Abstract
Adipocytes are key regulators of human metabolism, and their dysfunction in insulin signaling is central to metabolic diseases including type II diabetes mellitus (T2D). However, the progression of insulin resistance into T2D is still poorly understood. This limited understanding is due, in part, to the dearth of suitable models of insulin signaling in human adipocytes. Traditionally, adipocyte models fail to recapitulate in vivo insulin signaling, possibly due to exposure to supraphysiological nutrient and hormone conditions. We developed a protocol for human pluripotent stem cell–derived adipocytes that uses physiological nutrient conditions to produce a potent insulin response comparable to in vivo adipocytes. After systematic optimization, this protocol allows robust insulin-stimulated glucose uptake and transcriptional insulin response. Furthermore, exposure of sensitized adipocytes to physiological hyperinsulinemia dampens insulin-stimulated glucose uptake and dysregulates insulin-responsive transcription. Overall, our methodology provides a novel platform for the mechanistic study of insulin signaling and resistance using human pluripotent stem cell–derived adipocytes.
hPSC-adipocytes respond to physiological insulin levels and model insulin resistance pathophysiology after medium optimization.
INTRODUCTION
The increasing prevalence of metabolic diseases (1, 2) has led to a commensurate interest in mechanistically understanding how environment and nutrition affect metabolism. This is due to the evidence that these factors have been shown to regulate human metabolism and contribute to metabolic dysfunction, including insulin resistance and diabetes (3, 4). However, despite the significance of metabolic diseases in global public health, the tools for mechanistically studying the role of nongenetic factors in human metabolic dysfunction remain limited and insufficient. For example, current tools rely on either murine models or human in vitro cell culture systems, both of which poorly mimic in vivo human metabolism (5). A key limitation of in vitro cell culture systems is that the nutrient ingredients in culture media do not recapitulate in vivo human tissues (6). Despite multiple previous attempts to approximate human plasma with improved cell culture media, current in vitro culture models often still include specific nutrients at supraphysiological levels or have not yet been validated for insulin signaling response (7).
Adipose tissue is central to metabolic health and disease (8). The dysregulation of adipocyte function and loss of insulin sensitivity often observed in obesity is one of the key risk factors for the development of type II diabetes mellitus (T2D) (9). However, in vitro adipocyte culture models poorly mimic the functional capabilities of in vivo adipose tissue for the study of insulin resistance. Most notably, in vitro human adipocyte cultures lack the capacity to perform insulin-stimulated glucose uptake at sufficient levels to observe how environmental or nutritional factors might influence this process. Insulin-mediated glucose uptake and transcriptional response is primarily effectuated by AKT Serine/Threonine Kinase 2 (AKT2) after its phosphorylation downstream of the insulin receptor (10). Previous in vitro models have generally been able to show reasonable AKT2 phosphorylation induced by insulin, but this does not translate into physiologically relevant insulin-stimulated glucose uptake. Overall, these insufficiencies of in vitro culture models limit the ability to mechanistically study the dysregulation and loss of insulin sensitivity in human adipocytes.
Human pluripotent stem cells (hPSCs) are a powerful tool for pathobiology due to their ability to differentiate into almost any tissue to model various diseases, the ease of implementation of genome editing tools, and exquisite control over their culture conditions (11). They also have unlimited availability, which is a major advantage over primary tissues that can be notoriously hard to obtain. This previous limitation is especially true in the case of metabolically healthy adipose tissue. Previously, an established adipocyte differentiation protocol (12) recapitulated several key adipocyte phenotypes in terms of gene expression and lipid accumulation. While these adipocytes have been used to study obesity and their contribution to cardiovascular disease phenotypes (13, 14), they poorly recapitulate insulin signaling. Specifically, insulin-stimulated glucose uptake in hPSC-derived adipocytes has been previously performed at supraphysiological maximal insulin stimulation and portrayed after blocking and subtraction of basal glucose uptake, artificially inflating the observed fold changes. The lack of insulin response at physiologically relevant levels is a key hurdle to overcome to fully unlock the potential of hPSC-derived adipocytes in metabolic disease modeling.
Here, we hypothesized that supraphysiological concentrations of key nutrients in cell culture medium, especially glucose and insulin, negatively affect the function of in vitro adipocytes. Using an established hPSC white adipocyte differentiation protocol, we used a design of experiments (DoE) approach to identify optimal nutrient conditions to potentiate insulin-stimulated AKT2 phosphorylation and glucose uptake in hPSC adipocytes. This optimized sensitization protocol resulted in a physiologically relevant culture condition that improved the functional maturation of the hPSC-derived adipocytes. Adipocytes produced via this approach also demonstrated robust transcriptional response upon insulin stimulation in multiple relevant metabolic pathways. Furthermore, we generated hyperinsulinemia-induced insulin resistance in adipocytes by modulating the insulin concentration during the final differentiation step. These insulin-resistant adipocytes demonstrated impaired insulin-stimulated glucose uptake and transcriptional responses. Our physiologically relevant adipocyte culture model provides a new potential platform for mechanistic studies into how environmental and nutritional factors influence insulin signaling and resistance in human adipocytes.
