Abstract
BACE-1 is required for generating β-amyloid (Aβ) peptides in Alzheimer’s disease (AD). Here, we report that microglial BACE-1 regulates the transition of homeostatic to stage 1 disease-associated microglia (DAM-1) signature. BACE-1 deficiency elevated levels of transcription factors including Jun, Jund, Btg2, Erg1, Junb, Fos, and Fosb in the transition signature, which transition from more homeostatic to highly phagocytic DAM-1. Consistently, similar transition-state microglia in human AD brains correlated with lowered levels of BACE-1 expression. Targeted deletion of Bace-1 in adult 5xFAD mice microglia elevated these phagocytic microglia, correlated with significant reduction in amyloid plaques without synaptic toxicity. Silencing or pharmacologically inhibiting BACE-1 in cultured microglia-derived cells shows higher phagocytic function in microglia. Mechanistic exploration suggests that abolished cleavage of IL-1R2 and Toll-like receptors via BACE-1 inhibition contributes to the enhanced signaling via the PI3K and p38 MAPK kinase pathway. Together, targeted inhibition of BACE-1 in microglia may offer AD treatment.
BACE-1 acts as a trigger for transitioning from homeostatic to more efficiently phagocytic DAM-1 microglia.
INTRODUCTION
β-Amyloid (Aβ) peptide is produced through sequential cleavage of amyloid precursor protein (APP) by β- and γ-secretase, and its abnormal accumulation leads to amyloid deposition and senile plaques, which may drive the pathophysiology associated with synaptic dysfunction and memory loss in patients with Alzheimer’s disease (AD) (1). Elevated activity of neuronal BACE-1 (β-site APP cleaving enzyme–1) increases amyloidogenic processing of APP, as mouse genetic studies have established that BACE-1 is the sole enzyme for Aβ generation (2–4). Inhibiting this rate-limiting BACE-1 enzymatic activity by various pharmacological interventions suppresses Aβ production in humans (5, 6). However, despite a substantial reduction in Aβ load, human clinical trials have failed to demonstrate improvements in cognitive functions in patients with AD due to neuronal and synaptic toxicity (7).
Similar to the reduction of amyloid plaques in the human clinical trial, we showed that preexisting amyloid plaques can be removed in our previous study using Bace-1 conditional knockout (KO) mice to delete Bace-1 in the adult Alzheimer’s 5xFAD mouse model (8). To understand how preexisting amyloid plaques are removed in this mouse model, we assessed whether BACE-1 in microglia could have a role in removing amyloid plaques. This functional relevance was not fully understood due to the lack of systematic studies of BACE-1 in microglia.
The role of microglia in AD pathogenesis has recently gained considerable attention, mainly because of breakthrough genetic studies that have identified the microglial genes triggering receptor expressed on myeloid cells 2 (TREM2), clusterin (CLU), and CD33 as late-onset AD risk genes (9). Comparative transcriptomic analysis of human AD brains revealed genetic polymorphisms at the enhancer/promoter region of microglial genes that are related to phagocytosis, suggesting that altered functions of microglia are implicated in AD pathology (10). Recent studies have identified genetically distinct subtypes of microglia as they respond to changes in the brain microenvironment, namely, homeostatic and disease-associated microglia (DAM) (11). Stage 1 DAM (DAM-1) represents a more functional, phagocytic subtype of immune cell, while stage 2 DAM (DAM-2) may be dysfunctional and contribute to AD pathology (12, 13). We aimed to investigate the role of BACE-1 within microglial subtypes and asked whether BACE-1 in microglia can affect amyloid pathology in AD animal models.
To this purpose, we used an unbiased single-cell RNA sequencing (scRNA-seq) transcriptomic approach to analyze datasets obtained from various mouse models including Bace-1–null or Bace-1 conditional KO mice with and without breeding with 5xFAD mice as well as from human AD brains. We showed that specific deletion of Bace-1 in microglia in 5xFAD mice induced expression of a unique set of transcription factors (TFs) including Jun, Junb, Jund, and Fos. Induced expression of this unique set of TFs correlated with the transition from homeostatic microglia to DAM-1, and this was further validated using published data from human AD brains by single-nuclear RNA-seq (snRNA-seq). Moreover, conditional Bace-1 deletion from microglia in 5xFAD mice significantly reduced the expression of DAM-2 marker genes (Axl, Cst7, and Lpl), indicating a reduction of this dysfunctional subtype associated with reduced amyloid plaque deposition.
We also conducted in vitro studies using primary microglia and bone marrow–derived macrophages (BMDMs) and demonstrated enhanced phagocytosis of aggregated Aβ when Bace-1 was deleted or inhibited. This is in line with the observation that BACE-1 inhibition increased signaling pathways that are important for phagocytosis and degradation of Aβ, such as Toll-like receptor (TLR), LXR/RXR, Fc receptor (FcR), and Ras homolog family member A (RhoA) pathways. The up-regulation of these pathways was consistently seen in multiple Bace-1 deletion models. Mechanistic exploration suggests that abolished cleavage of interleukin-1R2 (IL-1R2) and TLR2 and TLR4 likely contributes to signaling changes via the p38 mitogen-activated protein kinase (MAPK) pathway. Together, our study demonstrates that targeted inhibition of BACE-1 in microglia will enhance phagocytosis and Aβ clearance through elevating genes that are unique to the DAM-1 signature and thus prevent transition to a more dysfunctional, DAM-2 state.
RESULTS
Targeted Bace-1 deletion in microglia induces a transition to DAM-1 signature
To understand the role of BACE-1 in microglia, we generated Bace-1fl/fl;Cx3cr1CreER mice by breeding Bace-1fl/fl mice with Cx3cr1CreER mice [B6.129P2(Cg)-Cx3cr1tm2.1(cre/ERT2)Litt/WganJ, purchased from JAX] for the purposes of deleting Bace-1 in microglia. Tamoxifen (TAM) treatment of 3-month-old Bace-1fl/fl;Cx3cr1CreER mice for 5 days selectively deleted Bace-1 in microglia, which were purified and examined by Western blot analysis at the age of 4 months (fig. S1B). We further crossed Bace-1fl/fl;Cx3cr1CreER mice with 5xFAD mice to obtain 5xFAD;Bace-1fl/fl;Cx3cr1CreER mice. Since Aβ accumulation in 5xFAD mice begins at around 2 months of age (14), Bace-1fl/fl;Cx3cr1CreER mice were treated with either TAM or vehicle for 5 days beginning at the age of 3 months old. Unbiased scRNA-seq experiments were conducted on CD11b+ immune cells sorted from cortical and hippocampal regions of TAM-treated 4-month-old 5xFAD;Bace-1fl/fl;Cx3cr1CreER mice (in the AD background) or Bace-1fl/fl;Cx3cr1CreER mice [in the wild-type (WT) background], compared to littermates with vehicle treatment.
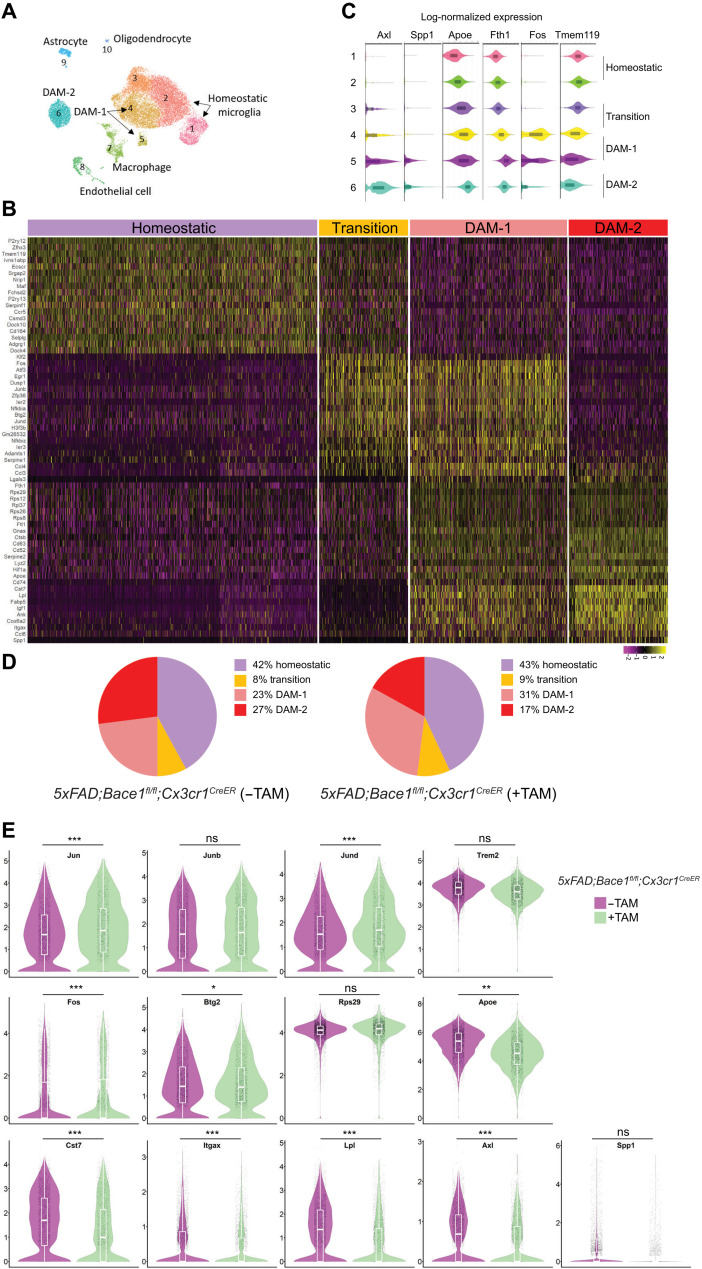
We then conducted RNA-seq analyses using 10x Genomics cloupe software and identified 10 clusters of cells in 4-month-old 5xFAD;Bace-1fl/fl;Cx3cr1CreER mice (Fig. 1A; additional cell type–specific signatures shown in fig. S1C), based on unique gene expression signatures. Clusters 1 to 6 belonged to microglia defined by microglia unique genes (Fig. 1A; signature genes listed in table S1). Clusters 1 and 2 expressed typical homeostatic genes (Tmem119 and P2ry12), while most of cluster 6 expressed typical DAM-2 genes (Fig. 1B). Clusters 4 and 5 expressed typical DAM-1 genes, while partial cluster 3– and cluster 4–expressed genes appeared in the transition state between homeostatic and DAM-1 signatures (Fig. 1C). These transitory genes—including a unique set of TFs such as Jun, Junb, Jund, Fos, Fosb, Btg2, and Egr1—were low in the homeostatic microglia and DAM-2 but overlapped with typical DAM-1 genes such as Fth1, Rps12, Rps26, Rps29, Rps36, Hif1a, and Serpin2. Some DAM-1 genes overlapped with DAM-2 genes, although typical DAM-2 genes such as Cst7, Lpl, Igf1, Ccl6, and Spp1 were expressed even more in DAM-2. Comparing these signatures in 5xFAD;Bace-1fl/fl;Cx3cr1CreER mice (+TAM) versus 5xFAD;Bace-1fl/fl;Cx3cr1CreER mice (−TAM), we noted a significant reduction of DAM-2 (~17% versus ~27%; Fig. 1D) and a clear increase in DAM-1 (~31% versus 23%). The increased DAM-1 population upon Bace-1 deletion in microglia correlated with elevated expression of transitory TFs in TAM-treated Bace-1fl/fl;Cx3cr1CreER mice (Fig. 1E); these TFs were expressed the highest in DAM-1 compared to homeostatic and DAM-2 signatures (fig. S1D). It is worth mentioning that expression of other major TFs such as P65, STAT1, and STAT6 was comparable. Typical DAM-2 genes—such as Cst7, Itgax, Lpl, and Apoe—were significantly reduced (Fig. 1E). Reduction in Trem2 expression was marginally insignificant. Deletion of microglia Bace-1 in 5xFAD;Bace-1fl/fl;Cx3cr1CreER mice (+TAM) caused changes in large sets of genes when compared to that in 5xFAD;Bace-1fl/fl;Cx3cr1CreER mice (−TAM) (see volcano plot in Fig. 5A).
