Abstract
Phosphoglycosyl transferases (PGTs) play a pivotal role at the inception of complex glycoconjugate biosynthesis pathways across all domains of life. PGTs promote the first membrane-committed step in the en bloc biosynthetic strategy by catalyzing the transfer of a phospho-sugar from a nucleoside diphospho-sugar to a membrane-resident polyprenol phosphate. Studies on the PGTs have been hampered because they are integral membrane proteins, and often prove to be recalcitrant to expression, purification and analysis. However, in recent years exciting new information has been derived on the structures and the mechanisms of PGTs, revealing the existence of two unique superfamilies of PGT enzymes that enact catalysis at the membrane interface. Genome neighborhood analysis shows that these superfamilies, the polytopic PGT (polyPGT) and monotopic PGT (monoPGT), may initiate different pathways within the same organism. Moreover, the same fundamental two-substrate reaction is enacted through two different chemical mechanisms with distinct modes of catalysis. This review highlights the structural and mechanistic divergence between the PGT enzyme superfamilies and how this is reflected in differences in regulation in their varied glycoconjugate biosynthesis pathways.
INTRODUCTION
Glycoconjugates serve essential functions in all domains of life. In prokaryotes, diverse biomolecules, such as the bacterial peptidoglycan (PG) and archaeal S-layer glycoproteins afford mechanical stability to unicellular organisms in rapidly changing environments. Also, complex glycolipids, including the O-antigen component of the lipopolysaccharide (LPS) of Gram-negative bacteria, function to mediate interactions amongst cells and propagate deleterious pathogenic processes (1, 2). In eukaryotes, glycosylation is highly significant in numerous aspects of biology. For example N-linked protein glycosylation contributes to myriad functions from protein folding to intercellular communication (3). With these examples of the central biological roles of glycoconjugates in mind, understanding the molecular logic of the biosynthesis of these complex non-templated biomolecules is an important and rich area for investigation.
Glycoconjugate biosynthesis occurs via two general strategies - designated as sequential and en bloc (2). The sequential mechanism is commonly associated with assembly of smaller, less complex glycans than the en bloc strategy. This review will focus on enzymes that catalyze the first key step in en bloc glycan assembly and glycoconjugate biosynthesis. En bloc glycoconjugate assembly is initiated on the cytoplasmic face of cellular membranes, including the outer and inner membranes of Gram-positive and Gram-negative bacteria, respectively, and at the endomembrane system of eukaryotic cells. These sites of biosynthesis are advantageous, as glycoconjugates commonly must be integrated into cell-surface structures or secreted from cells for function. For example, the cross-linked peptidoglycan polymer is localized to the periplasmic space of Gram-negative bacteria and provides a thick outer layer coating the cellular membranes of Gram-positive bacteria. A signature feature of the early steps in glycoconjugate biosynthesis is the stepwise assembly of glycans on membrane-resident long-chain linear polyprenol phosphates (PrenPs), such as undecaprenol phosphate (UndP), which is the most common PrenP in bacteria (Figure 1) (4). These terminal secondary metabolites are highly conserved in glycan assembly pathways. To date, a complete understanding of the role of the amphiphilic, long-chain PrenPs, which range from seven to 20 or more isoprene units, is unclear. The simplest view that these “superlipids” (5) serve as membrane-associated anchors upon which to assemble glycans is certainly a part of the story (5, 6). However, unique effects of extended PrenPs on membrane order and pathway assembly and flux may also be pivotal (7, 8).
Figure 1. Enzymes and stepwise pathways in glycoconjugate biosynthesis.
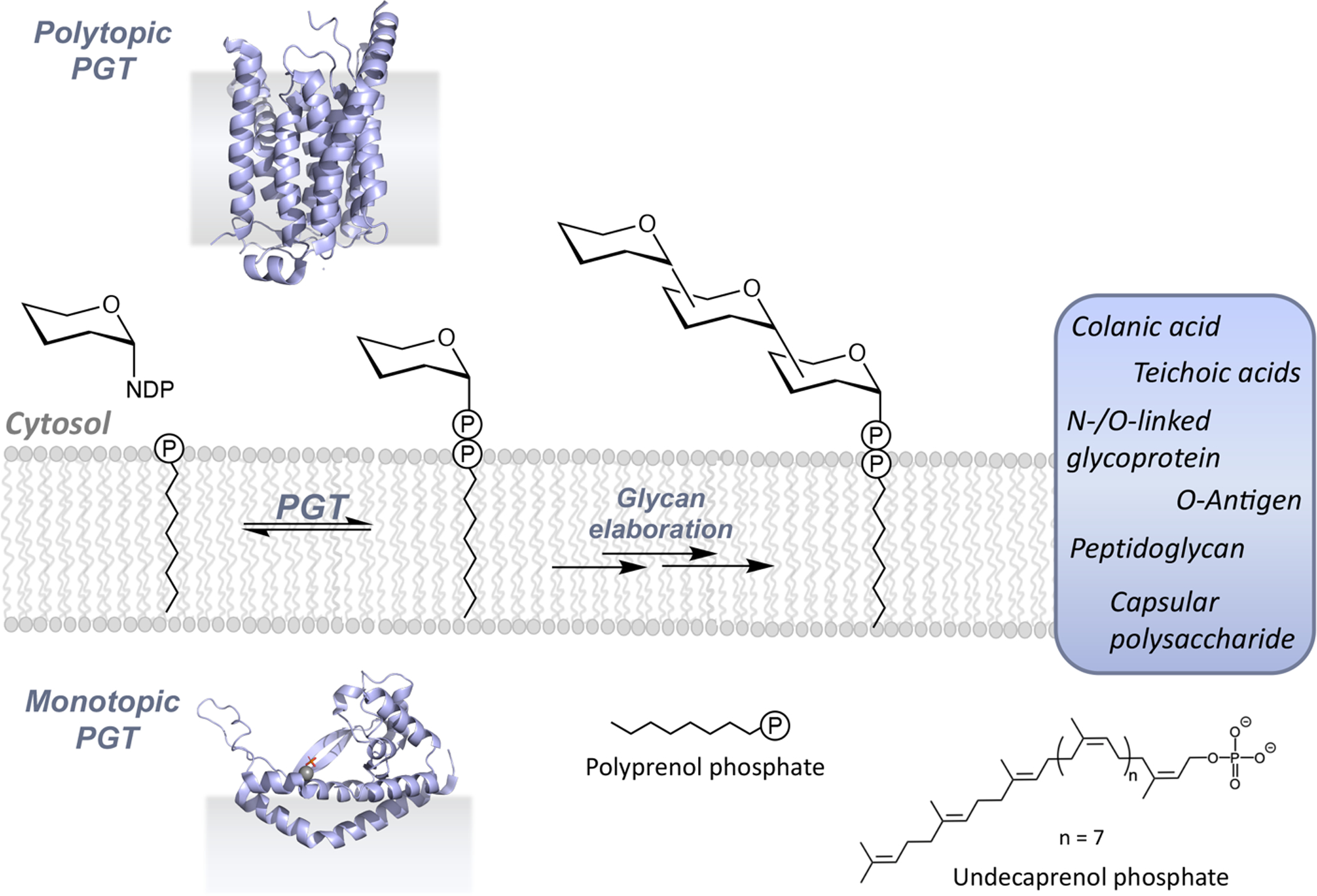
Polytopic and monotopic phosphoglycosyl transferases (PGTs) catalyze the initial, membrane-committed step in the en bloc mechanism of glycoconjugate biosynthesis. Inserts show ribbon diagrams of superfamily members PglC from Campylobacter concisus (PDB 5W7L) and MraY from Aquifex aeolicus (PDB 3CKR). Glycoconjugates are assembled via sequential modification of the PrenPP-linked product of the PGT reaction and ultimately polymerized and/or conjugated with lipids or proteins. PrenPs such as the bacterial undecaprenol phosphate are important lipid carriers in glycoconjugate assembly.
The first membrane-committed step in glycoconjugate biosynthesis is catalyzed by phosphoglycosyl transferases (PGTs) (Figure 1). This step is then followed by the action of a series of enzymes, acting at the membrane interface, which systematically elaborate the initial polyprenol diphosphate-linked carbohydrate (PrenPP-CHO). In recognition of the fact that PGTs catalyze the initial key step in the biosynthetic pathways in which they feature, these enzymes are often referred to as “priming glycosyl transferases”. However, this is an imprecise term as the enzymes are not formally glycosyl transferases and in fact, they catalyze a very distinctive phospho-sugar transfer reaction. PGTs have also been classified based on the nature of their nucleoside diphospho-sugar (NDP-sugar) substrates (9) – namely, as the polyisoprenol-phosphate N-acetyl hexosamine-1-phosphate transferase (PNPT) and polyisoprenol-phosphate hexose-1-phosphate transferase (PHPT) families. However, discovery of additional PGTs with greater structural diversity and increasing knowledge of the breadth of substrate specificity have blurred the lines of this latter definition (10–12).
Membrane-associated pathways involving the PGTs lead to a variety of glycoconjugates (Figure 1). In bacteria, the diversity is extraordinary due both to the wide range of different glycoconjugate scaffolds that are assembled and also to the remarkable array of prokaryote-specific carbohydrate building blocks (13). In eukaryotes, although the glycans are less diverse in terms of carbohydrate composition (14), these pathways, including the en bloc strategy N-linked protein glycosylation pathway, are essential for innumerable cellular processes (3). In en bloc glycoconjugate assembly, there are two PGT superfamilies that mediate phospho-sugar transfer from an activated NDP-sugar to a membrane-resident PrenP on the cytoplasmic face of cellular membranes. Representative structures of PGT superfamily members published in the last ten years reveal functional core structures that are either polytopic, including multiple membrane- spanning domains, or monotopic and resident of a single leaflet of the lipid bilayer (Figure 1). The fundamental basis for the prominent structural dichotomy is still unknown however, it is now clear that it results in the adoption of different mechanistic and catalytic strategies as well as opportunities for differential regulation. These themes with be discussed herein.
