Abstract
In Italy, serogroup C meningococci of the clonal complex cc11 (MenC/cc11) have caused several outbreaks of invasive meningococcal disease (IMD) during the past 20 years. Between December 2019 and January 2020, an outbreak of six cases of IMD infected with MenC/cc11 was identified in a limited area in the northern part of Italy. All cases presented a severe clinical picture, and two of them were fatal. This report is focused on the microbiological and molecular analysis of meningococcal isolates with the aim to reconstruct the chain of transmission. It further presents the vaccination strategy adopted to control the outbreak. The phylogenetic evaluation demonstrated the close genetic proximity between the strain involved in this outbreak and a strain responsible for a larger epidemic that had occurred in 2015 and 2016 in the Tuscany Region. The rapid identification and characterisation of IMD cases and an extensive vaccination campaign contributed to the successful control of this outbreak caused by a hyperinvasive meningococcal strain.
Keywords: Neisseria meningitidis, clonal complex 11, invasive meningocococcal disease, Italy, outbreak, serogroup C meningococcal vaccination
Background
The main pillars to manage an outbreak of meningococcal disease are rapid detection and whole genome sequencing to identify gene signature and possibly reconstruct the chain of transmission among cases, as well as public health interventions such as a targeted vaccination campaign.
Most cases of invasive meningococcal disease (IMD) are caused by serogroup B, C, W and Y meningococci [1]. However, serogroup C meningococci (MenC) are more commonly associated with outbreaks or epidemics [2]. Epidemiological analyses and molecular characterisations of meningococci have identified a few clonal complexes (cc) considered to be hyperinvasive. Among the main hyperinvasive cc, the cc11 [3] is frequently associated with serogroup C and W and, more rarely, with serogroups B and Y. Cases of IMD caused by MenC/cc11 are often associated with sepsis and with a high case fatality rate (CFR) and sequelae [4]. Several outbreaks and epidemics due to MenC/cc11 have been described worldwide since the mid-1980s [3,5]; since 2013, outbreaks have been reported also in European countries such as Belgium, France, Germany, Italy and the United Kingdom (UK) [6-9].
Italy is a low incidence country for IMD, with an average incidence around 0.31 cases per 100,000 inhabitants during the 3 years 2017 to 2019 [10]. Nevertheless, several outbreaks have been caused by cc11 strains during the past 15 years [11-13]. Notably, an outbreak of 62 cases in the Tuscany Region occurred in 2015 and 2016, with a CFR of ca 21% [9].
Sequencing of the entire genome of N. meningitidis is considered the most cost-effective typing method in routine meningococcal surveillance and in particular in outbreak investigations. Moreover, phylogenetic analysis permits to highlight similarity among the strains and the presence of specific clusters [2]. A targeted vaccination strategy is key to control an outbreak.
Outbreak detection
Between 2 December 2019 and 29 January 2020, an outbreak of six IMD cases caused by MenC occurred in a limited area of the Bergamo province, Lombardy region, northern Italy (Figure 1). The temporal sequence of cases is described in the Table.
Figure 1.
Geolocalisation of invasive meningococcal disease cases involved in the outbreak, Bergamo province, Italy, December 2019–January 2020 (n = 6)
ID: case identification number.
Panel A shows a map of Italy with the Lombardy Region highlighted.
Panel B shows a map of the Lombardy Region with the outbreak area highlighted.
Panel C shows the six IMD cases (ID1–6) by site of isolation and the percentage of population coverage for MenC or MenACWY vaccines in the 20 villages of the outbreak area following the extraordinary vaccination campaign conducted after the fourth case.
Blue stars: places of residence and/or work of the first three cases.
