Abstract
Oxytocin plays a vital role in social behavior and homeostatic processes, with animal models indicating that oxytocin receptor (OXTR) expression patterns in the brain influence behavior and physiology. However, the developmental trajectory of OXTR gene expression is unclear. By analyzing gene expression data in human post-mortem brain samples, from the prenatal period to late adulthood, we demonstrate distinct patterns of OXTR gene expression in the developing brain, with increasing OXTR expression along the course of the prenatal period culminating in a peak during early childhood. This early life OXTR expression peak pattern appears slightly earlier in a comparative macaque sample, which is consistent with the relative immaturity of the human brain during early life compared to macaques. We also show that a network of genes with strong spatiotemporal couplings with OXTR is enriched in several psychiatric illness and body composition phenotypes. Taken together, these results demonstrate that oxytocin signaling plays an important role in a diverse set of psychological and somatic processes across the lifespan.
Subject terms: Genetics of the nervous system, Gene expression
Introduction
Oxytocin is an evolutionarily ancient neuromodulator that facilitates mammalian social behavior and metabolic regulatory processes [1, 2]. Oxytocin is primarily produced in the hypothalamus, for both central and peripheral release [3], and binds to oxytocin receptors that are located throughout the brain and the periphery [1, 2]. Both the experimental manipulation of oxytocin receptor (OXTR) expression levels and OXTR gene knockout can have a dramatic effect on behavior and physiology [4, 5]. Thus, the identification of OXTR expression patterns in the human brain can help identify the functional relevance of the oxytocin signaling system. We recently mapped the voxel-wise density of OXTR expression across the adult human brain using donor tissue data, finding increased expression in subcortical and olfactory regions [6]. Moreover, the location of OXTR expression was highly correlated with dopaminergic gene expression and closely matched the neural activity patterns observed in anticipatory, appetitive, and aversive mental states, along with homeostatic regulation. Compared to neurotypical donors, OXTR expression in the dorsolateral prefrontal cortex is also increased in donors diagnosed with mood disorders [7] and reduced in the temporal cortex of donors diagnosed with schizophrenia [8].
While the OXTR gene expression pattern in the adult human brain has been described [6], little is known about both the functional relevance and evolution of OXTR gene expression and OXTR gene co-expression patterns across the lifespan. This is a critical knowledge gap, as genes are differentially regulated across the brain over development [9], disturbances in neural development contribute to the genesis of mental illness [10], and identifying the evolutionary history of a signaling system can help uncover its purpose [2]. Supporting the clinical relevance of spatially and temporally distinct aberrations, neurotypical postmortem brain tissue from the ventral pallidum (VP) and nucleus basalis of Maynert has been reported to have higher OXTR binding than postmortem VP tissue from autistic donors, and an early childhood peak of OXTR binding that was observed in VP tissue from neurotypical donors was absent in VP tissue from autistic donors [11].
Oxytocin signaling emerges very early in the course of mammalian development. Oxytocin has been detected prenatally during the neurogenesis period of fetal brain development [12] and magnocellular oxytocin neurons only mature a few weeks postnatally [13]. Oxytocin signaling dysregulation of receptor expression during early development also plays an important role in behavior later in life [14]. For example, oxytocin concentrations in cerebrospinal fluid (CSF) are significantly higher in mother-reared rhesus monkeys compared to nursery-reared monkeys [15] and increased maternal behaviors are associated with higher central OXTR expression levels in rat pups [16]. In humans, women with a history of child abuse also demonstrate reduced CSF levels of oxytocin [17], highlighting how the early environment can influence later oxytocin system functioning.
Psychiatric and somatic illnesses tend to follow specific developmental trajectories. Thus, the identification of a critical window of oxytocin receptor expression in the brain, together with associated genes, may illuminate our understanding of psychiatric illnesses. Research has identified patterns of increased OXTR expression in non-human mammals during either infancy, childhood, or adulthood, compared to other lifespan periods [18]. However, it is not known which of these infancy, childhood, or adulthood patterns, if any, are observed in humans and the functional significance of observed patterns. In addition, some aspects of oxytocin’s role in behavior are sexually dimorphic, but it is unclear whether sex differences in gene expression patterns are related to these effects. Therefore, there is a need to characterize typical oxytocin pathway system development, to determine whether expression patterns are stable over development, and to identify critical periods.
Here we identify the spatiotemporal distribution patterns of OXTR expression and gene expression interactions in humans, from the prenatal period to late adulthood, revealing that OXTR expression peaks in early childhood and late adulthood, and is highly correlated with dopaminergic signaling across the lifetime. We also examine the evolutionary conservation of OXTR expression patterns by replicating our first analysis using macaque OXTR expression data, which indicated that the human early life OXTR expression peak was observed temporally earlier in macaques, which is consistent with the relative immaturity of the neonate human brain compared to the neonate macaque. We extend this analysis by identifying the evolutionary origin of genes with strong spatiotemporal associations with OXTR expression, revealing that a number of these genes have undergone positive selection in humans, but not macaques. Moreover, exploring the functional significance of genes with a strong spatiotemporal association with the oxytocin receptor gene revealed enrichment of this geneset in various GWAS of psychiatric illnesses, cognitive processes, and somatic phenotypes. Collectively, these results provide novel insights into oxytocin signaling and its role in wellbeing.
Materials and methods
Spatiotemporal expression of the oxytocin pathway in humans
To better understand the functional relevance of spatiotemporal gene co-expression patterns across the lifespan and relevance to common human traits and diseases, we determined OXTR expression patterns, along with all available protein coding genes, across sixteen regions of the human brain from the prenatal stage (5.7 pre-conception weeks) to 82 years of age (i.e., spatiotemporally) in 57 donors (26 females, 31 males). Specimens were collected from clinically unremarkable donors without evidence of neurological or psychiatric illness (see Supplementary Note 1 for donor characteristics). Tissue specimens were collected from several institutions under approval of their respective institutional review boards [9]: Yale University, National Institute of Mental Health, Albert Einstein College of Medicine, University of Maryland, University of Washington, and University of Newcastle, UK. We used genome-wide exon-level transcriptome data available from the Gene Expression Omnibus database (https://www.ncbi.nlm.nih.gov/geo; series GSE25219) as a proxy of OXTR receptor density [19]. Gene expression data were converted from months and years into days for analyses.
Gene expression values for sixteen brain regions were visualized across the lifespan using two approaches. The first approach illustrates absolute change in gene expression patterns across the lifespan for sixteen brain regions: primary motor cortex (M1C), dorsolateral prefrontal cortex (DFC), ventrolateral prefrontal cortex (VFC), orbital frontal cortex (OFC), primary somatosensory cortex (S1C), inferior parietal cortex (IPC), primary auditory cortex (A1C), caudal superior temporal cortex (STC), inferolateral temporal cortex (ITC), primary visual cortex (V1C), medial prefrontal cortex (MFC), hippocampus (HIP), striatum (STR), amygdala (AMY), mediodorsal nucleus of the thalamus (MDT), and cerebellar cortex (CBC). Expression values were smoothed via a logarithmic scale using locally weighted least squares regression, then demeaned and scaled (i.e., divided by the standard deviation), yielding a dataset with expression values converted to Z-scores per brain region of interest. Thus, the mean expression of a given brain region across the lifespan is zero. In these “ribbon plots”, Z-values are stacked on top of each other and scaled again by number of regions, so when a single region (or group of regions) demonstrates increased expression relative to other life periods, the peaks will be higher. Our second visualization approach provides complementary information via a “heat plot”, with the key difference of age normalized expression values. This provides a stronger emphasis on expression changes in specific brain regions, rather than overall expression across the brain. For example, a value of 0.25 in the cerebellum during adolescence means that at this developmental stage 25% of gene expression occurs in the cerebellum compared to 75% in the remaining 15 regions at the same age period. Expression levels are illustrated by colors instead of ribbon height.
We also performed an out-of-sample validation of these gene expression patterns using data from the Allen human brain atlas (AHBA) [6]. While the AHBA has high spatial resolution, these samples are limited to adults, with most data available from the left hemisphere. Thus, to facilitate a more accurate comparison, we created a subset of the present lifespan gene expression data that was limited to donors above the age of twenty and from the left hemisphere. The number of donors used in this analysis varied per brain region, ranging from 12 to 16 (see Supplementary Note 2 for details). With this data, we calculated the mean expression for each of the 16 brain regions across the subset, for comparison with previously described research using the AHBA.
Lifespan correlation between genes
We have previously shown using a voxel-wise approach in the adult brain that OXTR expression is highly correlated with the expression of a selection of oxytocinergic, dopaminergic, muscarinic acetylcholine, and opioid pathway genes [6]. Thus, we assessed whether this same geneset also had a strong spatiotemporal relationship with OXTR (i.e., both across the brain and across the lifespan). To calculate the lifespan correlation for two given genes, we first calculated Spearman’s r correlation coefficient within each donor. Then, we interpolated calculated points on a logarithmic scale using locally weighted least squares regression. To evaluate the lifespan average, we calculated the mean of interpolated line, which we labeled the trajectory mean. By using this approach, each life period receives an equal weighting. To complement this measure, we also presented the arithmetic mean of all points, so that each donor was weighted equally. To assess the specificity of the correlation between two selected genes, we also plotted the distribution of the correlation between a gene of interest and bulk of all remaining genes (16659 for humans and 19049 for macaques), marking the location of the correlated genes of interest on the distribution.
