Abstract
A bacterium capable of utilizing fenitrothion (O,O-dimethyl O-4-nitro-m-tolyl phosphorothioate) as a sole carbon source was isolated from fenitrothion-treated soil. This bacterium was characterized taxonomically as being a member of the genus Burkholderia and was designated strain NF100. NF100 first hydrolyzed an organophosphate bond of fenitrothion, forming 3-methyl-4-nitrophenol, which was further metabolized to methylhydroquinone. The ability to degrade fenitrothion was found to be encoded on two plasmids, pNF1 and pNF2.
Organophosphorus insecticides such as fenitrothion (O,O-dimethyl O-p-nitro-m-tolyl phosphorothioate) and parathion (O,O-diethyl O-p-nitrophenyl phosphorothioate) are used all over the world for controlling a wide range of insects. These insecticides are potent inhibitors of cholinesterase and can thus be hazardous as a result of runoff from areas of application. Microbial degradation is considered to be a major factor determining the fate of organophosphorus insecticides in the environment. Studies of microbial degradation are useful in the development of strategies for the detoxification of the insecticides by microorganisms (14). While there have been many reports of isolation and characterization of bacterial species cometabolically hydrolyzing organophosphorus insecticides (3), reports of bacterial species that utilize an insecticide as a sole source of carbon and energy for growth have been limited to date (15–17). On the other hand, it is well known that plasmids can endow bacterial species with the ability to degrade various man-made organic compounds (18). Catabolic plasmids have been thought to play an important role in the evolution of pesticide-degrading ability in microorganisms (3, 18). A plasmid encoding the gene for hydrolysis of parathion to 4-nitrophenol has been found in Pseudomonas diminuta (19) and Flavobacterium sp. (13). However, there have been only a few studies of plasmid-associated organophosphorus insecticide degradation.
In the present study, we isolated and characterized a Burkholderia sp. strain capable of utilizing fenitrothion as a sole source of carbon. In addition, we demonstrated that the degradative capability of the isolate is associated with the two plasmids harbored by this bacterium.
Isolation and identification.
A fenitrothion-degrading bacterium was isolated from soil that had been exposed to fenitrothion for at least 2 years. The fenitrothion-exposed soil was suspended in sterilized distilled water, and its diluted suspensions were sprayed on plates of MMFF agar, which is minimal medium (MM) (8) containing 0.8% fenitrothion emulsion (consisting of 50% fenitrothion) and 2% agar. After a few days of incubation at 30°C, microbial colonies became visible, and a clear halo appeared around a colony capable of degrading fenitrothion. We selected and purified a colony that was able to use fenitrothion as a sole source of carbon, and this was designated strain NF100. Strain NF100 was identified on the basis of morphological, physiological, and biochemical characterizations. The guanine plus cytosine (G+C) content of bacterial DNA was determined as described by Tamaoka and Komagata (23). Quinone type was determined by high-performance liquid chromatography (HPLC). The 16S rRNA gene was amplified by PCR, and the nucleotide sequences of the purified PCR products were determined as described previously (9). Taxonomic properties of NF100 were as follows: cell morphology, a motile straight rod with dimensions of 1.2 to 2.0 μm in length and 0.5 to 0.8 μm in width and having a single polar flagellum; Gram stain, negative; oxidase and catalase production, positive; nitrate reduction, positive; urease production, negative; G+C content, 62.5% ± 2.5%; and major quinone type, ubiquinone Q8. The following additional tests or reactions were positive: acid production from glycerol, adonitol, l-arabinose, fructose, d-glucose, d-galactose, inositol, lactose, mannitol, mannose, melibiose, rhamnose, ribose, sorbitol, trehalose, xylose, and growth on succinate, gluconate, malonate, citrate, and malate. The following additional tests or reactions were negative: acid production from cellobiose, erythritol, inulin, maltose, raffinose, salicin, sorbose, sucrose, and starch; growth on maleate and propionate; production of H2S, indole, and acetoin; and hydrolysis of esculin, gelatin, starch, Tween 80, casein, and cellulose. The isolate grew at 37°C, but growth was negligible at 42°C; it did not require supplementation of vitamins in the growth medium. About 1,492 bases of 16S rRNA of NF100 were determined. A phylogenetic tree was constructed from evolutionary distances by the neighbor-joining method (data not shown). Following phylogetic analysis, strain NF100 was placed in a cluster making up the genus Burkholderia. The highest degree of similarity found, 97%, was obtained with the 16S rRNA genes of Burkholderia cepacia (DDBJ accession no. AF097532), B. vietnaminensis (AF097534), and B. multivorans (AF097531). Based on these observations (24), the isolate was identified as a Burkholderia sp. and designated strain NF100.
