Abstract
Very little is known about the contribution of surface appendages of Salmonella enterica serovar Enteritidis to pathogenesis in chickens. This study was designed to clarify the role of SEF14, SEF17, and SEF21 fimbriae in serovar Enteritidis pathogenesis. Stable, single, defined sefA (SEF14), agfA (SEF17), and fimA (SEF21) insertionally inactivated fimbrial gene mutants of serovar Enteritidis were constructed. All mutant strains invaded Caco-2 and HT-29 enterocytes at levels similar to that of the wild type. Both mutant and wild-type strains were ingested equally well by chicken macrophage cell lines HD11 and MQ-NCSU. There were no significant differences in the abilities of these strains to colonize chicken ceca. The SEF14− strain was isolated in lower numbers from the livers of infected chickens and was cleared from the spleens faster than other strains. No significant differences in fecal shedding of these strains were observed.
Salmonellosis is the third most commonly reported infectious disease in the United States. Since the middle 1980s, most human cases reported to the National Salmonella Surveillance System administered by the Centers for Disease Control and Prevention have been attributed to Salmonella enterica serovar Enteritidis. More than 80% of food-borne serovar Enteritidis outbreaks reported since 1985 are associated with the consumption of raw or undercooked eggs (16).
Salmonella species enter the host by invading the gastrointestinal mucosa, a step essential for pathogenesis (13). Fimbriae have been shown to be involved in colonization and in adherence to specific host target tissues in the early stages of infection (5, 14, 15). Studies involving fimbriae have drawn considerable interest because fimbriae can serve as potential immunogens against many pathogenic bacteria that colonize epithelial surfaces (3–5, 20). Currently, we are exploring the use of fimbrial antigens as vaccine candidates. However, the role of fimbriae in pathogenesis of serovar Enteritidis is poorly understood. More knowledge is necessary to clarify the role of serovar Enteritidis fimbriae in vaccines designed to reduce the colonization and prevalence of this bacterium in poultry.
Serovar Enteritidis produces at least five distinct fimbriae, namely SEF14 and SEF17 (type 1 fimbriae) and SEF21, LPF, and PEF (28). Of all serovar Enteritidis fimbriae, SEF14 has been studied the most. SEF14 has been shown to contribute to serovar Enteritidis adherence to mouse epithelial cells, and passive administration of SEF14 antibodies is protective in mice (26). However, in vitro and in vivo studies using isogenic mutants of serovar Enteritidis have demonstrated no role for SEF14 in pathogenesis (25, 28). Information regarding the role of SEF17 and SEF21 fimbriae in serovar Enteritidis pathogenesis is lacking. Because the factors which influence fimbrial phase variation in vivo and which influence variability in the fimbriation of cells carrying an intact fimbrial operon are not known, it is difficult to evaluate the role of fimbriae in pathogenesis. Therefore, a genetic approach was used in this study to investigate the role of the three known serovar Enteritidis fimbrial operons, sefA (7), agfA (10), and fimA (22), which encode SEF14, SEF17, and SEF21 fimbriae, respectively. The ability of these sefA, agfA, and fimA mutant strains to mediate colonization and invasion was assessed in vitro and in a chicken model.
Bacterial strains, plasmids, and growth conditions.
Serovar Enteritidis phage type 4 strain was originally isolated from the liver of a laying chicken (18). A spontaneous nalidixic-acid-resistant (Nalr) mutant was obtained by plating on a nalidixic-acid-containing Luria-Bertani (LB) agar plate and was used as the recipient strain for conjugation. Escherichia coli S17-1/λpir was used as the donor strain. Inactivation of fimbrial genes was carried out by using suicide plasmid vector PKNOCK-Km (2). pGEM-T plasmid vector (Promega, Madison, Wis.) was used for cloning PCR fragments. Bacterial strains were grown either in LB broth, colonization factor antigen (CFA) broth (9), or T medium (8).
Statistical analyses.
Statistical analyses were performed by using StatView 4.5 (Abacus Concepts, Berkeley, Calif.). Bacterial numbers were converted to log10 prior to statistical analyses, and differences were analyzed by a one-way analysis of variance followed by Fisher's test for significant difference. Fractional data were analyzed by a chi-square test with continuity correction.
