Abstract
Efficient expression of the dye-decolorizing peroxidase, DyP, from Geotrichum candidum Dec 1 in Aspergillus oryzae M-2-3 was achieved by fusing mature cDNA encoding dyp with the A. oryzae α-amylase promoter (amyB). The activity yield of the purified recombinant DyP (rDyP) was 42-fold compared with that of the purified native DyP from Dec 1. No exogenous heme was necessary for the expression of rDyP in A. oryzae. From the N-terminal amino acid sequence analyses of native DyP and rDyP, the absence of a histidine residue in both DyPs, which was considered to be important for heme binding of DyP, was confirmed. These results suggest that rDyP without a typical heme-binding region produced by A. oryzae exhibits a function similar to that of native DyP.
The newly isolated Geotrichum candidum Dec 1 was found to decolorize 21 kinds of synthetic dyes (13), and its degradation spectrum in relation to synthetic dyes is wider than that of any other decolorizing organisms reported so far. In our previous study, an extracellular enzyme, DyP (for dye-decolorizing peroxidase), was found to be responsible for the decolorization of dyes. DyP degraded phenolic compounds, such as 2,6-dimethoxyphenol and guaiacol, while it did not degrade nonphenolic veratryl alcohol (14). Considering its substrate specificity and molecular mass, DyP was found to be a novel peroxidase distinct from other peroxidases reported previously (25, 26, 28, 41). Furthermore, the absorption spectrum of DyP exhibited a Soret band at 406 nm corresponding to a hemoprotein, and its Na2S2O4-reduced form revealed a peak at 556 nm that indicates the presence of a protoheme as its prosthetic group (14). We also reported the cloning of cDNA of the dyp gene (32).
So far, several microorganisms capable of decolorizing some synthetic dyes have been reported (17–20, 22, 23). In particular, the white-rot fungus Phanerochaete chrysosporium was extensively studied as a dye-decolorizing fungus (5, 9, 19, 20, 21, 30) and several lignin peroxidases (LiPs) of P. chrysosporium have been reported to show decolorizing activity. However, their decolorizing spectrum toward dyes is not extensively investigated, mainly because research on their lignin-degrading reaction is focused. Furthermore, although cloning and expression of several LiPs have been reported (6, 11, 29), there is no report on the enhancement of the yield of these enzymes by heterologous expression.
Therefore, we focused on producing a large amount of DyP having dye-decolorizing activity in Aspergillus oryzae under the control of the amyB promoter. A. oryzae is known to exhibit a high growth rate and to be a safe host (1); furthermore, it can secrete gram-per-liter quantities of heterologous protein (4). From this study and our previous data, we show that DyP is a unique peroxidase.
Chemicals, enzymes, and other materials.
Ten kinds of synthetic dyes kindly provided by Nippon Kayaku Co., Ltd. (Tokyo, Japan), and Bayer Japan Co., Ltd. (Tokyo, Japan), were used. Cellulase and lysing enzymes were obtained from Sigma-Aldrich Japan (Tokyo, Japan). Restriction enzymes and all other reagents were of analytical grade and commercially available.
Strains, plasmids, and media.
G. candidum Dec 1, isolated in our laboratory (13), was grown in potato dextrose broth (20 g of potato infusion per liter and 20 g of dextrose per liter; Difco Laboratories, Detroit, Mich.). A. oryzae M-2-3 (10) and plasmid pTAex3 (8) were obtained from the National Research Institute of Brewing (Hiroshima, Japan). A. oryzae M-2-3 is an auxotroph for arginine, and pTAex3 has the argB gene from Aspergillus nidulans (10). Although pTAex3 is not an autonomously replicating plasmid in A. oryzae, it is designed for the expression of recombinant proteins by integration into the chromosome of A. oryzae. Therefore, a transformant of A. oryzae M-2-3 having pTAex3 was grown on an arginine-free medium. Furthermore, the plasmid can replicate in Escherichia coli because of the replication origin in pUC119. Construction, propagation, and amplification of the hybrid plasmids were performed with E. coli DH5α or JM109 (Takara Co., Ltd., Tokyo, Japan). E. coli was cultured in Luria-Bertani medium (1% tryptone, 0.5% yeast extract, 1% NaCl [pH 6.8]) according to the standard method (27).
Protein and enzyme assays.
