Abstract
The 35.5-kb ICESt1 element of Streptococcus thermophilus CNRZ368 is bordered by a 27-bp repeat and integrated into the 3′ end of a gene encoding a putative fructose-1,6-biphosphate aldolase. This element encodes site-specific integrase and excisionase enzymes related to those of conjugative transposons Tn5276 and Tn5252. The integrase was found to be involved in a site-specific excision of a circular form. ICESt1 also encodes putative conjugative transfer proteins related to those of the conjugative transposon Tn916. Therefore, ICESt1 could be or could be derived from an integrative conjugative element.
Cocultures of various lactic acid bacteria are used during the manufacture of dairy products. Sequence comparisons and hybridizations reveal that horizontal transfers between a large array of species of lactic acid bacteria have occurred, most likely during dairy cocultures (13, 32). The most convincing evidence indicates that insertion sequences IS1191, IS981, ISS1, and IS1194 (4, 5, 14) and some open reading frames (ORFs) involved in exopolysaccharide synthesis (6) or in restriction-modification (24) were transferred between the lactic acid bacteria Streptococcus thermophilus and Lactococcus lactis in cocultures used during cheese manufacture. However, the mechanism of genetic exchange between these two species remains unknown, and no conjugative element has been previously characterized in S. thermophilus.
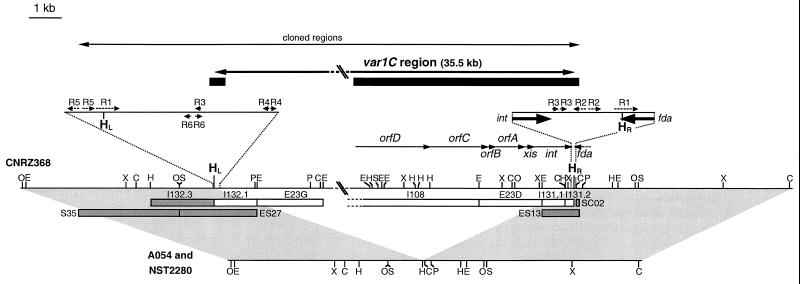
Cloning of var1C and localization of its limits.
The Sm4 fragment of the S. thermophilus CNRZ368 chromosome was previously found to contain the 35-kb variable region var1C, which was absent from the corresponding chromosomal fragments of strains A054 and NST2280 (28). A region containing an IS1191 copy inserted in a truncated IS981 element (14) was cloned and found to be included in var1C (28). Chromosome walking using a λGEM11 genomic library of CNRZ368 (25) was performed to isolate recombinant λ bacteriophages overlapping the var1C region. Their inserts were subcloned in pBC KS+ and used as hybridization probes on A054 and NST2280 DNAs. S35, ES27, I132.3, ES13, and SC02 fragments hybridized to A054 and NST2280 DNAs. On the contrary, all of the probes covering the 35.5-kb region (except IS1191 and IS981) and located between the HindIII sites HL and HR (Fig. 1) did not hybridize to A054 and NST2280 DNAs (data not shown). Furthermore, CNRZ368, A054, and NST2280 showed identical restriction maps in regions located to the left of the HindIII site HL and to the right of the HindIII site HR (Fig. 1). These data indicated that var1C limits are located near these HindIII sites. When ES27 including the left end and ES13 including the right end were hybridized to DNAs of the three strains digested by ClaI, EcoO109, EcoRI, PstI, or XbaI, they revealed the same fragment from A054 and NST2280, but two different fragments from CNRZ368. Thus, the flanking regions of var1C in CNRZ368 are adjacent to each other in strains A054 and NST2280 (Fig. 1).
FIG. 1.
Locations of var1C limits, ORFs, and repeated elements on a map comparison of the CNRZ368, A054, and NST2280 chromosomes. White boxes indicate probes hybridizing with CNRZ368 DNA only. Shaded boxes indicate probes hybridizing with the A054, NST2280, and CNRZ368 DNAs. Sequenced regions are shown by black boxes. Only the 3′ end of the ORF fda was cloned and sequenced. Arrows indicate ORF transcription direction. R1, R2, R3, R4, R5, and R6 repeated sequences are indicated by dotted arrows. HL and HR correspond to the HindIII sites included in the 27-bp repeated sequences delimiting var1C. C, ClaI; E, EcoRI; H, HindIII; O, EcoO109; P, PstI; S, SacI; X, XbaI.