PGT SUPERFAMILY TOPOLOGIES AND THE PATHWAYS TO WHICH THEY CONTRIBUTE
PGTs belong to two superfamilies, exhibiting strikingly different membrane-interaction modes of the functional cores (Figure 2A). The polytopic PGT (polyPGT) superfamily members are found in prokaryotes and eukaryotes. The common structural features of polyPGTs include a membrane-associated architecture comprising 10–11 transmembrane helices (TMHs) linked by soluble cytoplasmic loops that are involved in catalysis (15, 16). In contrast, the other PGT superfamily includes a monotopic functional core and thus is designated as the monotopic PGT (monoPGT) superfamily highlighting this topology. The monoPGT members are exclusively prokaryotic (17, 18). Extensive bioinformatic analysis reveals that the various monoPGT structures include a common monotopic functional core featuring a membrane-protein architecture that does not traverse the membrane but rather, engages only a single leaflet of the lipid bilayer via a re-entrant membrane helix (RMH) (12). The monoPGTs may simply comprise the signature functional core, as observed in the “small” monoPGT family exemplified by the Campylobacter jejuni PglC. Alternatively, the functional core may be elaborated at the N- or C-terminus (17). The most abundant monoPGT family, designated as the “large” monoPGT family, features an N-terminal extension that includes a series of four concatenated TMHs and a soluble cytoplasmic domain of unknown function. The topology of this monoPGT family was first elucidated experimentally by Valvano and coworkers for the Escherichia coli WcaJ (19). The third monoPGT family includes the “bifunctional” members, which are elaborated at either the N- or C-terminus with an additional functional domain. Recently, a sequence similarity network (SSN) analysis, representing over 38,000 non-redundant sequences, has been applied to the monoPGT superfamily. The analysis revealed the wide range of functional domains, including sugar-modifying enzymes, glycosyl transferases or regulatory domains, appended to the functional monoPGT core (17). Topology diagrams for representative polyPGTs (TarO and MraY also known as translocase 1) and the three families of monoPGTs including the minimal (PglC) and elaborated (PglB and CpsE) are illustrated in Figure 2A.
Figure 2. PGT topologies and genes associated with bacterial PGT-dependent pathways.
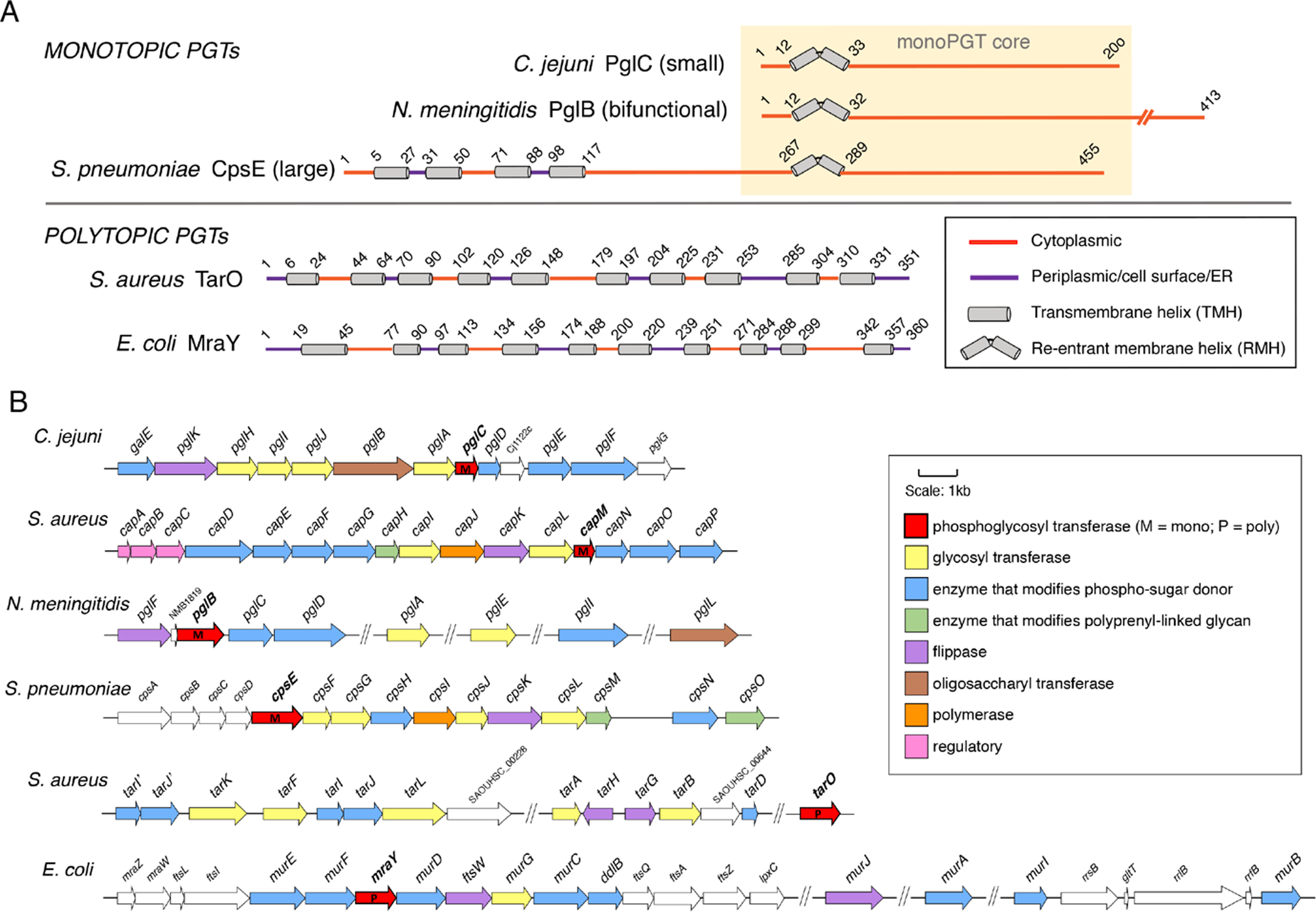
A. Topology diagrams showing the disposition of TMH and RMH motifs for poly- and monoPGTs. The topology of PglC (Campylobacter jejuni) is determined via biochemical analysis and by analogy with the structure of the Campylobacter concisus PglC (PDB 5W7L). The topology of the mono-PGT domain in PglB (Neisseria meningitidis) is inferred by homology, and the topology of CpsE (Streptococcus pneumoniae) is deduced by analogy with the experimentally determined structure of WcaJ (Escherichia coli). The topologies of the TarO (Staphylococcus aureus) and MraY (E. coli) are based on the experimental structures of homologous polyPGTs, (6FWZ and 3CKR respectively). B. Bacterial glycoconjugate assembly genes and pathways featuring mono- and polyPGTs including pathways from: C. jejuni (NCTC 11168), N-linked protein glycosylation; Staphylococcus aureus (NCTC 8325) - capsular polysaccharide; N. meningitidis (MC58) - O-linked protein glycosylation; S. pneumoniae (Serotype 9V) - capsular polysaccharide; S. aureus (NCTC 8325) – wall teichoic acid; E. coli (K-12 W3110) - peptidoglycan. Functionally-annotated genes related to the PGT-containing pathways are color coded by function (see insert).
The study of bacterial PGTs and the pathways that they initiate is particularly informative as genes encoding glycoconjugate biosynthesis enzymes are often organized into operons that provide insight into entire pathways. Alternatively, genes that are not in a single operon have been annotated in the context of other pathway components that may be in different genome locations (Figure 2B). Analysis of selected pathways shows that the different PGT superfamilies may be represented in separate pathways within the same organism. For example, in Staphylococcus aureus, the polyPGT, TarO, acts at the beginning of the wall teichoic acid (WTA) pathway (20) and a small monoPGT, CapM, features at the initiation of capsular polysaccharide (CPS) assembly (21). In some pathways, the PGT genes are located adjacent to genes that encode enzymes related to the function of the PGT. For example, the small monoPGT gene pglC (C. jejuni) is found adjacent to the pglF/pglE/pglD gene cluster, which encodes enzymes that convert UDP-GlcNAc into UDP-N,N’-diacetylbacillosamine (UDP-diNAcBac), the prokaryote-specific UDP-sugar substrate of PglC. Intriguingly, in N. meningitidis and N. gonorrhoeae, the gene that encodes the monoPGT (pglB in Neisseria) also encodes a domain that catalyzes AcCoA-dependent acetylation of UDP-2’-acetamido-4’-amino-bacillosamine, the precursor of UDP-diNAcBac, the substrate of the monoPGT domain of PglB. Therefore, the Neisseria PglB is bifunctional and catalyzes the same transformations as PglD and PglC from Campylobacter. Alternatively, pathway genes may encode enzymes for the biosynthesis of elaborate PGT substrates such as the Park’s nucleotide (UDP-MurNAc-pentapeptide) that is utilized by all bacterial MraYs for peptidoglycan biosynthesis. Such is the case with murE, murF, and murD, which are adjacent to MraY, in the E. coli pathway (Figure 2B) (22). By far the most common genome neighbors in the PGT-associated pathways are the glycosyl transferase (GT) genes (17). This is the case for cpsF and cpsG that are adjacent to the monoPGT gene cpsE from S. pneumoniae (23), capL, which is adjacent to capM in S. aureus (21) and pglA, which is adjacent to pglC in C. jejuni (24, 25). Some of the most important gene network components are those that are implicated in the regulation of glycoconjugate biosynthesis. Selected examples will be discussed in detail in a later section.
