Abstract
The aim of this study was to develop a reverse transcription-PCR assay and lateral flow detection protocol for specific identification of Cryptosporidium parvum. The method which we developed is sensitive and specific and has a low limit of detection. In our protocol a solid phase material, the Xtra Bind Capture System, was used for extraction and purification of double-stranded RNA (dsRNA) specific for C. parvum. The Xtra Bind Capture System interfaced with pellets concentrated from water samples collected with previously developed filtration devices. The pellets were resuspended in reagent water (final volume, 0.5 ml), and an equal amount of rupture buffer and the Xtra Bind Capture System was added to the resuspended pellet mixture. The dsRNA target sequences in a 0.5-ml portion were captured by the solid phase material via hybridization. The debris and potential inhibitors were removed by washing the Xtra Bind material several times with buffer. The Xtra Bind material with its bound dsRNA was added directly to an amplification reaction mixture, and the target was amplified without elution from the Xtra Bind material. A PCR was performed in the presence of the Xtra Bind Capture System, which resulted in robust amplification of the target. The detection system which we used was adapted from lateral flow chromatography methods typically used for antigen-antibody reactions. The result was a colored line that was visible if the organism was present. When this method was used, we were able to reproducibly and correctly identify 10 oocysts added to 0.5 ml of reagent water. When the protocol was evaluated with a small set of environmental samples, the level of detection was as low as 1 oocyst/liter. The total time from resuspension of the pellet to detection was about 3 h, which is considerably less than the 5 h required for immunomagnetic separation followed by an indirect immunofluorescence assay and microscopy.
In recent studies workers found that Cryptosporidium parvum oocysts were present in 65 to 97% of the surface water tested at locations throughout the United States (29, 30, 36, 37). Outbreaks have occurred in water systems ranging from simple chlorination systems to complete filtration and ozonation systems (5, 7, 9, 14, 15, 17–19, 32, 33, 38). An outbreak in Kitchner-Waterloo, Ontario, Canada, affected 1,000 people, and the water at this location was both filtered and ozonated. There have been more than 30 documented outbreaks of cryptosporidiosis in the United States. An outbreak during 1993 in Milwaukee, Wis., resulted in more than 400,000 cases of illness and 100 deaths (33). Filtration, which physically removes the parasite from contaminated water, is the only effective treatment since oocysts are resistant to chemical disinfectants. However, a filtration system that is not well maintained and operated may not provide absolute protection. Recent surveys in which the researchers examined the occurrence of Cryptosporidium oocysts in fully treated (disinfected and filtered) municipal water demonstrated that small numbers of oocysts were able to breach filters and were present in tap water in 27 to 54% of the communities evaluated (11, 31). All waterborne outbreaks of Cryptosporidium infection detected to date in the United States have occurred in communities in which the water utilities met state and federal standards for acceptable drinking water quality (22). Twenty-three million Americans reside in communities that do not filter municipal drinking water that comes from surface sources (12). Not all Cryptosporidium species are infectious to humans, so it is important that C. parvum is specifically identified before public health warnings are issued. The water industry needs a simple, easy, and rapid method for specific detection and identification of C. parvum.
The method now used to detect cryptosporidia requires passage of raw or finished water though filters with a nominal cutoff diameter of 1 μm. The filters are sent to laboratories for analysis with a indirect immunofluorescent assay (IFA) after a series of labor-intensive extraction and purification steps. The method is difficult, and the level of recovery and level of precision are low. The average level of recovery of oocysts is often less than 8% (8). False-positive results are common with IFA because of the lack of specificity of the antibody, which recognizes several diverse Cryptosporidium species. Only C. parvum is considered a human pathogen that requires species-specific detection in order to avoid reporting of false-positive results and unnecessary remediation. The limitations of the IFA method described above have been recognized, and a new method, Environmental Protection Agency method 1622 (2), has been developed. With this method recovery is more efficient, but the method still relies on an IFA for identification of oocysts. There is a need for a rapid and simple method that can be used to detect C. parvum with sensitivity and specificity unmatched by conventional testing procedures. The nucleic acid-based method described here is such a method.
There have been many previous descriptions of genetically based assays for Cryptosporidium spp. (1, 3, 4, 6, 10, 13, 20, 21, 27, 28, 34, 35, 41). These assays are most commonly PCR-based assays in which the small-subunit (18S) rRNA gene (19, 28), the heat shock protein (hsp70), or genomic fragments whose function is not known (6, 41) are used. While the ability of each of these assays to distinguish Cryptosporidium spp. from members of other pathogenic genera has been demonstrated, there is often a problem with species specificity. For example, the small-subunit rRNAs of three Cryptosporidium species have been sequenced, and the sequences exhibit approximately 97 to 98% homology. PCR assays in which these genes are used cannot distinguish among Cryptosporidium spp. or among subgroups of C. parvum (13, 28). To obtain species specificity, the assay methods may include a two-step nested set PCR (13), probing with cDNA or genomic DNA sequences specific for C. parvum (3, 6, 28, 41), analysis of the amplification product by restriction enzyme digestion (22), or other labor-intensive manipulations in order to confirm that C. parvum is present (6, 13, 21, 28, 41).
Molecular methods are very sensitive, but they have limitations when they are used with environmental samples. These limitations include (i) inhibition of PCR by environmental inhibitors, (ii) a limit on the sample size resulting from extraction that can be added to a PCR assay mixture, and (iii) time-consuming detection methods. Attempts have been made to overcome these limitations by magnetic cell sorting (34, 38), purification during nucleic acid extraction, addition of compounds to amplification reaction mixtures to overcome inhibition (35), hybridization to specific probes (3, 6, 28, 41), and restriction digestion (22, 42). Although these attempts have been successful and C. parvum has been detected, the processes are time-consuming, labor-intensive, and expensive.
The method described in this paper deals with the issues described above with a solid phase extraction and purification process, a single-tube nested set PCR, and rapid, visual detection. The extraction and purification process extracts and purifies C. parvum nucleic acid and easily interfaces with previously described sample collection techniques. Detection labeling and amplification of the target take place in a single-tube nested set PCR. Detection is visual and requires less than 5 min.