Table. Epidemiological and clinical characteristics of MenC invasive meningococcal disease cases involved in the outbreak, Bergamo province, December 2019–January 2020 (n = 6).
| ID | Day after the outbreak onset | Age group (years) | Clinical picture | Outcome |
|---|---|---|---|---|
| 1 | First day | 15–24 | Sepsis | Fatal |
| 2 | 1 day after | 15–24 | Sepsis | Recovered |
| 3 | 19 days after | 25–49 | Sepsis/meningitis | Recovered |
| 4 | 31 days after | 25–49 | Sepsis | Fatal |
| 5 | 39 days after | 50–64 | Sepsis/meningitis | Recovered |
| 6 | 58 days after | ≥ 65 | Sepsis | Recovered |
ID: case identification number.
Here we report the epidemiological and molecular analysis of the outbreak and its successful containment strategy.
Methods
Invasive meningococcal disease surveillance system
The IMD National Surveillance System (NSS) is coordinated by the national reference laboratory (NRL) of the Istituto Superiore di Sanità (ISS) with the support of the Italian Ministry of Health. The NRL receives bacterial isolates and/or clinical samples (blood and cerebrospinal fluid) from IMD cases, collected by the hospital laboratories, to perform serogroup identification/confirmation and molecular investigations. Meningococci isolated from the outbreak here described (five bacterial isolates and one clinical sample) were collected from hospital laboratories and sent to the NRL.
We used the outbreak definition by the ECDC: “The occurrence of more cases than expected in a particular population, in a specific geographical area and over a specified period of time” [14] and cases for this outbreak were defined as persons with confirmed IMD caused by MenC in the Bergamo province from December 2019 onwards.
Microbiological and molecular analyses
For each meningococcal isolate, the serogroup was determined by slide agglutination with commercial antisera (Thermo Scientific, Waltham, United States (US)) or, for the clinical sample, by multiplex PCR [15]. For the bacterial isolates, susceptibility to cefotaxime, ceftriaxone, ciprofloxacin, meropenem, penicillin G and rifampicin was determined on Mueller–Hinton agar (Thermo Scientific, Waltham, US) supplemented with 5% of sheep blood by the minimum inhibitory concentration (MIC) test strip method (Liofilchem, Roseto degli Abruzzi, Italy). The breakpoints were those recommended by the European Committee on Antimicrobial Susceptibility Testing [16].
Chromosomal DNA was extracted using the QIAamp mini kit (Qiagen, Hilden, Germany) from an overnight culture or directly from the clinical sample (blood). Molecular typing of the bacterial isolates was done through whole genome sequencing. For the clinical sample, multilocus sequence typing (MLST), PorA and FetA typing were performed by Sanger sequencing, referring to the PubMLST.org database (http://pubmlst.org/neisseria) [17]. The MLST defined the sequence type (ST) and the cc. PorA and FetA types together contribute to the finetype. For each sample, we adopted the standard typing nomenclature [18] comprising serogroup: PorA type:FetA type:ST (cc). Moreover, the Neis0430 gene, coding for a cytoplasmic axial filament protein (cafA), was also sequenced to compare the Neis0430 allele in our cases with the one characteristic of the meningococcal strain responsible for the Tuscany outbreak (Neis0430 allele 398) [19].
Whole genome sequencing
Cultivated isolates were analysed by whole genome sequencing. For each isolate, 1 ng of DNA was used to prepare the sequencing libraries following the Nextera XT DNA (Illumina) protocol (Document # 15031942 v05, May 2019). The Illumina MiSeq platform, with the reagent kit v3, 600 cycles, was used for the whole genome sequencing analysis. A first quality check of the raw sequence data was performed using FastQC [20]. Reads were trimmed using the software Sickle [21] to maintain a Q score > 25, and de novo assembly was carried out with the ABySS software version 1.5.2 (k parameter = 63) [22]. Contigs longer than 500 bp were selected using an ad hoc script and kept for further analysis. The final assembly ranged from 84 to 316 (median: 209) contigs per sample (N50: 10,999–59,092 bp; median: 19,790 bp), covering the ca 2.2 Mb of the N. meningitidis genome.