Ultimately, the extensive analysis of genomic data from different bacterial species and from genetic variants of the individual species, together with knowledge of the two PGT superfamilies that represent the hallmarks of PGT-dependent pathways will fuel new discovery. For example, informatics analysis will enable not only recognition of enzyme-interaction partnerships and regulation, but also provide the foundation for developing methods to predict the composition of glycoconjugates produced in selected pathways, and, potentially, the phenotypic effects of strain variation.
PHOSPHOGLYCOSYL TRANSFERASES – TWO ROUTES TO ONE DESTINATION
Historically, the first PGT superfamily to be defined included members showing the polytopic architecture. This was in part due to their importance as pharmacological targets and in part a result of the key role played by nucleoside natural product inhibitors in providing insight into the physiological significance of the enzymes (26). The core fold of the polytopic PGT superfamily (SCOP MraY-like family PF00953) includes members typified by the Clostridium bolteae MraY (PDB 5JNQ) (27), which has ten TMHs in which the four N-terminal helices have an antiparallel topology and flank the six C-terminal helices which form two interlocking tandem repeats of three helices each (28). The Homo sapiens polyPGT known as UDP-N-acetylglucosamine-dolichyl-phosphate N-acetylglucosamine phosphotransferase (DPAGT1: PDB 6FWZ) (29, 30) shares the core fold of MraY with an excursion preceding the ultimate helix. Despite the low sequence identity between these prokaryotic and eukaryotic polyPGTs (16% identity; 29.7% similarity) their structural homology is clear (RMSD 3.3 Å). Thus far, members of the polyPGT superfamily have been shown to be dimeric (27–30) with a common interface (although the residue composition differs), supporting enzyme stability within each of the active protomers (29).
Elucidating how the polyPGT active site supports the catalytic mechanism has been of significant interest to gain insight into inhibitor design. As with all two-substrate enzymes, the polyPGTs may proceed through either the formation of a ternary complex, in which both substrates are concurrently bound to the enzyme, or a ping-pong mechanism, in which reaction of the first substrate releases the first product and creates a covalent intermediate that is subsequently acted upon by the second substrate. Originally, there were conflicting reports regarding the mechanism of the polyPGT-catalyzed reaction, with some of the original studies hampered by the difficulty of obtaining purified samples of the challenging integral membrane enzymes. However, in 2016, several key experiments with pure enzyme elegantly and convincingly demonstrated that the polyPGT reaction follows the ternary complex model (Figure 3A). Liu et al. (31) probed the mechanism through isotope exchange studies, in which Bacillus subtilis MraY was incubated with [14N2]-UDP-MurNAc-pentapeptide and [15N2]-UMP, in the presence and absence of the PrenP substrate, followed by LC/MS analysis. In the presence of the PrenP, both [15N2]-UDP-MurNAc-pentapeptide and [14N2]-UMP were observed, indicating that isotope exchange had occurred. However, these products were not detected in the absence of PrenP, thus supporting a ternary complex. Additionally, steady-state kinetic analysis fits to a random binding model where complex formation with both UDP-MurNAc-pentapeptide and PrenP substrates are required for UMP and Lipid I product release. Al-Dabbagh et al. (32) used isotopic-exchange studies, with [14C]-labeled reactants and products, to draw similar conclusions. Furthermore, the potency of tunicamycin, the mureidomycins, and other bisubstrate analogs as inhibitors of polyPGTs supports the one-step, ternary-complex reaction mechanism (26).
Figure 3. Summary of mechanistic studies on poly- and monoPGT enzymes.
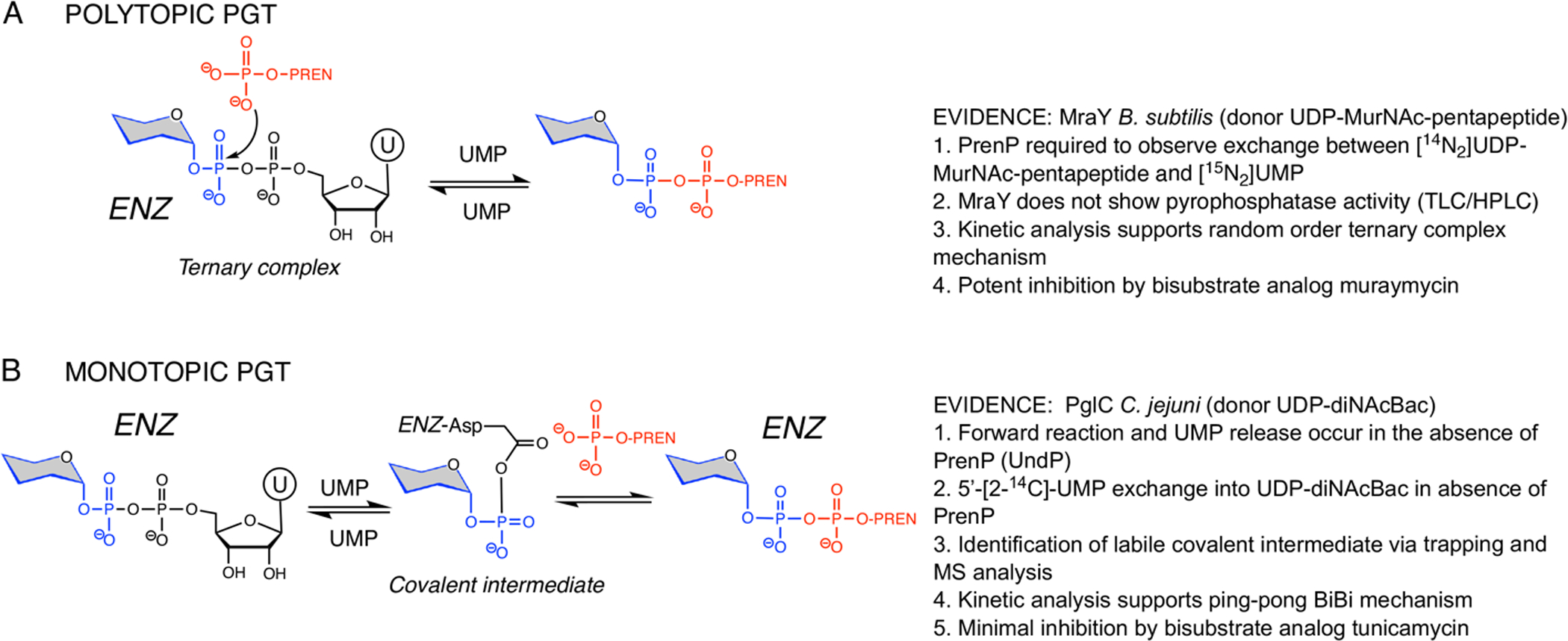
A. Experiments leading to mechanistic model and assignment of ternary complex (random binding) mechanism for the polyPGT MraY from Bacillus subtilis (31, 32). B. Experiments leading to mechanistic model and assignment of Bi-Bi ping-pong mechanism for PglC from Campylobacter jejuni (33).
In contrast to the polyPGTs, the monoPGT enzymes, typified by Campylobacter concisus PglC and Salmonella enterica WbaP differ dramatically in both structure and mechanism. The monoPGT enzymes share a core catalytic fold (11) where the signature structural motif is a α-helix-associated β-hairpin (designated the AHABh motif, PDB 5W7L) (12). The long β-hairpin of the AHABh motif is joined to two of the three amphipathic helices and to an extended loop structure and a double-twisted loop. These secondary structure elements make up the cytoplasmic module of the enzyme. At the N-terminus, two α-helices form a helix-break-helix motif comprising the RMH, which together with hydrophobic portions of the three amphipathic helices, make up the membrane-embedded module (thumbnail, Figure 1). The use of the RMH and amphipathic helices parallel to the membrane surface enforces interaction with a single leaflet and hence the monotopic membrane topology for which the superfamily is named.
Perhaps it is not surprising that the difference in molecular architecture of the polytopic and monotopic PGT superfamilies is mirrored by a divergence in the enzymatic mechanisms. In the monoPGT superfamily the same fundamental two-substrate reaction is catalyzed through a covalent phosphoglycosyl intermediate formed on the carboxyl group of an active site aspartyl residue (Figure 3B). The aspartyl-phosphoglycosyl intermediate was positively identified via reductive cleavage with sodium [3H]-borohydride (33), consistent with the known reactivity of the acyl-phosphate bond toward negatively charged nucleophiles (34). Moreover, in contrast to the findings in the case of the polyPGTs, Das et al. showed exchange of labeled 5’-[2-14C]-UMP into substrate sugar donor UDP-diNAcBac in the absence of the acceptor UndP, providing further support for a Bi-Bi ping-pong mechanism (33). It is also noteworthy that the monoPGTs are not inhibited by nucleoside antibiotics such as tunicamycin (35) which are most effective against the enzymes exhibiting a ternary complex mechanism.