Fig. 1. Bace-1 deletion facilitates microglial transition from a homeostatic to a DAM-like state.
(A) Uniform manifold approximation and projection (UMAP) clustering of CD11b-positive immune-sorted microglia derived from 4-month-old 5xFAD;Bace-1flfl; Cx3cr1CreER with and without TAM for 5 days at 3 months of age, N = 3 each genotype. (B) Heatmap showing unbiased top markers defining each microglia subtype. The color scale represents Z-score–transformed expression values (with yellow and purple representing up-regulated and down-regulated genes, respectively, compared with the mean expression value of a gene from all samples). Four distinct microglial cell populations represent homeostatic, transition, DAM-1–, and DAM-2–like states. (C) Violin plots showing selected genes associated with the homeostatic state and DAM-related microglial genes. On the basis of specific subsets of genes, subclusters 1 to 6 were composed of microglial cells. Tmem119 is concentrated in clusters 1 and 2 and lowered when transited to DAM-1 and DAM-2. Fos is the highest in transitory to DAM-1, while Axl is the highest in DAM-2. (D) Pie chart showing the percentages of microglia within homeostatic, transition, DAM-1, or DAM-2 cluster genes under the 5xFAD condition with and without conditional Bace-1 deletion in microglia [5xFAD;Bace-1fl/fl;Cx3cr1CreER mice (−TAM) versus 5xFAD;Bace-1fl/fl;Cx3cr1CreER mice (+TAM)]. (E) Violin plots compare the distribution of log-transformed normalized gene expression of transcriptions factors and genes strongly expressed in DAM-1 or DAM-2 in the 5xFAD mice with and without conditional microglial Bace-1 deletion. There was significant up-regulation of the AP-1 TF genes Jun, Junb, Jund, and Fos in Bace-1–deleted 5xFAD mice. ns, not significant.
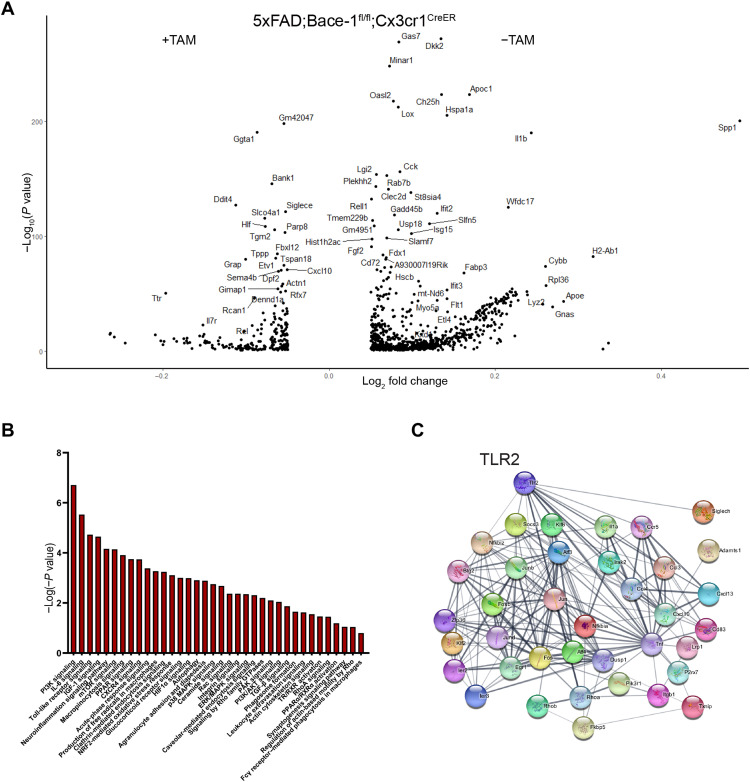
Fig. 5. Bace-1 deletion enhances signaling pathways associated with phagocytosis.
(A) Volcano plot shows significantly altered gene expression in 4-month-old 5xFAD;Bace-1fl/fl;Cx3cr1CreER mouse microglia with and without TAM treatment. (B) IPA-based canonical pathway analysis of significantly altered microglia genes from 5xFAD;Bace-1fl/fl;Cx3cr1CreER mice with or without Bace-1 deletion. (C) String-based analysis demonstrating potential protein-protein functional interactions and interconnected pathways enriched in statistically up-regulated genes after Bace-1 deletion in microglia. TLR2 is one of the top hit genes.
Targeted deletion of Bace-1 in microglia under normal conditions increases transition microglial signature
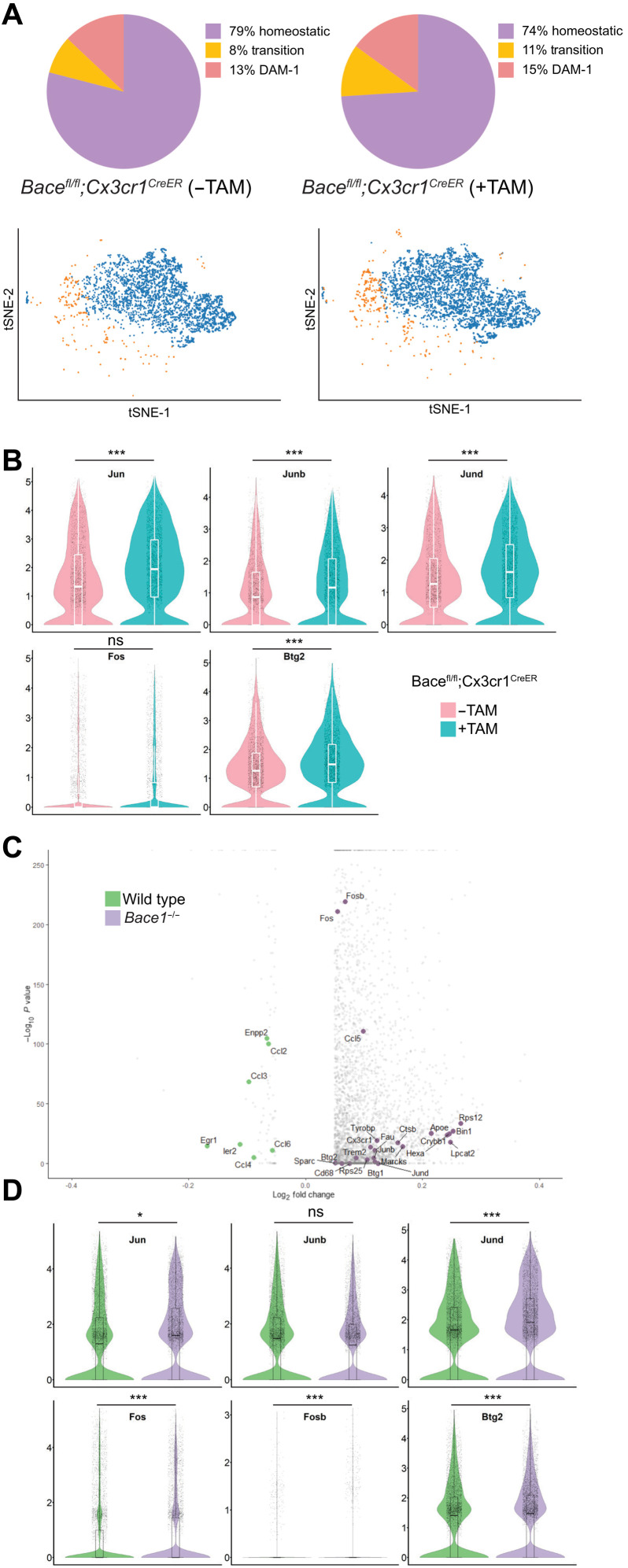
Since Bace-1 deletion enhances a unique set of TFs in 5xFAD mice, we asked whether this up-regulation is an intrinsic regulatory effect by BACE-1. We therefore analyzed scRNA-seq results of CD11b+ immune cells purified from 4-month-old Bace-1fl/fl;Cx3cr1CreER mice with and without TAM treatment. Again, we identified microglial cells based on canonical markers. Most cells fell into the homeostatic category since these mice were in the WT background (Fig. 2A). Bace-1 deletion in WT mouse microglia mainly increased the population of transitory microglia: ~11% versus ~8% (Fig. 2A). DAM-1 was slightly increased to 15% in TAM-treated Bace-1fl/fl;Cx3cr1CreER mice from ~13% in vehicle-treated Bace-1fl/fl;Cx3cr1CreER mice, while the DAM-2 population was minimal (0.2%) in both genotypes, consistent with the notion that DAM-2 is more related to amyloid deposition or the disease state. Among 74 significantly altered genes (fig. S2A; signature genes listed in table S2), TFs such as Jun, Junb, Jund, and Btg2 were significantly up-regulated, confirming that elevation of these genes correlates with the transition to activated microglia (Fig. 2B). This increased expression of TFs mentioned above was accompanied with significant increase in DAM-1 marker genes including Fth1, Rps12, Rps8, Rps29, and Lyz2. Moreover, we noted that mice with targeted Bace-1 deletion in microglia expressed higher levels of proinflammatory genes such as S100a8, S100a9, S100a6, Mmp-9, and Mmp-12 along with the significant reduction in homeostatic marker genes such as P2ry12, Hexb, and Sall1 (fig. S2B).
Fig. 2. Conditional Bace-1 deletion under a normal condition alters the microglial signature.
(A) tSNE clustering of CD11b-positive immune-sorted microglia derived from 4-month-old Bace-1flfl;Cx3cr1CreER with and without TAM for 5 days at 3 months of age (N = 3 each genotype). (B) Violin plots depict the distribution of log-transformed normalized gene expression of transcriptions factors under the control condition with and without conditional microglial Bace-1 deletion. There was significant up-regulation of the TF genes Jun, Junb, Jund, and Btg2 in Bace-1–targeted deletion mice, suggesting that Bace-1 deletion primes the microglia in an activated state in the WT background. (C) Volcano plot depicting the log2 fold changes in selected DEGs from scRNA-seq of microglia sorted from the brains of 2-month-old WT versus Bace-1–null mice (N = 3 mice per group). Elevated expression of TFs—such as Fosb, Fos, Ccl5, Jun, Junb, Jund, and Btg2—is shown in Bace-1–null mice. (D) Violin plots confirmed the elevated gene expression of transcriptions factors in the Bace-1–null microglia. The pattern of change in the microglial genes was consistent between conditional KO and Bace-1–null mice, and this excludes the potential artifact from the TAM treatment.
To exclude the effect of TAM treatment, we also conducted scRNA-seq of CD11b+ immune cells from Bace-1–null mice and WT control littermates at the age of 2 months (Fig. 2C). Again, expression of Fos, Fosb, Jun, Jund, and Btg2 was significantly elevated within the microglial population, and elevation of Jun and Jund was trending toward significance (Fig. 2C). Other significantly increased genes were Rps12, Rps25, S100a8, S100a9, Hspa1, Hba-a1, Retnlg, Mmp-9, Hif1α, PI3kK, Il-33, Il-1R2, Cd34, Cd74, and Clu (fig. S2C), also known as DAM-1 signature genes. While deletion of Bace-1 only in microglia caused no substantial increase in Apoe expression, its expression in Bace-1–null mice was significantly elevated (fig. S2C), likely resulting from effects of Bace-1 deletion in other cell types including neuronal Bace-1. Together, these scRNA-seq profiling experiments reveal the elevation of a unique set of TFs, intrinsically favoring a primed proinflammatory DAM-1 signature when Bace-1 in microglia is deleted.
