Abstract
Amphotericin B, an antifungal drug with a long history of use, forms fungicidal ion-permeable channels across cell membranes. Using solid-state nuclear magnetic resonance spectroscopy and molecular dynamics simulations, we experimentally elucidated the three-dimensional structure of the molecular assemblies formed by this drug in membranes in the presence of the fungal sterol ergosterol. A stable assembly consisting of seven drug molecules was observed to form an ion conductive channel. The structure is somewhat similar to the upper half of the barrel-stave model proposed in the 1970s but substantially different in the number of molecules and in their arrangement. The present structure explains many previous findings, including structure-activity relationships of the drug, which will be useful for improving drug efficacy and reducing adverse effects.
The structure of the amphotericin B ion channel was elucidated to be a seven-molecule assembly with a single molecular length.
INTRODUCTION
The antifungal antibiotic amphotericin B (AmB; Fig. 1A) is produced by the bacterium Streptomyces nodosus, which was isolated from the soil of the Orinoco River in Venezuela (1, 2). AmB has been used for over 60 years to treat systemic fungal infections and remains one of the most clinically important antibiotics (3–5). AmB penetrates the blood-brain barrier and thus serves as a standard therapeutic for most fungal infections of the central nervous system (6). However, serious adverse effects of AmB, such as nephrotoxicity, often cause discontinuation of therapy for life-threatening systemic fungal infections.
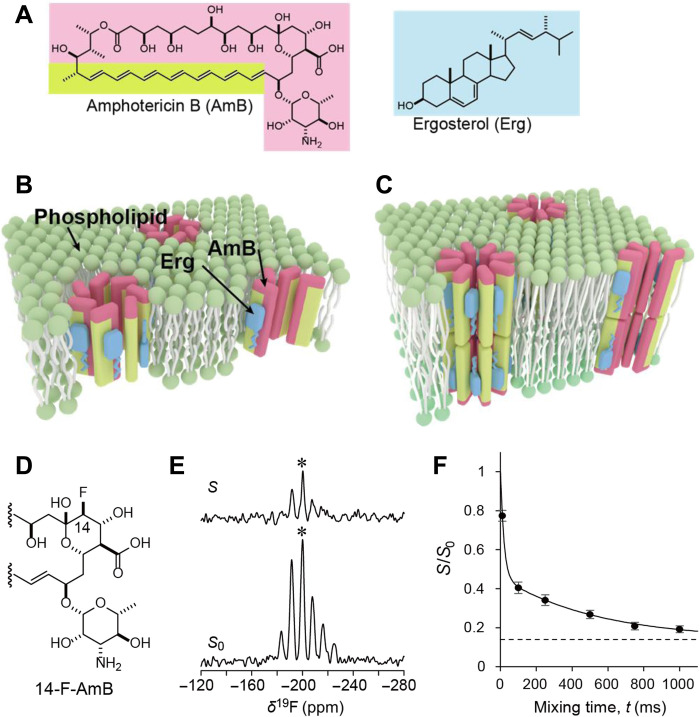
Fig. 1. Structures of AmB ion channel assemblies and the results of a solid-state NMR CODEX method used to determine the number of AmB molecules per assembly.
(A) Chemical structures of AmB and Erg. (B) The present model for channel assemblies with single molecular length. (C) The classic barrel-stave model of AmB ion channels with double molecular length proposed by de Kruijff et al. (13). The hydrophobic and hydrophilic regions of AmB are depicted in lime and pink, respectively. (D) Partial structure of 14-F-AmB. (E) S (top) and S0 (bottom) spectra of a 1:3:7 mixture of 14-F-AmB-Erg-POPC in a 19F-CODEX experiment (mixing time: 750 ms). The spectra of the sample were obtained at 0°C by applying the magic angle spinning method with a rotation speed of 3 kHz. The signal in the central band marked by an asterisk (*) corresponds to the signal of AmB that partly remained outside the membrane (see fig. S22). (F) The decay curve of S/S0, which denotes the fraction of the initial magnetization retained after mixing time. Error bars denote the noise level of the spectra obtained from integration of the baseline near the 19F signals. The intensity decayed according to the following biexponential equation: S/S0 = 0.14 + 0.54e(−t/19.1) + 0.32e(−t/541.1). This curve asymptotically approaches 0.14 (dotted line), indicating that 14-F-AmB forms a cluster of seven molecules.
The self-assembly of bioactive molecules, such as antimicrobial peptides, in cell membranes has long been a subject of research in structural biology (7, 8). However, despite their pharmacological importance, little is known about the self-assembled structures of nonpeptide natural products such as AmB mainly due to the lack of a suitable method for structure determination and the difficulties in isotope labeling of the aperiodic and diverse compounds necessary for nuclear magnetic resonance (NMR) measurements (9). AmB self-assembles into an oligomeric structure in ergosterol (Erg)–containing membranes of fungal and yeast cells; this oligomer acts as an ion channel and is thought to be responsible for the selective toxicity of the drug against eukaryotic microbes (4, 10–12). Even now, 50 years after the well-known barrel-stave model (13) was proposed (Fig. 1C), the structure of the AmB channel has not yet been determined. AmB-AmB interactions and functional groups involved in channel formation provide essential information for structural modifications, which are used to improve efficacy and reduce the toxicity of this important antibiotic. Thus, we aimed to determine the entire structure of the AmB channel assembly in Erg-containing membranes and its functionality by the following steps: (i) determine the number of AmB molecules per assembly by solid-state NMR spectroscopy; (ii) determine the interatomic distances between 13C-labeled AmB and 19F-labeled AmB in the assembly by 13C-19F rotational echo double resonance (REDOR) experiments (14, 15); (iii) determine how AmB molecules are arranged in the assembly by using 13C-19F distances to yield a time-averaged structure of the assembly; (iv) deduce the structural dynamics of the assembly with molecular dynamics (MD) simulations based on the time-averaged structure; and (v) estimate the ion permeability of the fluctuating assemblies by MD simulations and compare it with previous experimental results.
RESULTS
How many AmB molecules constitute one channel assembly
The average number of AmB molecules per channel assembly is crucial information for determining the three-dimensional (3D) structure of the AmB channel. In the past, this number has often been estimated by channel conductance measurements (16, 17) and MD calculations (18), but the results have been quite widely distributed from 4 to over 10. Fluorine-labeled AmB (Fig. 1D), which is easily detected in NMR measurements, shows almost the same biological activity as unlabeled AmB (19). Efficient preparation methods from natural products have been established for 19F- and 13C-labeled AmBs; the structural parameters and MD conditions of AmB and its derivatives used in this study are shown in figs. S1 to S6 and tables S1 and S2, their preparation methods are shown in figs. S7 to S18, and biological activities are shown in fig. S19 and tables S3 to S4. Using the 19F centerband-only detection of the exchange (CODEX) method, which is often used to determine the oligomer number of molecules in assemblies (20, 21), we attempted to directly determine the number of AmB molecules per channel assembly (Fig. 1E). After removing the influence of unbound AmB, we measured the S/S0 values. These experimental values were plotted as a function of mixing time and found to lie on a curve that asymptotically approached 0.14, which is the reciprocal of seven (Fig. 1F). These results implied that the average number of AmB molecules per assembly is seven because the magnetization of one AmB molecule diffuses equally toward the other neighboring molecules in the assembly during a long mixing time. However, the results do not rule out the possibility that assemblies with six and eight molecules coexist within the membrane.