A CATALYTIC CONUMDRUM
Is there an evolutionary driving force for the recruitment or invention of two different scaffolds to catalyze the same chemical reaction? As discussed below, this dissimilarity in scaffold could allow for differential regulation of two pathways with a shared membrane-embedded substrate (36) (Figures 4A and 5A). Parenthetically, the linear PrenPs comprise a very low percentage of the lipidic fraction of membranes, approximately 0.1 %, creating the need for strict regulation to control use of the precious secondary metabolite (6, 37). Furthermore, the particular scaffold may, consequentially, be more suited to a Bi-Bi ping-pong versus ternary-complex mechanism. Previous bioinformatic and mutagenesis studies of polytopic family members MraY and WecA identified a conserved motif of aspartyl residues across the superfamily, which is essential for catalysis and was proposed to provide the Mg2+-coordinating residues and nucleophile (Asp93, 94 and 231, respectively, C. bolteae numbering throughout) by analogy to prenyl transferases and terpene cyclases (38). Later, the Aquifex aeolicus structure of MraY (PDB 4J72) revealed that, of these residues, only Asp231 is within distance to coordinate to a bound Mg2+ ion (28). Overlay of the structure of MraY with that of human DPAGT1 (PDB 6FWZ, V264G variant (Figure 4A)) in complex with UDP-GlcNAc and Mg2+ ion shows that the other protein residue that coordinates to Mg2+ corresponds to Asn172 (29). Notably, these divalent cation-coordinating residues are two of only 12 amino acids conserved in all prokaryotic and eukaryotic sequences of the polyPGTs (38). The Mg2+ can act to neutralize the negative charge in the near-attack conformation and in the transition state. In ternary-complex formation, catalysis by approximation is expected to contribute significantly to reactivity and Mg2+ potentially plays a role in orienting the incoming UndP phosphoryl group with respect to the electrophilic phosphorus center (as judged by the relative positions in an overlay of UDP-GlcNAc and Mg2+ (PDB 6FWZ) and the branched chain acyl moiety of tunicamycin (PDB 6BW6) (Figure 4A, inset)). The co-positioning of two substrates is critical to the catalytic mechanism. Examination of the relative disposition of the polyprenyl moiety with respect to the electrophilic phosphate group of the activated sugar highlights catalytic residues. In terms of catalytic strategy, such SN2-type direct displacement reactions are enabled by an enzymatic general-acid catalyst activating the PrenP for attack, potentially with Lewis-acid activation of the departing UMP leaving group. Lys111 and Lys97, respectively, are positioned to potentially play these roles (Figure 4B). With a pKa of 6.4, the UMP leaving-group protonation is expected to be facile.
Figure 4. Structure and mechanism of the polyPGTs.
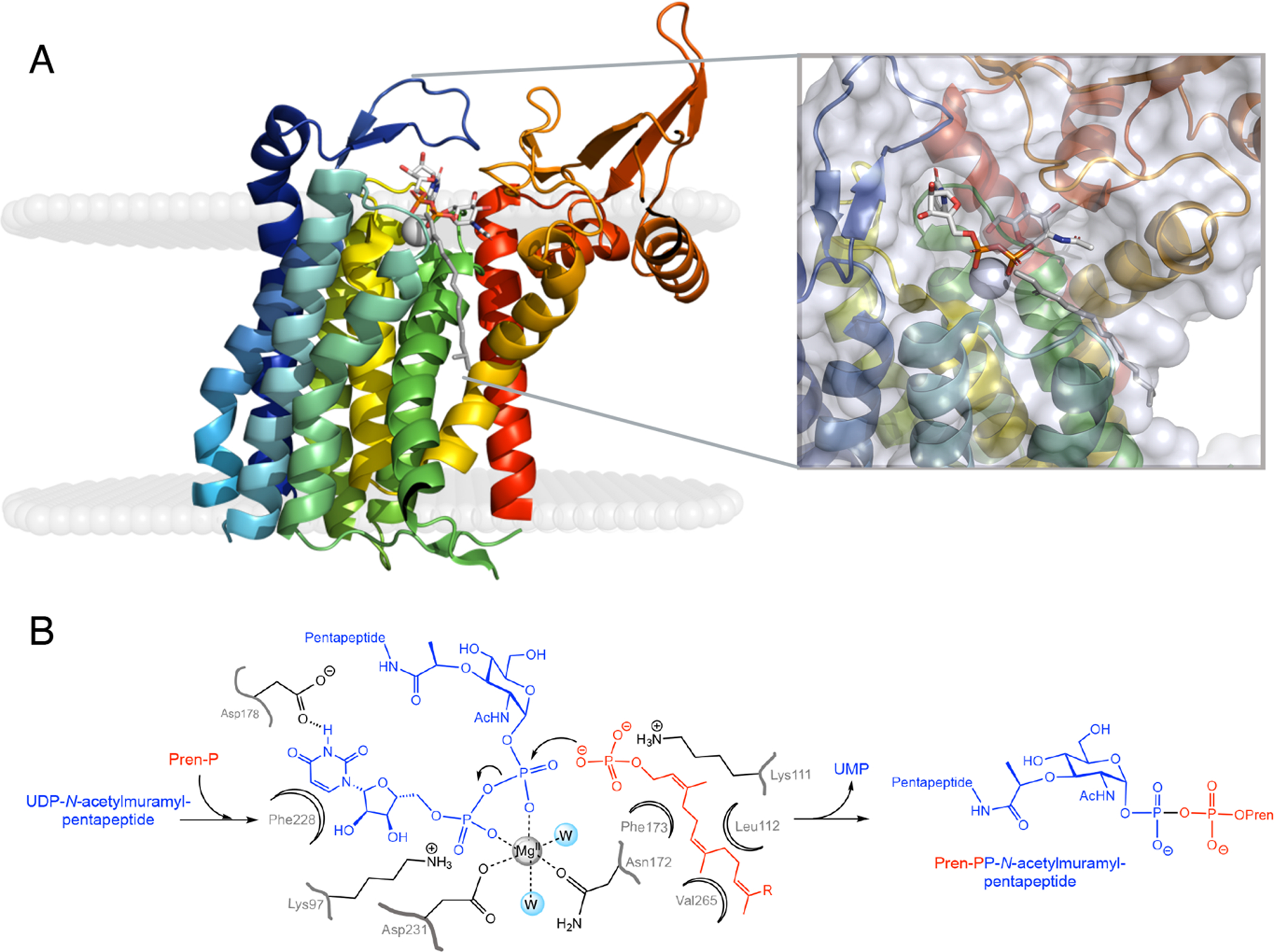
A. Ribbon diagram of polyPGT DPAGT1 (PDB 6FWZ) color ramped from N- to C- terminus (blue to red) shown in membrane as calculated using the OPM server (43) with UDP-GlcNAc (white stick) and Mg2+ ion (grey sphere). Also shown is polyprenyl moiety (grey stick) from superposition of tunicamycin bound complex (PDB 6BW6). Inset shows close-up view of active site with protein transparent surface. B. Ternary complex mechanism of polyPGTs with proposed roles for binding and catalytic residues (numbering from Clostridium bolteae MraY).
Figure 5. Structure and mechanism of the monoPGTs.