Using correlation data between genes, we generated a correlation matrix reflecting the spatiotemporal Spearman’s correlation for each donor for each gene expression map pair for 19 genes. In our correlation matrix, we presented correlations derived from the two different approaches that we described above: the trajectory mean and the arithmetic mean. The trajectory mean correlations were shown in the lower-left triangle of the correlation matrix, and the arithmetic mean correlations were shown in the upper-right triangle of the correlation matrix. As per our previous analysis [6], we used the following genes (Oxytocin pathway set: OXTR, CD38, OXT; Dopaminergic set: DRD1, DRD2, DRD3, DRD4, DRD5, COMT, and DAT1; muscarinic acetylcholine set: CRHM1, CRHM2, CRHM3, CRHM4, and CRHM5; opioid set: OPRM1, OPRD1, OPRK1, and AVPR1A, which encodes the vasopressin receptor 1 subtype). We used Ward’s hierarchical clustering to identify highly correlated groups of genes within this geneset.
The evolutionary origin of genes spatiotemporally associated with OXTR
To explore the evolutionary conservation of spatiotemporal patterns of oxytocin pathway expression across human development, rhesus macaque (macaca mulatta) gene expression data from post-mortem brain tissue was extracted from the National Institute of Health Blueprint Non-human primate (NIH Blueprint NHP) atlas (http://www.blueprintnhpatlas.org/). This atlas provides data for central gene expression across six prenatal stages in males and females (40, 50, 70, 80, 90, and 120 embryonic days) and four postnatal periods in males only (0, 3, 12, and 48 months), for which the human corresponding lifespan periods are birth, infant, childhood, and early adolescence [20]. We assumed pregnancy to last 40 weeks for humans and 166.5 embryonic days for macaques. Spatiotemporal OXTR expression patterns in macaques were generated using the same methods as described above as humans (i.e., a ribbon plot to best illustrate change across time and heat plot to best illustrate changes in specific brain regions). Equivalent human brain development ages, derived from the Translating Time resource [https://www.translatingtime.org]; [21, 22], were also included in these plots. Gene expression data from twenty brain regions that were available throughout the majority of the lifespan were used.
While comparing gene expression between humans and macaques is instructive, this only represents a comparison within a relatively short evolutionary period for two species. Thus, we used phylostratigraphy, which is a tool that can help pinpoint the origin of genes using genomic data from across the evolutionary tree [23]. Specifically, phylostratigraphy can identify the macroevolutionary period (i.e., phylogenetic stage) during which a gene’s first ancestor emerged. As gene modules with high co-expression in the brain are related to molecular functions [24], we created a geneset containing the genes with the 100 strongest spatiotemporal correlations with OXTR. For our phylostratigraphy analysis, a phylostratigraphic map with gene age inferences from Domazet-Lošo and Tautz [25] was combined with human gene expression data from five ontogenetic stages (prenatal, infant, child, adolescent, adult) extracted from the BrainSpan atlas (http://brainspan.org) for evolutionary transcriptomics via the “myTAI” R package [26]. For each brain region, a transcriptional age index (TAI) was calculated for each ontogenetic stage by calculating the average age of genes that contribute to the transcriptome [23]. To compute the TAI, the expression level for each gene is multiplied by its phylogenetic stage (i.e., gene age), and then the values for each gene in a geneset are averaged. As older phylostrata are associated with the genesis of a higher number of genes, the TAI provides more weight to genes from younger phylostrata. Altogether, lower values represent an older transcriptome age.
The evolution of protein coding genes in humans and macaques was assessed by comparing their genetic sequences to their common primate ancestor via the GenEvo tool [27]. From this data, the numbers of non-synonymous changes per non-synonymous sites (dN) and synonymous changes per synonymous sites (dS) were computed to calculate the dN/dS ratio to estimate gene conservation, with values closer to zero indicative of strong genetic conservation. Welch’s t-test was used to compare the dN/dS ratios (i.e., gene conservation) of the macaque and human genesets.
The functional significance of genes spatiotemporally associated with OXTR
To explore the functional significance of spatiotemporal expression patterns of genes strongly coupled with OXTR, a geneset with the top 100 genes with the strongest spatiotemporal correlations with OXTR was submitted to FUMA for annotation of genetic associations [28]. Hypergeometric tests (Benjamini-Hochberg adjusted) were performed to examine if this geneset was overrepresented in GWAS from the National Human Genome Research Institute-European Bioinformatics Institute (NHGRI-EBI) GWAS catalog (e96 2019-09-24; [29]) and biological processes from the Molecular Signatures database (MsigDB v7.0; [30]). We also calculated the top 20 correlated and anti-correlated genes between OXTR and the 16659 remaining genes across the lifespan, ordered by the lifespan trajectory mean (details provided above).
To assess if there was a significant difference in the association between the spatiotemporal expression of OXTR and genes that have been associated with psychiatric and physiological phenotypes of interest, we compared the distribution of correlation coefficients between phenotype genesets of interest (i.e., schizophrenia, major depressive disorder, IQ, general cognition, bone fracture, bone density, body mass index, bipolar disorder, autism spectrum disorder, and anorexia nervosa) against all remaining genes. To retrieve genesets associated with phenotypes of interest, we performed genome-wide gene-based association using MAGMA (v1.08) and functional mapping of variants to genes based on expression quantitative trait loci (eQTL) via FUMA on the complete GWAS input data available from public resources (Supplementary Table 1). All variants in the GWAS outside of the MHC region (chr6: 28,477,797-33,448,354) were included to estimate the significance value of that gene. The eQTL approach maps SNPs to genes that are likely to affect the expression of those genes up to 1 megabase away from the SNP of interest. To determine gene expression and assess eQTL functionality of likely regulatory SNPs, we used data from the eQTL Catalog [31], PsychENCODE [32], the xQTLServer [33], the CommonMind Consortium [34], GTEx v8 [35], and the Braineac eQTLs dataset [36]. MAGMA performs multiple linear regression to obtain gene-based p-values and the Bonferroni-corrected significant thresholds for each phenotype is listed in Supplementary Table 2.
After retrieving genes, we computed Spearman’s correlation coefficients to estimate the spatiotemporal relationship between OXTR and 16661 available genes. We then created a distribution for these correlation coefficients. Finally, genes were split into two groups, ones belonging to phenotype of interest and the remaining genes, and superimposed for comparison. We used non-parametric Mann–Whitney U tests to test if distributions had different medians. We adjusted reported p-values to the total number of brain phenotypes using a false discovery rate (FDR) threshold.
Donor-to-donor reproducibility of gene expression patterns
Differential stability measures were calculated, which index how closely related gene expression patterns are from donor-to-donor. If a gene has strong differential stability, this indicates that the gene’s expression patterns can be better generalized beyond the sample. Moreover, genes with strong differential stability tend to also have strong biological relevance and are associated with known pharmacological treatment targets [37]. To calculate spatiotemporal differential stability, each of the 57 human donors were matched to their three closest neighbors by age, with duplicates and matches with less than four regions in common removed, yielding a list with 93 matches. Each donor was matched 3.2 times, on average. We iterated through all the matched pairs, performing the following tasks for each protein coding gene in the database: (1) Extraction of gene expression for all available brain regions, (2) Calculation of Spearman’s correlation coefficient, (3) Calculation of the mean Spearman’s correlation coefficient.
