Abstract
Pin1 is a human peptidyl-prolyl cis-trans isomerase important for the regulation of phosphoproteins that are implicated in many diseases including cancer and Alzheimer’s. Further biophysical study of Pin1 will elucidate the importance of the two-domain system to regulate its own activity. Here, we report near-complete backbone and side-chain 1H, 13C and 15N NMR chemical shift assignments of full-length, apo Pin1 for the purpose of studying interdomain allostery and dynamics.
Keywords: Pin1, Prolyl isomerase, NMR, Chemical shift assignments, Allostery
Biological context
Phosphorylation and proline isomerization are critical signaling mechanisms that can change the structure and function of a protein. Enzymes that catalyze these reactions are thus necessary to maintain cellular homeostasis. Human Pin1 (protein interacting with NIMA) is a peptidyl-prolyl cis-trans isomerase that isomerizes the phosphorylated S/T-P motif, which results in a conformational change in the target protein (Lu et al. 1996). A quarter of all phosphorylated sites are on pSer-Pro or pThr-Pro motifs (Ubersax et al. 2007), and Pin1 has been shown to structurally regulate a large variety of these proteins, especially those involved in mitosis. Downstream effects of substrate isomerization by Pin1 include conformational changes, change in the phosphorylation state, and stability/turnover of the substrate (Shen et al. 1998; Liou et al. 2011). While misregulation and overexpression of Pin1 is common in various cancers (Bao et al. 2004; Rustighi et al. 2014), Pin1 protects against tauopathy and plaque formation that leads to Alzheimer’s disease neurodegeneration (Butterfield et al. 2006; Blair et al. 2015).
Pin1 is a small two-domain protein with a N-terminal Trp-Trp (WW) domain (residues 1–39), attached via a flexible linker to the peptidyl-prolyl isomerase (PPIase) domain (residues 50–163). The WW domain binds the ligand with high affinity, while the PPIase isomerizes the same pS/T-P motif (Ranganathan et al. 1997). Substantial evidence shows that the two domains tumble rather independently of one another, with occasional sampling of a compact conformation through an interdomain interface (Namanja et al. 2007; Matena et al. 2013). Interestingly, previous work suggests that the WW domain regulates catalysis in the PPIase through an interdomain allosteric mechanism (Wilson et al. 2013; Peng 2015). Besides the apparent implication of Pin1 in health and disease, Pin1 is also studied as a model to understand protein dynamics and allostery in the context of more than one domain within a protein, a feature found in more than two thirds of all mammalian proteins (Ekman et al. 2005).
The structure of apo Pin1 has been solved using both X-ray crystallography as well as NMR (Ranganathan et al. 1997; Bayer et al. 2003), yet there is much to still be discovered about the allosteric network and interdomain dynamics of Pin1. While some backbone and side-chain chemical shift assignments of the isolated apo domains along with the backbone assignment of full length Pin1 have been previously deposited (Kowalski et al. 2002; Jacobs et al. 2002; Xu et al. 2014), full-length Pin1 side-chain chemical shifts have yet to be deposited in BMRB. In addition, only proton chemical shifts have been deposited for the side chains of apo WW domain. Here, we report near-complete 1H, 13C and 15N backbone and side-chain chemical shift assignments and changes in chemical shifts from the isolated domains due to this interdomain communication.
Methods and experiments
Protein expression and purification
Full length, wild-type Pin1 was cloned into expression vector pET-28a(+) with a N-terminal 6xHistidine tag and kanamycin resistance. The plasmid was transformed and expressed in Escherichia coli strain BL21(DE3) as previously reported (Bayer et al. 2003; Born et al. 2018). Overnight culture plates were resuspended in M9 minimal media with 50 μg/mL kanamycin, and 1 g/L 15N-ammonium chloride and 2 g/L 13C-glucose for uniform labeling. Cultures were grown shaking at 37 °C until induction with 1 mM isopropyl-1-thio-D-galactopyranoside at optical density A600 = 0.8. After induction, cells were grown shaking at 25 °C overnight. To harvest, cells were centrifuged at 4 °C for 20 min at 4000 × g.
