Summary
We screened 47 subjects with DDX41 variants among 1529 subjects with myeloid neoplasms. The most common germline variants included Splice c.935 + 4A>T, p.T360Ifs*33, p.V152G, p.S217Ifs*4, p.R311* and p.R369*. Except for the p.R369*, no other variants have been previously reported. Clinical covariates of subjects with simple DDX41 somatic variants and germline/somatic biallelic variants are similar. The two-year overall survival (OS) of subjects with DDX41 variants was 85%. Overall response rate to demethylation therapy in subjects with DDX41 variants was 69%. The response did not correlate with the presence of a germline variant.
Keywords: DDX41 variants, genetic predisposition, myelodysplastic syndromes, demethylation therapy
Myeloid neoplasms with genetic predisposition (MNGP) is a new diagnostic category in the 2016 revised World Health Organisation (WHO) myeloid neoplasm classification. One subtype is a germline variant of DDX41.1 Many pedigrees with DDX41 germline variants are reported in diverse populations.2-6 Some data suggest that the DDX41 germline variants occur in 1–5% of myeloid neoplasms7-9 and are probably the most common germline susceptibility locus for myeloid neoplasms. In the present study, we report profiling of DDX41 variants in 1529 Han Chinese patients with diverse myeloid neoplasms.
Patients and methods
A total of 1529 patients with myeloid neoplasms, including 934 subjects with myelodysplastic syndrome (MDS), 174 with acute myeloid leukemia (AML), 335 with myeloproliferative neoplasm (MPN) and 86 MDS/MPN subjects, diagnosed according to the 2016 WHO criteria at our centre, were enrolled in the study from April 2017 to November 2019. All subjects provided informed consent in compliance with the Declaration of Helsinki.
Genomic DNA was extracted from bone marrow mononuclear cells and oral epithelial cells. Next-generation sequencing was performed on the coding sequence of the 114 genes closely related to haematologic neoplasms (see Table SI), as previously described.10 Details about targeted gene sequencing and mutation analysis are described in the supplemental methods, as shown in Data S1. The oral epithelial cells were taken as control samples to verify the mutations from possible germline sources (variant allele fraction >40%), and the specific sites were verified by Sanger sequencing.
Results and discussion
DDX41 variants were detected in 47 (3·1%) subjects, the median age was 65 years (33–78), and 44 (94%) were males. Forty-one patients had MDS [five with multilineage dysplasia (MLD), 20 with excess blasts 1 (EB1), 16 with excess blasts 2 (EB2)], four with AML (three cases secondary to MDS, one primary AML), and two with post-essential thrombocythaemia (ET) myelofibrosis (see Table 1. Cytogenetic data were available for 41 patients, of whom 40 (98%) had a normal karyotype and one had a t(17;17)(q21;q24). In contrast, 571 of the 1329 subjects with wild-type DDX41 had abnormal cytogenetics (P = 0·000). Thirty-two DDX41 variants were identified, including 17 germline variants and 15 somatic variants (Fig 1, Table SII). By investigating the family history, only one 62-year-old male patient (Patient No.38 in Fig 2) with the DDX41 germline variant had a relative with a myeloid neoplasm (an older brother died of MDS three years earlier). However, the actual prevalence of family members may be higher than in our survey. One possible explanation is that the young carriers were not yet showing signs of the disease.
Table 1.
Clinical and laboratory features of subjects with DDX41 variants.
| Parameter | Germline | Simple somatic |
|---|---|---|
| Sex | ||
| Male | 22 | 22 |
| Female | 3 | 0 |
| Age at diagnosis, y | ||
| Median | 67 | 65 |
| Range | 33–78 | 46–77 |
| Family history | 1 | 0 |
| BM evaluation | ||
| MDS-MLD | 3 | 2 |
| MDS-EB | 17 | 19 |
| AML | 3 | 1 |
| MPN | 2 | 0 |
| Cytogenetics | ||
| Normal | 21 | 19 |
| Abnormal | 1 | 0 |
| NA | 3 | 3 |
| Concomitant variants | ||
| ASXL1 | 7 | 3 |
| TP53 | 3 | 5 |
| EZH2 | 3 | 4 |
| SRSF2 | 3 | 3 |
| PHF6 | 4 | 2 |
| TET2 | 3 | 1 |
| Treatments of advanced phase | ||
| DMT | 8 | 8 |
| Lenalidomide | 1 | 0 |
| Transplantation | 1 | 1 |
| Others | 10 | 11 |
| Survival | ||
| MS | NR | NR |
| 2-year OS | 75 ± 11% | 88 ± 8% |
AML, acute myeloid leukemia; DMT, demethylation therapy; EB, excess blasts; MDS, myelodysplastic syndroms; MLD, multilineage dysplasia; MPN, myeloproliferative neoplasms; MS, median survival; NA, not available; OS, overall survival.