Because A054 and CNRZ368 are very closely related to each other, but distantly related to NST2280 (28), the absence of var1C in A054 and NST2280 probably results from an insertion in CNRZ368 rather than from two independent identical deletions in the two other strains.
var1C is bordered by a direct repeat and encodes an integrative system.
Sequencing of the var1C limits revealed that the element is bordered by a 27-bp direct repeated sequence (R1) containing a HindIII site (Fig. 2). A 362-bp fragment was obtained by PCRs performed with the DNA of S. thermophilus A054 by using the convergent primers O132.3 (GGACTACTAAGAGAACAT) and O131.2 (TGTTGCTGAATACGAAGC) (Fig. 3). The sequence of this fragment revealed a unique R1 copy identical to those found on either side of var1C in CNRZ368 (Fig. 2). Sequence comparison indicates that R1 direct repeats of CNRZ368 correspond to the boundaries of var1C (Fig. 2).
FIG. 2.
Comparison of the nucleotide sequences of the four attachment sites. attL and attR include, respectively, the left and right termini of var1C of strain CNRZ368. attB corresponds to the partial sequence of a PCR product obtained from strain A054 with the primers O132.3 and O131.2. attI corresponds to the partial sequence of a nested-PCR product obtained from strain CNRZ368 with the primer pairs O132.5-O131.5 and O132.4-O131.1 (Fig. 3). R1 sequences are written in capital letters. The italic letters correspond to the internal sequence of var1C. Underlined letters indicate the bases that are complementary to the 3′ end of the fda gene encoding fructose-1,6-biphosphate aldolase. Sequences underlined twice correspond to the HindIII restriction sites included in R1.
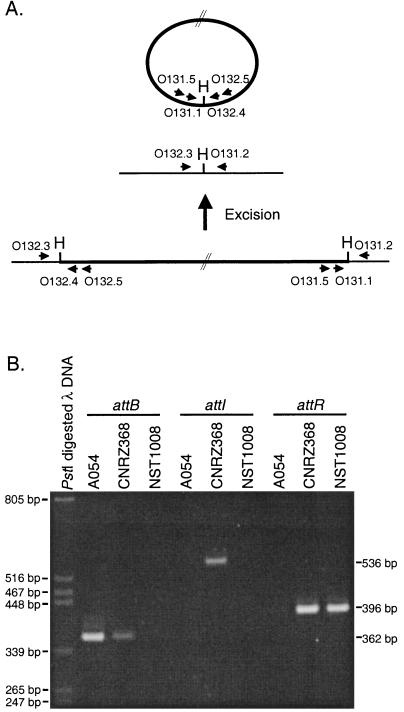
FIG. 3.
PCR amplification results. (A) Model of excision of ICESt1 and schematic localization of primer sets used to detect DNA molecules resulting from site-specific recombination events. H corresponds to the HindIII site included in the R1 cores of the attL, attR, attB, and attI attachment sites. (B) Electrophoresis of PCR products. attB, attI, and attR correspond to PCR with the O132.3 and O131.2 primers, nested PCR with the O131.5-O132.5 and O131.1-O132.4 primer pairs, or PCR with O131.1 and O131.2, respectively. The same volume (4 μl) of PCR product was loaded in each lane.
Two ORFs, int and xis, are located within var1C near the right copy of R1 (Fig. 1 and Table 1). The putative protein encoded by int shows significant similarities to site-specific recombinases belonging to the φLC3 subgroup of the integrase family (http://members.home.net/domespo/trhome.html). This subgroup includes a large array of integrases of temperate bacteriophages and conjugative transposons of lactic acid bacteria and other gram-positive low-G+C bacteria. The C terminus of Int contains the five amino acids which are perfectly conserved in this family (data not shown) (1, 3, 11). Furthermore, xis, located to the left of the int gene, encodes a small basic protein (pI 9.88) which show significant similarities to excisionases of two conjugative transposons, Tn5252 of Streptococcus pneumoniae and Tn5276 of L. lactis (Table 1). int and xis are located at comparable positions in many prophages and conjugative transposons.
TABLE 1.