RESULTS
Protocol optimization to generate insulin-responsive adipocytes
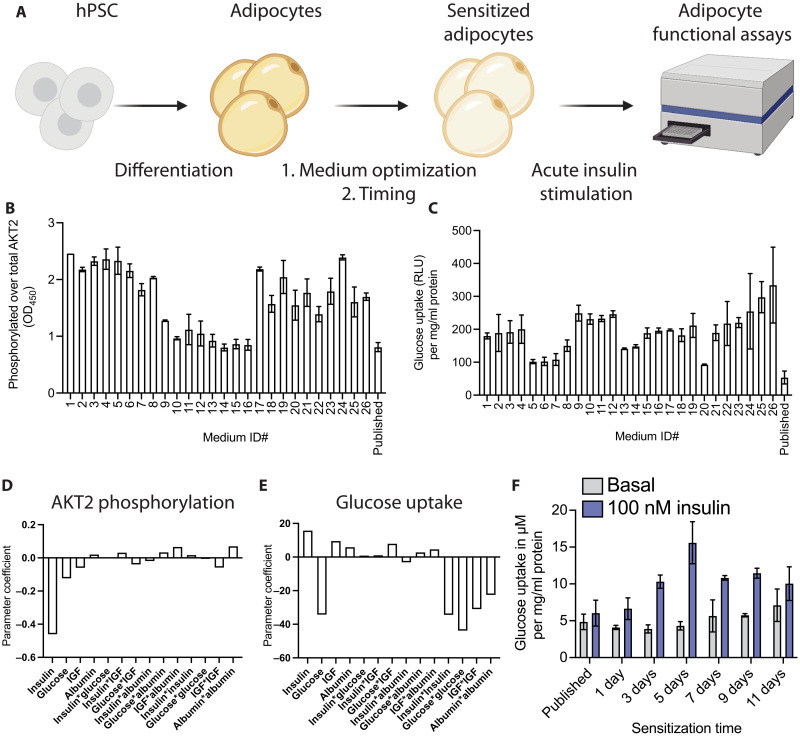
To enhance the insulin response of white hPSC adipocytes, we sought to optimize an established protocol (12) by adding a sensitization step with optimized nutrient and hormone conditions and length of culture (Fig. 1A). We started with a serum-free Dulbecco’s modified Eagle’s medium (DMEM) base medium and sought to determine the ideal concentrations of glucose, insulin, albumin, and insulin-like growth factor 1 (IGF-1) to maximize insulin-stimulated glucose uptake and AKT2 phosphorylation. We used a DoE approach to explore the relationship between these factors systematically and generated 26 different medium compositions (table S1), with which we treated adipocytes for 1 week, without loss of protein or total AKT2 (fig. S1, A and B). We compared these conditions to the results we obtained using the published differentiation medium in both AKT2 phosphorylation (Fig. 1B) and glucose uptake (Fig. 1C) and modeled the nutrient interactions to determine the principal components influencing the insulin responses and their desired concentrations. Our model indicated that the principal parameters driving both the AKT2 phosphorylation (Fig. 1D) and glucose uptake (Fig. 1E) are insulin and glucose, with IGF and albumin having negligible effects (fig. S1, C and D).
Fig. 1. Generation of an insulin-sensitive human adipocyte model.
(A) Schematic indicating the experimental setup to potentiate insulin response in functional assays. hPSCs are differentiated into adipocytes using a published protocol, after which an additional step was added and optimized to sensitize adipocytes. (B and C) Phosphorylation of AKT2 normalized to total AKT2 (B) and glucose uptake (C) after 100 nM insulin stimulation measured for all the medium compositions from our DoE model and the medium from a previously published protocol. RLU, relative light unit. (D and E) The parameter coefficient of each factor in our DoE model indicating its contribution to the AKT2 phosphorylation (D) and glucose uptake (E). (F) Time course of sensitization with the DoE-optimized media measuring glucose uptake at baseline and after insulin stimulation. Results are normalized to total protein for each sample. All bar graphs depict the mean with error bars representing SD, n = 2 biological replicates. OD450, optical density at 450 nm.
Informed by the DoE model, we generated a sensitization medium containing glucose (1 g/liter) and 100 pM insulin. Notably, these concentrations were in the range of what one would find in the plasma of a healthy human (15). We then optimized the exposure time to the sensitization medium, with a clear maximum in insulin-stimulated glucose uptake reached after 5 days of sensitization, with minimal effects on basal glucose uptake (Fig. 1F). This sensitization protocol also resulted in potent AKT2 phosphorylation (fig. S1E). Published literature frequently uses overnight serum starvation, which could roughly be compared to our 1-day sensitization time, indicating a present but weak insulin response in line with previous studies. No condition in the DoE screen or the exposure duration time points resulted in observable adverse effects on the adipocytes as measured by total protein (fig. S1, A and F). The finalized sensitization protocol also did not reduce adipocyte numbers or change their accumulated lipid content or lipid droplet number per cell (fig. S1, G, H, and I). Last, the important adipokine adiponectin is somewhat reduced in the sensitization medium but still secreted quite highly (fig. S1J). With this optimized media and 5-day sensitization protocol, the hPSC adipocytes displayed a >3-fold increase in glucose uptake upon insulin stimulation.
Optimized medium permits adipocyte response to physiological insulin levels
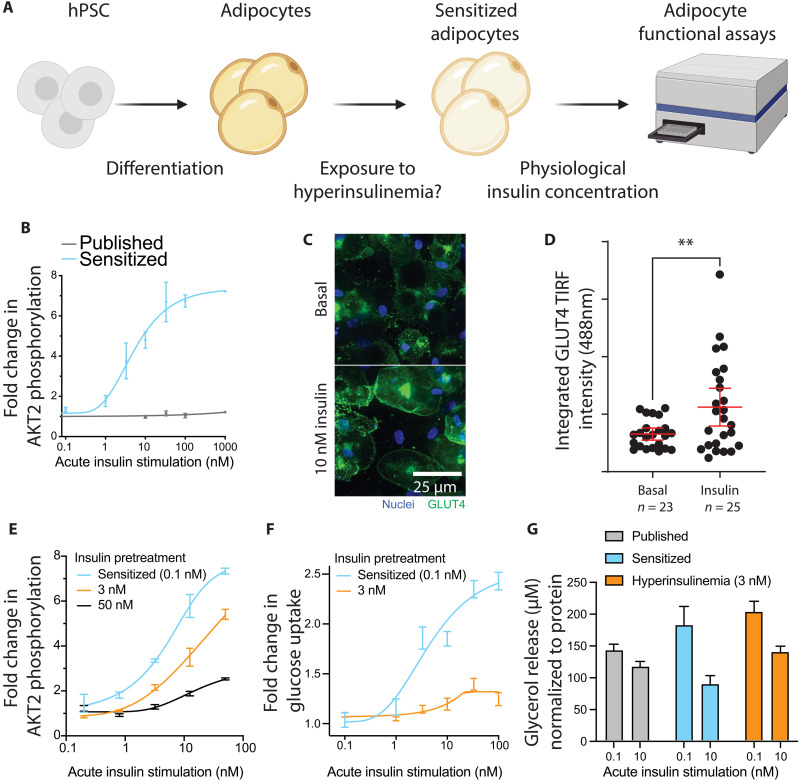
As shown in Fig. 1, the sensitized adipocytes responded strongly to maximal stimulation with 100 nM insulin, a standard concentration used in many previous publications. However, given our sensitization medium’s physiologically relevant baseline insulin concentration, we sought to identify whether insulin levels resembling human postprandial serum concentrations could also lead to insulin response in sensitized adipocytes (Fig. 2A). In an insulin dose-response assay, we demonstrated robust AKT2 phosphorylation at single-digit nanomolar concentrations of insulin in our sensitized adipocytes, with negligible response using previously published media (Fig. 2B). At a postprandial physiologically relevant stimulation of 10 nM insulin for 10 min (16), we observed translocation of Glucose transporter type 4 (GLUT4) from the intracellular space to the cell membrane (Fig. 2C and fig. S2A). Relative to baseline membrane GLUT4, we measured a 71% increase in membrane-resident signal via total internal reflection fluorescence (TIRF) imaging (Fig. 2D).