Interatomic distance gives a structural overview of AmB assembly in membranes
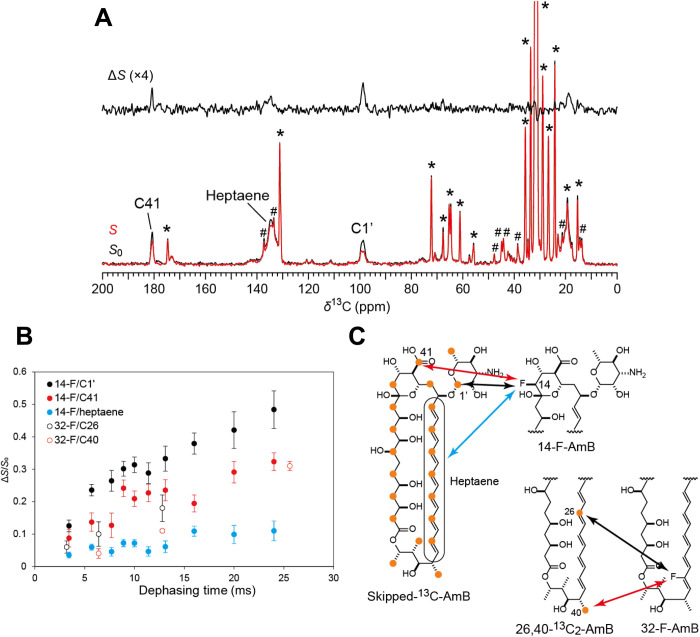
We next determined the interatomic distances between adjacent AmB molecules in the assembly using solid-state NMR (Fig. 2). Skipped 13C-labeled AmB was obtained by a biosynthetic method (Fig. 2C) (22). AmBs fluorinated at positions 14 or 32 and 26,40-13C2 AmB (Fig. 2C) were prepared by chemical synthesis (19, 23) (Supplementary Materials and Methods). Two lipid bilayer models with different lipid compositions and different Erg concentrations in palmitoyloleoylphosphatidylcholine (POPC) were used for 13C-19F REDOR (Fig. 2A) experiments because we used a low concentration of Erg to measure REDOR for 26,40-13C2 AmB due to signal overlap with Erg (Fig. 2A and fig. S20). Six parameters (Fig. 3, A and B, and figs. S21 and S23) had to be determined to elucidate the 3D structure of the AmB channel assembly: the number of molecules n per assembly, the Euler angles (α, β, γ), the channel inner diameter R, and the adjacent probability p, as described in fig. S23. Therefore, it was necessary to measure the distance by using as many labeled atom pairs as possible; we adopted five REDOR-derived distances (Fig. 2B) plus the CODEX result to determine the parameters. For the REDOR experiment, time-dependent increases in the ΔS/S0 values were observed for each 13C-19F pair (Fig. 2B), indicating that molecular motion was slow enough to allow accurate measurements of the 13C-19F distances in the membrane. With these distance constraints in hand, we investigated an assembly structure that could account for the NMR results without being constrained by previous models, such as the barrel-stave model and sterol-sponge model (24, 25). Recently, we found that the vertical penetration of AmB through the lipid bilayer corresponds to the length of a single drug molecule under experimental conditions similar to those used in this study (26, 27). The present REDOR experiment for the 14-F/C40 and 14-F/C26 distances showed that the AmB molecules in the assembly are arranged in a parallel manner, as assumed in the classic barrel-stave model (fig. S24). Together, the results strongly suggest that the AmB assembly resembles the upper half of the barrel-stave model (Fig. 1B and fig. S25) under the experimental conditions, and other possible structures, such as the sterol-sponge model, are considered highly unlikely.
Fig. 2. Solid-state NMR experiments used to determine intermolecular 13C-19F distances.
(A) 13C{19F}REDOR spectra (100 MHz) of 14-F-AmB/skipped-13C-AmB/Erg/POPC = 0.5:0.5:3:7 hydrated with 10 mM Hepes containing D2O at 25°C. The sample spinning speed was 7 kHz, and the dephasing time was 16 ms. The full echo (S0, bottom, black), REDOR-dephased (S, bottom, red), and difference (ΔS, top) spectra clearly showed a reduction in the peak intensity of the C41, heptaene, and C1′ signals; * and # indicate the signals attributed to POPC and Erg, respectively. (B) REDOR dephasing observed for C1′ (filled black), C41 (filled red), and heptaene (filled blue) upon irradiation at 14-F and for C26 (open black) and C40 (open red) upon irradiation at 32-F. The error bars denote the signal-to-noise ratios of ΔS signals. (C) 13C-labeled positions of the skipped labeled (22) and synthetic AmBs shown with orange circles. Arrows denote the interatomic distances determined by REDOR and are colored in the same manner as the dots in (B).
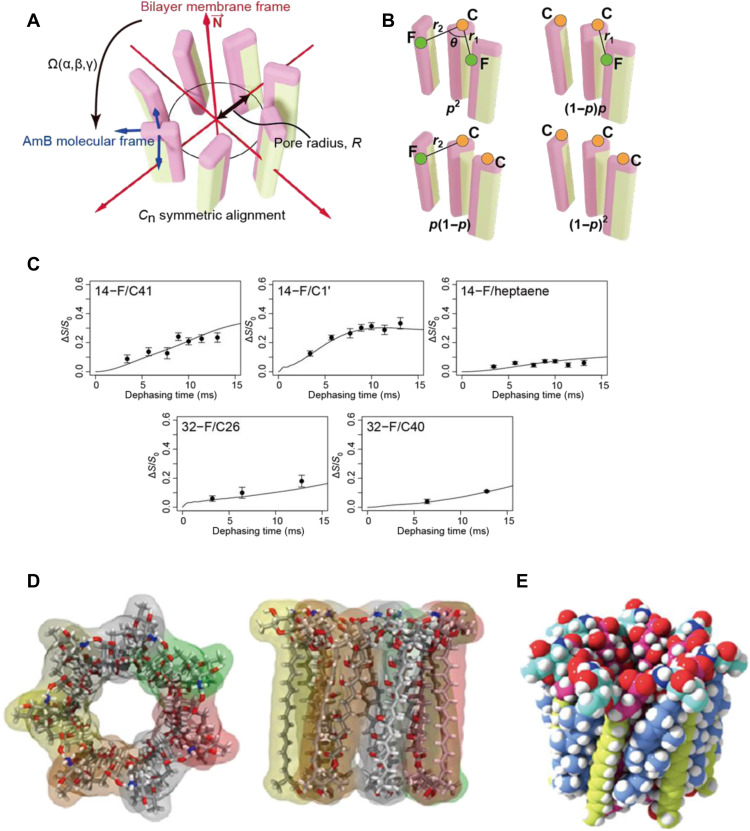
Fig. 3. Construction of a symmetric structure of the AmB assembly based on the interatomic distances obtained via NMR.
(A) Definition of the parameters α, β, γ, n, and R (see fig. S23). (B) Possible adjacent combinations of 19F-AmB and 13C-AmB and the abundance ratios for the possibility parameter p (fig. S21). (C) Simulated REDOR dephasing curves calculated from the best-fit parameters (α, β, γ, n, R, and p) = (−12°, 2°, −20°, 7, 5.4 Å, and 0.24) and experimental dephasing values (same as Fig. 2B). (D) Top and side views of the symmetric structure of the heptamer (seven-molecule) assembly. (E) Space-filling model of the seven AmB and seven Erg complex.
Time-averaged structure of AmB assembly revealed by NMR
We generated a time-averaged structure of the AmB assembly as follows: The upper half of the barrel-stave model was used as a starting point for an AmB self-assembled structure (table S1) (28, 29). We varied the six parameters that defined the structure to minimize the root mean square deviation (RMSD) between the interatomic distances in the assembly model and the REDOR-derived distances (figs. S23 and S26). Figure 3C shows curves of the experimental ΔS/S0 values obtained for each interatomic distance in the AmB assembly as a function of dephasing time. For the interatomic 14-F/C41, 14-F/C1′, and 32-F/C40 distances, two distances, r1 and r2 (Fig. 3B), were obtained for each labeled carbon atom; the 32-F/C26 distance deviated slightly from the measured value, presumably because of an interaction between assemblies as discussed later. This is the first experimentally validated structure of the AmB assembly in a membrane (Fig. 3D). In the structure, the molecular axis of AmB is nearly perpendicular to the central axis (the cone angle β is approximately 2°), and the channel radius at the oxygen atom at C8 is 5.4 Å.
Dynamic structure of AmB assembly deduced by MD simulation
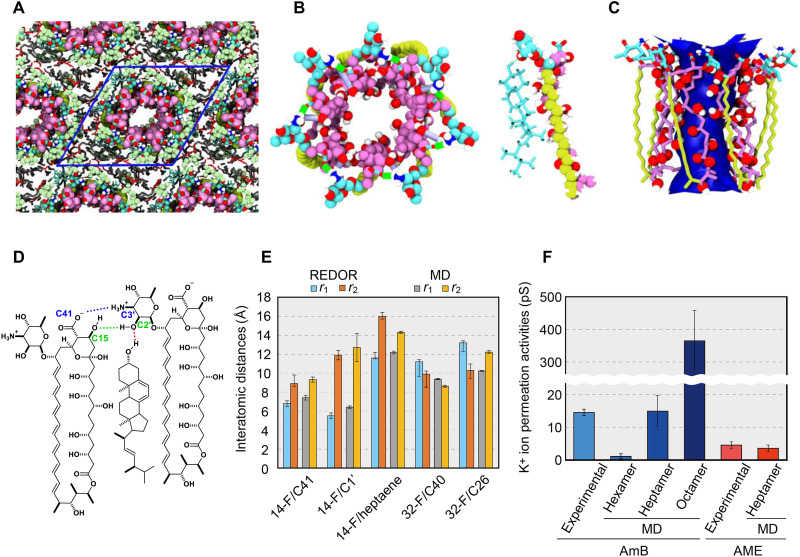
To clarify the structure of a functional ion channel from the AmB-AmB configuration derived from NMR, we next simulated the MD of the AmB assembly using MD calculations. The time scale of the REDOR experiment is in milliseconds, which is much slower than the molecular fluctuations occurring in the submicrosecond range in lipid bilayers. Therefore, the magnitude of REDOR dephasing reflects the time-averaged 13C-19F distances in the AmB assembly, and these distances correspond to a static structure with centrosymmetry (Fig. 3, D and E). It is important to know how the channel structure varies with time to more fully understand the molecular mechanisms of ion flux, which is responsible for the pharmacological activities of AmB. An accurate initial structure (Fig. 4A) is essential for simulating a dynamic structure because the simulation time is limited to microseconds with even the fastest computers, but the exchange of interacting molecules in the assembly occurs much more slowly than that in the simulation. In other words, the following events cannot occur during the simulation time: a change in the number of AmB molecules per assembly, an exchange between the single-length assembly (Fig. 1B) and the double-length assembly (Fig. 1B), or flipping of the AmB or Erg structure. Figure 4 (B and C) shows a typical snapshot and the average structure estimated from MD simulations, respectively. Each Erg molecule generally continues to interact with the same AmB molecule during the 1-μs simulation (Fig. 4, B and D, and fig. S29). These results are consistent with a previous report that the AmB-Erg complex has a lifetime longer than the NMR time scale (23). In addition, the averaged interatomic distances between isotope-labeled positions were calculated to directly compare the channel structures obtained from solid-state NMR to those obtained by MD simulations, and the results were in good agreement (Fig. 4E and table S5).
