Abstract
Cancer-associated cachexia (CAC) is a complex metabolic and behavioral syndrome with multiple manifestations that involve systemic inflammation, weight loss, and adipose lipolysis. It impacts the quality of life of patients and is the direct cause of death in 20–30% of cancer patients. The severity of fat loss and adipose tissue remodeling negatively correlate with patients’ survival outcomes. To address the mechanism of fat loss and design potential approaches to prevent the process, it will be essential to understand CAC pathophysiology through white adipose tissue models. In the present study, an engineered human white adipose tissue (eWAT) model based on three-dimensional (3D) bioprinting was developed and induced with pancreatic cancer cell-conditioned medium (CM) to mimic the status of CAC in vitro. We found that the CM induction significantly increased the lipolysis and accumulation of the extracellular matrix (ECM). The 3D eWATs were further vascularized to study the influence of vascularization on lipolysis and CAC progression, which was largely unknown. Results demonstrated that CM induction improved the angiogenesis of vascularized eWATs (veWATs), and veWATs demonstrated decreased glycerol release but increased UCP1 expression, compared to eWATs. Many unique inflammatory cytokines (IL-8, CXCL-1, GM-CSF, etc) from the CM were detected and supposed to contribute to eWAT lipolysis, UCP1 up-regulation, and ECM development. In response to CM induction, eWATs also secreted inflammatory adipokines related to the metastatic ability of cancer, muscle atrophy, and vascularization (NGAL, CD54, IGFBP-2, etc). Our work demonstrated that the eWAT is a robust model for studying cachectic fat loss and the accompanying remodeling of adipose tissue. It is therefore a useful tool for future research exploring CAC physiologies and developing potential therapies.
Keywords: Engineered 3D human white adipose tissue, Lipolysis, Browning, ECM remodeling, Vascularization
1. Introduction
Cancer-associated cachexia (CAC) is a multifactorial metabolic syndrome that is featured as systemic inflammation, weight loss, adipose tissue atrophy, and skeletal muscle wasting [1]. It frequently occurs in patients with pancreatic, lung, or colon cancer, during advanced stages [2]. CAC leads to reduced quality of life in patients and accounts for up to 30% of cancer deaths [3]. Currently, there are no effective early diagnosis methods for CAC and no effective treatments are available to reverse it once clinical signs are apparent. Fat loss is found to precede muscle wasting, and the intensity of fat depletion is reported to negatively correlate with the patients’ survival [4]. Fat remodeling caused by metabolic disorders, including elevated lipolysis, decreased lipogenesis, and white adipose browning, may happen in early stage CAC [1, 5]. Thus, fat loss and remodeling are important processes during CAC development. To better understand the underlying mechanisms of adipose tissue remodeling in CAC, it is necessary to generate a disease model that addresses the interactions between adipose tissue and cancer cells, identifies pathological responses, and discovers potential therapeutic targets.
At present, pre-clinical in vivo models that use orthotopic cancer cell inoculation are routinely applied to study adipose tissue remodeling in CAC [6]. For example, cancer cells were transplanted into mice to get tumor bearing mice, and fat tissue responses, including lipolysis and browning, were then analyzed [1, 7]. Although useful, these in vivo models are difficult to translate into CAC for humans, since either immunodeficient or immunocompromised mice models should be used to overcome immuno-incompatibility between humans and rodents. Therefore, in vitro human white adipose tissue models could be an effective tool to mimic native adipose tissues in CAC studies. Two-dimensional (2D) WAT models with adipocytes cultured in wells or dishes are simple and contribute to the majority of discoveries and research [8, 9]. Tumor-conditioned medium is used to induce the 2D cultured adipocytes, mimicking the cachexia process, and cell responses are determined. However, these 2D culture models cannot recapitulate the architectures of living tissues with abundant cell-cell interplay and cell-matrix interplay [10]. Three-dimensional (3D) WAT models have therefore been developed to better recapitulate the in vivo adipose environment with intensive cell-cell and cell-matrix interactions. Such models can be divided into two categories: scaffold-free and scaffold-based models [11]. For the scaffold-free WAT models, cells have more interactions with each other and can be fabricated by hanging droplets, magnetic levitation, or other cell assembly techniques [12–15]. Compared to scaffold-free models, scaffold-based WAT models provide a more well-established architecture for cells and have the advantages of easy handling, mimicking cell-matrix interactions, and providing tunable biophysical and biochemical microenvironments. The scaffolds can be composed of synthetic or natural materials, forming flexible fibers [16], hydrogels [17, 18], or membranes [19]. However, most of the current scaffold-based WAT models focus on model characterization and disease studies related to breast cancer and type II diabetes mellitus [20, 21], while their responses and effectiveness under CAC were largely undetermined.
Human WAT is vascularized, and its vasculature is essential for maintaining its mass, function, and homeostasis [22, 23]. Blood flow brings the nutrition, oxygen, and cytokines to adipocytes and also removes metabolic byproducts [24]. Hence, vascularization is crucial for adipose functions and needs to be taken into consideration when designing a functional adipose model for a CAC study. Previous studies have extensively demonstrated that co-cultures of adipocytes and adipose derived mesenchymal stem cells (ADMSCs) with endothelial cells promoted angiogenesis [25, 26], mitigated external radiation damage [27], and evoked physiological responses to hyperinsulinemia compared to monocultures [28]. However, the roles of vascularization on WAT remodeling during CAC and cancer progression are largely unknown.
In this study, we aimed to develop a scaffold-based engineered WAT (eWAT) through 3D bioprinting. We implemented methacrylated gelatin (GelMA) and methacrylated hyaluronic acid (HAMA) as bioinks, with encapsulated human white adipocyte progenitors (WAPs). After adipogenic differentiation, ADMSCs and human umbilical vein endothelial cells (HUVECs) were wrapped around eWATs and co-cultured with them to generate vascularized eWATs (veWATs) (figure 1(a)). Among all forms of malignancy, pancreatic ductal adenocarcinoma is the most highly associated with cachexia, with estimated 83% of patients suffering from CAC [29]. Conditioned cancer medium from human pancreatic cancer cells was used to induce CAC, and the remodeling of eWAT/veWAT, including lipolysis, browning, adipogenesis, and extracellular matrix (ECM) development, was studied (figure 1(a)). Cytokines from the cancer medium and adipokines released by the eWAT were also analyzed to identify potential CAC related molecules.
Figure 1.
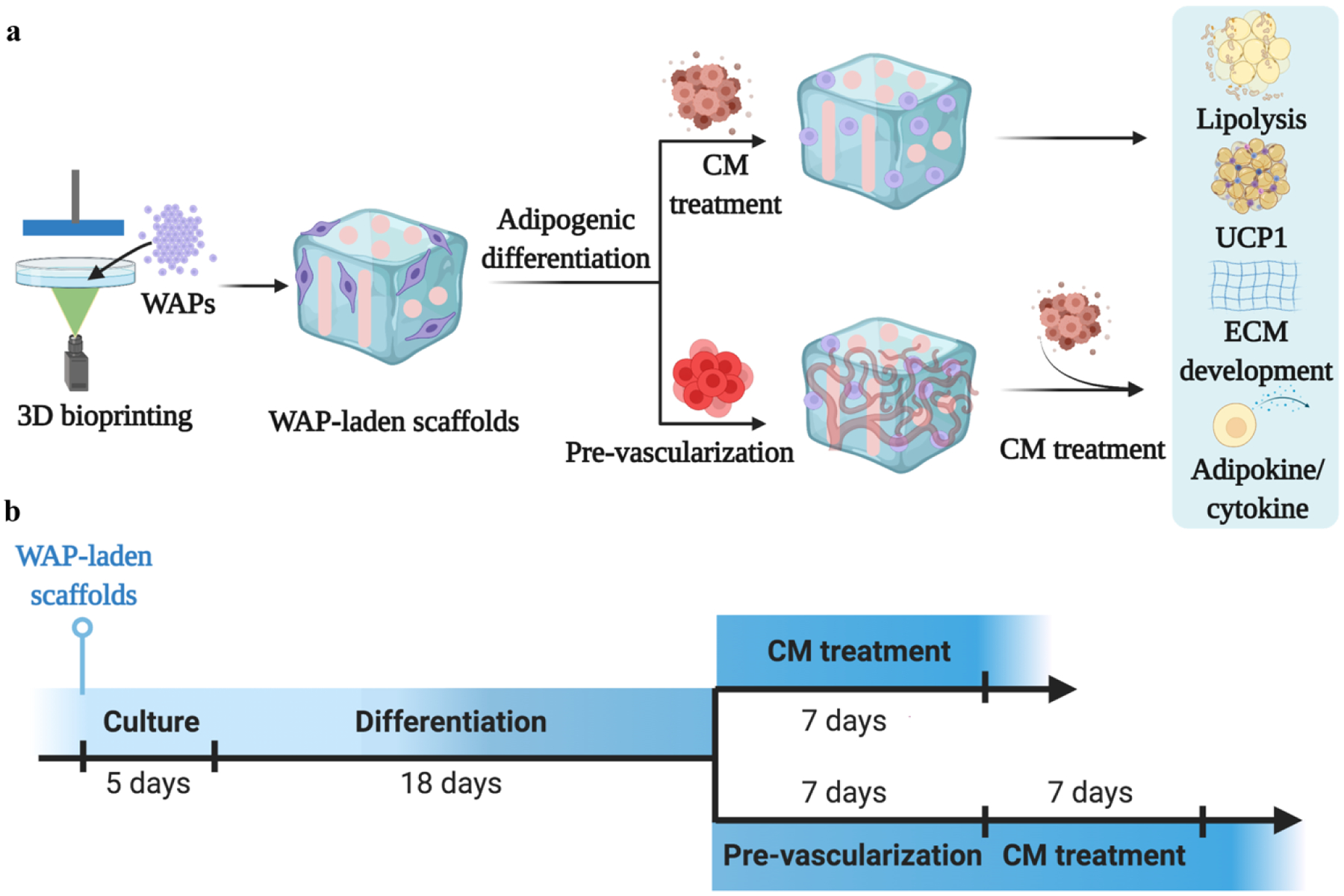
Schematic of the experimental design. (a) Schematic of human white adipocyte progenitor (WAP)-laden scaffold 3D printing, adipogenic differentiation, pre-vascularization, and conditioned medium (CM) induction. Adipose remodeling was evaluated in the aspects of lipolysis, browning gene expression, extracellular matrix (ECM) development, and cytokine/adipokine release. (b) Timeline of each process, with 5 days of WAP culture, 18 days of adipogenic differentiation, 7 days of pre-vascularization, and 7 days of CM induction.
2. Materials and Methods
2.1. Cell culture
Immortalized human WAPs were isolated from the subcutaneous fat from one anonymous female and infected with an hTERT expressing retrovirus [30]. The WAPs were cultured with a growth medium containing Dulbecco’s Modified Eagles Medium (DMEM/high glucose), 10% fetal bovine serum (FBS, Gibco), and 1% penicillin/streptomycin (P/S, Gibco) in 5% CO2 at 37 °C. Human ADMSCs (passage 3–5) were purchased from Lonza and were cultured in growth medium consisting of DMEM/F12 (Gibco), 10% FBS, and 1% P/S. HUVECs (passage 3–5) were also obtained from Lonza and were cultured in 100 μg/mL rat tail collagen (Advanced BioMatrix) coated flasks (3 h, at room temperature) and with Endothelial Cell Growth Medium v2 (ECGM v2, Cell Applications) and 1% P/S. BxPC-3 cells (ATCC, CRL-1678TM), a human pancreatic cancer cell line originally derived from a 61-year-old female with pancreas adenocarcinoma, at passages 5–6 were cultured in RPMI 1640 media supplemented with 10% FBS and 1% P/S.
2.2. 3D bioprinting of WAP-laden scaffolds
To prepare the bioink, methacrylated hyaluronic acid (HAMA) and methacrylated gelatin (GelMA) were synthesized as previously described [31]. The degree of methacrylation was determined to be around 22.5% for HAMA and 61.1% for GelMA. For the bioink, 7.5% wt GelMA and 1% wt HAMA were dissolved in WAP growth medium. 1 mM tartrazine and 0.01 g/mL lithium phenyl-2,4,6-trimethylbenzoylphosphinate (LAP) were added to act as the photoabsorber and photoinitiator, respectively.