Characteristics of the sequenced ORFs and encoded proteins examined in this study
| DNA sequence characteristic
|
Translation product characteristic
|
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| ORFa | % G+C | Start | Stop | RBSb | Dist1c | Dist2d | Length (no. of amino acids) | Related proteine | Origin | % Identity (no. of amino acids)f |
| fda | 38.3 | NDg | TAA | ND | ND | ND | ND | Fructose-1,6-biphosphate aldolase Fba (AJ005697) | Chromosome of S. pneumoniae | 80 (77) |
| int | 33.6 | ATG | TAA | TAAGGAGG | 7 | −1 | 448 | Integrase Int (U93688)h | Pathogenicity island SaPI1 of S. aureus | 26 |
| Integrase Int (M27965)h | Phage φL54a of S. aureus | 28 | ||||||||
| Integrase Int (M62697)h | Phage φadh of Lactobacillus gasseri | 32 (227) | ||||||||
| Integrase Int (L27649)h | Conjugative transposon Tn5276 of L. lactis | 24 | ||||||||
| Integrase Int (L29324)h | Conjugative transposon Tn5252 of S. pneumoniae | 26 (266) | ||||||||
| xis | 35.7 | ATG | TAA | AAAGGAGT | 5 | +13 | 82 | Excisionase Xis (L29324) | Conjugative transposon Tn5252 of S. pneumoniae | 41 |
| Excisionase Xis (L27649) | Conjugative transposon Tn5276 of L. lactis | 41 | ||||||||
| orfA | 43.3 | ATG | TAG | AAAGGAGA | 4 | +25 | 370 | Putative transfer protein TraG (AF051917) | Conjugative plasmid pSK41 of S. aureus | 30 |
| Putative transfer protein TrsG (L11998) | Conjugative plasmid pGO1 of S. aureus | 30 | ||||||||
| Immunogenic secreted protein Isp (U31811) | Chromosome of Streptococcus pyogenes D471 | 57 (325) | ||||||||
| orfB | 37.8 | ATG | TAA | AGAGGAGA | 5 | +1 | 74 | No similarity | ND | ND |
| orfC | 41.5 | ATG | TAG | TTAGGAGG | 7 | +11 | 626 | Putative membrane protein Orf15 (U09422) | Conjugative transposon Tn916 of E. faecalis | 18 (267) |
| orfD | 42.1 | ATG | TAG | AAAGGAGG | 4 | ND | 834 | Putative transfer protein Orf16 (U09422) | Conjugative transposon Tn916 of E. faecalis | 21 |
| Unknown protein YddE (AB001488) | Chromosome of B. subtilis 168 | 30 | ||||||||
ORFs are listed from the right to the left of the map.
The RBS consensus sequence of the gram-positive low-G+C bacterium Bacillus subtilis is AAAGGAGG.
Dist1, distance between the RBS and the start codon.
Dist2, distance between the start codon of an ORF and the stop codon of the previous ORF on the map (Fig. 1). A negative value indicates an overlapping of two ORFs.
Functions of proteins and GenBank accession numbers (in parentheses) of nucleotide sequences encoding proteins related to the product of the ORFs sequenced in this study are indicated.
Identities stretch over the entire length of each of the amino acid sequences of proteins encoded by ICESt1, except when indicated in parentheses.
ND, not determined.
Many other related integrases were found in databases, but in this table, we have only indicated a selection of the ones more related to the integrase encoded by var1C.
Therefore, these ORFs probably encode an integrative system which would mediate excision of var1C by site-specific recombination between the two R1 copies corresponding to the cores of the left and right attachment sites attL and attR. The unique R1 sequence found in A054 would be the attB attachment site used for var1C integration. fda, which flanks the right of var1C (Fig. 1), encodes a putative fructose-1,6-biphosphate aldolase (Table 1). The 3′ end of fda includes 20 bp of the R1 core of attR (Fig. 2). Thus, var1C integration does not change the sequence of fda. Numerous integrative elements (e.g., prophages or integrative conjugative elements) integrate into the 3′ end of genes encoding tRNAs, their sequences remaining unmodified by the integration (8, 15, 17, 23, 30, 31). Other integrative elements (e.g., most of the conjugative transposons) integrate into several or numerous sites (19, 26). Only a few elements site specifically integrate into the 3′ end of protein-encoding genes. The substitution sequence is then generally only similar to the original one (10, 18).