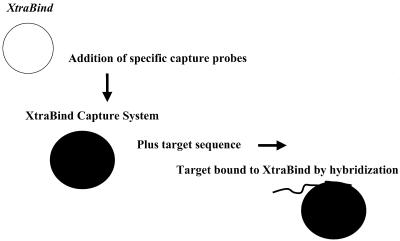
Our extraction method uses a solid phase material, Xtra Bind (Xtrana Inc., Denver, Colo.), which was supplied with directions for binding to single-stranded oligonucleotides and creates a capture system for a double-stranded RNA (dsRNA) target specific for C. parvum (23, 24). The specificity of the dsRNA was established by examining 16 human and 16 calf isolates of C. parvum (including genotype 1 and genotype 2 isolates), eight other members of the genus Cryptosporidium, and 12 unrelated species (24). The oligonucleotide used for the capture system was designed to complement sequences on the dsRNA which created a capture system for dsRNA via hybridization that facilitated binding to the Xtra Bind material. The concept is illustrated in Fig. 1. The capture system was used to establish the feasibility of purifying C. parvum dsRNA from environmental contaminants in finished water, groundwater, and raw water samples. Oocysts were chemically ruptured with a buffer that released the dsRNA from the sporozoites. The buffer not only released the dsRNA but also provided an environment that was conducive to hybridization of the dsRNA to the capture oligonucleotide on the Xtra Bind material. The dsRNA remained on the Xtra Bind material for washing, and the Xtra Bind material was added directly to a PCR mixture for amplification of the target sequences. With this method we were able to extract nucleic acids from volumes equivalent to 100-liter environmental samples without an immunomagnetic separation step.
FIG. 1.
Preparation of Xtra Bind Capture System. An oligonucleotide, termed a capture probe, is bound to the surface of solid phase Xtra Bind material to create an Xtra Bind Capture System. The capture probe contains sequences that are complementary to a region in the amplification target (but outside the amplification region). After lysis and release of nucleic acids, the target binds to the Xtra Bind Capture System through hybridization and is available for amplification.
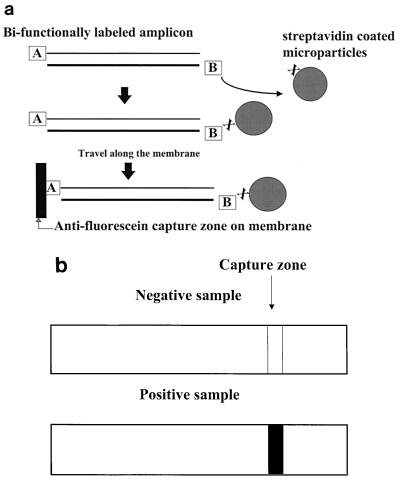
The PCR amplified the dsRNA in the presence of the Xtra Bind material and was designed to interface with the detection method. Typical PCR results are detected by gel electrophoresis alone or by gel electrophoresis in combination with blotting and probing. In this paper we describe a lateral flow format for detection in which we relied on bifunctional labeling of the product (amplicon) during the amplification reaction. The primers used for the PCR were designed with a hapten on the 5′ end of the reverse primer (hapten A) and a second hapten on the 5′ end of the forward primer (hapten B). Hapten A was fluorescein isothiocyanate (FITC), and hapten B was biotin. The bifunctionally labeled product reacted with streptavidin coupled to colored latex microparticles. The amplicon bound to the streptavidin-coated microparticles moved through the membrane by capillary action until the capture zone was reached. The capture zone contained antibody against the FITC hapten (anti-FITC). The antibody-antigen reaction between the capture zone (anti-FITC) and the FITC label on the amplicon resulted in visual detection if the target sequence was present in the sample. The amplicon created a “bridge” between the colored microparticles and the capture zone. If the target sequence was not present, a bridge between the microparticles and the capture zone was not created, the microparticles continued to move through the membrane without being captured, and the visual signal was not generated. These concepts are illustrated in Fig. 2.
FIG. 2.
Lateral flow chromatography detection with bifunctionally labeled amplicon. (a) The biotin moiety on the bifunctionally labeled amplicon binds to strepavidin conjugated to colored microparticles. As the microparticles travel along a nitrocellulose lateral flow chromatography strip, the FITC moiety binds to antibody in an anti-FITC capture zone, which creates a molecular bridge between the capture zone and the microparticles. This bridge causes the microparticles to accumulate and form a line. (b) Schematic representation of negative and positive lateral flow chromatography results when bifunctionally labeled amplicon was used. When the bifunctionally labeled amplicon is present, a molecular bridge between the microparticles and capture zone is formed, which results in a positive signal. No line is formed when microparticles do not accumulate in the absence of a molecular bridge between the capture zone and the microparticles.
One problem with detection by this method is the formation of primer-dimer artifacts. These artifacts would not be problematic with gel electrophoresis because it is possible to distinguish the correct band size of the product. With lateral flow detection, the visual line depends on the amplicon bridging the colored microparticles via biotin and the membrane via FITC. The formation of primer-dimer artifacts with labeled primers produces the same results without the amplicon bridge. To circumvent the primer-dimer problem, we designed a homogeneous nested set amplification reaction. Our nested set amplification procedure involved using four primers. Two unlabeled primers (the first primer set) extended at 70°C for 40 cycles created an unlabeled product. The second primer set was not functional at this high annealing temperature; these primers were located inside the first primer set and were labeled. The second primer set annealed at 55°C and used the product from the first round of amplification as the target for five cycles. At the lower temperature, the second amplification resulted in a bifunctionally labeled target. The amplification performed with the first primer set generated a high number of copies of unlabeled product specific for C. parvum. The second primer set labeled the amplicon already present in a few rounds of amplification. This approach eliminated the primer-dimer artifacts that were a potential cause of false-positive results during lateral flow chromatography. The concept of a homogeneous nested set PCR is illustrated in Fig. 3.
FIG. 3.