Protein concentrations were determined according to the Bradford method (3) using the Protein Assay Kit II (Bio-Rad, Tokyo, Japan) with bovine serum albumin as the standard protein. Reactive blue 5 (RB5), a representative anthraquinone dye, was used as the substrate. The substrate solution consists of 100 μg of RB5 per ml in 25 mM citrate buffer (pH 3.2). An appropriate amount of the enzyme solution was mixed with the substrate solution, and then H2O2 was added to give a final concentration of 0.2 mM. The total volume of the enzyme reaction mixture was adjusted to 3 ml. Enzyme activity was calculated from the decrease in absorbance at 600 nm (A600). One unit of enzyme activity was defined as the amount of enzyme that decolorized 1 μmol of RB5 at 30°C for 1 min.
Transformation of A. oryzae and dye-decolorizing activity.
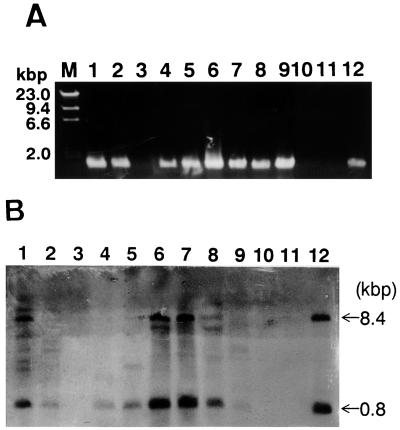
Plasmid pT-92 was constructed by the following method. The cDNA of dyp with the adapter (NotI-BamHI) at both its ends (1.6 kbp) was blunted with T4 DNA polymerase and ligated to the SmaI site which was located between the amyB promoter and amyB terminator of pTAex3 (7.6 kbp). Although the cDNA was ligated to forward or reverse orientation for the amyB promoter of pTAex3, only the forward-type hybrid plasmid was selected as pT-92 (9.2 kbp), which was used for transforming A. oryzae. The transformation of A. oryzae was performed according to a previously reported method with slight modification (10). Seventeen transformants were obtained by the transformation of A. oryzae M-2-3 with pT-92 on Czapek Dox medium plates. The efficiency of transformation was 0.85 clone per μg of pT-92. These transformants and A. oryzae M-2-3 (parent strain) were grown for 5 days in 5 ml of modified Czapek Dox medium (0.2% NaNO3, 0.1% K2HPO4, 0.05% MgSO4 · 7H2O, 0.05% KCl, 0.001% FeSO4, 3% maltose, 0.1% peptone [pH 5.5]) at 30°C with shaking at 120 spm. The culture was centrifuged, and the decolorizing activity of the supernatant was measured. Fifteen of these transformants decolorized RB5, but two did not decolorize RB5. A. oryzae M-2-3 (parent strain) also showed no dye-decolorizing activity. Eight of the 15 transformants which showed decolorizing activity (RD091, RD093, RD096, RD001, RD003, RD004, RD005, RD008), two nonactive strains (RD095, RD00A), A. oryzae M-2-3, and plasmid pT-92 were selected and PCR and Southern hybridization analyses were conducted according to standard methods (27). The results of the PCR analysis are shown in Fig. 1A. All the transformants possessing decolorizing activity, as well as pT-92, showed a positive band corresponding to dyp. No positive band corresponding to dyp appeared in the case of the two transformants (RD095 and RD00A) which do not have decolorizing activity and A. oryzae M-2-3. The result of Southern hybridization analysis was consistent with that of PCR analysis (Fig. 1B). Two fragments containing dyp digested with EcoRI are observed in Fig. 1B as 0.8- and 8.4-kb bands. All the transformants possessing decolorizing activity, as well as pT-92, showed a positive band corresponding to dyp. In contrast, the inactive transformants, RD095 (lane 3) and RD00A (lane 10), and A. oryzae M-2-3 (lane 11) showed no band. From these results, the dye-decolorizing activity of all transformants was derived from the expression of recombinant DyP (rDyP).
FIG. 1.
(A) PCR analysis of 10 A. oryzae transformants (RD series), A. oryzae M-2-3, and plasmid pT-92. The position of each fragment amplified with the primer of the 5′ coding region and the 3′ coding region of dyp is shown in kilobase pairs. Lanes: M, molecular marker; 1, A. oryzae RD091; 2, RD093; 3, RD095; 4, RD096; 5, RD001; 6, RD003; 7, RD004; 8, RD005; 9, RD008; 10, RD00A; 11, A. oryzae M-2-3 (parent strain); 12, pT-92. (B) Southern hybridization of the same samples as those subjected to PCR analysis. All templates were partially digested with EcoRI. Two fragments (0.8 and 8.4 kb) including dyp, obtained by EcoRI digestion of pT-92, are shown by arrows. The sample corresponding to each lane number is identical to that in panel A.