Fig. 2. Physiological insulin levels induce insulin response and resistance.
(A) Schematic indicating the experimental setup to measure insulin dose-response and induction of insulin resistance. (B) Insulin dose-response curve showing fold change in AKT2 phosphorylation compared to the unstimulated state. (C) Representative images of GLUT4 translocation to the cell membrane upon insulin stimulation. (D) TIRF measurement of GLUT4 signal intensity at the adipocyte cell membrane (**P < 0.01, unpaired two-tailed t test). (E) Insulin dose-response curve showing AKT2 phosphorylation fold change in three insulin preexposure conditions. Results are normalized to total AKT2 and plotted as fold change to unstimulated cells in that condition. (F) Insulin dose-response curve showing glucose uptake for sensitized and hyperinsulinemia-exposed adipocytes. Results are normalized to total protein content and plotted as fold change to unstimulated cells. (G) Glycerol release into the medium for published, sensitized, and hyperinsulinemia-exposed adipocytes showing basal or insulin-suppressed lipolysis, normalized to total protein. Bar graphs depict the mean with error bars representing SD, dose-response curves depict a nonlinear fit curve with error bars representing SD, and the scatterplot depicts individual cell values with mean and 95% confidence interval (CI) overlaid; n = 3 biological replicates unless otherwise indicated.
Hyperinsulinemia induces insulin resistance in adipocytes
After validating normal insulin responsiveness in the sensitized adipocytes, we sought to establish a model of hyperinsulinemia-induced insulin resistance. To induce insulin resistance, we exposed adipocytes throughout the sensitization period to various insulin concentrations. This exposure significantly dampened phosphorylation of AKT2 commensurate to the insulin level (fig. S2B). AKT2 phosphorylation fold changes were decreased across the entire insulin dose-response curve when exposed to a physiologically relevant chronic level of 3 nM insulin, mimicking systemic hyperinsulinemia in patients with T2D (Fig. 2E). At supraphysiological maximal (100 nM) insulin stimulation, the response was reduced by approximately 15%, while at 0.8 and 3.2 nM, acute insulin stimulation AKT2 phosphorylation was decreased by about 50 to 80%. Exposure to 3 nM insulin for the final 5 days of differentiation almost completely abrogated any insulin-responsive glucose uptake (Fig. 2F and fig. S2C). The degree of induced insulin resistance correlated with the concentration of insulin used during the sensitization period and was reproducible in one additional embryonic stem cell (ESC) and two induced pluripotent stem cell (iPSC) lines (fig. S2, D and E, and Table 1). As an additional functional readout, we also looked at adipocyte lipolysis, where we found that in line with previous results, sensitization allowed for insulin response. Glycerol release is suppressed in sensitized adipocytes by 10 nM insulin, while there is no effect in the published protocol (Fig. 2G). This response is blunted, but still present, in hyperinsulinemia-exposed adipocytes. In summary, these results demonstrate that the sensitized adipocytes are insulin sensitive with an appropriate dose-response to physiologically relevant nanomolar exposures and that this sensitivity can be blunted by exposure to hyperinsulinemia, leading to insulin resistance in both signaling and function.
Table 1. hPSC lines used in this study.
| hPSC identified | Cell line name | Source |
| ESC#1 | Hues9 | HSCI (Harvard Stem Cell Institute) iPS Core Facility, Harvard University |
| ESC#2 | WA09/H9 | WiCell |
| iPSC#1 | DiPS 1016 SevA | HSCI (Harvard Stem Cell Institute) iPS Core Facility, Harvard University |
| iPSC#2 | WIBR599 | Whitehead Institute for Biomedical Research |
Transcriptomic characterization of insulin-responsive and -resistant adipocytes
Having validated functional insulin responses in the sensitized adipocytes, we examined the transcriptional changes induced by insulin stimulation. We first verified the transcriptional adipocyte identity of the sensitized protocol developed here in comparison to adipocytes differentiated using a published protocol (12) and to primary adipocytes isolated from nonobese, nondiabetic humans (Table 2). On a global transcriptomic level, the sensitized hPSC-derived adipocytes were equivalent to cells derived using published protocols in their adipocyte identity, with similar principal component distances observed relative to the primary adipocytes (fig. S3, A and B). There was also strong concordance between all samples in their expression of core adipocyte genes as identified by the Human Protein Atlas (fig. S3C) (17).