Schematic representation of the homogeneous nested set PCR used to generate a bifunctionally labeled amplicon. Two unmodified primers (primer set 1) with a high melting temperature (Tm) are used to generate an unlabeled product that serves as the template for subsequent rounds of amplification with modified primers (primer set 2). The modified primers have a melting temperature that is 10°C lower than the melting temperature of the unmodified primers and do not function in early rounds of amplification. Amplification with primer set 2 results in labeling of the product with FITC and biotin for subsequent detection by lateral flow chromatography.
Our integrated extraction, amplification, and detection method was evaluated by using a variety of environmental water samples. The detection method was successful with groundwater, as well as finished water and raw water samples seeded with C. parvum oocysts. Oocysts were detected in 27 of the 32 environmental samples tested (84.4%). Our preliminary results demonstrate that our C. parvum detection method is compatible with environmental water samples. The simplicity, specificity, and rapidity of this method provide substantial advantages compared to previously described C. parvum detection methods.
MATERIALS AND METHODS
Oocyst stock preparation.
C. parvum oocysts (KSU-1 isolates) were obtained from calf fecal specimens supplied by Steve Upton (Kansas State University, Manhattan). The oocysts were purified from the feces of neonatal calves by sucrose-CsCl gradient centrifugation as previously described (25, 39) and were stored in distilled H2O (dH2O) at 4°C without preservatives. We noted that the oocysts spontaneously ruptured and released free nucleic acids. In order to determine an accurate limit of detection for oocyst enumeration, it was necessary to remove the free nucleic acids that might have been in the storage buffer prior to enumeration. A discontinuous gradient centrifugation technique was used to do this. Two sucrose layers were used; the density of one was 1.18 g/ml (41% sucrose), and the density of the other was 1.03 g/ml (9% sucrose). The oocysts were added to the gradients in Beckman Ultraclear centrifuge tubes and centrifuged with a Beckman model L 60 ultracentrifuge by using a type SW 28.1 swinging bucket rotor at 8,600 rpm for 10 min. The centrifuge was allowed to coast to a stop. The oocysts accumulated at the interface between the layers, and the free RNA was retained in the 1.03-g/ml layer. The oocyst layer was visualized, and an 18-gauge needle attached to a 5-ml syringe was used to remove the oocysts from the gradient.
Enumeration of oocysts and seeding studies.
The sucrose gradient-purified oocysts were initially enumerated by counting oocysts with a hemocytometer. The rough estimates obtained were used to prepare aliquots containing presumptive amounts of oocysts. These aliquots were sent to Clancy Environmental Consultants, Inc. (St. Albans, Vt.) for enumeration by a well slide method that is statistically significant (2). A Zeiss Axioskop fluorescence microscope equipped with a blue filter block (excitation wavelength, 490 nm; emission wavelength, 510 nm) was used for detection of FITC-monoclonal antibody-labeled oocysts at a magnification of ×200. The presence of oocysts was confirmed at a magnification of ×400 by using a UV filter block (excitation wavelength 400 nm; emission wavelength, 420 nm) for visualization of DAPI (4′,6′-diamidino-2-phenylindole), and the internal morphology of oocysts was observed by Nomarski- DIC microscopy. KSU-1 oocysts were added to preparations at the concentrations indicated below.
Detection method. (i) Preparation of the Xtra Bind Capture System.
An Xtra Bind Capture System kit (Xtrana Inc.) was used for extraction and purification of dsRNA from C. parvum. The extraction system was made specific for the target by adding an oligonucleotide (5′-CTATATCGTAATACGCTCTGATTA CGTAGGGAGTGGTACTCCTAACAGTAGGCCTCTGATTTGTCAGTCG ACATACCGCTGCGCTCAAATCCTTTTAGAA-3′) that was designed to bind C. parvum dsRNA to the Xtra Bind material as recommended by the manufacturer. The kit contained solid phase binding material (Xtra Bind), chemical rupture buffer, and washing buffer for making a capture system that could be used to specifically extract and purify C. parvum dsRNA.
(ii) Evaluation of chemical rupture buffer.
A 30-μl aliquot of a C. parvum oocyst preparation containing 9.5 × 105 oocysts was added to an equal volume of the Tris-HCl (pH 8.0)-based 2× lysis buffer provided with the kit. Three replicate tubes were prepared for this experiment. A fourth tube containing oocysts (30 μl) and an equal volume of deionized water was also analyzed. All of the tubes were vortexed and incubated at 95°C for 5 min. After incubation, the contents of the tubes were concentrated by centrifugation, the supernatants were aspirated, and the remaining pellets were loaded into a hemocytometer and analyzed to determine whether oocysts were present. The control concentrate was diluted in dH20 in order to enumerate the oocysts.
(iii) Extraction and purification.
The capture probe, a sequence specific for C. parvum dsRNA, was bound to the Xtra Bind material by following the instructions provided with the kit. dsRNA from C. parvum oocysts was extracted and prepared for PCR by using the following protocol for reagent water spikes. Ten microliters of the Xtra Bind Capture System was transferred to a 1.5-ml microcentrifuge tube for each sample analyzed. Once the Xtra Bind Capture System material settled to the bottom of the tube, the remaining buffer was removed carefully with a pipette. Reagents and a sample were added to each tube in the following order: 500 μl of 2× lysis buffer, 495 μl of an environmental water sample (or reagent water), and 5 μl of enumerated oocysts. The tubes were capped, mixed by vortexing, and placed in a 95°C heat block for 5 min. After 30 min of rotation at room temperature, the tubes were placed upright in a rack, which allowed the Xtra Bind Capture System material to settle. Most of the supernatant was removed and discarded. One milliliter of 1× washing buffer was added to each tube, and the mixture was gently pipetted up and down five times to wash the Xtra Bind Capture System material. Once the contents of the mixture settled, the washing buffer was removed carefully to ensure that the Xtra Bind Capture System material was not disturbed. This washing procedure was performed two more times. At the end of the last washing step, the washing buffer was completely removed, and 200 μl of additional washing buffer was added. PCR tubes for each of the reaction mixtures were appropriately labeled, and the Xtra Bind Capture System material and buffer were transferred into the PCR tubes. The Xtra Bind Capture System material was allowed to settle, the excess buffer was removed, and 5 μl of H2O was added immediately to each tube to keep the dsRNA from drying out. The extraction-purification procedure was complete, and the samples were ready for amplification. The positive amplification controls consisted of oocysts added directly to the amplification reaction mixtures. The positive extraction controls consisted of enumerated oocysts seeded into reagent water and subjected to the extraction protocol. The negative controls included no-template amplification controls and mock-extraction controls consisting of reagent water blanks without oocysts.