Cells were resuspended in 50 mM potassium phosphate buffer, 1 mM dithiothreitol, 0.3 mM phenylmethylsulfonyl fluoride (protease inhibitor), and 25 mM imidazole at pH 7.5 and lysed by sonication (4 × 30 seconds) at 4 °C. The cell lysate was centrifuged at 52,000 × g for 45 min at 4 °C, filtered through a 0.22 μm PVDF filter, and loaded on a pre-equilibrated nickel nitrilotriacetic acid column (GE Healthcare) on an Akta Pure. His-tagged Pin1 was eluted with 50 mM potassium phosphate buffer, 1 mM dithiothreitol, and 250 mM imidazole at pH 7.5. Protein was concentrated using a 3000 NMWL cutoff (Millipore), and then purified using Superdex 75 10/300 GL (GE Healthcare) size exclusion column using 50 mM sodium phosphate and 150 mM sodium chloride buffer at pH 6.5. The protein was concentrated to about 300 μl, and concentration was measured at 280 nm wavelength using a NanoDrop 2000 UV/Vis-spectrometer (Thermo Scientific) and calculated based on the predicted extinction coefficient of 20970 M−1 cm−1.
NMR spectroscopy
The 15N,13C-labeled apo sample contained 2 mM Pin1 in 20 mM sodium phosphate, 50 mM sodium chloride, 5 mM dithiothreitol, 0.03% sodium azide, and 3% D2O at pH 6.5 through buffer exchange. NMR experiments were performed at 298 K on a triple-resonance Varian 900 MHz spectrometer equipped with a cryoprobe. The sequence specific backbone assignment was determined using 15N-HSQC, HNCACB, HNCA, CBCA(CO)NH, and CCONH spectra. For side-chain assignments, 13C-resolved aliphatic and aromatic CT-HSQC, HBHA(CO)NH, and HCCH-TOCSY spectra were used. All data was processed with NMRPipe (Delaglio et al. 1995) and analyzed using CCPNMR (Vranken et al. 2005). We observed no systematic deviation from the previously published chemical shifts of the isolated domains. For comparison of the shifts, we corrected the isolated-domain shifts by the average deviation from the full-length Pin1 shifts. After excluding those peaks with the strongest deviation, we recalibrated the isolated-domain shifts again for the final comparison.
Assignments and data deposition
The purified full length Pin1 protein contains 163 amino acids, plus the residues in the His tag. The backbone and side-chain chemical shifts of apo Pin1 were assigned manually, with aid from previously published shift lists of the isolated WW (residues 1–39) and PPIase domains (residues 50–163), and the backbone assignment of full length Pin1 with BMRB accession codes 5248, 11559, and 5305, respectively. More WW domain chemical shifts were taken from our previous eNOE structure (Strotz 2016). Side-chain methyl chemical shifts were also previously reported in (Namanja et al. 2007). 152 out of 156 non-proline amide residues (97.4%) were assigned in the 15N-HSQC (Fig. 1). Missing amides are the N-terminal Met1, residues Arg17 and Ser19 in the dynamic WW binding loop, and Glu76 in the flexible PPIase catalytic loop, all likely due to conformational broadening or fast amide-solvent exchange. 100% of Cα and Cβ resonances were assigned. Excluding CγO resonances (which were not measured), 88% of Cγ resonances were assigned, with the remaining residues being all from aromatic residues. Not including CδO, 86% of Cδ resonances were assigned. For proton chemical shifts, 99.4% and 98.1% of the Hα and Hβ resonances were assigned. For side-chain protons, the completeness of Hγ, Hδ, Hε, HZ, and HH resonances was 95.1, 100, 79.7, 92.3, and 100%, respectively. Backbone and side-chain chemical shift assignments have been deposited to the Biological Magnetic Resonance Data Bank (BMRB) with the accession number 27579.
Fig. 1.