Ubiquitous Bace-1 deletion in the 5xFAD mouse model alters the balance between homeostatic and DAM
With the knowledge that Bace-1 regulates TFs favoring the DAM-1 signature, we further examined microglia signatures in 5xFAD;Bace-1fl/fl/UbcCreER mice, which deleted Bace-1 in adult 5xFAD mice via the ubiquitin promoter. We previously showed that preformed amyloid plaques were removed in this model after Bace-1 was sequentially deleted beginning at postnatal day 30 (P30) (8). To understand whether DAM-1 signature was promoted in this model, we conducted scRNA-seq on CD11b+ immune cells derived from 14-month-old 5xFAD;Bace-1fl/fl/UbcCreER, 5xFAD;Bace-1fl/fl, and aged-matched WT controls; amyloid plaques in 14-month-old 5xFAD;Bace-1fl/fl/UbcCreER were barely detectable, consistent with our prior report (8). Deletion of Bace-1 in 14-month-old 5xFAD;Bace-1fl/fl/UbcCreER significantly reduced overall DAM populations if only homeostatic and DAM signatures were plotted (fig. S3A).
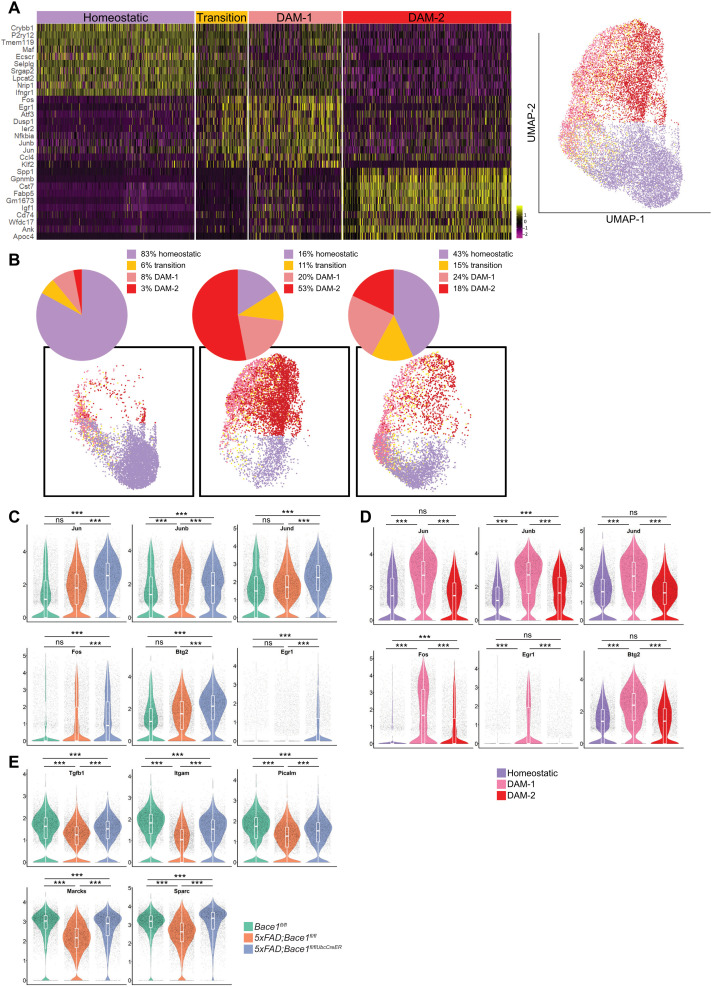
By further uniform manifold approximation and projection (UMAP) clustering of microglia signatures (Fig. 3A), we found that Bace-1 deletion in 5xFAD;Bace-1fl/fl/UbcCreER mice increased DAM-1 to ~24% and transition signature to ~15%, from ~20 and 11% in 5xFAD;Bace-1fl/fl mouse microglia, respectively. Note that both transition and DAM-1 were more in 5xFAD mice when compared to WT controls, which had no amyloid deposition (Fig. 3B). On the contrary, because of large accumulation of amyloid plaques, DAM-2 in 5xFAD;Bace-1fl/fl mice reached ~53%, while Bace-1 deletion reduced DAM-2 to ~18%. Considering the fact that WT controls had only ~3%, more DAM-2 in 5xFAD;Bace-1fl/fl/UbcCreER mice than in WT mice might reflect that either less DAM-1 transitioned to DAM-2 due to less amyloid deposition in the deletion model or a portion of DAM-2 was transitioned back to DAM-1 in response to clearance of amyloid plaques. Volcano plot showed large numbers of gene changes in 5xFAD with Bace-1 deletion (fig. S3B). In comparison to targeted deletion of Bace-1 in microglia (Fig. 1E), increased expression of transitory TFs such as Egr1, Fos, Btg2, Jun, Junb, and Jund was also evident in 5xFAD;Bace-1fl/fl/UbcCreER mouse microglia (Fig. 3C). Again, these genes were higher in DAM-1 compared to those in homeostatic microglia and were significantly higher compared to those in DAM-2 (Fig. 3D). Enhanced expression of this unique set of TFs in 5xFAD;Bace-1fl/fl/UbcCreER mice perhaps prevents the transition of DAM-1 to DAM-2. We also noted that anti-inflammatory genes and genes associated with phagocytosis such as Tgf-β1, Sparc, Picalm, Marcks, and Itgam were up-regulated in Bace-1–deleted 5xFAD;Bace-1fl/fl/UbcCreER mice compared to non–Bace-1 deletion and were comparable to those in WT mice (Fig. 3E). In line with the reduced DAM-2 signature, expression of DAM-2 genes such as Spp1, Gpnmb, Cst7, Axl, Lpl, and Igf1 was largely reduced, while homeostatic genes such as Cx3cr1, P2ry12, and Tmem119 were comparable to those in WT mice (fig. S3C). Expression of Trem2 and Apoe was reduced in DAM signatures, although total levels were elevated (fig. S3D).
Fig. 3. Ubiquitous BACE-1 deficiency promotes a pronounced microglial DAM-1 state during AD progression.
(A) Transcriptomic analysis, represented as UMAP, was performed on scRNA-seq of CD11b-positive microglia sorted from 14-month-old WT, 5xFAD, and 5xFAD;Bace-1fl/fl/UbcCreER mice (N = 3 per genotype). Heatmap showing four distinct microglia signatures based on subsets of genes. The color scale represents Z-score–transformed expression values (with yellow and purple representing up-regulated and down-regulated genes, respectively, compared with the mean expression value of a gene from all samples). (B) Pie chart showing the percentage of microglia expressing homeostatic, DAM-1, or DAM-2 signature genes in WT, 5xFAD, and 5xFAD;Bace-1fl/fl/UbcCreER mice. Four distinct colors represent each microglial cell cluster: homeostatic, transition, DAM-1–, and DAM-2–like states, depicted via combined UMAP clustering of CD11b-positive cells sorted from the cortex and hippocampus of WT, 5xFAD;Bace-1fl/fl, and 5xFAD;Bace-1fl/fl/UbcCreER mice. (C) Violin plots depict distribution of log-transformed normalized gene expression of unique sets of TFs in WT, 5xFAD;Bace-1fl/fl, and 5xFAD;Bace-1fl/fl/UbcCreER mice. (D) Violin plots show comparisons of specified TFs in the 5xFAD homeostatic, DAM-1, and DAM-2 states. (E) Violin plots compare the expression of homeostatic and phagocytosis genes such as Tgfb1, Itgam, Picalm, Marcks, and Sparc in three different genotypes of mice. These gene expressions were reduced in 5xFAD;Bace-1fl/fl while reversed when Bace-1 is deleted. Comparisons of expression levels of each specified gene between two groups were conducted (***P < 0.001).
We also compared TFs between microglia from 5xFAD mice at 2, 4, and 14 months of age and observed a significant increase in the abovementioned TFs at 2 months of age but reduced in the expression levels at 14 months (fig. S3, E and F), consistent with the notion that initial increase in these TFs in 2-month-old 5xFAD may favor the shift of homeostatic microglia to DAM-1. In contrast, lowered expression of TFs along with the expression of DAM-2 genes, such as Trem2 and Apoe, likely favors DAM-2 states, which suppress microglia to phagocytose amyloid plaques.
Lower expression of Bace-1 correlated with higher expression of TFs in human AD brains
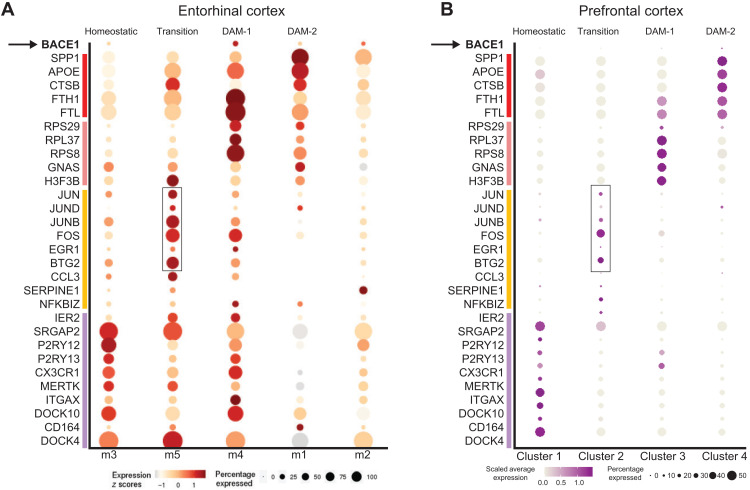
To examine the transitory microglia signature associated with the expression of the aforementioned TFs in human AD brains, we analyzed the open-source snRNA-seq data derived from the entorhinal cortex of 12 human samples (N = 6 AD patients with numerous diffuse and neuritic plaques, Braak stage VI, and N = 6 age-matched nondemented controls ranging from 72 to 90 years old) published by Grubman et al. (15). Using the originally analyzed dataset and Web interface provided (http://adsn.ddnetbio.com/), we examined the distribution pattern of the distinct sets of genes comparable to our scRNA-seq data derived from mouse models. In the original description by Grubman et al. (15), microglial cells were clustered into m1 to m5, with m1 and m2 expressing high levels of GWAS genes linked to late-onset AD such as APOE, APOC1, and STARD13 in m1 cluster while CSF3R and PTPRG in m2 cluster. We reanalyzed this dataset and found that the m1 cluster also expressed DAM-2–related genes such as GPNMB, SPP1, and LPL (Fig. 4A). The m2 cluster also expressed BIN1 and PICALM. The m3 cluster appeared to be more related to a homeostatic signature with the expression of well-defined homeostatic genes such as P2RY12, P2RY13, CX3CR1, and MERTK. Expression of BACE-1 was detectable in clusters m1 to m4 but not m5. m5 expressed high levels of transitory TFs such as BTG2, ERG1, FOS, JUN, JUNB, and JUND (as boxed in Fig. 4A), which were low in the DAM-2 clusters (m1 and m2). Early DAM-1 signature genes such as FTH1, FTL, RPS25, and RPS29 were specifically clustered into the m4 group (13). This human result is consistent with mouse dataset in Figs. 1 to 3, indicating that microglia may be favored in the transitory state when BACE-1 levels are low. Expression of BACE-1 is higher in microglia expressing DAM-2 like state (fig. S4).
Fig. 4. Low BACE-1 expression in human AD transitory microglia.
(A) Dot size and color are correlated with expression levels of each gene in various microglial clusters (1 to 5) derived from the single-nuclei (~13,000) postmortem entorhinal cortex tissue of AD and nondemented age-matched individuals (N = 12). Cluster m5 represents the transitioning state, having the highest expression of TFs (boxed) identified in our study. Correspondingly, BACE-1 levels were the lowest in the m5 cluster. (B) Dataset from snRNA-seq on single nuclei (~81,000) was extracted from the prefrontal cortices of 22 individuals (N = 11 patients with AD and N = 11 age-matched, nondemented controls). Typical genes for each of the four microglial subsets are highlighted with dots, with size and color in correlation with the expression level. Transitory microglia have the highest expression of these identified TFs.
We also examined another set of open-source AD human snRNA-seq data derived from prefrontal cortices of 22 individuals (N = 11 patients with AD and N = 11 age-matched, nondemented controls, the Rush AD Center) (12). Of 81,011 nuclei, we subclustered a total of 2625 microglia and were able to differentiate between homeostatic and DAM stage cells (Fig. 4B). We also identified a transitory microglia signature, which expressed higher levels of BTG2, ERG1, FOS, JUN, JUNB, and JUND as well as CCL3, CCL4, and SERPINE-1 (Fig. 4B), consistent with our aforementioned mouse results. Again, BACE-1 expression was only detectable in DAM-2, indicating that elevated BACE-1 activity is associated with DAM-2 signature.