Results
Spatiotemporal oxytocin receptor expression in humans
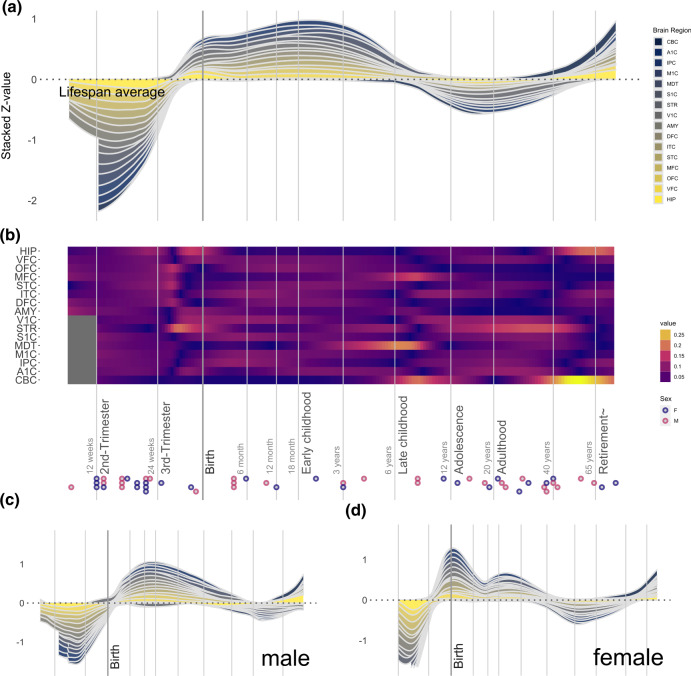
Our analysis revealed that OXTR expression across the brain in humans begins to accelerate just before birth, with a peak level of expression occurring during early childhood (Fig. 1a), consistent with previously reported OXTR receptor binding temporal patterns from the ventral pallidum [11]. There is also a general increase in OXTR expression, compared to the lifespan average, during late adulthood. Regional analyses demonstrated increased OXTR expression in the striatum during the third trimester and during a second period throughout adolescence and adulthood. There was also increased OXTR expression in the mediodorsal nucleus of the thalamus during early childhood (Fig. 1b) and in the cerebellar cortex and medial prefrontal cortex in later childhood. During adulthood, we observed increased OXTR expression in the striatum, cerebellar cortex, and hippocampus. Males exhibited a stronger early childhood peak in OXTR expression and more pronounced differentiation within brain regions, which reflected the trends described above (Fig. 1c, d; see Supplementary Fig. 1 for greater detail). In females, the strongest peak of OXTR expression was observed around birth, with a smaller childhood peak, and a dip during adolescence. The region with the most apparent increase in expression in females was the cerebellar cortex during adulthood. Mean OXTR expression patterns across these 16 regions from the adult donors in the present sample were comparable with OXTR expression patterns derived from the AHBA [6] sample of adult donors (Supplementary Note 1), which suggests that these gene expression patterns are generalizable.
Fig. 1. OXTR gene expression in sixteen regions of the human brain across different developmental phases.
a The ribbon plot illustrates normalized gene expression compared to the lifespan average, with expression of individual brain regions stacked (see methods for a key to all brain region abbreviations). The stacked Z-value is scaled to the number of regions (sixteen). b The heat plot illustrates gene expression across sixteen brain regions, normalized for each brain region across the lifespan. Each individual donor is represented at the bottom of the panel as a circle, with circle color representing donor sex (see legend). Time is presented using a log10 scale in both panels. Also presented are ribbon plots with data from only males (c) and females (d), with each panel illustrating normalized gene expression compared to the lifespan average.
We observed an increase of CD38 expression across the brain during the third trimester, which peaked around the first year of life (Supplementary Fig. 2). The regions with the largest increases of CD38 expression during this period was the cerebellar cortex and the striatum. There was also an increase in CD38 expression beginning in adolescence and peaking in adulthood in the cerebellar cortex, amygdala, and hippocampus. The strongest expression of OXT was observed during the prenatal stage, with a smaller peak around adolescence (Supplementary Fig. 3). There was also noticeable differentiation between brain regions, with the highest expression observed in the medial prefrontal cortex during the third trimester, the amygdala during adolescence, and the inferior parietal cortex during adulthood.
Lifespan correlation between genes
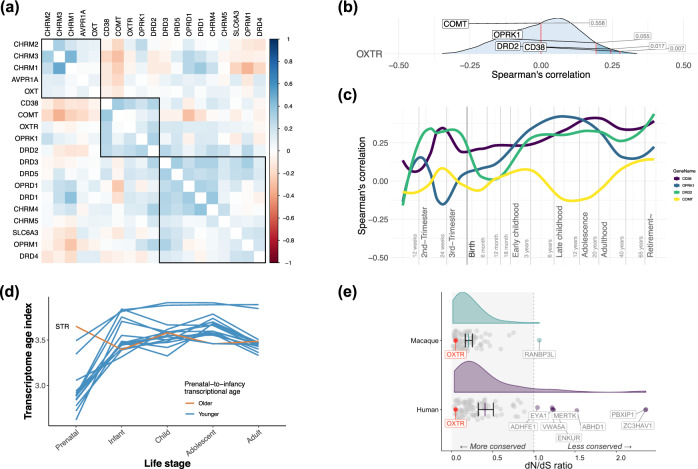
Hierarchical clustering identified a set of genes with particularly strong correlations for the spatiotemporal expression pattern of OXTR, which included Catechol-O-Methyltransferase (COMT), the CD38 molecule (CD38), opioid receptor kappa 1 (OPRK1), and the dopamine receptor (DRD2) (Fig. 2a). Figure 2b shows the distribution of the spatiotemporal correlations between OXTR and all remaining genes, marking the location of these four genes clustered with OXTR in Fig. 1a. This analysis revealed that DRD2 and CD38 were among the top 1.7% and top 0.7% of all correlated genes across the lifespan, respectively (total number of genes = 16,660). This high specificity was also observed when examining males and females separately (Supplementary Fig. 4). To complement lifespan correlation averages, which might disguise large differences between life periods, we additionally visualized how the correlation between OXTR these four genes of interest fluctuated across the lifespan. This revealed that the correlation between OXTR and DRD2 was relatively stable from early childhood onwards (Fig. 2c).
Fig. 2. OXTR spatiotemporal co-expression and the evolution of OXTR co-expression modules.
a The average spatiotemporal correlation between OXTR and a selection of oxytocinergic, dopaminergic, muscarinic acetylcholine, and opioid pathway genes. OXTR was part of a five-gene spatiotemporal co-expression module with DRD2, COMT, OPRK1, and CD38. b The percentage rank of the average spatiotemporal relationship between OXTR and DRD2, COMT, OPRK1, and CD38, compared with to the spatiotemporal relationship between OXTR and all protein coding genes (n = 16,659). Both DRD2 and CD38 were among with the top 5% of correlations (marked in dark blue). c The lifespan stability of the spatiotemporal correlation between OXTR and DRD2, COMT, OPRK1, CD38. d The transcriptional age of a gene co-expression module comprising OXTR and the 100 genes with the strongest spatiotemporal co-expression with OXTR across the lifespan in sixteen brain regions (see methods), with a smaller transcriptional age index representing an older transcriptome. Analysis suggested an older transcriptional age in infancy compared to the prenatal stage across the brain, except for the striatum (STR). e The top 100 gene module was more highly conserved in the macaque genome compared to the human genome (p = 0.0001), however, in both species OXTR (marked in red) is highly conserved. Several genes from this geneset had undergone positive selection in humans (i.e., dN/dS ratio values > 1).
The evolutionary origin of OXTR and spatiotemporally related genes
As we observed in our human data analysis, a late-prenatal acceleration of OXTR expression was observed in macaques, however, there was no evidence of an early childhood peak (Supplementary Fig. 5), which was observed in the human data. This prenatal peak was also observed when only including male donor samples (Supplementary Fig. 6). This temporal difference may reflect the immaturity of human neonates compared to macaques, as macaque brain development is faster than humans during the prenatal and postnatal period [20–22]. DRD2 was included in a cluster of genes with high spatiotemporal correlations with OXTR, but the relationship between OXTR and DRD2 was only in the top 81.7% of correlations of OXTR with all other protein coding genes (Supplementary Fig. 7), which indicates weak specificity in this dataset. Similar results were observed when only analyzing male macaque donor data (Supplementary Fig. 8).
A phylostratigraphic analysis revealed that most genes in a module containing genes with the 100 strongest spatiotemporal correlations with OXTR are evolutionary ancient, appearing among the first three phylostrata (Supplementary Fig. 9). According to this analysis, the ancestor of OXTR first emerged in the Eumetazoa phylostratum, about 600 million years ago. A transcriptional age index (TAI) was calculated for sixteen brain regions across five ontogenetic stages to better understand the role of older vs. newer genes from this geneset module in brain development. The TAI is derived by computing the average age of genes that contribute to a transcriptome [23], whereby lower TAI values represent an older transcriptome. For all brain regions except the striatum (STR), the transcriptome of the OXTR top 100 geneset was older during the prenatal stage, compared to later stages (Fig. 2d). This suggests that for most brain regions we investigated, genes in their transcriptome that are highly expressed from infancy onward evolved at a faster rate compared to genes that are highly expressed prenatally [38].
Next, we calculated the dN/dS ratio to assess whether genes in the top 100 geneset had experienced positive selection (dN/dS > 1), negative selection (dN/dS < 1), or if they have been evolving neutrally (dN/dS ~ 1), in both the human and macaque genome [27]. While OXTR is highly conserved in both humans and macaques, the total geneset on average had a significantly higher dN/dS ratio in humans, compared to macaques [t = 3.96 (123.3), p = 0.0001, d = 0.6; Fig. 2e]. This analysis also revealed that several genes show positive selection in humans, but not macaques, in which they were under selective constraint (ADHFE1, EYA1, MERTK, VWA5A, ENKUR, ABHD1, PBXIP1, and ZC3HAV1; See Supplementary Note 3 for full gene names). These genes have been associated with metabolism (ADHFE1) [39], bone density (EYA1, MERTK) [40, 41], brain volume (ENKUR) [42], intelligence (VWA5A) [43, 44], and body fat distribution (PBXIP1, ZC3HAV1) [45, 46].