Purification of rDyP.
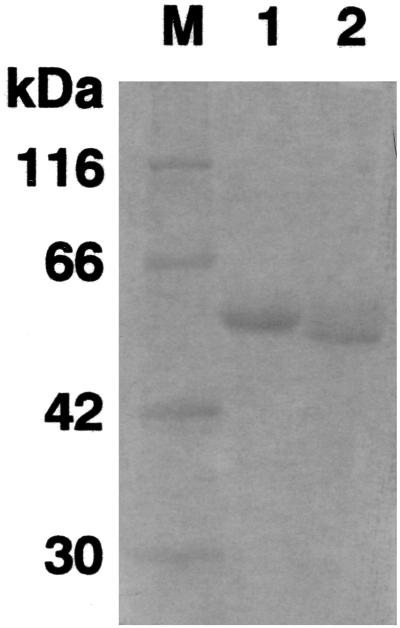
rDyP was purified from a culture of A. oryzae RD005 harboring pT-92, because RD005 showed strong dye-decolorizing activity. The strain was grown for 5 days in 900 ml of modified Czapek Dox medium at 30°C with shaking at 120 spm. All the following procedures were carried out at a temperature range of 0 to 4°C unless otherwise specified. The culture was filtered to remove insoluble components with filter paper 5A, and then the filtrate (622 ml) was concentrated to 70 ml by ultrafiltration through a YM-10 membrane (Amicon Grace Japan, Tokyo, Japan). Centriprep 10 (Amicon Grace Japan) was used in the buffer exchange treatment and concentration of the clear supernatant. Then, quaternary aminoethyl (QAE)-Toyopearl chromatography was carried out. A column (1 by 3 cm) of QAE-Toyopearl (Tosoh, Tokyo, Japan) was equilibrated with 20 mM acetate buffer (pH 5.5). Three milliliters of the supernatant treated as above was applied to the column and washed with 6 ml of the equilibration buffer. The column was eluted with a continuous linear gradient of 0 to 0.3 M NaCl in 20 mM acetate buffer (pH 5.5; total volume, 40 ml). The flow rate and the fraction volume were 1 ml/min and 2 ml, respectively. The active fractions (4 ml) were pooled and diluted to 16 ml with the equilibration buffer in order to reduce the concentration of salt. The fraction was then applied to a column (0.5 by 5 cm) of Mono-Q (Amersham-Pharmacia, Tokyo, Japan) equilibrated with 20 mM acetate buffer (pH 5.5) and washed with 3 ml of the same buffer. Elution was performed with a continuous linear gradient of 0 to 0.3 M NaCl in 20 mM acetate buffer (pH 5.5; total volume, 30 ml). The flow rate and the fraction volume were 0.5 ml/min and 1 ml, respectively. The active fractions were stored at 4°C for use in subsequent experiments. The specific activity and the percent recovery of the enzyme from each purification step are summarized in Table 1. Purified rDyP after Mono-Q chromatography is shown to migrate as a single band on sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) (Fig. 2). rDyP had a molecular mass of 58 kDa and was purified approximately 14.8-fold, with a yield of 39.8%. The rDyP expressed by A. oryzae revealed distinct decolorizing activity. The activity yield (8 × 102 U/liter of culture) of purified rDyP was 42-fold compared with that from Dec 1 (19 U/liter of culture), although rDyP production was not optimized here. Therefore, improvement of culture conditions or of the purification method could further increase the productivity of rDyP. As a solid culture of A. oryzae is applied to produce glucoamylase in Japanese fermentation industries (36), the solid culture may be a promising method of producing rDyP.
TABLE 1.
Purification of rDyP from A. oryzae RD005
| Purification step | Protein (mg) | Total activity (U) | Sp act (U/mg) | Yield (%) | Purification (fold) |
|---|---|---|---|---|---|
| Culture supernatant | 56.0 | 1,255 | 22.4 | 100 | 1 |
| Centriprep 10 | 25.7 | 5,109 | 199 | 407 | 8.9 |
| QAE-Toyopearl | 14.0 | 3,241 | 231 | 258 | 10.3 |
| Mono-Q | 1.51 | 500 | 331 | 39.8 | 14.8 |
FIG. 2.