(iv) Nested set RT-PCR amplification.
The primers used for PCR with dsRNA were designed by using primer design software (Oligo v5.0; Molecular Biology Insights, Casacade, Colo.). The primers were synthesized by Operon, Inc. (Foster City, Calif.). A single-tube, nested set reverse transcription-PCR (RT-PCR) was designed to be specific for C. parvum dsRNA. The outer set sequences were 40 bp long (primer CPOP716F [5′-CCG GAA GCA GTG CAA TCT GTT AGT CTC ACC TTC TAC TCA T-3′] and primer CPOP992R [5′-GAC CTA ATC TCA TTG TAT ATG GCG CGC ACG TAT ATC GGT A-3′]). These primers were not labeled. The primers of the inner primer set were labeled on their 5′ ends with FITC and biotin (primer CPSR805F-FITC [5′-GAG GAT AGA GGC ATT TGG TTG-3′] and primer CPSR948R-Biotin [5′-GTT TTG TAG GGG TCG CTC AT-3′], respectively). The PCR reagents and the Xtra Bind Capture System material were mixed well. A Perkin-Elmer model 2400 Gene Amp PCR system was used for direct amplification with the solid phase material. The nested set reaction was optimized with both primer sets in order to obtain a low limit of detection for oocysts. The reagents used in the nested set reaction mixture were (final concentrations) 1× EZ buffer (Perkin-Elmer, Foster City, Calif.), 1.25 mM manganese acetate, each deoxynucleoside triphosphate at a concentration of 160 μM, 500 nM CPOP716F, 500 nM CPOP992R, 100 nM CPOP805F-FITC, 100 nM CPOP948R-Biotin, and 5 U of rTth polymerase (Perkin-Elmer). The following PCR thermal profile was used: 95°C for 1 min; 60°C for 15 min; 40 cycles consisting of 95°C for 15 s and 65°C for 1 min; and then five cycles consisting of 95°C for 1 s and 55°C for 1 min. The nested set RT-PCR was performed with additional organisms to confirm that the primers were specific for C. parvum (Table 1).
TABLE 1.
Specificity of nested set RT-PCR for detection of C. parvum oocysts
| Organism (source) | Results of lateral flow detectiona |
|---|---|
| Cryptosporidium parvum KSU-1 (S. Upton, KSU)b | Positive |
| Cryptosporidium parvum KSU-4 (S. Upton, KSU) | Positive |
| Cryptosporidium muris (S. Upton, KSU) | Negative |
| Cryptosporidium serpentis (S. Upton, KSU) | Negative |
| Cryptosporidium sp. strain KSU-3 (S. Upton, KSU) | Negative |
| Cryptosporidium sp. (S. Upton, KSU) | Negative |
| Escherichia coli DH5α (U. Ochsner, UCHSC)c | Negative |
| Pseudomonas aeruginosa PA01 (U. Ochsner, UCHSC)c | Negative |
Lateral flow detection was performed after lysis and extraction of oocysts followed by amplification as described in Materials and Methods.
KSU, Kansas State University, Manhattan; UCHSC, University of Colorado Health Sciences Center, Denver.
Purified chromosomal DNA of this organism was tested directly by using the nested set RT-PCR.
(v) Lateral flow detection. (a) Microparticle preparation.
Blue latex microparticles (diameter, 0.293 μm) from Seradyn Inc. (Indianapolis, Ind.) were covalently coupled to streptavidin. Covalent coupling was accomplished by using carbodiiamide chemistry (16), a 1.0-mg/ml solution of streptavidin, and a 0.75% suspension of microparticles. The binding buffer was 500 mM MES (morpholineethanesulfonic acid) buffer (pH 5.5).
(b) Membrane preparation.
Nitrocellulose (Millipore Corp., Bedford, Mass.) was “striped” with a Linear Striper instrument (IVEK Corp., North Springfield, Vt.). Striping consisted of depositing antibody (1 mg/ml) directed against the FITC hapten label (Dako A/S, Glostrup, Denmark) onto the membrane at a rate of 1 ml/s. The membrane was dried and blocked with a 1% casein solution.
(c) Detection of amplified products by lateral flow chromatography.
For lateral flow detection we added 2.5 μl of amplified product to a mixture containing 0.1% microparticles and 20 μl of detection buffer (50 mM Tris-HCl [pH 8.0], 8 mM MgCl2, 0.025% Triton X-100). The product and reaction components were vortexed and allowed to migrate through a previously prepared nitrocellulose membrane by capillary action.
Limit of detection with reagent water spiked with enumerated oocysts.
The lowest number of oocysts that could be detected with the lysis, extraction, amplification, and detection procedures used was determined by adding enumerated oocysts to 0.5 ml of sterile dH2O. The numbers of oocysts added were 1 × 103 and 1 × 102. Dilutions containing lower oocyst densities were prepared by using enumerated stock preparations, and they presumptively contained certain numbers of oocysts. The seeded oocyst preparations were subjected to the procedure described above.
Comparison of 50-μl reaction mixtures with 500-μl integrated extraction-amplification reaction mixtures.
The amplification sensitivity of a 50-μl reaction mixture that was seeded directly with oocysts was compared to the amplification sensitivity of a 500-μl sample that was seeded with oocysts and subjected to the integrated extraction-amplification protocol. To ensure that equivalent amounts of target were available for amplification, the nucleic acid from oocysts that were seeded directly into the PCR mixture was released by rupturing the oocysts with repeated freeze-thaw cycles as follows. The oocysts were incubated for 10 min at 95°C and then frozen at −70°C for 10 min. Then they were thawed at 65°C for 5 min. The freeze-thaw cycles were repeated eight times. We verified that the oocysts were ruptured by microscopic examination. The PCR conditions used were the conditions described above. A parallel amplification experiment was performed with a 500-μl sample that was subjected to the extraction procedure described above.