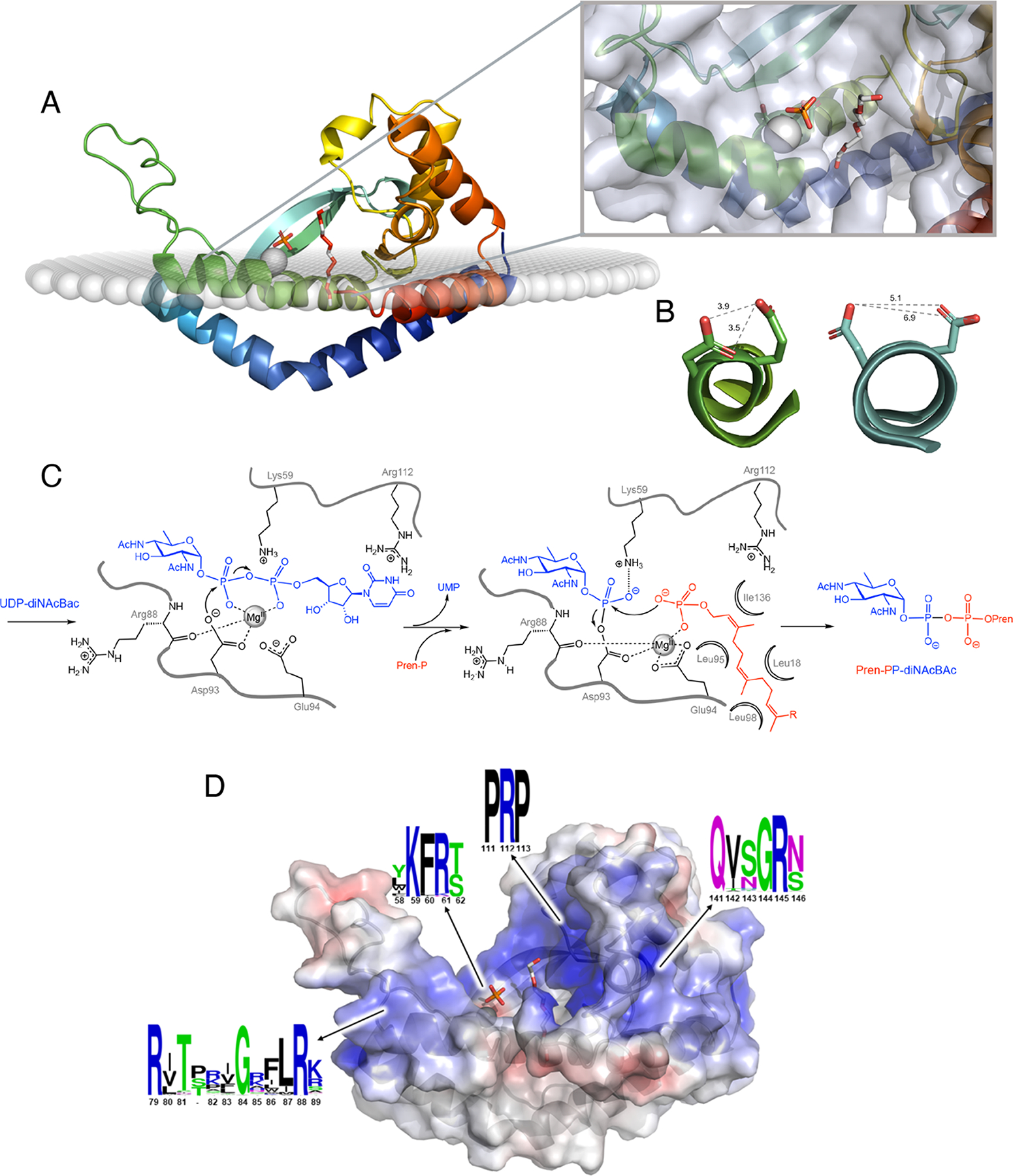
A. Ribbon diagram of monoPGT PglC (PDB 5W7L) color ramped from N- to C- terminus (blue to red shown in membrane as calculated using the OPM server (43) with Mg2+ ion (grey sphere), phosphate and PEG (white stick; PEG molecule shows proposed position for UndP binding). Inset shows close-up view of active site with protein transparent surface. B. Active-site helix 310-geometry allows for co-facial positioning of Asp-Glu catalytic dyad. MonoPGT PglC residues 91–99 shown in ribbon representations colored as in panel A with side chains of the catalytic dyad Asp93-Glu94 shown as stick. PolyPGT DPAGT1 (PDB 6FWZ) residues 100–118 shown in ribbon representation colored as in Figure 4A with residues Asp115 and Asp116 from the conserved aspartyl motif shown as stick. In polyPGTs these residues do not play an analogous role in catalysis (see text). Shortest distances between oxygen atoms on proximal residues (gray dashed lines). C. Bi-Bi ping-pong mechanism of monoPGTs with proposed roles for binding and catalytic residues (numbering from Campylobacter concisus PglC). D. Electrostatic mapping of PglC revealing positive funnel for substrate binding. Ribbon diagram of PglC with transparent surface colored by electrostatic potential ramped from −5 kT/e (red) to 5 kT/e (blue). Ligands shown as in panel A. The sequence conservation of positive pockets was calculated using sequences of all three classes of monoPGT (small, large, and bifunctional), visualized using WebLogo (C. concisus PglC numbering).
In the monoPGTs, a mixed carboxylic–phosphoric anhydride intermediate kinetically and thermodynamically activates the phosphoryl group towards nucleophilic substitution, allowing the observed stepwise chemical transformation (33). In the second half-reaction, the electron-withdrawing aspartyl acyl group increases the electrophilicity of the phosphorus atom and lowers the energy of the transition state by stabilizing the developing negative charge on the aspartyl leaving group. The thermodynamic stability of such intermediates has been explained, in part, by protection of the active site from bulk solvent (39). Within the Asp93-Glu94 dyad (C. concisus PglC numbering), Asp has lower side-chain entropy than Glu (40) and this may be a driving force for its selection as the nucleophile, to attain greater catalytic efficiency (41). Additional orientation and activation of the attacking aspartyl carboxylate is afforded by positioning of Asp93 as the “cap” of a 310 helix. A kink formed by Pro96 in concert with residues with low helical propensity (Ser91, Asp93, Glu94) distorts the N-terminal end of the helix (residues 92–96) into this 310-helix geometry. In such a helix, each turn, defined by the hydrogen-bonding partners, comprises ten atoms, making it more tightly-wound than a canonical α-helix. As a result, the catalytic Asp93-Glu94 dyad is brought into closer proximity than would be possible for two residues that are adjacent in primary structure in a typical α-helix (see Figure 5B for distances and comparison with a canonical α-helix). This proximity allows the proposed shift in coordination of the Mg2+ cofactor, facilitating the attack of the PrenP phosphoryl oxygen on the phosphoglycosyl aspartyl intermediate (Figure 5C). Moreover, participation of the Asp93 side-chain carboxylate oxygen in a helix-capping hydrogen bond, in addition to coordination of the catalytically-required Mg2+ ion, contributes to the nucleophilic reactivity of the non-coordinating oxygen via a disruption of resonance stabilization.
As it does for the polyPGTs, the Mg2+ cofactor in the monoPGTs can enhance electrophilicity at the phosphorus center by neutralizing the negative charge in the transition state and can serve to co-position the nucleophile and electrophile and to increase the acidity of the leaving group. The charge on the nucleophilic Asp carboxylate is shielded from the electrophilic center by the Mg2+ cofactor and side-chain functionality. Moreover, the divalent cation shields electrostatic repulsion of the attacking PrenP phosphate from the phosphorus center in the second half-reaction. Model studies using isotopic labeling on acetyl phosphate show that the Mg2+ ion favors P-O cleavage whereas with Ca2+, C-O cleavage predominates (42). Thus, the metal ion cofactor is critical to both reactivity and selectivity of the second half reaction.
THE SCAFFOLD AS A STAGE FOR REACTION AT THE INTERFACE
The TMHs in the polyPGT scaffold culminate in loops that serve to form the active site, positioning the binding site for the water-soluble nucleotide-activated substrate at the interface for reaction with the membrane-resident PrenP substrate. Rotational and translational calculation of the placement of DPAGT1 (PDB 6FWZ) in the membrane environment using the OPM server (43) places the electrophilic phosphoryl moiety of the bound UDP-GlcNAc coincident with the lipid headgroups (Figure 4A). This allows the co-positioning of this group with the nucleophilic PrenP phosphoryl moiety in the ternary complex without the energetically unfavorable removal of the PrenP prenyl chain from the hydrophobic lipid-tail environment (44, 45). In the monoPGTs, positioning of the Asp-Glu catalytic dyad on an amphipathic helix, likewise, positions the active site at the cytoplasmic interface with the membrane, estimated to be ~3Å from the membrane surface in C. concisus PglC (44) (Figure 5A). This “leave in place” strategy, describing the location of active sites, either at the interface or at a distance from the interface that does not necessitate extraction of the hydrophobic moiety of the membrane-resident substrate, is also utilized in other non-homologous monotopic enzymes such as WaaA (46) and PglH (47).
A difference in strategy for substrate colocalization is revealed by the analysis of the conservation of cationic residues within the mono- and polyPGT superfamilies. In the monoPGTs there is conservation of motifs containing arginine across all three families which act to frame and control the electrostatic environment of the active site. Mapping of the electrostatic charge onto the molecular structure of C. concisus PglC reveals numerous basic residues forming a positive “electrostatic funnel” poised for binding and orienting the negatively-charged phosphate-rich UDP-diNAcBac substrate (Figure 5D). Such cation-rich motifs are not revealed in the analysis of the polyPGTs and therefore, substrate binding may be more dependent upon colocation of these metabolites.
REGULATION - LEVELING THE PLAYING FIELD
Control of specific enzyme catalysts becomes more critical in pathways that share a common substrate. Indeed, studies using variants of enzymes in the outer membrane glycolipid pathway in E. coli suggest that there is competition for the pool of PrenP, wherein appropriate distribution among competing metabolic pathways is required to maintain cell shape (36). Examination of the steady-state kinetic constants for prototypes from the two PGT superfamilies highlights the necessity for control. A global approach utilizing omics data confirms that in vitro enzyme assays generally accurately reflect the maximal rates of enzyme-catalyzed reactions in vivo (48). A comparison of either kcat, which defines the reaction rate of the PGTs under saturating substrate concentrations or of kcat/Km, which defines the reaction rate under low substrate conditions, finds that, unchecked, the monoPGT-catalyzed reaction would be faster than that of the polyPGT. For C. jejuni PglC kcat is 3.7-fold greater than that of B. subtilis MraY and the kcat/KMUndP is 50-fold greater (31, 33). Thus, differential regulation of the two pathways is critical, lest the rare PrenP substrate be preferentially partitioned into glycoconjugate pathways initiated by the monoPGT reactions. Having different scaffolds associated with each pathway facilitates such regulation. Using similar arguments, availability of sugar substrates could also affect pathway flux.
Glycoconjugate biosynthesis pathways may be regulated either at the transcriptional level or at the enzyme level via post-translational modification (see Table 1). Transcriptional regulation of genes encoding enzymes in glycoconjugate assembly is employed in numerous pathways as an effective means of glycoconjugate regulation in response to environmental and cellular cues. Pathway regulation in bacteria is also contingent on a common pool of the essential, but limiting, PrenP lipid carrier substrate (36). In this context, the PGT-catalyzed step itself is reversible, therefore problematic sequestration of the PrenPP-linked substrates occurs from dysregulation of subsequent steps further downstream of the PGT reaction in the pathway. In vivo studies of wall teichoic acid biosynthesis in B. subtilis, initiated by the polyPGT TagO (homologous to S. aureus TarO) have shown that inactivation of later genes in the biosynthetic pathway leads to buildup of dead-end PrenPP-linked products, which is lethal because the PrenP is sequestered from the accessible pool (49). In contrast, suppressor mutations of the CpsE (large monoPGT) gene in the Staphylococcus pneumoniae CPS biosynthesis pathway, prevent buildup of PrenPP-linked products but results in reduction of CPS levels on the outer membrane (50, 51). The suppressor mutations occur in both the PGT core domain and the cytoplasmic domain of unknown function, emphasizing the functional importance of the cytoplasmic domain.