Fig. 4. Construction of asymmetric assembly and ion channel activity based on MD simulations.
(A) Initial structure of the AmB assemblies with periodic boundary conditions constructed on the basis of NMR results. (B) Snapshots of all-atom MD simulations of an AmB assembly without Erg (left) and an AmB-Erg complex (right). Bright blue shows the mycosamine moiety of AmB (both) and Erg (right). (C) Average structure of an AmB assembly showing the water-accessible region inside the pore. (D) Major intermolecular interactions stabilizing the AmB-Erg channel assembly (see figs. S29 and S38). (E) Interatomic distances measured by solid-state NMR experiments and the values from MD simulations. The distances differ between the front and back of the molecule and correspond to r1 and r2 (Fig. 3B); the structure obtained by MD is not rotationally symmetric, so the average ± SD are shown, where SD obtained from 10 runs of the simulation were less than 5%. (F) Experimental and MD-derived values of K+ permeability (30) (fig. S34 and table S12); the experiments in the literature carried out by using the membranes of DPhPC-Erg 5:1 (2 M KCl, at 100 mV) (30) and egg yolk lecithin (43). This clearly indicates that the channel that shows a value close to those of past single-channel measurements is the heptamer assembly.
Ion-conducting activity of AmB assembly evaluated by MD simulation
To evaluate how well the structure of the AmB assembly constructed on the basis of NMR and MD results emulates the real ion channel responsible for drug activity (fig. S30), we measured the conductance of the assembly under conditions similar to those used for single-channel recording experiments (tables S6 to S10) (30). The channel activity of the dynamic AmB assembly was estimated in Erg-containing diphytanoylphosphatidylcholine (DPhPC) membranes with MD simulations (figs. S31 and S39 to S42 and tables S8 to S10). As shown in Fig. 4F and movies S1 and S2, the heptamer channel best reproduced the experimental ion permeability (30), and the formation of a water column in the channel pore was deduced to significantly stabilize the channel structure (Fig. 4C). Assemblies consisting of six or fewer AmB molecules did not allow K+ passage (movie S3), whereas assemblies of eight molecules allowed much more K+ to pass through than was observed experimentally (Fig. 4F and movies S4 and S5).
DISCUSSION
In this study, we investigated the ion channel structure of AmB using solid-state NMR spectroscopy and MD simulations. Although the resulting channel structure shares similarities with the classic structure (13) proposed in the 1970s, significant differences were observed: The number of AmB molecules lining a channel pore is seven rather than eight, the macrocycle plane is arranged not radially but in a spiral configuration (Fig. 3, D and E), and the length of the conductive AmB channel that penetrates the lipid bilayer corresponds to the length of a single molecule rather than the length of two molecules (Fig. 1, B and C, and fig. S30), although the single-length assembly was assumed to be too short to function as an ion channel (18).
Regarding the length of the ion channels, previous studies (31–34) have shown that AmB forms single-length or double-length channels, depending on the membrane environment such as bilayer thickness. Some of these studies have described whether or not the single-length assembly can penetrate lipid bilayers. In bilayers formed in the liquid-crystalline phase by phospholipids with C14-C18 fatty chains, single-length channels tend to form (31, 32), while in the gel phase bilayers of saturated phospholipids or in the liquid-crystalline bilayers with very long chains of C20:1 or C22:1, single-length channels cannot span the membrane (33, 34). The POPC-Erg bilayer used in this study is in the liquid-crystalline phase under the NMR conditions. Therefore, considering the bilayer thickness, the previous studies suggest that AmB assemblies in this study were formed under the single-length condition; MD simulations (fig. S30) imply that the double-length octamer channels (Fig. 1C) (13) are unstable compared with the single-length heptamer channels in POPC-Erg bilayers. Since the objective of this study is to deduce the channel structure formed in fungal membranes, we used POPC, which has a chain length and unsaturation similar to those of the target biological system; e.g., in Candida albicans, phosphatidylcholines, mainly consisting of saturated and unsaturated C16-C18 chains, are the most abundant phospholipid and Erg accounts for 28 to 35 weight % of total neutral lipids (35).
From the viewpoint of ion channel conductance as a pharmacological activity of AmB, it is important to know which channel contributes the most to the activity (figs. S35 to S37). Although we cannot rule out the possibility that a significant percentage of hexamer assemblies exists, MD results reveal that the AmB hexamers exhibit little conductance (Fig. 4F). On the other hand, the presence of octamer channels, even a small number of them, would have a significant impact on ion conductance so that the proportion of octamer channels, if any, is considered to be small. Thus, our results indicate that, in the POPC-Erg model membrane, the ion channel activity is largely attributed to single-length heptamer channels.
We also found that intermolecular hydrogen bonds from the amino sugar moieties in AmB-AmB and AmB-Erg provide an important force that stabilizes these ion channels in an active form (Fig. 4D); the AmB-AmB contact is further stabilized by a salt bridge between the carboxy group of C41 and the amino group at C3′. In addition, the hydrogen bond formed between the C2′-OH of one AmB and the C15-OH of an adjacent AmB stabilizes the channel complex (table S7).
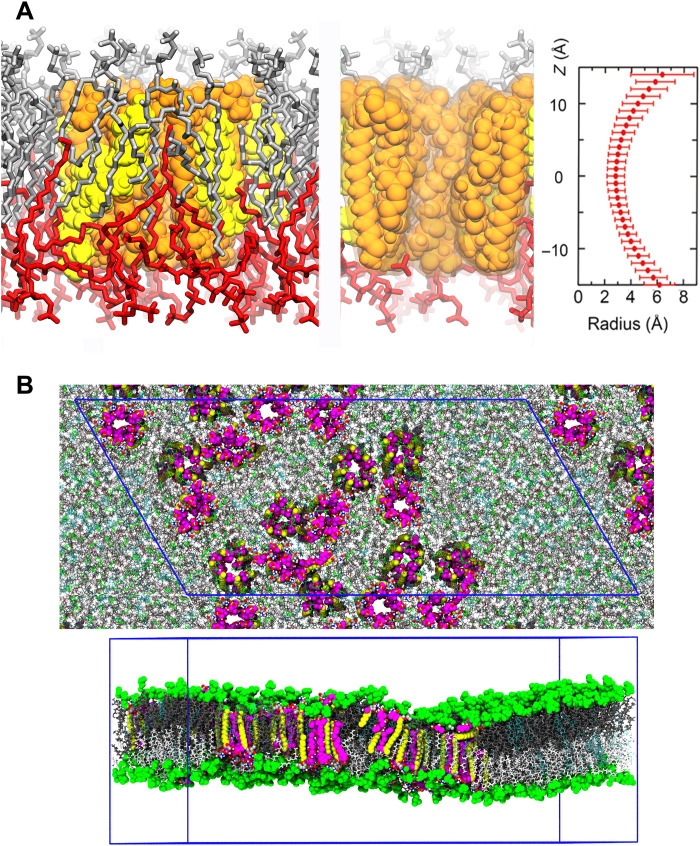
We have to consider how similar the present AmB channel formed in the model bilayers is to the real ion channels that exert antifungal activity in the human body. The apparent difference in the conditions of channel formation involves the concentration of AmB in the membranes since the AmB concentration for NMR measurements was approximately 100 times that found in real fungal membranes (5). First, we investigated using ultraviolet-visible (UV-VIS) and circular dichroism (CD) spectroscopy to determine whether the 3D structure of the channel assembly changed with changes in the concentration of AmB in the membrane. The UV-VIS spectra (fig. S27) revealed that the heptaene moiety of AmB in the Erg-containing membrane showed typical absorbance bands with sharper and red-shifted peaks, which were distinct from those of cholesterol (Cho)–containing or sterol-free membranes. These features were unaffected by the concentration of AmB, indicating that the heptaene moiety resided stably in the membrane interior at concentrations ranging from 0.1 to 10 mole percent (mol %) of those of total lipids (36). The CD spectra of AmB, which sensitively reflect the molecular arrangements of AmB in membranes (37), showed that AmB formed essentially the same assembly over the same concentration range used for UV-VIS measurements (fig. S28). Therefore, we presume that the structure of AmB assemblies elucidated by solid-state NMR is basically the same as that of ion channels occurring in fungal membranes at low concentrations of the drug. Usually, peptides remain in a monomeric form at very low membrane concentrations and tend to form assemblies with a larger number of molecules at higher concentrations (38). Why do AmB molecules form a similar assembly despite the large concentration difference? We speculate that this similarity can be explained by the stability of the heptamer assembly (fig. S26) and the resulting aggregation of the channel assemblies (figs. S32 and S33). As deduced from MD simulation in Fig. 5 (also figs. S32, S43, and S44), the number of unstable interdigitating lipids surrounding the assemblies is expected to be reduced by aggregation of the channels. Even if the overall concentration of AmB is low, as in the case of antifungal assays, the AmB concentration is assumed to increase locally as a result of aggregation of the assemblies.