SDS-PAGE of DyP. Purified DyP was subjected to electrophoresis on a 10% polyacrylamide gel at pH 8.0 in a Tris-glycine buffer, and the protein was stained with Coomassie brilliant blue R-250. β-Galactosidase (116 kDa), bovine serum albumin (66 kDa), aldolase (42 kDa), and carbonic anhydrase (30 kDa) were used as the standards for molecular mass determination. Lanes: M, molecular mass standards; 1, 5 μg of native DyP from G. candidum Dec 1; 2, 5 μg of rDyP from A. oryzae RD005.
Surprisingly, exogenous heme, which was reported to be necessary and important for heterologous peroxidase expression in A. oryzae (31), was unnecessary for the expression of rDyP. The purified rDyP showed a Soret band at 407 nm, corresponding to a hemoprotein. This reveals that DyP can use the same heme as that produced by A. oryzae and also suggests that the heme-binding site of DyP adapts to the heme which is produced by A. oryzae. This unique characteristic of DyP is reported for the first time in this work.
Analyses of thermostability and pH profile.
Fifteen microliters of rDyP (0.2 mg/ml) was placed in a water bath at a temperature of 30, 40, 50, 60, or 70°C for a period of 0, 5, 15, 30, 60, or 120 min. The enzyme activity for RB5 after heat treatment was measured according to the method described above in “Protein and enzyme assays.” After heating at 30, 40, and 50°C for 120 min, rDyP retained more than 90% of its activity when measured at 30°C for 1 min, but the enzyme gradually became inactive after treatment at 60°C and was rapidly inactivated at 70°C.
The pH profile of the RB5-decolorizing activity of rDyP was determined in a substrate solution in 25 mM citrate buffer adjusted to different pHs. Fifteen microliters of rDyP solution (0.4 mg/ml) was used, and the enzyme activity was measured according to the method described in “Protein and enzyme assays.” The optimum pH for this enzyme was found to lie between 3.0 and 3.2.
Amino acid sequence analysis.
SDS-PAGE of native DyP and rDyP (10 μg of each) was performed according to the method of Laemmli (15). Subsequently, both native DyP and rDyP bands were transferred from the gel to a polyvinyl difluoride membrane according to the standard method (35), and then amino acid sequencing analysis of each was performed with an amino acid sequencing apparatus (PPSQ-21; Shimadzu, Kyoto, Japan) according to the conventional method (7).
The first 14 residues of the N-terminal sequence of native DyP and rDyP were determined to be AXDTILPLNNIQGD in the single-letter amino acid code.
Substrate specificity.
Ten dyes which were decolorized by Dec 1 were selected. The concentration of each was adjusted so that the initial absorbance at each maximum absorption wavelength (λmax) was around 1 in 25 mM citrate buffer (pH 3.2) (14). Fifteen microliters of rDyP (0.15 mg/ml) was added to the substrate solution and mixed with H2O2 to give a final concentration of 0.2 mM. The total volume of the enzyme reaction mixture was adjusted to 3 ml. The decolorizing rate was calculated from the decrease in the absorbance at λmax. The decolorizing rates of RB5 (2.2 × 102 μmol/min), reactive blue 19 (2.2 × 102 μM/min), and reactive blue 21 (70 μM/min) were higher than those of the other dyes. Reactive red 33 (4.0 μM/min), reactive black 5 (1.9 μM/min), and reactive violet 23 (10 μM/min) were weakly decolorized. On the other hand, reactive red 120 was decolorized only slightly (0.7 μM/min), and reactive red 123, reactive orange 13, and reactive yellow 2 were not decolorized at all.
Moreover, veratryl alcohol, which is a well-known substrate of LiP, and guaiacol and 2,6-dimethoxyphenol, which are widely used as standard substrates of MnP, were tested according to the previously reported method (14, 34). Veratryl alcohol was not oxidized by rDyP. In contrast, the oxidation of 2,6-dimethoxyphenol and guaiacol by rDyP occurred without the addition of Mn2+, and no enhancement of activity by the addition of Mn2+ was observed.
Comparison between native DyP and rDyP.