Bace-1 deletion induces elevation of multiple signaling pathways toward more efficient phagocytosis
Since we conducted scRNA-seq on several mouse models to delete microglia Bace-1 in the 5xFAD background (4-month-old Bace-1fl/fl;Cx3cr1CreER with and without TAM treatment), we were able to determine a common set of differentially expressed genes (DEGs) within the microglial population under this condition. We found 45 genes down-regulated and 181 genes up-regulated (Fig. 5A). To determine which specific signaling pathways are altered as a result of Bace-1 deletion in microglia, we analyzed these DEGs using the Ingenuity Pathway Analysis (IPA) tool. We observed up-regulation of phosphatidylinositol 3-kinase (PI3K)/AKT, IL-6, TLR, p38 MAPK, and Rho, Rac, and downstream PAK family pathways (Fig. 5B).
We also conducted IPA of DEGs in 2-month-old Bace-1–null mice and found 45 canonical pathways (P < 0.05) significantly altered in comparison to WT littermates (fig. S5). Among these up-regulated pathways, we found that up-regulation of PI3K/AKT, TLR, p38 MAPK, and IL-6 signaling pathways was common under two different conditions. The commonly elevated pathways in both Bace-1–null mice and targeted deletion of Bace-1 in microglia indicate the intrinsic function of BACE-1 in microglia no matter whether it is under the WT or 5xFAD conditions.
In addition, we also noted commonly elevated pathways such as LXR/RXR, FXR/RXR activation, clathrin-mediated endocytosis, FcR signaling, and Rho guanosine triphosphatase (GTPase) signaling pathways. These elevations likely facilitated actin remodeling, phagosome maturation, phagocytosis, and clearance. Bace-1 deletion also up-regulated FcR-mediated phagocytosis and phagosome maturation, favoring efficient phagocytosis. Together, unbiased scRNA-seq experiments reveal changes in microglial genes and pathways in response to Bace-1 deletion, and these changes likely lead to more efficient microglial membrane ruffling and motility, phagosome maturation, and phagocytosis. In the protein functional network, we noted that TLR2 was the top gene that potentially mediated the expression of TFs such as Junb, Fos, and Erg1 (Fig. 5C).
Bace-1 upregulates phagocytosis of Aβ in primary microglia
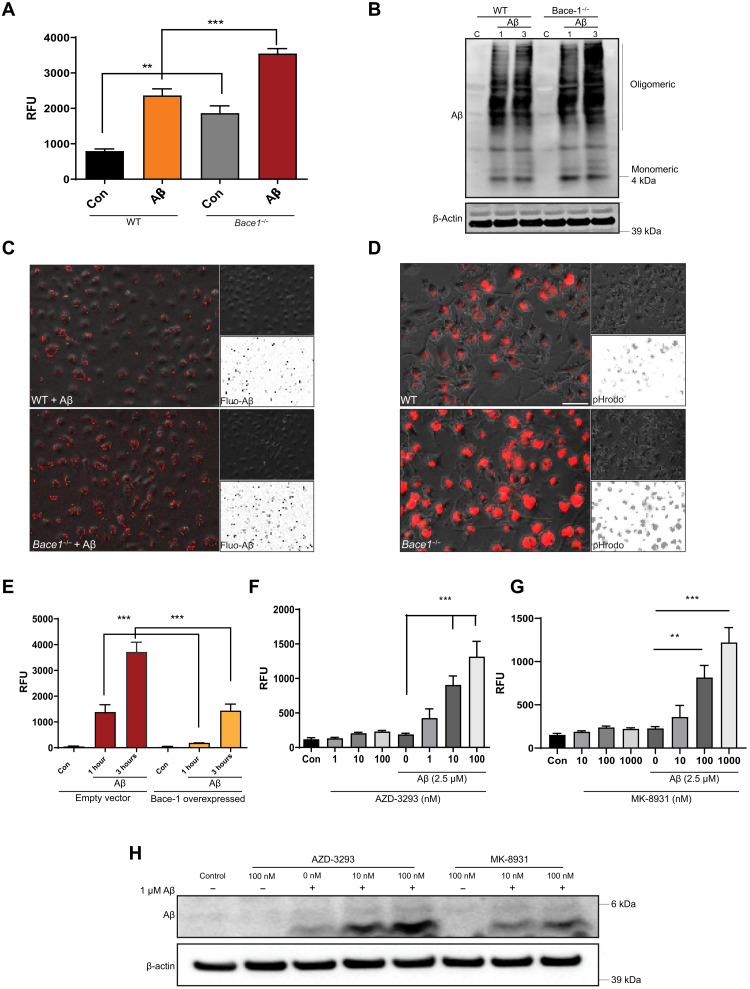
To determine whether Bace-1 deletion in microglia would enhance phagocytosis, we prepared primary microglial cultures from WT and Bace-1–null mice to test their uptake of fluorescent-tagged Aβ (fluo-Aβ) or pHrodo Red Escherichia coli BioParticles dyes (pHrodo E. coli, Thermo Fisher Scientific). A set of these cells was also pretreated with Aβ oligomers (nonfluorescently tagged) to mimic activation of microglia by aggregated Aβ in mice. After treatment, the phagocytized fluo-Aβ or pH-sensitive pHrodo E. coli was fluorometrically quantified on the basis of relative fluorescent units (RFU). In comparison to WT, Bace-1 deletion alone enhanced pHrodo E. coli uptake by ~2-fold (794 ± 53 versus 1866 ± 158 RFU; Fig. 6A); uptake of fluo-Aβ was increased by ~30% (111 ± 2.7 versus 143 ± 2.3 RFU; fig. S6A). After Aβ treatment, WT microglial cells were more active and visibly engulfed more pHrodo E. coli (2363 ± 187 RFU; Fig. 6A); Bace-1 deficiency led to increased uptake of ~50% more pHrodo E. coli (3550 ± 137 RFU; ***P < 0.001, Student’s t test). Immunoblotting results revealed that Bace-1 deletion visibly increased the uptake of not only monomeric but also high–molecular weight oligomeric Aβ1–42 (Fig. 6B). Quantification showed that Bace-1 deletion significantly enhanced Aβ1–42 uptake by ~33% at 1 hour and ~40% at 3 hours.
Fig. 6. Bace-1 deletion enhances phagocytic functions in microglia.
(A) Cultured primary microglia from WT and Bace-1–null (Bace-1−/−) microglia, with or without pretreatment with oligomerized Aβ1–42 (Aβ), were incubated with pHrodo E. coli particles for 1 to 3 hours, and the amount of phagocytized particles was fluorometrically determined (N = 6 independent experiments, ***P < 0.001 and **P < 0.01, Student’s t test). (B) Primary microglia were treated with Aβ for 1 and 3 hours, and the extent of Aβ phagocytosis was quantified via immunoblotting. Monoclonal 6E10 antibody was used to detect both monomeric and oligomeric Aβ. (C) WT and Bace-1–null BMDMs or Bace-1 KD BV-2 cells (D) were treated with fluorescent-tagged HiLyte 555–Aβ1–42 [fluo-Aβ in (C)] or pHrodo E. coli particles (D). Pictorial representation of fluorescence-emitting fluo-Aβ or phagocytized E. coli particles in lysosomes. Insets show black-white phase-contrast images for clearer comparisons. (E) BACE-1 overexpression suppresses fluorescently tagged HiLyte 555–Aβ1–42 uptake in BV-2 cells (N = 6, ***P < 0.001, Student’s t test). (F to H) BMDM cells were pretreated with BACE-1 inhibitor AZD-3293 (F) or MK-8931 (G) followed by incubation with pHrodo E. coli particles (N = 6, ***P < 0.001 and **P < 0.01, Student’s t test). The amount of phagocytized Aβ1–42 was quantified from the Western blot (H).
We also replicated this uptake experiment using BMDMs isolated and differentiated from WT and Bace-1–null mice (KO) and BV-2 cells. Expression of BACE-1 in BV-2 cells was knocked down (KD) using a CRISPR-Cas9–mediated ablation approach, and marked KD was demonstrated in fig. S6B. In these two cell types, Bace-1 KO or Bace-1 KD clearly increased uptake of either fluo-Aβ per unit area in BMDM (Fig. 6C) or BV-2 (Fig. 6D), respectively. Quantification is shown in fig. S6C (785 ± 130 versus 1892 ± 195 RFU) and fig. S6D (710 ± 102 versus 2087 ± 284 RFU). LysoTracker staining showed that more engulfed fluo-Aβ colocalized with acidic lysosomal compartments in Bace-1 KD BV-2 cells than in WT controls (fig. S6E). Conversely, increased expression of BACE-1 via lentivirus-mediated delivery in BV-2 cells delayed fluorescent Aβ1–42 uptake (Fig. 6E); uptake was slowed by ~7-fold (1385 ± 267 versus 190 ± 7 RFU) and ~2.5-fold (3718 ± 350 versus 1438 ± 235 RFU) at 1 and 3 hours after incubation, respectively.
To examine whether pharmacological inhibition of BACE-1 would also enhance microglial phagocytosis, we treated BMDM with the clinically used BACE-1 inhibitors lanabecestat (AZD-3293) (16) and verubecestat (MK-8931) (17) to evaluate phagocytosis. We first conducted a cell viability assay after treatment with AZD-3293 or MK-8931 in BMDM to determine the safe dose range. No significant toxicity was observed with up to 100 nM AZD-3293 (fig. S6F). The effect of BACE-1 inhibition on phagocytosis was therefore tested by preincubation of BMDM with 1 to 100 nM AZD-3293 for 2 hours, followed by cellular challenges with or without 2.5 μM Aβ for 1 hour. Following these treatments, cells were incubated with fresh medium containing pHrodo E. coli particles. Similar to the aforementioned Aβ treatment in inducing phagocytosis, AZD-3293 treatment increased the uptake of pHrodo E. coli particles in a dose-dependent manner: 1 nM by ~1-fold, 10 nM by ~4-fold, and 100 nM by ~6-fold (***P < 0.001; Fig. 6F). This increase was also validated in BV-2 cells pretreated with either 10 or 100 nM AZD-3293 (**P < 0.01 and ***P < 0.001; fig. S6G). Pretreatment with 1 μM or more of AZD-3293 suppressed pHrodo intake, mainly due to higher toxicity (fig. S6F).
In addition, BV-2 cell lysates treated with AZD-3293 or MK-8931 were immunoblotted to examine engulfed Aβ1–42. Consistent with our fluorometric assay, both BACE-1 inhibitors significantly increased Aβ1–42 uptake, as evidenced by the higher levels of 4.5-kDa monomeric Aβ1–42 present in cellular lysates (Fig. 6H). Together, these results indicate that higher microglial BACE-1 levels in AD brains may reduce Aβ phagocytosis, while reduced BACE-1 activity may enhance Aβ phagocytosis.