Detection when environmental samples were used.
A variety of concentrated pellets resulting from passage of raw or finished water through a filter and subsequent elution of the material collected were analyzed. The environmental water sample sources included groundwater, raw water, and finished water. The samples were collected from different geographical locations and exhibited a range of turbidities and a range of biological and organic compound contents, as well as a range of mineral and inorganic compound contents. For finished water, the turbidities of the samples ranged from 0.03 to 0.263 nephelometric turbidity units, while the pH values ranged from 5 to 7.4. For raw water, the turbidities of the samples ranged from 0.19 to 1.30 nephelometric turbidity units, and the pH values ranged from 4 to 7.5. Algae were present in most of the samples, while some samples also contained amoebae, rotifers, nematodes, and protozoans. Giardia sp. was detected by indirect immunofluorescence in some of the environmental samples; however, Cryptosporidium sp. was not identified in any of the samples before the seeding studies were performed. Samples were stored at 4°C and were typically used within 2 weeks of preparation.
Preparation of environmental water samples.
The following modifications to the protocol described above were made for interfacing with environmental water samples. The pellet material obtained from filtered environmental water samples was typically received in 1.5- to 15-ml tubes, and the equivalent initial volume was given for each sample. For each sample, the exact volume received was measured and recorded. To assay a 100-liter equivalent volume, the following calculation was performed: volume received (in milliliters)/initial filtered volume (in liters) × 100. Duplicates were prepared for each sample. The volume calculated as described above was removed and placed in a 1.5-ml microcentrifuge tube. The samples were centrifuged for 10 min at 13,500 × g. Each supernatant was carefully removed, and 1 ml of sterile water was added to wash away potential amplification inhibitors. The samples were resuspended by vigorous pipetting and then were centrifuged again. Each pellet was resuspended in 1 ml of sterile dH2O as described above. A total of three washing steps were performed. After the last washing step, the samples were centrifuged again, and the supernatants were discarded. Each pellet was resuspended in 500 μl of sterile water and thoroughly mixed by pipetting. The sample was then placed directly into a tube containing Xtra Bind Capture System material and chemical rupture buffer, seeded with oocysts, and subjected to the extraction protocol.
RESULTS
Chemical rupture.
To demonstrate that the chemical rupture buffer effectively lysed C. parvum oocysts, treated oocysts were examined and enumerated by microscopy and compared to untreated controls. After the oocysts were heated in dH2O at 95°C for 5 min, the number of oocytes was reduced from 8.5 × 105 to 1.4 × 104, and the remaining oocysts appeared to be round and refractile. These oocysts were not noticeably different from unexposed oocysts (oocysts examined before the experiment). In the samples containing lysis buffer, the numbers of oocysts were reduced by more than 4 logs, from 8.5 × 105 to an average of 18 oocysts in three experiments. Although small numbers of oocysts remained, these oocysts appeared to be damaged and to have lost some or all of their contents. The 2× lysis buffer released and lysed the sporozoites simultaneously in less than 5 min. The efficiency of the buffer at 95 to 100°C was 99.8%.
Comparison of 50-μl amplification reaction mixture with 500-μl extracted and amplified sample.
A 50-μl amplification reaction mixture that was directly seeded with oocysts was compared with a 500-μl extracted and amplified sample in order to demonstrate that the extraction procedure can deal with relatively large sample volumes. Since oocysts subjected to the extraction procedure were chemically lysed to release the amplification target, oocysts seeded directly into the amplification reaction mixture were first ruptured by freeze-thaw cycles to ensure that equivalent amounts of target were available for amplification. Figure 4 shows that equivalent numbers of oocysts were detected in the 50-μl reaction mixture and the 500-μl extracted sample. For the 50-μl amplification mixture spiked directly with oocysts, a positive signal was obtained with the lateral flow method for all oocyst spikes (103, 102, and 101). Similar results were obtained with the 500-μl extracted samples. The signals observed with samples that had been subjected to the extraction procedure and then amplified were stronger overall than the signals observed with reaction mixtures prepared with oocysts that were directly seeded into the 50-μl amplification mixtures, suggesting that more product was generated in the amplification reactions.
FIG. 4.
Comparison of a 50-μl amplification reaction mixture with a 500-μl extracted and amplified sample: lateral flow chromatography detection results for 50-μl amplification reaction mixtures that were directly spiked with target obtained from freeze-thaw-ruptured oocysts and results for 500-μl extracted and amplified samples. Similar results were observed for both conditions, and approximately 10 oocysts were detected.
Limit of detection of the assay with reagent water spiked with enumerated oocysts.
To demonstrate the sensitivity of the method, the limit of detection when the integrated assay was used was determined by adding enumerated oocysts or dilutions prepared by using enumerated oocysts to reagent water. Figure 5 shows that oocysts prepared by using enumerated oocyst dilutions were detected at a level of approximately 101. Lower numbers of oocysts were not tested due to the difficulty of accurately enumerating low numbers of oocysts.
FIG. 5.
Limit of detection when reagent water was used: lateral flow chromatography detection results used to determine the limit of detection when oocysts were seeded into reagent water and preparations were subjected to the lysis, extraction, and amplification procedures. The limit of detection was found to be 100 oocysts. When we used dilutions prepared from enumerated stock preparations containing approximately 10 oocysts, oocysts were also detected.
Specificity of the nested set RT-PCR.
To demonstrate the specificity of the RT-PCR, two C. parvum strains and several Cryptosporidium species were tested. DNA from Escherichia coli and Pseudomonas aeruginosa, two bacterial species commonly found in water and soil, were also tested. The results showed that the nested set RT-PCR was specific for the dsRNA target found in C. parvum, as the two C. parvum strains were the only organisms detected (Table 1).