Prior to 3D bioprinting, WAPs were digested by TrypLE Reagents (Gibco) and resuspended in the bioinks at a density of 5×106 cells/mL. A 3D digital light processing (DLP)-based bioprinter (Lumen X, CELLINK) was used for the 3D bioprinting. The temperature of bioink support platform was set around 37 °C, at which the bioinks were maintained in solution state. The bioprinting was conducted layer-by-layer through a UV projector that crosslinked each layer under a sterile environment. The WAP-laden scaffolds were designed to be 4 mm × 4 mm × 2 mm (height), with four channels through them from the top view and two channels through them from the side view. The diameter of the channels was 0.8 mm, which is advantageous for nutrient infiltration and cell migration and growth. Although most hydrogels illustrate pores defined by polymer chains comprising the hydrogel network, their sizes are limited to less than ten micrometers. Microchannels were designed here to meet nutrition and molecule transport requirements for the large number of cells. Once the bioprinting was finished, the constructs were washed in WAP growth medium to remove excess un-crosslinked solution.
2.3. Characterization of 3D printed scaffolds
The overall structure of the 3D printed acellular scaffolds was observed by an optical microscope (Zeiss). After lyophilization, the scaffolds were sputter coated with gold, and the morphology was observed under a scanning electron microscope (SEM, FEI Quanta 200) at 25 kV.
The rheological performance of the bioinks, as well as the 3D printed acellular scaffolds, was evaluated by a rheometer (HR-2, TA Instruments). The plate diameter was set as 8 mm with a gap of 1 mm, and the plate temperature was 37 °C. Both the oscillation strain sweep (0.01–10% strain, 1 rad/s) and frequency sweep (0.1–100 rad/s, 1% strain) were conducted to evaluate the mechanical performances of the bioinks and printed scaffolds. Their storage moduli (G’) and loss moduli (G”) were recorded.
The compressive mechanical performance of scaffolds was conducted by the UniVert mechanical testing system (CellScale). The scaffolds were compressed to 50% of their height, at a speed of 0.5 mm/min. Five loading-unloading cycles were applied to investigate scaffold compression resilience property. Compressive modulus of scaffolds was calculated according to the stress-strain curve.
2.4. Differentiation of WAP-laden scaffolds
To differentiate WAPs into adipocytes, 3D bioprinted WAP-laden scaffolds were first cultured in growth medium for 5 days and then cultured with a differentiation medium consisting of DMEM/high glucose with 2% FBS, 1% P/S, 0.5 μM recombinant human insulin (Alfa Aesar), and 2 nM tri-iodothyronine (T3, Sigma) for 6 days, with the medium changed every 3 days. Induction was further conducted for another 12 days with an induction medium that consisted of the differentiation medium plus 33 μM biotin (Sigma), 17 μM pantothenate (Sigma), 0.1 μM dexamethasone (Sigma), 500 μM 3-isobutyl-1-methyl xanthine (IBMX, Sigma), and 30 μM indomethacin (Sigma). The medium was changed every 3 days. After adipogenic differentiation, we obtained eWATs, and they were maintained in growth medium before further vascularization or cancer medium application.
Bright field images of WAP-laden scaffolds with or without adipogenic differentiation were taken. The cell viability was examined by a Live/Dead assay (Invitrogen). Images of Live/Dead assays were taken by a confocal laser scanning microscope (CLSM, LSM 710, Zeiss). For immunofluorescent (IF) staining, samples were fixed in 4% paraformaldehyde (PFA) for 4 h, dehydrated by 30% sucrose overnight, and cryo-sectioned. Slides were then blocked by 1% bovine serum albumin (BSA) overnight at 4 °C. 10 μg/mL BODIPY™ 493/503 (D3922, Thermo Fisher Scientific) was added and incubated for 30 min at room temperature. After nuclear staining with DAPI (1:1000 in PBS), the slides were imaged by the 710 CLSM. The lipid droplet size for WAP-laden scaffolds with or without adipogenic differentiation was quantified by ImageJ software.
For quantitative real time polymerase chain reaction (qPCR) analysis, constructs were homogenized by a lysis buffer using a bead miller (Fisher Scientific). The total RNA was extracted through RNeasy mini-kits (QIAgen) and reverse transcribed to cDNA through iScript cDNA synthesis kits (BioRad Laboratories). QPCR was performed by a StepOnePlusTM PCR System (Thermo Fisher Scientific) with SsoAdvanced SYBR Green Supermix (Bio-Rad). The relative expressions of the genes of interest, like peroxisome proliferator-activated receptor gamma (PPARG) and CCAAT/enhancer binding protein epsilon (CEBPE), were quantified with by normalizing them to housekeeping gene 18S. Their sequences are shown in Table S1.
2.5. Induction of eWATs with cancer-cell-conditioned medium
When BxPC-3 cells reached 100% confluence, the medium was refreshed with fresh growth medium and maintained for 48 hours. Then, the supernatants were collected and filtered by a 0.02 μm filter to remove cells and debris. To avoid the risk of nutrient deprivation of eWATs and over dilution of cancer cell supernatants, the obtained supernatants were mixed with WAP growth medium in a ratio of 1:1 (v/v), and this mixture was used as the cancer-cell-conditioned medium (CM) for inducing eWATs. BxPC-3 fresh growth medium mixed with WAP growth medium (1:1 v/v) worked as the control growth medium (GM) for inducing eWATs. 3D printed scaffolds without cells were also induced with CM and served as the baseline.
At days 3, 5, and 7, the culture media in all groups were collected. Lipolysis levels were determined by measuring the amount of glycerol released into the media by a glycerol assay (Sigma). QPCR was conducted after 7 days as mentioned in Section 2.4. Genes related to adipose browning, like Uncoupling Protein 1 (UCP1), Cell Death Inducing DFFA Like Effector A (CIDEA), and PR/SET Domain 16 (PRDM16), were analyzed. Lipoprotein lipase (LPL), along with PPARG and CEBPE, were also evaluated (Table S1).
After CM application for 7 days, eWATs were fixed in 4% PFA, dehydrated, and cryo-sectioned as mentioned in 2.4. For IF staining, slides were blocked by 1% BSA and cultured overnight with primary antibodies to UCP1 (1:100, Abcam). Secondary Alexa Fluor™ 568 goat anti-rabbit antibodies (1:100 in 1% BSA) and 10 μg/mL BODIPY were cultured for another 2 h at room temperature. After nuclear staining with DAPI, the slides were imaged by a 710 CLSM. Oil Red O, hematoxylin and eosin (H&E), and immunohistochemical (IHC) staining of collagen I were also conducted and imaged by a microscope (Leica). Briefly, slides were incubated in Oil Red O working solutions for around 10 min. For H&E staining, slides were incubated in hematoxylin solution, to stain the nuclei, and eosin solution subsequently. To stain collagen I, slides were pre-incubated with 3% hydrogen peroxide in methanol for 15 min to deactivate the endogenous peroxidase and then blocked with blocking serum for 1 hour at room temperature.
The sections were then incubated overnight at 4 °C with anti-collagen type I primary antibodies (Col I, NB600–408, Novus Biologicals). After washing three times with PBS, biotinylated goat anti-rabbit IgG (1:150, Vector Laboratories) was added and cultured for 30 min at room temperature. After washing with PBS, avidin-biotin complex conjugated to peroxidase (Vector Laboratories) was applied for 30 min. The bound complexes were visualized by adding DAB substrate (3,3’-diaminobenzidine, Vector Laboratories) to the sections for 5 minutes. Then, counterstaining in hematoxylin was finally conducted. Semi-quantitative analysis of the eosin-stained intensity in H&E images and collagen I in IHC images were evaluated by Fiji software. The mean optical density was calculated as log (255/mean intensity).
2.6. Induction of HUVECs with CM
Proliferation, tube formation, and wound healing assays were conducted to evaluate the influence of CM on 2D cultured HUVEC behaviors in vitro. For the proliferation test, a 96-well plate was coated with 100 μg/mL of rat tail collagen prior to the experiments. HUVECs were seeded at a density of 1×104 cells/100 μL into each well. The CM consisted of BxPC-3 supernatants, prepared as described in Section 2.5, and fresh Endothelial Cell Growth Medium (ECGM) (1:1, v/v), while fresh BxPC-3 growth medium mixed with ECGM served as the control GM. The media were changed every 2 days. HUVEC proliferation was measured by a Cell Counting Kit 8 (CCK8, Abcam) at days 3 and 7, following manufacturer’s instructions.
For the tube formation, 50 μL of Matrigel was added into each well of a 96-well plate and incubated at 37 °C for 1 h. HUVECs (2×104 cells/100 μL) were then seeded into the wells and cultured with CM or GM. After 24 and 48 h, the formed HUVEC tubes were observed, and three images of each well were randomly taken by an inverted microscope (Leica). The tube nodes were counted afterwards [32].
For the wound healing assay, a 24-well plate was first coated with 100 μg/mL of rat tail collagen. HUVECs were then seeded (3×104 cells/well) and grew until 70% confluency. A 200 μL pipette tip was used to make a scratch among the cells. After washing three times with PBS, CM and GM were added. Images at t= 0 h and t= 24 h were taken by an inverted microscope. The wound healing rate (%) was calculated as: [(wound length at t=0 h)− (wound length at t=24 h)/wound length at t= 0 h] × 100% [32]. Five samples of each group and three images from each sample were included.
2.7. Development of veWATs and induced with CM
Mixed collagen and fibrin solutions were used to encapsulate HUVECs (2×106 cells/mL) and ADMSCs (5×105 cells/mL) for vascularization of eWATs. ADMSCs were used to support HUVEC growth. The pH of type I human collagen solution (3.1 mg/mL, Advanced BioMatrix) was adjusted to 7.5, and its ionic strength was neutralized by adding 1/10 final volume of 10×PBS. Human fibrinogen (EMD Millipore Corporation) was dissolved in PBS at a concentration of 20 mg/mL. HUVECs and ADMSCs were suspended in 2 units/mL of thrombin (Thermo Fisher Scientific) in ECGM v2 medium. To pre-vascularize eWATs, type I human collagen solution, fibrinogen, and cell-encapsulated thrombin were mixed in equal volume, wrapped around the eWAT (50 μL per scaffold), and cultured at 37 °C for 1 h [33]. ECGM and WAP growth medium (1:1, v/v) were then added and changed every 2 days. After 7 days of culture, CM consisting of BxPC-3 supernatants, ECGM, and WAP growth medium (2:1:1, v/v/v) was used to induce vascularized eWAT (veWAT) for another 7 days (timeline is shown in figure 1(b)). A mixture of fresh BxPC-3 growth medium, ECGM, and WAP growth medium (2:1:1, v/v/v) was used as the control GM. IF staining was conducted after scaffold fixation (4% PFA, 4 h) and blocking to determine the CM influence on veWAT angiogenesis. A primary antibody to CD31 (1:100, eBioscience™) was incubated overnight at 4 °C. Secondary Alexa Fluor™ 568 goat anti-mouse antibodies (1:100 in 1% BSA) and 10 μg/mL BODIPY were applied for another 2 h at room temperature. After nuclear staining with DAPI, the constructs were imaged by a 710 CLSM.
To compare lipolysis and browning related gene expressions of eWAT and veWAT after CM induction, the media from eWAT, veWAT, and vascularized 3D bioprinted acellular scaffolds (vScaffolds) were collected, and the released glycerol was tested as described in Section 2.5. At day 7, the expressions of adipocyte browning related genes (UCP1, CIDEA, PRDM16) were also evaluated by qPCR.
2.8. Human cytokine and adipokine array analyses
The relative amounts of cytokines in BxPC-3 fresh growth medium and supernatants were evaluated by a Human Cytokine Array Kit (ARY005B, R&D Systems) by following the manufacturer’s protocol. Briefly, 1 mL of fresh BxPC-3 growth medium or BxPC-3 supernatants was mixed with a cocktail of biotinylated detection antibodies and incubated with the array membrane overnight at 4 °C. After washing three times with washing buffer (10 min each time), streptavidin-horseradish peroxidase solution and chemiluminescent detection reagents were added to the membrane in sequence. Then, the array membranes were imaged and scanned. The pixel density of each dot was analyzed with ImageJ software.