BACE-1 regulates PI3K-AKT-Rac1 activity through TLR/IL-1 signaling in microglia
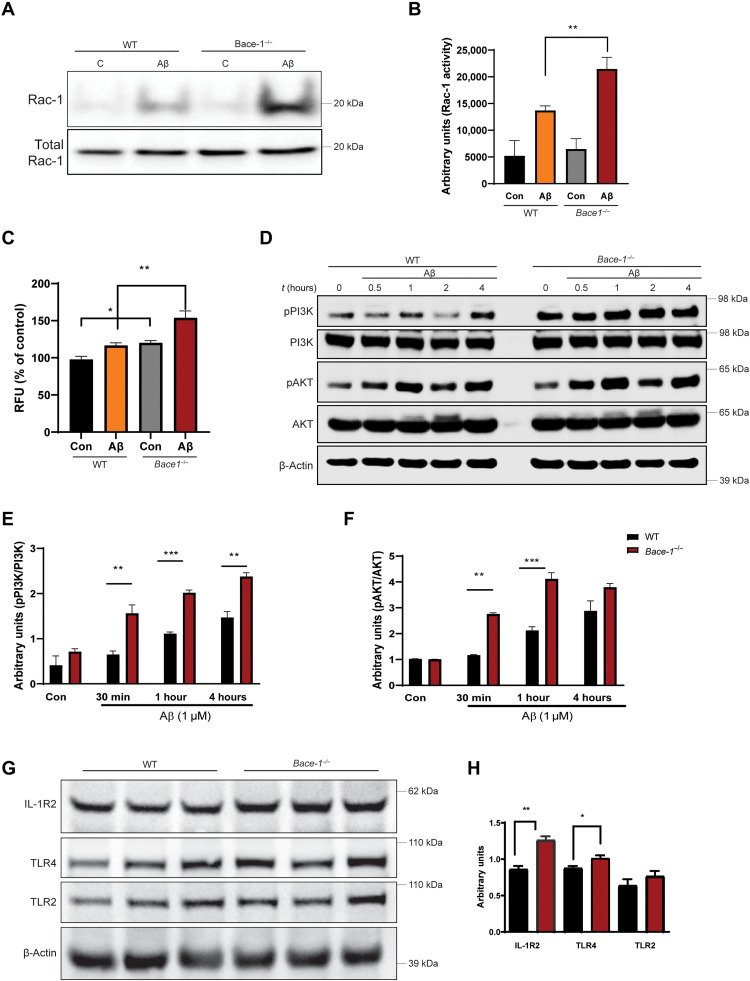
To explore the mechanism underlying enhanced phagocytosis by BACE-1–inhibited microglia, we asked whether BACE-1 regulates phagocytosis through actin remodeling. One noted feature of micro/macropinocytosis or phagocytosis is the dynamic extension and retraction of actin filaments, resulting in the formation of a phagocytic cup for internalization of particles (18). For example, FcR-mediated phagocytosis of Aβ oligomers and aggregates requires Rac-1, Rho, and CDC42-dependent actin remodeling and phagocytic cup formation (19). As shown in our pathway analysis (Fig. 5), BACE-1 deficiency increases FcγR, RhoA, actin cytoskeleton, and clathrin-mediated endocytosis signaling pathways. To test whether Bace-1 deletion would alter Rac-1 activity, we used BMDM cells derived from WT and Bace-1–null mice to measure Rac-1 protein levels and activity; Rac-1 activity was measured on the basis of Rac-1 pulldown activation assay (Cytoskeleton Inc). Compared to WT, Bace-1 deletion enhanced Rac-1 activity levels by ~57%; Aβ treatment induced significant up-regulation in Rac-1 levels as early as 30 min after Aβ treatment (13,698 ± 1337 versus 21,470 ± 3375 arbitrary units; Fig. 7, A and B). During phagocytosis, Rac-1 activity is known to regulate the generation of reactive oxygen species (ROS) by modulating the reduced form of nicotinamide adenine dinucleotide phosphate oxidase machinery (20). We therefore measured ROS generation in both WT and Bace-1–null BMDM. There was significant up-regulation in ROS levels in Bace-1–null BMDM (Fig. 7C), correlating with the elevated Rac-1 activity.
Fig. 7. BACE-1 signaling regulates Aβ uptake by modulating the PI3K-AKT signaling pathway.
(A and B) WT and Bace-1–null BMDM were treated with Aβ for 30 min, and Rac-1 activity was determined using the PBD pulldown assay, followed by immunoblotting with an anti–Rac-1 antibody. The bar graph in (B) shows elevation of Rac-1 activity in Bace-1–null BMDM (N = 3, **P < 0.01, Student’s t test). (C) Elevated ROS levels in WT and Bace-1–null BMDM measured spectrophotometrically by changes in DCFDA fluorescence 30 min after Aβ treatment. (D to F) Immunoblot panel and quantification for phosphorylated PI3K (pPI3K) and pAKT, as performed on lysates from WT and Bace-1–null BMDM treated with Aβ (1 μM) for the indicated time points. There was significant up-regulation of pPI3K and pAKT as early as 30 min after Aβ treatment (N = 3, **P < 0.01, Student’s t test). (G and H) Levels of full-length IL-1R2 and TLR4 in the cortex of Bace-1–null brains were elevated, but TLR2 levels were slightly increased (N = 3 independent experiments, **P < 0.01 and *P < 0.05, Student’s t test), likely related to abolished cleavage of these proteins by BACE-1.
PI3K-AKT signaling has been previously shown to activate Rac-1 in vivo (21, 22), and this pathway is elevated in Bace-1–deleted microglia (Fig. 5). We indeed observed significantly increased phosphorylation of both PI3K and AKT in Bace-1–null BMDM as early as 30 min after Aβ treatment (Fig. 7, D to F), in line with previous work showing PI3K-AKT as the upstream signal to Rac-1 activation. This elevated PI3K-AKT signaling was also seen in Bace-1 KD BV-2 cells (fig. S7, A to D). We further confirmed this PI3K-AKT activation in BV-2 cells pretreated with 100 nM AZD-3293 (fig. S7E). BACE-1 inhibition (1 hour of treatment) adequately induced elevation of pPI3K and peaked at 2.5 hours, independent of whether Aβ challenge was applied (**P < 0.01; fig. S7F). Consistently, the phosphorylated downstream molecule AKT (pAKT) was subsequently elevated, peaking at 5 hours after AZD-3293 treatment (fig. S7G). P38 activation appeared to peak at 2.5 hours after treatment (fig. S7H).
To understand how BACE-1 deficiency increases PI3K-AKT signaling in microglia, we examined changes in potential BACE-1 substrates that are richly expressed by microglia, including IL-1R (IL-1R2) and TLRs, as described in a previous publication (23). We reasoned that BACE-1 deficiency or inhibition would suppress cleavage of these substrates by potentially augmenting their signal transduction activities. Specifically, the type 1 transmembrane substrate IL-1R2 was shown to be expressed by immune cells in response to proinflammatory stimuli such as Aβ oligomers (24) and processed at extracellular sites by α-, β-, and γ-secretases (25). We found a significant increase in IL-1R2 levels in the cortical lysates from Bace-1–null mice compared to WT controls (**P < 0.01; Fig. 7, G and H). A smaller increase in another potential BACE-1 substrate, TLR4, was also observed (*P < 0.05; Fig. 7, G and H). TLR2 protein levels were slightly increased. Considering that TLR2 is one of the top proteins in the functional network (Fig. 5C) and IL-1r2 and Tlr4 signaling have long been shown to activate PI3K signaling in various cells (26, 27), we speculated that abolished cleavages of IL-1r2 and Tlr4 might contribute to Bace1-mediated activation of PI3K-AKT-Rac1 in microglia from Bace-1–null mice or inhibited cells.
To support this further, we examined the effects of Bace-1 deletion on downstream signaling in response to proinflammatory IL-1β and Aβ treatments. While the p65 and ERK MAPK pathways remained unaltered, BACE-1 deficiency significantly induced p38 MAPK phosphorylation as early as 5 min after IL-1β treatment in Bace-1−/− microglia (*P < 0.05 and **P < 0.01; fig. S7, I and J). Likewise, BACE-1 deficiency significantly augmented Aβ-induced p38 MAPK phosphorylation (*P < 0.05 and **P < 0.01; fig. S7, K and L). Together, our study highlights BACE-1 as a critical signaling protein in regulating microglial functions through the PI3K and P38 pathways.
DISCUSSION
BACE-1 is widely recognized as a neuronal protein due to its robust expression by neurons, and its role in microglia has therefore been underexplored. In this study, we took advantage of scRNA-seq to analyze gene changes in microglia when Bace-1 is deleted under normal conditions or in the 5xFAD background. Our comprehensive analysis was able to reveal distinct populations of microglia as they transformed from homeostatic to DAM-1 and DAM-2 states. We demonstrated that genetic deletion of microglial Bace-1 in mice up-regulated a set of TFs that are important for maintaining or promoting the DAM-1 signature. This observation is novel and has not explicitly been revealed in previous studies. Our results show that BACE-1 normally controls the balanced expression of these genes in microglia and that targeted inhibition of BACE-1 is a viable approach to induce their up-regulation for enhancing the phagocytic functions of microglia.
In WT mouse brains, microglia are mostly in the homeostatic stage, which is recognized as cluster 1 by scRNA-seq (fig. S3A). Microglial Bace-1 deletion under the WT condition increased DAM-1 from ~13 to ~15%, and an obvious change was the transitory microglia, which increased from 8 to 11% (Fig. 2A). Under the 5xFAD condition, amyloid deposition would increase the overall DAM signatures of 14-month-old 5xFAD;Bace-1fl/fl mice (fig. S3B), mostly the DAM-2 signature (3 to ~53% in Fig. 3A). Notably, Bace-1 deletion in 5xFAD;Bace-1fl/fl;UbcCreER microglia had ~18% DAM-2, while it increased DAM-1 from ~20 to ~24%. Similarly, specific deletion of Bace-1 in microglia increased DAM-1 from ~23 to ~31% (Fig. 1D). The emergence of transitory and DAM-1 may reflect a protective mechanism, presumably to have higher capacity of phagocytosis or to remove an insult like Aβ deposition. This shift to transitory signature is more obvious in 14-month-old 5xFAD;Bace-1fl/fl mouse brains, in which amyloid deposition may facilitate transitory signature to around 11%, compared to ~6% in WT mice (Fig. 3B); Bace-1 deletion further increased this population to ~15% in these 14-month-old 5xFAD;Bace-1fl/fl;UbcCreER mice.
High levels of the transitory and DAM-1 signatures are correlative to increased expression of a set of transcriptional factors such as Jun, Junb, Jund, Egr1, Fos, and Btg2 (Figs. 1E, 2C, and 3E). We noted that in young 5xFAD mice (i.e., 2-month-old 5xFAD;Bace-1fl/fl mice), these TFs began to be up-regulated in microglia, likely in response to the initial amyloid deposition; nadirs of amyloid plaques were just beginning to form at this time point (14). This is consistent with the shift of homeostatic microglia to DAM-1 for phagocytosing the produced amyloid plaques, which are morphologically detectable in contacting Aβ plaques (28–30). As amyloid pathology progresses, elevated Trem2 may induce a transition from DAM-1 to DAM-2 (11, 31). DAM-2 likely becomes dysfunctional in phagocytosis and cannot arrest the growth of amyloid plaques as they increase in number and size in 5xFAD mice. We inferred that enhanced transitory/DAM-1 signatures while reduced DAM-2 will reduce amyloid plaques because more healthy microglia are able to clear away amyloid plaques. In support of this notion, we also noted increased expression of proinflammatory genes such as S100a8, S00a9, and Hspa1a (fig. S2), which may also facilitate phagocytosis.
Trem2 has been viewed as an inducer for the transition from DAM-1 to DAM-2 (11). We found that Trem2 expression was significantly increased in Bace-1–null microglia but slightly reduced if it was deleted only in 5xFAD microglia fig. S3D. Comparing 14-month-old 5xFAD;Bace-1fl/fl;UbcCreER mice with 5xFAD;Bace-1fl/fl mice, Trem2 expression was reduced, mainly in DAM signature (fig. S3D), indicating that Bace-1 deletion induces less DAM-1 being transited to DAM-2. Expression of Apoe had a largely similar pattern to Trem2, reduced mainly in DAM signature but higher in the homeostatic state (fig. S3D). Apoe expression was the highest in DAM-2 (Fig. 1B). It is likely that slightly higher APOE may facilitate microglia phagocytic function, while significantly high level of APOE correlated with impaired DAM-2. Bace-1 deletion slightly reduced Apoe, and this reduction was mainly seen in DAM while elevated in homeostatic cluster 1 (fig. S3D). Increased APOE in homeostatic and DAM-1 microglia is likely a beneficial event as previously implied (32). In non-AD background, the scRNA-seq results also support such speculation, as Bace-1–null mice show elevated expression of microglial genes such as Apoe (fig. S2C). Elevated APOE expression promotes protein clearance. In AD brains, more APOE in microglia will facilitate Aβ trafficking to lysosomes for degradation (33), and functional APOE isoform, ApoE3 in induced pluripotent stem cells (iPSC)–derived microglia, can attenuate multiple AD-related pathologies (34).
