Abstract
Whole-genome duplications (WGDs) have occurred in many eukaryotic lineages. However, the underlying evolutionary forces and molecular mechanisms responsible for the long-term retention of gene duplicates created by WGDs are not well understood. We employ a population-genomic approach to understand the selective forces acting on paralogs and investigate ongoing duplicate-gene loss in multiple species of Paramecium that share an ancient WGD. We show that mutations that abolish protein function are more likely to be segregating in retained WGD paralogs than in single-copy genes, most likely because of ongoing nonfunctionalization post-WGD. This relaxation of purifying selection occurs in only one WGD paralog, accompanied by the gradual fixation of nonsynonymous mutations and reduction in levels of expression, and occurs over a long period of evolutionary time, “marking” one locus for future loss. Concordantly, the fitness effects of new nonsynonymous mutations and frameshift-causing indels are significantly more deleterious in the highly expressed copy compared with their paralogs with lower expression. Our results provide a novel mechanistic model of gene duplicate loss following WGDs, wherein selection acts on the sum of functional activity of both duplicate genes, allowing the two to wander in expression and functional space, until one duplicate locus eventually degenerates enough in functional efficiency or expression that its contribution to total activity is too insignificant to be retained by purifying selection. Retention of duplicates by such mechanisms predicts long times to duplicate-gene loss, which should not be falsely attributed to retention due to gain/change in function.
Keywords: distribution of fitness effects, loss-of-function mutations, nonfunctionalization, Paramecium, whole-genome duplications
Introduction
Gene duplications are a potentially important source of new genes (Ohno 1970). Although segmental duplications encompassing small numbers of genes are extremely common (Lynch and Conery 2000; Zhang 2003), duplications of entire genomes, that is whole-genome duplications (WGDs) have also occurred in multiple eukaryotic lineages. For example, the model organisms Saccharomyces cerevisiae (Shields and Wolfe 1997) and Xenopus laevis (Morin et al. 2006) have each experienced an ancestral WGD. Two additional WGDs preceded the radiation of vertebrate lineages (Dehal and Boore 2005; Van de Peer et al. 2010) with an additional third round of WGD at the base of the teleost fish lineage (Postlethwait et al. 2000; Jaillon et al. 2004). At least two successive WGDs have occurred below the base of the Paramecium aurelia complex (Aury et al. 2006; McGrath, Gout, Johri, et al. 2014), and WGDs have occurred many times independently in various plant lineages (Jiao et al. 2011, 2014), including the model organism Arabidopsis thaliana (Simillion et al. 2002).
Although WGDs are an important source of new gene duplicates across eukaryotes, the short- and long-term evolutionary forces responsible for the maintenance and loss of resulting duplicates are not well understood. Many models of preservation and loss of duplicates have been proposed (Lynch 2007; Hahn 2009; Innan and Kondrashov 2010), but it has been difficult to understand the relative contributions of alternative mechanisms of retention of paralogs. This issue is even more challenging in the case of WGDs, which increase the scale of the study and introduce novel considerations (such as preservation of dosage) relative to single-gene duplicates. Studies in different model organisms have repeatedly shown a bias toward the post-WGD retention of genes that encode subunits of protein complexes or are involved in many different complexes (Blanc and Wolfe 2004; Maere et al. 2005; Aury et al. 2006; Hakes et al. 2007), as well as genes with high expression levels and slow rates of evolution (Davis and Petrov 2004; Aury et al. 2006; Gout et al. 2010; McGrath, Gout, Doak, et al. 2014). These observations have largely pointed toward the roles of dosage and dosage-balance in maintaining gene duplicates post-WGD (Gout and Lynch 2015).
Despite these advances in our understanding of gene-duplicate loss and retention, the evolutionary mechanisms responsible for the process of gene loss following WGD remain unclear. If the paralogs start off being identical and are largely preserved for their ancestral function (because of dosage or dosage-balance), then why does one get lost eventually, and which evolutionary forces determine which paralog will be lost? Do both members of a duplicate pair experience similar or different selective forces? How long does it take to lose a gene duplicate? The initial phases of duplicate-gene loss must involve the increase in frequency and eventual fixation of mutations that result in loss-of-function or complete deletion of one of the two duplicate-gene sequences. Thus, the probability and time to fixation of such mutations depend on population-genetic parameters such as the effective population size, the strength of selection against or for the loss of gene duplicate, and the rate of input of loss-of-function mutations into the population. Therefore, understanding evolutionary parameters that govern the fixation of loss-of-function mutations or null alleles in a paralog and lead to its removal is ultimately a population-genetic question. However, such a perspective has been lacking, in that most previous studies have only examined gene duplicate evolution via between-species comparative and phylogenetic approaches (e.g., Scannell and Wolfe 2008; Inoue et al. 2015; Braasch et al. 2016).