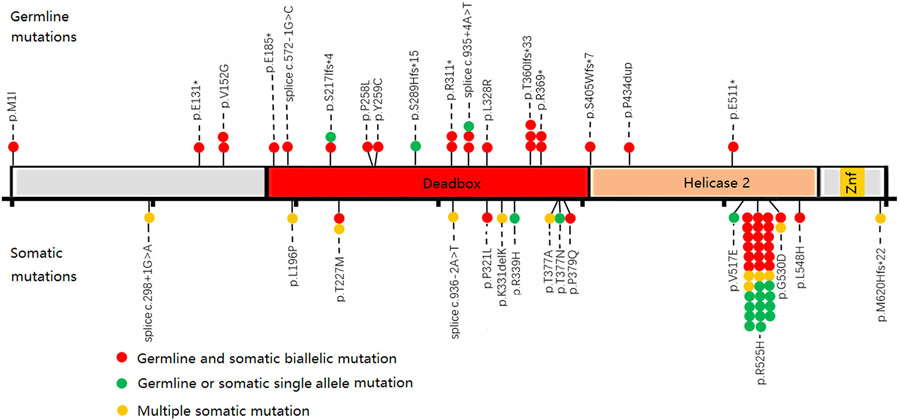
Fig 1.
Distribution of DDX41 variants. There were 32 DDX41 variants in 47 subjects with myeloid neoplasms, including 17 germline variants and 15 somatic variants. The upper part of the gene structure map represents the germline variant site, and the lower part represents the somatic variant site. The red round marker represents the presence of both germline and somatic variants concurrently, the green round marker represents the detection of only germline or somatic variants, and the yellow round marker represents the presence of multiple somatic variants.
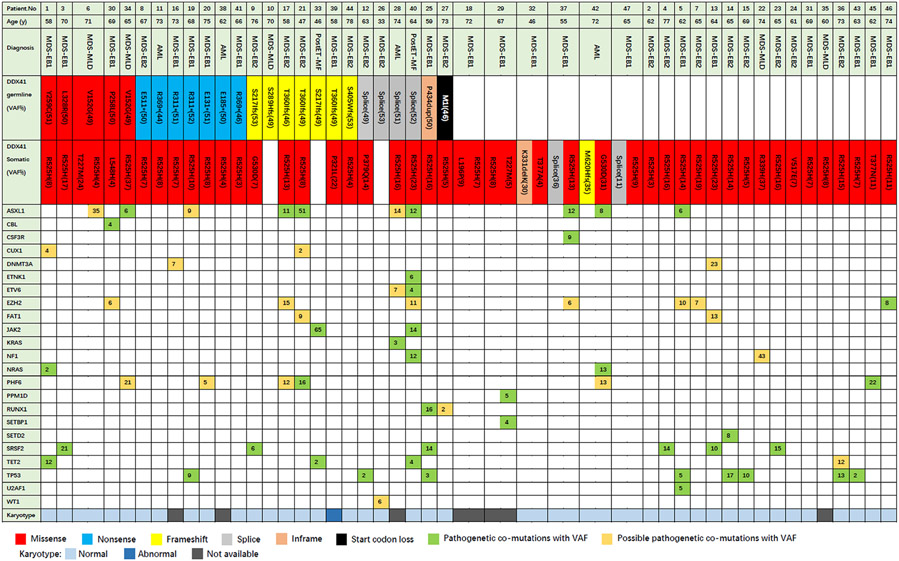
Fig 2.
The oncoplot represented the diagnosis of each patient, the DDX41 variant site and concomitant mutations. The number displayed in the box represents the mutant gene variant allele fraction (%).