Fig. 5. MD simulations of phospholipid interaction with and aggregate formation of AmB assemblies in membrane.
(A) Left, the heptamer channel of AmB (tan) and Erg (yellow) surrounded by partly interdigitating POPC molecules (silver and red). Middle, a slice of the left image at the channel center. Right, channel radius profile (distance from the center of a water molecule) along the pore axis. The horizontal bars denote the SD of the channel radius during the simulation (fig. S30). (B) The snapshot of the partial aggregation of AmB-Erg assemblies after 2-μs MD simulation under similar conditions to those of NMR experiments with the antiparallel channel-channel arrangement. The blue frame shows the periodic boundary box (also see figs. S43 and S44).
In this study, since we mix AmB and lipids before preparing vesicles, AmB assemblies are randomly oriented as shown in Fig. 5B. There is a long-standing debate as to whether or not the orientation of AmB ion channels is aligned (39–41). When liposomal AmB injected intravenously is taken up by lipoproteins or other substances and absorbed directly into cells (39), the channels are unlikely oriented in the same way. On the other hand, there is a model in which AmB is adsorbed as a single molecule on fungal membranes (40). In this case, the channel orientation can be aligned in parallel. There is another study that AmB adsorbs on yeast membranes as a large aggregate state (41). In this case, the orientation may not be aligned. The assembly structure elucidated in this study may be somewhat affected by the association direction of the channel. MD simulation based on the NMR-derived channel structure, however, reproduced the experimental K+ conductance of a single channel (Fig. 4F), implying that the NMR structure obtained from aggregated channels should be similar to that of the single channel. Thus, it is reasonable to assume that basically the same structure is maintained in the aggregated state. The orientation of channel aggregates in vivo needs to be clarified by future studies.
Accurately elucidating the channel structure enables us to explain some previously reported structure-activity relationships. For example, the destructive effect of N-acylation on the antifungal activity of AmB and the retained activity of AmB methyl ester (AME), which was subjected to human clinical trials, were demonstrated by MD simulations (figs. S34 and S45 and tables S11 and S12) (42); the simulated channel activity of AME also reproduced previous experimental values (Fig. 4F) (43). In mammalian cell membranes containing Cho, the interaction between AmB and Cho is too weak to form stable ion channels (figs. S29 and S38), often resulting in unstructured aggregates of AmB molecules outside membranes, as revealed by the UV-VIS spectra (fig. S27) (26). Nevertheless, AmB channels occurring in mammalian cells could be very similar in structure to those in Erg-containing membranes (fig. S28G and table S7) and thus weakly increase the ion permeability of the membrane by allowing a small number of ions to pass, which results in the adverse effects of the drug.
We elucidated the dynamic structure of the AmB assembly in model bilayers of fungal membranes using NMR experiments and MD simulations. To investigate whether this molecular assembly functions as an ion channel, the value for K+ conductance obtained from MD simulation was compared with the experimental values from single-channel recording. The two values agreed well, and it is suggested that the present channel structure in POPC-Erg bilayers is more accurate in terms of the number of molecules per channel and the mode of AmB-AmB interaction than those previously proposed. Since it is still very challenging to investigate the channel structure under biologically relevant conditions, it remains to be confirmed whether this channel is formed in the fungal membrane. Nevertheless, the present 3D structure of the AmB channel could be useful for improving drug efficacy and reducing adverse effects.
MATERIALS AND METHODS
Materials
AmB was purchased from Nacalai Tesque (Kyoto, Japan). Erg, Cho, and aliquat 336 were purchased from Tokyo Kasei (Tokyo, Japan). POPC was purchased from NOF Corp (Tokyo, Japan). [1-13C] glucose was purchased from Cambridge Isotope Laboratory (Massachusetts, USA). The AmB-producing microorganism, S. nodosus, was provided by the RIKEN BRC through the National Bio Resource Project of the MEXT/AMED, Japan. All other analytical-grade chemicals were purchased from standard sources. 14-F-AmB was chemically derivatized from AmB (19); a high-performance liquid chromatogram is shown in fig. S8. Skipped-13C-AmB and 26,40-13C2-AmB were prepared as described below. Solution NMR spectra were measured by JEOL ECS400 or ECA500 (Tokyo, Japan). Mass spectra were recorded on Thermo Fisher Scientific Orbitrap XL (Massachusetts, USA). Synthesis of 26,40-13C2-AmB:26,40-13C2-AmB was prepared according to scheme S1. The details of the synthesis are provided in the Supplementary Materials.
Preparation of skipped-13C-AmB
Skipped-13C-AmB was prepared as follows (22). S. nodosus was cultured in the production media (50 ml) in a 500-ml Erlenmeyer flask at 26°C with shaking at 200 rpm. The production medium contains (1-13C) glucose (5.0 g/liter), dextrin (15 g/liter), soy bean flour (7.5 g/liter), and CaCO3 (10 g/liter). Additional (1-13C) glucose (111 mg in 200 μl of water) was pulse-fed to the medium 7, 15, 24, 31, 38, 48, 55, 62, and 72 hours after incubation. After incubation for 122 hours, 7 w/v % of aliquot 336 containing ethyl acetate (25 ml) was added to the production medium, and the pH of the mixture was adjusted to 10.5 with 5 M NaOH. The broth was shaken vigorously at 200 rpm for 1 hour, and the pH of the broth was adjusted to 10.5 with 5 M NaOH. The organic layer was collected and stored at 4°C for 7 days. The crude skipped-13C-AmB was precipitated as yellow solids, and the crudes were collected by centrifugation, washed with acetone (3 ml × 2) and MeOH (2 ml), and dried with Ar flow. The yellow solid was dissolved in N,N′-dimethylformamide (1 ml) containing 0.2 ml of AcOH and precipitated by adding diethyl ether (10 ml). The precipitate was filtered using Celite and dissolved in CHCl3/MeOH/H2O = 10:6:1 to afford purified skipped-13C-AmB (8.9 mg). The purity was checked with 13C NMR. The average labeling rate of 13C was estimated to be 26% according to the isotope pattern of electrospray ionization mass spectra (fig. S7, see the Supplementary Materials).
Membrane sample preparation
Preparation of MLVs for 19F CODEX
14-F-AmB (1.98 mg, 2.1 μmol), Erg (2.50 mg, 6.3 μmol), and POPC (11.2 mg, 14.7 μmol) were dissolved in CHCl3/MeOH (1:2, v/v). The solvent was removed under reduced pressure and dried in vacuo for 8 hours. The lipid film was hydrated with H2O (500 μl) and 10 mM Hepes buffer (14.0 μl) by vortexing, and the lipid dispersion was freeze-thawed for five cycles. After transferring to an Eppendorf tube, water was removed by freeze-drying. After the powder was hydrated with D2O (14.0 μl, 50% hydration), 300 μl of H2O was added, followed by centrifugation at 15,000 rpm for 10 min, and then supernatant was removed by decantation. This operation was repeated three times. The obtained hydrated multi-lamellar vesicles (MLVs) were transferred into a 4-mm magic angle spinning (MAS) rotor (Bruker).
Preparation of liposome sample for 13C{19F}REDOR of 14-F-AmB and skipped-13C-AmB in POPC-Erg MLVs
14-F-AmB, skipped-13C-AmB, Erg, and POPC (0.5:0.5:3:7, total 15 mg) were dissolved in CHCl3/MeOH. Then, the solvent was removed by evaporation to make thin film, which was then dried over in vacuo for 18 hours. After hydrating with H2O (1 ml) and 10 mM Hepes buffer (pH 7.0, 15 μl), the lipid dispersion was freeze-thawed five times and then lyophilized. The powder thus obtained was rehydrated with 15 μl of D2O and packed into a high-resolution MAS insert (Bruker, Germany). Then, the insert was placed into a 4-mm-diameter ϕ MAS rotor (Bruker).