We take a population-genomic approach to investigate the evolutionary forces responsible for duplicate-gene loss and retention by observing the process of ongoing loss of gene duplicates in a large complex of Paramecium species, following an ancient WGD event. The P. aurelia complex (Sonneborn 1975) is a promising system for examining the process of ongoing loss, as about 40–60% of all gene duplicates from the most recent WGD are retained on a phylogeny of 15 morphologically cryptic post-WGD species. Because even the most recent WGD is extremely ancient (∼320 Ma), the WGD paralogs are easily distinguishable by sequence from one another, with extremely high average divergence (∼1.8) at synonymous sites (McGrath, Gout, Johri, et al. 2014). This then allows for the possibility of population-genetic studies of the two paralogs separately, providing a unique opportunity for understanding the evolutionary mechanisms and consequences of retention of gene duplications.
Notably, whereas two rounds of WGDs at the base of the vertebrates have been hypothesized to facilitate morphological diversification (referred to as the 2R hypothesis; Meyer and Van de Peer 2005; Freeling and Thomas 2006), the WGDs in Paramecium species have instead been accompanied by morphological stasis: all P. aurelia species are morphologically indistinguishable from each other (Sonneborn 1975). The Paramecium complex thus also provides an interesting counterexample to the 2R hypothesis.
To investigate the evolutionary forces acting on WGD duplicates, we use population-genomic data in three species of the P. aurelia complex and an outgroup species P. caudatum that predates the WGD. By examining mutations that are likely to abolish protein function (referred to as loss-of-function mutations or null alleles) segregating in populations, we identify WGD paralogs that may be headed toward loss. Using population-genetic methods, we infer the distribution of fitness effects (DFE) of new base-substitution mutations at nonsynonymous sites and of frameshift-causing indels for WGD paralogs, and demonstrate how mutations in the lowly expressed paralog are much less deleterious than those in the highly expressed paralog. Using the inferred DFE allows us to more precisely estimate the expectation of time taken for fixation of null alleles in WGD paralogs. This unique approach of combining comparative and population-genomics along with transcriptomics sheds new light on the mechanism of duplicate-gene loss following a WGD.
Results
Detection of Loss-of Function Variants in Paramecium
The P. aurelia complex consists of about 15 species that share two and possibly three rounds of WGDs, with most species retaining between 40% and 60% of the duplicates created by the most recent WGD (Aury et al. 2006; McGrath, Gout, Doak, et al. 2014; Gout et al. 2019), which is the focus of this study. To investigate the process of ongoing loss of gene duplicates in the P. aurelia species, we used within-species genomic variation for three P. aurelia species (P. tetraurelia, P. biaurelia, and P. sexaurelia) that share the most recent WGD as well as for one outgroup species, P. caudatum (fig. 1a), which predates the WGD. The population-genomic data utilized in this project, which are publicly available (see Materials and Methods; Johri et al. 2017), are based on whole-genome sequences of 10–13 individuals sampled globally for each species and sequenced to high-depth (with ∼40–80× coverage for each individual).
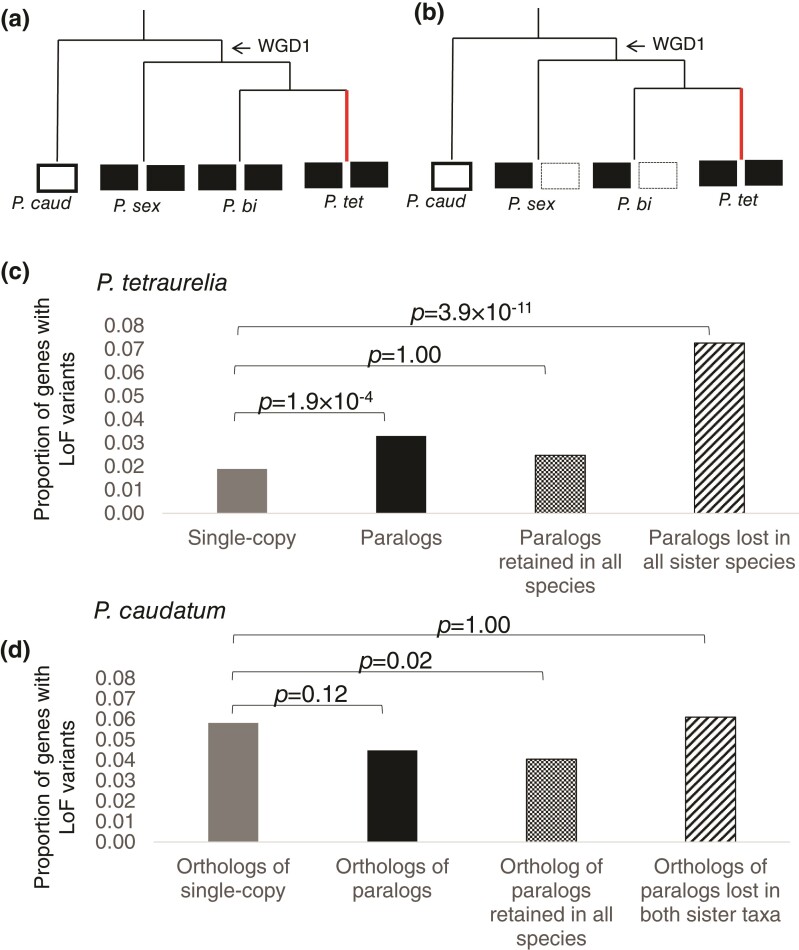
Fig. 1.