Metabolism of fenitrothion.
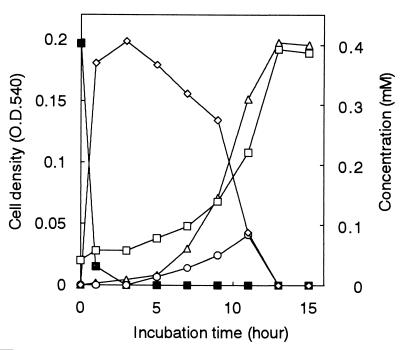
To determine the degradation pathway of fenitrothion, NF100 was inoculated into MM containing 0.4 mM fenitrothion and incubated at 30°C on a reciprocal shaker. Samples of culture medium were periodically withdrawn and analyzed. Growth of NF100 was measured as the increase in absorbance at 540 nm by spectrophotometry. Fenitrothion and its metabolites were identified and measured by HPLC, using a Shimazu model 6A high-performance liquid chromatograph with a diode array detector. The samples were injected into a C18 column (Develosil ODS, 0.6-cm internal diameter, 20 cm long; Nomurakagaku Co., Aichi, Japan) at a flow rate of 1.0 ml/min. Retention times and UV spectra of the sample peaks were compared with those of known standards. Nitrite was determined by the modified Griess-Ilosvay method (11). The concentrations of fenitrothion and its metabolites and culture growth were observed for 15 h after inoculation of the medium (Fig. 1). NF100 completely hydrolyzed the added fenitrothion in the initial 3 h without detectable growth. 3-Methyl-4-nitrophenol levels increased and then disappeared completely at between 1 and 13 h of growth, and methylhydroquinone was detected in the culture during the logarithmic growth phase. Nitrite production continued during the logarithmic phase and ceased upon entry into stationary phase. These observations indicated that the metabolism of fenitrothion in NF100 was initiated by its hydrolysis to 3-methyl-4-nitrophenol, which was metabolized to nitrite and methylhydroquinone, the substrate for oxygenase-catalyzed ring fission.
FIG. 1.
Utilization of fenitrothion as a sole source of carbon for growth by Burkholderia sp. strain NF100. Symbols: □, optical density of the culture at 540 nm (O.D. 540); ■, fenitrothion; ◊, 3-methyl-4-nitrophenol; ○, methylhydroquinone; ▵, nitrite.
Hydrolysis and utilization of organophosphate insecticides by NF100 were examined on the basis of growth in MM containing an organophosphate insecticide as the carbon source. NF100 was inoculated into MM containing a 0.5 mM concentration of an organophosphate insecticide and incubated at 30°C on a reciprocal shaker for 24 h. The concentration of the insecticide and growth of NF100 in the culture were measured as described above. The following insecticides were used: methyl parathion (O,O-dimethyl O-p-nitrophenyl phosphorothioate), parathion, EPN (o-ethyl O-p-nitrophenyl phenylphosphonothioate), diazinone [O,O-diethyl O-2-isopropyl-4-methyl-6-pyrimidinylthiophosphate, and malathion [S-1,2-bis (ethoxycarbonyl)ethyl O,O-dimethyl phosphorodithioate]. NF100 hydrolyzed methyl parathion, parathion, and ENP to 4-nitrophenol, which was further metabolized as a carbon source. The strain also hydrolyzed diazinone but could not utilize its metabolites. Malathion was not hydrolyzed by NF100.
Plasmid curing.