Preparation of liposome sample for 13C{19F}REDOR of 32-F-AmB, 26,40-13C2-AmB in POPC-Erg MLVs
32-F-AmB, 26,40-13C2-AmB, Erg, and POPC (0.5:0.5:1:9, total 20 mg) were dissolved in CHCl3/MeOH. Then, the solvent was removed by evaporation to make thin film, which was then dried in vacuo for 18 hours. After hydrating using H2O (0.5 ml) and 10 mM Hepes buffer (pH 7.0, 20 μl), the lipid dispersion was freeze-thawed five times to prepare MLVs and lyophilized. The powder thus obtained was rehydrated with 20 μl of D2O and packed into a glass tube and sealed with epoxy glue. Then, the tube was inserted into a 5-mm MAS rotor (Varian).
NMR spectral measurements
Solid-state NMR measurements
Solid-state NMR spectra were recorded using a Bruker Avance III spectrometer equipped with a 4-mm HFX triple resonance probe or CMX300 (Chemagnetics) equipped with a 5-mm HFC triple resonance probe (Varian). The 19F CODEX (20, 21) experiments were carried out with 3 kHz of MAS at 0°C. The chemical shift anisotropy (CSA) evolution period was set to 0.67 ms, and 66 kHz of 1H-decoupling field strength was applied during the CSA evolution and free induction decay (FID) acquisition period. The 90° pulse width of 19F was 4.43 μs. The recycle delay was set to 10 s, and 4000 to 16,000 scans were acquired. A Gaussian (300 Hz) window function was applied before the Fourier transformation. The 13C{19F}REDOR spectra were recorded with 5 or 7 kHz MAS at 25° or 30°C. The 90° pulse width of 1H, the 180° pulse width of 19F, and 180° pulse width of 13C were 3.78, 10.8, and 7.86 μs, respectively. 1H decoupling field strength at 66 kHz was applied during the dipole evolution and FID acquisition period. The recycle delay was set to 2 or 3 s, and 32,000 to 64,000 scans were acquired. A Gaussian (30 or 40 Hz) window function was applied before the Fourier transformation.
UV-VIS/CD spectral measurements
AmB and lipids were mixed into vials from the stock solution to a molar ratio of approximately 1:10, 1:100, and 1:1000 (10 or 30 mol % of POPC was replaced with sterols in sterol-containing samples). The solvent was distilled off (V-10, Biotage, Sweden), and the lipid film obtained was dried in vacuum overnight. After hydration with 2 ml of H2O (MilliQ), freeze-thawing was repeated three times to obtain MLV. The MLV was passed through a polycarbonate filter (Liposofast) with a pore size of 800 nm to produce large uni-lamellar vesicles (LUVs). To determine the composition ratio after extrusion through the filter, AmB was quantified by dissolving 100 μl of the LUV suspension in 900 μl of dimethyl sulfoxide and subjected to colorimetric determination by UV-VIS spectra. The POPC content was quantified by using the phospholipid C-test Wako kit (FUJIFILM Wako Pure Chemical Corporation, Japan), and the Cho and Erg contents were quantified using the cholesterol E-test Wako kit (FUJIFILM Wako Pure Chemical Corporation, Japan). From these quantitative values, the final ratio of AmB/sterol/POPC of each LUV sample was determined as described in the legends of figs. S27 and S28. UV-VIS absorption spectra were recorded on a V-730BIO (JASCO Corporation, Japan) at room temperature (22° to 25°C) using a cell with an optical path length of 10 mm. CD spectra were measured with a J-720W spectrophotometer (JASCO Corporation, Japan). The measurements were performed at 25°C using a cell with an optical path length of 2 mm.
Construction of AmB assembly based on REDOR dephasing curves
The xyz coordinate of AmB was constructed on the basis of an x-ray structure (28). Mycosamine orientation was modified according to our report (table S4) (29). The coordinates are summarized in table S1. The AmB coordinates were then rotated by the Euler angle (α, β, γ) as follows
where (x, y, z) and (x′, y′, z′) are coordinates of AmB before and after Euler rotation, respectively. AmB coordinates were moved in parallel with the x axis by R (see fig. S23 for the definition of R as a pore radius)
and duplicated by rotating around the z axis to form the Cn symmetric channel assembly
where () is the coordinate of the kth duplicate of AmB, and n is the oligomeric number. The interatomic distance between kth and (k + 1)th, and between kth and (k − 1)th AmB molecules were then calculated.
The REDOR curves were calculated by considering both 14-F-AmB intramolecular natural abundance 13C contribution and 14-F-AmB/skipped-13C-AmB intermolecular contribution. The intramolecular REDOR dephasings were calculated on the basis of the coordinates shown in table S1. In the case of 14-F/C41 combination, the overall REDOR dephasing, (∆S/S0)C41, was calculated as follows
where Xn, Xe, and p are a 13C ratio of natural abundance, a 13C-enriched ratio, and the probability parameter (Fig. 3B), respectively. The (∆S/S0)intra, (∆S/S0)r1, and (∆S/S0)r2 are intramolecular REDOR dephasing and intermolecular REDOR dephasings depending on two neighboring distance r1 and r2, respectively (see Fig. 3B). They were calculated as two-spin systems. The (∆S/S0)r12 is the REDOR dephasing depending on both r1 and r2, which was calculated as a three-spin system. Bessel function approximation (44) was used for the REDOR dephasing curves of the two-spin system, and 144 angle sets were used for the REDOR dephasing curves of the three-spin system.
In the case of the REDOR dephasing curve observed on C1′ carbon, the natural abundance 13C signal of the C13 position of skipped-13C-AmB overlapped with that of C1′. Therefore, both C1′ and C13 contributions were taken into account. Similarly, in the case of the heptaene moiety, the signals from C20 to C33 positions largely overlapped. Therefore, each REDOR curve for overlapping carbons was calculated and summed up. To eliminate the REDOR dephasings caused by longer 13C-19F spin pairs such as interchannel interaction, the experimental data observed at less than 15-ms dephasing time (seven data points for each labeled position) were taken into account for channel structure screening. The best-fit parameters were obtained by RMSD analysis.
where ,, and are the calculated REDOR dephasing ratios of the ith dephasing time, respectively. ,, and are the observed REDOR dephasing ratios at the ith dephasing time, respectively. RMSD for 32-F-AmB was calculated by the same method with fewer dephasing (∆S/S0) values (fig. S23).
Methods of MD simulations
We have carried out a series of MD simulations of channels composed of different numbers of AmB monomers (fig. S1A and table S2; see the Supplementary Materials), where the initial configuration of the channel complexed with Erg (fig. S1D) was generated using the results of solid-state NMR (23, 45) in a POPC (fig. S1F) lipid membrane. We also performed MD simulations of channels made by AmB derivatives; i.e., AME and N-acetyl-AmB (AmB-NHCOCH3) in fig. S1 (B and C). To examine the effect of orientation of AmB channels on the assembling behavior of AmB channels, we simulated the multiple AmB channels with parallel and antiparallel arrangements in POPC membrane. We also examined the ion channel activity of hexamer, heptamer, and octamer AmB channels in the DPhPC lipid bilayer (fig. S1G).
For the simulated systems, we examined the ion permeation through channels consisting of different numbers of AmB monomers. The details of the simulated systems are provided in table S2. The parallel orientation of the AmB/Erg or AmB/Cho channel structure is considered to be consistent with previous experimental data (45). On the basis of the solid-state NMR measurements (Fig. 3), AmB molecules were arranged in an axisymmetric manner to build a channel as shown in fig. S2. The 3β-hydroxyl group of Erg or Cho was placed toward the 2′-OH group of AmBs, which was reported as “head-to-head” binding conformation (23, 46). The generated channel was embedded in a lipid bilayer for systems V, VI-1, VII-1–4, and VIII-1 (table S2), as shown in fig. S3. We inserted a single AmB channel in the partially “interdigitated” lipid membrane composed of 16 POPC molecules (AmB5, AmB6, and AmB7) or 32 POPC (1-palmitoyl-2-oleoyl-sn-glycero-3-phosphocholine) (AmB8) molecules in a triclinic unit cell. The interdigitated lipid membrane was generated by arranging POPC molecules that were partially interdigitated. When the AmB channels form the denser domain, the channels should be best placed in a hexagonal crystal to minimize the energy penalty of the partial interdigitation. By taking into account this hypothesis, we adopted a triclinic unit cell to mimic the single part of the hexagonal AmB7 channel domain in case of 32 or 16 POPC molecules. We have also inserted the single AmB7 channel POPC-Erg lipid bilayer (system VII-8; table S2) by using the CHARMM-GUI web server. A simulation system (system VIII-3; table-S2) is composed of 16:48:112 (1:3:7) AmB/Erg/POPC molecules, hydrated with 8430 water molecules. The double-length assembly of AmB8 is embedded in lipid membrane using the CHARMM-GUI.