(a) Phylogenetic relationships between postduplication (P. biaurelia, P. tetraurelia, and P. sexaurelia) and outgroup species (P. caudatum) considered in this study, where all species retain paralogs (filled black boxes) for the same gene, whereas the outgroup species has the corresponding single-copy orthologs (white boxes). (b) Schematic representation of a scenario where P. tetraurelia retains both WGD paralogs but paralogs in P. biaurelia and P. sexaurelia have returned to single-copy genes. In both cases, the focal lineage is depicted by the red line. (c) Proportion of genes that have segregating loss-of-function (LoF) mutations in P. tetraurelia in different categories of genes—single-copy genes, paralogs, paralogs that have been retained in all species (as shown in a), and paralogs that have been lost in P. biaurelia and P. sexaurelia (as shown in b). (d) Proportions of P. caudatum genes orthologous to the ones in figure 1c that have segregating loss-of-function mutations. In (c) and (d), statistical significance of the difference in the proportions of genes with loss-of-function variants between the set of paralogs and single-copy genes is shown as Bonferroni-corrected P-values obtained via Fisher’s exact test and this analysis is restricted to genes that have orthologs in all four species.
In order to identify gene duplicates that might be heading toward pseudogenization (also known as nonfunctionalization), the population-genomic data were used to detect segregating mutations that could abolish protein function, also referred to as loss-of-function mutations or null alleles, which are not necessarily lethal and might result in only partial loss-of-function. As potential loss-of-function variants, we considered single nucleotide polymorphisms (SNPs) and indels that cause premature termination codons (PTCs), frameshifts that cause mistranslation, or missing start and stop codons (see Materials and Methods). We also detected larger deletion polymorphisms (up to 2,000 bp) in protein-coding genes using CNVnator (Abyzov et al. 2011), which uses read depth to search for deletions. Because it is difficult to precisely detect insertions/deletions (indels) from population-genomic data, reduction of false positives of loss-of-function variants was achieved by performing several checks and by including a rigorous set of filters (see details in supplemental Methods, Supplementary Material online).
Consistent with previous studies in humans (MacArthur 2012) and Drosophila (Lee and Reinhardt 2012), an elevated fraction of frameshift-causing indels were observed at the 3′ ends of genes (supplementary fig. S1, Supplementary Material online), a pattern that is likely due to relaxation of purifying selection against frameshift-causing indels toward the downstream ends of genes. In addition, a slightly higher proportion of frameshift-causing indels at the 5′ end of genes was also observed, which was found to be primarily due to the presence of downstream alternative start codons that can rescue the transcription of most of the rest of the protein (rendering the indels before the alternate start codons to be less deleterious; supplementary fig. S1, Supplementary Material online). Very similar patterns have been found in protein-coding genes in previous population-genomic studies (Lee and Reinhardt 2012; MacArthur 2012), strongly indicating that the majority of detected variants in this study are real.