Preparation of gene knockout constructs.
Internal fragments (lacking sequences on 5′ and 3′ ends) of the sefA, agfA, and fimA genes were amplified from the serovar Enteritidis genomic DNA by PCR by using the following primers: for sefA, forward, 5′ GGGCTCGAGCTTGCTTAAATTGCATGTGGC and reverse, 5′ GGGCTCGAGGG TTGTGACAGGGACATTTAGC; for agfA, forward, 5′ GGCTCGAGATCGTAGTT TCTGGCAGTGC and reverse, 5′ GGGCTCGAGCCTGACGCACCATTACGC; and for fimA, forward, 5′ GGGCTCGAGGTCTGATGTTTGCTGGC and reverse, 5′ GGGCTCGAGATTAGCCTGGCCTGGCG. Additional bases were added to the 5′ end of each primer to confer a recognition sequence for XhoI (underlined above). The amplification conditions were 94°C for 1.5 min, 56°C for 1 min, and 72°C for 2 min, for 30 cycles. PCR products were purified from 1% agarose gel and cloned into pGEM-T vector. Sequences of the inserts were confirmed by automated DNA sequencing. Finally, internal sequences of sefA, agfA, and fimA were subcloned into XhoI-digested pKNOCK-Km suicide vector, thus creating pKNOCK-Km/ΔsefA, pKNOCK-Km/ΔagfA, and pKNOCK-Km/ΔfimA, respectively. Resulting plasmids were electroporated into E. coli S17-1/λpir donor cells. Electroporation was performed in a Gene Pulser apparatus according to the manufacturer's instructions (Bio-Rad Laboratories, Richmond, Calif.). Recombinant clones were selected based on restriction enzyme analysis of the plasmid DNA extracted from kanamycin-resistant (Kmr) colonies.
Construction of fimbrial gene knockouts.
SEF14−, SEF17−, and SEF21− strains of serovar Enteritidis were constructed by inactivation of the respective genes through homologous exchange of DNA material between the wild-type fimbrial genes and 5′, 3′-truncated fimbrial genes on pKNOCK. Suicide vector constructs pKNOCK-Km/ΔsefA, pKNOCK-Km/ΔagfA, and pKNOCK-Km/ΔfimA were mobilized from donor E. coli S17-1/λpir into spontaneous Nalr serovar Enteritidis by conjugation as described (1). The Kmr and Nalr bacterial colonies were selected and analyzed by PCR, nucleotide sequence analysis, and Western blotting. For each conjugation experiment, at least 20 exconjugants were tested for the presence of autonomously replicating pKNOCK plasmid. All exconjugants tested had integrated the plasmid DNA into their genome (results not shown). For PCR, regions of chromosomal DNA flanking the insertion site were amplified by using two pairs of primers for each exconjugant tested. One primer in each pair was specific to the chromosomal DNA flanking the insertion site (5′ or 3′ sequences of the fimbrial gene absent in the pKNOCK construct) and another was specific to the Kmr gene (forward, 5′ TTGGGTGGAGAGGCTATTCG and reverse, 5′ CACCATGATATTCGGCAAGC). The chromosomal-DNA-specific primer sequences were as follows: for sefA, forward, 5′ ATGCGTAAATCAGCAT CTGCA and reverse, 5′ TTAGTTTTGATA CTGCTGAACGTAG; for agfA, forward, 5′ ATGAAACTCCTAAAAGTGGCAGC and reverse, 5′ AGCGCAGACGCTAAA TTAATACTG; and for fimA, forward, 5′ ATGAAACATAAATTAATGACCTCTAC and reverse, 5′ TTATTCGTATTTCATGATAAAGGTGG. PCR amplification was performed as described above, except that annealing was performed at 57°C for 1 min. PCR products were purified from a 1% agarose gel with identity confirmed by nucleotide sequence analysis. More than 90% of the clones tested from each conjugation had integrated the plasmid DNA site specifically into their genome. To further support the PCR results, the PCR products of two exconjugants from each conjugation reaction were sequenced by automated sequencing, and the identities of products were confirmed. However, when the same sets of primers were used, no DNA amplification was observed with the wild-type counterpart. No evidence for the presence of an unaltered sefA, agfA, or fimA allele in SEF14−, SEF17−, and SEF21− strains was observed, and PCR with wild-type-gene-specific primers failed to amplify specific DNA fragments from these strains.