The functional significance of spatiotemporally expression pattern
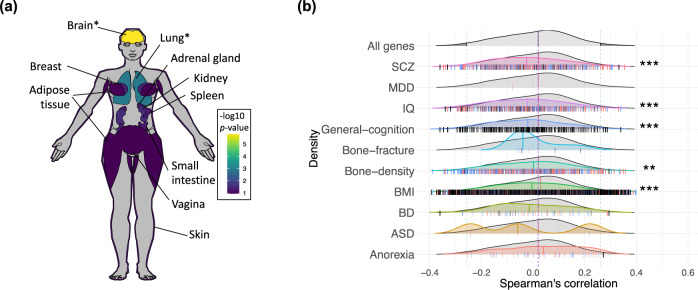
Annotating the associations of a geneset including 100 genes with the strongest spatiotemporal associations with OXTR revealed enrichment in GWAS-derived genes for bone fracture in osteoporosis (p = 8.293 × 10−3). In addition, there was enrichment with genes associated with age-related macular degeneration GWAS (p = 8.293 × 10−3), as well as gene ontology genesets associated with reproduction (p = 5.83 × 10−3) and penile erection (p = 5.83 × 10−3). We also examined the enrichment of this geneset in thirty tissue types across the body using the GTEx database (version 8; [35]) in FUMA, discovering up-regulated differentially expressed genes in brain and lung tissue (p < 0.05, Bonferroni corrected; Fig. 3a; Supplementary Fig. 10). The twenty genes showing the strongest spatiotemporal correlations with OXTR gene expression are presented in Table 1. The most positively correlated genes have been associated with type 2 diabetes (FGFR3) [47], obesity (INHBB) [48], bone density (DIRAS3) [49], and body morphology (KCNN3) [45]. The strongest negatively correlated genes have been associated with intelligence (DEPDC5, MORC2) [44, 50], body morphology (MAP3K12, KLC2) [51, 52], and cholesterol levels (CTCF) [53]. Notably, some of the highest correlated genes in this spatiotemporal analysis (e.g., NTSR2, GLUD1, SRPK1, MTCL1, KCNJ3, PAK7, THBS4, HEYL, ZC3HAV1) were also among the highest correlated in a previous analysis of OXTR expression in human adults using the AHBA gene expression dataset [6].
Fig. 3. Networks of genes with strong spatiotemporal couplings with OXTR are enriched in brain and lung tissue, body composition phenotypes, and psychiatric illnesses.
a Top ten highest levels of differential expression in human tissue types of a geneset containing the top 100 genes correlated with OXTR. The -log10 p-values for upregulation of this geneset in various tissue types represent the probability of the hypergeometric tests. See Supplementary Fig. 7 for a list of results with 30 tissue types. *p < 0.05 (Bonferroni corrected). b Comparisons between the correlation of OXTR with genes related to disorders based on eQTL mapping and phenotypes and the correlation of OXTR with remaining genes. Vertical dashes in each distribution represent correlation between genes involved in each phenotype and OXTR, and the direction of the effect. Blue notches indicate risk increasing alleles which increase the expression of the gene, red notches indicate when risk increasing alleles decrease the expression of the gene, and black notches indicate when direction is unknown, under their respective distributions. ***p < 0.001 (FDR corrected).
Table 1.
Genes with the strongest positive and negative spatiotemporal correlations with OXTR.
| Rank | Top correlated | Top anti-correlated | ||||
|---|---|---|---|---|---|---|
| Gene | Trajectory | Arithmetic | Gene | Trajectory | Arithmetic | |
| 1 | FGFR3 | 0.368 | 0.365 | DEPDC5 | −0.381 | −0.365 |
| 2 | GRAMD1C | 0.358 | 0.363 | CACNA1A | −0.368 | −0.382 |
| 3 | C1orf88 | 0.356 | 0.358 | KLC2 | −0.363 | −0.373 |
| 4 | NTSR2 | 0.35 | 0.318 | KCNC3 | −0.363 | −0.313 |
| 5 | INHBB | 0.348 | 0.328 | C2CD3 | −0.361 | −0.319 |
| 6 | CXorf59 | 0.343 | 0.348 | HDAC8 | −0.359 | −0.345 |
| 7 | KCNN3 | 0.341 | 0.336 | CTCF | −0.349 | −0.3 |
| 8 | DIRAS3 | 0.34 | 0.286 | MAP3K12 | −0.347 | −0.356 |
| 9 | STON2 | 0.338 | 0.345 | MICAL2 | −0.345 | −0.313 |
| 10 | RDH10 | 0.335 | 0.338 | APBA2 | −0.345 | −0.285 |
| 11 | AGXT2L1 | 0.328 | 0.301 | MORC2 | −0.343 | −0.334 |
| 12 | SIL1 | 0.327 | 0.293 | NOVA1 | −0.341 | −0.318 |
| 13 | HRSP12 | 0.326 | 0.316 | NFIX | −0.34 | −0.321 |
| 14 | GLUD1 | 0.323 | 0.309 | KCNJ3 | −0.338 | −0.333 |
| 15 | GNG12 | 0.323 | 0.32 | KDM5C | −0.337 | −0.345 |
| 16 | PBXIP1 | 0.321 | 0.304 | SEMA4C | −0.333 | −0.315 |
| 17 | RUNX3 | 0.319 | 0.289 | EPS15L1 | −0.332 | −0.33 |
| 18 | F3 | 0.317 | 0.312 | TMEM25 | −0.33 | −0.325 |
| 19 | ACSBG1 | 0.317 | 0.303 | DHX8 | −0.33 | −0.307 |
| 20 | GJA1 | 0.316 | 0.301 | ADAM11 | −0.329 | −0.275 |
Two approaches for this analysis are presented. “Trajectory” represents a mean correlation across the lifespan trajectory, in which each life period receives an equal weighting. “Arithmetic” represents the mean of individual correlations between donors, in which each donor is weighted equally.
To assess the relationship between the spatiotemporal expression of OXTR and genes that have been associated with psychological and physiological phenotypes of interest, we extracted phenotype genesets by using expression quantitative trait loci (eQTL) mapping of GWAS data (Supplementary Table 1). We selected a range of psychiatric and psychological phenotypes that have been previously associated with oxytocin signaling dysfunction [i.e., schizophrenia (SCZ), major depressive disorder (MDD), IQ, general cognition, bipolar disorder (BD), autism spectrum disorder (ASD), and anorexia nervosa], and a set of physiological phenotypes based on results from analysis above (i.e., bone fracture, bone density, and BMI). To determine specificity, we compared these results with the relationship between spatiotemporal expression of OXTR and genes not included in the phenotype geneset of interest. The correlations with the genesets of interest and the background genes were both plotted as density distributions and non-parametric Mann–Whitney U tests were performed to compare the distributions. There was a significantly lower density median for the relationship of OXTR with genesets enriched in schizophrenia, IQ, general cognition, bone density, and BMI, compared to the median of the density distribution with background genes, adjusting tests via a false discovery rate threshold (p < 0.001; Fig. 3b; Supplementary Table 3). Genes related to phenotypes falling within top or bottom 2.5% of OXTR correlation’s distribution are provided in Supplementary Tables 4 and 5. Similar results were found using a MAGMA analysis (Supplementary Fig 11; Supplementary Tables 6, 7, and 8).
Donor-to-donor reproducibility of gene expression patterns
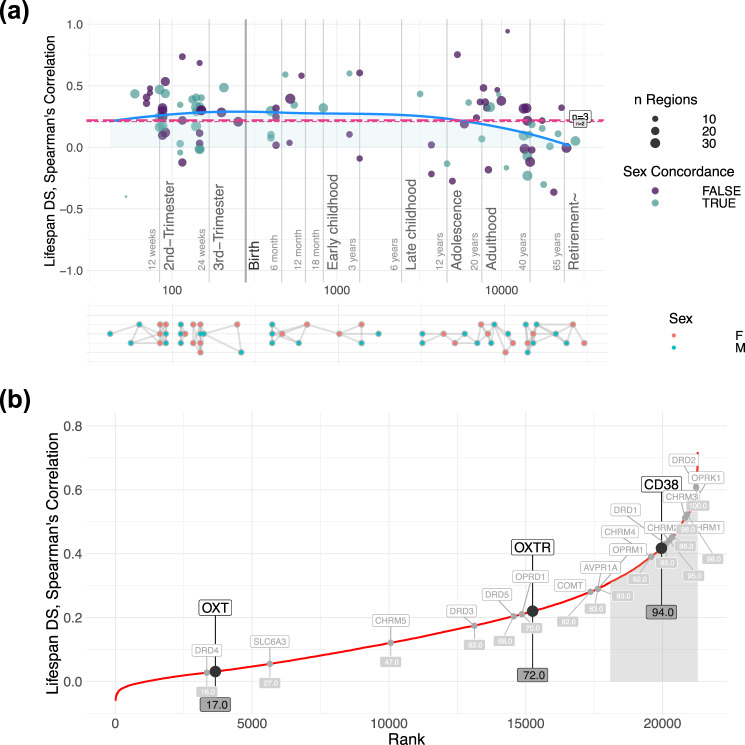
As the number of donor samples in these analyses is relatively small, it is important to confirm the stability of spatiotemporal gene expression patterns to make meaningful inferences beyond the sample. Thus, we calculated a measure of donor-to-donor spatiotemporal differential stability. Genes with a differential stability score in the top 50% of all genes are considered to be conserved [37]. Previous work has demonstrated that genes with high differential stability have strong biological relevance [37], and that OXTR expression patterns have high person-to-person stability in an adult sample [6]. We found that differential stability of OXTR remains relatively constant across the lifespan, except for a modest decrease during adulthood (Fig. 4a). Moreover, both OXTR and CD38 were among the top 30% of all protein coding genes in terms of spatiotemporal differential stability (Fig. 4b). Notably, DRD2 was among the top 1% of protein coding genes for spatiotemporal differential stability. Differential stability values were relatively similar when analyzing males and females separately (Supplementary Fig. 12). This indicates that brain expression patterns of these genes are relatively stable in donors of a similar age. Differential stability of OXTR expression across development also remains relatively constant across macaque development (Supplementary Fig. 13).