Overall, a total of 2,218 protein-coding gene loci in P. tetraurelia, 2,949 in P. biaurelia, 4,247 in P. sexaurelia, and 1,296 in P. caudatum were found to harbor potential loss-of-function variants at a high enough frequency to be detected in our sample (supplementary table S1 and methods, Supplementary Material online), representing 5.5%, 7.9%, 12.2%, and 7.0% of all genes in each species, respectively. After including larger deletions detected using CNVnator, these proportions increase to 6.5%, 10.6%, 14.7%, and 11.9% in P. tetraurelia, P. biaurelia, P. sexaurelia, and P. caudatum, respectively. On average, we estimate that ∼700 genes in P. tetraurelia, ∼1,000 in P. biaurelia, ∼1,100 in P. sexaurelia, and ∼500 in P. caudatum are homozygous null in at least one individual in our samples. Interestingly, whereas the nucleotide diversity at 4-fold degenerate sites in the P. aurelia species (0.006, 0.009, and 0.027 in P. tetraurelia, P. biaurelia, and P. sexaurelia) is much lower than in P. caudatum (0.069; Johri et al. 2017), the proportion of genes with loss-of-function variants is similar or higher in P. aurelia species than in P. caudatum. Although factors such as specific demographic histories (e.g., the extent of population structure, presence of recent bottlenecks reducing the efficacy of selection), sample sizes, and different criteria for sampling are likely to contribute to the between-species differences in the prevalence of null alleles, the higher proportion of genes with null alleles in the P. aurelia species could also be due to ongoing duplicate-gene loss post-WGD. Moreover, because the approximate minimal observed frequency in a sample of size n is 1/n, many of the loss-of-function variants detected are likely relatively common, and could thus represent ongoing nonfunctionalization.
Loss-of-Function Variants are Overrepresented in WGD Paralogs versus Single-Copy Genes Due to Ongoing Nonfunctionalization
If a significant number of the loss-of-function variants in the P. aurelia species are due to progression toward post-WGD nonfunctionalization, there should be observable differences between genes that have lost their duplicate (referred to as single-copy genes) versus those that still retain their duplicate created by the WGD. Only 1.9, 3.3, and 7.1% of single-copy genes in P. tetraurelia, P. biaurelia, and P. sexaurelia have loss-of-function variants, whereas much larger proportions, ∼3.3%, 5.8%, and 10.6%, of retained WGD paralogs have segregating loss-of-function variants respectively (fig. 1c; supplementary figs. S2 and S3, Supplementary Material online). Thus, paralogous genes are 1.5–1.7× more likely to harbor loss-of-function variants than are single-copy genes in the postduplication species (p = 5.38 × 10−6, p = 1.10 × 10−9, p = 1.08 × 10−11 in P. tetraurelia, P. biaurelia, and P. sexaurelia respectively).
Interestingly, in the subset of genes whose WGD paralogs are only retained in one of the three P. aurelia species (i.e., have been lost in the other two species; fig. 1b), the species with retained duplicates has an even higher proportion of genes with segregating loss-of-function variants—7.3%, 11.3%, and 13.3% in P. tetraurelia, P. biaurelia, and P. sexaurelia, respectively (fig. 1c; supplementary figs. S2 and S3, Supplementary Material online). Thus, WGD paralogs that are retained in only one of the three P. aurelia species are 3.8× (P. tetraurelia), 3.4× (P. biaurelia), and 1.9× (P. sexaurelia) more likely than single-copy genes to harbor segregating loss-of-function variants, strongly supporting the idea that the majority of segregating loss-of-function variants in WGD paralogs in the P. aurelia species are due to ongoing nonfunctionalization. Concordantly, among genes whose WGD paralogs are retained in all three postduplication species (fig. 1a), a sign of selection for joint retention, only 2.5%, 4.6%, and 10.1% of them in P. tetraurelia, P. biaurelia, and P. sexaurelia, respectively, have loss-of-function variants (fig. 1b; supplementary figs. S2 and S3, Supplementary Material online).
One could argue that all of the observed trends above are simply due to differences in evolutionary constraints (predating the WGD) among the sets of genes tested. In other words, it is possible that genes under weak selective constraints are more likely to be lost and more likely to harbor loss-of-function variants than genes under stronger purifying selection. We therefore looked at loss-of-function variants in orthologous genes from P. caudatum (the outgroup species not sharing the WGD). In P. caudatum, there is no significant difference between the proportion of loss-of-function variants in genes whose orthologs still retain their WGD paralogs in P. aurelia species versus those that have reverted to single copies in P. aurelia species (fig. 1d; supplementary figs. S2 and S3, Supplementary Material online). Thus, differences in the proportion of genes harboring null alleles between WGD paralogs and single-copy genes do not predate the WGD, reinforcing our hypothesis that retained paralogs are undergoing pseudogenization. Because comparisons of prevalence of loss-of-function variants in the above analyses were conducted entirely within-species, our results should not be overly sensitive to differences in sample sizes and genome-wide levels of variation across species due to differences in population-genetic parameters such as demographic histories.