Adipokines released from the eWAT after being induced with CM and control GM were also evaluated. The relative amount of adipokines was calculated as the ratio of the adipokine pixel density/positive control pixel density from the CM induced samples to the adipokine pixel density/positive control pixel density from the GM.
2.9. Statistical analysis
All quantitative data were expressed as the mean ± standard deviation (SD). Statistical analysis was also performed. In experiments with two groups, student t-tests were used, while in experiments with more than two groups, one-way analysis of variance ANOVA was used with Tukey post-hoc tests for statistical analysis. Differences with p<0.05 were denoted as *, while p<0.01 was denoted as **, p<0.001 was denoted as ***, p<0.0001 was denoted as ****, and ns indicated not significant.
3. Results
3.1. 3D printing and characterization of scaffolds
We implemented HAMA and GelMA based bioinks to 3D bioprint WAP-laden scaffolds. The printed scaffolds had four channels through them from the top view and two from the side view (figure 2(a)), which were designed to help nutrition transportation and keep high cell viability. SEM images of the scaffolds showed an interconnected and porous structure (figure 2(b)). Under oscillation strain sweep, bioinks before crosslinking and printed scaffolds showed almost constant G’ and G” during a 1–10% strain range (figure 2(c)). 1% strain was chosen for the frequency sweep later. The bioinks before crosslinking demonstrated similar storage and loss modulus values (figure 2(c)). After crosslinking during the 3D printing process, scaffolds showed an around ten times higher storage modulus compared to loss modulus, indicating successful hydrogel formation (figure 2(c)). The compressive stress of printed scaffolds at 50% strain decreased from 54±1.8 kPa to 34±2.3 kPa after 5 cycles of compression-release. The modulus of scaffolds was calculated to be 0.9±0.3 kPa in the first cycle of compression process, which is comparable to that of the native fat tissue tested in the previous literature [34].
Figure 2.
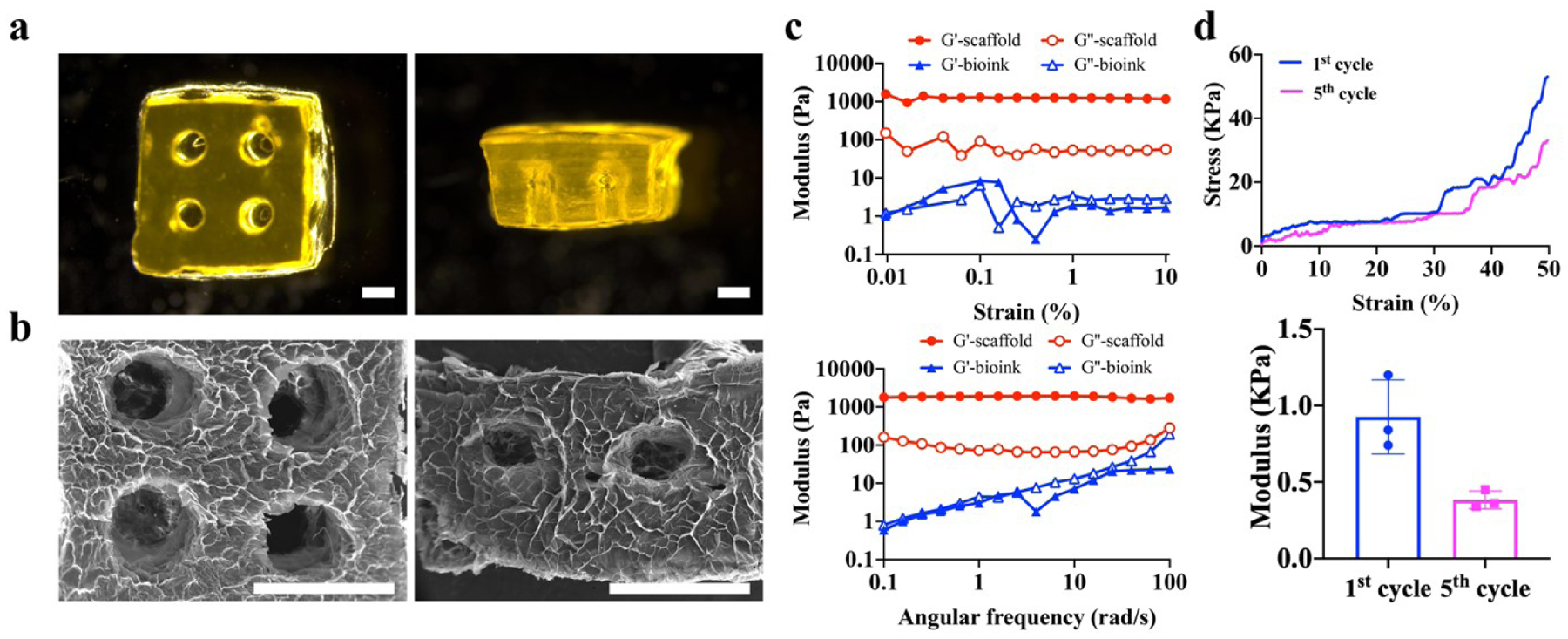
Characterization of 3D printed acellular scaffolds. (a) Microscopic images of 3D printed acellular scaffolds. Scale bar=1 mm. (b) Scanning electron microscope (SEM) images of 3D printed acellular scaffolds. Scale bar=2 mm. (c) Oscillation strain sweep (top) and frequency sweep performance (bottom) of the bioink and acellular scaffolds. (d) Stress-strain curve (top) and modulus (bottom) of acellular scaffolds under compression.
3.2. Adipogenic differentiation of WAP-laden scaffolds
After adipogenic differentiation, the eWATs accumulated more cytoplasmic lipids, compared to undifferentiated constructs (figure 3(a). The WAPs encapsulated within 3D bioprinted constructs showed high cell viability with or without adipogenic differentiation (figure 3(a)). Undifferentiated WAPs had a spindle-like morphology and were weakly stained by BODIPY, which is a lipophilic bright green dye for non-polar lipids (figure 3(a)). In contrast, WAPs after adipogenic differentiation showed a round shape and had much more and brighter BODIPY-stained lipid droplets localized around the nuclei (figure 3(a)). In addition, the adipogenic differentiation process significantly increased the lipid droplet size, compared to WAPs without differentiation (figure 3(b)). QPCR results also illustrated significant upregulation of PPARG and CEBPE (figure 3(c)), which are two major transcription factors that drive terminal adipocyte differentiation [35, 36].
Figure 3.
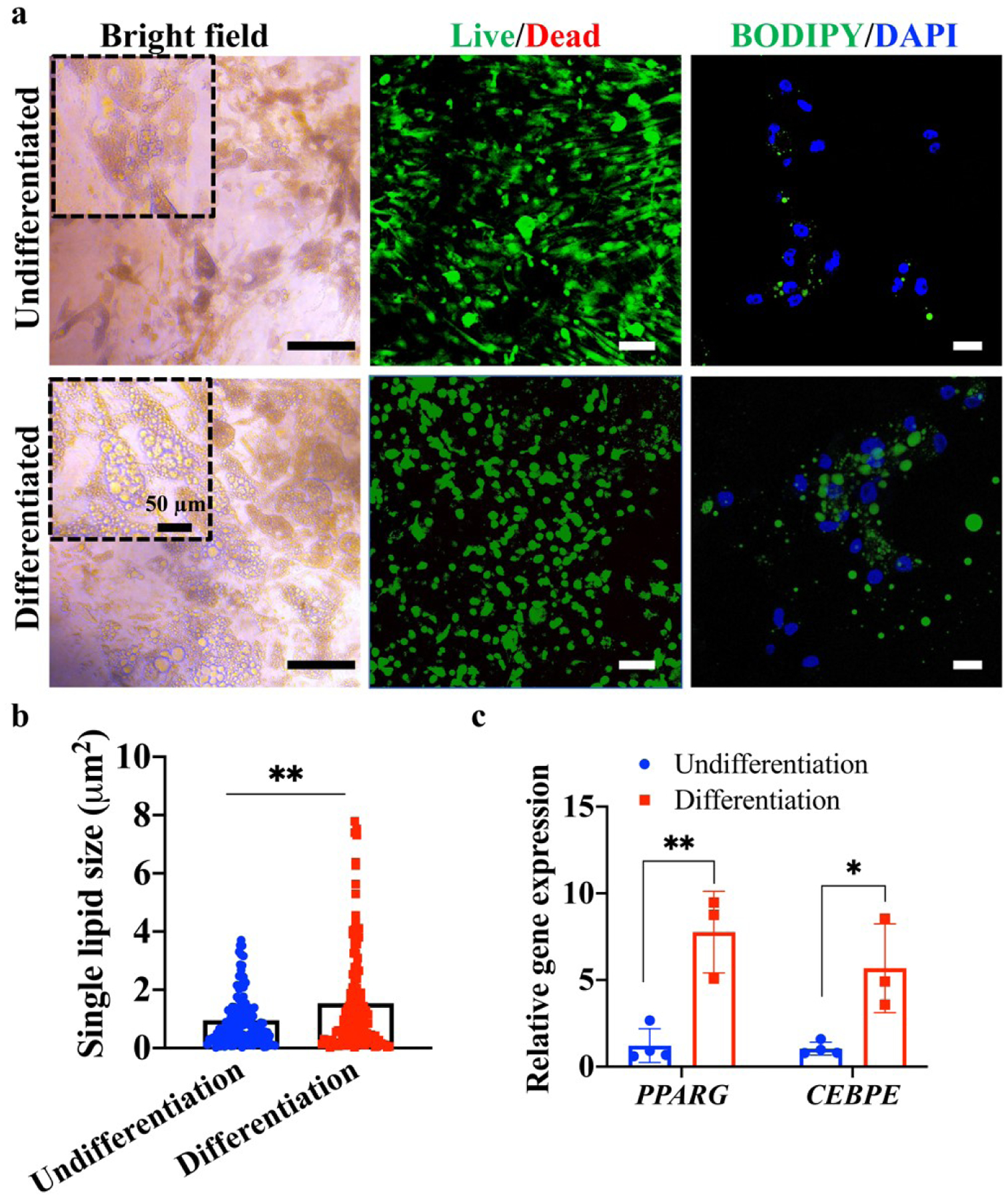
Characterization of the adipogenic differentiation of white adipocyte progenitors (WAPs) within 3D bioprinted constructs. (a) Optical images, Live/Dead staining, and lipid staining of WAP-laden constructs after differentiation and without differentiation. Scale bar=50 μm, 100 μm, and 20 μm for bright field, Live/Dead, and BODIPY/DAPI images, respectively. (b) Single lipid size in differentiated and undifferentiated constructs (n=3, 20–30 lipids from each sample were evaluated). (c) Relative adipogenic gene expression (PPARG and CEBPE) of differentiated and undifferentiated constructs (n=3–4).
3.3. eWAT remodeling induced by CM induction
The cachexia process was mimicked in vitro by using CM to induce eWATs. The 3D printed acellular scaffolds induced with CM (scaffold/CM) and eWATs induced with growth medium (eWAT/GM) served as controls. Most cells within eWATs were alive after 7 days of treatment, as shown in figure S1. The lipolysis of eWATs in response to CM was evaluated by quantifying the free glycerol released in the media at days 3, 5, and 7. As shown in figure 4(a), at each time interval, eWATs induced with CM had a significantly higher degree of glycerol release than other groups. The cumulative release of glycerol was also evaluated, and the amount of glycerol was the highest in CM-induced eWATs (figure 4(b)).
Figure 4.
