-2,8-sialyltransferase 1
- mTOR
Mammalian target of rapamycin
- TGFBR2
Transforming growth factor-beta receptor type II
- TAZ
Tafazzin, phospholipid-lysophospholipid transacylase
- ROCK2
Rho‑associated protein kinase 2
- MSI2
Musashi RNA binding protein 2
- KLF6
Kruppel like factor 6
- ceRNA
Competing endogenous RNA
- RBM3
RNA binding motif protein 3
- PTEN
Phosphatase and tensing homology
- P1NP
Procollagen-1 N-terminal peptide
- TRACP-5b
Tartrate-resistant acid phosphatase 5b
- RUNX2
Runt‑related transcription factor 2
- MSDs
Mesenchymal stem cells
- HMSC-bm
Human Mesenchymal Stem Cells-Bone Marrow
- RPTOR
Regulatory associated protein of MTOR complex 1
- ROC
Receiver operating characteristic
- CRC
Colorectal cancer
- GC
Gastric cancer
- HCC
Hepatocellular carcinoma
- NSCLC
Non‑small cell lung cancer
- OC
Ovarian cancer
- PC
Prostate cancer
- GBM
Glioblastoma
- T‑ALL
T‑cell acute lymphoblastic leukemia
- AML
Acute myeloid leukemia
- CC
Cervical cancer
- ccRCC
Clear cell renal cell carcinoma
- ESCC
Esophageal squamous cell
- HNSCC
Head and neck squamous cell carcinoma
- BCa
Bladder cancer
- NPC
Nasopharyngeal carcinoma
- BC
Breast cancer
- LC
Lung cancer
Author contributions
MG, US, SS collected the related literatures and article writing. MM drew the figures and prepared tables. VH participated in editing the manuscript. RM and MR initiated the study and revised and finalized the manuscript. All authors read and approved the final manuscript.
Funding
Not applicable.
Availability of data and materials
Not applicable.
Declarations
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no conflict of interests.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Masoumeh Rajabibazl, Email: rajabi_m@sbmu.ac.ir.
Reza Mirfakhraie, Email: reza_mirfakhraie@yahoo.com.
References
- 1.Huarte M. The emerging role of lncrnas in cancer. Nat Med. 2015;21:1253–1261. doi: 10.1038/nm.3981. [DOI] [PubMed] [Google Scholar]
- 2.Chakravarthi BV, Nepal S, Varambally S. Genomic and epigenomic alterations in cancer. Am J Pathol. 2016;186:1724–1735. doi: 10.1016/j.ajpath.2016.02.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Armaghany T, Wilson JD, Chu Q, Mills G. Genetic alterations in colorectal cancer. Gastrointest Cancer Res: GCR. 2012;5:19. [PMC free article] [PubMed] [Google Scholar]
- 4.Xu S-p, Zhang J-f, Sui S-y, Bai N-x, Gao S, Zhang G-w, et al. Downregulation of the long noncoding rna egot correlates with malignant status and poor prognosis in breast cancer. Tumor Biol. 2015;36:9807–12. doi: 10.1007/s13277-015-3746-y. [DOI] [PubMed] [Google Scholar]
- 5.Kangarlouei R, Irani S, Noormohammadi Z, Memari F, Mirfakhraie R. Anril and anrassf1 long noncoding rnas are upregulated in gastric cancer. J Cell Biochem. 2019;120:12544–12548. doi: 10.1002/jcb.28520. [DOI] [PubMed] [Google Scholar]
- 6.Sadeghi H, Nazemalhosseini-Mojarad E, Sahebi U, Fazeli E, Azizi-Tabesh G, Yassaee VR, et al. Novel long noncoding rnas upregulation may have synergistic effects on the cyp24a1 and pfdn4 biomarker role in human colorectal cancer. J Cell Physiol. 2021;236:2051–2057. doi: 10.1002/jcp.29992. [DOI] [PubMed] [Google Scholar]
- 7.Esmkhani S, Sadeghi H, Ghasemian M, Pirjani R, Amin-Beidokhti M, Gholami M, et al. Contribution of long noncoding rna hotair variants to preeclampsia susceptibility in iranian women. Hypertens Pregnancy. 2021;40:29–35. doi: 10.1080/10641955.2020.1855192. [DOI] [PubMed] [Google Scholar]