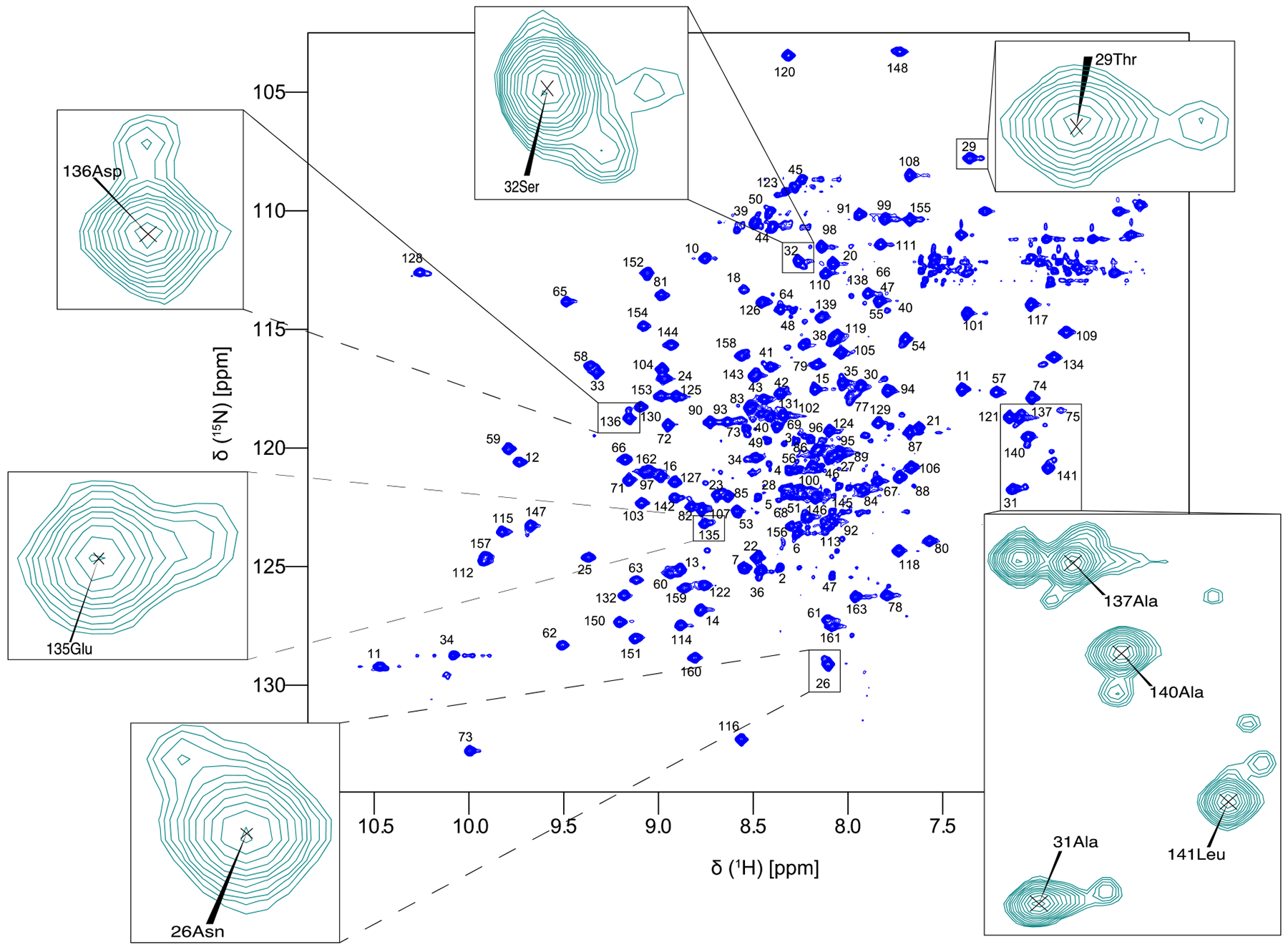
Assigned 15N-HSQC spectrum of apo full-length Pin1 recorded at 900 MHz with peaks in the interdomain interface zoomed to show the minor peaks only present in the full-length protein.
While the structure of Pin1 has been solved using both X-ray crystallography and NMR, minor peaks present in the 15N-HSQC of the full-length protein have not been fully discussed. As can been seen in the zoomed in regions of Fig. 1, many peaks from the interdomain interface have a nearby minor peak. The minor peak in WW domain residues Thr29 and Ala31 have previously been investigated in (Matena et al. 2013), and the minor peak is indicative of the structure populating a compact state, while the larger, main peak is the residue in an extended state. We see the same phenomenon in multiple residues in the interdomain interface of the WW side (between residues 26 and 32) and on the PPIase side (between 135 and 141). In 15N-HSQC of the isolated WW and PPIase domain, these minor peaks were not noticed, possibly due to differences in the signal-to-noise ratio or to differences in resolution (note previous work has been carried out at B0 fields smaller than 900 MHz). These minor peaks are further evidence for the existence of extended and compact states of the two domains of Pin1.