Genome comparison
Genomes, uploaded to the PubMLST.org database (http://pubmlst.org/neisseria), were analysed and compared using the BIGSdb Genome Comparator through gene-by-gene analysis [23]. Through core genome MLST (cgMLST), a phylogenetic analysis was carried out [24]. Incomplete loci were automatically removed from the distance matrix calculation for the NeighbourNet graphs. Based on the distance matrices, a NeighbourNet network was generated by SplitsTree4 (version 4.13.1) [25].
Results
Invasive meningococcal disease surveillance data
During the year 2019, 189 IMD cases were reported in Italy, with an incidence of 0.31 cases per 100,000 inhabitants. Thirty-eight cases occurred in Lombardy Region (18 serogroup B, nine serogroup C, eight serogroup Y, one capsule null locus (cnl) strain and two non-determined), with an incidence of 0.38 cases per 100,000 inhabitants (ISS database https://w3.iss.it/site/mabi, last accessed: 30 April 2021), in line with what had been observed in the previous years (0.35 in 2018 and 0.32 in 2017). However, 19 IMD cases occurred in Lombardy (10 serogroup B, seven serogroup C and two serogroup Y) in December 2019 and January 2020 vs only four cases during the same period of the previous year (two serogroup B, one serogroup C and one serogroup Y) (https://w3.iss.it/site/mabi/, last accessed: 30 April 2021). Of the seven MenC cases, six (four female and two male cases) occurred in a limited geographical area of the Bergamo province. The clinical picture was characterised by a rapidly evolving sepsis (n = 4) or meningitis and sepsis (n = 2). The median age was 40 years (range: 15–70 years). Two of the six cases were fatal (Table). None of the patients had been vaccinated against MenC.
Microbiological analyses
The following samples were analysed by the NRL: five meningococcal isolates (ID 1, 2, 4, 5, 6) and one clinical sample (blood; ID 3). Infection with serogroup C was confirmed through serological tests and/or molecular analysis.
All five meningococcal isolates were found susceptible to cefotaxime, ceftriaxone, ciprofloxacin, meropenem, penicillin G and rifampicin. All isolates showed MIC values to penicillin G ranging from 0.125 to 0.25 mg/L.
Molecular analyses
The molecular analyses showed that the six meningococci belonged to ST-11/cc11 and shared the identical finetype P1.5–1,10–8:F3–6. The 398 Neis0430 allele was found in all of them.
Genome comparison
For all isolates (except for the ID 3 case), we analysed the genomes for a high-resolution comparison using the gene-by-gene approach of cgMLST. A total of 1,581 of the 1,605 core genome loci were included in the cgMLST analysis (we excluded 24 loci that were missing in all): of those, 1,552 core genome loci were identical in all the isolates, with a mean distance of 14 loci with allelic differences (data not shown).
We carried out an additional cgMLST comparison of the five outbreak genomes and representative genomes of meningococci with identical genotypic designation: 13 genomes from Italian sporadic cases from the period 2019 to 2020, four genomes from an outbreak on a cruise ship sailing along the Italian coast in 2012 [12], 11 genomes from an outbreak in Tuscany in 2015 and 2016 [9,19] and five genomes from an outbreak on Sardinia in 2018 caused by a strain with switched capsule B:P1.5–1,10–8:F3–6:ST-11(cc11) [13]. Among the genomes obtained from sporadic cases, 10 presented the Neis0430 allele 398 and three the allele 6.
A total of 1,597 of the 1,605 core genome loci were included in the cgMLST analysis (we excluded eight loci that were missing in all). A total of 1,039 core genome loci were identical in all the isolates, with a mean distance of 73 loci with allelic differences.