Asymmetric Relaxation of Purifying Selection and Reduction of Expression in One WGD Paralog
To further distinguish between the possibility of ongoing nonfunctionalization post-WGD versus relaxed evolutionary constraints on both WGD paralogs, other population-genetic and transcriptomic signatures of selective constraints were evaluated. Although in the latter scenario, both paralogs would show evidence of relatively weak purifying selection, asymmetric relaxation of selective constraints on only one paralog would be expected if just one of the two paralogs is on its way toward nonfunctionalization. We therefore compared other measures of evolutionary constraints between WGD paralogs where one paralog had segregating loss-of-function variants, whereas the other did not, referred to as intact (fig. 2). As a comparison, we also show the same measures for (1) a set of well-conserved paralogs retained in all three postduplication species, and with identifiable orthologs in the outgroup and without loss-of-function variants and (2) WGD paralogs that were not observed to have any loss-of-function variants.
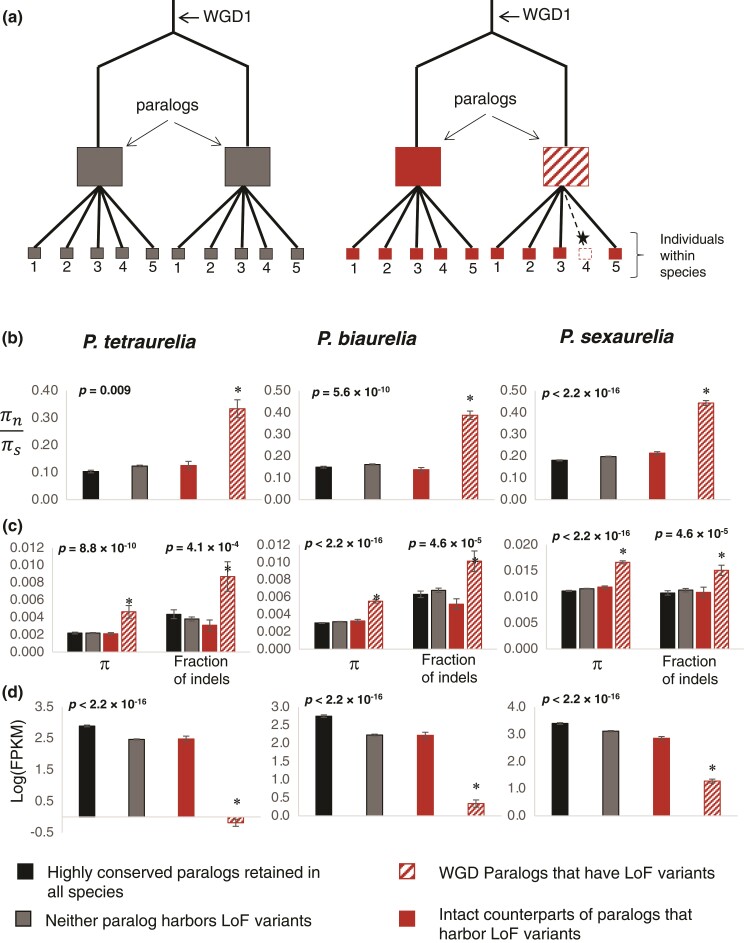
Fig. 2.
Relaxation of purifying selection in only one of the two paralogs. (a) Schematic of WGD paralogs analyzed in this figure. Dark gray squares represent paralogous loci where neither of the two were found to harbor loss-of-function polymorphisms. Red squares represent WGD paralogs where one paralog harbors loss-of-function (LoF) polymorphism (patterned squares) and the other is intact (solid square). Black star represents a loss-of-function mutation. (b) Nucleotide diversity at nonsynonymous sites (πn) relative to that at synonymous sites (πs) in WGD paralogs. (c) Nucleotide diversity and fraction of indels per base in 150 bp upstream of genes. (d) Expression levels shown in logarithm of FPKM values. For (b), (c), and (d), patterned red bars represent WGD paralogs that harbor segregating loss-of-function mutations, solid red bars represent their corresponding intact paralogs that do not have segregating loss-of-function mutations, gray bars represent all WGD paralogs where both copies were intact, and black bars represent a set of highly conserved WGD paralogs that have been retained by all three P. aurelia species. Asterisks represent significant differences between the paralogs that have loss-of-function polymorphisms (patterned red bars) and their respective WGD paralogs that did not have loss-of-function polymorphisms (solid red bars).