- 8.Aprile M, Katopodi V, Leucci E, Costa V. Lncrnas in cancer: From garbage to junk. Cancers. 2020;12:3220. doi: 10.3390/cancers12113220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Xiong J, Liu Y, Luo S, Jiang L, Zeng Y, Chen Z, et al. High expression of the long non-coding rna heircc promotes renal cell carcinoma metastasis by inducing epithelial-mesenchymal transition. Oncotarget. 2017;8:6555. doi: 10.18632/oncotarget.14149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cheetham S, Gruhl F, Mattick J, Dinger M. Long noncoding rnas and the genetics of cancer. Br J Cancer. 2013;108:2419–2425. doi: 10.1038/bjc.2013.233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Carlevaro-Fita J, Lanzós A, Feuerbach L, Hong C, Mas-Ponte D, Pedersen JS, et al. Cancer lncrna census reveals evidence for deep functional conservation of long noncoding rnas in tumorigenesis. Commun Biol. 2020;3:1–16. doi: 10.1038/s42003-019-0741-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tang T, Wang B. Clinical significance of lncrna-awpph in coronary artery diseases. Eur Rev Med Pharmacol Sci. 2020;24:11747–11751. doi: 10.26355/eurrev_202011_23826. [DOI] [PubMed] [Google Scholar]
- 13.Chen X, Li J, Liang D, Zhang L, Wang Q. Lncrna awpph participates in the development of non-traumatic osteonecrosis of femoral head by upregulating runx2. Exp Ther Med. 2020;19:153–159. doi: 10.3892/etm.2019.8185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Xiao Y, Bao Y, Tang L, Wang L. Lncrna mir4435-2hg is downregulated in osteoarthritis and regulates chondrocyte cell proliferation and apoptosis. J Orthop Surg Res. 2019;14:1–5. doi: 10.1186/s13018-019-1278-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Qian G, Yu Y, Dong Y, Hong Y, Wang M. Lncrna awpph is downregulated in osteoporosis and regulates type i collagen α1 and α2 ratio. Archives Physiol Biochem. 2020 doi: 10.1080/13813455.2020.1767150. [DOI] [PubMed] [Google Scholar]
- 16.Wang X, Ma F, Jia P. Lncrna awpph overexpression predicts the recurrence of periodontitis. Biosci Rep. 2019;39:BSR20190636. doi: 10.1042/BSR20190636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ouyang W, Ren L, Liu G, Chi X, Wei H. Lncrna mir4435-2hg predicts poor prognosis in patients with colorectal cancer. PeerJ. 2019;7:e6683. doi: 10.7717/peerj.6683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dong X, Yang Z, Yang H, Li D, Qiu X. Long non-coding rna mir4435-2hg promotes colorectal cancer proliferation and metastasis through mir-206/yap1 axis. Front Oncol. 2020;10:160. doi: 10.3389/fonc.2020.00160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Luo P, Wu S, Ji K, Yuan X, Li H, Chen J, et al. Lncrna mir4435-2hg mediates cisplatin resistance in hct116 cells by regulating nrf2 and ho-1. PLoS ONE. 2020;15:e0223035. doi: 10.1371/journal.pone.0223035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bai J, Xu J, Zhao J, Zhang R. Downregulation of lncrna awpph inhibits colon cancer cell proliferation by downregulating glut-1. Oncol Lett. 2019;18:2007–2012. doi: 10.3892/ol.2019.10515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ghasemian M, Rajabibazl M, Mirfakhraie R, Razavi AE, Sadeghi H. Long noncoding rna linc00978 acts as a potential diagnostic biomarker in patients with colorectal cancer. Exp Mol Pathol. 2021;122:104666. doi: 10.1016/j.yexmp.2021.104666. [DOI] [PubMed] [Google Scholar]
- 22.Shen M, Zhou G, Zhang Z. Lncrna mir4435-2hg contributes into colorectal cancer development and predicts poor prognosis. Eur Rev Med Pharmacol Sci. 2020;24:1771–1777. doi: 10.26355/eurrev_202002_20354. [DOI] [PubMed] [Google Scholar]