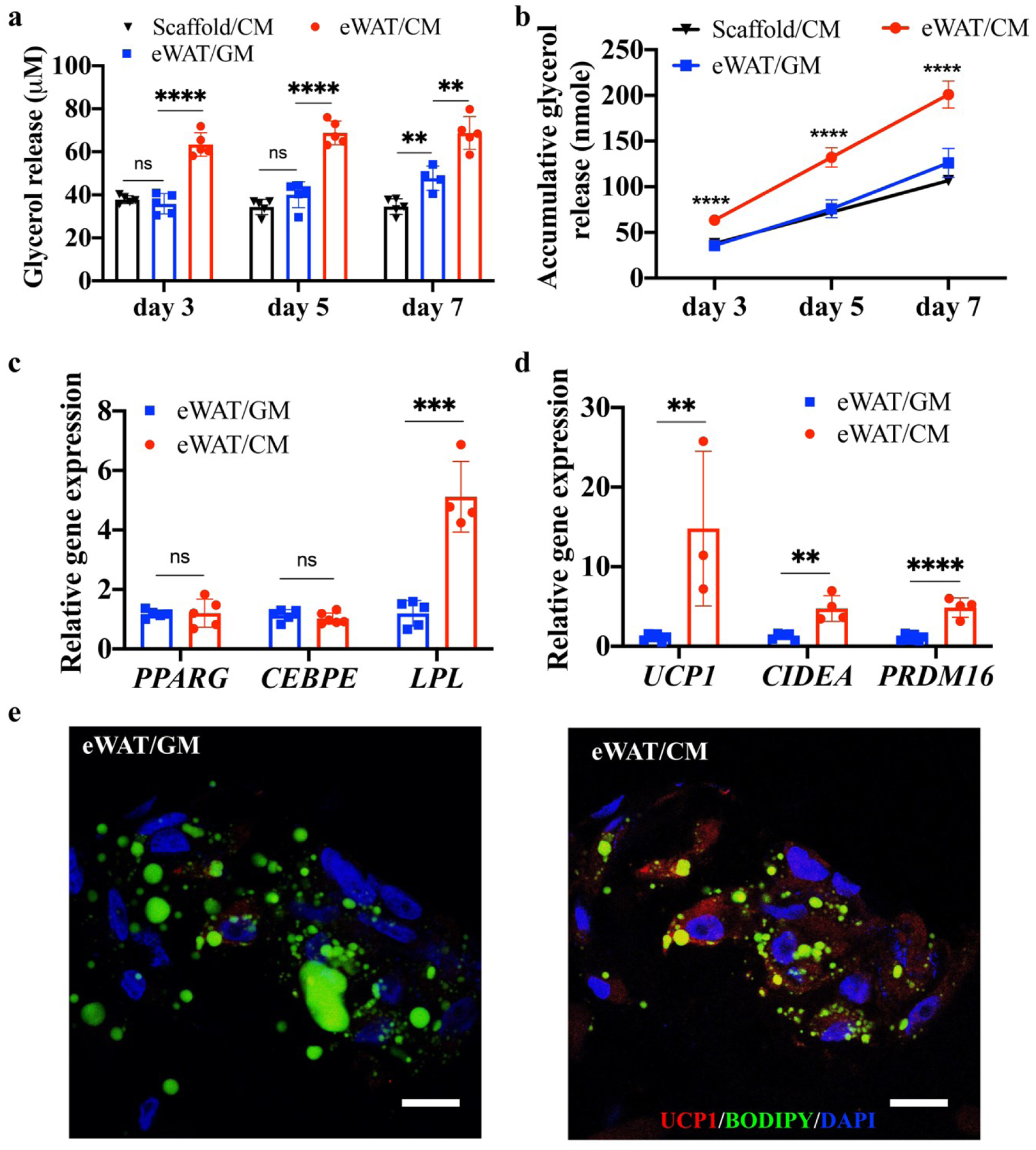
The effects of conditioned medium (CM) on glycerol release and browning of engineered human white adipose tissues (eWATs). (a) Glycerol release of acellular scaffolds and eWATs under treatments with CM and growth medium (GM) at different time intervals (n=5). (b) Accumulative glycerol release of acellular scaffolds and eWATs (n=5). (c) Gene expressions of eWATs after induction with CM and GM (n=3–4). (d) Adipocyte browning gene expressions (UCP1, CIDEA, PRDM16) of eWATs after induction with CM and GM (n=3–4). (e) Immunofluorescent staining of eWATs induced with CM and GM. Scale bar=20 μm.
Adipogenic differentiation-related genes, PPARG and CEBPE, did not show significant changes after CM induction, while the gene related to triacylglycerol hydrolysis, LPL, increased significantly in the eWAT/CM group (figure 4(c)). The induction of eWATs with CM significantly upregulated the expressions of browning genes (i.e., UCP1, CIDEA, PRDM16) (figure 4(d)). The expression of UCP1 was increased by about 15-fold after the induction with CM (figure 4(d)). IF staining demonstrated that the eWATs induced with CM showed more positive UCP1 expression than those induced by GM (figure 4(e)), which was consistent with the qPCR results. CM-induced eWATs did illustrate a weaker signal and less and smaller lipids.
To further evaluate the eWAT remodeling, Oil Red O, H&E, and collagen I staining were examined (figure 5). According to the Oil Red O staining images, a limited distribution of lipids (red) was shown in CM-induced eWAT groups, which confirmed that CM contributed to the adipocytes’ lipid loss (figure 5(a)). The hematoxylin stains cell nuclei blue, and eosin stains the ECM and cytoplasm pink [37]. We found that CM-induced eWATs presented a higher distribution of pink stained ECM (figure 5(a)), and semi-quantitative analysis of the mean optical density of the H&E images also confirmed that the eWATs induced with CM had higher eosin-stained optical density than their counterparts with GM induction (figure 5(b)). The stained ECM was composed of crosslinked HAMA and GelMA in the 3D printed scaffolds, as well as adipocyte-secreted collagens, fibronectin, glycoproteins, and proteoglycans. Since the same material recipe was applied in all groups, we speculated that CM might induce extensive ECM development. To further confirm our hypothesis, collagen I was stained by IHC. CM-induced eWATs exhibited a higher distribution of collagen I and a higher collagen I optical density than the GM-induced eWAT group (figure 5(a) and 5(c)).
Figure 5.
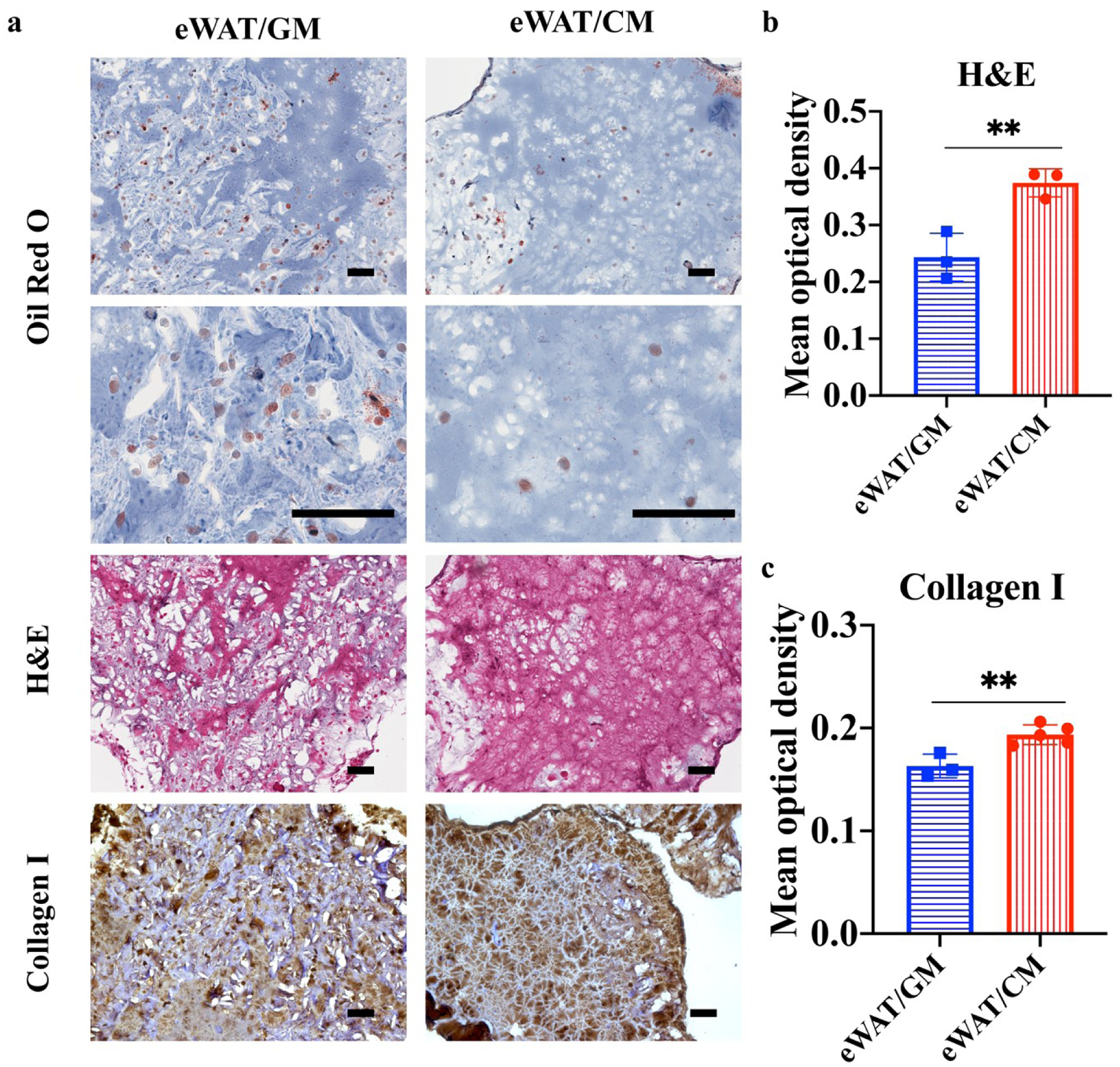
Oil Red O, H&E, and Collagen I staining of engineered human white adipose tissues (eWATs) induced by growth medium (GM) and conditioned medium (CM). (a) Microscopical images of Oil Red O, H&E, and Collagen I staining of eWATs induced with CM and GM. Scale bar=100 μm. (b) Mean optical density of eosin in H&E staining (n=3). (c) Mean optical density of collagen I (n=3–5).
3.4. Influence of CM induction on HUVECs and veWATs
Angiogenesis is a crucial event in pancreatic cancer development, progression, and metastasis [38]. Previous studies have demonstrated that a co-culture of BxPC-3 and HUVECs significantly enhanced angiogenesis and vascular endothelial growth factor (VEGF) secretion [39, 40]. However, it is largely unknown how CAC affects adipose tissue angiogenesis and crosstalk between endothelial cells and adipocytes. We first investigated the influence of CM induction on 2D HUVEC proliferation, migration, and cellular angiogenesis capacity. As shown in figure 6(a), CM induction was found to significantly enhance HUVEC proliferation after 3 and 7-day cultures (figure 6(a)). A tube formation assay was conducted to evaluate the in vitro angiogenesis of HUVECs in response to CM. HUVECs that were seeded onto Matrigel coated wells developed vascular tubular structures (figure 6(b)). Although no statistical difference was observed after 24 h culture (figure 6(c)), the node density of the HUVECs was significantly increased after CM induction for 48 h. HUVEC migration was assessed by a scratch wound assay (figure 6(d)). HUVECs migrated to heal the gap, and CM induced HUVECs showed significantly higher healing rate than cells cultured in GM (figure 6(e)).
Figure 6.