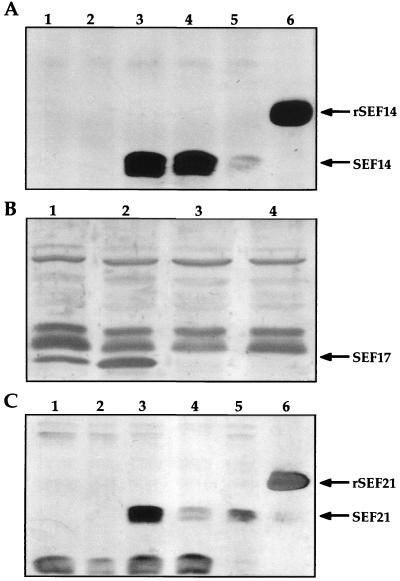
For Western blot analyses, whole-cell lysates from serovar Enteritidis exconjugants were prepared as previously described (8, 9). The samples were separated on 12% polyacrylamide gels, were transferred onto a nitrocellulose membrane, and were stained with monospecific polyclonal antibodies to SEF14, SEF17, and SEF21 (9, 10, 22). No SEF14, SEF17, or SEF21 proteins were observed in extracts obtained from SEF14−, SEF17−, or SEF21− strains, respectively; however, all three antigens were detected in extracts of the wild-type serovar Enteritidis (Fig. 1).
FIG. 1.
Western immunoblot analysis of serovar Enteritidis wild-type and mutant strains. (A) Sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) sample buffer-glycine extracts of serovar Enteritidis wild-type and SEF14− strains. Lanes 1 and 2, serovar Enteritidis SEF14−; lanes 3 to 5, wild-type serovar Enteritidis; lane 6, rSEF14 control. (B) SDS-PAGE sample buffer-glycine insoluble formic acid treated extract of serovar Enteritidis wild-type and SEF17− strains. Lanes 1 and 2, wild-type serovar Enteritidis; lanes 3 and 4, SEF17− serovar Enteritidis. (C) SDS-PAGE sample buffer-glycine extracts of serovar Enteritidis wild-type and SEF21− strains. Lanes 1 and 2, SEF21− serovar Enteritidis; lanes 3 to 5, serovar Enteritidis; lane 6, rSEF21 control. rSEF14 and rSEF21 are expressed as fusion proteins.
The mutation induced through the pKNOCK plasmid was structurally stable. Mutant strains were able to grow on LB agar supplemented with kanamycin and nalidixic acid even after 100 serial passages on LB agar without antibiotic. Stability was further confirmed by PCR with primers specific for the integrated plasmid and flanking DNA. Mutant serovar Enteritidis strains were also biochemically identical to wild-type serovar Enteritidis on standard biochemical media (19).
Invasion of HT-29 and Caco-2 enterocytes.