We embedded nine modeled AmB-Erg channel complexes in the lipid membrane composed of 450 POPC molecules. This resulted in the same molecular ratio as in the experimental setup (AmB/Erg/POPC, 1:3:7) (system VII-5 in table S2; fig. S43A). The configuration of the additional lipid membrane was generated using the CHARMM-GUI (47–50). In the initial structures of systems VI-2, VII-7, and VIII-2 (table S2), single AmB6, AmB7, and AmB8 channels were embedded in DPhPC lipid membrane, respectively, and surrounded by ~14,500 water molecules and ions (~550 K+, ~550 Cl−), resulting in the salt concentration of 2 M, using the CHARMM-GUI web server. The initial structures are shown in fig. S4. A similar salt concentration was chosen for the single AmB channel conductance measurement in DPhPC (30). The 10 different initial structures of system VI-2, 16 different initial structures of system VII-7, and 10 different initial structures of system VIII-2 were constructed by using CHARMM-GUI (47–50). In the case of AmB derivatives, we prepared systems by replacing all AmB molecules of system VII-1 (table S2) to each AmB derivative shown in figs. S1 (B and C), respectively (system VII-3–4). We also set up the system by placing the antiparallel arrangement of AmB channels (system VII-6) (figs. S5 and S44A).
To check the dependency on the initial conditions, we constructed “10” different lipid membrane structures including the AmB-sterol complex for systems V, VI-1, VII-1–4, and VIII-1 (table S2). For systems VII-3 and VII-4, seven Cl− counter-ions and seven K+ counter-ions were added to neutralize AmB derivatives, respectively. In the case of systems V, VI-1, VII-1–4, and VIII-1, 7 K+ and 7 Cl− ions were added to the equilibrated systems after 100-ns MD runs to realize physiological salt concentration. The GROMACS “genion” tool was used to replace bulk water molecules with K+ and Cl− ions. The final number of molecules in each system is given in table S2.
Statistical analysis
All experimental data except NMR results were basically expressed with mean and SD. For NMR data, error bars denote the noise level of the spectra obtained from integration of the baseline near the signals. For ion conductance data derived from MD simulation, error bars denote SD from 10 simulations. In MD simulations, error bars of the channel diameter denote SD.
Acknowledgments
The calculations were performed using the facilities of the supercomputer center at the Research Center for Computational Science, Okazaki, Japan (project codes: 20-IMS-C029 and 21-IMS-C106) and the Institute for Solid State Physics, The University of Tokyo (Tokyo, Japan).
Funding: This work was funded by JSPS KAKENHI grant numbers 16H06315 (U.Y., M.D., W.S., and M.M.); JSPS KAKENHI grant numbers 17H06406 (H.T. and M.M.); and JST ERATO Lipid active structure project JPMJER1005, (U.Y., S.M., and M.M.).
Author contributions: Project conception by M.M., N.M., and W.S.; writing of the original manuscript by Y.U., T.Y., M.D.,W.S., and M.M.; syntheses of labeled AmBs by Y.U., T.Y., Y.N., T.S., H.T., and S.M.; synthetic strategy by T.Y., H.T., S.H., and T.O.; NMR measurements by Y.U., T.Y., Y.N., T.S., and S.M.; NMR data interpretation by Y.U., T.Y., T.S., S.M., N.M., and M.M.; MD simulations by M.D., K.F., S.S., and W. S.; and comprehensive analysis of ion channel structure and function by Y.U., T.Y., M.D., Y.N., W.S., and M.M.
Competing interests: The authors declare that they have no competing interests.
Data and materials availability: All data needed to evaluate the conclusions in the paper are present in the paper and/or the Supplementary Materials.
Supplementary Materials
This PDF file includes:
Figs. S1 to S47
Tables S1 to S12
References
Other Supplementary Material for this manuscript includes the following:
Videos S1 to S5
REFERENCES AND NOTES
- 1.Donovick R., Gold W., Pagano J. F., Stout H. A., Amphotericins A and B, antifungal antibiotics produced by a streptomycete. I. In vitro studies. Antibiot. Annu. 3, 579–586 (1956). [PubMed] [Google Scholar]
- 2.Stiller E. T., Vandeputte J., Wachtel J. L., Amphotericins A and B, antifungal antibiotics produced by a streptomycete. II. The isolation and properties of the crystalline amphotericins. Antibiot. Annu. 3, 587–591 (1956). [PubMed] [Google Scholar]
- 3.Hartsel S., Bolard J., Amphotericin B: New life for an old drug. Trends Pharmacol. Sci. 17, 445–449 (1996). [DOI] [PubMed] [Google Scholar]
- 4.Bolard J., How do the polyene macrolide antibiotics affect the cellular membrane properties? Biochim. Biophys. Acta 864, 257–304 (1986). [DOI] [PubMed] [Google Scholar]
- 5.Ellis D., Amphotericin B: Spectrum and resistance. J. Antimicrob. Chemother. 49, 7–10 (2002). [DOI] [PubMed] [Google Scholar]
- 6.Petraitis V., Petraitiene R., Valdez J. M., Pyrgos V., Lizak M. J., Klaunberg B. A., Kalasauskas D., Basevicius A., Bacher J. D., Benjamin D. K. Jr., Hope W. W., Walsh T. J., Amphotericin B penetrates into the central nervous system through focal disruption of the blood-brain barrier in experimental hematogenous Candida meningoencephalitis. Antimicrob. Agents Chemother. 63, e01626-19 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Luo W., Hong M., Determination of the oligomeric number and intermolecular distances of membrane protein assemblies by anisotropic 1H-driven spin diffusion NMR spectroscopy. J. Am. Chem. Soc. 128, 7242–7251 (2006). [DOI] [PubMed] [Google Scholar]
- 8.G. Gröbner, P. Williamson, Solid-state NMR methods for studying membrane systems, in Solid-State NMR (IOP Publising, 2020), pp. 1–30. [Google Scholar]
- 9.Matsuoka S., Inoue M., Application of REDOR NMR in natural product chemistry. Chem. Commun. 5664–5675 (2009). [DOI] [PubMed] [Google Scholar]
- 10.Vertut-Croquin A., Bolard J., Gary-Bobo C. M., Enhancement of amphotericin B selectivity by antibiotic incorporation into gel state vesicles. A circular dichroism and permeability study. Biochem. Biophys. Res. Commun. 125, 360–366 (1984). [DOI] [PubMed] [Google Scholar]
- 11.Davis S. A., Vincent B. M., Endo M. M., Whitesell L., Marchillo K., Andes D. R., Lindquist S., Burke M. D., Nontoxic antimicrobials that evade drug resistance. Nat. Chem. Biol. 11, 481–487 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dong P., Zong C., Dagher Z., Hui J., Li J., Zhan Y., Polarization-sensitive stimulated Raman scattering imaging resolves amphotericin B orientation in Candida membrane. Sci. Adv. 7, eabd5230 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.de Kruijff B., Demel R. A., Polyene antibiotic-sterol interactions in membranes of Acholeplasma laidlawii cells and lecithin liposomes. 3. Molecular structure of the polyene antibiotic-cholesterol complexes. Biochim. Biophys. Acta 339, 57–70 (1974). [DOI] [PubMed] [Google Scholar]
- 14.Gullion T., Schaefer J., Rotational-echo double-resonance NMR. J. Magn. Reson. 81, 196–200 (1989). [DOI] [PubMed] [Google Scholar]
- 15.Gehman J. D., Separovic F., Lu K., Mehta A. K., Boltzmann statistics rotational-echo double-resonance analysis. J. Phys. Chem. B 111, 7802–7811 (2007). [DOI] [PubMed] [Google Scholar]
- 16.Venegas B., González-Damián J., Celis H., Ortega-Blake I., Amphotericin B channels in the bacterial membrane: Role of sterol and temperature. Biophys. J. 85, 2323–2332 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gabrielska J., Gagoś M., Gubernator J., Gruszecki W. I., Binding of antibiotic amphotericin B to lipid membranes: A 1H NMR study. FEBS Lett. 580, 2677–2685 (2006). [DOI] [PubMed] [Google Scholar]
- 18.Zielińska J., Wieczor M., Chodnicki P., Grela E., Luchowski R., Nierzwicki Ł., Bączek T., Gruszecki W., Czub J., Self-assembly, stability and conductance of amphotericin B channels: Bridging the gap between structure and function. Nanoscale 13, 3686–3697 (2021). [DOI] [PubMed] [Google Scholar]