Detection when environmental samples were used.
To demonstrate that the integrated extraction-amplification-detection method could be used with environmental samples, pellets from filtered water samples were seeded with oocysts and subjected to the procedure. The target was washed three times with buffer to remove possible amplification inhibitors while it remained bound to the Xtra Bind Capture System. Table 2 shows that the oocysts were detected in 27 of 32 environmental samples (84.4%). Samples that gave negative results were not reinvestigated, as the samples were usually consumed in the assay. However, polyacrylamide was present as a treatment chemical in one of the finished water samples that gave negative results and was not present in any of the samples in which oocysts were detected. The presence of Giardia sp. and other organisms did not inhibit the assay, as C. parvum was detected in all samples that contained Giardia sp., including samples that also contained nematodes, rotifers, amoebae, and other protozoans. These results show that our integrated method is compatible with a variety of environmental water samples, including groundwater, finished water, and raw water samples.
TABLE 2.
Environmental water sample extraction and detection after PCR amplification
| Sample type | Vol. equivalent (liters)a | No. of oocysts added | No. of positive samples/ no. of samples testedb |
|---|---|---|---|
| Finished water | 100 | 1 × 103 | 6/7 |
| Finished water | 100 | 1 × 102 | 9/10 |
| Finished water | 10 | 1 × 102 | 0/2 |
| Groundwater | 100 | 1 × 103 | 2/2 |
| Raw water | 100 | 1 × 104 | 2/2 |
| Raw water | 100 | 1 × 102 | 2/2 |
| Raw water | 10 | 1 × 102 | 6/7 |
An aliquot of an environmental water concentrate representing the volume indicated was processed and seeded with the number of oocysts indicated as described in Materials and Methods.
Water concentrates were seeded with the numbers of oocysts indicated and subjected to the lysis, extraction, amplification, and detection procedures described in Materials and Methods.
DISCUSSION
There is a need for a sensitive and specific nucleic acid-based test for detection of C. parvum in water. The previously described microbiological methods are labor-intensive, costly, and inaccurate. The number of oocysts present decreases at each step in the lengthy process. Since the antibody used in the IFA step can cross-react with organisms other than Cryptosporidium sp., a trained microscopist is required to correctly identify cryptosporidia.
More recent molecular methods are accurate and sensitive but are labor-intensive and generally require postamplification manipulation to determine that C. parvum is present. The protocol described here eliminates postamplification manipulations to specifically determine if any oocysts present are C. parvum oocysts. The target which we used was chosen because the dsRNA are species specific and multiple copies are present in each oocyst (23, 24). We expected that PCR-based detection assays targeting these dsRNA would be very sensitive due to the abundance of these sequences in the cells. We expected that these sequences would provide the lowest limit of oocyst detection and robust PCR assays.
The 2× lysis buffer very effectively denatured oocysts and sporozoites. The few oocysts that remained in the buffer after incubation were damaged and appeared to have lost some or all of their contents. This result demonstrates that this buffer effectively ruptured the oocysts and subsequently released the dsRNA amplification target.
Using the Xtra Bind Capture System was an effective method for extracting dsRNA. For reagent water samples spiked with enumerated oocysts the limit of detection after PCR was 10 oocysts in 500 μl of extraction preparation. This finding shows that low numbers of oocysts can be detected in preparations whose volumes are 10 to 20 times larger than the volume of a typical PCR mixture (25 to 50 μl). This protocol overcomes the limitation of the amount of sample that can be added to a PCR mixture. Since the Xtra Bind Capture System is added directly to an amplification reaction mixture, target from a concentrated sample as large as 0.5 ml can be evaluated by this method. We anticipate that the volumes of the pellets resulting from concentration of filtered water samples (10 to 1,000 liters) should be less than 0.5 ml in most cases.
With the Xtra Bind Capture System it is feasible to purify the target sequence from organic and inorganic debris that may be present in an environmental sample. Our hypothesis is that the target sequences hybridized to the Xtra Bind Capture System, which immobilizes the target on the solid phase. This allows the debris and other material to be decanted or aspirated from the target.
One objective of this study was to eliminate the use of gel electrophoresis product analysis and replace it with a lateral flow detection format. Using the nested set PCR provided an amplicon that was ready for rapid analysis by lateral flow chromatography detection. This detection method is advantageous because it is simple to perform, takes less than 5 min, and requires no washing steps and no instruments for interpreting the results. The nested set PCR was the foundation for evaluating the limit of detection of the dsRNA system and for evaluating the extraction-lysis-purification protocol.
The limit of detection when our method was evaluated with enumerated doses in reagent water was determined to be 10 oocysts. Note that this dose was prepared from a dilution of an enumerated oocyst stock preparation, so the actual number of oocysts that were present is presumptive. The data obtained established the basis for evaluating the method with environmental samples. Environmental samples are expected to behave quite differently than oocysts in reagent water. Environmental samples may contain algae and organic and inorganic components, as well as various kinds of debris. Any one of these is a potential inhibitor of PCR. The feasibility of using the method described here for detecting C. parvum in environmental samples was evaluated with a limited number of samples. Our results indicated that the test could correctly identify 84.4% of the positive samples after a known number of oocysts was added. The sample size was 32, and the samples included 19 finished water samples, 2 groundwater samples, and 11 raw water samples. The reasons why negative results were obtained with five samples were not determined, and these samples were not reinvestigated. However, treatment chemicals, such as polyacrylamide, may have contributed to the negative results in some of the finished water sample experiments, as this compound was used for one of the finished water samples that were negative but for none of the samples in which oocysts were detected. The sample size was small, and this was just a preliminary study, but the possibility that this technology could be used for a rapid and easy detection of C. parvum in environmental samples was established. This method compares favorably with other recently described C. parvum detection methods. For example, similar numbers of oocysts were detected in environmental samples with this protocol and with the protocol of Kostrzynska et al. (26), but an immunomagnetic separation step or nucleic acid precipitation was not necessary when our protocol was used. Our method is also specific for C. parvum but does not require postamplification manipulations, such as restriction enzyme digestion, which is necessary for specific identification of C. parvum by methods described in other recent reports (42). Our method is a promising alternative to other C. parvum detection protocols.