Table 1.
Regulation of PGT-initiated pathways
| Regulatory Protein/Receiver Domain | PGT | Organism | Target protein | Regulated step | Effect of Regulation | Pathway |
|---|---|---|---|---|---|---|
| Polytopic Pathways | ||||||
| PknB (PASTA-eSTKs) | MraY | Mycobacterium tuberculosis | GlmU | UDP-GlcNAc synthesis | reduced activity | PG |
| MviN | glycan translocation | inhibited by FhaA | PG | |||
| PonA1 | transglycosylation/transpeptidation | activation | PG | |||
| CwlM | lipid II export via MurJ | cellular localization, activation of MurA | PG | |||
| PrkA (PASTA-eSTKs) | MraY | Listeria monocytgenes | YvcK | cell-wall homeostasis | unknown | unknown |
| GpsB | transglycosylation/transpeptidation | ClpCP-dependent degradation of MurA | PG | |||
| Wnt/β-catenin | DPAGT1 | Homo sapiens | --- | gene expression | increased N-glycosylation of E-cadherin | N-glycosylation |
| Monotopic Pathways | ||||||
| RcsB/RcsC | WcaJ | Escherichia coli | RcsA | gene expression | RcsA regulates gene expression | colanic acid |
| CapB/CapA | CapM | Staphylococcus aureus | CapO | UDP-ManNAc synthesis | activation | CPS |
| CapM | phosphoglycosyl transfer | increases activity | CPS | |||
| CapE | UDP-FucNAc synthesis | increases activity | CPS | |||
| Wzc | WcaJ | Escherichia coli | Ugd | UDP-glucuronic acid synthesis | increases activity | colanic acid |
| CpsD/CpsC | CpsE | Streptococcus pneumoniae | unknown | unknown | regulates CPS length | CPS |
| EpsD/EpsC | EpsE | Streptococcus thermophilus | EpsE | phosphoglycosyl transfer | activation | EPS |
In another example from E. coli, the two-component system that controls biosynthesis of colanic acid, an exopolysaccharide (EPS), initiated by the monoPGT WcaJ, shows regulation at the transcriptional level. In this case, a transmembrane sensor domain (RcsC), a response regulator (RcsB), and a positive regulator (RcsA) function to modulate the transcription and subsequent expression of colanic acid biosynthetic genes (52–54). At 37 °C, RcsA protein levels are low, and thus colanic acid is not synthesized. However, at lower temperatures or when under stress, the RcsC sensor domain phosphorylates the RscB regulator, which in turn activates RscA, leading to upregulation of genes in the colanic acid biosynthetic pathway. In the yeast Saccharomyces cerevisiae, there is considerable evidence that the entire dolichol pathway is transcriptionally regulated at the alg7 gene (55), which encodes a polyPGT. Also, the mammalian homolog of Alg7, DPAGT1, is regulated at the transcriptional level by the Wnt/β-catenin signaling pathway and upregulation of dpagt1 gene transcription leads to increased N-linked protein glycosylation of E-cadherin (56)
Although there is no evidence of direct regulation of the polyPGT MraY or its orthologs, there are numerous examples of regulation of the downstream enzymes in peptidoglycan biosynthesis. In the Gram-negative exopolysaccharide pathway two-component systems, PASTA-eSTKs are integral membrane proteins with a cytoplasmic kinase domain fused to numerous PASTA receiver domains (57). Stimulated by Lipid II or free muropeptides, PASTA-eSTKs phosphorylate specific enzymes, modulating their respective activities. The action of PknB, a PASTA-eSTK from Mycobacterium tuberculosis includes modulation of flippase, uridyltransferase/acetyltransferase and export activities (58–63). Listeria monocytogenes PrkA, an M. tuberculosis PknB homolog, phosphorylates YvcK and GpsB (64, 65), which are involved in cell-wall homeostasis and assembly of class A penicillin-binding proteins, respectively. Phosphorylation of these targets in M. tuberculosis and L. monocytogenes lead to regulation of peptidoglycan at numerous steps and via processes including inhibition of activity and changes in cellular localization.
There are only a few definitive studies on systems showing direct regulation of glycoconjugate biosynthesis at the enzyme level, although this has been proposed in many cases. However, it is likely that regulation specifically at the PGT step would be favorable as it would control use of a limited pool of PrenP substrate across numerous pathways. In fact, the exopolysaccharide biosynthesis tyrosine kinase superfamily (InterPro IPR005702) comprises bacterial protein kinases which specifically phosphorylate enzymes in the monoPGT-associated exopolysaccharide (EPS) biosynthetic pathways (see Table 1) (21, 66–68). Gram-negative bacteria may express one protein with both the receiver domain and the kinase domain, as exemplified by the E. coli tyrosine kinase Wzc. Alternatively, Gram-positive bacteria, such as S. aureus, have two distinct proteins acting as the intrinsic membrane receiver domain (CapA) and the tyrosine kinase (CapB). The targets of these kinases include the monoPGTs, CapM (21) and EpsE (68), as well as the sugar-modifying enzymes, UDP-Glc dehydrogenase (Ugd) (67), UDP-ManNAc dehydrogenase (CapO) (69), and UDP-GlcNAc dehydratase (CapE) (21), where phosphorylation activates or increases activity. In addition to the kinase, a complementary phosphatase is often encoded in the same operon, to fine-tune pathway activity. In the case of S. aureus CPS, the phosphatase CapC dephosphorylates both CapE and CapM (21). Notably, in vitro analyses of CapM also identified it as a substrate of the PASTA-eSTK PknB at a conserved threonine. This phosphorylation serves to negatively regulate CapM, based on the decrease in lipid Icap production measured in vitro, allowing for cross-pathway regulation of peptidoglycan and capsular polysaccharide biosynthesis (21).
Together, the S. aureus CPS pathway employs positive and negative regulation of glycoconjugate biosynthesis at key steps throughout the pathway. These are the first membrane-committed, monoPGT-dependent steps and two sugar-modifying steps that control NDP-sugar substrate production. Such regulation allows for tight control of common substrate pools (PrenP and UDP-GlcNAc, respectively). Notably, these regulatory events in pathways producing essential cellular wall components or necessary virulence factors in bacteria may serve to control the pathogenesis of these organisms (70). Further, in commensal bacteria, regulation of CPS or EPS production may modulate their respective host-cell interactions (71).
CONCLUSIONS
In nature, the polyprenol phosphate phosphoglycosyl transferases carry out a strategic transformation at the inception of membrane-associated, en bloc glycoconjugate biosynthesis pathways. Until relatively recently, the polyPGTs, which are found in eukaryotes and prokaryotes, were believed to represent the principal enzymes catalyzing phospho-sugar transfer to PrenPs. However, biochemical and biophysical evidence has emerged revealing that the polyPGT superfamily is juxtaposed with a distinct monoPGT superfamily that is exclusive to prokaryotes. Therefore, two PGT superfamilies, the polyPGTs and monoPGTs, have evolved to carry out an identical chemical transformation using completely different structural scaffolds and chemical mechanisms. This evolutionary “sleight of hand” is particularly remarkable given the significant physical constraints on the transformation that they catalyze. Poly- and monoPGTs are integral membrane proteins that catalyze phospho-sugar transfer from a soluble negatively-charged NDP-sugar substrate to an amphiphilic linear polyprenol phosphate, which is most favorably positioned with the nucleophilic phosphate directly at the membrane interface. Extracting the PrenP from the membrane represents a formidable task and therefore, both PGT superfamilies have simply elected a “leave in place” strategy. But, this is where the similarity ends.
The difference between the mechanisms, with intermediacy of a ternary complex with NDP-sugar substrate and PrenP versus a phosphoglycosyl aspartyl intermediate, leads to very different affinity profiles for inhibitory nucleoside natural product ligands. The exclusive occurrence of the monoPGTs in prokaryotes, coupled to the opportunities afforded by the dissimilar scaffolds, opens the way to design inhibitors that distinguish these PGTs, in ways that substrates do not. Moreover, each PGT superfamily makes use of the low-abundance, but highly-conserved PrenP substrate, which anchors the product facilitating subsequent steps at the membrane. However, the binding mode through potential interactions with a transmembrane versus a re-entrant membrane helix must differ. This dichotomy offers an embarrassment of riches for the study of evolutionary pressures on the scaffolds themselves and local interplay of these fascinating enzymes with the membrane environment and in the presence of the membrane-associated PrenP substrate.
PERSPECTIVES.
The existence of structurally- and mechanistically-distinct PGT superfamilies at the inception of glycoconjugate biosynthesis raises important questions related to the evolution of each superfamily and the biological imperative of this dichotomy.
There is a need for additional high-resolution structures of mono- and polyPGTs in native-like membrane environments with bound ligands to reveal aspects of catalysis and regulation and for structure-guided inhibitor design. The definition of the monoPGT superfamily as prokaryote-specific, offers opportunities for therapeutic targeting of monoPGTs in bacterial survival and virulence-associated pathways.
Although the catalytic mechanisms of the two PGT superfamilies are divergent, both employ linear polyprenol phosphate substrates and a “leave-in-place” strategy for action at the interface with this membrane-embedded substrate, pointing to evolution-driven necessity for scaffold recruitment and the possibility of constraints imposed by co-evolution with the membrane environment.