An imperfect 14-bp inverted repeat, R2, is located 29 bp to the right of the 3′ end of the int gene and 21 bp to the left of the R1 core of attR (Fig. 1). The potential stem-loop structure (ΔG = −14.8 kcal · mol−1) (33), preceded by a stretch of A's and followed by a stretch of T's, could be used as a ρ-independent transcription terminator for both int and fda. A perfect 13-bp inverted repeat, R5 (ΔG = −18.8 kcal · mol−1), preceded by a stretch of A's, is located 2 bp to the left of the core of attL (Fig. 1) and could be used as a transcription termination signal for fda prior to the var1C integration. Therefore, these data suggest that the expression of fda would not be changed after var1C integration.
R3, a perfect 9-bp direct repeat, was found 2 bp downstream from the stop codon of int (Fig. 1). A copy of this 9-bp sequence was also found 148 bp to the right of the R1 core of attL. R6, an imperfect 12-bp inverted repeat, and R4, an imperfect 9-bp inverted repeat, are located 123 and 229 bp to the right of the core of attL, respectively. R2, R3, R4, and R6 could be binding sites for integrase or host-encoded proteins involved in the recombination.
Detection of site-specific recombination products.
A nested PCR was performed to amplify the putative junction between the var1C termini, which could result from a site-specific recombination event between the R1 cores of attL and attR. Nested-PCR amplification was performed with the O132.5 (GATGAAATTCACATCATC)-O131.5 (CAGGAATCGATATTGACA) outer primer pair and the O132.4 (AGTTGAAACTAGACTCAG)-O131.1 (TTCCGACATACGCATATC) inner primer pair (Fig. 3A) according to the method described by Manganelli et al. (21). As expected, no product was identified in strain A054 (Fig. 3B), which does not contain var1C. The sequence of the 536-bp PCR product obtained in CNRZ368 (attI, Fig. 2) is identical to the expected sequence resulting from site-specific recombination between the R1 cores of attL and attR. The PCR product was digoxigenin labelled and hybridized to EcoRI-digested A054 and CNRZ368 chromosomal DNA. As expected, this probe hybridizes with the two fragments containing the var1C termini in CNRZ368, but not with A054 DNA (data not shown). Site-specific excision of var1C in CNRZ368 should also lead to a junction between sequences flanking var1C, identical to that observed in A054. PCR amplification using the O132.3-O131.2 primer pair (Fig. 3A) was performed to detect this junction. PCR products obtained for A054 and CNRZ368 show the same size (Fig. 3B) and restriction map (data not shown).
Detection of these two junction fragments implies in CNRZ368 the excision of a covalent circular molecule in some cells of the population. The R1 sequences found in the chromosome of A054, in the circular form of var1C, and in the ends of integrated var1C probably constitute the core of the attB, attI, attL, and attR attachment sites: the strand exchange reaction probably takes place by crossover events similar to those involved in λ integration and excision. The length of the core of attachment sites suggests that this element would show very strong insertional site specificity.
Disruption of the int gene prevents var1C excision.
The ORF int was disrupted in order to prove its involvement in var1C excision. The thermosensitive plasmid pNST152 was constructed by subcloning the 754-bp HindIII fragment of pNST131.1 containing a fragment of int (region encoding residues 137 to 383 of the integrase) into pG+Host9 (20). pNST152 was used to transform S. thermophilus CNRZ368 by electroporation according to the method of Marciset and Mollet (22). Integration of pNST152 into the int gene was promoted by homologous recombination at a nonpermissive temperature (42°C). The integration site and the number of integrated copies were verified by hybridization of probe I131.1 to PstI patterns of integrants (data not shown). The recombinant strain NST1008 contains two truncated copies of int resulting from the integration of a unique copy of pNST152 within the int gene of CNRZ368. Junction fragments containing attB or attI were not detected in NST1008 by PCR experiments (Fig. 3B), whereas a fragment bearing attR was amplified from NST1008 by using the O131.1 and O131.2 primers (Fig. 3). Therefore, int gene disruption causes the disappearance of the two junction fragments and, therefore, of the covalent circular molecule, showing that this gene is actually involved in var1C excision.
var1C encodes proteins related to those of some conjugative system.
The 5,881-bp region located to the left of the xis ORF start codon was sequenced. Four ORFs have been identified by GeneMark (http://genemark.biology.gatech.edu/GeneMark/) and/or by comparison of the putative translation products with proteins from the EMBL/GenBank databases by using BLASTX and BLASTP (2) (Fig. 1 and Table 1). All of these ORFs are preceded by a suitably located ribosome binding site (RBS) (27), have the same orientation as xis and int, and are spaced by very short sequences (Table 1). Therefore, orfDCBA, xis, and int could be translated from a unique transcript.