ACKNOWLEDGMENTS
This work was supported by grant 98-33610-6345 from the U.S. Department of Agriculture.
We thank Steve Upton for providing oocysts and Clancey Environmental Consultants for performing enumeration and rupture studies.
REFERENCES
- 1.Abbaszadegan M, DiGiovanni G, Czajka J, LeChevallier M. Water Quality Technical Conference Proceedings. American Water Works Association, Denver, Colo. [on CD-ROM.] 1998. Development of a PCR based kit for the detection of Cryptosporidium using a fluorescent homogeneous format. [Google Scholar]
- 2.Anonymous. Method 1622: Cryptosporidium in water by filtration/IMS/FA. EPA publication 821-R-97-021. U.S. Washington, D.C.: Environmental Protection Agency; 1997. [Google Scholar]
- 3.Awad-el-Kariem F M, Warhurst D C, McDonald V. Detection and species identification of Cryptosporidium oocysts using a system based on PCR and endonuclease restriction. Parasitology. 1994;109:19–22. doi: 10.1017/s0031182000077714. [DOI] [PubMed] [Google Scholar]
- 4.Balatbat A B, Jordan G W, Tang Y J, Silva J. Detection of Cryptosporidium parvum DNA in human feces by nested PCR. J Clin Microbiol. 1996;34:1769–1772. doi: 10.1128/jcm.34.7.1769-1772.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bell A, Guasparini R, Meeds D, Mathias R G, Farley J D. A swimming pool-associated outbreak of cryptosporidiosis in British Columbia. Can J Public Health. 1993;84:334–337. [PubMed] [Google Scholar]
- 6.Carraway M, Tzipori S, Widmer G. Identification of genetic heterogeneity in the Cryptosporidium parvum ribosomal repeat. Appl Environ Microbiol. 1996;62:712–716. doi: 10.1128/aem.62.2.712-716.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Centers for Disease Control and Prevention. Cryptosporidium infections associated with swimming pools—Dane County, Wisconsin, 1993. Morbid Mortal Weekly Rep. 1994;43(31):561–563. [PubMed] [Google Scholar]
- 8.Centers for Disease Control and Prevention. CDC assessing the public health threat associated with waterborne cryptosporidiosis: report of a workshop. Morbid Mortal Weekly Rep. 1995;44(6):1–18. [PubMed] [Google Scholar]
- 9.D'Antonio R G, Winn R E, Taylor J P, Gustafson T L, Current W L, Rhodes M M, Gary G W, Jr, Zajac R A. A waterborne outbreak of cryptosporidiosis in normal hosts. Ann Intern Med. 1985;103:886–888. doi: 10.7326/0003-4819-103-6-886. [DOI] [PubMed] [Google Scholar]
- 10.Deng M Q, Cliver D O, Mariam T W. Immunomagnetic capture PCR to detect viable Cryptosporidium parvum oocysts from environmental samples. Appl Environ Microbiol. 1997;63:3134–3138. doi: 10.1128/aem.63.8.3134-3138.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Federal Register. National primary drinking water regulations: filtration, disinfection, turbidity, Giardia lamblia, viruses, legionella and heterotrophic bacteria. Proposed rule 40CFR, parts 141 and 142. Fed Reg. 1987;52(212):42718. [Google Scholar]
- 12.Federal Register. National primary drinking regulations: monitoring requirements for public drinking water supplies. Final rule. Fed Reg. 1996;61(94):24353–24387. [Google Scholar]
- 13.Filkhorn R, Wiedenmann A, Botzenhart K. Selective detection of viable Cryptosporidium oocysts by PCR. Zentralbl Hyg. 1994;195:489–494. [PubMed] [Google Scholar]
- 14.Gallaher M M, Herndon J L, Nims L J, Sterling C R, Grabowski D J, Hull H F. Cryptosporidiosis and surface water. Am J Public Health. 1989;79:39–42. doi: 10.2105/ajph.79.1.39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hayes E B, Matte T D, O'Brien T R, McKinley T W, Logsdon G S, Rose J B, Ungar B L, Word D M, Pinsky P F, Cummings M L, et al. Large community outbreak of cryptosporidiosis due to contamination of a filtered public water supply. N Engl J Med. 1989;320:1372–1376. doi: 10.1056/NEJM198905253202103. [DOI] [PubMed] [Google Scholar]
- 16.Hermanson G T. Bioconjugate techniques. New York, N.Y: Academic Press; 1996. pp. 169–173. [Google Scholar]
- 17.Herwaldt B L, Craun G F, Calderon R L, Highsmith A R, Juarnek D D. Waterborne disease outbreaks, 1991–1992. Morbid Mortal Weekly Rep CDC Surveill Summ. 1993;42(5):1–22. [PubMed] [Google Scholar]
- 18.Herwaldt B L, Craun G F, Stokes S L, Juarnek D D. Waterborne disease outbreaks, 1989–1990. Morbid Mortal Weekly Rep CDC Surveill Summ. 1991;40(3):1–21. [PubMed] [Google Scholar]
- 19.Joce R E, Bruce J, Kiely D, Noah N D, Dempster W B, Stalker R, Gumsley P, Chapman P A, Norman P, Watkins J, et al. An outbreak of cryptosporidiosis associated with a swimming pool. Epidemiol Infect. 1991;107:497–508. doi: 10.1017/s0950268800049190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Johnson D W, Pieniazek N J, Griffin D W, Misener L, Rose J B. Development of a PCR protocol for sensitive detection of Cryptosporidium oocysts in water samples. Appl Environ Microbiol. 1995;61:3849–3855. doi: 10.1128/aem.61.11.3849-3855.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Johnson D W, Pieniazck N, Rose J B. DNA probe hybridization and PCR detection of Cryptosporidium compared to immunofluorescence assay. Water Sci Technol. 1993;27:77–84. [Google Scholar]