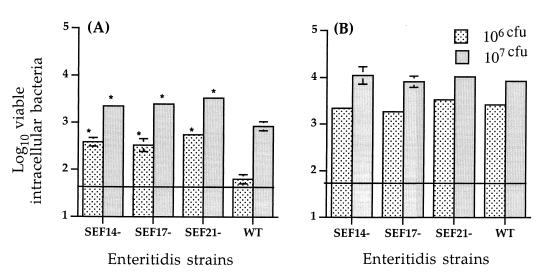
The abilities of wild-type serovar Enteritidis and fimbrial mutants to invade HT-29 and Caco-2 enterocytes were compared. Individual bacterial strains were grown in static CFA or T medium for 48 to 72 h (9, 22) at 37°C, were washed, and were diluted to the appropriate concentration in tissue culture medium. Bacterial concentrations were determined by densitometry and were confirmed by serial dilution followed by viable plate counts on MacConkey agar. One milliliter of bacterial suspension containing either 106 or 107 viable bacteria was added to each tissue culture well (15-mm-diameter, 24-well microtiter plates; Nunc) containing approximately 106 enterocytes, and invasion assays were performed as previously described (31). Each bacterial strain was tested in at least three separate assays, each assay representing the average of triplicate tissue culture wells. The lower limit of assay detection was 50 bacteria; for statistical analysis, values below this limit were assigned a value equal to the lower limit of assay detection. Figure 2 shows the number of viable Salmonella cells internalized by HT-29 and Caco-2 enterocytes. In general, serovar Enteritidis strains were more invasive in Caco-2 cells than in HT-29 cells. The mutant strains were internalized by Caco-2 and HT-29 enterocytes as efficiently as the wild-type serovar Enteritidis. Quantitative recovery of viable intracellular bacteria was reproducible, as indicated by small standard error bars (Fig. 2). Previous studies involving SEF14 and epithelial cell monolayers have produced variable results. SEF14 fimbriae have been shown to contribute to serovar Enteritidis adherence to mouse epithelial cells (25, 26) but not to adherence and invasion of human HEp-2, INT-407, Caco-2, and HeLa cells (12, 25, 28), although a SEF14− mutant derived from avirulent serovar Enteritidis invaded HeLa cells more poorly than the wild type (25). Because HeLa cells are of cervical epithelial origin, they may have unique receptors for SEF14 compared to receptors on enterocytes. It has been suggested that SEF17 and SEF21 fimbriae of serovar Enteritidis bind basement membrane proteins (9, 21) and may thus provide a mechanism for intestinal colonization. Recent studies have shown that serovar Enteritidis cells lacking either SEF17 or SEF21 are less invasive in INT-407 (12). However, data from our study suggest that neither SEF14, SEF17, nor SEF21 augmented serovar Enteritidis internalization by enterocytes. It is possible that cell types used in our study may not express the appropriate receptor for fimbrial attachment or that bacteria might utilize multiple adhesins to attach. It has recently been shown that multiple fimbrial adhesins are required for full virulence of Salmonella enterica serovar Typhimurium in mice (29).
FIG. 2.
Internalization of HT-29 (A) and Caco-2 (B) enterocytes by wild-type and mutant serovar Enteritidis strains. Each bacterial strain was tested in at least three separate assays. Horizontal line indicates lower limit of assay detection. Asterisks indicate values significantly different from that of the wild type at the corresponding concentration (P < 0.05).
Ingestion by chicken macrophage cell lines.
Macrophage cell lines HD-11 and MQ-NCSU (6, 27) were cultivated at 41°C in 5% CO2 in either RPMI medium supplemented with 5% fetal bovine serum (HD-11) or Liebovitz-McCoy medium supplemented with 10% fetal bovine serum (MQ-NCSU). Macrophage cell suspension (107 cells/ml) was prepared in a mixture of 1% gelatin in Hank's balanced salt solution, and viability was determined by trypan blue exclusion. Wild-type serovar Enteritidis and mutant strains were cultivated in CFA broth for 24 h at 37°C with gentle shaking, were washed, and were resuspended in phosphate-buffered saline to a concentration of 108 CFU/ml. Bacterial opsonization was carried out by mixing equal volumes of bacteria with 20% (vol/vol) normal chicken serum for 15 min at 37°C with shaking. Bacteria were then centrifuged at 6,000 × g for 15 min and were resuspended to the original concentration in 1% gelatin in Hank's balanced salt solution. Ingestion of the preopsonized bacteria was measured as described (30) with modifications. Five hundred microliters of bacteria was added to 500 μl of the macrophage cell suspension, and the mixture was incubated with shaking for 45 min to compare the efficacy of bacterial ingestion of each strain. The number of bacteria in the supernatant after ingestion was verified to be at least 1 log10 fewer than the number of internalized bacteria. Each bacterial strain was tested in at least three separate assays, each assay representing the average of triplicate samples. No significant differences in the numbers of internalized bacteria were noted with both macrophage cell lines (values ranging from 6.3 to 6.8 log10 CFU/5 × 106 NCSU cells or 6.0 to 6.4 log10 CFU/5 × 106 HD-11 cells for mutant strains and 6.3 log10 CFU/5 × 106 NCSU cells or 6.0 log10 CFU/5 × 106 HD-11 cells for the wild type). Total bacterial numbers before and after phagocytosis were similar, indicating that the bacteria did not multiply during phagocytosis. Type 1 fimbriae of serovar Typhimurium have been shown to mediate attachment, internalization, and intracellular survival in murine and porcine phagocytes (17, 23). However, in the present study, wild-type serovar Enteritidis and serovar Enteritidis lacking SEF21, SEF14, or SEF17 were internalized equally well by chicken macrophages. Thorns et al. (28) have also reported that serovar Enteritidis lacking SEF14 is internalized and persists in murine peritoneal macrophages similar to the wild-type strain.