Table 2. Primary adipocyte RNA source.
| Catalog # | Lot # | Description |
| RNA-A10-1 | LM082310A | Human culture adipocyte RNA < 24.9 BMI |
| RNA-A10-2 | L071498 | Human culture adipocyte RNA 25–29.9 BMI |
| RNA-A10-2 | L090198 | Human culture adipocyte RNA 25–29.9 BMI |
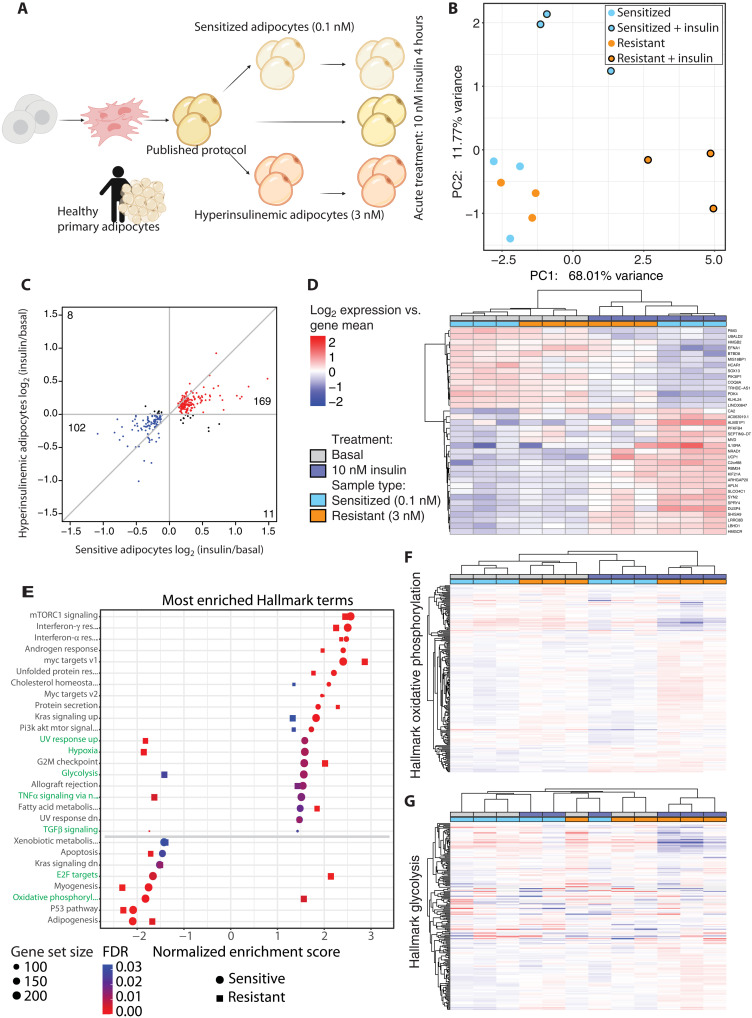
Despite similar transcriptomic adipocyte identities during basal culture, we explored how transcriptional responses after insulin stimulation might differ between adipocytes derived using published methods and the cells described here. We generated three groups of adipocytes; using the published protocol, as well as the sensitized (0.1 nM), and chronic hyperinsulinemia (3 nM) insulin-resistant cells described above. We observed that adipocytes derived using a standard published protocol show little to no transcriptional response upon insulin stimulation, while both the sensitized and insulin-resistant adipocytes responded potently to stimulation with 10 nM insulin for 4 hours (fig. S3D). There are few changes between the sensitized and hyperinsulinemic adipocytes in their basal state (Fig. 3B and fig. S3E), but the 50 significantly differentially expressed genes are enriched in gene sets annotated by DAVID (18) as primarily restricted to lipid and metabolism-related processes (table S2). However, as opposed to the basal state, insulin-responsive transcriptional changes did display numerous differences between the sensitive and resistant adipocytes (Fig. 3, B and C). The top differentially regulated genes after insulin stimulation in the sensitized adipocytes were also largely regulated in the resistant adipocytes, albeit with a diminished fold change (Fig. 3D).
Fig. 3. Induction of insulin resistance perturbs insulin-stimulated transcription.
(A) Schematic indicating the experimental setup for the transcriptomic experiments. (B) PCA plot showing the sensitized and resistant adipocyte in their basal or insulin-stimulated state. (C) Scatterplot comparing fold change in sensitive and resistant adipocytes upon insulin stimulation, filtered on false discovery rate (FDR) ≤ 0.05 in the sensitive adipocytes. Genes with a positive or negative fold change in both comparisons are colored red and blue, respectively, and genes with opposite fold changes appear in black. (D) Heatmap of the genes most changed upon insulin stimulation, filtered by FDR ≤ 0.05 and log2 fold change > 0.5 in the sensitive adipocytes. (E) GSEA results of Hallmark terms that are enriched upon insulin stimulation in both sensitive and resistant adipocytes. Ranked by normalized enrichment score (NES) in the sensitive adipocytes. (F) Heatmap of expression of genes in the oxidative phosphorylation Hallmark gene set. (G) Heatmap of expression of genes in the glycolysis Hallmark gene set. (D), (F), and (G) use the same legend.
Given the lack of clear differences between the sensitized and insulin-resistant adipocytes in the basal state, we performed gene set enrichment analysis (GSEA) (19) on insulin-responsive genes in both of these conditions. We observed a large overlap in the most enriched Hallmark terms (Fig. 3E and fig. S3F). As expected, many of the top enriched gene sets affected by insulin were related to metabolism [mechanistic target of rapamycin complex 1 (mTORC1) signaling, cholesterol homeostasis, phosphatidylinositol 3-kinase AKT MTOR signaling, glycolysis, fatty acid metabolism, and oxidative phosphorylation]. Curiously, several gene sets were enriched in the opposite direction between sensitized and resistant adipocytes, with the most notable examples being the “glycolysis” and “oxidative phosphorylation” gene sets (Fig. 3E). In the sensitized adipocytes, we observed the expected metabolic response to insulin exposure; the expression of genes involved in glycolysis increased while oxidative phosphorylation-related gene expression was decreased. In contrast, the resistant adipocytes exhibited the opposite response to insulin exposure—expression of glycolysis genes was decreased, and oxidative phosphorylation genes increased with insulin—indicating a divergent regulation of metabolism. Gene expression changes in the oxidative phosphorylation gene set were generally mirrored between sensitized and resistant adipocytes, although there were several genes that were increased in the basal insulin-resistant state, which aberrantly further increased with insulin stimulation (Fig. 3F). Meanwhile, the resistant adipocytes in their basal state most closely resembled insulin-stimulated sensitive adipocytes with respect to glycolysis-related genes, while insulin stimulation in the resistant adipocytes wholly dysregulates the glycolytic transcriptional program (Fig. 3G). These results indicate that the sensitization protocol described here produced adipocytes similar in identity to published protocols with respect to primary tissue but that our approach bolstered the transcriptional response to insulin. As expected, the most potentiated transcriptional pathways were related to metabolism, and these were adversely affected in the resistant adipocytes compared to the sensitive protocol.