Funding
This work was funded by National Institutes of Health grant R01 GM131627 (to K.N.A and B.I.) and GM039334 (B.I.).
Abbreviations
- CPS
capsular polysaccharide
- EPS
exopolysaccharide
- LPS
Lipopolysaccharide
- PASTA-eSTKs
penicillin-binding protein and serine/threonine kinase associated domain-containing eukaryotic-like serine/threonine protein kinases
- monoPGT
monotopic phosphoglycosyl transferase
- PG
peptidoglycan
- PGT
phosphoglycosyl transferase
- polyPGT
polytopic phosphoglycosyl transferase
- PrenP
polyprenol phosphate
- RMH
reentrant membrane helix
- SSN
sequence similarity network
- TMH
transmembrane helix
- UndP
undecaprenol phosphate
- WTA
wall teichoic acid
Footnotes
Competing Interests
The authors declare that there are no competing interests associated with the manuscript.
REFERENCES
- 1.Kay E, Lesk VI, Tamaddoni-Nezhad A, Hitchen PG, Dell A, Sternberg MJ, et al. Systems analysis of bacterial glycomes. Biochem Soc Trans. 2010;38(5):1290–3. [DOI] [PubMed] [Google Scholar]
- 2.Tytgat HL, Lebeer S. The sweet tooth of bacteria: common themes in bacterial glycoconjugates. Microbiol Mol Biol Rev. 2014;78(3):372–417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Varki A Biological roles of glycans. Glycobiology. 2017;27(1):3–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Jones MB, Rosenberg JN, Betenbaugh MJ, Krag SS. Structure and synthesis of polyisoprenoids used in N-glycosylation across the three domains of life. Biochim Biophys Acta. 2009;1790(6):485–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Surmacz L, Swiezewska E. Polyisoprenoids - Secondary metabolites or physiologically important superlipids? Biochem Biophys Res Commun. 2011;407(4):627–32. [DOI] [PubMed] [Google Scholar]
- 6.Hartley MD, Imperiali B. At the membrane frontier: a prospectus on the remarkable evolutionary conservation of polyprenols and polyprenyl-phosphates. Arch Biochem Biophys. 2012;517(2):83–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Valtersson C, van Duyn G, Verkleij AJ, Chojnacki T, de Kruijff B, Dallner G. The influence of dolichol, dolichol esters, and dolichyl phosphate on phospholipid polymorphism and fluidity in model membranes. J Biol Chem. 1985;260(5):2742–51. [PubMed] [Google Scholar]
- 8.Janas T, Walinska K, Chojnacki T, Swiezewska E, Janas T. Modulation of properties of phospholipid membranes by the long-chain polyprenol (C(160)). Chem Phys Lipids. 2000;106(1):31–40. [DOI] [PubMed] [Google Scholar]
- 9.Valvano MA. Export of O-specific lipopolysaccharide. Front Biosci. 2003;8:s452–71. [DOI] [PubMed] [Google Scholar]
- 10.Hug I, Feldman MF. Analogies and homologies in lipopolysaccharide and glycoprotein biosynthesis in bacteria. Glycobiology. 2011;21(2):138–51. [DOI] [PubMed] [Google Scholar]
- 11.Lukose V, Walvoort MTC, Imperiali B. Bacterial phosphoglycosyl transferases: Initiators of glycan biosynthesis at the membrane interface. Glycobiology. 2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ray LC, Das D, Entova S, Lukose V, Lynch AJ, Imperiali B, et al. Membrane association of monotopic phosphoglycosyl transferase underpins function. Nat Chem Biol. 2018;14(6):538–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Imperiali B Bacterial carbohydrate diversity - a Brave New World. Curr Opin Chem Biol. 2019;53:1–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Stanley P, Cummings RD. Structures Common to Different Glycans. In: Varki A,; Cummings RD; Esko JD, et al. , editor. Essentials of Glycobiology. 3rd ed. New York: Cold Spring Harbor Laboratory Press; 2017. [Google Scholar]
- 15.Price NP, Momany FA. Modeling bacterial UDP-HexNAc: polyprenol-P HexNAc-1-P transferases. Glycobiology. 2005;15(9):29R–42R. [DOI] [PubMed] [Google Scholar]
- 16.Lehrer J, Vigeant KA, Tatar LD, Valvano MA. Functional characterization and membrane topology of Escherichia coli WecA, a sugar-phosphate transferase initiating the biosynthesis of enterobacterial common antigen and O-antigen lipopolysaccharide. J Bacteriol. 2007;189(7):2618–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.O’Toole KH, Imperiali B, Allen KN. Glycoconjugate pathway connections revealed by sequence similarity network analysis of the monotopic phosphoglycosyl transferases. Proc Natl Acad Sci U S A. 2021;118(4). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lukose V, Luo L, Kozakov D, Vajda S, Allen KN, Imperiali B. Conservation and Covariance in Small Bacterial Phosphoglycosyltransferases Identify the Functional Catalytic Core. Biochemistry. 2015;54(50):7326–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Furlong SE, Ford A, Albarnez-Rodriguez L, Valvano MA. Topological analysis of the Escherichia coli WcaJ protein reveals a new conserved configuration for the polyisoprenyl-phosphate hexose-1-phosphate transferase family. Sci Rep. 2015;5:9178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Brown S, Zhang YH, Walker S. A revised pathway proposed for Staphylococcus aureus wall teichoic acid biosynthesis based on in vitro reconstitution of the intracellular steps. Chem Biol. 2008;15(1):12–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rausch M, Deisinger JP, Ulm H, Muller A, Li W, Hardt P, et al. Coordination of capsule assembly and cell wall biosynthesis in Staphylococcus aureus. Nat Commun. 2019;10(1):1404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bugg TD, Walsh CT. Intracellular steps of bacterial cell wall peptidoglycan biosynthesis: enzymology, antibiotics, and antibiotic resistance. Nat Prod Rep. 1992;9(3):199–215. [DOI] [PubMed] [Google Scholar]
- 23.van Selm S, Kolkman MAB, van der Zeijst BAM, Zwaagstra KA, Gaastra W, van Putten JPM. Organization and characterization of the capsule biosynthesis locus of Streptococcus pneumoniae serotype 9 V. Microbiology (Reading) 2002;148(Pt 6):1747–55. [DOI] [PubMed] [Google Scholar]
- 24.Linton D, Dorrell N, Hitchen PG, Amber S, Karlyshev AV, Morris HR, et al. Functional analysis of the Campylobacter jejuni N-linked protein glycosylation pathway. Mol Microbiol. 2005;55(6):1695–703. [DOI] [PubMed] [Google Scholar]
- 25.Glover KJ, Weerapana E, Imperiali B. In vitro assembly of the undecaprenylpyrophosphate-linked heptasaccharide for prokaryotic N-linked glycosylation. Proc Natl Acad Sci U S A. 2005;102(40):14255–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Brandish PE, Kimura KI, Inukai M, Southgate R, Lonsdale JT, Bugg TD. Modes of action of tunicamycin, liposidomycin B, and mureidomycin A: inhibition of phospho-N-acetylmuramyl-pentapeptide translocase from Escherichia coli. Antimicrob Agents Chemother. 1996;40(7):1640–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hakulinen JK, Hering J, Branden G, Chen H, Snijder A, Ek M, et al. MraY-antibiotic complex reveals details of tunicamycin mode of action. Nat Chem Biol. 2017;13(3):265–7. [DOI] [PubMed] [Google Scholar]
- 28.Chung BC, Zhao J, Gillespie RA, Kwon DY, Guan Z, Hong J, et al. Crystal structure of MraY, an essential membrane enzyme for bacterial cell wall synthesis. Science. 2013;341(6149):1012–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Dong YY, Wang H, Pike ACW, Cochrane SA, Hamedzadeh S, Wyszynski FJ, et al. Structures of DPAGT1 Explain Glycosylation Disease Mechanisms and Advance TB Antibiotic Design. Cell. 2018;175(4):1045–58 e16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yoo J, Mashalidis EH, Kuk ACY, Yamamoto K, Kaeser B, Ichikawa S, et al. GlcNAc-1-P-transferase-tunicamycin complex structure reveals basis for inhibition of N-glycosylation. Nat Struct Mol Biol. 2018;25(3):217–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Liu Y, Rodrigues JP, Bonvin AM, Zaal EA, Berkers CR, Heger M, et al. New Insight into the Catalytic Mechanism of Bacterial MraY from Enzyme Kinetics and Docking Studies. J Biol Chem. 2016;291(29):15057–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Al-Dabbagh B, Olatunji S, Crouvoisier M, El Ghachi M, Blanot D, Mengin-Lecreulx D, et al. Catalytic mechanism of MraY and WecA, two paralogues of the polyprenyl-phosphate N-acetylhexosamine 1-phosphate transferase superfamily. Biochimie. 2016;127:249–57. [DOI] [PubMed] [Google Scholar]
- 33.Das D, Kuzmic P, Imperiali B. Analysis of a dual domain phosphoglycosyl transferase reveals a ping-pong mechanism with a covalent enzyme intermediate. Proc Natl Acad Sci U S A. 2017;114(27):7019–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Degani C, Boyer PD. A borohydride reduction method for characterization of the acyl phosphate linkage in proteins and its application to sarcoplasmic reticulum adenosine triphosphatase. J Biol Chem. 1973;248(23):8222–6. [PubMed] [Google Scholar]
- 35.Glover KJ, Weerapana E, Chen MM, Imperiali B. Direct biochemical evidence for the utilization of UDP-bacillosamine by PglC, an essential glycosyl-1-phosphate transferase in the Campylobacter jejuni N-linked glycosylation pathway. Biochemistry. 2006;45(16):5343–50. [DOI] [PubMed] [Google Scholar]