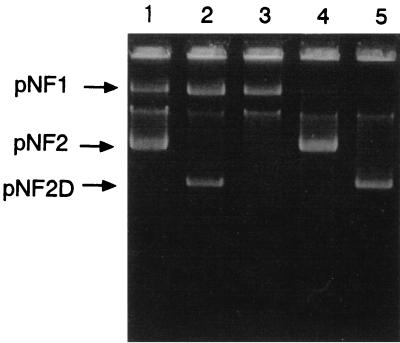
The ability of strain NF100 to utilize fenitrothion was unstable and was lost irreversibly. The ability of bacteria to degrade some pesticides is known to be controlled often by plasmids (18). Thus, NF100 was examined for its plasmid content by the alkaline sodium dodecyl sulfate plasmid extraction method (2) and agarose gel electrophoresis. The molecular sizes of the plasmids were determined by analysis of their digestion patterns after treatment with restriction enzymes BamHI and PstI. The total size of each plasmid was estimated by comparing the electrophoretic mobilities of the fragments with those of lambda DNA digestion products of known sizes. NF100 was found to harbor two large-molecular-size plasmids, designated pNF1 (105 kb) and pNF2 (33 kb) (Fig. 2, lane 1). To confirm that fenitrothion metabolism is controlled by pNF1 and pNF2, we examined the correlation of loss of fenitrothion degradative ability with plasmid removal. A curing experiment with mitomycin C, which is the agent used for curing Pseudomonas and related genera of plasmids, was conducted as described previously (9). When NF100 was treated with mitomycin C, 2% of the treated cells lost their fenitrothion-hydrolyzing activity. The cells unable to hydrolyze fenitrothion either were carrying a smaller plasmid termed pNFD2 or had lost pNF2 (Fig. 2, lanes 2 and 3). These cured strains, NF101(pNF1, pNF2D) and NF102(pNF1), simultaneously lost their ability to hydrolyze methyl parathion (Table 1). Restriction analysis of pNF2 and pNF2D revealed that a portion of approximately 13 kb was deleted from pNF2, with the concomitant loss of fenitrothion hydrolase activity. A phenotypic property correlating with the presence of pNF2 was fenitrothion-hydrolyzing activity (Fed+). The ability to utilize 3-methyl-4-nitrophenol and methylhydroquinone was lost in 0.5% of cells treated with mitomycin C. These cured strains, NF103 and NF104, had lost pNF1 (Fig. 2, lanes 4 and 5), indicating that a phenotypic property correlating with the presence of pNF1 is utilization of methylhydroquinone (Mhq+) (Table 1). Both of these cured strains, NF102(pNF1) and NF103(pNF2), retained their ability to liberate nitrite from 3-methyl-4-nitrophenol and to utilize 4-nitrophenol and hydroquinone as sole sources of carbon (Table 1), indicating that the oxidative pathway for removal of the nitro group and the hydroquinone-degrading pathway are encoded by the chromosome.
FIG. 2.
Agarose gel electrophoresis of plasmids from strain NF100 and its plasmid-cured derivatives. Lanes: 1, NF100(pNF1, pNF2); 2, NF101(pNF1, pNFD2); 3, NF102(pNF1); 4, NF103(pNF2); 5, NF104(pNFD2).
TABLE 1.
Growth responses and plasmid contents of NF100 and its derivatives
| Straina | Plasmid(s) contained | Growthb on carbon sourcec
|
|||||
|---|---|---|---|---|---|---|---|
| FNT | MP | 3M4N | 4N | MHQ | HQ | ||
| NF100 | pNF1, pNF2 | + | + | + | + | + | + |
| NF101 | pNF1, pNFD2 | − | − | + | + | + | + |
| NF102 | pNF1 | − | − | + | + | + | + |
| NF103 | pNF2 | − | + | − | + | − | + |
| NF104 | pNFD2 | − | − | − | + | − | + |
| NF1023 | pNF2 | − | + | − | + | − | + |
| NF1033 | pNF1 | − | − | + | + | + | + |
| NF1023E | pNF1, pNF2 | + | + | + | + | + | + |
| NF1033T | pNF1, pNF2 | + | + | + | + | + | + |
Strain descriptions: NF100, wild type; NF101 to NF104, cured strains; NF1023 and NF1033, spontaneous mutants (Rifr Strr) of NF102 and NF103, respectively; NF1023E, transformant of NF1023 (via electroporation with pNF2); NF1033T, transconjugant of NF1033 (via mating with NF100).
+, growth; −, no growth.
Carbon source abbreviations: FNT, fenitrothion; MP, methyl parathion; 3M4N, 3-methyl-4-nitrophenol; 4N, 4-nitrophenol; MHQ, methylhydroquinone; HQ, hydroquinone.
Mating and electroporation.