From the above results, the characteristics of rDyP and native DyP were compared, as shown in Table 2. The substrate specificities in relation to dyes of rDyP and native DyP were almost the same. In addition, the activities of rDyP toward veratryl alcohol, guaiacol, and 2,6-dimethoxyphenol were almost the same as that of native DyP. These results suggest that the substrate specificities of DyP and rDyP were different from those of well-known peroxidases such as LiP and manganese peroxidase. The thermostability and molecular mass of rDyP were slightly different from those of native DyP. The N-terminal sequences of rDyP and native DyP were identical to that of residues 57 to 70 of the amino acid sequence of DyP deduced from the cDNA (32). Therefore, the processing site of the N-terminal region of native DyP and rDyP was found to be between the residues 56 and 57. As DyP is an extracellular protein, the N-terminal hydrophobic region is considered to function as a secretion signal peptide. However, the N-terminal hydrophobic region of DyP was deduced from our previous report to be around 20 amino acid residues from the initiation residue (32), and the potential cleavage site was considered to be between residues 22 and 23. If this is the case, the sequence from residue 23 to 56 was considered to be ruled out by secondary processing to form mature DyP. Interestingly, this deduced processing occurred in G. candidum and A. oryzae, although these two fungi belong to different genera. This suggests that the maturation process of DyP depends not on the genus of fungi but on DyP itself. The maturation of DyP might be critical for its function as a unique peroxidase. The glycosylation ratio and the sugar chain structures of the polypeptides are generally considered to differ between recombinant and native enzymes. Therefore, the slight difference in molecular mass between rDyP (58 kDa) and native DyP (60 kDa) was considered to depend on the difference in the glycosylation ratio between them; this was also reflected in the difference in migration on SDS-PAGE. If this is the case, the slight difference in thermostability between rDyP and native DyP could be explained because the glycosylation ratio is generally related to the thermostability or structural stability of an enzyme (16, 33, 37). However, the difference in glycosylation ratio was not critical for the decolorizing activity of the enzyme, as shown in Table 2. In a recent report, recombinant LiPH2, a glycoprotein cloned from P. chrysosporium, was expressed in E. coli, and its peroxidase activity was not found to be affected by the glycosylation ratio (11).
TABLE 2.
Comparison of characteristics of rDyP and native DyP
| Test or characteristic | Value for:
|
|
|---|---|---|
| Native DyPa | rDyPb | |
| Substrate specificity (%)c | ||
| Reactive blue 5 | 100 | 100 |
| Reactive blue 19 | 92 | 100 |
| Reactive black 5 | 0.5 | 0.9 |
| Reactive red 33 | 2.0 | 1.8 |
| Reactive yellow 2 | 2.6 | 0 |
| Veratryl alcohol | 0 | 0 |
| Guaiacol | 0.31g | 0.17g |
| 2,6-Dimethoxyphenol | 0.61g | 0.54g |
| Optimum pHd | 3.2 | 3.0–3.2 |
| Thermostability (%)e at: | ||
| 40°C | 97 | 100 |
| 50°C | 93 | 93 |
| 60°C | 79 | 12 |
| Molecular mass (kDa)f | 60 | 58 |
Data were summarized from our previous report (14).
Data were summarized from this work.
Substrate specificity was defined as relative activity toward RB5.
RB5 was used as the substrate.
Remaining activity at 30°C after 2 h of treatment at each temperature.
Molecular mass was estimated from data shown in Fig. 2.
Each datum was the increase in absorbance at 470 nm per min at pH 3.2 and at 30°C with 0.6 U of rDyP or native DyP.
The most unique characteristic of DyP as a peroxidase.
Peroxidases are classified into two superfamilies, animal peroxidases and plant peroxidases. The plant peroxidase superfamily is further categorized into three classes according to origin (40). Class I peroxidases are from the procaryotic lineage (12, 39). Class II peroxidases are secretory fungal peroxidases (25, 26, 28). Class III peroxidases are classical, secretory plant peroxidases (38, 41). According to this classification, DyP was expected to belong to the class II peroxidases. However, in our study, the characteristics of DyP were not similar to those of any other known peroxidases (14, 32). Especially, the nucleotide sequence of dyp and its primary translation product have no homology with any other reported peroxidases, except one peroxidase (cpop21) from a Polyporaceae sp., which has been registered under accession no. U77073 in GenBank, EMBL, and DDBJ. Although the characteristics of cpop21 are unclear because its data have been unpublished, we have already discussed the relation between the primary structure of DyP and that of cpop21 in our previous report (32). Therefore, we have omitted the discussion here.