- 23.Zanconato F, Cordenonsi M, Piccolo S. Yap/taz at the roots of cancer. Cancer Cell. 2016;29:783–803. doi: 10.1016/j.ccell.2016.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Barrera G, Cucci MA, Grattarola M, Dianzani C, Muzio G, Pizzimenti S. Control of oxidative stress in cancer chemoresistance: spotlight on nrf2 role. Antioxidants. 2021;10:510. doi: 10.3390/antiox10040510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wang X-J, Sun Z, Villeneuve NF, Zhang S, Zhao F, Li Y, et al. Nrf2 enhances resistance of cancer cells to chemotherapeutic drugs, the dark side of nrf2. Carcinogenesis. 2008;29:1235–1243. doi: 10.1093/carcin/bgn095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ke D, Li H, Zhang Y, An Y, Fu H, Fang X, et al. The combination of circulating long noncoding rnas ak001058, inhba-as1, mir4435-2hg, and cebpa-as1 fragments in plasma serve as diagnostic markers for gastric cancer. Oncotarget. 2017;8:21516. doi: 10.18632/oncotarget.15628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Fu M, Huang Z, Zang X, Pan L, Liang W, Chen J, et al. Long noncoding rna linc 00978 promotes cancer growth and acts as a diagnostic biomarker in gastric cancer. Cell Prolif. 2018;51:e12425. doi: 10.1111/cpr.12425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bu J-Y, Lv W-Z, Liao Y-F, Xiao X-Y, Lv B-J. Long non-coding rna linc00978 promotes cell proliferation and tumorigenesis via regulating microrna-497/ntrk3 axis in gastric cancer. Int J Biol Macromol. 2019;123:1106–1114. doi: 10.1016/j.ijbiomac.2018.11.162. [DOI] [PubMed] [Google Scholar]
- 29.Gao L-F, Li W, Liu Y-G, Zhang C, Gao W-N, Wang L. Inhibition of mir4435–2hg on invasion, migration, and emt of gastric carcinoma cells by mediating mir-138–5p/sox4 axis. Front Oncol. 2021;11:3057. doi: 10.3389/fonc.2021.661288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhao M, Mishra L, Deng C-X. The role of tgf-β/smad4 signaling in cancer. Int J Biol Sci. 2018;14:111. doi: 10.7150/ijbs.23230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sabbadini F, Bertolini M, De Matteis S, Mangiameli D, Contarelli S, Pietrobono S, et al. The multifaceted role of tgf-β in gastrointestinal tumors. Cancers. 2021;13:3960. doi: 10.3390/cancers13163960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Liu S, Ren J, Ten Dijke P. Targeting tgfβ signal transduction for cancer therapy. Signal Transduct Target Ther. 2021;6:1–20. doi: 10.1038/s41392-020-00451-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Han Q, Chen B, Zhang K, Xia S, Zhong W, Zhao Z. The long non-coding rna ak001796 contributes to poor prognosis and tumor progression in hepatocellular carcinoma. Eur Rev Med Pharmacol Sci. 2019;23:2013–2019. doi: 10.26355/eurrev_201903_17240. [DOI] [PubMed] [Google Scholar]
- 34.Zhao X, Liu Y, Yu S. Long noncoding rna awpph promotes hepatocellular carcinoma progression through ybx1 and serves as a prognostic biomarker. Biochimica et Biophysica Acta (BBA)-Molecular Basis of Disease. 2017;1863:1805–16. doi: 10.1016/j.bbadis.2017.04.014. [DOI] [PubMed] [Google Scholar]
- 35.Kong Q, Liang C, Jin Y, Pan Y, Tong D, Kong Q, et al. The lncrna mir4435-2hg is upregulated in hepatocellular carcinoma and promotes cancer cell proliferation by upregulating mirna-487a. Cell Mol Biol Lett. 2019;24:1–7. doi: 10.1186/s11658-019-0148-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Xu X, Gu J, Ding X, Ge G, Zang X, Ji R, et al. Linc00978 promotes the progression of hepatocellular carcinoma by regulating ezh2-mediated silencing of p21 and e-cadherin expression. Cell Death Dis. 2019;10:1–15. doi: 10.1038/s41419-018-1236-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zhang Q, Cheng S, Cao L, Yang J, Wang Y, Chen Y. Linc00978 promotes hepatocellular carcinoma carcinogenesis partly via activating the mapk/erk pathway. Biosci Rep. 2020;40:BSR20192790. doi: 10.1042/BSR20192790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zhu Y, Li B, Xu G, Han C, Xing G. Lncrna mir4435-2hg promotes the progression of liver cancer by upregulating b3gnt5 expression. Mol Med Rep. 2022;25:1–11. doi: 10.3892/mmr.2022.12695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Evdokimova V, Tognon C, Ng T, Ruzanov P, Melnyk N, Fink D, et al. Translational activation of snail1 and other developmentally regulated transcription factors by yb-1 promotes an epithelial-mesenchymal transition. Cancer Cell. 2009;15:402–415. doi: 10.1016/j.ccr.2009.03.017. [DOI] [PubMed] [Google Scholar]