Wider applications of our insulin sensitization protocol
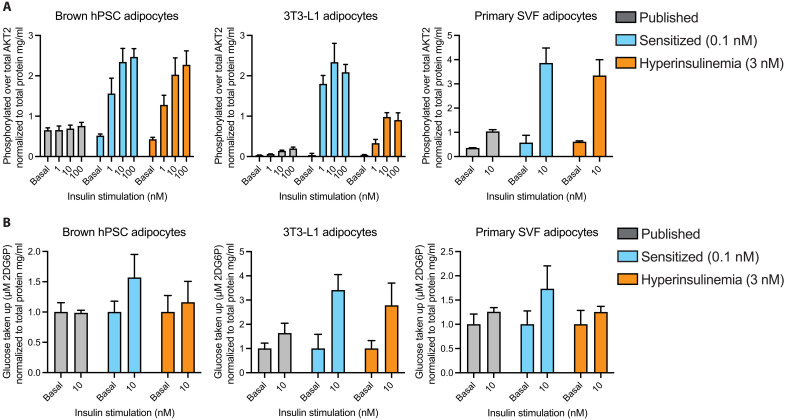
We applied our sensitization method to several other methods of deriving adipocytes, hPSC-derived brown adipocytes (12), mouse 3T3-L1 differentiated adipocytes, and human primary stromal vascular fraction differentiated adipocytes, to explore whether the insulin sensitization strategy described here could be applied to other adipocyte models and thus broaden the applicability of our approach. Without further optimization, we applied our protocol to each of these three adipocyte types and measured their insulin response to AKT2 phosphorylation and glucose uptake. Compared to these cells’ standard culture medium, all three adipocyte models demonstrated exceptional improvement with sensitization with respect to increased insulin response to both phosphorylation of AKT2 and insulin-stimulated glucose uptake at physiological acute stimulation levels (Fig. 4, A and B). Hyperinsulinemia-induced insulin resistance also blunted 3T3-L1 adipocyte AKT2 phosphorylation and insulin-stimulated glucose uptake in brown hPSC adipocytes and primary stromal vascular fraction (SVF) adipocytes. Last, we explored whether additional methods of inducing insulin resistance other than hyperinsulinemia would be possible with our model. We exposed our hPSC adipocytes to tumor necrosis factor–α (TNFα), a known inducer of insulin resistance in adipose tissue (20) and observed severely dampened AKT2 phosphorylation and insulin-stimulated glucose uptake (fig. S4, A and B). While further optimization, similar to the systematic approach described in this manuscript, could be done to further improve each of these models, these results demonstrate the broad applicability of the methods developed here for in vitro adipocyte models at large.
Fig. 4. Application in other adipocyte models.
(A) Phosphorylated AKT2 measurements as a fraction of total AKT2 normalized to total protein per well for brown hPSC adipocytes, mouse 3T3-L1 differentiated adipocytes, and primary human SVF adipocytes. (B) Glucose uptake measurements normalized to total protein per well and displayed as fold change in glucose uptake versus the unstimulated condition for brown hPSC adipocytes, mouse 3T3-L1 differentiated adipocytes, and primary human SVF adipocytes. All bar graphs depict the mean with error bars representing SD, n = 4 biological replicates.
DISCUSSION
The protocol developed here results in insulin-sensitive human adipocytes. By optimizing the timing and medium composition through a statistically driven DoE approach, we developed a simple, easily adoptable media formulation for producing adipocytes from hPSCs that show exquisite sensitivity to insulin signaling. This protocol can also be applied to other adipocyte models. Moreover, we have shown that insulin sensitivity can be dampened by exposing these cells to hyperinsulinemic conditions during the sensitization period, thus providing a new and physiologically relevant model of insulin resistance in human adipocytes. In addition, our induction of insulin resistance did not produce extensive basal changes in transcription but did lead to a diverging response in gene expression upon insulin stimulation. This finding may suggest that this model resembles progression toward insulin resistance in adipose tissue during hyperinsulinemia in humans. It also informs future research, as the lack of transcriptional changes between sensitive and resistant adipocytes strongly implies that the insulin resistance we observe in signaling and function is mediated at a protein or metabolic level. In addition, these observed insulin-stimulated transcriptional differences in sensitive and resistant adipocytes are primarily in metabolic pathways, hinting that metabolism-related protein posttranslation modifications such as oxidation or lipidation could be studied in our model system. Ultimately, this is the first human stem cell–derived adipocyte model that is insulin-responsive at physiologically relevant insulin concentrations and thus better mimics in vivo conditions. This unlocks a plethora of future research possibilities with all the advantages of human stem cell disease models.
MATERIALS AND METHODS
hPSC maintenance
hPSCs were maintained feeder-free on Matrigel (Corning, 354234) in StemFlex medium (Thermo Fisher Scientific, A3349401) and passaged as clumps using Versene solution (Thermo Fisher Scientific, 15040066).
Adipocyte differentiation
Differentiation of hPSCs into adipocytes was performed as previously published (12). Briefly, hPSCs were passaged and grown in suspension culture to form embryoid bodies. After 1 week, the embryoid bodies were plated on tissue culture–treated plastic and grown in DMEM + 10% fetal bovine serum. The outgrowing mesenchymal progenitor cells (MPCs) were used from passage 4 through 8. For adipocyte differentiation, MPCs were infected with previously described lentivirus to express the transactivator reverse tetracycline-controlled transactivator (rtTA) and inducible expression of peroxisome proliferator-activated receptor gamma isoform-2 (PPARg2). MPCs were passaged and grown to confluence and then exposed to doxycycline (700 ng/ml) for 2 weeks in previously established A2 medium and then 1 week without doxycycline in A2 medium. Afterward, adipocytes were exposed to the experimental treatment media for 5 days unless otherwise indicated.