Paralogs with segregating loss-of-function variants have significantly higher ratios of nonsynonymous to synonymous diversity (πN/πS) compared with their intact paralogs (0.33 vs. 0.13 in P. tetraurelia; 0.39 vs. 0.14 in P. biaurelia; 0.44 vs. 0.21 in P. sexaurelia; fig. 2), whereas intact paralogs have similar levels of πN/πS as the set of well-conserved paralogs (0.13 vs. 0.10 in P. tetraurelia; 0.14 vs. 0.15 in P. biaurelia; 0.21 vs. 0.18 in P. sexaurelia; fig. 2). A caveat of this analysis is that indels, which are responsible for many loss-of-function variants, can cause misalignments of nearby sites and result in false-positive SNP calls. To account for this issue, πN/πS was also calculated by excluding individuals or haplotypes with the loss-of-function variant, revealing that for paralogs with segregating loss-of-function variants, even haplotypes without loss-of-function variants exhibit an elevated level of diversity at nonsynonymous sites (supplementary fig. S4, Supplementary Material online). In addition, nonsynonymous polymorphisms in paralogs with segregating loss-of-function variants typically caused more radical amino-acid changes (as scored by the BLOSUM62 matrix) than those in their respective intact paralogs (supplementary fig. S5, Supplementary Material online).
Because there is also a significantly higher density of both SNPs and indels in upstream (5′) intergenic regions of duplicate loci with loss-of-function variants (fig. 2), suggesting relaxed selection on the regulatory elements of these genes, we examined expression levels of these paralogs, which had been measured in the reference genome strains (Gout and Lynch 2015). As the reference genome by definition has the intact version of most genes that were detected to have loss-of-function variants, expression levels should not have been affected by the presence of loss-of-function variants and/or PTCs in their sequences (which might induce nonsense-mediated decay and thus reduce expression).
Duplicate loci harboring loss-of-function variants have significantly lower expression levels (given in logarithm of FPKM values; see Materials and Methods) than their intact paralogs in all postduplication species (−0.19 vs. 2.48 in P. tetraurelia; 0.34 vs. 2.22 in P. biaurelia; 1.28 vs. 2.85 in P. sexaurelia; fig. 2), whereas expression levels of the intact paralogs were close to those of the set of conserved paralogs (2.48 vs. 2.90 in P. tetraurelia; 2.22 vs. 2.75 in P. biaurelia; 2.85 vs. 3.39 in P. sexaurelia). It is possible that in a minority of cases, the reference genome has the loss-of-function variant and would thus result in lowered expression level due to nonsense-mediated mRNA decay. For instance, if a PTC was present in the reference genome, it would be identified as a missing stop codon in the resequenced individuals. On excluding all cases where the presence of termination codons was polymorphic in the population (which would exclude any scenarios where the reference genome might have loss-of-function mutations), there continues to be a drastic difference in levels of expression between paralogs with and without loss-of-function variants (−0.20 vs. 2.47 in P. tetraurelia; 0.45 vs. 2.23 in P. biaurelia; 1.49 vs. 3.09 in P. sexaurelia), suggesting that this observation is not a technical artifact and that duplicate loci exhibiting reduced selective constraints indeed have lower levels of expression. Because the average expression levels could be biased by a few extreme outliers, we also looked at the proportion of cases where the paralog with segregating loss-of-function variants has lower expression than the intact paralog in the reference genome and found this to be the case in ∼79%, ∼71%, and ∼69% of WGD-derived paralogs in P. tetraurelia, P. biaurelia, and P. sexaurelia, respectively. We therefore conclude that segregating loss-of-function variants at a retained WGD duplicate-gene harbor a signature of relaxed selective constraints associated with both protein function and expression.
It has been demonstrated that the rate of evolution at coding sites is higher for lowly expressed genes (Drummond et al. 2005). We tested whether the observation of reduced selective constraints at paralogs with lower levels of expression was more than expected when controlling for the levels of expression. We find that both the paralog with the lower expression and single-copy genes that no longer retain their WGD paralogs follow a very similar trend of being more likely to segregate with loss-of-function variants when they are lowly expressed (supplementary fig. S6, Supplementary Material online). Thus, genes with low levels of expression, whether they have retained or lost their WGD paralogs, exhibit relaxed selective constraints and are more likely to have segregating null alleles. Interestingly, however, on restricting this analysis to well-conserved genes (i.e., single-copy and WGD paralogs that have an identifiable ortholog in P. caudatum), we find that for similar expression levels, WGD paralogs are much more likely than single-copy genes to have segregating loss-of-function variants (supplementary fig. S7, Supplementary Material online). This suggests that when all else is equal, the WGD paralog with lower expression might be more likely to harbor null alleles even after controlling for lower levels of expression. Future work in other species might help confirm this observation.