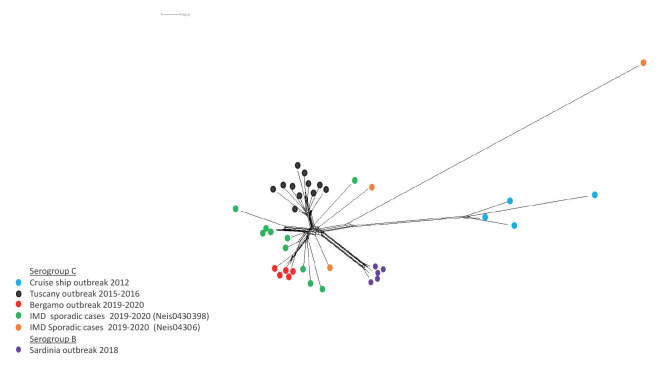
In the NeighbourNet network (Figure 2), reconstructed on the estimated allelic distances, we identified several genomes that were more closely related. In particular, genomes belonging to the same outbreak showed high proximity.
Figure 2.
NeighbourNet phylogenetic network based on a comparison of 1,597 core genome loci (cgMLST) of meningococcal genomes C:P1.5–1,10–8:F3–6:ST-11(cc11) (n = 33) and B:P1.5–1,10–8:F3–6:ST-11(cc11) (n = 5) obtained from invasive meningococcal disease cases, Italy, 2012–2020
IMD: invasive meningococcal disease.
Sequences relate to the following outbreaks: Outbreak in a cruise ship sailing along the Italian coast of Tuscany, 2012 (n = 4) [12]; outbreak in Tuscany, 2015–2016 (n = 11) [9,19]; outbreak in Sardinia, 2018 (n = 5) [13]; outbreak in Bergamo province, 2019–2020 (n = 5); IMD sporadic cases 2019–20 Neis0430 allele 398 (n = 10); IMD sporadic cases 2019–2020 Neis0430 allele 6 (n = 3). The scale bar indicates the number of differences among the compared loci.
The five genomes isolated in the outbreak in Bergamo clustered together. They were in proximity of MenC:cc11 genomes deriving from representative samples of the outbreak in Tuscany and from more recent (2019–2020) sporadic cases, all characterised by the Neis0430 allele 398. Genomes belonging to the outbreak on Sardinia and the outbreak on the cruise ship, as well as three sporadic cases, all presenting the Neis0430 allele 6, were positioned further away from the genomes from Bergamo.
Outbreak control measures
Epidemiological investigations, contact tracing and other public health measures such as chemoprophylaxis of close contacts were conducted by the regional Agency for the Protection of Health (Agenzia di Tutela della Salute), together with the local Agency for Territorial Social-Health (Azienda Socio Sanitaria Territoriale), the Directorate General Welfare, and the Organisational Unit Prevention. Vaccination strategies were designed by local health authorities in collaboration with the ISS and the Italian Ministry of Health.
After the first two cases, chemoprophylaxis with rifampicin (600 mg every 12 h for 2 days) and vaccination with a conjugate vaccine against serogroup C (MenC) or a quadrivalent conjugate vaccine against serogroups A, C, W and Y (MenACWY) were administrated to all close contacts of the six IMD cases. After identification of the third case, because it was determined by the same meningococcal serogroup and the geographical area affected by the outbreak was small, vaccination was extended to the overall population up to 50 years in the towns where the cases resided or worked (Figure 1, blue stars). After the fourth case, the vaccination campaign was extended to a wider geographical area, comprising places visited by the cases, contacts' places of residence and some villages geographically adjacent (Figure 1, green shading) and including the population up to 60 years of age.
Finally, of 67,571 inhabitants representing the entire population of the outbreak area, 28,070 were vaccinated with MenC or MenACWY within 9 weeks, (from 24 December 2019 to the first week of February 2020). After this prompt and extensive reactive vaccination campaign, no further MenC cases were notified in the outbreak area from February 2020 until the end of the year.
Discussion
We identified an outbreak caused by a highly transmissible and hypervirulent MenC/cc11 strain. The same strain was has caused several outbreaks worldwide [2,3,5-8].