Design of experiments
Media for the DoE experiments were prepared using no-glucose DMEM (Thermo Fisher Scientific, 11966025) and adding the components outlined in table S1. The selection of these formulations was made using JMP software (SAS) to maximize the experimental space created by the four factors. The design was a rotatable surface around a center point for the four factors. Factor levels were selected as ±1 step sizes with log bases 10, 2, 5, and 2 for insulin, glucose, IGF-1, and albumin, respectively (table S1). Adipocytes were fed every other day with these media for 7 days before assays were performed. For modeling, the results of duplicate treatments for each condition were entered into JMP software. We performed least-squares fitting of linear dependencies for each factor and squared dependencies via self- and interfactor crosses to determine any interacting effects on both glucose uptake and AKT2 phosphorylation, normalized to total protein for each condition. The model dependence for each of these linear and secondary factor interactions for both glucose uptake and AKT2 phosphorylation was reported as the parameter coefficient, and the significance of each contribution to the model was reported as the t-ratio P value.
Insulin stimulation assays
All experiments were performed with a no-glucose DMEM as the assay medium (Thermo Fisher Scientific, A1443001). At the end point of differentiation and medium treatment, adipocytes were rinsed with assay medium and then washed for 10 min at 37°C. For enzyme-linked immunosorbent assays (ELISAs), adipocytes were treated with assay medium ± insulin for 10 min and harvested in cell lysis buffer (Cell Signaling Technology, #9803) with phosphatase inhibitor (Thermo Fisher Scientific, 78442). For glucose uptake assays, adipocytes were treated with assay medium ± insulin for 40 min and 2 μM 2-deoxy-glucose and processed as per the manufacturer’s instructions. For lipolysis assays, cells were treated with assay medium ± insulin for 3 hours, and medium was collected.
AKT2 ELISA
After cell lysis, protein samples were quantified and diluted as appropriate for the ELISA (~100 to 200 ng/ml protein). PathScan Phospho-Akt2 (Ser474) and Total Akt2 Sandwich ELISA kits were used to quantify AKT2 levels (Cell Signaling Technology, #7048 and #7046, respectively) as per the manufacturer’s instructions, by colorimetric reading at 450 nm on a Thermo Fisher Multiskan Go plate reader. Results are plotted as the OD450 (optical density at 450 nm) ratio of phosphorylated over total AKT2 or as fold change compared to the basal unstimulated state.
Glucose uptake
Glucose uptake was measured using the Glucose Uptake-Glo Assay (Promega, J1342) per the manufacturer’s instructions. Briefly, after assay medium treatment, 0.5 volume equivalent of lysis buffer was added, followed by 0.5 volume neutralization buffer and 1 volume of the prepared 2-deoxy-D-glucose-6-phosphate (2DG6P) detection reagent. Luminescence was measured to quantify the amount of 2DG taken up. Results are normalized by total protein per well and plotted as relative luminescence units or as fold change compared to the basal unstimulated state.
Lipolysis
Glycerol release into the medium was measured with the Cultured Human Adipocyte Lipolysis Assay Kit–Glycerol (colorimetric) (Zenbio LIP-1) per the manufacturer’s instructions. Briefly, after assay medium treatment, 100 μl of medium was collected and transferred into a new assay plate. Glycerol reagent A was added and total glycerol was measured as colorimetric reading at 450 nm. Results are displayed normalized to total protein content per well.
Protein quantification
Total protein was quantified for all samples using the Pierce BCA Protein Assay Kit (Thermo Fisher Scientific, 23225) as per the manufacturer’s instructions.
Immunofluorescence and TIRF
Differentiated adipocytes were fixed with 10% neutral buffered formalin for 15 min and then washed twice with phosphate-buffered saline (PBS). Cells were permeabilized for 10 min with 0.1% Triton X-100 and incubated in blocking solution consisting of 2% bovine serum albumin (BSA) in PBS for 1 hour at room temperature. After blocking, the cells were incubated in primary antibodies [anti-glucose transporter GLUT4 (Abcam, ab654; 1:500 dilution) or rabbit anti-C/EBPα (CCAAT Enhancer Binding Protein Alpha) (Cell Signaling Technology, D56F10; 1:200 dilution)] for 1 hour at room temperature in 0.2% BSA and 0.1% Triton X-100 in PBS. After primary incubation, samples were washed three times for 5 min each in 0.05% Tween-20 in PBS. After washing, cells were incubated in secondary antibodies [donkey anti-rabbit 488 (Life Technologies, A-21206; 1:500)] for 30 min at room temperature in 0.2% BSA and 0.1% Triton X-100 in PBS. In addition, 4′,6-diamidino-2-phenylindole (DAPI) (Life Technologies, A-62248; 1:500) and HCS LipidTOX Deep Red Neutral Lipid Stain (Thermo Fisher Scientific, H34477; 1:500) were added to the secondary stain solution for counterstains of the nuclei and lipid droplets, respectively. After secondary and counterstain incubation, samples were washed three times for 5 min each in 0.05% Tween-20 in PBS. GLUT4 TIRF imaging was performed on a Nikon Ti-E inverted microscope with a Yokogawa CSU-X1 spinning disk confocal scan head, an Andor iXon 897E EM-CCD camera, and an Andor FRAPPA TIRF photomanipulation system using a 60× plan apo TIRF objective. C/EBPα epifluorescence imaging was performed on a Nikon TE2000 inverted microscope