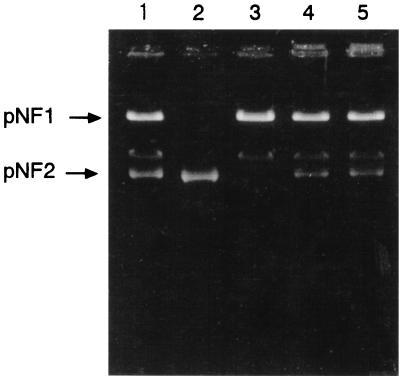
To further establish the roles of pNF1 and pNF2 in fenitrothion metabolism, a mating experiment was performed by the filter mating method as described previously (9). Spontaneous rifampin-resistant (Rifr) and streptomycin-resistant (Strr) mutants of NF102 and NF103 were isolated and designated NF1023 and NF1033, respectively. Conjugal transfer of pNF2 into NF1023(pNF1) (Fed− Mhq+ Strr Rifr) was not successful. Therefore, NF1023 was transformed with pNF2 by electroporation. The pNF2 DNA purified from NF103 was transformed into NF1023 by using a Gene Pulser (Bio-Rad, Richmond, Calif.). Electrocompetent cells prepared by extensive washing with a 10% glycerol solution were mixed with the pNF2, and the mixture was electrically pulsed at 400 Ω, 25 μF, and 2.5 kV in accordance with the manufacturer's instructions. The electroporation enabled NF1023 to hydrolyze fenitrothion. No differences in the fenitrothion-hydrolyzing abilities of wild-type NF100 and NF1023 transformed with pNF2 (NF1023E) were observed (Table 1). A plasmid similar to pNF2 was recovered from the transformant NF1023E (Fig. 3, lane 4). On the other hand, the Mhq+ phenotype was successfully introduced via filter mating into NF1033(pNF2) (Fed+ Mhq− Strr Rifr), using NF100 donors (Table 1). The frequency of plasmid transfer from NF100 to NF1033 ranged from 10−6 to 10−5 per donor cell. All of the NF1033T transconjugants (Table 1) were able to utilize methylhydroquinone and harbored pNF1 (Fig. 3, lane 5), indicating that methylhydroquinone degradation is mediated by pNF1. These observations indicated that both pNF1 and pNF2 were needed to metabolize fenitrothion and that pNF2 was needed to metabolize methyl parathion (Fig. 4). We concluded that pNF2 encodes a fenitrothion-hydrolyzing activity, that pNF1 encodes a methylhydroquinone-degrading activity, and that the oxidative pathway for removal of the nitro group from nitrophenol is not mediated by the two plasmids. These conclusions were supported by the following observations: (i) the loss of pNF1 and pNF2 from NF100 via mitomycin C treatment coincided with the loss of methylhydroquinone-degrading and fenitrothion-hydrolyzing abilities, respectively; (ii) the subsequent introduction of pNF1 into NF1033 and pNF2 into NF1023 resulted in reestablishment of the ability to degrade methylhydroquinone and the ability to hydrolyze fenitrothion, respectively; (iii) both cured strains NF102(pNF2) and NF103(pNF1) retained the capability of oxidative removal of the nitro group from nitrophenol; and (iv) the plasmid profiles in strain NF100 and other derivatives used in the experiments (points i to iii above) were confirmed by agarose gel electrophoresis (Fig. 2 and 3).
FIG. 3.
Agarose gel electrophoresis of plasmids from recipient strains, transconjugants, and transformants. Lanes: 1, NF100 (wild-type strain); 2, NF1033 (recipient strain); 3, NF1023 (recipient strain); 4, NF1023E (transformant of NF1023 [via electroporation with pNF2]); 5, NF1033T (transconjugant of NF1033 [via mating with NF100]).
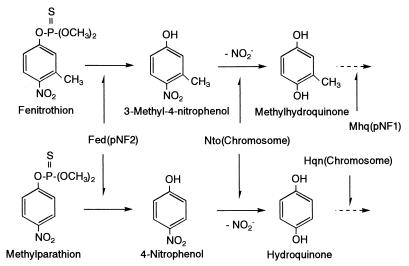
FIG. 4.
Proposed pathways of fenitrothion and methyl parathion degradation by Burkholderia sp. strain NF100. Fed(pNF2), fenitrothion-hydrolyzing activity encoded on pNF2; Nto(chromosome), nitro group-removing oxidative pathway encoded on the chromosome; Mhq(pNF1), methylhydroquinone-degrading ability encoded on pNF1; Hqn(chromosome), hydroquinone-degrading ability encoded on the chromosome.