How BACE-1 controls the expression of TFs such as Jun, Fos, and Junb is intriguing. Pathway analyses showed increased TLRs, p38 MAPK, and PI3K signaling pathways (Fig. 5, B and C). We indeed found that BACE-1 increased protein levels of TLR proteins TLR2 and TLR4, as well as IL-1R2 (Fig. 7G). These three receptor molecules are type I transmembrane receptors and are likely cleaved by transmembrane BACE-1 (25, 35). BACE-1 inhibition or deficiency will decrease their cleavages, and more full-length receptor molecules will be available for inducing signaling to the downstream molecules, including TFs such as Jun, Junb, and Fos as illustrated in Fig. 8. More relevantly, we found that deletion of microglial Bace-1 also induced LXR/RXR, Fcγ, RhoA, and Rac signaling pathways in either WT or 5xFAD background. We measured Rac-1 activity and protein levels and found only a slight increase in cultured Bace-1–null microglia compared to the WT controls (Fig. 7, A and B). Rac-1 activity was induced under the AD condition, as Aβ could mediate this induction. Under this induced condition, Bace-1 deficiency appeared to augment its expression. This also appeared to be consistent with IPA pathway analyses: Rac signaling is up-regulated in TAM-treated 4-month-old 5xFAD;Bace-1fl/fl;Cx3cr1CreER mice, compared to TAM-treated 4-month-old Bace-1fl/fl;Cx3cr1CreER mice. This up-regulation may facilitate the phagocytic function of microglia. Our biochemical assays confirmed that Bace-1 deficiency or inhibition in cultured microglia enhances phagocytosis (Fig. 6).
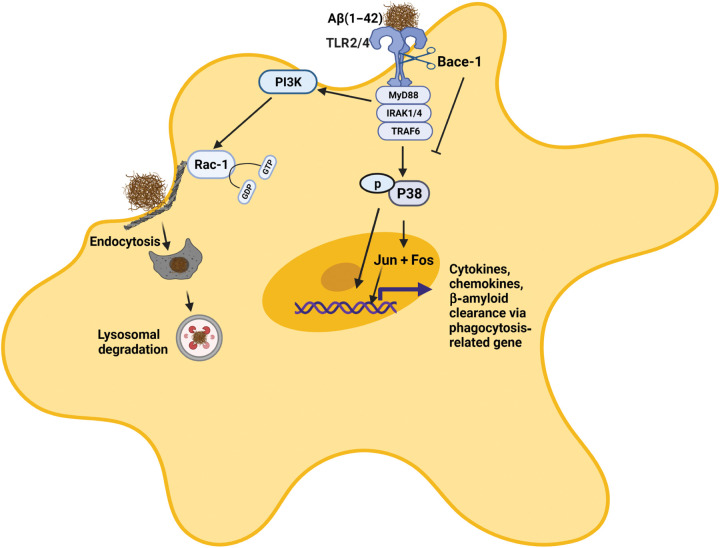
Fig. 8. BACE-1 regulates expression of TFs through TLR2/4 and IL-1R2.
BACE-1 regulates the neuroinflammatory response through signaling from TLRs and IL-1R2, which are a potential BACE-1 substrate. The abolished or inhibited cleavage of these type 1 transmembrane proteins by BACE-1 will likely increase its signaling activity. Therefore, BACE-1 deletion or inhibition enhances TLR2 or TLR4 and IL-1β signaling activity, which further increases the subsequent phosphorylation of the downstream molecule p38 MAPK. Activated MAPK will increase phosphorylation and nuclear translocation of TFs such as Jun and Fos. In addition, BACE-1 also regulates Aβ-induced PI3K and downstream Rac-1 signaling associated with actin remodeling, endocytosis, and phagocytosis. Activation of these signaling pathways may also facilitate lysosomal functions.
Bace-1–null mice exhibited phenotypes such as hypomyelination, spontaneous seizures, decreased neurogenesis, and reduced LTP (36). Expression of these molecules is not detectable in microglia (based on our RNA-seq results), and the effects of these molecules in microglia are minimally affected. Microglia have been shown to regulate synaptic plasticity in multiple ways (37). It will be exciting to find out whether increased transitory and DAM-1 will prime microglia for enhancing synaptic function. Collectively, our study demonstrates a unique role of BACE-1 in regulating microglial gene expressions, likely through the cleavage of its substrates TLR2, TLR4 and IL-1R2 (illustration in Fig. 8). Targeted inhibition or deletion of BACE-1 in microglia has the potential to change microglial functions toward more beneficial states in patients with AD.
MATERIALS AND METHODS
Cell culture
The immortalized BV-2 microglial cell line and primary microglia were grown in RPMI 1640 and Dulbecco’s modified Eagle’s medium (DMEM)–F12 medium, respectively, containing 10% fetal bovine serum, 2 mM l-glutamine, 50 U of penicillin, and streptomycin (50 μg/ml). Cells were grown in a humidified atmosphere of 5% CO2 at 37°C. The cells were then monitored for growth, and the medium was replaced at least every 2 days.
Primary microglial culture and isolation
Primary mixed neuroglial cultures were prepared from P1 to P2 mouse pups as described previously (38). Briefly, meninges and blood vessels were removed from the cortex. Brain tissue was gently triturated, and a single-cell suspension was cultured in a poly-d-lysine–coated T75 flask in DMEM-F12 containing 10% heat-inactivated fetal bovine serum, 2 mM l-glutamine, and 1% penicillin/streptomycin (Life Technologies) for 12 to 14 days. Microglia were separated from mixed glial cell cultures using either a shake-off method (180 rpm for 2 hours) or via a magnetic separation kit (EasyStep Mouse Cd11b-positive selection kit) from STEMCELL Technologies.
Preparation of fibrillar and soluble Aβ peptides
Peptides were solubilized in 0.1% NH4OH containing 0.01% (w/v) NaN3 and further resuspended in sodium phosphate buffer (pH 7.4). Peptides were oligomerized by incubation with constant rotation for 1 to 7 days at 37°C using an Innova 40 incubator shaker (New Brunswick). To ensure the presence of predominantly monomeric form, lyophilized fluorescence peptides were suspended in 0.01% NH4OH. These solutions were stored at −20°C. Each aliquot was thawed only once immediately before the experiment.
ROS measurement
Intracellular ROS levels were detected by the fluorescent probe chloromethyl-2′,7′-dichlorofluorescein diacetate (CM-H2DCFDA; Molecular Probes, Eugene, OR). Microglial cells were seeded in 96-well plates at a confluence of 40,000 cells per well. Cells were loaded with 10 μM CM-DCFDA dye at 37°C for 30 min in the dark, then extracellular dye was washed, and cells were treated with Aβ for 30 min. The formation of fluorescent product, dichlorofluorescin (DCF), was analyzed using a fluorescence spectrometer with excitation and emission wavelengths of 488 and 525 nm, respectively. Data are represented as RFU.
Cytotoxicity assay
Cell viability was determined using the MTS [3-(4,5-dimethylthiazol-2-yl)-5-(3-carboxymethoxyphenyl)-2-(4-sulfophenyl)-2H-tetrazolium] assay protocol (39). BV-2 cells were grown on a 96-well plate with approximately 20,000 cells per well. The cells were treated with AZD-3293 for another 20 hours. MTS was added in all the treatment groups for another 45 min at 37°C. The amount of soluble formazan, an indicator of cell viability, was quantified spectrophotometrically at an absorbance wavelength of 490 nm.
Aβ detection
To quantify intracellular HiLyte Fluor 555 Aβ1–42 uptake after incubation, the cells were washed and fixed with 4% paraformaldehyde. Images were taken via a BZ-X810 Keyence fluorescence microscope displaying the intracellular Aβ42 in transfected BV-2 or mouse primary microglial cells. The area corresponding to intracellular Aβ42 was traced using a bright-field overlay image, and the average intensity of the signal was determined within this area via ImageJ software. Data are represented as percentage of area covered with Aβ1–42.
Western blotting
After Aβ or compound treatment, the BV-2 or microglia were washed twice with ice-cold phosphate-buffered saline (PBS) and lysed on ice in radioimmunoprecipitation assay (RIPA) lysis buffer containing 50 mM tris-HCl (pH 7.4), 1 mM EDTA, 100 mM NaCl, 0.1% SDS, 1 mM phenylmethylsulfonyl fluoride (PMSF), 1 mM sodium orthovanadate, leupeptin (1 μg/ml), pepstatin (1 μg/ml), and aprotinin (10 μg/ml), for 5 min. The lysate was collected and further sonicated on ice for 30 s on and off cycle for 5 min and then centrifuged at 15,000g for 15 min at 4°C. Protein concentrations were determined using a BCA assay kit (Pierce). Equal amounts of protein from each sample were loaded and electrophoretically resolved on 4 to 12% SDS–polyacrylamide gel electrophoresis (NuPAGE system, Life Technologies) gels. After electrophoresis, proteins were transferred to nitrocellulose membranes at 100 V for 2 hours. The membranes were blocked with 5% bovine serum albumin for 1 hour at room temperature. The membranes were probed with primary antibody (see the list in Table 1), followed by incubation with secondary fluorescence-labeled antibody (1:3000). The antibody-bound proteins were detected by iBright 1500 imaging system (Invitrogen). To ensure equal loading, the blots were reprobed with monoclonal anti-actin (1:1000). For quantification purposes, band intensities of immunoblots were analyzed using ImageJ software.
Table 1. The list of antibodies used for the study.
| Antibody name | Catalog no. | RRID: | |
| P-AKT(Ser473) | 9271 | AB_329825 | Cell Signaling |
| β-Amyloid (1–42 specific) (D9A3A) | 14974 | AB_2798671 | Cell Signaling |
| β-Actin | A5441 | AB_476744 | Sigma-Aldrich |
| β-Amyloid 1–16, 6E10 | 803003 | AB_2564652 | BioLegend |
| TREM2 | ab86491 | AB_1925525 | Abcam |
| TLR4 | sc-293072 | AB_10611320 | Santa Cruz |
| BACE1 (D10E5) | 5606 | AB_1903900 | Cell Signaling Inc. |
| Iba1 | 019-19741 | AB_839504 | Wako |
| IL-1 RII | AF563 | AB_2125055 | R and D Systems |
| Rac-1 | ARC03 | AB_2721173 | Cytoskeleton Inc. |
| p38-P-MAPK(Thr180/Tyr182)(D3F9) | 4511 | AB_2139682 | Cell Signaling Inc. |
| p38 MAPK | 8690 | AB_10999090 | Cell Signaling Inc. |
| PI3-Kinase-pp85 | 4228 | 659940 | Cell Signaling Inc. |
| TLR2 | 12276 | AB_2797867 | Cell Signaling Inc. |
| AKT | 4691 | AB_329827 | Cell Signaling Inc. |
Single-cell RNA sequencing
Microglia were isolated from mouse brains using adult brain dissociation kits (MiltenyiBiotec, catalog no. 130-107-677). Briefly, forebrain was dissociated into a single-cell suspension; myelin, cell debris, and erythrocytes were removed subsequently; and cells were immunolabeled with CD11b microbeads (catalog no. 130-093-634). The cell suspension was allowed to pass though the magnetic column and retained CD11b-positive cells from the column, which were flushed out and used for analysis. Microglia were loaded into capture plates (a droplet-based 10x Chromium controller) to perform single-cell partitioning, and mRNA was barcoded and subsequently converted into cDNA. After library quality inspection, libraries were sequenced using an Illumina Nextseq 500 sequencer. Raw sequencing was aligned and annotated using the CellRanger v3.1.0 pipeline. During FASTQ generation, reads with more than one mismatch in the 8-bp i7 index were excluded. During alignment using STAR (40), only reads with MAPping Quality (MAPQ) scores greater than 255 aligned to annotated transcripts were retained. Reads containing bases with Q30 scores below 3 were also excluded. After alignment, cell barcodes were filtered up to one mismatch against a whitelist of 737,500 barcodes provided by 10x Genomics. Barcodes associated with cells were distinguished from ambient mRNA using an adaptively computed Unique Molecular Identifier (UMI) threshold. The raw count matrix was filtered using cutoff values of mitochondrial transcripts below 10% and 600 to 6500 unique features.