We next compared our full-length, apo Pin1 chemical shifts to those of the isolated WW and PPIase domains (Fig. 2). The WT isolated WW domain chemical shifts were taken from a BMRB accession code 5248 (Kowalski et al. 2002) and from a previous project (Zhang et al. 2018), and the PPIase chemical shifts were from BMRB 11559 (Xu et al. 2014). Due to lack of WW domain heavy atoms previously deposited in the BMRB, 13C shifts were also taken from a mutant, isolated WW domain (S18N and W34F) (Strotz 2016). Similar to tracking chemical shift perturbations, we scaled the C and N heavy atoms by 0.2 so their deviations are comparable to protons. The average deviation for the WW domain and PPIase domains were 0.05 ± 0.07 ppm and 0.02 ± 0.03 ppm, respectively. In contrast to the PPIase domain, the much smaller WW domain is affected almost over the entire sequence. Large chemical shift differences between full-length Pin1 WW and isolated WW have also been previously reported (Jacobs et al. 2003). We are considering deviations that are over one standard deviation over the average chemical shift deviation of the individual domains to be significant. The termini of the domains were ignored, since different construct termini cause additional shifts that would mask the deviation of interest. Interestingly, the largest deviations when the termini from the domains are excluded are in ligand binding regions and the interdomain interface of the PPIase domain. While deviations in the interface have been reported previously (Bayer et al. 2003; Jacobs et al. 2003; Namanja et al. 2011), the variance in the ligand binding sites are novel. In the WW domain, residues 11–24 are responsible for ligand binding in full-length Pin1, and appear to be in a slightly different chemical environment in this region compared to the isolated WW domain. In the PPIase domain of full-length Pin1, we observe significant chemical shift deviation for the catalytic loop (residues ~63–76) as well as in the PPIase active site (residues 54, 110–115,125–134, and 151–159). The deviations in the ligand binding sites of both domains indicates that the other domain does indeed influence the structure, and therefore potentially the dynamics and activity, of the binding sites. One of the most distinct regions of deviation comes from the putative interdomain interface on the PPIase domain, residues 136–143 and 147. This suggests that full-length Pin1 does sample both an extended and compact state even without ligand present, and thus the isolated PPIase experiences a different chemical environment than when the WW domain is present. While the deviations in the interdomain interface of the WW and PPIase domains were a similar value, the WW domain had much larger deviations throughout the whole sequence therefore making the interdomain interface deviations less distinct. Nonetheless, this highlights the need to study full-length Pin1, not just the domains independently in order to understand this complex system. Our chemical shift assignments will be used for multi-state ensemble calculations to understand the allosteric network of Pin1.
Fig. 2.
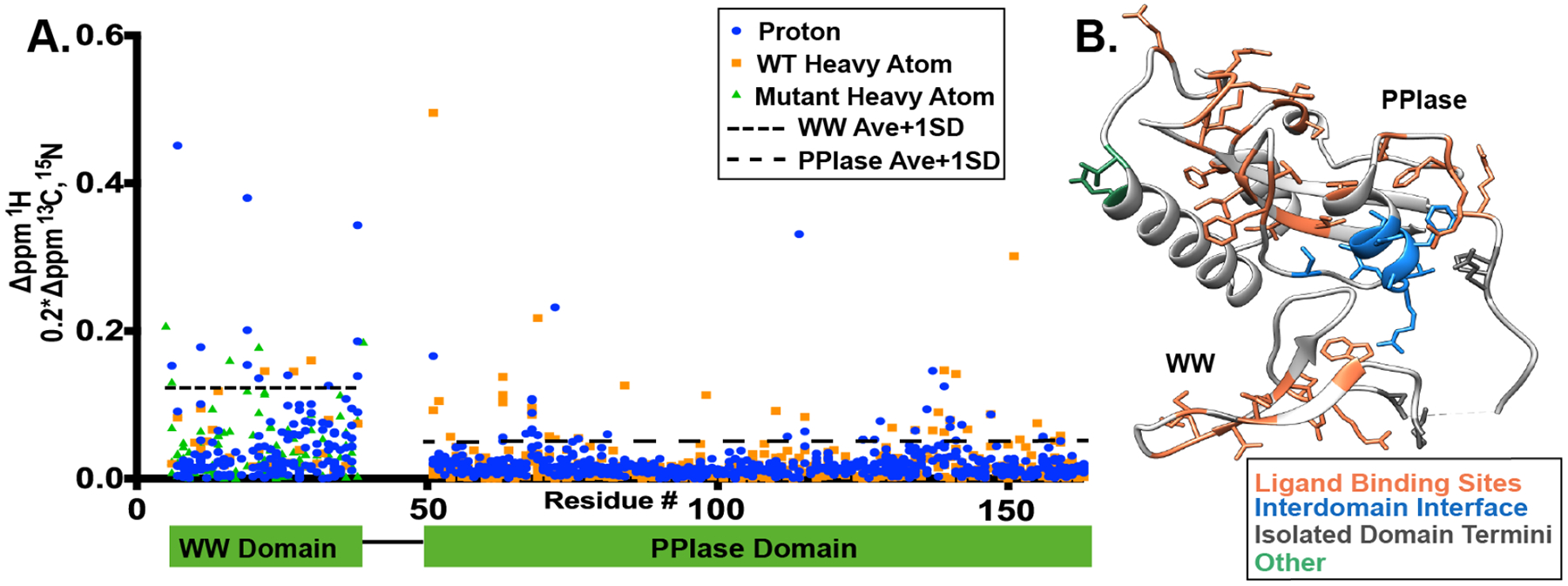