A germline variant of DDX41 was detected in 25 (1·6%) subjects, 22 (88%) of whom had a DDX41 somatic mutation (Fig 2). Seventeen causal germline variants were detected (Table SIII), the most common variants including Splice c.935 + 4A>T (n = 3), p.T360Ifs*33 (n = 3), p.V152G (n = 2), p.S217Ifs*4 (n = 2), p.R311* (n = 2) and p.R369* (n = 2). Except for the p.R369*, no other variants have previously been reported. Fifteen somatic variants were detected, with p.R525H (n = 35) the most common DDX41 somatic variant. Other recurrent variants included p.T227M (n = 2) and p.G530D (n = 2). The most common germline variants in persons of predominantly European descent were p.D140fs*22 and p.M1I7. Only one of our subjects had a germline p.M1I variant and none had a germline p.D140fs*2 variant. Our data reflect those reported by Takeda et al. in Japan, but we did not detect the A500Cfs*9 variant, which is common in their population9. In contrast, results of several studies on somatic variants are consistent, with p.R525H variant being the most common variant, usually with a low variant allele frequency (VAF) (0·05-0·25). The mechanism of the diversity of germline variants versus the consistency of somatic variants is still not clear, but obtaining p.R525H will lead to abnormal growth and differentiation of haematopoietic cells.11
There are a few studies on the characteristics of simple DDX41 somatic variants in myeloid neoplasms. In our cohort, simple DDX41 somatic variations were detected in 22 (1·4%) patients. We compared the clinical covariates of germline/somatic biallelic variants (n = 22) and simple somatic variants (n = 22). There was no significant difference between the two groups in age, sex or cytogenetic abnormalities. Subjects with DDX41 variants were typically older, predominantly male, had a normal karyotype, and were more likely to have advanced MDS and AML. These data are consistent with previous reports2,7,8. Interestingly, in 22 patients with simple DDX41 somatic variants, six had variants at two loci. These subjects often had one missense variant accompanied by another non-missense variant. Generally, non-missense mutations have higher VAF than missense mutations, 0·33 (0·11–0·36) versus 0·11 (0·04–0·31, P = 0·147). These data suggest that double hits caused by DDX41 biallelic variants may lead to the development of myeloid neoplasms. We cannot exclude the possibility that in some subjects with a seeming single hit we failed to detect a variant in an intronic regulatory region, or insertion or deletion of a large fragment.
We also detected 23 mutations other than germline and somatic DDX41 variants. The most common mutations were in ASXL1 (n = 10), (TP53 (n = 8), EZH2 (n = 7), SRSF2 (n = 6), PHF6 (n = 6) and TET2 (n = 4). Other gene abnormalities were rare, as shown in Fig 2. These concomitant mutations are similar to the previous report,7 but PHF6 variants are more common in our cohort. Simple DDX41 somatic variants have similar concomitant mutations with germline/somatic biallelic variants.
DDX41 is mainly involved in the biological functions of pre-mRNA splicing, innate immunity and ribosome biosynthesis.12 SRSF2 and PHF6 are similarly involved in the splicing of mRNAs and ribosomal RNA transcription. Median VAF of SRSF2 and PHF6 are 0·14 (0·06–0·21) and 0·15 (0·05–0·22), similar to the VAF of 0·11 (0·03–0·37) of the somatic DDX41 variants. We speculate that these mutations may interact with DDX41 somatic variants in the development of myeloid neoplasms.
The overall response rate of advanced MDS/AML patients who received chemotherapy or azacytidine was 100% and 73%, respectively.8 In the present study, 16 subjects with advanced phase (AML/MDS-EB) received at least two courses of hypomethylating drugs (Table SIV) for a median 3·5 (2–10) courses and had an overall response rate of 69%. The eight subjects with germline and somatic DDX41 variants had an overall response rate of 63%, including four complete remissions (CR) and one marrow remission (mCR). The eight subjects with simple somatic variants had an overall response rate of 75%, including four CR, one mCR and one haematologic improvement. These data indicate that subjects with somatic DDX41 variants have a high response rate to hypomethylating drugs, but there was no correlation with the presence of a germline variant. Previous reports suggest subjects with DDX41 germline variants have a good response to lenalidomide.13,14 The one subject (Patient No.12) in our study with MDS-EB2 and a germline DDX41 variant responded to lenalidomide.
A recent study showed that the two-year overall survival (OS) of subjects with DDX41 germline variants and MDS/AML was 90%.8 Median interval from diagnosis to DDX41 screening in our study was 15 months (range, 1–72 months). Median follow-up was 27 (1–81) months. The two-year OS of subjects with DDX41 variants was 85% (95% CI 79–91%). Subjects with germline variants had similar OS to those with simple somatic variants, 75% (95% CI 64–86%) versus 88% (95% CI 80–96%) (P = 0·688). Median survival (MS) time has not yet been reached in the two groups (Fig S1).
There are several limitations to our study. For example, some subjects died early, before the DDX41 screening was done. Consequently, exclusion of these subjects artificially extends estimated survival. Also, our depth of coverage was only 800x and our method failed to detect variants in intronic regulatory regions, and insertion or deletion of a large fragment. Lastly, oral epithelial cells are not strict germline samples, but choosing >40% VAF in these samples likely indicates germline.
In conclusion, the features of germline DDX41 variant in our population were different from the studies of other populations. In common with other studies, we found that myeloid neoplasms with somatic DDX41 variants have a good prognosis and could be considered as an independent entity.
Supplementary Material
Data S1. Supplemental methods.
Figure S1. The survival curve of subjects with DDX41 variants.
Table SI. Gene list of the 114-gene Next Generation Sequencing (NGS) panel.
Table SII. High-confidence mutations of 47 subjects with DDX41 variant.
Table SIII. American College of Medical Genetics and Genomics (ACMG) classification of germline DDX41 variants.
Table SIV. Data of demethylation treatment for advanced phase.