Many bacteria that metabolize organophosphorus insecticides have been isolated from soils. However, only a few of these bacteria were found to carry a plasmid encoding an insecticide-hydrolyzing enzyme. Two bacterial strains from the closely related genera Pseudomonas and Flavobacterium encode organophosphorus insecticide hydrolase (opd) genes on large plasmids (13, 19). Characteristics of the opd gene and properties of the hydrolase have been extensively studied (6, 7, 10, 12, 20). In the present study, NF100 was found to carry a plasmid encoding a fenitrothion-hydrolyzing enzyme. The few microorganisms capable of hydrolyzing organophosphorus insecticides can utilize one or more hydrolysis products as carbon or nutrient sources (15–17). Formation of 4-nitrophenol via hydrolysis of methyl parathion has been reported by several authors (1, 4, 16, 17). NF100 was able to utilize 3-methyl-4-nitrophenol and 4-nitrophenol, which are hydrolysis products of fenitrothion and methyl parathion, respectively. A variety of species able to utilize nitrophenol as a carbon source have been isolated by researchers (4, 5, 14, 16, 17, 21). The initial step in the bacterial degradation of 4-nitrophenol was shown to be oxidative removal of the nitro group, resulting in the formation of hydroquinone (17, 21, 22). Hydroquinone is further hydroxylated to 1,2,4-benzenetriol prior to ring fission in Pseudomonas putida (17). However, Spain and Gibson (22) indicated that hydroquinone was directly cleaved to β-ketoadipic acid via τ-hydroxymuconic semialdehyde and maleylacetic acid in a Moraxella species. Nitrite and methylhydroquinone were detected in fenitrothion-treated cultures of NF100, indicating that NF100 has the oxidative nitrite group-removing pathway. Methylhydroquinone may be further metabolized through either Moraxella (22)- or P. putida (17)-type ring fission. A number of plasmids encoding the degradation pathway of hydrocarbons such as toluene and naphthalene were isolated from natural habitats or in selective enrichment studies. However, there are very few plasmids encoding methylhydroquinone-degrading enzymes as does the plasmid pNF1 isolated in the present study.
Although a number of plasmid-mediated xenobiotic-compound degradations have been described in the literature (18), such degradation pathways are in most cases encoded on a single plasmid in a microbial cell. Burkholderia sp. strain NF100, isolated in the present study, has a fenitrothion degradation pathway encoded on two plasmids, pNF1 and pNF2 (Fig. 4). In the fenitrothion-treated soil used in the present study, dissemination of the catabolic plasmid may have led to the rapid evolution of a strain capable of utilizing fenitrothion as a carbon source. The combination of pNF1 and pNF2 leading to total degradation of fenitrothion is an example of the role of various degradative plasmids in the total mineralization of some pesticides.
Nucleotide sequence accession number.
The 16S rRNA gene sequence of strain NF100 has been deposited in the DDBJ database under accession no. AB025790.
Acknowledgments
This work was supported by a program for promotion of basic research activities for innovative biosciences of the Bio-oriented Technology Research Advancement Institution (JAPAN).
REFERENCES
- 1.Adhya T K, Sudhakar B, Sethunatha N. Hydrolysis of selected organophosphorus insecticides by two bacteria isolated from flooded soil. J Appl Bacteriol. 1981;50:167–172. doi: 10.1111/j.1365-2672.1981.tb00881.x. [DOI] [PubMed] [Google Scholar]
- 2.Casse F, Boucher C, Julliot J S, Michel M, Denarie J. Identification and characterization of large plasmids in Rhizobium meliloti using agarose gel electrophoresis. J Gen Microbiol. 1979;113:229–242. [Google Scholar]
- 3.Chapalamadugu S, Chaudhry G R. Microbiological and biotechnological aspects of metabolism of carbamates and organophosphates. Crit Rev Biotechnol. 1992;12:357–389. doi: 10.3109/07388559209114232. [DOI] [PubMed] [Google Scholar]
- 4.Chaudhry G R, Ali A N, Wheeler W B. Isolation of a methyl parathion-degrading Pseudomonas sp. that possesses DNA homologous to the opd gene from a Flavobacterium sp. Appl Environ Microbiol. 1988;54:288–293. doi: 10.1128/aem.54.2.288-293.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Daughton C G, Hsieh D P H. Parathion utilization by bacterial symbionts in a chemostat. Appl Environ Microbiol. 1977;34:175–184. doi: 10.1128/aem.34.2.175-184.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dumas D P, Caldwell S R, Wild J R, Raushel F R. Purification and properties of the phosphotriesterase from Pseudomonas diminuta. J Biol Chem. 1989;264:19659–19665. [PubMed] [Google Scholar]