Fig. 4. Differential stability for OXTR, CD38, and OXT across development.
a The differential stability of OXTR is relatively constant across the lifespan, with a modest dip during adulthood. The blue line represents lifespan trajectory and the dashed horizontal red lines illustrate the mean of all points (long dashes = 3 pair comparisons; short dashes = 4 pair comparisons). Purple dots show the correlation between two donors of the same sex, whereas turquoise dots show the correlations of opposite sexes. The size of the dots corresponds to the number of brain regions. The bottom section illustrates a map of donor comparisons, where pairs used for differential stability estimation are connected with edges. b The differential stability of OXTR, CD38, and OXT were compared against 21,323 genes, across development. The top 15% of differential stability values are shaded in light gray. Both CD38 and OXTR can be considered to be highly conserved as their scores are in the top 50% [37].
Discussion
Here, we characterized OXTR spatiotemporal expression patterns in humans along with gene-gene co-expression patterns, revealing increased OXTR expression during early childhood and a strong association between the spatiotemporal expression of OXTR and a key gene regulating dopaminergic signaling (DRD2). These temporal gene expression patterns were not observed in a macaque sample, whereby OXTR expression peaks were observed prenatally and perinatally, which may reflect the altricial nature of human young compared to macaques. We also found that a geneset including genes with the strongest 100 associations with the OXTR spatiotemporal expression pattern is enriched in several psychiatric, psychological, and physiological phenotypes, such as schizophrenia, IQ, general cognition, osteoporosis, reproductive processes, and BMI.
Our analyses revealed a distinct pattern of OXTR expression in the human brain across the lifespan. We observed an OXTR expression peak during childhood, which was especially pronounced in males. These sex differences, thought to be influenced by gonadal steroids [54–57], may contribute to reported sex differences in neurodevelopmental disorder diagnoses [58, 59]. The data also indicated an increase in OXTR expression across the brain in late adulthood in both sexes. However, it should be noted that the differential stability of OXTR expression during this life period was lower compared to the rest of the lifespan, which is indicative of greater interindividual differences in OXTR expression patterns. While the production and distribution of oxytocin in the brain are relatively similar across mammalian species [60], the location of oxytocin receptors in the brain varies between mammalian species both spatially [61, 62] and temporally [18]. Indeed, differences in temporal expression peaks may reveal dissimilar critical periods for experience-dependent development mediated by oxytocin signaling [63]. For example, peak OXTR expression in mice occurs during the post-natal period [64, 65], which has been shown to be a critical period for oxytocin-mediated cortical plasticity [66]. Altogether, our observation of increased expression during early childhood and late adulthood points to a critical role for oxytocin signaling during these life periods in humans [11].
Calculating OXTR gene expression in specific brain regions revealed increased OXTR expression in the striatum in the third trimester, as well during adolescence and adulthood. Oxytocin expression levels in the striatum has been associated with parental care [67] and general affiliative behavior [68] in animal models. We speculate that the increase in receptor expression in the striatum just before birth may help prepare the neonate for caregiver bonding. We also discovered increased OXTR expression in the mediodorsal nucleus of the thalamus during childhood, which regulates cognitive tasks associated with the prefrontal cortex, such as attention and memory [69, 70]. As oxytocin signaling has been shown to regulate social recognition and memory [71, 72], these results suggest that this process might be especially critical during childhood. Relatedly, Watanabe and colleagues [73] reported that improved social cognition after intranasal oxytocin treatment in ASD was associated with increased activity in the medial prefrontal cortex. There was an increase in OXTR expression in the cerebellar cortex in both early childhood and late adulthood. Postmortem samples from the cerebellar vermis from donors diagnosed with schizophrenia, which is characterized by social dysfunction, demonstrate reduced OXTR-like binding compared to controls [8], which points to the potential role of oxytocin signaling dysfunction in the cerebellar cortex in the pathophysiology of severe psychiatric illness. There was also an increase in OXTR expression in the hippocampus in late adulthood, which is critical for the recognition of social stimuli in a rodent model [74]. This may also play a role in the adult hippocampal neurogenesis [75, 76], which appears to be facilitated via oxytocin receptors that are expressed in hippocampal CA3 pyramidal neurons [77]. While these region-specific patterns are certainly informative, it is likely that OXTR expression patterns temporally shift in other regions that were not sampled.
We have previously demonstrated the strong co-expression of OXTR and dopaminergic signaling genes and the link between the OXTR expression pattern and brain regions associated with learning in the adult human brain [6]. Moreover, animal models indicate that oxytocin and dopamine signaling may also interact in both bonding [78] and sexual behavior [79]. The strong spatiotemporal correlation between OXTR and DRD2 gene expression observed in the present study suggests that the oxytocin signaling works synergistically with the dopaminergic signaling to support learning also throughout development [80], especially during critical developmental periods [2]. In addition, the spatiotemporal expression patterns of DRD2 from donor-to-donor was among the most stable of all protein coding genes, which is indicative of its strong biological relevance and generalizability beyond the sample, along with its strongly conserved function.
We also demonstrated that OXTR is evolutionarily ancient, with its homolog emerging around the Bilateria phylostratum during which basic nervous systems first appeared [81], and that OXTR has been conserved in both humans and macaques, as indicated by low dN/dS ratios (i.e., negative selection). However, several genes among those with the strongest spatiotemporal gene expression associations with OXTR have experienced recent positive selection in humans, but not macaques (ADHFE1, EYA1, MERTK, VWA5A, ENKUR, ABHD1, PBXIP1, and ZC3HAV1; See Supplementary Note 3 for full gene names). Our results also point to a stronger integration of oxytocin signaling with dopaminergic signaling in humans compared to macaques, as operationalized by the comparison of the specificity of OXTR and DRD2 spatiotemporal co-expression. However, it is important to recognize that regardless of this difference in specificity, the integration between oxytocin-like signaling and dopamine signaling is still evolutionarily conserved, as it has been demonstrated in both non-primate vertebrate [82] and invertebrate [83] species. Regarding the developing brain, we found that genes with the strongest spatiotemporal associations with OXTR that are highly expressed in most brain regions prenatally are evolutionarily older compared to genes that are highly expressed later in life. More recently evolved genes during infancy, and beyond, is indicative of a response to changed ecological conditions. While this is consistent with the analogy of ontogeny recapitulating phylogeny [38], this is perhaps better explained by the fetus’ relative isolation from changes in ecological conditions [23].
In terms of the functional relevance of the OXTR spatiotemporal expression pattern, we found that this was highly correlated with a set of genes enriched for bone density, derived from a GWAS of over 120,000 individuals [40]. These results are especially notable considering the increased expression of OXTR during late adulthood, which is a life period associated with the increased risk of bone fracture [84]. This is certainly not the first time the link between oxytocin signaling and bone remodeling has been highlighted [85], however, the present results suggest that gene-gene co-expression in the brain may contribute to this association. Intriguingly, bone remodeling issues have been identified in autism [86], which has been associated with oxytocin signaling dysfunction [59], and oxytocin receptor knockout mice have been shown to develop osteoporosis [87]. Moreover, peripheral oxytocin levels have been linked to bone mineral density in men with hypopituitarism [88] and post-menopausal women [89]. Leptin release has been proposed as the primary central mediator of bone remodeling [90], but our results suggest that oxytocin may also play some role in this process. Although speculative, this finding, along with the increase in OXTR expression during late adulthood, points to a possible pleiotropic effect of oxytocin dysfunction on social difficulties and bone remodeling, which warrants further investigation.