- 40.Astanehe A, Finkbeiner M, Hojabrpour P, To K, Fotovati A, Shadeo A, et al. The transcriptional induction of pik3ca in tumor cells is dependent on the oncoprotein y-box binding protein-1. Oncogene. 2009;28:2406–2418. doi: 10.1038/onc.2009.81. [DOI] [PubMed] [Google Scholar]
- 41.Moritz LE, Trievel RC. Structure, mechanism, and regulation of polycomb-repressive complex 2. J Biol Chem. 2018;293:13805–13814. doi: 10.1074/jbc.R117.800367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kim KH, Roberts CW. Targeting ezh2 in cancer. Nat Med. 2016;22:128–134. doi: 10.1038/nm.4036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Yang Q, Xu E, Dai J, Liu B, Han Z, Wu J, et al. A novel long noncoding rna ak001796 acts as an oncogene and is involved in cell growth inhibition by resveratrol in lung cancer. Toxicol Appl Pharmacol. 2015;285:79–88. doi: 10.1016/j.taap.2015.04.003. [DOI] [PubMed] [Google Scholar]
- 44.Qian H, Chen L, Huang J, Wang X, Ma S, Cui F, et al. The lncrna mir4435-2hg promotes lung cancer progression by activating β-catenin signalling. J Mol Med. 2018;96:753–764. doi: 10.1007/s00109-018-1654-5. [DOI] [PubMed] [Google Scholar]
- 45.Li X, Ren Y, Zuo T. Long noncoding rna linc00978 promotes cell proliferation and invasion in non-small cell lung cancer by inhibiting mir-6754-5p. Mol Med Rep. 2018;18:4725–4732. doi: 10.3892/mmr.2018.9463. [DOI] [PubMed] [Google Scholar]
- 46.Wu D, Qin B, Qi X, Hong L, Zhong H, Huang J. Lncrna awpph accelerates the progression of non-small cell lung cancer by sponging mirna-204 to upregulate cdk6. Eur Rev Med Pharmacol Sci. 2020;24:4281–4287. doi: 10.26355/eurrev_202004_21008. [DOI] [PubMed] [Google Scholar]
- 47.Deng L-l, Chi Y-y, Liu L, Huang N-s, Wang L, Wu J. Linc00978 predicts poor prognosis in breast cancer patients. Sci Rep. 2016;6:1–7. doi: 10.1038/s41598-016-0001-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kea J, Wang Q, Zhang W, Mei H. Lncrna mir4435–2hg promotes proliferation, migration, invasion and emt via targeting mir-22–3p/tmem9b in breast cancer. Res Square. 2021 doi: 10.21203/rs.3.rs-860533/v1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Liu AN, Qu HJ, Gong WJ, Xiang JY, Yang MM, Zhang W. Lncrna awpph and mirna-21 regulates cancer cell proliferation and chemosensitivity in triple-negative breast cancer by interacting with each other. J Cell Biochem. 2019;120:14860–14866. doi: 10.1002/jcb.28747. [DOI] [PubMed] [Google Scholar]
- 50.Gong J, Xu X, Zhang X, Zhou Y. Lncrna mir4435-2hg is a potential early diagnostic marker for ovarian carcinoma. Acta Biochim Biophys Sin. 2019;51:953–959. doi: 10.1093/abbs/gmz085. [DOI] [PubMed] [Google Scholar]
- 51.Zhu L, Wang A, Gao M, Duan X, Li Z. Lncrna mir4435-2hg triggers ovarian cancer progression by regulating mir-128-3p/ckd14 axis. Cancer Cell Int. 2020;20:1–16. doi: 10.1186/s12935-019-1086-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Xing P, Wang Y, Zhang L, Ma C, Lu J. Knockdown of lncrna mir4435-2hg and st8sia1 expression inhibits the proliferation, invasion and migration of prostate cancer cells in vitro and in vivo by blocking the activation of the fak/akt/β-catenin signaling pathway. Int J Mol Med. 2021;47:1–13. doi: 10.3892/ijmm.2021.4926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Zhang H, Meng H, Huang X, Tong W, Liang X, Li J, et al. Lncrna mir4435-2hg