- 7.Harper L L, McDaniel C S, Miller C E, Wild J R. Dissimilar plasmids isolated from Pseudomonas diminuta MG and a Flavobacterium sp. (ATCC 27551) contain identical opd genes. Appl Environ Microbiol. 1988;54:2586–2589. doi: 10.1128/aem.54.10.2586-2589.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hayatsu M, Nagata T. Purification and characterization of carbaryl hydrolase from Blastobacter sp. strain M501. Appl Environ Microbiol. 1993;59:2121–2125. doi: 10.1128/aem.59.7.2121-2125.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hayatsu M, Hirano M, Nagata T. Involvement of two plasmids in the degradation of carbaryl by Arthrobacter sp. strain RC100. Appl Environ Microbiol. 1998;65:1015–1019. doi: 10.1128/aem.65.3.1015-1019.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.McDaniel C S, Harper L L, Wild J R. Cloning and sequencing of a plasmid-borne gene (opd) encoding a phosphotriesterase. J Bacteriol. 1988;170:2306–2311. doi: 10.1128/jb.170.5.2306-2311.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Montgomery H A C, Dymock J F. The determination of nitrite in water. Analyst. 1961;86:414–416. [Google Scholar]
- 12.Mulbry W W, Karns J S. Parathion hydrolase specified by the Flavobacterium opd gene: relationship between the gene and protein. J Bacteriol. 1989;171:6740–6746. doi: 10.1128/jb.171.12.6740-6746.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mulbry W W, Karns J S, Kearney P C, Nelson J O, McDaniel C S, Wild J R. Identification of a plasmid-borne parathion hydrolase gene from Flavobacterium sp. by Southern hybridization with opd from Pseudomonas diminuta. Appl Environ Microbiol. 1986;51:926–930. doi: 10.1128/aem.51.5.926-930.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Munnecke D M, Hsieh D P H. Microbial decontamination of parathion and p-nitrophenol in aqueous media. Appl Microbiol. 1974;28:212–217. doi: 10.1128/am.28.2.212-217.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Nelson L M. Biologically induced hydrolysis of parathion in soil: isolation of hydrolyzing bacteria. Soil Biol Biochem. 1982;44:219–222. [Google Scholar]
- 16.Ou L T, Sharma A. Degradation of methyl parathion by a mixed bacterial culture and a Bacillus sp. isolated from different soils. J Agric Food Chem. 1989;37:1514–1518. [Google Scholar]
- 17.Rani N L, Lalithakumari D. Degradation of methylparathion by Pseudomonas putida. Can J Microbiol. 1994;40:1000–1006. doi: 10.1139/m94-160. [DOI] [PubMed] [Google Scholar]
- 18.Sayler G S, Hooper S W, Layton A C, Henry King J M. Catabolic plasmids of environmental and ecological significance. Microb Ecol. 1990;19:1–20. doi: 10.1007/BF02015050. [DOI] [PubMed] [Google Scholar]
- 19.Serdar C M, Gibson D T, Munnecke D M, Lancaster J H. Plasmid involvement in parathion hydrolysis by Pseudomonas diminuta. Appl Environ Microbiol. 1982;44:246–249. doi: 10.1128/aem.44.1.246-249.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Serdar C M, Murdock D C, Rohde M F. Parathion hydrolase gene from Pseudomonas diminuta MG: subcloning, complete nucleotide sequence, and expression of the mature portion of the enzyme in Escherichia coli. Bio/Technology. 1989;7:1151–1155. [Google Scholar]
- 21.Simpson J R, Evans W C. The metabolism of nitrophenols by certain bacteria. Biochem J. 1953;55:24. [PubMed] [Google Scholar]
- 22.Spain J C, Gibson D T. Pathway for biodegradation of p-nitrophenol in a Moraxella sp. Appl Environ Microbiol. 1991;57:812–819. doi: 10.1128/aem.57.3.812-819.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tamaoka J, Komagata K. Determination of DNA base composition by reversed-phase high-performance liquid chromatography. FEMS Microbiol Lett. 1984;25:125–128. [Google Scholar]
- 24.Yabuuchi E, Kosako Y, Oyaizu H, Yano I, Hotta H, Hashimoto Y, Ezaki T, Arakawa M. Proposal of Burkholderia gen. nov. and transfer of seven species of the genus Pseudomonas homology group II to the new genus, with the type species Burkholdria cepacia (Palleroni and Holmes 1981) comb. nov. Microbiol Immunol. 1992;36:1251–1275. doi: 10.1111/j.1348-0421.1992.tb02129.x. [DOI] [PubMed] [Google Scholar]