Invasion, persistence, and excretion of serovar Enteritidis fimbrial mutants in SPF chickens.
Groups of 25 5-day-old, specific-pathogen-free chickens (SPAFAS Inc., Roanoke, Ill.) were orally inoculated with a pure culture of approximately 107 wild-type serovar Enteritidis cells or fimbrial mutant suspended in 1 ml of phosphate-buffered saline. At 1, 3, 7, 14, and 21 days postinoculation (p.i.), five chickens from each group were killed, and the liver, spleen, and cecum were aseptically excised. The number of viable bacteria per gram of tissue was determined as previously described (11). The lower limit of assay detection was 12 bacteria or 1.09 log10 cells per g of tissue. For statistical analysis, values below this limit were assigned a value equal to the lower limit of assay detection. Fecal excretion of Salmonella cells was monitored by culturing cloacal swabs. Stability of the mutants following reisolation from chickens was determined by plating onto LB agar with and without antibiotics and by PCR on genomic DNA from five individual colonies from each plate.
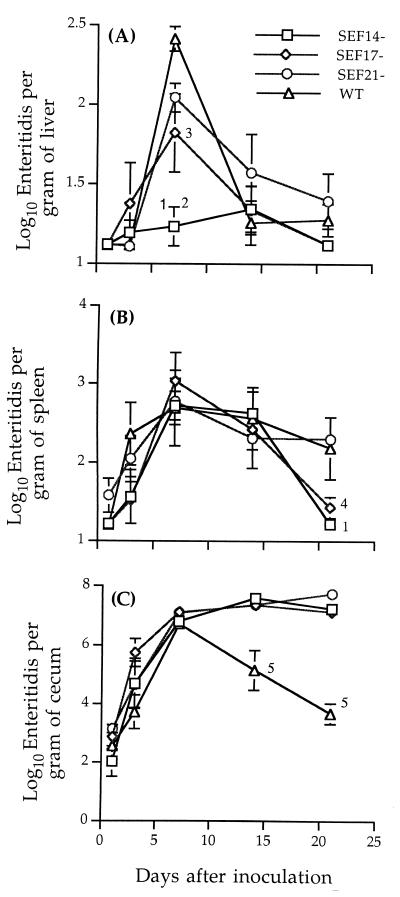
In general, no differences were observed among the serovar Enteritidis strains in their ability to colonize chicken ceca, to invade liver and spleen (Fig. 3), and to be shed in feces (data not shown). The only significant finding was that in birds inoculated with SEF14− serovar Enteritidis, fewer Salmonella cells were isolated from the liver at all time points compared to the other treatment groups, and this number was significantly reduced at day 7 p.i. (Fig. 3A). Birds inoculated with SEF17− serovar Enteritidis also had a low number of Salmonella cells in the liver at day 7 p.i. compared to birds inoculated with SEF21− bacteria or the wild type. In addition, in birds inoculated with SEF14− serovar Enteritidis, no Salmonella cells were recovered from the spleen at day 21 p.i. Although there was no difference in colonization of chicken ceca by the SEF14− strain compared to other strains, the lower numbers of SEF14− bacteria recovered from livers and spleens of infected chickens suggested that SEF14 might contribute to the persistence of serovar Enteritidis in extraintestinal tissues. Although it was also possible that these differences were due to an indirect effect of sefA gene disruption, clarification of this mechanism was beyond the scope of this study. The SEF14−, SEF17−, and SEF21− mutants were stable even after reisolation from chickens.