with a Hamamatsu Orca-ER charge-coupled device (CCD) camera using a 10× plan fluor objective. For GLUT4 TIRF imaging quantification, single adipocytes in the stimulated and unstimulated condition were selected at random using the lipid droplet and then centered around the nuclei. Approximately 30 individual cells were selected and imaged between two biological replicates. The Alexa Fluor 488 GLUT4 area was outlined in ImageJ [National Institutes of Health (NIH)], and the integrated intensity was measured and plotted. For C/EBPα imaging and quantification, three representative areas were selected from three replicate differentiations using the DAPI counterstain. The differentiation efficacy for each cell line in using the previously published medium and the sensitization medium was reported as C/EBPα per unit area by averaging the three representative areas together for a single n and generating n = 3 for the replicate differentiations. For lipid droplet analysis, cells were stained for 20 min at room temperature using HCS CellMask Green (Thermo Fisher Scientific; 1:5000) for the cell bodies, DAPI (Thermo Fisher Scientific; 1 μg/ml) for the nuclei, and HCS LipidTOX Deep Red (Thermo Fisher Scientific; 1:1000) for the lipid droplets. The cells were imaged on a Nikon Ti Eclipse inverted microscope equipped with a Lumencor Sola Light Engine light-emitting diode fluorescence lamp and a Nikon DS-QiMc camera using a Nikon CFI Plan Fluor 10× objective with DAPI, GFP, and Far Red single-channel fluorescence filter cubes. The lipid droplets were analyzed on a per-cell basis using an automated image analysis pipeline generated in Cell Profiler (21). Briefly, the DAPI-stained nuclei were used to identify individual cell locations as primary objects, and then secondary objects were propagated outward using the green HCS cell mask to identify and segment cell bodies. Lipid droplets were separately analyzed by subtracting the HCS LipidTOX deep red stain from the HCS CellMask green stain to generate dark circular reliefs. These reliefs were automatically detected and then converted to a binary image, inverted, and identified as tertiary objects. These tertiary objects were recursively eroded and filled to enhance droplet segmentation, declumping, and precise identification of lipid droplets. The identified lipid droplets were then overlaid with the segmented cell bodies measured for number and area within each cell body. The analyses presented are from analyzed images of two experimental replicates from each culture condition with n = 281 to 326 cells per condition.
Primary adipocytes
Primary adipocyte RNA was procured from Zenbio. The samples listed in Table 2 were used.
Other adipocyte sources
Brown hPSC adipocytes were differentiated as previously described (12) with the only difference with the white hPSC adipocyte protocol being CCAAT Enhancer Binding Protein Beta (CEBPB) overexpression in addition to PPARg2. Primary SVF adipocytes were procured as differentiated adipocytes from Zenbio (OA-1048-3) and maintained in Omental/Visceral Adipocyte Maintenance Medium (Zenbio OM-AM). Mouse 3T3-L1 preadipocytes were procured from Zenbio (SP-L1-F), maintained in 3T3-L1 Preadipocyte Medium (Zenbio PM-1-L1), and differentiated per Zenbio instructions in 3T3-L1 Differentiation Medium (Zen-Bio DM-2-L1-500) and 3T3-L1 Adipocyte Medium (Zen-Bio AM-1-L1).
TNFα treatment
hPSC adipocytes were treated for 1 day at the end of the sensitization period with 2.5 nM TNFα (Abcam, ab9642).
RNA sequencing
We mapped the reads to the hg38 version of the human genome, containing only canonical chromosomes, with STAR (22) using a general transfer format (GTF) file downloaded from ENSEMBL version GRCh38.99. We set the overhang parameter to 50 and the “alignIntronMax” parameter to 95,000. We assigned reads to genes with featureCounts (23) with parameters “-p -s 2” and the same GTF file used on the mapping step. We normalized the counts and performed principal components analysis (PCA) with DESeq2 (24) using “vst” on the variance stabilization step. Samples were contrasted in PCA space by Euclidean distance. We performed differential expression (DE) analysis with DESeq2 without log fold change shrinkage. We used statistics from the DE analysis to rank the genes and run the preranked GSEA (19), with the Hallmark gene sets version 7.4. We used Cluster 3.0 (25) to cluster the genes and samples displayed on the heatmaps.
Acknowledgments
We thank the Whitehead Institute genome technology core for the RNA sequencing. Some of the figures were made with the help of biorender.com.
Funding: This study was supported by Novo Nordisk to M.F. and R.J., the NIH to R.J. (1U19AI131135-01 and 5R01MH104610-21), and the National Institute of Biomedical Imaging and Bioengineering at the NIH (T32 EB016652) to A.S.K. and D.J.M.
Author contributions: M.F., A.S.K., J.F.J., and R.J. designed the study. M.F. and A.S.K. performed the experiments. M.F., A.S.K., and M.I.B. analyzed the data. M.F., A.S.K., M.I.B., J.F.J., D.J.M., and R.J. contributed to the manuscript.
Competing interests: J.F.J. is a shareholder of Novo Nordisk A/S and R.J. is a cofounder of Fate, Fulcrum and Omega Therapeutics and an advisor to Dewpoint and Camp4 Therapeutics.
Data and materials availability: All data needed to evaluate the conclusions in the paper are present in the paper and/or the Supplementary Materials. The RNA-sequencing data are available in the Gene Expression Omnibus series GSE201908.