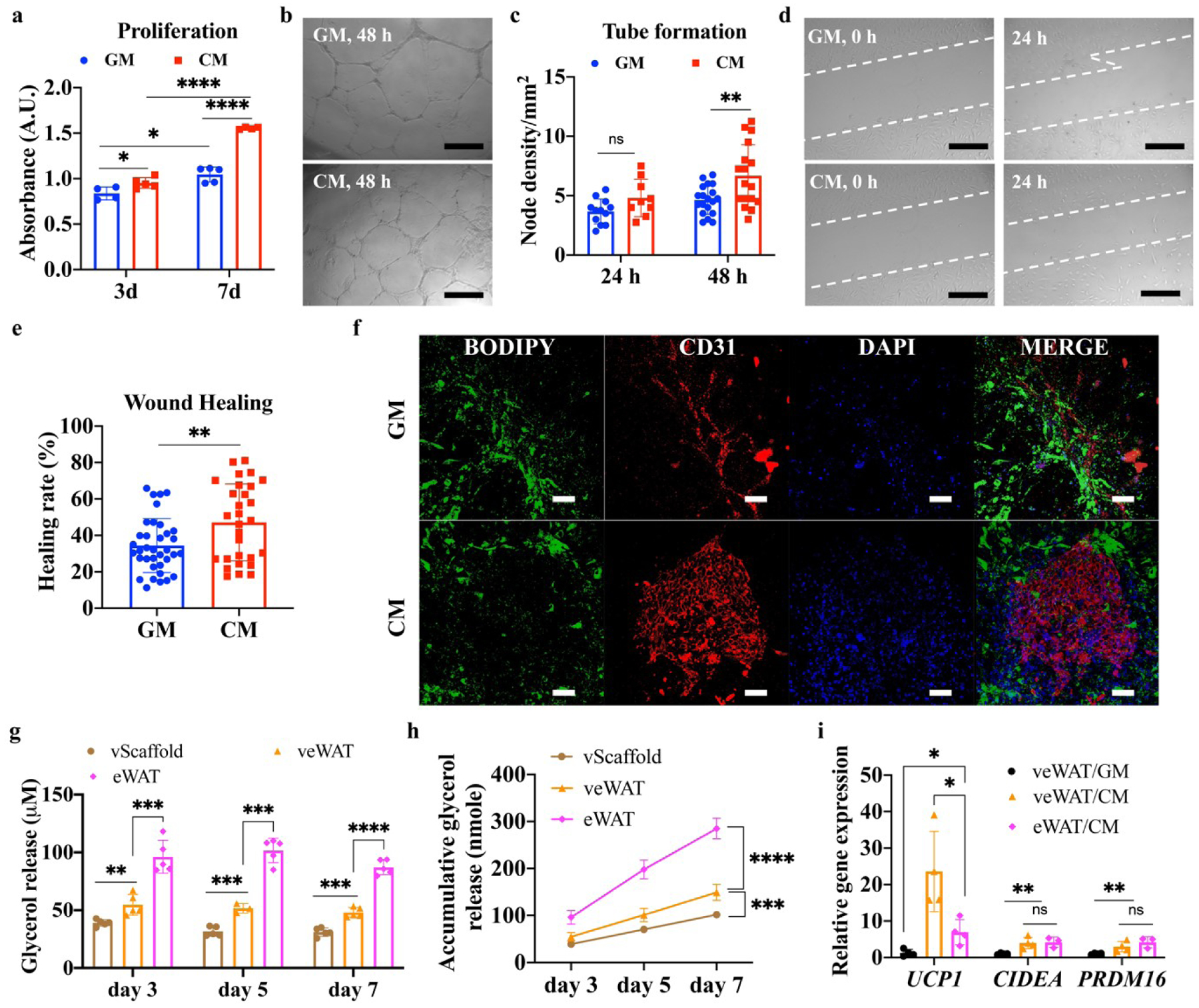
The effects of growth medium (GM) and conditioned medium (CM) on 2D cultured human umbilical vein endothelial cells (HUVECs) and vascularized engineered human white adipose tissues (veWATs). (a) Proliferation of HUVECs after CM and GM induction (n=4–5). (b) Tubular structures of HUVECs after CM and GM induction. Scale bar=500 μm. (c) Node density of HUVECs after CM and GM induction for 24 h and 48 h (n=5, 3 images from each sample were evaluated). (d) Wound healing images of HUVECs after CM and GM induction for 24 h. Scale bar=500 μm. (e) Healing rate of HUVECs after CM and GM induction for 24 h (n=5, 3 images from each sample were evaluated). (f) Immunofluorescent staining of veWAT after induction with CM and GM for 7 days. Scale bar=100 μm. (g) Glycerol release of vascularized acellular scaffolds (vScaffolds), engineered human white adipose tissue (eWATs), and veWATs under CM induction at different time intervals (n=5). (h) Cumulative glycerol release of vScaffold, eWATs, and veWATs under CM treatment (n=5). (i) Browning gene expressions (UCP1, CIDEA, PRDM16) of eWATs and veWATs under CM and GM induction for 7 days (n=3–4).
To better simulate native vascularized adipose tissue, HUVECs and ADMSCs in mixed collagen and fibrin hydrogels were wrapped around 3D eWATs and cultured for 7 days to undergo angiogenesis. The obtained veWATs were further induced with CM or GM for another 7 days. Their viabilities are shown in figure S2. Consistent with the 2D results, we found that CM induction enhanced angiogenesis within veWATs. As shown in figure 6(f), pre-vascularized constructs induced by CM illustrated denser and more interconnected capillaries, while veWATs in GM showed weak and partial CD31 distribution. In addition, less BODIPY stained lipids were detected in CM induced veWATs, indicating augmented fat loss.
Quantitative evaluation of glycerol release and lipolysis showed that both eWATs and veWATs responded to CM induction with increased glycerol release after 3, 5, and 7-day culture, compared to the vScaffold (figure 6(g) and 6(h)). Interestingly, we also found that veWATs had a significantly lower glycerol release but an upregulated expression of UCP1, compared to their counterparts without pre-vascularization (i.e., eWATs) (figure 6(i)). The expressions of CIDEA and PRDM16 were comparable in both eWATs and veWATs with CM induction but were significantly downregulated in veWATs under GM conditions (figure 6(i)). This indicated that the pre-vascularization process might decrease the lipolysis but increase the browning process in response to CM induction.
3.5. Pro-inflammatory cytokines in CM and alteration of adipokines released from eWATs after CM induction
We hypothesized that cytokines in CM contributed to the lipolysis, UCP1 up-regulation, and ECM enhancement in eWATs and veWATs. A cytokine array assay was performed with BxPC-3 fresh growth medium and supernatants. As expected, no cytokines were detected in BxPC-3 growth medium per se (figure 7(a)). In contrast, 8 different cytokines were identified in the supernatant of BxPC-3 cells. Among them, interleukin (IL)-8 and C-X-C motif chemokine ligand (CXCL) 1 were the two most abundant cytokines, as reflected by dot pixel intensity (figure 7(b)). IL-8 is an inflammatory cytokine, and its level is reported to have stronger positive correlation with cachexia status, weight loss, and sarcopenia in pancreatic patients than other pro-inflammatory cytokines like IL-1β, IL-6 and tumor necrosis factor alpha (TNF-α) [41]. IL-8 was also found to promote HUVEC proliferation and tube formation [42]. Thus, IL-8 can be used as a prognostic indicator in pancreatic CAC [43]. CXCL1, a major member of the chemokine family, was also identified and reported to promote diverse biological activities, including tumor growth and angiogenesis, in former research [39]. Other present cytokines included plasminogen activator inhibitor-1 (PAI-1), granulocyte-macrophage colony stimulating factor (GM-CSF), macrophage migration inhibitory factor (MIF), IL-18, granulocyte colony-stimulating factor (G-CSF), and cluster of differentiation (CD) 54 (figure 7(a) and 7(b)). GM-CSF and G-CSF are known to induce myeloid progenitor differentiation and myeloid-derived suppressor cell (MDSC) expansion [44, 45]. MDSCs can suppress antitumor T cell responses and promote tumor angiogenesis and metastasis [46]. While IL-6 and IL-8 have been extensively studied in CAC, the effects of IL-18 were largely undiscovered [47]. Andrew et. al. reported that IL-18 was activated by Caspase-1 in inflammasome complexes and had anti-obesity effects. They found that mice with increased plasma IL-18 concentrations strikingly led to adipose tissue lipolysis, which could be prevented by genetic deletion of IL-18 [48]. PAI-1, MIF, and CD54 also play roles in the maintenance of an inflammatory state and tumor aggressiveness in cancer disease [49, 50].
Figure 7.
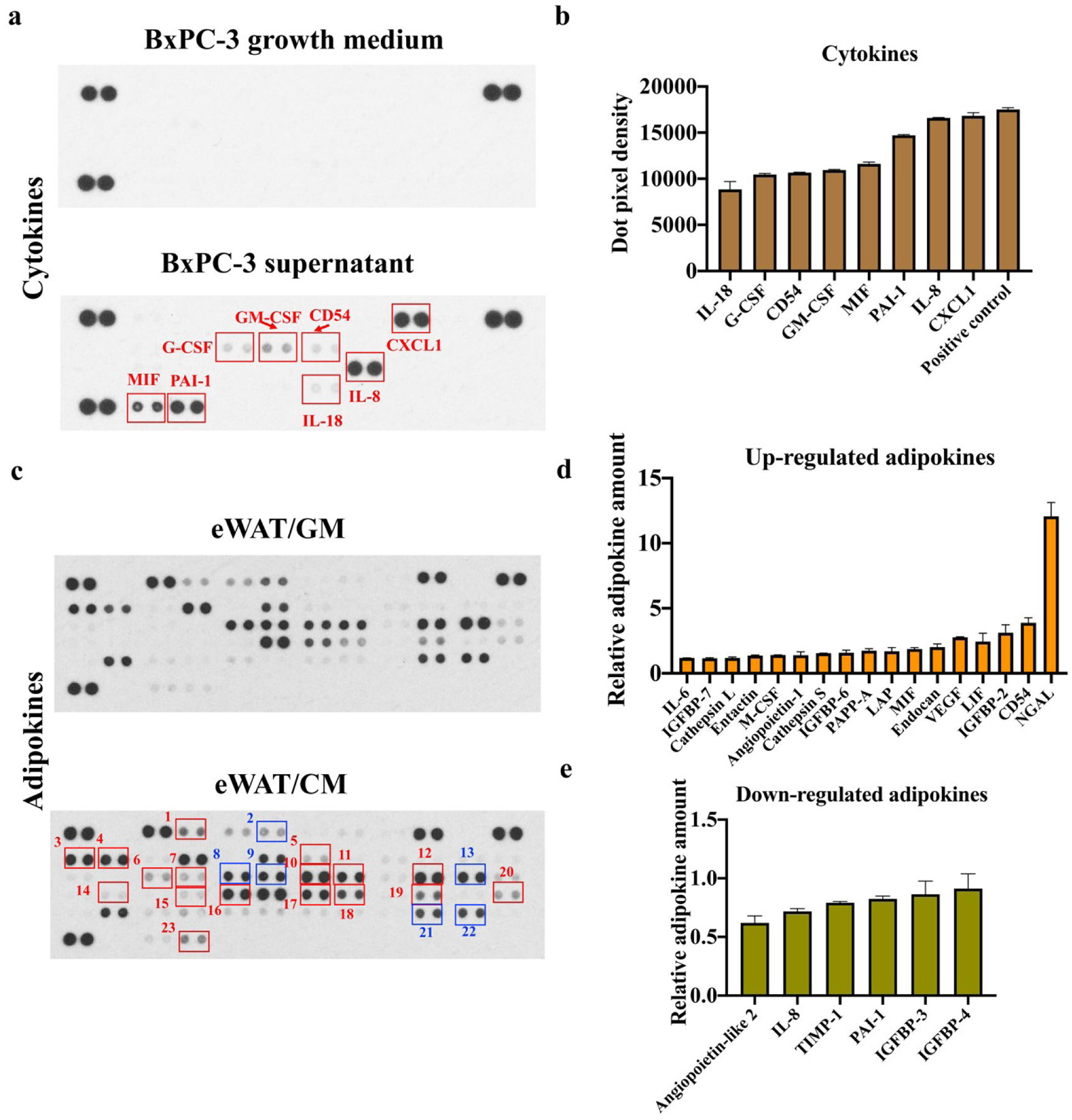
Human cytokine and adipokine arrays. (a) Cytokine array of BxPC-3 growth medium and BxPC-3 supernatants. (b) Dot pixel density of cytokines in BxPC-3 supernatants. (c) Adipokine arrays of engineered human white adipose tissue (eWATs) induced with growth medium (GM) or conditioned medium (CM). Red rectangles represent up-regulated adipokines, and blue rectangles represent down-regulated adipokines (1: Angiopoietin-1; 2: Angiopoietin-like 2; 3: Cathepsin L; 4: Cathepsin S; 5: Endocan; 6: CD54; 7: IGFBP-2; 8: IGFBP-3; 9: IGFBP-4; 10: IGFBP-6; 11: IGFBP-7; 12: IL-6; 13: IL-8; 14: LAP; 15: LIF; 16: Lipocalin-2/NGAL; 17: M-CSF; 18: MIF; 19: Nidogen-1/Entactin; 20: Pappalysin-1/PAPP-A; 21: PAI-1; 22: TIMP-1; 23: VEGF). (d) Relative amount of up-regulated adipokines in eWATs induced with CM. (e) Relative amount of down-regulated adipokines in eWATs induced with CM.