Differences in Divergence and Expression Between Paralogs Precede the Acquisition of Null Alleles
Although relaxation of constraints was found to be selectively experienced by one of the two WGD paralogs, the length of time to reach this point may provide insight into the mechanism of loss and retention of gene duplicates. We therefore investigated how long ago WGD paralogs with loss-of-function variants started experiencing divergent selective constraints and expression levels, in particular, whether this preceded the acquisition of null alleles. Because the WGD paralogs are extremely old (∼320 My), the divergence in sequence and expression level between WGD paralogs can be investigated phylogenetically using additional P. aurelia species with high-quality genome assemblies (Gout et al. 2019). Because the clade comprising the closely related species, P. tetraurelia, P. octaurelia, P. decaurelia, P. dodecaurelia, and P. biaurelia, offers the best resolution to track small changes in selective constraints over evolutionary time (fig. 3), we focused on the WGD paralogs retained in these five species. By evaluating the divergence in levels of expression and rates of amino-acid substitutions between the WGD paralogs retained in these five species, it is possible to approximately date how long ago the paralogs started diverging.
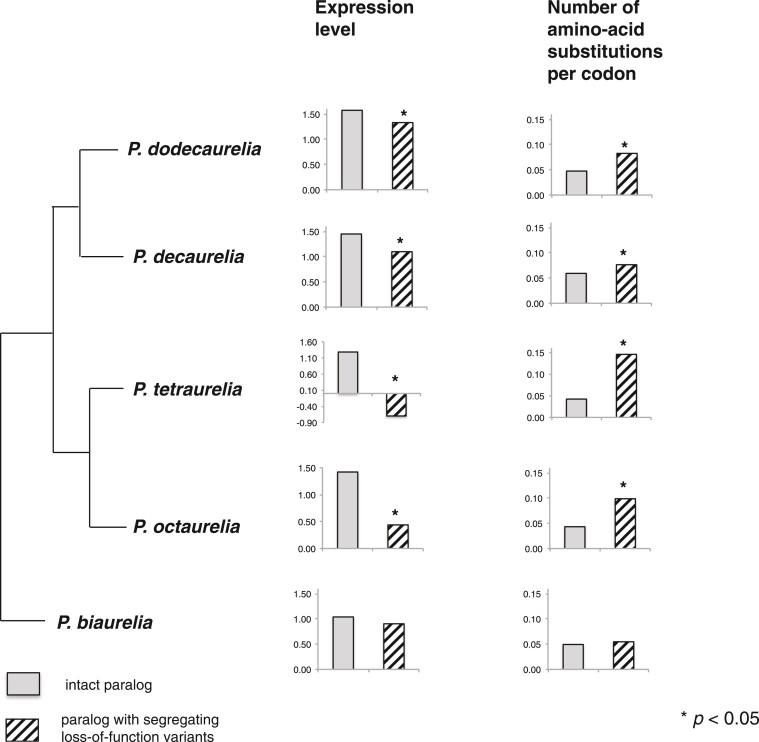
Fig. 3.
Lowering of expression level and accumulation of amino-acid substitutions in the nonfunctionalizing paralog in P. tetraurelia, as observed phylogenetically in orthologs in sister species. The patterned bar in P. tetraurelia corresponds to the duplicate-gene loci that were observed to have segregating loss-of-function mutations in P. tetraurelia, whereas the gray bar corresponds to their intact WGD paralogs (without loss-of-function variants). For all other species—P. octaurelia, P. decaurelia, P. dodecaurelia, and P. biaurelia—the gray and patterned bars show expression levels and rates of amino-acid substitution for the orthologs of the corresponding genes in P. tetraurelia. The number of amino-acid substitutions per codon was calculated as the unique number of amino-acid differences (divided by total length of the gene) obtained for each WGD paralog with respect to the ortholog in the outgroup P. caudatum (using Tajima’s relative rate test).