Supplementary Materials
This PDF file includes:
Figs. S1 to S4
Other Supplementary Material for this manuscript includes the following:
Tables S1 and S2
REFERENCES AND NOTES
- 1.DeFronzo R. A., Ferrannini E., Groop L., Henry R. R., Herman W. H., Holst J. J., Hu F. B., Kahn C. R., Raz I., Shulman G. I., Simonson D. C., Testa M. A., Weiss R., Type 2 diabetes mellitus. Nat. Rev. Dis. Primers. 1, 15019 (2015). [DOI] [PubMed] [Google Scholar]
- 2.González-Muniesa P., Mártinez-González M.-A., Hu F. B., Després J.-P., Matsuzawa Y., Loos R. J. F., Moreno L. A., Bray G. A., Martinez J. A., Obesity. Nat. Rev. Dis. Primers. 3, 17034 (2017). [DOI] [PubMed] [Google Scholar]
- 3.Alonso-Magdalena P., Quesada I., Nadal A., Endocrine disruptors in the etiology of type 2 diabetes mellitus. Nat. Rev. Endocrinol. 7, 346–353 (2011). [DOI] [PubMed] [Google Scholar]
- 4.San-Cristobal R., Navas-Carretero S., Martinez-Gonzalez M. A., Ordovas J. M., Martinez J. A., Contribution of macronutrients to obesity: Implications for precision nutrition. Nat. Rev. Endocrinol. 16, 305–320 (2020). [DOI] [PubMed] [Google Scholar]
- 5.Chandrasekera P. C., Pippin J. J., Of rodents and men: Species-specific glucose regulation and type 2 diabetes research. ALTEX 31, 157–176 (2014). [DOI] [PubMed] [Google Scholar]
- 6.R. I. Freshney, Culture of Animal Cells: A Manual of Basic Technique and Specialized Applications (John Wiley & Sons, 2015). [Google Scholar]
- 7.Cantor J. R., Abu-Remaileh M., Kanarek N., Freinkman E., Gao X., Louissaint A. Jr., Lewis C. A., Sabatini D. M., Physiologic medium rewires cellular metabolism and reveals uric acid as an endogenous inhibitor of UMP synthase. Cell 169, 258–272.e17 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Morigny P., Boucher J., Arner P., Langin D., Lipid and glucose metabolism in white adipocytes: Pathways, dysfunction and therapeutics. Nat. Rev. Endocrinol. 17, 276–295 (2021). [DOI] [PubMed] [Google Scholar]
- 9.Friesen M., Cowan C. A., Adipocyte metabolism and insulin signaling perturbations: Insights from genetics. Trends Endocrinol. Metab. 30, 396–406 (2019). [DOI] [PubMed] [Google Scholar]
- 10.Haeusler R. A., McGraw T. E., Accili D., Biochemical and cellular properties of insulin receptor signalling. Nat. Rev. Mol. Cell Biol. 19, 31–44 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Donovan P. J., Gearhart J., The end of the beginning for pluripotent stem cells. Nature 414, 92–97 (2001). [DOI] [PubMed] [Google Scholar]
- 12.Ahfeldt T., Schinzel R. T., Lee Y.-K., Hendrickson D., Kaplan A., Lum D. H., Camahort R., Xia F., Shay J., Rhee E. P., Clish C. B., Deo R. C., Shen T., Lau F. H., Cowley A., Mowrer G., al-Siddiqi H., Nahrendorf M., Musunuru K., Gerszten R. E., Rinn J. L., Cowan C. A., Programming human pluripotent stem cells into white and brown adipocytes. Nat. Cell Biol. 14, 209–219 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Friesen M., Camahort R., Lee Y.-K., Xia F., Gerszten R. E., Rhee E. P., Deo R. C., Cowan C. A., Activation of IRF1 in human adipocytes leads to phenotypes associated with metabolic disease. Stem Cell Rep. 8, 1164–1173 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Warren C. R., O’Sullivan J. F., Friesen M., Becker C. E., Zhang X., Liu P., Wakabayashi Y., Morningstar J. E., Shi X., Choi J., Xia F., Peters D. T., Florido M. H. C., Tsankov A. M., Duberow E., Comisar L., Shay J., Jiang X., Meissner A., Musunuru K., Kathiresan S., Daheron L., Zhu J., Gerszten R. E., Deo R. C., Vasan R. S., O’Donnell C. J., Cowan C. A., Induced pluripotent stem cell differentiation enables functional validation of gwas variants in metabolic disease. Cell Stem Cell 20, 547–557.e7 (2017). [DOI] [PubMed] [Google Scholar]
- 15.Gutt M., Davis C. L., Spitzer S. B., Llabre M. M., Kumar M., Czarnecki E. M., Schneiderman N., Skyler J. S., Marks J. B., Validation of the insulin sensitivity index (ISI0,120): Comparison with other measures. Diabetes Res. Clin. Pract. 47, 177–184 (2000). [DOI] [PubMed] [Google Scholar]
- 16.Song S. H., McIntyre S. S., Shah H., Veldhuis J. D., Hayes P. C., Butler P. C., Direct measurement of pulsatile insulin secretion from the portal vein in human subjects1. J. Clin. Endocrinol. Metab. 85, 4491–4499 (2000). [DOI] [PubMed] [Google Scholar]
- 17.Uhlen M., Oksvold P., Fagerberg L., Lundberg E., Jonasson K., Forsberg M., Zwahlen M., Kampf C., Wester K., Hober S., Wernerus H., Björling L., Ponten F., Towards a knowledge-based human protein Atlas. Nat. Biotechnol. 28, 1248–1250 (2010). [DOI] [PubMed] [Google Scholar]
- 18.Huang D. W., Sherman B. T., Lempicki R. A., Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat. Protoc. 4, 44–57 (2009). [DOI] [PubMed] [Google Scholar]
- 19.Subramanian A., Tamayo P., Mootha V. K., Mukherjee S., Ebert B. L., Gillette M. A., Paulovich A., Pomeroy S. L., Golub T. R., Lander E. S., Mesirov J. P., Gene set enrichment analysis: A knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl. Acad. Sci. 102, 15545–15550 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hotamisligil G. S., Molecular mechanisms of insulin resistance and the role of the adipocyte. Int. J. Obes. (Lond) 24, S23–S27 (2000). [DOI] [PubMed] [Google Scholar]
- 21.Carpenter A. E., Jones T. R., Lamprecht M. R., Clarke C., Kang I., Friman O., Guertin D. A., Chang J., Lindquist R. A., Moffat J., Golland P., Sabatini D. M., CellProfiler: Image analysis software for identifying and quantifying cell phenotypes. Genome Biol. 7, R100 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Li H., Durbin R., Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 25, 1754–1760 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Liao Y., Smyth G. K., Shi W., featureCounts: An efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics 30, 923–930 (2014). [DOI] [PubMed] [Google Scholar]
- 24.Love M. I., Huber W., Anders S., Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 15, 550 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.de Hoon M. J. L., Imoto S., Nolan J., Miyano S., Open source clustering software. Bioinformatics 20, 1453–1454 (2004). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figs. S1 to S4
Tables S1 and S2