- 19.Matsumori N., Umegawa Y., Oishi T., Murata M., Bioactive fluorinated derivative of amphotericin B. Bioorg. Med. Chem. Lett. 15, 3565–3567 (2005). [DOI] [PubMed] [Google Scholar]
- 20.deAzevedo E. R., Hu W.-G., Bonagamba T. J., Schmidt-Rohr K., Centerband-only detection of exchange: Efficient analysis of dynamics in solids by NMR. J. Am. Chem. Soc. 121, 8411–8412 (1999). [Google Scholar]
- 21.Mani R., Cady S. D., Tang M., Waring A. J., Lehrer R. I., Hong M., Membrane-dependent oligomeric structure and pore formation of a β-hairpin antimicrobial peptide in lipid bilayers from solid-state NMR. Proc. Natl. Acad. Sci. U.S.A. 103, 16242–16247 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.McNamara C. M., Box S., Crawforth J. M., Hickman B. S., Norwood T. J., Rawlings B. J., Biosynthesis of amphotericin B. J. Chem. Soc. Perkin Trans. 1 1998, 83–87 (1998). [Google Scholar]
- 23.Nakagawa Y., Umegawa Y., Matsushita N., Yamamoto T., Tsuchikawa H., Hanashima S., Oishi T., Matsumori N., Murata M., The structure of the bimolecular complex between amphotericin B and ergosterol in membranes is stabilized by face-to-face van der Waals interaction with their rigid cyclic cores. Biochemistry 55, 3392–3402 (2016). [DOI] [PubMed] [Google Scholar]
- 24.Anderson T. M., Clay M. C., Cioffi A. G., Diaz K. A., Hisao G. S., Tuttle M. D., Nieuwkoop A. J., Comellas G., Maryum N., Wang S., Uno B. E., Wildeman E. L., Gonen T., Rienstra C. M., Burke M. D., Amphotericin forms an extramembranous and fungicidal sterol sponge. Nat. Chem. Biol. 10, 400–406 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lewandowska A., Soutar C. P., Greenwood A. I., Nimerovsky E., De Lio A. M., Holler J. T., Hisao G. S., Khandelwal A., Zhang J., SantaMaria A. M., Schwieters C. D., Pogorelov T. V., Burke M. D., Rienstra C. M., Fungicidal amphotericin B sponges are assemblies of staggered asymmetric homodimers encasing large void volumes. Nat. Struc. Mol. Biol. 28, 972–981 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yamamoto T., Umegawa Y., Tsuchikawa H., Hanashima S., Matsumori N., Funahashi K., Seo S., Shinoda W., Murata M., The Amphotericin B-ergosterol complex spans a lipid bilayer as a single-length assembly. Biochemistry 58, 5188–5196 (2019). [DOI] [PubMed] [Google Scholar]
- 27.Yamamoto T., Umegawa Y., Yamagami M., Suzuki T., Tsuchikawa H., Hanashima S., Matsumori N., Murata M., The perpendicular orientation of amphotericin B methyl ester in hydrated lipid bilayers supports the barrel-stave model. Biochemistry 58, 2282–2291 (2019). [DOI] [PubMed] [Google Scholar]
- 28.Jarzembska K. N., Kamiński D., Hoser A. A., Malińska M., Senczyna B., Woźniak K., Gagoś M., Controlled crystallization, structure, and molecular properties of iodoacetylamphotericin B. Cryst. Growth Des. 12, 2336–2345 (2012). [Google Scholar]
- 29.Matsumori N., Sawada Y., Murata M., Mycosamine orientation of amphotericin B controlling interaction with ergosterol: Sterol-dependent activity of conformation-restricted derivatives with an amino-carbonyl bridge. J. Am. Chem. Soc. 127, 10667–10675 (2005). [DOI] [PubMed] [Google Scholar]
- 30.Brutyan R. A., McPhie P., On the one-sided action of amphotericin B on lipid bilayer membranes. J. Gen. Phisiol. 107, 69–78 (1996). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Matsuoka S., Murata M., Membrane permeabilizing activity of amphotericin B is affected by chain length of phosphatidylcholine added as minor constituent. Biochim. Biophys. Acta 1617, 109–115 (2003). [DOI] [PubMed] [Google Scholar]
- 32.Van Hoogevest P., De Kruijff B., Effect of amphotericin B on cholesterol-containing liposomes of egg phosphatidylcholine and didocosenoyl phosphatidylcholine. A refinement of the model for the formation of pores by amphotericin B in membranes. Biochim. Biophys. Acta 511, 397–407 (1978). [DOI] [PubMed] [Google Scholar]
- 33.Kleinberg M. E., Finkelstain A., Single-length and double-length channels formed by nystatin in lipid bilayer membranes. J. Membr. Biol. 80, 257–269 (1984). [DOI] [PubMed] [Google Scholar]
- 34.Ruckwardt T., Scott A., Scott J., Mikulecky P., Hartsel S. C., Lipid and stress dependence of amphotericin B ion selective channels in sterol free membranes. Biochim. Biophys. Acta 1372, 283–288 (1998). [DOI] [PubMed] [Google Scholar]
- 35.Hitchcock C. A., Barrett-Bee K. J., Russell N. J., The lipid composition of azole-sensitive and azole-resistant strains of Candida albicans. J. Gen. Microbiol. 132, 2421–2431 (1986). [DOI] [PubMed] [Google Scholar]
- 36.Fujii G., Chang J. E., Coley T., Steere B., The formation of amphotericin B ion channels in lipid bilayers. Biochemistry 36, 4959–4968 (1997). [DOI] [PubMed] [Google Scholar]
- 37.Vertut-Croquin A., Bolard J., Chabbert M., Gary-Bobo C., Differences in the interaction of the polyene antibiotic amphotericin B with cholesterol- or ergosterol-containing phospholipid vesicles. A circular dichroism and permeability study. Biochemistry 22, 2939–2944 (1983). [DOI] [PubMed] [Google Scholar]
- 38.Huang H. W., Molecular mechanism of antimicrobial peptides: The origin of cooperativity. Biochim. Biophys. Acta 1758, 1292–1302 (2006). [DOI] [PubMed] [Google Scholar]
- 39.Hong Y., Shaw P. J., Tattam B. N., Nath C. E., Earl J. W., Stephen K. R., McLachlan A. J., Plasma protein distribution and its impact on pharmacokinetics of liposomal amphotericin B in paediatric patients with malignant diseases. Eur. J. Clin. Pharmacol. 63, 165–172 (2007). [DOI] [PubMed] [Google Scholar]
- 40.Bolard J., Legrand P., Heitz F., Cybulska B., One-sided action of amphotericin B on cholesterol-containing membranes is determined by its self-association in the medium. Biochemistry 30, 5707–5715 (1991). [DOI] [PubMed] [Google Scholar]
- 41.Walker-Caprioglio H. M., MacKenzie J. M., Parks L. W., Antibodies to nystatin demonstrate polyene sterol specificity and allow immunolabeling of sterols in Saccharomyces cerevisiae. Antimicrob. Agents Chemother. 33, 2092–2095 (1989). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Chéron M., Cybulska B., Mazerski J., Grzybowska J., CzerwiŃski A., Borowski E., Quantitative structure-activity relationships in amphotericin B derivatives. Biochem. Pharmacol. 37, 827–836 (1988). [DOI] [PubMed] [Google Scholar]
- 43.Hartsel S. C., Benz S. K., Ayenew W., Bolard J., Na+, K+ and Cl− selectivity of the permeability pathways induced through sterol-containing membrane vesicles by amphotericin B and other polyene antibiotics. Eur. Biophys. J. 23, 125–132 (1994). [DOI] [PubMed] [Google Scholar]
- 44.Mueller K. T., Analytic solution for the time evolution of dipolar-dephasing NMR signals. J. Magn. Reason. Ser. A A113, 81–93 (1995). [Google Scholar]
- 45.Umegawa Y., Matsumori N., Oishi T., Murata M., Ergosterol increases the intermolecular distance of amphotericin B in the membrane-bound assembly as evidenced by solid-state NMR. Biochemistry 47, 13463–13469 (2008). [DOI] [PubMed] [Google Scholar]
- 46.Umegawa Y., Nakagawa Y., Tahara K., Tsuchikawa H., Matsumori N., Oishi T., Murata M., Head-to-tail interaction between amphotericin B and ergosterol occurs in hydrated phospholipid membrane. Biochemistry 51, 83–89 (2012). [DOI] [PubMed] [Google Scholar]
- 47.Jo S., Lim J. B., Klauda J. B., Im W., CHARMM-GUI membrane builder for mixed bilayers and its application to yeast membranes. Biophys. J. 97, 50–58 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wu E. L., Cheng X., Jo S., Rui H., Song K. C., Dávila-Contreras E. M., Qi Y., Lee J., Monje-Galvan V., Venable R. M., Klauda J. B., Im W., CHARMM-GUI membrane builder toward realistic biological membrane simulations. J. Comput. Chem. 35, 1997–2004 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Lee J., Cheng X., Swails J. M., Yeom M. S., Eastman P. K., Lemkul J. A., Wei S., Buckner J., Jeong J. C., Qi Y., Jo S., Pande V. S., Case D. A., Brooks C. L. III, MacKerell A. D. Jr., Klauda J. B., Im W., CHARMM-GUI input generator for NAMD, GROMACS, AMBER, OpenMM, and CHARMM/OpenMM simulations using the CHARMM36 additive force field. J. Chem. Theory Comput. 12, 405–413 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Jo S., Kim T., Iyer V. G., Im W., CHARMM-GUI: A web-based graphical user interface for CHARMM. J. Comput. Chem. 29, 1859–1865 (2008). [DOI] [PubMed] [Google Scholar]