There are two limitations to the study worth noting. First, the donor sample sizes for both humans and macaques were relatively small and participant life history details (e.g., stressors), which can influence oxytocin receptor expression [91], were also not available for human donors. However, high differential stability values demonstrate that OXTR expression patterns were relatively stable from donor-to-donor across the lifespan. Moreover, genes with high spatiotemporal co-expression with OXTR in the present study were also found to be highly co-expressed in our previous study in human adults, which used a different dataset for analysis [6]. Second, we used transcriptome data as a proxy for gene expression density [19]. While other methods can directly measure OXTR expression density (e.g., competitive-binding receptor autoradiography), it is not feasible to assess co-expression for more than a few receptors at a time using such approaches. While transcriptome measures are a less direct method, this facilitates the analysis of gene-gene co-expression patterns for thousands of receptor and non-receptor genes, which can help unravel the functional organization of the brain [37], for which oxytocin signaling was the focus in the present paper.
Here, we provide evidence for distinct OXTR expression patterns that are enriched in psychiatric disorders and metabolic regulation processes across development. These findings are consistent with the allostatic theory of oxytocin, which uniquely accounts for oxytocin’s effects on both behavioral and non-behavioral traits and highlights the importance of oxytocin signaling function changes across the lifespan to adapt to shifting environmental challenges [2]. By mapping the spatiotemporal OXTR gene expression pattern and identifying co-expressed genes, we provide evidence that oxytocin signaling is implicated in a broad suite of psychological and somatic functions across the lifespan.
Supplementary information
Author contributions
JR, LTW, and DSQ conceived the study. JR analyzed the data, with contributions from DSQ and AMS. All authors contributed to the interpretation of results. JR and DSQ wrote the first draft of the paper and all authors contributed to the final manuscript.
Funding
This research was funded by the Research Council of Norway (301767), the Novo Nordisk Foundation (NNF16OC0019856), the Kavli Trust, and the ERA-Net Cofund through the ERA PerMed project “IMPLEMENT”.
Code availability
The R code to recreate our analyses and figures, along links to the public data used in our analyses, are available at https://gitlab.com/jarek.rokicki/spatio-temporal-oxytocin/.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
The online version contains supplementary material available at 10.1038/s41386-022-01305-5.
References
- 1.Jurek B, Neumann ID. The oxytocin receptor: from intracellular signaling to behavior. Physiol Rev. 2018;98:1805–908. doi: 10.1152/physrev.00031.2017. [DOI] [PubMed] [Google Scholar]
- 2.Quintana DS, Guastella AJ. An allostatic theory of oxytocin. Trends Cogn Sci. 2020;24:515–28. doi: 10.1016/j.tics.2020.03.008. [DOI] [PubMed] [Google Scholar]
- 3.Busnelli M, Chini B. Molecular basis of oxytocin receptor signalling in the brain: what we know and what we need to know. Curr Top Behav Neurosci. 2018;35:3–29. doi: 10.1007/7854_2017_6. [DOI] [PubMed] [Google Scholar]
- 4.Keebaugh AC, Barrett CE, Laprairie JL, Jenkins JJ, Young LJ. RNAi knockdown of oxytocin receptor in the nucleus accumbens inhibits social attachment and parental care in monogamous female prairie voles. Soc Neurosci. 2015;10:561–70. doi: 10.1080/17470919.2015.1040893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Nishimori K, Takayanagi Y, Yoshida M, Kasahara Y, Young LJ, Kawamata M. New aspects of oxytocin receptor function revealed by knockout mice: sociosexual behaviour and control of energy balance. Prog Brain Res. 2008;170:79–90. doi: 10.1016/S0079-6123(08)00408-1. [DOI] [PubMed] [Google Scholar]
- 6.Quintana DS, Rokicki J, van der Meer D, Alnæs D, Kaufmann T, Córdova-Palomera A, et al. Oxytocin pathway gene networks in the human brain. Nat Commun. 2019;10:668. doi: 10.1038/s41467-019-08503-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lee MR, Sheskier MB, Farokhnia M, Feng N, Marenco S, Lipska BK, et al. Oxytocin receptor mRNA expression in dorsolateral prefrontal cortex in major psychiatric disorders: A human post-mortem study. Psychoneuroendocrinology. 2018;96:143–7. doi: 10.1016/j.psyneuen.2018.05.039. [DOI] [PubMed] [Google Scholar]
- 8.Uhrig S, Hirth N, Broccoli L, von Wilmsdorff M, Bauer M, Sommer C, et al. Reduced oxytocin receptor gene expression and binding sites in different brain regions in schizophrenia: A post-mortem study. Schizophr Res. 2016. 2016. 10.1016/j.schres.2016.04.019. [DOI] [PubMed]
- 9.Kang HJ, Kawasawa YI, Cheng F, Zhu Y, Xu X, Li M, et al. Spatiotemporal transcriptome of the human brain. Nature. 2011;478:483–9. doi: 10.1038/nature10523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kim DR, Bale TL, Epperson CN. Prenatal programming of mental illness: current understanding of relationship and mechanisms. Curr Psychiatry Rep. 2015;17:5. doi: 10.1007/s11920-014-0546-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Freeman SM, Palumbo MC, Lawrence RH, Smith AL, Goodman MM, Bales KL. Effect of age and autism spectrum disorder on oxytocin receptor density in the human basal forebrain and midbrain. Transl Psychiatry. 2018;8:1–11.. doi: 10.1038/s41398-018-0315-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Boer GJ, Swaab DF, Uylings HBM, Boer K, Buijs RM, Velis DN. Neuropeptides in Rat Brain Development. In: McConnell PS, Boer GJ, Romijn HJ, Van De Poll NE, Corner MA, editors. Prog. Brain Res., vol. 53, Elsevier; 1980. p. 207–27. [DOI] [PubMed]
- 13.Swaab DF. Development of the human hypothalamus. Neurochem Res. 1995;20:509–19. doi: 10.1007/BF01694533. [DOI] [PubMed] [Google Scholar]
- 14.Pedersen CA, Boccia ML. Oxytocin links mothering received, mothering bestowed and adult stress responses. Stress Amst Neth. 2002;5:259–67. doi: 10.1080/1025389021000037586. [DOI] [PubMed] [Google Scholar]
- 15.Winslow JT, Noble PL, Lyons CK, Sterk SM, Insel TR. Rearing effects on cerebrospinal fluid oxytocin concentration and social buffering in rhesus monkeys. Neuropsychopharmacology. 2003;28:910–8. doi: 10.1038/sj.npp.1300128. [DOI] [PubMed] [Google Scholar]
- 16.Francis DD, Champagne FC, Meaney MJ. Variations in maternal behaviour are associated with differences in oxytocin receptor levels in the rat. J Neuroendocrinol. 2000;12:1145–8. doi: 10.1046/j.1365-2826.2000.00599.x. [DOI] [PubMed] [Google Scholar]
- 17.Heim C, Young LJ, Newport DJ, Mletzko T, Miller AH, Nemeroff CB. Lower CSF oxytocin concentrations in women with a history of childhood abuse. Mol Psychiatry. 2009;14:954–8. doi: 10.1038/mp.2008.112. [DOI] [PubMed] [Google Scholar]
- 18.Vaidyanathan R, Hammock EA. Oxytocin receptor dynamics in the brain across development and species. Dev Neurobiol. 2017;77:143–57. doi: 10.1002/dneu.22403. [DOI] [PubMed] [Google Scholar]
- 19.Young LJ, Muns S, Wang Z, Insel TR. Changes in oxytocin receptor mRNA in rat brain during pregnancy and the effects of estrogen and interleukin‐6. J Neuroendocrinol. 1997;9:859–65. doi: 10.1046/j.1365-2826.1997.00654.x. [DOI] [PubMed] [Google Scholar]
- 20.Zhu Y, Sousa AMM, Gao T, Skarica M, Li M, Santpere G, et al. Spatiotemporal transcriptomic divergence across human and macaque brain development. Science. 2018;362:eaat8077. doi: 10.1126/science.aat8077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Workman AD, Charvet CJ, Clancy B, Darlington RB, Finlay BL. Modeling transformations of neurodevelopmental sequences across mammalian species. J Neurosci. 2013;33:7368–83. doi: 10.1523/JNEUROSCI.5746-12.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Clancy B, Finlay BL, Darlington RB, Anand KJS. Extrapolating brain development from experimental species to humans. NeuroToxicology. 2007;28:931–7. doi: 10.1016/j.neuro.2007.01.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Domazet-Lošo T, Tautz D. A phylogenetically based transcriptome age index mirrors ontogenetic divergence patterns. Nature. 2010;468:815–8. doi: 10.1038/nature09632. [DOI] [PubMed] [Google Scholar]