Outbreaks involving the C:P1.5–1,10–8:F3–6:ST-11(cc11) strain had already been identified in previous outbreaks in Italy [9,11-13]. In fact, the phylogenetic analysis highlighted the close genetic proximity between the strain responsible for the outbreak in Bergamo and that of a larger outbreak in Tuscany in 2015 and 2016 [9,19]. Both outbreaks were characterised by cases with severe disease and a CFR of more than 20%. The genetic relationship between the strains from Bergamo and Tuscany is also supported by the presence of Neis0430 allele 398. Of note, the C:P1.5–1,10–8:F3–6:ST-11(cc11) isolates with Neis0430 allele 398 responsible for sporadic cases throughout the country in the same period showed a genetic proximity with the genomes from Bergamo in the phylogenetic analysis. These cases were characterised by a high CFR (50%, data not shown).
In this outbreak, all patients except one were older than the recommended vaccination target age groups in Italy. The national immunisation plan for the period 2017 to 2019 recommended the use of MenC vaccine during the second year of life and the MenACWY vaccine from 12 to 18 years of age [26].The immunisation campaign has provided the MenC vaccine for children up to 10 years of age, MenACWY for adolescents, and MenC or MenACWY for adults up to 60 years of age.
Conclusion
Meningococcal outbreaks caused by hypervirulent strains require rapid identification and response with specific public health measures. In parallel with a prompt post-exposure prophylaxis, also a reactive vaccination campaign proved necessary in order to contain the outbreak, preventing new cases and deaths. Rapid microbiological identification and characterisation of the outbreak strain is also important to plan a timely vaccination campaign.
Acknowledgements
The authors thank Carlo Tersalvi (Health Protection Agency of Bergamo, Bergamo, Italy), Giancarlo Malchiodi (Health Protection Agency of Bergamo, Bergamo, Italy), Pietro Imbrogno (Health Protection Agency of Bergamo, Bergamo, Italy), Lucia Antonioli (Health Protection Agency of Bergamo, Bergamo, Italy), Carla Masia (MS Medical Laboratory of Clinical Chemistry and Microbiology, ASST Melegnano and Martesana, Milano, Italy), Chiara Vignati (MS Medical Laboratory of Clinical Chemistry and Microbiology, ASST Melegnano and Martesana, Milano, Italy), Claudio Farina (UOC Microbiology, Papa Giovanni XXIII Hospital, Bergamo, Italy), Antonella Dodaro (MS SIMT Melzo e Cernusco, ASST Melegnano and Martesana, Milano, Italy) and Stefania Bellino, Stefano Boros, Roberta Urciuoli and Flavia Riccardo (Department of Infectious Diseases, Istituto Superiore di Sanità, Rome, Italy). This publication made use of the Neisseria Multi Locus Sequence Typing website (http://pubmlst.org/neisseria/) developed by Keith Jolley and sited at the University of Oxford. The development of this site has been funded by the Wellcome Trust and European Union.
Ethical statement
No ethical approval was required for this study, as it was part of the outbreak control measures.
Conflict of interest: None declared.
Authors’ contributions: Paola Stefanelli conceived the study and revised the manuscript. Cecilia Fazio provided insight on microbiological investigation and drafted the manuscript. Arianna Neri, Paola Vacca, Luigina Ambrosio, Annapina Palmieri and Anna Carannante carried out the laboratory analyses, contributed in the molecular analyses and provided insight into the interpretation of results. Andrea Ciammaruconi, Silvia Fillo, Antonella Fortunato and Florigio Lista carried out whole genome sequencing. Giovanni Rezza was involved in the public health measures management, in the definition of vaccination strategies and in the revision of the manuscript. Laura Daprai, Marcello Tirani, Milena Arghittu, Danilo Cereda, Maria Gramegna, Maria Rosa Bertoli, Lucia Crottogini, Giorgio Gennati, Eugenia Quinz, Livia Trezzi were involved in the invasive meningococcal diseases surveillance at the local level: they were in charge of the data collection and management of invasive meningococcal disease cases. Paola Stefanelli revised the results. All authors participated in the drafting and revision of this manuscript and gave their final approval of this version.