Adipose tissue is a crucial endocrine organ that produces and releases a variety of adipokines and bioactive factors [51]. Previous studies suggested that during CAC progression, pathological alterations of WAT may precede muscle wasting [4, 52]. This indicates that WAT may not only be a victim of the CAC but also an important contributing factor to muscle atrophy and further vicious CAC cycle formation. Therefore, inflammatory cytokines in the CM may trigger adipocytes to express and secrete various adipokines. The adipokine array analysis was conducted to evaluate eWATs in response to CM and GM induction (figure 7(c)). As shown in figure 7(d), neutrophil gelatinase-associated lipocalin (NGAL), a glycoprotein belonging to the lipocalin family, was the most upregulated adipokine after CM induction and has been demonstrated to be an inflammatory marker closely interrelated with insulin resistance and hyperglycemia [53]. The secretions of several insulin-like growth factor-binding protein (IGFBP) family members were also increased. The proteins from the IGFBP family serve as transport proteins for insulin-like growth factor, and their increased expressions were reported to correlate with cancer’s metastatic ability and muscle atrophy [54, 55]. The synthesis and secretion of pro-inflammatory adipokines, like leukemia inhibitory factor (LIF) and IL-6, have been observed, and those molecules may promote lipid depletion, inhibit lipid synthesis, and drive the expression of UCP1 [51, 56]. The increased secretions of VEGF, endocan, and angiopoietin 1 contribute to the regulation of angiogenesis [57]. Enhanced secretion of Entactin after CM induction may help collagen synthesis and ECM remodeling [58]. Among six down-regulated adipokines, angiopoietin-like (ANGPTL) 2 was determined to be the most downregulated (figure 7(e)). ANGPTL2 maintains tissue homeostasis and displays both physiological and pathological functions [59]. A reduction of the ANGPTL2 expression was reported to inhibit adipogenesis and induce insulin resistance [60]. Despite the upregulation of several inflammatory adipokines in eWAT/CM samples, IL-8 and PAI-1, which were highly expressed in CM, were downregulated (figure 7(e)). Most current studies have been restricted to assessing systemic inflammation by investigating the parameters of the plasma, while neglecting local tissue inflammation [61]. It is speculated that IL-8 and PAI-1 in CM may bind with the receptors on adipocytes within eWATs and thus showed the decreased secretion.
4. Discussion
In this study, we applied 3D bioprinting technology to develop scaffold-based WAT models with interconnected channels. The human WAPs were encapsulated in HAMA and GelMA bioinks and underwent UV crosslinking to obtain WAP-laden constructs. The interconnected channel structure was designed to facilitate the nutrient transportation, and it was generated in a single 3D bioprinting process, rather than involving any sacrificial materials or further washing step in traditional molding method. After adipogenic differentiation, WAPs exhibited increased lipid droplet size and augmented PPARG and CEBPE expression, establishing a human 3D eWATs system in vitro. Then, a pancreatic cancer-cell-conditioned medium was used to induce eWATs to study their remodeling during cancer-associated cachexia in vitro.
After CM induction, a series of functional, gene expression, and morphological alterations happened in the eWAT models. Lipolysis, with increased free glycerol release, was detected in the media of eWATs after CM induction (figure 4(a) and 4(b)). Morphologically, a smaller lipid size and fewer lipids in CM-induced eWATs were shown by IF staining and Oil Red O images, compared to control groups (figure 4(e) and figure 5). This all indicated adipose lipolysis, where lipid triglycerides were hydrolyzed into glycerol and three fatty acids [62], under CM inducement. The underlying mechanisms that control the adipose atrophy and lipolysis were found to be related to adipose triglyceride lipase (ATGL) and hormone-sensitive lipase (HSL), according to Das [63]. ATGL or HSL deficient mice showed resistance to WAT lipolysis and proteasomal muscle degradation by CAC. Adipose browning-related genes (UCP1, CIDEA, PRDM16) and proteins (UCP1) were also up-regulated in CM-induced eWATs (figure 4(c) and 4(e)). Browning is the conversion of white adipocytes into brown adipocytes and has been found to be associated with CAC [1, 64]. WAT browning will lead to lipid mobilization and increased energy expenditure, through the characteristic protein, UCP1 [65]. Inflammatory molecules were found to cause an increase in UCP1 expression in CAC [1]. In addition, eWATs illustrated obvious ECM development after CM induction (figure 5). The ECM of WAT is mainly composed of collagens, fibronectin, glycoproteins, and proteoglycans, and its remodeling happens under physiological or pathological conditions [64]. During adipogenic differentiation, there is a reduction of the fibronectin matrix and a formation of basal lamina [66]. Under CAC, collagen fiber content augmentation, elastic fiber synthesis, and fibrosis were reported [67, 68]. In our study, we also found an enhancement of ECM distribution in eWATs after CM induction, especially with the increase of collagen I deposition, demonstrated by IHC staining (figure 5).
It is currently believed that CAC is a chronic inflammatory syndrome related to cytokines from the tumor and the host, including IL-8, CXCL-1, IL-18, PAI-1, MIF, and CD54, as we found in BxPC-3 supernatants (figure 7(a) and 7(b)) [69]. Those pro-inflammatory cytokines could promote tumor growth, angiogenesis, and tissue inflammation [39]. As a complex tissue, WAT is susceptible to those cytokines and remodels itself through lipolysis, browning, ECM accumulation, and adipose inflammation [70, 71]. Adipose tissue inflammation is characterized by the infiltration of macrophages and neutrophils, as reported by previous studies [72]. The dysfunctional WAT also secretes various adipokines, like NGAL, CD54, IGFBP-2, etc., as we reported (figure 7(c) and 7(d)), to promote tumorigenesis and cancer progression via oncogenic signaling or indirect mechanisms, such as angiogenesis and immunomodulation [73, 74]. Crosstalk between inflamed WAT and muscle tissue may also further promote muscle atrophy and wasting [75].
WAT is highly vascularized, with adipocytes close to capillaries in vivo [76]. A change of the vasculature can be induced by numerous physiological and pathological stimuli and is an important step in disease progression [77]. CM induction was applied then [78]. For the 2D cultured cells, we found that CM could improve the proliferation, tube formation, and wound healing rate of HUVECs, compared to GM (figure 6(a)-(e)). The angiogenesis of 3D veWATs was also promoted under CM induction (figure 6(f)). The results were highly related to pro-inflammatory cytokines in the CM and correspondingly released adipokines from adipocytes, like IL-8, GM-CSF, G-CSF, and VEGF, which could improve HUVEC growth and capillaries. Interestingly, pre-vascularization of eWATs provoked a decrease of glycerol release and an increase of UCP1 gene expression under CM induction, compared to eWATs without vascularization (figure 6(g)-(i)). Previous studies also reported that the incorporation of HUVECs in the coculture models decreased adipogenesis and isoproterenol-stimulated lipolysis [79, 80]. In addition, endothelial cells and angiogenesis were also reported to promote the browning of WATs [81, 82]. All these results indicate that endothelial cells play a complicated role in the pathological remodeling of WATs. Meanwhile, the adipokines secreted by adipocytes can also affect angiogenesis and endothelial cell functions [83, 84]. The underlying process includes not only the crosstalk between endothelial cells and adipocytes but also their interactions and remodeling under a cancerous environment, like CM induction in this study. Tissue crosstalk during CAC remains a subject of debate and is not well understood. We speculate that both cytokines in CM and adipokines from adipocytes increased WAT vasculature, and they also alter the inflammatory states of adipocytes and HUVECs [85].
Thus, we successfully developed an engineered WAT model that could recapitulate typical CAC manifestations to study adipose tissue loss in vitro. The eWATs were also combined with HUVECs to evaluate the effects of vascularization on adipose remodeling under a cancer medium environment. The eWAT is therefore an ideal model system for future research to uncover the biology of CAC pathophysiology and to develop potential therapies. One limitation of the current eWAT model is that the adipocyte density is low compared to that of adipose tissue in vivo, and the adipocytes may not have enough interactions within the scaffolds, which is hoped to be resolved in our future research. Besides, cancer cells cultured in different environments, like 3D instead of 2D conditions, may have a different secretome composition and level. It can thus lead to altered cachexia remodeling of the adipose tissue, which can be studied in the future. In this study, BxPC-3 was used as an example to investigate the model feasibility in cachexia related research, and more cancer cell lines or primary pancreatic cancer cells can be investigated in the future.
5. Conclusion
In this study, we developed scaffold-based 3D bioprinted eWATs for studying the underlying mechanisms of adipose tissue atrophy during the development of cancer cachexia. The GelMA and HAMA-based bioink and 3D printed scaffolds successfully supported the differentiation of adipocytes. The developed 3D eWATs demonstrated the biology of cancer cachexia, exhibiting an increase of glycerol release, upregulation of browning-related gene and protein expressions, and enhancement of ECM deposition after CM induction. To further mimic native WATs and understand the role of vascularization in adipose remodeling under CAC, eWATs were vascularized into veWATs. CM was found to promote the angiogenesis of 2D cultured HUVECs as well as 3D veWATs. In addition, compared to eWATs, veWATs had decreased glycerol release but increased UCP1 expression under CM induction. Inflammatory cytokines, like IL-8, CXCL-1, GM-CSF, etc., were found in CM and are thought to induce the remodeling of eWATs/veWATs. Inflammatory cytokines in the CM also altered the adipokine secretion of eWATs and increased the secretion of various cancer metastasis and vascularization related adipokines.
Supplementary Material
Acknowledgement
This work has been supported by Mary & Dick Holland Regenerative Medicine Program start-up grant and pilot grant, University of Nebraska Collaboration Initiative to Dr. Bin Duan, and 5R01HD096042-02 and University of Nebraska Collaboration Initiative NRI2132270010 to Dr. So-Youn Kim.
Abbreviations:
- 2D
two-dimensional
- 3D
three-dimensional
- ADMSCs
adipose derived mesenchymal stem cells
- BSA
bovine serum albumin
- CAC
cancer-associated cachexia
- CEBPE
CCAAT/enhancer binding protein epsilon
- CIDEA
Cell Death Inducing DFFA Like Effector A
- CLSM
confocal laser scanning microscope
- CM
conditioned medium
- DLP
digital light processing
- ECGM
Endothelial Cell Growth Medium
- ECM
extracellular matrix
- GelMA
methacrylated gelatin
- GM
growth medium
- HAMA
methacrylated hyaluronic acid
- H&E
hematoxylin and eosin
- HUVECs
human umbilical vein endothelial cells
- IF
immunofluorescent
- IHC
immunohistochemical
- LAP
lithium phenyl-2,4,6-trimethylbenzoylphosphinate
- LPL
lipoprotein lipase
- PFA
paraformaldehyde
- PPARG
peroxisome proliferator-activated receptor gamma
- PRDM16
PR/SET Domain 16
- qPCR
quantitative polymerase chain reaction
- SEM
scanning electron microscope
- UCP1
uncoupling Protein 1
- vScaffold
vascularized acellular scaffolds
- WAPs
human white adipocyte progenitors
- WAT
human white adipose tissue
- eWAT
engineered human white adipose tissue
- veWATs
vascularized eWATs
Footnotes
Conflict of Interest
The authors declare no competing financial interest.