- 24.Hawrylycz M, Lein ES, Guillozet-Bongaarts AL, Shen EH, Ng L, Miller JA, et al. An anatomically comprehensive atlas of the adult human brain transcriptome. Nature. 2012;489:391. doi: 10.1038/nature11405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Domazet-Lošo T, Tautz D. An ancient evolutionary origin of genes associated with human genetic diseases. Mol Biol Evol. 2008;25:2699–707. doi: 10.1093/molbev/msn214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Drost H-G, Gabel A, Liu J, Quint M, Grosse I. myTAI: evolutionary transcriptomics with R. Bioinformatics. 2018;34:1589–90. doi: 10.1093/bioinformatics/btx835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Dumas G, Malesys S, Bourgeron T. Systematic detection of brain protein-coding genes under positive selection during primate evolution and their roles in cognition. Genome Res. 2021;31:484–96. doi: 10.1101/gr.262113.120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Watanabe K, Taskesen E, Bochoven A, Posthuma D. Functional mapping and annotation of genetic associations with FUMA. Nat Commun. 2017;8:1826. doi: 10.1038/s41467-017-01261-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.MacArthur J, Bowler E, Cerezo M, Gil L, Hall P, Hastings E, et al. The new NHGRI-EBI Catalog of published genome-wide association studies (GWAS Catalog) Nucleic Acids Res. 2017;45:D896–D901.. doi: 10.1093/nar/gkw1133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Liberzon A, Subramanian A, Pinchback R, Thorvaldsdóttir H, Tamayo P, Mesirov JP. Molecular signatures database (MSigDB) 3.0. Bioinformatics. 2011;27:1739–40. doi: 10.1093/bioinformatics/btr260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kerimov N, Hayhurst JD, Peikova K, Manning JR, Walter P, Kolberg L, et al. eQTL Catalogue: a compendium of uniformly processed human gene expression and splicing QTLs. BioRxiv. 2021:2020.01.29.924266. [DOI] [PMC free article] [PubMed]
- 32.Akbarian S, Liu C, Knowles JA, Vaccarino FM, Farnham PJ, Crawford GE, et al. The PsychENCODE project. Nat Neurosci. 2015;18:1707–12. doi: 10.1038/nn.4156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ng B, White CC, Klein H-U, Sieberts SK, McCabe C, Patrick E, et al. An xQTL map integrates the genetic architecture of the human brain’s transcriptome and epigenome. Nat Neurosci. 2017;20:1418–26. doi: 10.1038/nn.4632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hoffman GE, Bendl J, Voloudakis G, Montgomery KS, Sloofman L, Wang Y-C, et al. CommonMind Consortium provides transcriptomic and epigenomic data for Schizophrenia and Bipolar Disorder. Sci Data. 2019;6:180. doi: 10.1038/s41597-019-0183-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Aguet F, Brown AA, Castel SE, Davis JR, He Y, Jo B, et al. Genetic effects on gene expression across human tissues. Nature. 2017;550:204–13. doi: 10.1038/nature24277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ramasamy A, Trabzuni D, Guelfi S, Varghese V, Smith C, Walker R, et al. Genetic variability in the regulation of gene expression in ten regions of the human brain. Nat Neurosci. 2014;17:1418–28. doi: 10.1038/nn.3801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hawrylycz M, Miller JA, Menon V, Feng D, Dolbeare T, Guillozet-Bongaarts AL, et al. Canonical genetic signatures of the adult human brain. Nat Neurosci. 2015;18:1832. doi: 10.1038/nn.4171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kuratani S, Uesaka M, Irie N. How can recapitulation be reconciled with modern concepts of evolution? J Exp Zoolog B Mol Dev Evol. 2022;338:28–35. [DOI] [PubMed]
- 39.Shin S-Y, Fauman EB, Petersen A-K, Krumsiek J, Santos R, Huang J, et al. An atlas of genetic influences on human blood metabolites. Nat Genet. 2014;46:543–50. doi: 10.1038/ng.2982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kemp JP, Morris JA, Medina-Gomez C, Forgetta V, Warrington NM, Youlten SE, et al. Identification of 153 new loci associated with heel bone mineral density and functional involvement of GPC6 in osteoporosis. Nat Genet. 2017;49:1468–75. doi: 10.1038/ng.3949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Morris JA, Kemp JP, Youlten SE, Laurent L, Logan JG, Chai RC, et al. An atlas of genetic influences on osteoporosis in humans and mice. Nat Genet. 2019;51:258–66. doi: 10.1038/s41588-018-0302-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zhao B, Luo T, Li T, Li Y, Zhang J, Shan Y, et al. Genome-wide association analysis of 19,629 individuals identifies variants influencing regional brain volumes and refines their genetic co-architecture with cognitive and mental health traits. Nat Genet. 2019;51:1637–44. doi: 10.1038/s41588-019-0516-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Davies G, Lam M, Harris SE, Trampush JW, Luciano M, Hill WD, et al. Study of 300,486 individuals identifies 148 independent genetic loci influencing general cognitive function. Nat Commun. 2018;9:2098. doi: 10.1038/s41467-018-04362-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Savage JE, Jansen PR, Stringer S, Watanabe K, Bryois J, de Leeuw CA, et al. Genome-wide association meta-analysis in 269,867 individuals identifies new genetic and functional links to intelligence. Nat Genet. 2018;50:912–9. doi: 10.1038/s41588-018-0152-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zhu Z, Guo Y, Shi H, Liu C-L, Panganiban RA, Chung W, et al. Shared genetic and experimental links between obesity-related traits and asthma subtypes in UK Biobank. J Allergy Clin Immunol. 2020;145:537–49. doi: 10.1016/j.jaci.2019.09.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Pulit SL, Stoneman C, Morris AP, Wood AR, Glastonbury CA, Tyrrell J, et al. Meta-analysis of genome-wide association studies for body fat distribution in 694 649 individuals of European ancestry. Hum Mol Genet. 2019;28:166–74. doi: 10.1093/hmg/ddy327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Vujkovic M, Keaton JM, Lynch JA, Miller DR, Zhou J, Tcheandjieu C, et al. Discovery of 318 new risk loci for type 2 diabetes and related vascular outcomes among 1.4 million participants in a multi-ancestry meta-analysis. Nat Genet. 2020;52:680–91. doi: 10.1038/s41588-020-0637-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wang K, Li W-D, Zhang CK, Wang Z, Glessner JT, Grant SFA, et al. A genome-wide association study on obesity and obesity-related traits. PloS One. 2011;6:e18939. doi: 10.1371/journal.pone.0018939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kim SK. Identification of 613 new loci associated with heel bone mineral density and a polygenic risk score for bone mineral density, osteoporosis and fracture. PloS One. 2018;13:e0200785. doi: 10.1371/journal.pone.0200785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Loo SK, Shtir C, Doyle AE, Mick E, McGough JJ, McCracken J, et al. Genome-wide association study of intelligence: additive effects of novel brain expressed genes. J Am Acad Child Adolesc Psychiatry. 2012;51:432–.e2. doi: 10.1016/j.jaac.2012.01.006. [DOI] [PubMed] [Google Scholar]
- 51.Akiyama M, Okada Y, Kanai M, Takahashi A, Momozawa Y, Ikeda M, et al. Genome-wide association study identifies 112 new loci for body mass index in the Japanese population. Nat Genet. 2017;49:1458–67. doi: 10.1038/ng.3951. [DOI] [PubMed] [Google Scholar]
- 52.Christakoudi S, Evangelou E, Riboli E, Tsilidis KK. GWAS of allometric body-shape indices in UK Biobank identifies loci suggesting associations with morphogenesis, organogenesis, adrenal cell renewal and cancer. Sci Rep. 2021;11:10688. doi: 10.1038/s41598-021-89176-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Sinnott-Armstrong N, Tanigawa Y, Amar D, Mars N, Benner C, Aguirre M, et al. Genetics of 35 blood and urine biomarkers in the UK Biobank. Nat Genet. 2021;53:185–94. doi: 10.1038/s41588-020-00757-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Bale TL, Pedersen CA, Dorsa DM. CNS oxytocin receptor mRNA expression and regulation by gonadal steroids. Adv Exp Med Biol. 1995;395:269–80. [PubMed] [Google Scholar]
- 55.Tribollet E, Audigier S, Dubois-Dauphin M, Dreifuss JJ. Gonadal steroids regulate oxytocin receptors but not vasopressin receptors in the brain of male and female rats. Autoradiographical Study Brain Res. 1990;511:129–40. doi: 10.1016/0006-8993(90)90232-Z. [DOI] [PubMed] [Google Scholar]
- 56.Carter CS, Perkeybile AM. The monogamy paradox: what do love and sex have to do with it? Front Ecol Evol. 2018;6:202. doi: 10.3389/fevo.2018.00202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Jirikowski GF, Ochs SD, Caldwell JD Oxytocin and Steroid Actions. In: Hurlemann R, Grinevich V, editors. Behav. Pharmacol. Neuropept. Oxytocin, Cham: Springer International Publishing; 2018. 77–95. [DOI] [PubMed]