promotes cancer cell migration and invasion in prostate carcinoma by upregulating tgf-β1. Oncol Lett. 2019;18:4016–4021. doi: 10.3892/ol.2019.10757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Nguyen K, Yan Y, Yuan B, Dasgupta A, Sun J, Mu H, et al. St8sia1 regulates tumor growth and metastasis in tnbc by activating the fak–akt–mtor signaling pathway. Mol Cancer Ther. 2018;17:2689–2701. doi: 10.1158/1535-7163.MCT-18-0399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Shan Y, Liu Y, Zhao L, Liu B, Li Y, Jia L. Microrna-33a and let-7e inhibit human colorectal cancer progression by targeting st8sia1. Int J Biochem Cell Biol. 2017;90:48–58. doi: 10.1016/j.biocel.2017.07.016. [DOI] [PubMed] [Google Scholar]
- 56.Yu S, Wang S, Sun X, Wu Y, Zhao J, Liu J, et al. St8sia1 inhibits the proliferation, migration and invasion of bladder cancer cells by blocking the jak/stat signaling pathway. Oncol Lett. 2021;22:1–11. doi: 10.3892/ol.2021.12997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Drabsch Y, Ten Dijke P. Tgf-β signalling and its role in cancer progression and metastasis. Cancer Metastasis Rev. 2012;31:553–568. doi: 10.1007/s10555-012-9375-7. [DOI] [PubMed] [Google Scholar]
- 58.Xu H, Zhang B, Yang Y, Li Z, Zhao P, Wu W, et al. Lncrna mir4435-2hg potentiates the proliferation and invasion of glioblastoma cells via modulating mir-1224-5p/tgfbr2 axis. J Cell Mol Med. 2020;24:6362–6372. doi: 10.1111/jcmm.15280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Dai B, Xiao Z, Mao B, Zhu G, Huang H, Guan F, et al. Lncrna awpph promotes the migration and invasion of glioma cells by activating the tgf-β pathway. Oncol Lett. 2019;18:5923–5929. doi: 10.3892/ol.2019.10918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Elbediwy A, Thompson BJ. Evolution of mechanotransduction via yap/taz in animal epithelia. Curr Opin Cell Biol. 2018;51:117–123. doi: 10.1016/j.ceb.2018.02.003. [DOI] [PubMed] [Google Scholar]
- 61.Shen W, Zhang J, Pan Y, Jin Y. Lncrna mir4435-2hg functions as a cerna against mir-125a-5p and promotes neuroglioma development by upregulating taz. J Clin Lab Anal. 2021;35:e24066. doi: 10.1002/jcla.24066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Wei L, Surma M, Shi S, Lambert-Cheatham N, Shi J. Novel insights into the roles of rho kinase in cancer. Arch Immunol Ther Exp. 2016;64:259–278. doi: 10.1007/s00005-015-0382-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Mali RS, Kapur R. Targeting rho associated kinases in leukemia and myeloproliferative neoplasms. Oncotarget. 2012;3:909. doi: 10.18632/oncotarget.664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Li X, Song F, Sun H. Long non-coding rna awpph interacts with rock2 and regulates the proliferation and apoptosis of cancer cells in pediatric t-cell acute lymphoblastic leukemia. Oncol Lett. 2020;20:1. doi: 10.3892/ol.2020.12102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Cai Z, Aguilera F, Ramdas B, Daulatabad SV, Srivastava R, Kotzin JJ, et al. Targeting bim via a lncrna morrbid regulates the survival of preleukemic and leukemic cells. Cell Rep. 2020;31:107816. doi: 10.1016/j.celrep.2020.107816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Wang R, Liu L, Jiao J, Gao D. Knockdown of mir4435-2hg suppresses the proliferation, migration and invasion of cervical cancer cells via regulating the mir-128-3p/msi2 axis in vitro. Cancer Manage Res. 2020;12:8745. doi: 10.2147/CMAR.S265545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Wu K, Hu L, Lv X, Chen J, Yan Z, Jiang J, et al. Long non-coding rna mir4435-1hg promotes cancer growth in clear cell renal cell carcinoma. Cancer Biomark. 2020;29:39–50. doi: 10.3233/CBM-201451. [DOI] [PubMed] [Google Scholar]