FIG. 3.
Recovery of wild-type and mutant serovar Enteritidis strains in liver (A), spleen (B), and cecum (C) of chickens orally inoculated with 107 cells of each strain. Data represent the means ± standard errors of five tested chickens. 1, significantly different from the SEF21− and wild-type strains (P < 0.01); 2, significantly different from the SEF17− strain (P < 0.05); 3, significantly different from the wild-type strain (P < 0.05); 4, significantly different from the wild-type and SEF21− strains (P < 0.05); 5, significantly different from the SEF14−, SEF17−, and SEF21− strains (P < 0.01).
Studies involving chickens vaccinated parenterally with recombinant SEF14 as well as with recombinant SEF17 or SEF21 and subsequently challenged with serovar Enteritidis have shown no major differences in cecal colonization or persistence of serovar Enteritidis (G. Rajashekara, S. Munir, D. A. Halvorson, and K. V. Nagaraja, unpublished data). These observations support the results of our in vitro studies using enterocytes or chicken macrophage cell lines. There is evidence that SEF14 is a T-cell immunogen (24) and that oral administration of hen egg yolk antibodies specific to SEF14 provides passive protection against experimental salmonellosis (26); using a variety of antigen delivery systems, similar results have been obtained following immunization of BALB/c mice with SEF14 (25). Differences observed in the present study could be due to differences in host species and in tissue tropism. Recent evidence that serovar Typhimurium fimbrial types contribute to tissue tropism for murine small intestinal villi (3) and murine Peyer's patches (5), as well as attachment to and invasion of epithelial cell lines (4), supports this view. Interestingly, from day 14 p.i. onwards, all three serovar Enteritidis mutant strains were recovered from chicken ceca in higher numbers than the wild type (Fig. 3C). This suggested that in the absence of these fimbrial antigens, the host might be unable to efficiently clear the organisms from the cecum. Perhaps fimbrial antigens might act in concert to establish infection; thus, the host immune response to multiple fimbrial antigens might be necessary for clearance from the cecum. In conclusion, our data suggest no major role for SEF14, SEF17, or SEF21 fimbriae under the conditions tested. Because distinct fimbrial types contribute to tissue tropism in serovar Typhimurium (3–5) and multiple fimbrial adhesins are required for full virulence of serovar Typhimurium (29), investigations using serovar Enteritidis strains lacking multiple fimbriae may provide additional information regarding the role of fimbriae in serovar Enteritidis pathogenesis.
Acknowledgments
This work was supported in part by Minnesota Agricultural Experiment Station grant AES6325 and by Public Health Service grant AI23484 from the National Institutes of Health.
We thank W. W. Kay, University of Victoria, British Columbia, Canada, for providing anti-SEF14, anti-SEF17, and anti-SEF21 antibodies and J. M. Sharma, University of Minnesota, St. Paul, for providing macrophage cell lines.