- 58.Ferri SL, Abel T, Brodkin ES. Sex differences in autism spectrum disorder: a review. Curr Psychiatry Rep. 2018;20:9. doi: 10.1007/s11920-018-0874-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Guastella AJ, Hickie IB. Oxytocin treatment, circuitry and autism: a critical review of the literature placing oxytocin into the autism context. Biol Psychiatry. 2016;79:234–42. doi: 10.1016/j.biopsych.2015.06.028. [DOI] [PubMed] [Google Scholar]
- 60.Knobloch HS, Grinevich V. Evolution of oxytocin pathways in the brain of vertebrates. Front Behav Neurosci. 2014;8:31. doi: 10.3389/fnbeh.2014.00031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Leung CH, Abebe DF, Earp SE, Goode CT, Grozhik AV, Mididoddi P, et al. Neural distribution of vasotocin receptor mRNA in two species of songbird. Endocrinology. 2011;152:4865–81. doi: 10.1210/en.2011-1394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Freeman SM, Young LJ. Comparative perspectives on oxytocin and vasopressin receptor research in rodents and primates: translational implications. J Neuroendocrinol. 2016;28. [DOI] [PMC free article] [PubMed]
- 63.Hammock E. Developmental perspectives on oxytocin and vasopressin. Neuropsychopharmacology. 2015;40:24. doi: 10.1038/npp.2014.120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Mitre M, Marlin BJ, Schiavo JK, Morina E, Norden SE, Hackett TA, et al. A distributed network for social cognition enriched for oxytocin receptors. J Neurosci. 2016;36:2517–35. doi: 10.1523/JNEUROSCI.2409-15.2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Hammock E, Levitt P. Oxytocin receptor ligand binding in embryonic tissue and postnatal brain development of the C57BL/6J mouse. Front Behav Neurosci. 2013;7:195. doi: 10.3389/fnbeh.2013.00195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Zheng J-J, Li S-J, Zhang X-D, Miao W-Y, Zhang D, Yao H, et al. Oxytocin mediates early experience–dependent cross-modal plasticity in the sensory cortices. Nat Neurosci. 2014;17:391–9. doi: 10.1038/nn.3634. [DOI] [PubMed] [Google Scholar]
- 67.Olazábal DE, Young LJ. Species and individual differences in juvenile female alloparental care are associated with oxytocin receptor density in the striatum and the lateral septum. Horm Behav. 2006;49:681–7. doi: 10.1016/j.yhbeh.2005.12.010. [DOI] [PubMed] [Google Scholar]
- 68.Ross HE, Freeman SM, Spiegel LL, Ren X, Terwilliger EF, Young LJ. Variation in oxytocin receptor density in the nucleus accumbens has differential effects on affiliative behaviors in monogamous and polygamous voles. J Neurosci J Soc Neurosci. 2009;29:1312–8. doi: 10.1523/JNEUROSCI.5039-08.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Pergola G, Danet L, Pitel A-L, Carlesimo GA, Segobin S, Pariente J, et al. The regulatory role of the human mediodorsal thalamus. Trends Cogn Sci. 2018;22:1011–25. doi: 10.1016/j.tics.2018.08.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Xing B, Mack NR, Guo K-M, Zhang Y-X, Ramirez B, Yang S-S, et al. A subpopulation of prefrontal cortical neurons is required for social memory. Biol Psychiatry. 2021;89:521–31. doi: 10.1016/j.biopsych.2020.08.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Ferguson JN, Young LJ, Hearn EF, Matzuk MM, Insel TR, Winslow JT. Social amnesia in mice lacking the oxytocin gene. Nat Genet. 2000;25:284. doi: 10.1038/77040. [DOI] [PubMed] [Google Scholar]
- 72.Maroun M, Wagner S. Oxytocin and memory of emotional stimuli: some dance to remember, some dance to forget. Biol Psychiatry. 2016;79:203–12. doi: 10.1016/j.biopsych.2015.07.016. [DOI] [PubMed] [Google Scholar]
- 73.Watanabe T, Abe O, Kuwabara H, Yahata N, Takano Y, Iwashiro N, et al. Mitigation of sociocommunicational deficits of autism through oxytocin-induced recovery of medial prefrontal activity: a randomized trial. JAMA Psychiatry. 2014;71:166–75. doi: 10.1001/jamapsychiatry.2013.3181. [DOI] [PubMed] [Google Scholar]
- 74.Raam T, McAvoy KM, Besnard A, Veenema AH, Sahay A. Hippocampal oxytocin receptors are necessary for discrimination of social stimuli. Nat Commun. 2017;8:2001. doi: 10.1038/s41467-017-02173-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Leuner B, Caponiti JM, Gould E. Oxytocin stimulates adult neurogenesis even under conditions of stress and elevated glucocorticoids. Hippocampus. 2012;22:861–8. doi: 10.1002/hipo.20947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Boldrini M, Fulmore CA, Tartt AN, Simeon LR, Pavlova I, Poposka V, et al. Human hippocampal neurogenesis persists throughout aging. Cell Stem Cell. 2018;22:589–99. doi: 10.1016/j.stem.2018.03.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Lin Y-T, Chen C-C, Huang C-C, Nishimori K, Hsu K-S. Oxytocin stimulates hippocampal neurogenesis via oxytocin receptor expressed in CA3 pyramidal neurons. Nat Commun. 2017;8:537. doi: 10.1038/s41467-017-00675-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Liu Y, Wang ZX. Nucleus accumbens oxytocin and dopamine interact to regulate pair bond formation in female prairie voles. Neuroscience. 2003;121:537–44. doi: 10.1016/S0306-4522(03)00555-4. [DOI] [PubMed] [Google Scholar]
- 79.Baskerville TA, Allard J, Wayman C, Douglas AJ. Dopamine–oxytocin interactions in penile erection. Eur J Neurosci. 2009;30:2151–64. doi: 10.1111/j.1460-9568.2009.06999.x. [DOI] [PubMed] [Google Scholar]
- 80.Love TM. Oxytocin, motivation and the role of dopamine. Pharm Biochem Behav. 2014;119:49–60. doi: 10.1016/j.pbb.2013.06.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Kelava I, Rentzsch F, Technau U. Evolution of eumetazoan nervous systems: insights from cnidarians. Philos Trans R Soc B Biol Sci. 2015;370:20150065. doi: 10.1098/rstb.2015.0065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Hung LW, Neuner S, Polepalli JS, Beier KT, Wright M, Walsh JJ, et al. Gating of social reward by oxytocin in the ventral tegmental area. Science. 2017. 29 September 2017. 10.1126/science.aan4994. [DOI] [PMC free article] [PubMed]
- 83.Beets I, Janssen T, Meelkop E, Temmerman L, Suetens N, Rademakers S, et al. Vasopressin/oxytocin-related signaling regulates gustatory associative learning in C. elegans. Science. 2012;338:543–5. doi: 10.1126/science.1226860. [DOI] [PubMed] [Google Scholar]
- 84.Hui SL, Slemenda CW, Johnston CC. Age and bone mass as predictors of fracture in a prospective study. J Clin Invest. 1988;81:1804–9. doi: 10.1172/JCI113523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Copland JA, Ives KL, Simmons DJ, Soloff MS. Functional oxytocin receptors discovered in human osteoblasts. Endocrinology. 1999;140:4371–4. doi: 10.1210/endo.140.9.7130. [DOI] [PubMed] [Google Scholar]
- 86.Hediger ML, England LJ, Molloy CA, Yu KF, Manning-Courtney P, Mills JL. Reduced bone cortical thickness in boys with autism or autism spectrum disorder. J Autism Dev Disord. 2008;38:848–56. doi: 10.1007/s10803-007-0453-6. [DOI] [PubMed] [Google Scholar]
- 87.Tamma R, Colaianni G, Zhu L, DiBenedetto A, Greco G, Montemurro G, et al. Oxytocin is an anabolic bone hormone. Proc Natl Acad Sci. 2009;106:7149–54. doi: 10.1073/pnas.0901890106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Aulinas A, Guarda FJ, Yu EW, Haines MS, Asanza E, Silva L, et al. Lower oxytocin levels are associated with lower bone mineral density and less favorable hip geometry in hypopituitary men. Neuroendocrinology. 2021;111:87–98. doi: 10.1159/000506638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Breuil V, Panaia-Ferrari P, Fontas E, Roux C, Kolta S, Eastell R, et al. Oxytocin, a new determinant of bone mineral density in post-menopausal women: analysis of the OPUS cohort. J Clin Endocrinol Metab. 2014;99:E634–E641.. doi: 10.1210/jc.2013-4126. [DOI] [PubMed] [Google Scholar]
- 90.Ducy P, Amling M, Takeda S, Priemel M, Schilling AF, Beil FT, et al. Leptin inhibits bone formation through a hypothalamic relay: a central control of bone mass. Cell. 2000;100:197–207. doi: 10.1016/S0092-8674(00)81558-5. [DOI] [PubMed] [Google Scholar]
- 91.Perkeybile AM, Carter CS, Wroblewski KL, Puglia MH, Kenkel WM, Lillard TS, et al. Early nurture epigenetically tunes the oxytocin receptor. Psychoneuroendocrinology. 2019;99:128–36. doi: 10.1016/j.psyneuen.2018.08.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The R code to recreate our analyses and figures, along links to the public data used in our analyses, are available at https://gitlab.com/jarek.rokicki/spatio-temporal-oxytocin/.