Reference:
- [1].Petruzzelli M, Schweiger M, Schreiber R, Campos-Olivas R, Tsoli M, Allen J, Swarbrick M, Rose-John S, Rincon M, Robertson G, A switch from white to brown fat increases energy expenditure in cancer-associated cachexia, Cell metabolism 20(3) (2014) 433–447. [DOI] [PubMed] [Google Scholar]
- [2].Hu W, Ru Z, Xiao W, Xiong Z, Wang C, Yuan C, Zhang X, Yang H, Adipose tissue browning in cancer-associated cachexia can be attenuated by inhibition of exosome generation, Biochemical and biophysical research communications 506(1) (2018) 122–129. [DOI] [PubMed] [Google Scholar]
- [3].Palesty J, Dudrick S, What we have learned about cachexia in gastrointestinal cancer, Digestive Diseases 21(3) (2003) 198–213. [DOI] [PubMed] [Google Scholar]
- [4].Kliewer KL, Ke J-Y, Tian M, Cole RM, Andridge RR, Belury MA, Adipose tissue lipolysis and energy metabolism in early cancer cachexia in mice, Cancer biology & therapy 16(6) (2015) 886–897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Agustsson T, Rydén M, Hoffstedt J, van Harmelen V, Dicker A, Laurencikiene J, Isaksson B, Permert J, Arner P, Mechanism of increased lipolysis in cancer cachexia, Cancer research 67(11) (2007) 5531–5537. [DOI] [PubMed] [Google Scholar]
- [6].Visavadiya NP, Pena GS, Khamoui AV, Mitochondrial dynamics and quality control are altered in a hepatic cell culture model of cancer cachexia, Molecular and Cellular Biochemistry 476(1) (2021) 23–34. [DOI] [PubMed] [Google Scholar]
- [7].Bonetto A, Rupert JE, Barreto R, Zimmers TA, The colon-26 carcinoma tumor-bearing mouse as a model for the study of cancer cachexia, JoVE (Journal of Visualized Experiments) (117) (2016) e54893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Han Y-H, Mun J-G, Jeon HD, Yoon DH, Choi B-M, Kee J-Y, Hong S-H, The extract of Arctium lappa L. fruit (Arctii Fructus) improves cancer-induced cachexia by inhibiting weight loss of skeletal muscle and adipose tissue, Nutrients 12(10) (2020) 3195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Larian N, Ensor M, Thatcher SE, English V, Morris AJ, Stromberg A, Cassis LA, Pseudomonas aeruginosa-derived pyocyanin reduces adipocyte differentiation, body weight, and fat mass as mechanisms contributing to septic cachexia, Food and Chemical Toxicology 130 (2019) 219–230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Murphy CS, Liaw L, Reagan MR, In vitro tissue-engineered adipose constructs for modeling disease, BMC biomedical engineering 1(1) (2019) 1–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Taylor J, Sellin J, Kuerschner L, Krähl L, Majlesain Y, Förster I, Thiele C, Weighardt H, Weber E, Generation of immune cell containing adipose organoids for in vitro analysis of immune metabolism, Scientific reports 10(1) (2020) 1–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Nzou G, Wicks R, Wicks E, Seale S, Sane C, Chen A, Murphy S, Jackson J, Atala A, Human cortex spheroid with a functional blood brain barrier for high-throughput neurotoxicity screening and disease modeling, Scientific reports 8(1) (2018) 1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Haisler WL, Timm DM, Gage JA, Tseng H, Killian T, Souza GR, Three-dimensional cell culturing by magnetic levitation, Nature protocols 8(10) (2013) 1940–1949. [DOI] [PubMed] [Google Scholar]
- [14].Bartosh TJ, Ylöstalo JH, Mohammadipoor A, Bazhanov N, Coble K, Claypool K, Lee RH, Choi H, Prockop DJ, Aggregation of human mesenchymal stromal cells (MSCs) into 3D spheroids enhances their antiinflammatory properties, Proceedings of the National Academy of Sciences 107(31) (2010) 13724–13729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Batista ML Jr, Meshulam T, Desevin K, Rabhi N, Farmer SR, Three-Dimensional Adipocyte Culture as a Model to Study Cachexia-Induced White Adipose Tissue Remodeling, JoVE (Journal of Visualized Experiments) (167) (2021) e61853. [DOI] [PubMed] [Google Scholar]
- [16].Hsiao AY, Okitsu T, Teramae H, Takeuchi S, 3D tissue formation of unilocular adipocytes in hydrogel microfibers, Advanced healthcare materials 5(5) (2016) 548–556. [DOI] [PubMed] [Google Scholar]
- [17].Qi D, Wu S, Kuss MA, Shi W, Chung S, Deegan PT, Kamenskiy A, He Y, Duan B, Mechanically robust cryogels with injectability and bioprinting supportability for adipose tissue engineering, Acta biomaterialia 74 (2018) 131–142. [DOI] [PubMed] [Google Scholar]
- [18].Mauney JR, Nguyen T, Gillen K, Kirker-Head C, Gimble JM, Kaplan DL, Engineering adipose-like tissue in vitro and in vivo utilizing human bone marrow and adipose-derived mesenchymal stem cells with silk fibroin 3D scaffolds, Biomaterials 28(35) (2007) 5280–5290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Aubin K, Safoine M, Proulx M, Audet-Casgrain M-A, Côté J-F, Têtu F-A, Roy A, Fradette J, Characterization of in vitro engineered human adipose tissues: relevant adipokine secretion and impact of TNF-α, PloS one 10(9) (2015) e0137612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Klingelhutz AJ, Gourronc FA, Chaly A, Wadkins DA, Burand AJ, Markan KR, Idiga SO, Wu M, Potthoff MJ, Ankrum JA, Scaffold-free generation of uniform adipose spheroids for metabolism research and drug discovery, Scientific reports 8(1) (2018) 1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Delort L, Lequeux C, Dubois V, Dubouloz A, Billard H, Mojallal A, Damour O, Vasson M-P, Caldefie-Chezet F, Reciprocal interactions between breast tumor and its adipose microenvironment based on a 3D adipose equivalent model, PLoS One 8(6) (2013) e66284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Rupnick MA, Panigrahy D, Zhang C-Y, Dallabrida SM, Lowell BB, Langer R, Folkman MJ, Adipose tissue mass can be regulated through the vasculature, Proceedings of the National Academy of Sciences 99(16) (2002) 10730–10735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Cao Y, Angiogenesis and vascular functions in modulation of obesity,adipose metabolism, and insulin sensitivity, Cell metabolism 18(4) (2013) 478–489. [DOI] [PubMed] [Google Scholar]
- [24].Muller S, Ader I, Creff J, Leménager H, Achard P, Casteilla L, Sensebé L, Carrière A, Deschaseaux F, Human adipose stromal-vascular fraction self-organizes to form vascularized adipose tissue in 3D cultures, Scientific reports 9(1) (2019) 1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Strassburg S, Nienhueser H, Björn Stark G, Finkenzeller G, Torio-Padron N, Co-culture of adipose-derived stem cells and endothelial cells in fibrin induces angiogenesis and vasculogenesis in a chorioallantoic membrane model, Journal of tissue engineering and regenerative medicine 10(6) (2016) 496–506. [DOI] [PubMed] [Google Scholar]
- [26].Kuss MA, Wu S, Wang Y, Untrauer JB, Li W, Lim JY, Duan B, Prevascularization of 3D printed bone scaffolds by bioactive hydrogels and cell co-culture, Journal of Biomedical Materials Research Part B: Applied Biomaterials 106(5) (2018) 1788–1798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Haubner F, Leyh M, Ohmann E, Pohl F, Prantl L, Gassner HG, Effects of external radiation in a co-culture model of endothelial cells and adipose-derived stem cells, Radiation Oncology 8(1) (2013) 1–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Choi JH, Gimble JM, Vunjak-Novakovic G, Kaplan DL, Effects of hyperinsulinemia on lipolytic function of three-dimensional adipocyte/endothelial co-cultures, Tissue Engineering Part C: Methods 16(5) (2010) 1157–1165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Michaelis KA, Zhu X, Burfeind KG, Krasnow SM, Levasseur PR, Morgan TK, Marks DL, Establishment and characterization of a novel murine model of pancreatic cancer cachexia, Journal of cachexia, sarcopenia and muscle 8(5) (2017) 824–838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Xue R, Lynes MD, Dreyfuss JM, Shamsi F, Schulz TJ, Zhang H, Huang TL, Townsend KL, Li Y, Takahashi H, Clonal analyses and gene profiling identify genetic biomarkers of the thermogenic potential of human brown and white preadipocytes, Nature medicine 21(7) (2015) 760–768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Duan B, Yin Z, Kang LH, Magin RL, Butcher JT, Active tissue stiffness modulation controls valve interstitial cell phenotype and osteogenic potential in 3D culture, Acta biomaterialia 36 (2016) 42–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Zhang W, Shi W, Wu S, Kuss M, Jiang X, Untrauer JB, Reid SP, Duan B, 3D printed composite scaffolds with dual small molecule delivery for mandibular bone regeneration, Biofabrication 12(3) (2020) 035020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Jiang X, Wu S, Kuss M, Kong Y, Shi W, Streubel PN, Li T, Duan B, 3D printing of multilayered scaffolds for rotator cuff tendon regeneration, Bioactive materials 5(3) (2020) 636–643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Alkhouli N, Mansfield J, Green E, Bell J, Knight B, Liversedge N, Tham JC, Welbourn R, Shore AC, Kos K, The mechanical properties of human adipose tissues and their relationships to the structure and composition of the extracellular matrix, American Journal of Physiology-Endocrinology and Metabolism 305(12) (2013) E1427–E1435. [DOI] [PubMed] [Google Scholar]
- [35].Park HS, Kim SH, Kim YS, Ryu SY, Hwang JT, Yang HJ, Kim GH, Kwon DY, Kim MS, Luteolin inhibits adipogenic differentiation by regulating PPARγ activation, Biofactors 35(4) (2009) 373–379. [DOI] [PubMed] [Google Scholar]
- [36].Hadrich F, Sayadi S, Apigetrin inhibits adipogenesis in 3T3-L1 cells by downregulating PPARγ and CEBP-α, Lipids in health and disease 17(1) (2018) 1–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Chan JK, The wonderful colors of the hematoxylin–eosin stain in diagnostic surgical pathology, International journal of surgical pathology 22(1) (2014) 12–32. [DOI] [PubMed] [Google Scholar]
- [38].Annese T, Tamma R, Ruggieri S, Ribatti D, Angiogenesis in pancreatic cancer: pre-clinical and clinical studies, Cancers 11(3) (2019) 381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Matsuo Y, Raimondo M, Woodward TA, Wallace MB, Gill KR, Tong Z, Burdick MD, Yang Z, Strieter RM, Hoffman RM, CXC-chemokine/CXCR2 biological axis promotes angiogenesis in vitro and in vivo in pancreatic cancer, International journal of cancer 125(5) (2009) 1027–1037. [DOI] [PubMed] [Google Scholar]
- [40].Ma J, Sawai H, Ochi N, Matsuo Y, Xu D, Yasuda A, Takahashi H, Wakasugi T, Takeyama H, PTEN regulate angiogenesis through PI3K/Akt/VEGF signaling pathway in human pancreatic cancer cells, Molecular and cellular biochemistry 331(1) (2009) 161–171. [DOI] [PubMed] [Google Scholar]
- [41].Hou Y-C, Wang C-J, Chao Y-J, Chen H-Y, Wang H-C, Tung H-L, Lin J-T, Shan Y-S, Elevated serum interleukin-8 level correlates with cancer-related cachexia and sarcopenia: An indicator for pancreatic cancer outcomes, Journal of clinical medicine 7(12) (2018) 502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Matsuo Y, Sawai H, Funahashi H, Takahashi H, Sakamoto M, Yamamoto M, Okada Y, Hayakawa T, Manabe T, Enhanced angiogenesis due to inflammatory cytokines from pancreatic cancer cell lines and relation to metastatic potential, Pancreas 28(3) (2004) 344–352. [DOI] [PubMed] [Google Scholar]