Dimensionality reduction and clustering
The expression profiles of each cell using the 2000 most variable genes as measured by dispersion (41, 42) were used for neighborhood graph generation and dimensionality reduction with t-distributed stochastic neighbour embedding (tSNE) or UMAP (43). Clustering was performed on this neighborhood graph using the Leiden community detection algorithm (44). Because the experiments consisted of multiple samples, the neighborhood graph was batch-corrected using the batch correction software BBKNN (45). Subclustering and differential expression were performed ad hoc on a per-cluster basis using the Seurat R toolkit v4.0 (46).
Generation of microglial Bace-1 conditional KO mice
TAM-inducible microglia-specific Bace-1 deletion was achieved by breeding microglia-specific B6.129P2(Cg)Cx3cr1tm2.1(cre/ERT2)Litt/WganJ (JAX stock #020940, The Jackson Laboratory) with Bace-1 conditional mice (Bace-1fl/fl) carrying loxP-flanked genes as previously described (8). The heterozygous mice were further bred to obtain a colony with the following genotype: Cx3cr1Cre/ER;Bace-1fl/fl. At the age of 2 months, the Bace-1 deletion in microglia was initiated by injecting TAM intraperitoneally at 100 mg/kg for five consecutive days. The level of Bace-1 deletion was confirmed by isolating microglia, and the extent of Bace-1 deletion was examined by immunoblotting.
Rac-1 GTPase activation assay
The GTPase activity of Rac1 was measured using a Rac1 Activation Assay Biochem Kit (Cytoskeleton Inc., catalog no. BK035). After AB treatment, BMDMs were washed with ice-cold PBS and lysed in RIPA buffer containing protease inhibitors. Lysates were cleared by centrifugation (10,000g for 1 min), and protein concentration was measured via a BCA assay kit. Equal amounts of protein (500 μg) were incubated with 50 μg of glutathione S-transferase (GST) fusion protein of Cdc42- and Rac-interactive binding (CRIB)–binding domain of p21-activated kinase (PAK) [GST-Rac/Cdc42 (p21) binding domain (PBD)] bound to glutathione-Sepharose beads for 60 min at 4°C. Active Rac1 was pulled down/precipitated with beads following brief centrifugation. Beads were gently washed three times in ice-cold buffer [50 mM tris-HCl (pH 7.6), 500 mM NaCl, 1% Triton X-100, 0.5 mM MgCl2, 1 mM PMSF, leupeptin (10 μg/ml), and aprotinin (10 μg/ml)] to remove unbound guanosine diphosphate–tagged inactive Rac-1 proteins. Rac-1 proteins were eluted with 2× Laemmli sample buffer. The amount of active Rac1 was analyzed by immunoblotting.
CRISPR null of BACE1 in BV-2 cells
Guide RNA (gRNA) was designed to specifically cut in the second exon of the Bace-1 gene in the mouse genome (Ensembl sequences ENSMUSG00000032086). gRNA was designed for high specificity and to guard against genome-wide off-target effects using https://chopchop.cbu.uib.no/. Oligos containing the gRNA sequence were cloned into lentiCRISPR v2 (Addgene plasmid) (47). Scramble gRNA was also designed to be used as a control in this study. gRNAs were designed to bind to the genome in a position flanked by a NGG at the 3′ end. Oligos were ordered and analyzed by the OligoAnalyzer tool Integrated DNA Technologies (IDT) and phosphorylated/annealed. The annealed gRNA oligos were cloned into the lentiCRISPRv2 plasmid digested with BSMB1 to create the appropriate overhangs.
gRNA for Bace-1 was 5′-TCCTGCATCGCTACTACCAG. gRNA for scramble was 5′-CAGTCGGGCGTCATCATGAT.
Plasmids were sequenced to verify the correct insertion of gRNAs. Lentivirus was made using the packaging plasmids psPAX2 and pMD2.G (Addgene plasmids #12260 and #12259; gift of D. Trono) into human embryonic kidney 293FT cells using Lipofectamine 2000 (Thermo Fisher Scientific). Virus was harvested at 48 hours after transfection.
BV-2 cells were plated and transduced by the lentivirus. Forty-eight hours after transduction, puromycin was added to the culture at 4 ng/μl to select for lentiCRISPR integration. After selection, a group of bulk cells was tested using a T7E1 assay to verify the creation of insertions or deletions (INDELs). Once INDELs were verified, cells were plated sparsely as single cells and allowed to create colonies. Colonies were isolated into 24-well plates and allowed to expand. The plate was duplicated, one set of wells was lysed, and genomic DNA was isolated. The region of CRISPR binding was amplified by polymerase chain reaction (PCR) and sequenced by Sanger sequencing. Sequences were analyzed to identify frameshifts in exon 2. Cells with different INDELs on each allele were analyzed using TIDE analysis 2 using chromatograms of the edited samples comparing to the WT cells.
Clones that showed frameshift INDELs on both alleles leading to nonsense and early termination were expanded for use in this study. Primers used for analysis were as follows: Bace-1 mouse ex2 PCR forward, GACGATCAGGTGACAGGAAA; BACE mouse ex2 pcr reverse; BACE1 mouse exon 2 PCR forward, 5-GACGATCAGGTGACAGGAAA-3; BACE mouse ex 2 PCR reverse, 5-TGGTTCATGTTCTGCTCTGG-3; BACE mouse exon2 seq forward, 5-ACAGACAGACGCAAGTGCAG-3.
Statistical analysis
Results are expressed as means ± SEM. The statistical analyses were performed using GraphPad Prism 6.0 software (GraphPad Software, San Diego). Student’s t tests were used to compare between two groups. Multiple group analyses were performed by one-way analysis of variance followed by Tukey’s post hoc test. Differential expression between conditions in the scRNA-seq dataset was analyzed via Wilcoxon rank sum test. Differences with *P < 0.05, **P < 0.01, and ***P < 0.001 were considered significant.
Acknowledgments
We thank the Bioinformatics Core at UConn Health and P. Robson at The Jackson Laboratory for support in RNA-seq experiment and analyses.
Funding: R.Y. is supported by grants RF1AG058261, AG025493, NS074256, and AG046929 from the NIH. Yan’s laboratory is also supported by the Cure Alzheimer’s Fund. B.D. is supported by a postdoctoral fellowship from BrightFocus Foundation (A20201729F).
Author contributions: N.S. and R.Y. designed and initiated this study. N.S. conducted most of the experiments and wrote the draft; N.S. and M.R.B. conducted the analysis of RNA-seq results; J.Z. and B.D. conducted partial experiments; X.H. helped in some data analysis; J.D.-V., L.-H.T., and M.K. helped with data analysis of human samples; M.R.B. also helped in data submission and wrote the data analysis part; R.Y. supervised the project and wrote the manuscript.
Competing interests: The authors declare that they have no competing interests.
Data and materials availability: All data needed to evaluate the conclusions in the paper are present in the paper and/or the Supplementary Materials. All mouse RNA-seq data present in the paper have been uploaded to the NCBI Gene Expression Omnibus (GEO) at the following accession number GSE199027.
Supplementary Materials
This PDF file includes:
Figs. S1 to S7
Other Supplementary Material for this manuscript includes the following:
Tables S1 and S2
Data S1
REFERENCES AND NOTES
- 1.Corriveau R. A., Koroshetz W. J., Gladman J. T., Jeon S., Babcock D., Bennett D. A., Carmichael S. T., Dickinson S. L., Dickson D. W., Emr M., Fillit H., Greenberg S. M., Hutton M. L., Knopman D. S., Manly J. J., Marder K. S., Moy C. S., Phelps C. H., Scott P. A., Seeley W. W., Sieber B. A., Silverberg N. B., Sutherland M. L., Taylor A., Torborg C. L., Waddy S. P., Gubitz A. K., Holtzman D. M., Alzheimer’s disease–related dementias summit 2016: National research priorities. Neurology 89, 2381–2391 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Cai H., Wang Y., McCarthy D., Wen H., Borchelt D. R., Price D. L., Wong P. C., BACE1 is the major β-secretase for generation of Aβ peptides by neurons. Nat. Neurosci. 4, 233–234 (2001). [DOI] [PubMed] [Google Scholar]
- 3.Luo Y., Bolon B., Kahn S., Bennett B. D., Babu-Khan S., Denis P., Fan W., Kha H., Zhang J., Gong Y., Martin L., Louis J. C., Yan Q., Richards W. G., Citron M., Vassar R., Mice deficient in BACE1, the Alzheimer’s beta-secretase, have normal phenotype and abolished beta-amyloid generation. Nat. Neurosci. 4, 231–232 (2001). [DOI] [PubMed] [Google Scholar]
- 4.Roberds S. L., Anderson J., Basi G., Bienkowski M. J., Branstetter D. G., Chen K. S., Freedman S. B., Frigon N. L., Games D., Hu K., Johnson-Wood K., Kappenman K. E., Kawabe T. T., Kola I., Kuehn R., Lee M., Liu W., Motter R., Nichols N. F., Power M., Robertson D. W., Schenk D., Schoor M., Shopp G. M., Shuck M. E., Sinha S., Svensson K. A., Tatsuno G., Tintrup H., Wijsman J., Wright S., McConlogue L., BACE knockout mice are healthy despite lacking the primary beta-secretase activity in brain: Implications for Alzheimer’s disease therapeutics. Hum. Mol. Genet. 10, 1317–1324 (2001). [DOI] [PubMed] [Google Scholar]
- 5.Egan M. F., Kost J., Voss T., Mukai Y., Aisen P. S., Cummings J. L., Tariot P. N., Vellas B., van Dyck C. H., Boada M., Zhang Y., Li W., Furtek C., Mahoney E., Harper Mozley L., Mo Y., Sur C., Michelson D., Randomized trial of verubecestat for prodromal alzheimer’s disease. N. Engl. J. Med. 380, 1408–1420 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wessels A. M., Tariot P. N., Zimmer J. A., Selzler K. J., Bragg S. M., Andersen S. W., Landry J., Krull J. H., Downing A. M., Willis B. A., Shcherbinin S., Mullen J., Barker P., Schumi J., Shering C., Matthews B. R., Stern R. A., Vellas B., Cohen S., MacSweeney E., Boada M., Sims J. R., Efficacy and safety of lanabecestat for treatment of early and mild alzheimer disease: The AMARANTH and DAYBREAK-ALZ randomized clinical trials. JAMA Neurol. 77, 199–209 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Imbimbo B. P., Watling M., Investigational BACE inhibitors for the treatment of Alzheimer’s disease. Expert Opin. Investig. Drugs 28, 967–975 (2019). [DOI] [PubMed] [Google Scholar]
- 8.Hu X., Das B., Hou H., He W., Yan R., BACE1 deletion in the adult mouse reverses preformed amyloid deposition and improves cognitive functions. J. Exp. Med. 215, 927–940 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tansey K. E., Cameron D., Hill M. J., Genetic risk for Alzheimer’s disease is concentrated in specific macrophage and microglial transcriptional networks. Genome Med. 10, 14 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Nott A., Holtman I. R., Coufal N. G., Schlachetzki J. C. M., Yu M., Hu R., Han C. Z., Pena M., Xiao J., Wu Y., Keulen Z., Pasillas M. P., O’Connor C., Nickl C. K., Schafer S. T., Shen Z., Rissman R. A., Brewer J. B., Gosselin D., Gonda D. D., Levy M. L., Rosenfeld M. G., McVicker G., Gage F. H., Ren B., Glass C. K., Brain cell type-specific enhancer-promoter interactome maps and disease-risk association. Science 366, 1134–1139 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Deczkowska A., Keren-Shaul H., Weiner A., Colonna M., Schwartz M., Amit I., Disease-associated microglia: A universal immune sensor of neurodegeneration. Cell 173, 1073–1081 (2018). [DOI] [PubMed] [Google Scholar]