Acknowledgements
Supported, in part, by grants from the National Natural Science Funds (No. 81870104, No.81530008, No.81470297), CAMS Initiative Fund for Medical Sciences (No. 2016-I2M-1-001), Tianjin Key Natural Science Funds (18JCZDJC34900), Tianjin Natural Science Funds (19JCQNJC09400).
R.P.G. is a part-time employee of Celgene Corp.
Footnotes
Supporting Information
Additional supporting information may be found online in the Supporting Information section at the end of the article.
References
- 1.Arber DA, Orazi A, Hasserjian R, Thiele J, Borowitz MJ, Le Beau MM, et al. The 2016 revision to the World Health Organization classification of myeloid neoplasms and acute leukemia. Blood. 2016;127:2391–405. [DOI] [PubMed] [Google Scholar]
- 2.Polprasert C, Schulze I, Sekeres MA, Makishima H, Przychodzen B, Hosono N, et al. Inherited and somatic defects in DDX41 in myeloid neoplasms. Cancer Cell. 2015;27:658–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cardoso SR, Ryan G, Walne AJ, Ellison A, Lowe R, Tummala H, et al. Germline heterozygous DDX41 variants in a subset of familial myelodysplasia and acute myeloid leukemia. Leukemia 2016;30:2083–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lewinsohn M, Brown AL, Weinel LM, Phung C, Rafidi G, Lee MK, et al. Novel germ line DDX41 mutations define families with a lower age of MDS/AML onset and lymphoid malignancies. Blood. 2016;127:1017–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Li R, Sobreira N, Witmer PD, Pratz KW, Braunstein EM. (2016) Two novel germline DDX41 mutations in a family with inherited myelodysplasia/acute myeloid leukemia. Haematologica. 2016;101:e228–231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Berger G, van den Berg E, Sikkema-Raddatz B, Abbott KM, Sinke RJ, Bungener LB, et al. Re-emergence of acute myeloid leukemia in donor cells following allogeneic transplantation in a family with a germline DDX41 mutation. Leukemia. 2017;31:520–2. [DOI] [PubMed] [Google Scholar]
- 7.Quesada AE, Routbort MJ, DiNardo CD, Bueso-Ramos CE, Kanagal-Shamanna R, Khoury JD, et al. DDX41 variants in myeloid neoplasms are associated with male gender, TP53 variants and high-risk disease. Am J Hematol. 2019;94:757–66. [DOI] [PubMed] [Google Scholar]
- 8.Sébert M, Passet M, Raimbault A, Rahmé R, Raffoux E, Sicre de Fontbrune F, et al. Germline DDX41 variants define a significant entity within adult MDS/AML patients. Blood. 2019;134:1441–4. [DOI] [PubMed] [Google Scholar]
- 9.Takeda J, Yoshida K, Makishima H, Yoshizato T, Shiozawa Y, Shiraishi Y, et al. (2015) Genetic predispositions to myeloid neoplasms caused by germline DDX41 variants [abstract]. Blood, 126, Abstract 2843. [Google Scholar]
- 10.Li B, Liu J, Jia Y, Wang J, Xu Z, Qin T, et al. Clinical features and biological implications of different U2AF1 mutation types in myelodysplastic syndromes. Genes Chromosomes Cancer. 2018;57:80–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kadono M, Kanai A, Nagamachi A, Shinriki S, Kawata J, Iwato K, et al. Biological implications of somatic DDX41 p. R525H mutation in acute myeloid leukemia. Exp Hematol. 2016;44:745–54. [DOI] [PubMed] [Google Scholar]
- 12.Cheah JJC, Hahn CN, Hiwase DK, Scott HS, Brown AL. Myeloid neoplasms with germline DDX41 variant. Int J Hematol. 2017;106:163–74. [DOI] [PubMed] [Google Scholar]
- 13.Abou Dalle I, Kantarjian H, Bannon SA, Kanagal-Shamanna R, Routbort M, Patel KP, et al. Successful lenalidomide treatment in high risk myelodysplastic syndrome with germline DDX41 mutation. Am J Hematol. 2020;95:227–9. [DOI] [PubMed] [Google Scholar]
- 14.Negoro E, Radivoyevitch T, Polprasert C, Adema V, Hosono N, Makishima H, et al. Molecular predictors of response in patients with myeloid neoplasms treated with lenalidomide. Leukemia. 2016; 30:2405–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data S1. Supplemental methods.
Figure S1. The survival curve of subjects with DDX41 variants.
Table SI. Gene list of the 114-gene Next Generation Sequencing (NGS) panel.
Table SII. High-confidence mutations of 47 subjects with DDX41 variant.
Table SIII. American College of Medical Genetics and Genomics (ACMG) classification of germline DDX41 variants.
Table SIV. Data of demethylation treatment for advanced phase.