The orfA and orfD products share significant sequence similarities with proteins involved in conjugative transfer of plasmids from Staphylococcus aureus and Tn916 from Enterococcus faecalis (Table 1). orfC encodes a putative protein weakly related to the translational product of orf15 of the conjugative transposon Tn916. Topology predictions using the HMMTOP server (http://www.enzim.hu/hmmtop/) indicate that the proteins encoded by these two ORFs would be transmembrane proteins with similar tridimensional structures, suggesting that they are actually related. Thus, this region of var1C could encode conjugative functions. Various recently identified elements excise by forming a circular intermediate, promote self-transfer by conjugation into the recipient cell, and integrate by recombination between the specific site of the circular molecule and another site (17, 26, 29, 31). Therefore, the entire var1C sequence could be or could be derived from a site-specific integrative conjugative element. This possible conjugative element, which would be the first isolated in S. thermophilus, was named ICESt1, for integrative conjugative element of S. thermophilus no. 1.
The possible conjugative system of ICESt1 is related to that of Tn916, but not to the system encoded by Tn5252. On the contrary, the ICESt1 excisionase is related only to those of Tn5276 and Tn5252. Moreover, the integrases of ICESt1, Tn5276, and Tn5252 belong to the φLC3 integrase subfamily, whereas the integrase of Tn916 belongs to another subfamily (http://members.home.net/domespo/trhome.html). Furthermore, differences in G+C content between the xis and int genes (about 34%) and orfABCD (about 42%) of ICESt1 also suggest that the integration-excision system and the possible conjugative system have different origins or have undergone very different evolutions. A similar structure is observed in Tn916 (about 36% G+C for the xis and int genes versus about 40% G+C for the conjugative system). This suggests that ICESt1 and Tn916 possess a modular structure which results from exchanges or acquisitions of sequences from different sources. This modular structure and evolution are similar to those of bacteriophages (9, 16) and enterobacterial plasmids (7).
The large size of ICESt1 (35 kb) suggests that this element, like Tn5276, which encodes nisin synthesis (26), could carry industrially attractive genes. The ICESt1 element contains a complete copy of IS1191, an insertion sequence probably transferred from S. thermophilus to L. lactis, and a truncated copy of IS981, which was probably transferred from L. lactis to S. thermophilus, most likely in cocultures of these species used during the manufacture of cheese (14). Furthermore, conjugative transposons related to ICESt1, like Tn916 of Enterococcus faecalis and Tn5252 of S. pneumoniae, are broad-host-range elements (12, 34). Therefore, ICESt1 or elements related to ICESt1 could be involved not only in intraspecific but also in interspecific horizontal transfers between S. thermophilus and other lactic acid bacteria.
Nucleotide sequence accession numbers.
The GenBank accession numbers of the nucleotide sequences reported in this paper are AJ243105 (left terminus of var1C) and AJ243106 (right terminus of var1C).
Acknowledgments
We thank E. Maguin for providing the thermosensitive plasmid pG+Host9.
This work was supported by grants from the Institut National de la Recherche Agronomique, the University of Nancy 1, and the Ministère de l'Education Nationale, de la Recherche et de la Technologie, France.