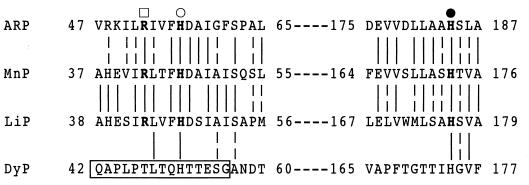
Heme-containing peroxidases have two conserved His residues and one conserved Arg residue. One His residue (proximal histidine) serves as the axial ligand for the heme, and the other His (distal histidine) and Arg (essential arginine) residues are considered to be involved in charge stabilization during the reaction between heme and H2O2 (2, 23, 24). Therefore, these conserved residues are considered to be essential for peroxidase activity. All these aforementioned residues are conserved in LiP, manganese peroxidase, and Arthromyces ramosus peroxidase, which are classified as class II peroxidases, as shown in Fig. 3. So far, it is believed that there are no exceptions to this rule. However, DyP has no heme-binding region which is common to the plant peroxidase superfamily, as described in our previous report (32). Furthermore, in this work, a probable His residue for heme binding, located at residue 51 of the amino acid sequence deduced from the cDNA of dyp, could have been lost by N-terminal processing, as shown in Fig. 3. It was predicted that DyP had a heme-binding site quite different from those of other peroxidases. In this case, the heme binding of DyP was considered to be specific for that in G. candidum. Therefore, the rDyP obtained would have no enzyme activity, since it lacks heme, even though the heterologous expression was successful. However, we obtained active rDyP from A. oryzae. This suggests that there is a novel heme-binding region other than the well-characterized region in the plant peroxidase superfamily and that this region is not specific to G. candidum. This is the most unique point of DyP as a heme-containing peroxidase. To clarify this interesting finding, crystallization and analysis of DyP are necessary.
FIG. 3.
Amino acid sequence comparison of DyP and other peroxidases in regions surrounding the conserved motif for the plant peroxidase superfamily. Vertical lines indicate points of identity between two enzymes. In addition, broken vertical lines indicate the matches between the residues, classified according to the functional property of each amino acid. The amino acid residues critical for heme binding are shown in boldface type. The numbers at both ends of each sequence indicate the number of residues from the initiation codon. The boxed region shown here is absent in mature DyP. ARP, peroxidase from A. ramosus (28); MnP, manganese peroxidase from P. chrysosporium (31); LiP, lignin peroxidase from P. chrysosporium (30); ●, proximal histidine; ○, distal histidine; □, essential arginine.
Acknowledgments
We are grateful to Katsuya Gomi for helpful advice and Hideki Taguchi for the N-terminal analysis of rDyP.
REFERENCES
- 1.Barvesgaard P, Heldt-Hansen H P, Diderichen B. On the safety of Aspergillus oryzae: a review. Appl Microbiol Biotechnol. 1992;36:569–572. doi: 10.1007/BF00183230. [DOI] [PubMed] [Google Scholar]
- 2.Bosshard H R, Banziger J, Hasler T, Poulos T L. The cytochrome c peroxidase-cytochrome c electron transfer complex. The role of histidine residues. J Biol Chem. 1984;259:5683–5690. [PubMed] [Google Scholar]
- 3.Bradford M M. A rapid and sensitive method for the quantification of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1976;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- 4.Christensen T, Woeldike H, Boel E, Motensen S B, Hjortsjoej K, Thim L, Hansen M T. High level expression of recombinant genes in Aspergillus oryzae. Bio/Technology. 1988;6:1419–1422. [Google Scholar]
- 5.Cripps C, Bumpus J S, Aust S D. Biodegradation of azo and heterocyclic dyes by Phanerochaete chrysosporium. Appl Environ Microbiol. 1990;56:1114–1118. doi: 10.1128/aem.56.4.1114-1118.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cullen D. Recent advances on the molecular genetics of ligninolytic fungi. J Biotechnol. 1997;53:273–289. doi: 10.1016/s0168-1656(97)01684-2. [DOI] [PubMed] [Google Scholar]
- 7.Edman P. A method for the determination of the amino acid sequence in peptides. Arch Biochem Biophys. 1949;22:475–476. [PubMed] [Google Scholar]