Chemical shift deviations of full-length Pin1 compared to isolated WW and PPIase domains. A) Deviations in units of ppm plotted for protons (blue) and heavy atoms (orange for WT and green for S18N/W34F mutant). One standard deviation above the average is indicated by the broken lines. *Amide of Gly 39 lies outside our scale. B) Deviation values one standard deviation over the average deviation for each domain plotted on the X-ray crystal structure 1pin (Ranganathan et al. 1997).
Acknowledgments
We would like to thank Dr. David Jones at Univ. of Colorado for helping set up initial experiments. This work was supported by a start-up package from the University of Colorado at Denver to B.V.
Footnotes
Conflict of interest. The authors declare no conflict of interest.
References
- Apic G, Gough J, Teichmann SA (2001) Domain combinations in archaeal, eubacterial and eukaryotic proteomes. J Mol Biol 310:311–325. doi: 10.1006/JMBI.2001.4776 [DOI] [PubMed] [Google Scholar]
- Bao L, Kimzey A, Sauter G, et al. (2004) Prevalent Overexpression of Prolyl Isomerase Pin1 in Human Cancers. Am J Pathol 164:1727–1737. doi: 10.1016/S0002-9440(10)63731-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bayer E, Goettsch S, Mueller JW, et al. (2003) Structural analysis of the mitotic regulator hPin1 in solution: Insights into domain architecture and substrate binding. J Biol Chem 278:26183–26193. doi: 10.1074/jbc.M300721200 [DOI] [PubMed] [Google Scholar]
- Blair LJ, Baker JD, Sabbagh JJ, Dickey CA (2015) The emerging role of peptidyl-prolyl isomerase chaperones in tau oligomerization, amyloid processing, and Alzheimer’s disease. J Neurochem 133:1–13. doi: 10.1111/jnc.13033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Born A, Henen M, Nichols P, et al. (2018) Efficient Stereospecific Hβ2/3 NMR Assignment Strategy for Mid-Size Proteins. Magnetochemistry 4:25. doi: 10.3390/magnetochemistry4020025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Butterfield DA, Abdul HM, Opii W, et al. (2006) REVIEW: Pin1 in Alzheimer’s disease. J Neurochem 98:1697–1706. doi: 10.1111/j.1471-4159.2006.03995.x [DOI] [PubMed] [Google Scholar]
- Delaglio F, Grzesiek S, Vuister GW, et al. (1995) NMRPipe: a multidimensional spectral processing system based on UNIX pipes. J Biomol NMR 6:277–93 [DOI] [PubMed] [Google Scholar]
- Ekman D, Björklund ÅK, Frey-Skött J, Elofsson A (2005) Multi-domain Proteins in the Three Kingdoms of Life: Orphan Domains and Other Unassigned Regions. J Mol Biol 348:231–243. doi: 10.1016/J.JMB.2005.02.007 [DOI] [PubMed] [Google Scholar]
- Jacobs DM, Saxena K, Grimme S, et al. (2002) 1H, 13C and 15N backbone resonance assignment of the peptidyl-prolyl cis-trans isomerase Pin1. J Biomol NMR 23:163–4 [DOI] [PubMed] [Google Scholar]
- Jacobs DM, Saxena K, Vogtherr M, et al. (2003) Peptide binding induces large scale changes in inter-domain mobility in human Pin1. J Biol Chem 278:26174–26182. doi: 10.1074/jbc.M300796200 [DOI] [PubMed] [Google Scholar]
- Kowalski JA, Liu K, Kelly JW (2002) NMR solution structure of the isolated Apo Pin1 WW domain: Comparison to the x-ray crystal structures of Pin1. Biopolymers 63:111–121. doi: 10.1002/bip.10020 [DOI] [PubMed] [Google Scholar]