REFERENCES
- 1.Abremski K E, Hoess R H. Evidence for a second conserved arginine residue in the integrase family of recombination proteins. Protein Eng. 1992;5:87–91. doi: 10.1093/protein/5.1.87. [DOI] [PubMed] [Google Scholar]
- 2.Altschul S F, Madden T L, Schäffer A A, Zhang J, Zhang Z, Miller W, Lipman D J. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Argos P, Landy A, Abremski K, Egan J B, Haggard-Ljungquist E, Hoess R H, Kahn M L, Kalionis B, Narayana S V L, Pierson III L S, Sternberg N, Leong J M. The integrase family of site-specific recombinases: regional similarities and global diversity. EMBO J. 1986;5:433–440. doi: 10.1002/j.1460-2075.1986.tb04229.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bourgoin F, Guédon G, Gintz B, Decaris B. Characterization of a novel insertion sequence, IS1194, in Streptococcus thermophilus. Plasmid. 1998;40:44–49. doi: 10.1006/plas.1998.1337. [DOI] [PubMed] [Google Scholar]
- 5.Bourgoin F, Guédon G, Pébay M, Roussel Y, Panis C, Decaris B. Characterization of a mosaic ISS1 element and evidence for the recent horizontal transfer of two different types of ISS1 between Streptococcus thermophilus and Lactococcus lactis. Gene. 1996;178:15–23. doi: 10.1016/0378-1119(96)00316-2. [DOI] [PubMed] [Google Scholar]
- 6.Bourgoin F, Pluvinet A, Gintz B, Decaris B, Guédon G. Are horizontal transfers involved in the evolution of the Streptococcus thermophilus exopolysaccharide synthesis loci? Gene. 1999;233:151–161. doi: 10.1016/s0378-1119(99)00144-4. [DOI] [PubMed] [Google Scholar]
- 7.Boyd E F, Hill C W, Rich S M, Hartl D L. Mosaic structure of plasmids from natural population of Escherichia coli. Genetics. 1996;143:1091–1100. doi: 10.1093/genetics/143.3.1091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Brown D P, Idler K B, Katz L. Characterization of the genetic elements required for site-specific integration of plasmid pSE211 in Saccharopolyspora erythraea. J Bacteriol. 1990;172:1877–1888. doi: 10.1128/jb.172.4.1877-1888.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Brüssow H, Bruttin A, Desiere F, Lucchini S, Foley S. Molecular ecology and evolution of Streptococcus thermophilus bacteriophages—a review. Virus Genes. 1998;16:95–109. doi: 10.1023/a:1007957911848. [DOI] [PubMed] [Google Scholar]
- 10.Campbell A M. Chromosomal insertion sites for phages and plasmids. J Bacteriol. 1992;174:7495–7499. doi: 10.1128/jb.174.23.7495-7499.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Caroll D, Kehoe M A, Cavanagh D, Coleman D C. Novel organization of the site-specific integration and excision recombination functions of the Staphylococcus aureus serotype F virulence-converting phages φ13 and φ42. Mol Microbiol. 1995;16:877–893. doi: 10.1111/j.1365-2958.1995.tb02315.x. [DOI] [PubMed] [Google Scholar]
- 12.Celli J, Poyart C, Trieu-Cuot P. Use of an excision reporter plasmid to study the intracellular mobility of the conjugative transposon Tn916 in Gram-positive bacteria. Microbiology. 1997;143:1253–1261. doi: 10.1099/00221287-143-4-1253. [DOI] [PubMed] [Google Scholar]
- 13.Guédon G, Bourgoin F, Decaris B. Does gene horizontal transfer occur in lactic acid bacteria co-cultures? Lait. 1998;78:53–58. [Google Scholar]
- 14.Guédon G, Bourgoin F, Pébay M, Roussel Y, Colmin C, Simonet J M, Decaris B. Characterization and distribution of two insertion sequences, IS1191 and iso-IS981, in Streptococcus thermophilus: does intergeneric transfer of insertion sequences occur in lactic acid bacteria co-cultures? Mol Microbiol. 1995;16:69–78. doi: 10.1111/j.1365-2958.1995.tb02392.x. [DOI] [PubMed] [Google Scholar]
- 15.Hacker J, Blum-Oehler G, Mühldorfer I, Tschäpe H. Pathogenicity islands of virulent bacteria: structure, function and impact on microbial evolution. Mol Microbiol. 1997;23:1089–1097. doi: 10.1046/j.1365-2958.1997.3101672.x. [DOI] [PubMed] [Google Scholar]