References
- 1. Peterson ME, Li Y, Bita A, Moureau A, Nair H, Kyaw MH, et al. Meningococcal serogroups and surveillance: a systematic review and survey. J Glob Health. 2019;9(1):010409. 10.7189/jogh.09.010409 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Lucidarme J, Hill DM, Bratcher HB, Gray SJ, du Plessis M, Tsang RS, et al. Genomic resolution of an aggressive, widespread, diverse and expanding meningococcal serogroup B, C and W lineage. J Infect. 2015;71(5):544-52. 10.1016/j.jinf.2015.07.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Waśko I, Hryniewicz W, Skoczyńska A. Significance of meningococcal hyperinvasive clonal complexes and their influence on vaccines development. Pol J Microbiol. 2015;64(4):313-21. 10.5604/17331331.1185912 [DOI] [PubMed] [Google Scholar]
- 4. Rosenstein NE, Perkins BA, Stephens DS, Popovic T, Hughes JM. Meningococcal disease. N Engl J Med. 2001;344(18):1378-88. 10.1056/NEJM200105033441807 [DOI] [PubMed] [Google Scholar]
- 5. Ezeoke I, Galac MR, Lin Y, Liem AT, Roth PA, Kilianski A, et al. Tracking a serial killer: Integrating phylogenetic relationships, epidemiology, and geography for two invasive meningococcal disease outbreaks. PLoS One. 2018;13(11):e0202615. 10.1371/journal.pone.0202615 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Ladhani SN, Lucidarme J, Parikh SR, Campbell H, Borrow R, Ramsay ME. Meningococcal disease and sexual transmission: urogenital and anorectal infections and invasive disease due to Neisseria meningitidis. Lancet. 2020;395(10240):1865-77. 10.1016/S0140-6736(20)30913-2 [DOI] [PubMed] [Google Scholar]
- 7. Taha MK, Claus H, Lappann M, Veyrier FJ, Otto A, Becher D, et al. Evolutionary events associated with an outbreak of meningococcal disease in men who have sex with men. PLoS One. 2016;11(5):e0154047. 10.1371/journal.pone.0154047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. De Schrijver K, Maes I. An outbreak of serogroup C meningococcal disease in the province of Antwerp (Belgium) in 2001-2002. Eur J Epidemiol. 2003;18(11):1073-7. 10.1023/A:1026100321871 [DOI] [PubMed] [Google Scholar]
- 9. Miglietta A, Fazio C, Neri A, Pezzotti P, Innocenti F, Azzari C, et al. Interconnected clusters of invasive meningococcal disease due to Neisseria meningitidis serogroup C ST-11 (cc11), involving bisexuals and men who have sex with men, with discos and gay-venues hotspots of transmission, Tuscany, Italy, 2015 to 2016. Euro Surveill. 2018;23(34):1700636. 10.2807/1560-7917.ES.2018.23.34.1700636 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Italian National Health Institute (Istituto Superiore di Sanità, ISS). Sorveglianza delle malattie batteriche invasive in Italia. Rapporto consolidato MIB 2019 . [Surveillance data on invasive bacterial diseases, consolidated report 2019]. Rome: ISS; 2020. Italian. Abvailable from: https://www.iss.it/-/rapporto-consolidato-mib-2019
- 11. Fazio C, Neri A, Tonino S, Carannante A, Caporali MG, Salmaso S, et al. Characterisation of Neisseria meningitidis C strains causing two clusters in the north of Italy in 2007 and 2008. Euro Surveill. 2009;14(16):19179. 10.2807/ese.14.16.19179-en [DOI] [PubMed] [Google Scholar]
- 12. Stefanelli P, Fazio C, Neri A, Isola P, Sani S, Marelli P, et al. Cluster of invasive Neisseria meningitidis infections on a cruise ship, Italy, October 2012. Euro Surveill. 2012;17(50):20336. 10.2807/ese.17.50.20336-en [DOI] [PubMed] [Google Scholar]