- 51.Abraham M. J., Murtola T., Schulz R., Páll S., Smith J. C., Hess B., Lindah E., Gromacs: High performance molecular simulations through multi-level parallelism from laptops to supercomputers. SoftwareX 1–2, 19–25 (2015). [Google Scholar]
- 52.Klauda J. B., Venable R. M., Freites J. A., O’Connor J. W., Tobias D. J., Mondragon-Ramirez C., Vorobyov I., MacKerell A. D., Pastor R. W., Update of the CHARMM all-atom additive force field for lipids: Validation on six lipid types. J. Phys. Chem. 114, 7830–7843 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Lim J. B., Rogaski B., Klauda J. B., Update of the cholesterol force field parameters in CHARMM. J. Phys. Chem. 116, 203–210 (2012). [DOI] [PubMed] [Google Scholar]
- 54.Vanommeslaeghe K., Hatcher E., Acharya C., Kundu S., Zhong S., Shim J., Darian E., Guvench O., Lopes P., Vorobyov I., MacKerell A. D. Jr., CHARMM general force field: A force field for drug-like molecules compatible with the CHARMM all-atom additive biological force fields. J. Comput. Chem. 31, 671–690 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Vanommeslaeghe K., MacKerell A. D. Jr., Automation of the CHARMM general force field (CGenFF) I: Bond perception and atom typing. J. Chem. Inf. Model. 52, 3144–3154 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Vanommeslaeghe K., Prabhu Raman E., MacKerell A. D. Jr., Automation of the CHARMM general force field (CGenFF)II: Assignment of bonded parameters and partial atomic charges. J. Chem. Inf. Model. 52, 3155–3168 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Soteras Gutiérrez I., Lin F. Y., Vanommeslaeghe K., Lemkul J. A., Armacost K. A., Brooks C. L. III, MacKerell A. D. Jr., Parametrization of halogen bonds in the CHARMM general force field: Improved treatment of ligand–protein interactions. Bioorg. Med. Chem. 24, 4812–4825 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Jorgensen W. L., Chandrasekhar J., Madura J. D., Impey R. W., Klein M. L., Comparison of simple potential functions for simulating liquid water. J. Chem. Phys. 79, 926–935 (1983). [Google Scholar]
- 59.Nosé S., A unified formulation of the constant temperature molecular dynamics methods. J. Chem. Phys. 81, 511–519 (1984). [Google Scholar]
- 60.Hoover W. G., Canonical dynamics: Equilibrium phase-space distributions. Phys. Rev. 31, 1695–1697 (1985). [DOI] [PubMed] [Google Scholar]
- 61.Parrinello M., Rahman A., Polymorphic transitions in single crystals: A new molecular dynamics method. J. Appl. Phys. 52, 7182–7190 (1981). [Google Scholar]
- 62.Essmann U., Perera L., Berkowitz M. L., Darden T., Lee H., Pedersen L. G., A smooth particle mesh Ewald method. J. Chem. Phys. 103, 8577–8593 (1995). [Google Scholar]
- 63.Darden T., York D., Pedersen L., Particle mesh Ewald: An N·log(N) method for Ewald sums in large systems. J. Chem. Phys. 98, 10089–10092 (1993). [Google Scholar]
- 64.Hess B., Bekker H., Berendsen H. J. C., Fraaije J. G. E. M., LINCS: A linear constraint solver for molecular simulations. J. Comput. Chem. 18, 1463–1472 (1997). [Google Scholar]
- 65.Roux B., The membrane potential and its representation by a constant electric field in computer simulations. Biophys. J. 95, 4205–4216 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Smart O. S., Neduvelil J. G., Wang X., Wallace B. A., Sansom M. S. P., HOLE: A program for the analysis of the pore dimensions of ion channel structural models. J. Mol. Graph. 14, 354–360 (1996). [DOI] [PubMed] [Google Scholar]
- 67.Rui H., Lee K. I., Pastor R. W., Im W., Molecular dynamics studies of ion permeation in VDAC. Biophys. J. 100, 602–610 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Tieleman D. P., The molecular basis of electroporation. BMC Biochem. 5, 1–12 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Böckmann R. A., De Groot B. L., Kakorin S., Neumann E., Grubmüller H., Kinetics, statistics, and energetics of lipid membrane electroporation studied by molecular dynamics simulations. Biophys. J. 95, 1837–1850 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Dasa S., Goswami R. K., Total synthesis of marine natural products separacenes A and B. Org. Biomol. Chem. 15, 4842–4850 (2017). [DOI] [PubMed] [Google Scholar]
- 71.Paquette L. A., Pissarnitski D., Barriault L., A modular enantioselective approach to construction of the macrolactone core of polycavernoside A. J. Org. Chem. 63, 7389–7398 (1998). [DOI] [PubMed] [Google Scholar]
- 72.Hervé M., Cybulska B., Gary-Bobo C. M., Cation permeability induced by valinomycin, gramicidin D and amphotericin B in large lipidic unilamellar vesicles studied by 31P-NMR. Eur. Biophys. J. 12, 121–128 (1985). [Google Scholar]
- 73.Baginski M., Resat H., Borowski E., Comparative molecular dynamics simulations of amphotericin B–cholesterol/ergosterol membrane channels. Biochim. Biophys. Acta Biomembr. 1567, 63–78 (2002). [DOI] [PubMed] [Google Scholar]
- 74.Katsu T., Imamura T., Komagoe K., Masuda K., Mizushima T., Simultaneous measurements of K+ and calcein release from liposomes and the determination of pore size formed in a membrane. Anal. Sci. 23, 517–522 (2007). [DOI] [PubMed] [Google Scholar]
- 75.Houdai T., Matsuoka S., Matsumori N., Murata M., Membrane-permeabilizing activities of amphidinol 3, polyene-polyhydroxy antifungal from a marine dinoflagellate. Biochim. Biophys. Acta Biomembr. 1667, 91–100 (2004). [DOI] [PubMed] [Google Scholar]
- 76.Nakagawa Y., Umegawa Y., Nonomura K., Matsushita N., Takano T., Tsuchikawa H., Hanashima S., Oishi T., Matsumori N., Murata M., Axial hydrogen at C7 position and bumpy tetracyclic core markedly reduce sterol’s affinity to amphotericin B in membrane. Biochemistry 54, 303–312 (2015). [DOI] [PubMed] [Google Scholar]
- 77.Ernst C., Grange J., Rinnert H., Dupont G., Lematre J., Structure of amphotericin B aggregates as revealed by UV and CD spectroscopies. Biopolymers 20, 1575–1588 (1981). [Google Scholar]
- 78.Milhaud J., Hartmann M. A., Bolard J., Interaction of the polyene antibiotic amphotericin B with model membranes: Differences between small and large unilamellar vesicles. Biochimie 71, 49–56 (1989). [DOI] [PubMed] [Google Scholar]
- 79.Matsuoka S., Ikeuchi H., Matsumori N., Murata M., Dominant formation of a single-length channel by amphotericin B in dimyristoylphosphatidylcholine membrane evidenced by 13C-31P rotational echo double resonance. Biochemistry 44, 704–710 (2005). [DOI] [PubMed] [Google Scholar]
- 80.Mouri R., Konoki K., Matsumori N., Oishi T., Murata M., Complex formation of amphotericin B in sterol-containing membranes as evidenced by surface plasmon resonance. Biochemistry 47, 7807–7815 (2008). [DOI] [PubMed] [Google Scholar]
- 81.Shvinka N., Caffier G., Cation conductance and efflux induced by polyene antibiotics in the membrane of skeletal muscle fiber. Biophys. J. 67, 143–152 (1994). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Mariusz B., Mazerski J., Molecular modelling of amphotericin B–ergosterol primary complex in water. Biophys. Chem. 95, 125–133 (2002). [DOI] [PubMed] [Google Scholar]
- 83.Nezil F. A., Bloom M., Combined influence of cholesterol and synthetic amphiphillic peptides upon bilayer thickness in model membranes. Biophys. J. 61, 1176–1183 (1992). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figs. S1 to S47
Tables S1 to S12
References
Videos S1 to S5