- 8.Fujii T, Yamaoka H, Gomi K, Kitamikado K, Kumagai C. Cloning and nucleotide sequence of the ribonuclease T1 gene (rntA) from Aspergillus oryzae and its expression in Saccharomyces cerevisiae and Aspergillus oryzae. Biosci Biotechnol Biochem. 1995;59:1869–1874. doi: 10.1271/bbb.59.1869. [DOI] [PubMed] [Google Scholar]
- 9.Glenn J K, Gold M H. Decolorization of several polymeric dyes by the lignin-degrading basidiomycete Phanerochaete chrysosporium. Appl Environ Microbiol. 1983;45:1741–1747. doi: 10.1128/aem.45.6.1741-1747.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gomi K, Iimura Y, Hara S. Integrative transformation of Aspergillus oryzae with a plasmid containing the Aspergillus nidulans argB gene. Agric Biol Chem. 1987;51:2549–2555. [Google Scholar]
- 11.Guojun N, Reading N S, Aust D. Expression of the lignin peroxidase H2 gene from Phanerochaete chrysosporium in Escherichia coli. Biochem Biophys Res Commun. 1998;249:146–150. doi: 10.1006/bbrc.1998.9106. [DOI] [PubMed] [Google Scholar]
- 12.Kaput J, Goltz S, Blobel G. Nucleotide sequence of the yeast nuclear gene for cytochrome c peroxidase precursor: functional implications of the presequence for protein transport into mitochondria. J Biol Chem. 1982;257:15054–15058. [PubMed] [Google Scholar]
- 13.Kim S J, Ishikawa K, Hirai M, Shoda M. Characteristics of a newly isolated fungus, Geotrichum candidum Dec 1, which decolorizes various dyes. J Ferment Bioeng. 1995;79:601–607. [Google Scholar]
- 14.Kim S J, Shoda M. Purification and characterization of a novel peroxidase from Geotrichum candidum Dec 1 involved in decolorization of dyes. Appl Environ Microbiol. 1999;65:1029–1035. doi: 10.1128/aem.65.3.1029-1035.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Laemmli U K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 16.Lis H, Sharon N. Protein glycosylation: structural and functional aspects. Eur J Biochem. 1993;218:1–27. doi: 10.1111/j.1432-1033.1993.tb18347.x. [DOI] [PubMed] [Google Scholar]
- 17.Livernoche D, Jurasek L, Desrochers M, Dorica J. Removal of color from kraft mill wastewaters with cultures of white-rot fungi and with immobilized mycelium of Coriolus versicolor. Biotechnol Bioeng. 1983;25:2055–2065. doi: 10.1002/bit.260250814. [DOI] [PubMed] [Google Scholar]
- 18.Pasti M B, Crawford D L. Relationship between the abilities of streptomycetes to decolorize three anthron-type dyes and to degrade lignocellulose. Can J Microbiol. 1991;37:902–907. [Google Scholar]
- 19.Pasti-Grigsby M B, Paszczynski A, Goszczynski S, Crawford D L, Crawford R L. Influence of aromatic substitution patterns on azo dye degradability by Streptomyces spp. and Phanerochaete chrysosporium. Appl Environ Microbiol. 1992;58:3605–3613. doi: 10.1128/aem.58.11.3605-3613.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Paszczynski A, Pasti-Grigsby M B, Goszczynski S, Crawford R L, Crawford D L. Mineralization of sulfonated azo dyes and sulfanilic acid by Phanerochaete chrysosporium and Streptomyces chromofuscus. Appl Environ Microbiol. 1992;58:3598–3604. doi: 10.1128/aem.58.11.3598-3604.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Paszczynski A, Pasti M B, Goszczynski S, Crawford D L, Crawford R L. New approach to improve degradation of recalcitrant azo dyes by Streptomyces spp. and Phanerochaete chrysosporium. Enzyme Microb Technol. 1991;13:378–384. [Google Scholar]
- 22.Platt M W, Hadar Y, Chet I. The decolorization of the polymeric dye poly-blue (polyvinylamine sulfonate-anthraquinone) by lignin-degrading fungi. Appl Microbiol Biotechnol. 1985;21:394–396. [Google Scholar]
- 23.Poulos T L, Edwards S L, Wariishi H, Gold M H. Crystallographic refinement of lignin peroxidase at 2 Å. J Biol Chem. 1993;268:4429–4440. doi: 10.2210/pdb1lga/pdb. [DOI] [PubMed] [Google Scholar]
- 24.Poulos T L, Kraut J. The stereochemistry of peroxidase catalysis. J Biol Chem. 1980;255:8199–8205. [PubMed] [Google Scholar]