REFERENCES
- 1.Alexeyev M F, Shokolenko I N. Mini-Tn10 transposon derivatives for insertion mutagenesis and gene delivery in the chromosome of Gram-negative bacteria. Gene. 1995;160:59–62. doi: 10.1016/0378-1119(95)00141-r. [DOI] [PubMed] [Google Scholar]
- 2.Alexeyev M F. The pKNOCK series of broad-host-range mobilizable suicide vectors for gene knockout and targeted DNA insertion into the chromosome of Gram's negative bacteria. BioTechniques. 1999;26:824–828. doi: 10.2144/99265bm05. [DOI] [PubMed] [Google Scholar]
- 3.Baumler A J, Tsolis R M, Bowe F A, Kusters J G, Hoffmann S, Heffron F. The pef fimbrial operon of Salmonella typhimurium mediates adhesion to murine small intestine and is necessary for fluid accumulation in the infant mouse. Infect Immun. 1996;64:61–68. doi: 10.1128/iai.64.1.61-68.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Baumler A J, Tsolis R M, Heffron F. Contribution of fimbrial operons to attachments to and invasion of epithelial cell lines by Salmonella typhimurium. Infect Immun. 1996;64:1862–1865. doi: 10.1128/iai.64.5.1862-1865.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Baumler A J, Tsolis R M, Heffron F. The lpf fimbrial operon mediates adhesion of Salmonella typhimurium to murine Peyer's patches. Proc Natl Acad Sci USA. 1996;93:279–283. doi: 10.1073/pnas.93.1.279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Beug H, von Kirchback A, Doderlein G, Conscience J F, Graf T. Chicken hematopoietic cells transformed by seven strains of defective avian leukemia viruses display three distinct phenotypes of differentiation. Cell. 1979;18:375–382. doi: 10.1016/0092-8674(79)90057-6. [DOI] [PubMed] [Google Scholar]
- 7.Clouthier S C, Muller K H, Doran J L, Collinson S K, Kay W W. Characterization of three fimbrial genes of sefABC, of Salmonella enteritidis. J Bacteriol. 1993;175:2523–2533. doi: 10.1128/jb.175.9.2523-2533.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Collinson S K, Emody L, Muler K H, Trust T J, Kay W W. Purification and characterization of thin, aggregative fimbriae from Salmonella enteritidis. J Bacteriol. 1991;173:4773–4781. doi: 10.1128/jb.173.15.4773-4781.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Collinson S K, Doig P C, Doran J L, Clouthier S, Trust T J, Kay W W. Thin aggregative fimbriae mediate binding of Salmonella enteritidis to fibronectin. J Bacteriol. 1993;175:12–18. doi: 10.1128/jb.175.1.12-18.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Collinson S K, Clouthier S C, Doran J L, Banser P A, Kay W W. Salmonella enteritidis agfBAC operon encoding thin, aggregative fimbriae. J Bacteriol. 1996;178:662–667. doi: 10.1128/jb.178.3.662-667.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cooper G L, Nicholas R A J, Cullen G A, Hormaeche C E. Vaccination of chickens with a Salmonella enteritidis aroA− live oral Salmonella vaccine. Microb Pathog. 1990;9:255–265. doi: 10.1016/0882-4010(90)90014-h. [DOI] [PubMed] [Google Scholar]
- 12.Dibb-Fuller M, Allen-Vercoe E, Thorns C J, Woodward M J. Fimbriae- and flagella-mediated association with and invasion of cultured epithelial cells by Salmonella enteritidis. Microbiology (New York) 1999;145:1023–1031. doi: 10.1099/13500872-145-5-1023. [DOI] [PubMed] [Google Scholar]
- 13.Finlay B B, Falkow S. Common themes in microbial pathogenicity. Microbiol Rev. 1997;61:136–169. doi: 10.1128/mmbr.61.2.136-169.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hohmann A W, Schmidt G, Rowley D. Intestinal colonization and virulence of Salmonella in mice. Infect Immun. 1978;22:763–770. doi: 10.1128/iai.22.3.763-770.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jones G W, Richardson L A. The attachment to and invasion of HeLa cells by Salmonella typhimurium: the contribution of mannose-sensitive and mannose-resistant hemagglutinating activities. J Gen Microbiol. 1981;127:361–370. doi: 10.1099/00221287-127-2-361. [DOI] [PubMed] [Google Scholar]
- 16.Jordan Lin C T, Morales R A, Ralston K. Raw and undercooked eggs: a danger of salmonellosis. Food Rev. 1997;20:27–32. [Google Scholar]