- 12.Zhou Y., Song W. M., Andhey P. S., Swain A., Levy T., Miller K. R., Poliani P. L., Cominelli M., Grover S., Gilfillan S., Cella M., Ulland T. K., Zaitsev K., Miyashita A., Ikeuchi T., Sainouchi M., Kakita A., Bennett D. A., Schneider J. A., Nichols M. R., Beausoleil S. A., Ulrich J. D., Holtzman D. M., Artyomov M. N., Colonna M., Human and mouse single-nucleus transcriptomics reveal TREM2-dependent and TREM2-independent cellular responses in Alzheimer’s disease. Nat. Med. 26, 131–142 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Huang Y., Happonen K. E., Burrola P. G., O’Connor C., Hah N., Huang L., Nimmerjahn A., Lemke G., Microglia use TAM receptors to detect and engulf amyloid β plaques. Nat. Immunol. 22, 586–594 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Oakley H., Cole S. L., Logan S., Maus E., Shao P., Craft J., Guillozet-Bongaarts A., Ohno M., Disterhoft J., Van E. L., Berry R., Vassar R., Intraneuronal beta-amyloid aggregates, neurodegeneration, and neuron loss in transgenic mice with five familial Alzheimer’s disease mutations: Potential factors in amyloid plaque formation. J. Neurosci. 26, 10129–10140 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Grubman A., Chew G., Ouyang J. F., Sun G., Choo X. Y., McLean C., Simmons R. K., Buckberry S., Vargas-Landin D. B., Poppe D., Pflueger J., Lister R., Rackham O. J. L., Petretto E., Polo J. M., A single-cell atlas of entorhinal cortex from individuals with Alzheimer’s disease reveals cell-type-specific gene expression regulation. Nat. Neurosci. 22, 2087–2097 (2019). [DOI] [PubMed] [Google Scholar]
- 16.Eketjall S., Janson J., Kaspersson K., Bogstedt A., Jeppsson F., Falting J., Haeberlein S. B., Kugler A. R., Alexander R. C., Cebers G., AZD3293: A novel, orally active BACE1 inhibitor with high potency and permeability and markedly slow off-rate kinetics. J. Alzheimers Dis. 50, 1109–1123 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kennedy M. E., Stamford A. W., Chen X., Cox K., Cumming J. N., Dockendorf M. F., Egan M., Ereshefsky L., Hodgson R. A., Hyde L. A., Jhee S., Kleijn H. J., Kuvelkar R., Li W., Mattson B. A., Mei H., Palcza J., Scott J. D., Tanen M., Troyer M. D., Tseng J. L., Stone J. A., Parker E. M., Forman M. S., The BACE1 inhibitor verubecestat (MK-8931) reduces CNS β-amyloid in animal models and in Alzheimer’s disease patients. Sci. Transl. Med. 8, 363ra150 (2016). [DOI] [PubMed] [Google Scholar]
- 18.Groves E., Dart A. E., Covarelli V., Caron E., Molecular mechanisms of phagocytic uptake in mammalian cells. Cell. Mol. Life Sci. 65, 1957–1976 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Massol P., Montcourrier P., Guillemot J. C., Chavrier P., Fc receptor-mediated phagocytosis requires CDC42 and Rac1. EMBO J. 17, 6219–6229 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wilkinson B., Koenigsknecht-Talboo J., Grommes C., Lee C. Y., Landreth G., Fibrillar beta-amyloid-stimulated intracellular signaling cascades require Vav for induction of respiratory burst and phagocytosis in monocytes and microglia. J. Biol. Chem. 281, 20842–20850 (2006). [DOI] [PubMed] [Google Scholar]
- 21.Pan J., Kao Y. L., Joshi S., Jeetendran S., Dipette D., Singh U. S., Activation of Rac1 by phosphatidylinositol 3-kinase in vivo: Role in activation of mitogen-activated protein kinase (MAPK) pathways and retinoic acid-induced neuronal differentiation of SH-SY5Y cells. J. Neurochem. 93, 571–583 (2005). [DOI] [PubMed] [Google Scholar]
- 22.Koyasu S., The role of PI3K in immune cells. Nat. Immunol. 4, 313–319 (2003). [DOI] [PubMed] [Google Scholar]
- 23.Dislich B., Wohlrab F., Bachhuber T., Mueller S., Kuhn P. H., Hogl S., Meyer-Luehmann M., Lichtenthaler S. F., Label-free quantitative proteomics of mouse cerebrospinal fluid detects β-site APP cleaving enzyme (BACE1) protease substrates in vivo. Mol. Cell. Proteomics 14, 2550–2563 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Boraschi D., Italiani P., Weil S., Martin M. U., The family of the interleukin-1 receptors. Immunol. Rev. 281, 197–232 (2018). [DOI] [PubMed] [Google Scholar]
- 25.Kuhn P. H., Marjaux E., Imhof A., De Strooper B., Haass C., Lichtenthaler S. F., Regulated intramembrane proteolysis of the interleukin-1 receptor II by alpha-, beta-, and gamma-secretase. J. Biol. Chem. 282, 11982–11995 (2007). [DOI] [PubMed] [Google Scholar]
- 26.Sizemore N., Leung S., Stark G. R., Activation of phosphatidylinositol 3-kinase in response to interleukin-1 leads to phosphorylation and activation of the NF-kappaB p65/RelA subunit. Mol. Cell. Biol. 19, 4798–4805 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Doan H. Q., Bowen K. A., Jackson L. A., Evers B. M., Toll-like receptor 4 activation increases Akt phosphorylation in colon cancer cells. Anticancer Res 29, 2473–2478 (2009). [PMC free article] [PubMed] [Google Scholar]
- 28.Keren-Shaul H., Spinrad A., Weiner A., Matcovitch-Natan O., Dvir-Szternfeld R., Ulland T. K., David E., Baruch K., Lara-Astaiso D., Toth B., Itzkovitz S., Colonna M., Schwartz M., Amit I., A unique microglia type associated with restricting development of alzheimer’s disease. Cell 169, 1276–1290.e17 (2017). [DOI] [PubMed] [Google Scholar]
- 29.Mathys H., Adaikkan C., Gao F., Young J. Z., Manet E., Hemberg M., De Jager P. L., Ransohoff R. M., Regev A., Tsai L. H., Temporal tracking of microglia activation in neurodegeneration at single-cell resolution. Cell Rep. 21, 366–380 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mrdjen D., Pavlovic A., Hartmann F. J., Schreiner B., Utz S. G., Leung B. P., Lelios I., Heppner F. L., Kipnis J., Merkler D., Greter M., Becher B., High-dimensional single-cell mapping of central nervous system immune cells reveals distinct myeloid subsets in health, aging, and disease. Immunity 48, 599 (2018). [DOI] [PubMed] [Google Scholar]
- 31.Song W. M., Colonna M., The identity and function of microglia in neurodegeneration. Nat. Immunol. 19, 1048–1058 (2018). [DOI] [PubMed] [Google Scholar]
- 32.Krasemann S., Madore C., Cialic R., Baufeld C., Calcagno N., El Fatimy R., Beckers L., O’Loughlin E., Xu Y., Fanek Z., Greco D. J., Smith S. T., Tweet G., Humulock Z., Zrzavy T., Conde-Sanroman P., Gacias M., Weng Z., Chen H., Tjon E., Mazaheri F., Hartmann K., Madi A., Ulrich J. D., Glatzel M., Worthmann A., Heeren J., Budnik B., Lemere C., Ikezu T., Heppner F. L., Litvak V., Holtzman D. M., Lassmann H., Weiner H. L., Ochando J., Haass C., Butovsky O., The TREM2-APOE pathway drives the transcriptional phenotype of dysfunctional microglia in neurodegenerative diseases. Immunity 47, 566–581.e9 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lee C. Y., Tse W., Smith J. D., Landreth G. E., Apolipoprotein E promotes β-amyloid trafficking and degradation by modulating microglial cholesterol levels. J. Biol. Chem. 287, 2032–2044 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lin Y. T., Seo J., Gao F., Feldman H. M., Wen H. L., Penney J., Cam H. P., Gjoneska E., Raja W. K., Cheng J., Rueda R., Kritskiy O., Abdurrob F., Peng Z., Milo B., Yu C. J., Elmsaouri S., Dey D., Ko T., Yankner B. A., Tsai L. H., APOE4 causes widespread molecular and cellular alterations associated with alzheimer’s disease phenotypes in human iPSC-derived brain cell types. Neuron 98, 1141–1154.e7 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Johnson J. L., Chambers E., Jayasundera K., Application of a bioinformatics-based approach to identify novel putative in vivo BACE1 substrates. Biomed. Eng. Comput. Biol. 5, 1–15 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hampel H., Lista S., Vanmechelen E., Zetterberg H., Giorgi F. S., Galgani A., Blennow K., Caraci F., Das B., Yan R., Vergallo A., β-Secretase1 biological markers for Alzheimer’s disease: State-of-art of validation and qualification. Alzheimers Res. Ther. 12, 130 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Rajendran L., Paolicelli R. C., Microglia-mediated synapse loss in Alzheimer’s disease. J. Neurosci. 38, 2911–2919 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Chen X., Stoeck A., Lee S. J., Shih I., Wang M. M., Wang T. L., Jagged1 expression regulated by Notch3 and Wnt/β-catenin signaling pathways in ovarian cancer. Oncotarget 1, 210–218 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Cory A. H., Owen T. C., Barltrop J. A., Cory J. G., Use of an aqueous soluble tetrazolium/formazan assay for cell growth assays in culture. Cancer Commun. 3, 207–212 (1991). [DOI] [PubMed] [Google Scholar]
- 40.Dobin A., Davis C. A., Schlesinger F., Drenkow J., Zaleski C., Jha S., Batut P., Chaisson M., Gingeras T. R., STAR: Ultrafast universal RNA-seq aligner. Bioinformatics 29, 15–21 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Satija R., Farrell J. A., Gennert D., Schier A. F., Regev A., Spatial reconstruction of single-cell gene expression data. Nat. Biotechnol. 33, 495–502 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zheng G. X., Terry J. M., Belgrader P., Ryvkin P., Bent Z. W., Wilson R., Ziraldo S. B., Wheeler T. D., McDermott G. P., Zhu J., Gregory M. T., Shuga J., Montesclaros L., Underwood J. G., Masquelier D. A., Nishimura S. Y., Schnall-Levin M., Wyatt P. W., Hindson C. M., Bharadwaj R., Wong A., Ness K. D., Beppu L. W., Deeg H. J., McFarland C., Loeb K. R., Valente W. J., Ericson N. G., Stevens E. A., Radich J. P., Mikkelsen T. S., Hindson B. J., Bielas J. H., Massively parallel digital transcriptional profiling of single cells. Nat. Commun. 8, 14049 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wang M., Song W. M., Ming C., Wang Q., Zhou X., Xu P., Krek A., Yoon Y., Ho L., Orr M. E., Yuan G. C., Zhang B., Guidelines for bioinformatics of single-cell sequencing data analysis in Alzheimer’s disease: Review, recommendation, implementation and application. Mol. Neurodegener. 17, 17 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Traag V. A., Waltman L., van Eck N. J., From Louvain to Leiden: Guaranteeing well-connected communities. Sci. Rep. 9, 5233 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Polański K., Young M. D., Miao Z., Meyer K. B., Teichmann S. A., Park J. E., BBKNN: Fast batch alignment of single cell transcriptomes. Bioinformatics 36, 964–965 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Hao Y., Hao S., Andersen-Nissen E., Mauck W. M. III, Zheng S., Butler A., Lee M. J., Wilk A. J., Darby C., Zager M., Hoffman P., Stoeckius M., Papalexi E., Mimitou E. P., Jain J., Srivastava A., Stuart T., Fleming L. M., Yeung B., Rogers A. J., McElrath J. M., Blish C. A., Gottardo R., Smibert P., Satija R., Integrated analysis of multimodal single-cell data. Cell 184, 3573–3587.e29 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Shalem O., Sanjana N. E., Zhang F., High-throughput functional genomics using CRISPR-Cas9. Nat. Rev. Genet. 16, 299–311 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figs. S1 to S7
Tables S1 and S2
Data S1