- 13. Stefanelli P, Fazio C, Vacca P, Palmieri A, Ambrosio L, Neri A, et al. An outbreak of severe invasive meningococcal disease due to a capsular switched Neisseria meningitidis hypervirulent strain B:cc11. Clin Microbiol Infect. 2019;25(1):111.e1-4. 10.1016/j.cmi.2018.07.014 [DOI] [PubMed] [Google Scholar]
- 14.European Centre for Disease Prevention and Control (ECDC). Field epidemiology manual wiki. Outbreak investigations. Stockholm: ECDC. [Accessed: 26 May 2022]. Available from: https://wiki.ecdc.europa.eu/fem/Pages/Outbreak%20Investigations.aspx
- 15. Zhu H, Wang Q, Wen L, Xu J, Shao Z, Chen M, et al. Development of a multiplex PCR assay for detection and genogrouping of Neisseria meningitidis. J Clin Microbiol. 2012;50(1):46-51. 10.1128/JCM.00918-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.The European Committee on Antimicrobial Susceptibility Testing (EUCAST). Breakpoint tables for interpretation of MICs and zone diameters. Version 12.0, 2022. Växjö: EUCAST. Available from: http://www.eucast.org/clinical_breakpoints
- 17. Jolley KA, Bray JE, Maiden MCJ. Open-access bacterial population genomics: BIGSdb software, the PubMLST.org website and their applications. Wellcome Open Res. 2018;3:124. 10.12688/wellcomeopenres.14826.1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Jolley KA, Brehony C, Maiden MCJ. Molecular typing of meningococci: recommendations for target choice and nomenclature. FEMS Microbiol Rev. 2007;31(1):89-96. 10.1111/j.1574-6976.2006.00057.x [DOI] [PubMed] [Google Scholar]
- 19. Stefanelli P, Fazio C, Neri A, Ciammaruconi A, Balocchini E, Anselmo A, et al. Genome-based study of a spatio-temporal cluster of invasive meningococcal disease due to Neisseria meningitidis serogroup C, clonal complex 11. J Infect. 2016;73(2):136-44. 10.1016/j.jinf.2016.05.003 [DOI] [PubMed] [Google Scholar]
- 20.Andrews S. FastQC. A quality control tool for high throughput sequence data. Cambridge: Babraham Institute; 2010. [Google Scholar]
- 21.Joshi NA, Fass JN. Sickle: A sliding-window, adaptive, quality-based trimming tool for FastQ files using quality (Version 1.33). 2011. Available from https://github.com/najoshi/sickle
- 22. Simpson JT, Wong K, Jackman SD, Schein JE, Jones SJ, Birol I. ABySS: a parallel assembler for short read sequence data. Genome Res. 2009;19(6):1117-23. 10.1101/gr.089532.108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Jolley KA, Maiden MC. BIGSdb: Scalable analysis of bacterial genome variation at the population level. BMC Bioinformatics. 2010;11(1):595. 10.1186/1471-2105-11-595 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Bratcher HB, Corton C, Jolley KA, Parkhill J, Maiden MC. A gene-by-gene population genomics platform: de novo assembly, annotation and genealogical analysis of 108 representative Neisseria meningitidis genomes. BMC Genomics. 2014;15(1):1138. 10.1186/1471-2164-15-1138 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Huson DH, Bryant D. Application of phylogenetic networks in evolutionary studies. Mol Biol Evol. 2006;23(2):254-67. 10.1093/molbev/msj030 [DOI] [PubMed] [Google Scholar]
- 26.Ministero della Salute. Piano Nazionale Prevenzione Vaccinale PNPV 2017-2019. [National plan for vaccine prevention NPVP 2017-2019]. Rome: Ministero della Salute; 2017. Italian. Available from: http://www.salute.gov.it/imgs/C_17_pubblicazioni_2571_allegato.pdf