- Liou Y-C, Zhou XZ, Lu KP (2011) Prolyl isomerase Pin1 as a molecular switch to determine the fate of phosphoproteins. Trends Biochem Sci 36:501–14. doi: 10.1016/j.tibs.2011.07.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matena A, Sinnen C, Van Den Boom J, et al. (2013) Transient domain interactions enhance the affinity of the mitotic regulator pin1 toward phosphorylated peptide ligands. Structure 21:1769–1777. doi: 10.1016/j.str.2013.07.016 [DOI] [PubMed] [Google Scholar]
- Namanja AT, Peng T, Zintsmaster JS, et al. (2007) Substrate Recognition Reduces Side-Chain Flexibility for Conserved Hydrophobic Residues in Human Pin1. Structure 15:313–327. doi: 10.1016/j.str.2007.01.014 [DOI] [PubMed] [Google Scholar]
- Namanja AT, Wang XJ, Xu B, et al. (2011) Stereospecific gating of functional motions in Pin1. Proc Natl Acad Sci 108:12289–12294. doi: 10.1073/pnas.1019382108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng JW (2015) Investigating dynamic interdomain allostery in Pin1. Biophys Rev 7:239–249. doi: 10.1007/s12551-015-0171-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ping Lu K, Hanes SD, Hunter T (1996) A human peptidyl–prolyl isomerase essential for regulation of mitosis. Nature 380:544–547. doi: 10.1038/380544a0 [DOI] [PubMed] [Google Scholar]
- Ranganathan R, Lu KP, Hunter T, Noel JP (1997) Structural and Functional Analysis of the Mitotic Rotamase Pin1 suggest substrate recognition is phosphorylation dependent_Noel 1997.pdf. Cell 89:875–886 [DOI] [PubMed] [Google Scholar]
- Rustighi A, Zannini A, Tiberi L, et al. (2014) Prolyl-isomerase Pin1 controls normal and cancer stem cells of the breast. EMBO Mol Med 6:99–119. doi: 10.1002/emmm.201302909 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen M, Stukenberg PT, Kirschner MW, Lu KP (1998) The essential mitotic peptidyl-prolyl isomerase Pin1 binds and regulates mitosis-specific phosphoproteins. Genes Dev 12:706–20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strotz D (2016) eNOE Method Development And Applications To Protein Allostery. PhD Thesis Diss No 23867 ETH Zurich [Google Scholar]
- Ubersax JA, Ferrell JE Jr (2007) Mechanisms of specificity in protein phosphorylation. Nat Rev Mol Cell Biol 8:530–541. doi: 10.1038/nrm2203 [DOI] [PubMed] [Google Scholar]
- Vranken WF, Boucher W, Stevens TJ, et al. (2005) The CCPN data model for NMR spectroscopy: Development of a software pipeline. Proteins Struct Funct Bioinforma 59:687–696. doi: 10.1002/prot.20449 [DOI] [PubMed] [Google Scholar]
- Wilson KA, Bouchard JJ, Peng JW (2013) Interdomain interactions support interdomain communication in human pin1. Biochemistry 52:6968–6981. doi: 10.1021/bi401057x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu N, Tochio N, Wang J, et al. (2014) The C113D mutation in human Pin1 causes allosteric structural changes in the phosphate binding pocket of the ppiase domain through the tug of war in the dual-histidine motif. Biochemistry 53:5568–5578. doi: 10.1021/bi5007817 [DOI] [PubMed] [Google Scholar]
- Zhang M, Case DA, Peng Correspondence JW, Peng JW (2018) Propagated Perturbations from a Peripheral Mutation Show Interactions Supporting WW Domain Thermostability. Struct Des 26:. doi: 10.1016/j.str.2018.07.014 [DOI] [PubMed] [Google Scholar]