- 17.Keith B R, Harris S L, Walker P R, Orndorff P E. Effect of type 1 piliation on in vitro killing of Escherichia coli by mouse peritoneal macrophages. Infect Immun. 1990;58:3448–3454. doi: 10.1128/iai.58.10.3448-3454.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kinde H, Read D H, Ardans A, Little H, Keri D, Gireesh R, Nagaraja K V. Sewage effluent: source of Salmonella enteritidis phage type 4 infection and disease in a commercial chicken layer flock in Southern California. Avian Dis. 1996;40:672–676. [PubMed] [Google Scholar]
- 19.Koneman E W, Allen S D, Janda W M, Schreckenberger P C, Winn W C. Color atlas and textbook of diagnostic microbiology. 5th ed. Philadelphia, Pa: Lippincott; 1997. [Google Scholar]
- 20.Krogfelt K A. Bacterial adhesion: genetics, biogenesis and role in pathogenesis of fimbrial adhesion of E. coli. Rev Infect Dis. 1991;13:721–735. doi: 10.1093/clinids/13.4.721. [DOI] [PubMed] [Google Scholar]
- 21.Kukkonen M, Raunio T, Virkola R, Lahteenmaki K, Makela P H, Klemm P, Clegg S, Korhonen T K. Basement membrane carbohydrate as a target for bacterial adhesion: binding of type 1 fimbriae of Salmonella enterica and Escherichia coli to laminin. Mol Microbiol. 1993;7:229–237. doi: 10.1111/j.1365-2958.1993.tb01114.x. [DOI] [PubMed] [Google Scholar]
- 22.Muller K H, Collinson S K, Trust T J, Kay W W. Type 1 fimbriae of Salmonella enteritidis. J Bacteriol. 1991;173:4765–4772. doi: 10.1128/jb.173.15.4765-4772.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ngeleka M, Marineau-Doize B, Fairbrother J M. Septicemia-inducing Escherichia coli O115:K′V165′F1651 resists killing by porcine polymorphonuclear leukocytes in vitro: role of F1651 fimbriae and K′V165′ O-antigen capsule. Infect Immun. 1994;62:398–404. doi: 10.1128/iai.62.2.398-404.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ogunniyi A D, Manning P A, Kotlarski I. A Salmonella enteritidis 11RX pilin induces strong T-lymphocyte responses. Infect Immun. 1994;62:5376–5383. doi: 10.1128/iai.62.12.5376-5383.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ogunniyi A D, Kotlarski I, Morona R, Manning P A. Role of SefA subunit protein of SEF14 fimbriae in the pathogenesis of Salmonella enterica serovar enteritidis. Infect Immun. 1997;65:708–717. doi: 10.1128/iai.65.2.708-717.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pertala R C, Yokoyama H, Ikemori Y, Kuroki M, Kodama Y. Passive immunization against experimental salmonellosis in mice by orally administered hen egg yolk antibodies specific for 14kDa fimbriae of Salmonella enteritidis. J Med Microbiol. 1994;41:29–35. doi: 10.1099/00222615-41-1-29. [DOI] [PubMed] [Google Scholar]
- 27.Qureshi M A, Miller L, Lillehoj H S, Ficken M D. Establishment and characterization of a chicken mononuclear cell line. Vet Immunol Immunopathol. 1990;26:237–250. doi: 10.1016/0165-2427(90)90094-9. [DOI] [PubMed] [Google Scholar]
- 28.Thorns C J, Turcotte C, Gemmell C G, Woodward M J. Studies into the role of the SEF14 fimbrial antigen in the pathogenesis of Salmonella enteritidis. Microb Pathog. 1996;20:235–246. doi: 10.1006/mpat.1996.0022. [DOI] [PubMed] [Google Scholar]
- 29.Van der Velden A W M, Baumler A J, Tsolis R M, Heffron F. Multiple fimbrial adhesins are required for full virulence of Salmonella typhimurium in mice. Infect Immun. 1998;66:2803–2808. doi: 10.1128/iai.66.6.2803-2808.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Verhoef J, Peterson P K, Quie P G. Kinetics of staphylococcal opsonization, attachment, ingestion and killing by human polymorphonuclear leukocytes: a quantitative assay using [3H] thymidine labeled bacteria. J Immunol Methods. 1977;14:303–311. doi: 10.1016/0022-1759(77)90141-7. [DOI] [PubMed] [Google Scholar]
- 31.Wells C L, Elisabeth M A, De Westerlo V, Jechorek R P, Feltis B A, Wilkins T D, Erlandsen S L. Bacteroides fragilis enterotoxin modulates permeability and bacterial internalization by HT-29 enterocytes. Gastroenterology. 1996;110:1429–1437. doi: 10.1053/gast.1996.v110.pm8613048. [DOI] [PubMed] [Google Scholar]