- 68.Zhu K, Miao C, Tian Y, Qin Z, Xue J, Xia J, et al. Lncrna mir4435-2hg promoted clear cell renal cell carcinoma malignant progression via mir-513a-5p/klf6 axis. J Cell Mol Med. 2020;24:10013–10026. doi: 10.1111/jcmm.15609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Liu B, Pan C-F, Yao G-L, Wei K, Xia Y, Chen Y-J. The long non-coding rna ak001796 contributes to tumor growth via regulating expression of p53 in esophageal squamous cell carcinoma. Cancer Cell Int. 2018;18:1–8. doi: 10.1186/s12935-017-0498-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Wang S, Chen X, Qiao T. Long non-coding rna mir4435-2hg promotes the progression of head and neck squamous cell carcinoma by regulating the mir-383-5p/rbm3 axis. Oncol Rep. 2021;45:1–10. doi: 10.3892/or.2021.7952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Wang W, Xu Z, Wang J, Chen R. Linc00978 promotes bladder cancer cell proliferation, migration and invasion by sponging mir-4288. Mol Med Rep. 2019;20:1866–1872. doi: 10.3892/mmr.2019.10395. [DOI] [PubMed] [Google Scholar]
- 72.Ma D, Sun D, Wang J, Jin D, Li Y, Han Y. Long non-coding rna mir4435-2hg recruits mir-802 from flot2 to promote melanoma progression. Eur Rev Med Pharmacol Sci. 2020;24:2616–2624. doi: 10.26355/eurrev_202003_20530. [DOI] [PubMed] [Google Scholar]
- 73.Guo D, Liu F, Zhang L, Bian N, Liu L, Kong L, et al. Long non-coding rna awpph enhances malignant phenotypes in nasopharyngeal carcinoma via silencing pten through interacting with lsd1 and ezh2. Biochem Cell Biol. 2021;99:195–202. doi: 10.1139/bcb-2019-0497. [DOI] [PubMed] [Google Scholar]
- 74.Zhao L, Lin L, Pan C, Shi M, Liao Y, Bin J, et al. Flotillin-2 promotes nasopharyngeal carcinoma metastasis and is necessary for the epithelial-mesenchymal transition induced by transforming growth factor-β. Oncotarget. 2015;6:9781. doi: 10.18632/oncotarget.3382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Huang S, Zheng S, Huang S, Cheng H, Lin Y, Wen Y, et al. Flot2 targeted by mir-449 acts as a prognostic biomarker in glioma. Artif Cells Nanomed Biotechnol. 2019;47:250–255. doi: 10.1080/21691401.2018.1549062. [DOI] [PubMed] [Google Scholar]
- 76.Zhang X, Yang M, Lin L, Chen P, Ma K, Zhou C, et al. Runx2 overexpression enhances osteoblastic differentiation and mineralization in adipose-derived stem cells in vitro and in vivo. Calcif Tissue Int. 2006;79:169–178. doi: 10.1007/s00223-006-0083-6. [DOI] [PubMed] [Google Scholar]
- 77.Matsubara T, Kida K, Yamaguchi A, Hata K, Ichida F, Meguro H, et al. Bmp2 regulates osterix through msx2 and runx2 during osteoblast differentiation∗. J Biol Chem. 2008;283:29119–29125. doi: 10.1074/jbc.M801774200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Pengde K, Fuxing P, Bin S, Jing Y, Jingqiu C. Lovastatin inhibits adipogenesis and prevents osteonecrosis in steroid-treated rabbits. Joint Bone Spine. 2008;75:696–701. doi: 10.1016/j.jbspin.2007.12.008. [DOI] [PubMed] [Google Scholar]
- 79.Hartana CA, Rassadkina Y, Gao C, Martin-Gayo E, Walker BD, Lichterfeld M, et al. Long noncoding rna mir4435-2hg enhances metabolic function of myeloid dendritic cells from hiv-1 elite controllers. J Clin Investig. 2021;131:e146136. doi: 10.1172/JCI146136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Zheng Y, Song D, Xiao K, Yang C, Ding Y, Deng W, et al. Lncrna gas5 contributes to lymphatic metastasis in colorectal cancer. Oncotarget. 2016;7:83727. doi: 10.18632/oncotarget.13384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Xie X, Tang B, Xiao Y-F, Xie R, Li B-S, Dong H, et al. Long non-coding rnas in colorectal cancer. Oncotarget. 2016;7:5226. doi: 10.18632/oncotarget.6446. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Not applicable.