- 36.Jorgenson MA, Kannan S, Laubacher ME, Young KD. Dead-end intermediates in the enterobacterial common antigen pathway induce morphological defects in Escherichia coli by competing for undecaprenyl phosphate. Mol Microbiol. 2016;100(1):1–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Entova S, Guan Z, Imperiali B. Investigation of the conserved reentrant membrane helix in the monotopic phosphoglycosyl transferase superfamily supports key molecular interactions with polyprenol phosphate substrates. Arch Biochem Biophys. 2019;675:108111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lloyd AJ, Brandish PE, Gilbey AM, Bugg TD. Phospho-N-acetyl-muramyl-pentapeptide translocase from Escherichia coli: catalytic role of conserved aspartic acid residues. J Bacteriol. 2004;186(6):1747–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wolfenden R, Liang YL. Influences of solvent on group transfer potentials and biochemical recognition of carbohydrates. Anomalous solvation of the anomeric hydroxyl group. J Biol Chem. 1988;263(17):8022–6. [PubMed] [Google Scholar]
- 40.Pickett SD, Sternberg MJ. Empirical scale of side-chain conformational entropy in protein folding. J Mol Biol. 1993;231(3):825–39. [DOI] [PubMed] [Google Scholar]
- 41.Allen KN, Dunaway-Mariano D. Catalytic scaffolds for phosphoryl group transfer. Curr Opin Struct Biol. 2016;41:172–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Klinman JP, Samuel D. Oxygen-18 studies to determine the position of bond cleavage of acetyl phosphate in the presence of divalent metal ions. Biochemistry. 1971;10(11):2126–31. [DOI] [PubMed] [Google Scholar]
- 43.Lomize MA, Pogozheva ID, Joo H, Mosberg HI, Lomize AL. OPM database and PPM web server: resources for positioning of proteins in membranes. Nucleic Acids Res. 2012;40(Database issue):D370–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Allen KN, Entova S, Ray LC, Imperiali B. Monotopic Membrane Proteins Join the Fold. Trends Biochem Sci. 2019;44(1):7–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Allen KN, Imperiali B. Structural and mechanistic themes in glycoconjugate biosynthesis at membrane interfaces. Curr Opin Struct Biol. 2019;59:81–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Schmidt H, Hansen G, Singh S, Hanuszkiewicz A, Lindner B, Fukase K, et al. Structural and mechanistic analysis of the membrane-embedded glycosyltransferase WaaA required for lipopolysaccharide synthesis. Proc Natl Acad Sci U S A. 2012;109(16):6253–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ramirez AS, Boilevin J, Mehdipour AR, Hummer G, Darbre T, Reymond JL, et al. Structural basis of the molecular ruler mechanism of a bacterial glycosyltransferase. Nat Commun. 2018;9(1):445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Davidi D, Noor E, Liebermeister W, Bar-Even A, Flamholz A, Tummler K, et al. Global characterization of in vivo enzyme catalytic rates and their correspondence to in vitro kcat measurements. Proc Natl Acad Sci U S A. 2016;113(12):3401–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.D’Elia MA, Millar KE, Bhavsar AP, Tomljenovic AM, Hutter B, Schaab C, et al. Probing teichoic acid genetics with bioactive molecules reveals new interactions among diverse processes in bacterial cell wall biogenesis. Chem Biol. 2009;16(5):548–56. [DOI] [PubMed] [Google Scholar]
- 50.Xayarath B, Yother J. Mutations blocking side chain assembly, polymerization, or transport of a Wzy-dependent Streptococcus pneumoniae capsule are lethal in the absence of suppressor mutations and can affect polymer transfer to the cell wall. J Bacteriol. 2007;189(9):3369–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.James DB, Gupta K, Hauser JR, Yother J. Biochemical activities of Streptococcus pneumoniae serotype 2 capsular glycosyltransferases and significance of suppressor mutations affecting the initiating glycosyltransferase Cps2E. J Bacteriol. 2013;195(24):5469–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Stout V Identification of the promoter region for the colanic acid polysaccharide biosynthetic genes in Escherichia coli K-12. J Bacteriol. 1996;178(14):4273–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Whitfield C, Roberts IS. Structure, assembly and regulation of expression of capsules in Escherichia coli. Mol Microbiol. 1999;31(5):1307–19. [DOI] [PubMed] [Google Scholar]
- 54.Whitfield C, Paiment A. Biosynthesis and assembly of Group 1 capsular polysaccharides in Escherichia coli and related extracellular polysaccharides in other bacteria. Carbohydr Res. 2003;338(23):2491–502. [DOI] [PubMed] [Google Scholar]
- 55.Kukuruzinska MA, Lennon-Hopkins K. ALG gene expression and cell cycle progression. Biochim Biophys Acta. 1999;1426(2):359–72. [DOI] [PubMed] [Google Scholar]
- 56.Sengupta PK, Bouchie MP, Kukuruzinska MA. N-glycosylation gene DPAGT1 is a target of the Wnt/beta-catenin signaling pathway. J Biol Chem. 2010;285(41):31164–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Manuse S, Fleurie A, Zucchini L, Lesterlin C, Grangeasse C. Role of eukaryotic-like serine/threonine kinases in bacterial cell division and morphogenesis. FEMS Microbiol Rev. 2016;40(1):41–56. [DOI] [PubMed] [Google Scholar]
- 58.Deng LL, Humphries DE, Arbeit RD, Carlton LE, Smole SC, Carroll JD. Identification of a novel peptidoglycan hydrolase CwlM in Mycobacterium tuberculosis. Biochim Biophys Acta. 2005;1747(1):57–66. [DOI] [PubMed] [Google Scholar]
- 59.Boutte CC, Baer CE, Papavinasasundaram K, Liu W, Chase MR, Meniche X, et al. A cytoplasmic peptidoglycan amidase homologue controls mycobacterial cell wall synthesis. Elife. 2016;5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Turapov O, Forti F, Kadhim B, Ghisotti D, Sassine J, Straatman-Iwanowska A, et al. Two Faces of CwlM, an Essential PknB Substrate, in Mycobacterium tuberculosis. Cell Rep. 2018;25(1):57–67 e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Gee CL, Papavinasasundaram KG, Blair SR, Baer CE, Falick AM, King DS, et al. A phosphorylated pseudokinase complex controls cell wall synthesis in mycobacteria. Sci Signal. 2012;5(208):ra7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Parikh A, Verma SK, Khan S, Prakash B, Nandicoori VK. PknB-mediated phosphorylation of a novel substrate, N-acetylglucosamine-1-phosphate uridyltransferase, modulates its acetyltransferase activity. J Mol Biol. 2009;386(2):451–64. [DOI] [PubMed] [Google Scholar]
- 63.Kieser KJ, Boutte CC, Kester JC, Baer CE, Barczak AK, Meniche X, et al. Phosphorylation of the Peptidoglycan Synthase PonA1 Governs the Rate of Polar Elongation in Mycobacteria. PLoS Pathog. 2015;11(6):e1005010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Pensinger DA, Boldon KM, Chen GY, Vincent WJ, Sherman K, Xiong M, et al. The Listeria monocytogenes PASTA Kinase PrkA and Its Substrate YvcK Are Required for Cell Wall Homeostasis, Metabolism, and Virulence. PLoS Pathog. 2016;12(11):e1006001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Wamp S, Rutter ZJ, Rismondo J, Jennings CE, Moller L, Lewis RJ, et al. PrkA controls peptidoglycan biosynthesis through the essential phosphorylation of ReoM. Elife. 2020;9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Toniolo C, Balducci E, Romano MR, Proietti D, Ferlenghi I, Grandi G, et al. Streptococcus agalactiae capsule polymer length and attachment is determined by the proteins CpsABCD. J Biol Chem. 2015;290(15):9521–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Grangeasse C, Obadia B, Mijakovic I, Deutscher J, Cozzone AJ, Doublet P. Autophosphorylation of the Escherichia coli protein kinase Wzc regulates tyrosine phosphorylation of Ugd, a UDP-glucose dehydrogenase. J Biol Chem. 2003;278(41):39323–9. [DOI] [PubMed] [Google Scholar]
- 68.Minic Z, Marie C, Delorme C, Faurie JM, Mercier G, Ehrlich D, et al. Control of EpsE, the phosphoglycosyltransferase initiating exopolysaccharide synthesis in Streptococcus thermophilus, by EpsD tyrosine kinase. J Bacteriol. 2007;189(4):1351–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Soulat D, Grangeasse C, Vaganay E, Cozzone AJ, Duclos B. UDP-acetyl-mannosamine dehydrogenase is an endogenous protein substrate of Staphylococcus aureus protein-tyrosine kinase activity. J Mol Microbiol Biotechnol. 2007;13(1–3):45–54. [DOI] [PubMed] [Google Scholar]
- 70.Wugeditsch T, Paiment A, Hocking J, Drummelsmith J, Forrester C, Whitfield C. Phosphorylation of Wzc, a tyrosine autokinase, is essential for assembly of group 1 capsular polysaccharides in Escherichia coli. J Biol Chem. 2001;276(4):2361–71. [DOI] [PubMed] [Google Scholar]
- 71.Hsieh SA, Allen PM. Immunomodulatory Roles of Polysaccharide Capsules in the Intestine. Front Immunol. 2020;11:690. [DOI] [PMC free article] [PubMed] [Google Scholar]