- 25.Pribnow D, Mayfield M B, Nipper V J, Brown J A, Gold M H. Characterization of a cDNA encoding a manganese peroxidase, from the lignin-degrading basidiomycete Phanerochaete chrysosporium. J Biol Chem. 1989;264:5036–5040. [PubMed] [Google Scholar]
- 26.Ritch T G, Jr, Nipper V J, Akireswaran L, Smith A J, Pribnow D G, Gold M H. Lignin peroxidase from the basidiomycete Phanerochaete chrysosporium is synthesized as a preproenzyme. Gene. 1991;107:119–126. doi: 10.1016/0378-1119(91)90304-t. [DOI] [PubMed] [Google Scholar]
- 27.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 28.Sawai-Hatanaka H, Ashikari T, Tanaka Y, Asada Y, Nakayama T, Minakata H, Kunishima N, Yamada H, Shibano Y, Amachi T. Cloning, sequencing, and heterologous expression of a gene coding for Arthromyces ramosus peroxidase. Biosci Biotechnol Biochem. 1995;59:1221–1228. doi: 10.1271/bbb.59.1221. [DOI] [PubMed] [Google Scholar]
- 29.Sollewijin M D, Mayfield-Gambill M, Lin Cereghino G P, Gold M H. Homologous expression of recombinant lignin peroxidase in Phanerochaete chrysosporium. Appl Environ Microbiol. 1999;65:1670–1674. doi: 10.1128/aem.65.4.1670-1674.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Spadaro J T, Gold M H, Renganathan V. Degradation of azo dyes by the lignin-degrading fungus Phanerochaete chrysosporium. Appl Environ Microbiol. 1992;58:2397–2401. doi: 10.1128/aem.58.8.2397-2401.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Stewart P, Whitwam R E, Kersten P J, Cullen D, Tien M. Efficient expression of a Phanerochaete chrysosporium manganese peroxidase gene in Aspergillus oryzae. Appl Environ Microbiol. 1996;62:860–864. doi: 10.1128/aem.62.3.860-864.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sugano Y, Sasaki K, Shoda M. cDNA cloning and genetic analysis of a novel decolorizing enzyme, peroxidase gene dyp from Geotrichum candidum Dec 1. J Biosci Bioeng. 1999;87:411–417. doi: 10.1016/s1389-1723(99)80087-5. [DOI] [PubMed] [Google Scholar]
- 33.Tams J W, Welinder K G. Glycosylation and thermodynamics versus kinetic stability of horseradish peroxidase. FEBS Lett. 1998;421:234–236. doi: 10.1016/s0014-5793(97)01573-1. [DOI] [PubMed] [Google Scholar]
- 34.Tien M, Kirk T K. Lignin-degrading enzyme from Phanerochaete chrysosporium: purification, characterization, and catalytic properties of a unique H2O2-requiring oxygenase. Proc Natl Acad Sci USA. 1984;81:2280–2284. doi: 10.1073/pnas.81.8.2280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Towbin H, Staehelin T, Gordon J. Electrophoretic transfer of proteins from polyacrylamide gels to nitrocellulose sheets: procedure and some applications. Proc Natl Acad Sci USA. 1979;76:4350–4354. doi: 10.1073/pnas.76.9.4350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Tsuchiya K, Nagashima T, Yamamoto Y, Gomi K, Kitamikado K, Kumagai C, Tamura G. High level secretion of calf chymosin using a glucoamylase-prochymosin fusion gene in Aspergillus oryzae. Biosci Biotechnol Biochem. 1994;58:895–899. doi: 10.1271/bbb.58.895. [DOI] [PubMed] [Google Scholar]
- 37.Wang C, Eufemi M, Turano C, Giartosio A. Influence of the carbohydrate moiety on the stability of glycoproteins. Biochemistry. 1996;35:7299–7307. doi: 10.1021/bi9517704. [DOI] [PubMed] [Google Scholar]
- 38.Welinder K G. Covalent structure of the glycoprotein horseradish peroxidase (EC 1.11.1.7) FEBS Lett. 1976;72:19–23. doi: 10.1016/0014-5793(76)80804-6. [DOI] [PubMed] [Google Scholar]
- 39.Welinder K G. Bacterial catalase-peroxidases are gene duplicated members of the plant peroxidase superfamily. Biochim Biophys Acta. 1991;1080:215–220. doi: 10.1016/0167-4838(91)90004-j. [DOI] [PubMed] [Google Scholar]
- 40.Welinder K G. Superfamily of plant, fungal, and bacterial peroxidases. Curr Opin Struct Biol. 1992;2:388–393. [Google Scholar]
- 41.Welinder K G, Mazza G. Amino-acid sequences of heme-linked, histidine-containing peptides of five peroxidases from horseradish and turnip. Eur J Biochem. 1977;73:353–358. doi: 10.1111/j.1432-1033.1977.tb11325.x. [DOI] [PubMed] [Google Scholar]