- [43].Vaes RD, van Dijk DP, Welbers TT, Blok MJ, Aberle MR, Heij L, Boj SF, Olde Damink SW, Rensen SS, Generation and initial characterization of novel tumour organoid models to study human pancreatic cancer-induced cachexia, Journal of cachexia, sarcopenia and muscle 11(6) (2020) 1509–1524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Wu W-C, Sun H-W, Chen H-T, Liang J, Yu X-J, Wu C, Wang Z, Zheng L, Circulating hematopoietic stem and progenitor cells are myeloid-biased in cancer patients, Proceedings of the National Academy of Sciences 111(11) (2014) 4221–4226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].Dolcetti L, Peranzoni E, Ugel S, Marigo I, Fernandez Gomez A, Mesa C, Geilich M, Winkels G, Traggiai E, Casati A, Hierarchy of immunosuppressive strength among myeloid‐derived suppressor cell subsets is determined by GM-CSF, European journal of immunology 40(1) (2010) 22–35. [DOI] [PubMed] [Google Scholar]
- [46].Sun H-W, Wu W-C, Chen H-T, Xu Y-T, Yang Y-Y, Chen J, Yu X-J, Wang Z, Shuang Z-Y, Zheng L, Glutamine deprivation promotes the generation and mobilization of MDSCs by enhancing expression of G-CSF and GM-CSF, Frontiers in immunology 11 (2020) 3759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47].Penafuerte CA, Gagnon B, Sirois J, Murphy J, MacDonald N, Tremblay ML, Identification of neutrophil-derived proteases and angiotensin II as biomarkers of cancer cachexia, British journal of cancer 114(6) (2016) 680–687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [48].Murphy AJ, Kraakman MJ, Kammoun HL, Dragoljevic D, Lee MK, Lawlor KE, Wentworth JM, Vasanthakumar A, Gerlic M, Whitehead LW, IL-18 production from the NLRP1 inflammasome prevents obesity and metabolic syndrome, Cell metabolism 23(1) (2016) 155–164. [DOI] [PubMed] [Google Scholar]
- [49].Serag Esmat RF, Rashed L, Effect of exercise on plasminogen activator inhibitor-1 (PAI-1) level in patients with metabolic syndrome, Journal of American Science 6(12) (2010) 1374–1380. [Google Scholar]
- [50].Manukyan G, Papajik T, Gajdos P, Mikulkova Z, Urbanova R, Gabcova G, Kudelka M, Turcsányi P, Ryznerova P, Prochazka V, Neutrophils in chronic lymphocytic leukemia are permanently activated and have functional defects, Oncotarget 8(49) (2017) 84889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [51].Mannelli M, Gamberi T, Magherini F, Fiaschi T, The adipokines in cancer cachexia, International Journal of Molecular Sciences 21(14) (2020) 4860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [52].Byerley LO, Lee SH, Redmann S, Culberson C, Clemens M, Lively MO, Evidence for a novel serum factor distinct from zinc alpha-2 glycoprotein that promotes body fat loss early in the development of cachexia, Nutrition and cancer 62(4) (2010) 484–494. [DOI] [PubMed] [Google Scholar]
- [53].Abella V, Scotece M, Conde J, Gómez R, Lois A, Pino J, Gómez-Reino JJ, Lago F, Mobasheri A, Gualillo O, The potential of lipocalin-2/NGAL as biomarker for inflammatory and metabolic diseases, Biomarkers 20(8) (2015) 565–571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [54].Dong J, Yu J, Li Z, Gao S, Wang H, Yang S, Wu L, Lan C, Zhao T, Gao C, Serum insulin-like growth factor binding protein 2 levels as biomarker for pancreatic ductal adenocarcinoma-associated malnutrition and muscle wasting, Journal of cachexia, sarcopenia and muscle 12 (2021) 704–716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [55].Wang C, Gao C, Meng K, Qiao H, Wang Y, Human adipocytes stimulate invasion of breast cancer MCF-7 cells by secreting IGFBP-2, PloS one 10(3) (2015) e0119348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [56].Terawaki K, Sawada Y, Kashiwase Y, Hashimoto H, Yoshimura M, Suzuki M, Miyano K, Sudo Y, Shiraishi S, Higami Y, New cancer cachexia rat model generated by implantation of a peritoneal dissemination-derived human stomach cancer cell line, American Journal of Physiology-Endocrinology and Metabolism 306(4) (2014) E373–E387. [DOI] [PubMed] [Google Scholar]
- [57].Poon PY-K, Ng JK-C, Fung WW-S, Chow K-M, Kwan BC-H, Li PK-T, Szeto C-C, Relationship between plasma endocan level and clinical outcome of Chinese peritoneal dialysis patients, Kidney and Blood Pressure Research 44(5) (2019) 1259–1270. [DOI] [PubMed] [Google Scholar]
- [58].Aratani Y, Kitagawa Y, Enhanced synthesis and secretion of type IV collagen and entactin during adipose conversion of 3T3-L1 cells and production of unorthodox laminin complex, Journal of Biological Chemistry 263(31) (1988) 16163–16169. [PubMed] [Google Scholar]
- [59].Thorin-Trescases N, Thorin E, Angiopoietin-like-2: a multifaceted protein with physiological and pathophysiological properties, Expert Reviews in Molecular Medicine 16 (2014) e17. [DOI] [PubMed] [Google Scholar]
- [60].Kitazawa M, Nagano M, Masumoto K.-h., Shigeyoshi Y, Natsume T, Hashimoto S, Angiopoietin-like 2, a circadian gene, improves type 2 diabetes through potentiation of insulin sensitivity in mice adipocytes, Endocrinology 152(7) (2011) 2558–2567. [DOI] [PubMed] [Google Scholar]
- [61].Henriques F, Júnior MLB, Adipose tissue remodeling during cancer-associated cachexia: translational features from adipose tissue dysfunction, Immunometabolism 4(3) (2020) e200032. [Google Scholar]
- [62].Arner P, Lipases in cachexia, Science 333(6039) (2011) 163–164. [DOI] [PubMed] [Google Scholar]
- [63].Das SK, Eder S, Schauer S, Diwoky C, Temmel H, Guertl B, Gorkiewicz G, Tamilarasan KP, Kumari P, Trauner M, Adipose triglyceride lipase contributes to cancer-associated cachexia, Science 333(6039) (2011) 233–238. [DOI] [PubMed] [Google Scholar]
- [64].Kir S, White JP, Kleiner S, Kazak L, Cohen P, Baracos VE, Spiegelman BM, Tumour-derived PTH-related protein triggers adipose tissue browning and cancer cachexia, Nature 513(7516) (2014) 100–104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [65].Bae S, Park J, Kim J-S, Cas-OFFinder: a fast and versatile algorithm that searches for potential off-target sites of Cas9 RNA-guided endonucleases, Bioinformatics 30(10) (2014) 1473–1475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [66].Spiegelman BM, Ginty CA, Fibronectin modulation of cell shape and lipogenic gene expression in 3T3-adipocytes, Cell 35(3) (1983) 657–666. [DOI] [PubMed] [Google Scholar]
- [67].Alves MJ, Figuerêdo RG, Azevedo FF, Cavallaro DA, Neto NIP, Lima JDC, Matos-Neto E, Radloff K, Riccardi DM, Camargo RG, Adipose tissue fibrosis in human cancer cachexia: the role of TGFβ pathway, BMC cancer 17(1) (2017) 1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [68].Batista ML Jr, Henriques FS, Neves RX, Olivan MR, Matos-Neto EM, Alcântara PS, Maximiano LF, Otoch JP, Alves MJ, Seelaender M, Cachexia-associated adipose tissue morphological rearrangement in gastrointestinal cancer patients, Journal of cachexia, sarcopenia and muscle 7(1) (2016) 37–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [69].Fearon K, Strasser F, Anker SD, Bosaeus I, Bruera E, Fainsinger RL, Jatoi A, Loprinzi C, MacDonald N, Mantovani G, Definition and classification of cancer cachexia: an international consensus, The lancet oncology 12(5) (2011) 489–495. [DOI] [PubMed] [Google Scholar]
- [70].Batista M Jr, Olivan M, Alcantara P, Sandoval R, Peres S, Neves R, Silverio R, Maximiano L, Otoch J, Seelaender M, Adipose tissue-derived factors as potential biomarkers in cachectic cancer patients, Cytokine 61(2) (2013) 532–539. [DOI] [PubMed] [Google Scholar]
- [71].Batista M Jr, Peres S, McDonald M, Alcântara P.S.M.d., Olivan M, Otoch JP, Farmer S, Seelaender M, Adipose tissue inflammation and cancer cachexia: possible role of nuclear transcription factors, Cytokine 57(1) (2012) 9–16. [DOI] [PubMed] [Google Scholar]
- [72].Henriques FS, Antônio Laurato Sertié R, Franco FO, Knobl P, Neves RX, Andreotti S, Lima FB, Guilherme A, Seelaender M, Batista ML Jr, Early suppression of adipocyte lipid turnover induces immunometabolic modulation in cancer cachexia syndrome, The FASEB Journal 31(5) (2017) 1976–1986. [DOI] [PubMed] [Google Scholar]
- [73].Paz-Filho G, Lim EL, Wong M-L, Licinio J, Associations between adipokines and obesity-related cancer, Front Biosci 16(1) (2011) 1634–1650. [DOI] [PubMed] [Google Scholar]
- [74].Khandekar MJ, Cohen P, Spiegelman BM, Molecular mechanisms of cancer development in obesity, Nature Reviews Cancer 11(12) (2011) 886–895. [DOI] [PubMed] [Google Scholar]
- [75].Aguirre G, De Ita JR, De La Garza R, Castilla-Cortazar I, Insulin-like growth factor-1 deficiency and metabolic syndrome, Journal of translational medicine 14(1) (2016) 1–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [76].Yang F, Cohen RN, Brey EM, Optimization of Co-Culture Conditions for a Human Vascularized Adipose Tissue Model, Bioengineering 7(3) (2020) 114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [77].Huttala O, Sarkanen J-R, Heinonen T, Ylikomi T, Presence of Vasculature Results in Faster Insulin Response in Adipocytes in Novel In Vitro Vascularized Adipose Tissue Model, ALTEX - Alternatives to animal experimentation, 36(3) (2019) 419–434. [DOI] [PubMed] [Google Scholar]
- [78].Alves da Silva ML, Costa-Pinto AR, Martins A, Correlo VM,Sol P, Bhattacharya M,Faria S, Reis RL, Neves Nuno M., Conditioned medium as a strategy for human stem cells chondrogenic differentiation, Journal of tissue engineering and regenerative medicine, 2015, 9(6) 714–723. [DOI] [PubMed] [Google Scholar]
- [79].Choi JH, Bellas E, Gimble JM, Vunjak-Novakovic G, Kaplan DL, Lipolytic function of adipocyte/endothelial cocultures, Tissue Engineering Part A 17(9–10) (2011) 1437–1444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [80].Pellegrinelli V, Rouault C, Veyrie N, Clément K, Lacasa D, Endothelial cells from visceral adipose tissue disrupt adipocyte functions in a three-dimensional setting: partial rescue by angiopoietin-1, Diabetes 63(2) (2014) 535–549. [DOI] [PubMed] [Google Scholar]
- [81].Hammel JH, Bellas E, Endothelial cell crosstalk improves browning but hinders white adipocyte maturation in 3D engineered adipose tissue, Integrative Biology 12(4) (2020) 81–89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [82].Herold J, Kalucka J, Angiogenesis in Adipose Tissue: the interplay between adipose and endothelial cells, Frontiers in Physiology 11 (2020) 624903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [83].Singh M, Benencia F, Inflammatory processes in obesity: focus on endothelial dysfunction and the role of adipokines as inflammatory mediators: We reviewed obesity-induced metabolic and immunological changes at the level of vasculature and emphasize on the importance of adipokines, International reviews of immunology 38(4) (2019) 157–171. [DOI] [PubMed] [Google Scholar]
- [84].Gu P, Xu A, Interplay between adipose tissue and blood vessels in obesity and vascular dysfunction, Reviews in Endocrine and Metabolic Disorders 14(1) (2013) 49–58. [DOI] [PubMed] [Google Scholar]
- [85].Robciuc MR, Kivelä R, Williams IM, de Boer JF, van Dijk TH, Elamaa H, Tigistu-Sahle F, Molotkov D, Leppänen V-M, Käkelä R, VEGFB/VEGFR1-induced expansion of adipose vasculature counteracts obesity and related metabolic complications, Cell metabolism 23(4) (2016) 712–724. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