- 16.Hendrix R W, Smith M C M, Burns R N, Ford M E, Hatfull G F. Evolutionary relationships among diverse bacteriophages and prophages: all the world's a phage. Proc Natl Acad Sci USA. 1999;96:2192–2197. doi: 10.1073/pnas.96.5.2192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hochhut B, Jahreis K, Lengeler J W, Schmid K. CTnscr94, a conjugative transposon found in enterobacteria. J Bacteriol. 1997;179:2097–2102. doi: 10.1128/jb.179.7.2097-2102.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lillehaug D, Birkeland N-K. Characterization of genetic elements required for site-specific integration of the temperate lactococcal bacteriophage φLC3 and construction of integration-negative φLC3 mutants. J Bacteriol. 1993;175:1745–1755. doi: 10.1128/jb.175.6.1745-1755.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lu F, Churchward G. Tn916 target DNA sequences bind the C-terminal domain of integrase protein with different affinities that correlate with transposon insertion frequency. J Bacteriol. 1995;177:1938–1946. doi: 10.1128/jb.177.8.1938-1946.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Maguin E, Prévost H, Ehrlich S D, Gruss A. Efficient insertional mutagenesis in lactococci and other gram-positive bacteria. J Bacteriol. 1996;178:931–935. doi: 10.1128/jb.178.3.931-935.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Manganelli R, Romano L, Ricci S, Zazzi M, Pozzi G. Dosage of Tn916 circular intermediates in Enterococcus faecalis. Plasmid. 1995;34:48–57. doi: 10.1006/plas.1995.1032. [DOI] [PubMed] [Google Scholar]
- 22.Marciset O, Mollet B. Multifactorial experimental designs for optimizing transformation: electroporation of Streptococcus thermophilus. Biotechnol Bioeng. 1994;43:490–496. doi: 10.1002/bit.260430609. [DOI] [PubMed] [Google Scholar]
- 23.McShan W M, Tang Y F, Ferretti J J. Bacteriophage T12 of Streptococcus pyogenes integrates into the gene encoding a serine tRNA. Mol Microbiol. 1997;23:719–728. doi: 10.1046/j.1365-2958.1997.2591616.x. [DOI] [PubMed] [Google Scholar]
- 24.O'Sullivan T, Van Sinderen D, Fitzgerald G. Structural and functional analysis of pCI65st, a 6.5 kb plasmid from Streptococcus thermophilus NDI-6. Microbiology. 1999;145:127–134. doi: 10.1099/13500872-145-1-127. [DOI] [PubMed] [Google Scholar]
- 25.Pébay M, Roussel Y, Simonet J-M, Decaris B. High-frequency deletion involving closely spaced rRNA gene sets in Streptococcus thermophilus. FEMS Microbiol Lett. 1992;98:51–56. [Google Scholar]
- 26.Rauch P J G, De Vos W M. Characterization of the novel nisin-sucrose conjugative transposon Tn5276 and its insertion in Lactococcus lactis. J Bacteriol. 1992;174:1280–1287. doi: 10.1128/jb.174.4.1280-1287.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rocha E P C, Danchin A, Viari A. Translation in Bacillus subtilis: roles and trends of initiation and termination, insights from a genome analysis. Nucleic Acids Res. 1999;27:3567–3576. doi: 10.1093/nar/27.17.3567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Roussel Y, Bourgoin F, Guédon G, Pébay M, Decaris B. Analysis of the genetic polymorphism between three Streptococcus thermophilus strains by comparing their physical and genetic organization. Microbiology. 1997;143:1335–1343. doi: 10.1099/00221287-143-4-1335. [DOI] [PubMed] [Google Scholar]
- 29.Scott J R, Churchward G G. Conjugative transposition. Annu Rev Microbiol. 1995;49:367–397. doi: 10.1146/annurev.mi.49.100195.002055. [DOI] [PubMed] [Google Scholar]
- 30.Shoemaker N B, Wang G-R, Salyers A A. The Bacteroides mobilizable insertion element, NBU1, integrates into the 3′ end of a Leu-tRNA gene and has an integrase that is a member of the lambda integrase family. J Bacteriol. 1996;178:3594–3600. doi: 10.1128/jb.178.12.3594-3600.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sullivan J T, Ronson C W. Evolution of rhizobia by acquisition of a 500-kb symbiosis island that integrates into a phe-tRNA gene. Proc Natl Acad Sci USA. 1998;95:5145–5149. doi: 10.1073/pnas.95.9.5145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Teuber M, Meile L, Schwarz F. Acquired antibiotic resistance in lactic acid bacteria from food. Antonie Leeuwenhoek. 1999;76:115–137. [PubMed] [Google Scholar]
- 33.Tinoco I, Borer P N, Dengler B, Levin M D, Uhlenbeck O C, Crothers D M, Bralla J. Improved estimation of secondary structure in ribonucleic acids. Nature. 1973;246:40–41. doi: 10.1038/newbio246040a0. [DOI] [PubMed] [Google Scholar]
- 34.Vijayakumar M N, Ayalew S. Nucleotide sequence analysis of the termini and chromosomal locus involved in site-specific integration of the streptococcal conjugative transposon Tn5252. J Bacteriol. 1993;175:2713–2719. doi: 10.1128/jb.175.9.2713-2719.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]