- 22.Juranek D D. Cryptosporidiosis: sources of infection and guidelines for prevention. Clin Infect Dis Suppl. 1995;1:S57–S61. doi: 10.1093/clinids/21.supplement_1.s57. [DOI] [PubMed] [Google Scholar]
- 23.Khramtsov N V, Woods K M, Nesterenko M V, Dykstra C C, Upton S J. Virus-like, double-stranded RNAs in the parasitic protozoan Cryptosporidium parvum. Mol Microbiol. 1997;26:289–300. doi: 10.1046/j.1365-2958.1997.5721933.x. [DOI] [PubMed] [Google Scholar]
- 24.Khramtsov N V, Chung P A, Dykstra C C, Griffiths J K, Morgan U M, Arrowood M J, Upton S J. Presence of dsRNAs in human and calf isolates of Cryptosporidium parvum. J Parasitol. 2000;86:275–282. doi: 10.1645/0022-3395(2000)086[0275:PODSRI]2.0.CO;2. [DOI] [PubMed] [Google Scholar]
- 25.Kilani R T, Sekla L. Purification of Cryptosporidium oocysts and sporozoites by cesium chloride and Percoll gradients. Am J Trop Med Hyg. 1987;36:505–508. doi: 10.4269/ajtmh.1987.36.505. [DOI] [PubMed] [Google Scholar]
- 26.Kostrzynska M, Sankey M, Haack E, Power C, Aldom J E, Chagla A H, Unger S, Palmateer G, Lee H, Trevors J T, De Grandis S A. Three sample preparation protocols for polymerase chain reaction based detection of Cryptosporidium parvum in environmental samples. J Microbiol Methods. 1999;35:65–71. doi: 10.1016/s0167-7012(98)00106-7. [DOI] [PubMed] [Google Scholar]
- 27.Laxer M A, D'Nicuola M E, Patel R J. Detection of Cryptosporidium parvum DNA in fixed, paraffin-embedded tissue by the polymerase chain reaction. Am J Trop Med Hyg. 1992;47:450–455. doi: 10.4269/ajtmh.1992.47.450. [DOI] [PubMed] [Google Scholar]
- 28.Laxer M A, Timblin B K, Patel R J. DNA sequences for the specific detection of Cryptosporidium parvum by the polymerase chain reaction. Am J Trop Med Hyg. 1991;45:688–694. doi: 10.4269/ajtmh.1991.45.688. [DOI] [PubMed] [Google Scholar]
- 29.LeChevallier M W, Norton W D. Giardia and Cryptosporidium in raw and finished water. J Am Water Works Assoc. 1995;87:54. [Google Scholar]
- 30.LeChevallier M W, Norton W D, Lee R G. Occurrence of Giardia and Cryptosporidium spp. in surface water supplies. Appl Environ Microbiol. 1991;57:2610–2616. doi: 10.1128/aem.57.9.2610-2616.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.LeChevallier M W, Norton W D, Lee R G. Giardia and Cryptosporidium spp. in filtered drinking water supplies. Appl Environ Microbiol. 1991;57:2617–2621. doi: 10.1128/aem.57.9.2617-2621.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Levine W C, Stephenson W T, Craun G F. Waterborne disease outbreaks, 1986–1988. Morbid Mortal Weekly Rep. 1990;39:1–13. [PubMed] [Google Scholar]
- 33.MacKenzie W R, Hoxie N J, Proctor M E, Gradus M S, Blair K A, Peterson D E, Kazmierczak J J, Addiss D G, Fox K R, Rose J B, et al. A massive outbreak in Milwaukee of Cryptosporidium infection transmitted through the public water supply. N Engl J Med. 1994;331:161–167. doi: 10.1056/NEJM199407213310304. [DOI] [PubMed] [Google Scholar]
- 34.Mayer C L, Palmer C J. Evaluation of PCR, nested PCR, and fluorescent antibodies for detection of Giardia and Cryptosporidium species in wastewater. Appl Environ Microbiol. 1996;62:2081–2085. doi: 10.1128/aem.62.6.2081-2085.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Rochelle P, De Leon R, Stewart M, Wolfe R. Comparison of primers and optimization of PCR conditions for detection of Cryptosporidium parvum and Giardia lamblia in water. Appl Environ Microbiol. 1997;63:106–114. doi: 10.1128/aem.63.1.106-114.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Rose J B. Occurrence and significance of Cryptosporidium in water. J Am Water Works Assoc. 1988;80:53–58. [Google Scholar]
- 37.Rose J B, Gerba C P, Jakubowski W. Survey of potable water supplies for Cryptosporidium and Giardia. Environ Sci Technol. 1991;25:1393–1400. [Google Scholar]
- 38.Sorvillo F J, Fujioka K, Nahlen B, Tormey M P, Kebabjian R, Mascola L. Swimming-associated cryptosporidiosis. Am J Public Health. 1992;82:742–744. doi: 10.2105/ajph.82.5.742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Upton S J, Tilley M, Nesterenko M V, Brillhart D B. A simple and reliable method of producing in vitro infections of Cryptosporidium parvum (Apicomplexa) FEMS Microbiol Lett. 1994;118:45–49. doi: 10.1111/j.1574-6968.1994.tb06801.x. [DOI] [PubMed] [Google Scholar]
- 40.Wagner-Wiening C, Kimmig P. Detection of viable Cryptosporidium parvum oocysts by PCR. Appl Environ Microbiol. 1995;61:4514–4516. doi: 10.1128/aem.61.12.4514-4516.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Webster K, Pow J D E, Giles M, Catchpole J, Woodward M J. Detection of Cryptosporidium parvum using a specific polymerase chain reaction. Vet Parasitol. 1993;50:35–44. doi: 10.1016/0304-4017(93)90005-8. [DOI] [PubMed] [Google Scholar]
- 42.Xiao L, Escalante L, Yang C, Sulaiman I, Escalante A A, Montali R J, Fayer R, Lal A A. Phylogenetic analysis of Cryptosporidium parasites based on the small-subunit rRNA gene locus. Appl Environ Microbiol. 1999;65:1578–1583. doi: 10.1128/aem.65.4.1578-1583.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]