Overall, it appears that the decrease in levels of expression and concomitant increase in amino-acid substitution rates in one paralog can be tracked phylogenetically and appear to have started at the time when the ancestor of P. tetraurelia and P. biaurelia split. More specifically, when the samples for neither WGD paralog in P. tetraurelia exhibit loss-of-function variants (left panel of fig. 2a), ∼11-14% of the corresponding WGD orthologous paralogs in P. octaurelia, P. decaurelia, P. dodecaurelia, and P. biaurelia exhibit asymmetry in amino-acid substitutions (tested using the relative rate test by Tajima 1993). However, when one of the two WGD paralogs in P. tetraurelia has segregating loss-of-function variants (right panel of fig. 2a), a significantly higher fraction of the corresponding duplicate pairs in all four related species exhibit asymmetry in amino-acid substitutions (fig. 3): 29% in P. octaurelia (p = 3.2 × 10−10 for comparison to cases with no loss-of-function variant), 27% in P. decaurelia (p = 5.9 × 10−12), 24% in P. dodecaurelia (p = 1.5 × 10−9), and 16% in P. biaurelia (p = 0.04). Moreover, of the WGD paralogs with significantly asymmetric rates of amino-acid substitution in P. octaurelia, in 85% of the cases the increased substitution rate was found to be in the ortholog of the P. tetraurelia locus with segregating loss-of-function variants (p= 1.6 × 10−6 in comparison to 50:50 expectation). This fraction decreases with increasing phylogenetic distance to P. tetraurelia, with about 77% in P. dodecaurelia (p = 1.5 × 10−3), 64% in P. decaurelia (p= 0.081), and 50% duplicates in P. biaurelia.
Similar trends are observed with expression levels (fig. 3). Specifically, we find that in P. octaurelia, P. dodecaurelia, and P. decaurelia the copy orthologous to that with loss-of-function variants in P. tetraurelia is typically less expressed than the copy orthologous to the intact copy in P. tetraurelia (0.44 vs. 1.42 in P. octaurelia, p < 2.2 × 10−16; 1.33 vs. 1.57 in P. dodecaurelia, p = 0.02; 1.1 vs. 1.45 in P. decaurelia, p = 2.12×10−4, Wilcoxon rank sum test). These observations strongly suggest that between-paralog changes in both protein-coding sequence and expression levels started occurring around the time when the ancestors of P. tetraurelia and P. biaurelia diverged (∼236Ne generations; see Materials and Methods) or perhaps shortly before, and therefore predate the acquisition of loss-of-function variants (which would on average be younger than 4Ne generations, the approximate maximum age of alleles within populations). Moreover, both the lowering of expression level and accumulation of amino-acid changing substitutions started occurring preferentially in one paralog.
Paralogs with Loss-of-Function Variants are Not Likely to have Undergone Expression Neofunctionalization
The reduced expression of one paralog could also reflect cases of expression neofunctionalization. For instance, it is possible that paralogs undergoing expression neofunctionalization have gained expression in different conditions or life stages at the expense of their expression during vegetative growth, which is the state during which expression levels were measured in this study. First, we asked whether expression neofunctionalization is prevalent, by looking at the difference in levels of expression between paralogs across different environmental conditions—starvation and four different time points during the process of autogamy in P. tetraurelia (Arnaiz et al. 2017). Interestingly, we found that differences in expression levels between paralogs are largely highly correlated across various environmental conditions (supplementary fig. S8, Supplementary Material online), suggesting that expression neofunctionalization is not extremely prevalent in paralogs created by the most recent WGD, at least under the measured conditions. We then obtained a set of paralogs (resulting from the most recent WGD) for which one of the two loci could be a putative candidate for expression neofunctionalization using a very nonconservative outlier approach (see Materials and Methods). Interestingly, we observe that WGD paralogs with putative candidates for expression neofunctionalization have lower mean levels of expression (0.666 in log[FPKM]; supplementary fig. S9, Supplementary Material online) compared with WGD paralogs that do not possess such candidates (2.461 in log[FPKM]), suggesting that lowly expressed paralogs might be more likely to be neofunctionalized. Fortunately, we find that the putative candidates of expression neofunctionalization only comprise of 8% of the WGD paralogs that were found to have loss-of-function variants. Thus, whereas it is possible that a minority of the paralogs segregating with null alleles have undergone expression neofunctionalization instead of pseudogenization, neofunctionalization does not appear to be the common outcome and the majority of paralogs we have identified are indeed heading toward nonfunctionalization.
New Mutations in WGD Paralogs with Higher Levels of Expression are More Deleterious Than in Paralogs With Lower Expression Levels
We hypothesized that mutations might be less deleterious in the paralog with a lower level of expression because of its prior descendance down a path of nonfunctionalization. To test this hypothesis, we contrasted the DFE of new mutations between highly and lowly expressed WGD paralogs in the P. aurelia species. Because mutations of different selection coefficients (s) are expected to segregate at different allele frequencies in populations, the distribution of allele frequencies of variants can be used to infer the most likely distribution of selection coefficients. We inferred the DFE of single base-pair mutations at 0-fold degenerate sites (i.e., amino-acid changing mutations) and those of 1- and 2-bp indels that result in frameshifts, using the site frequency spectrum (SFS) of these polymorphisms (fig. 4). To this end, we employed the method of DFE-α (Keightley and Eyre-Walker 2007), which uses the SFS of putatively neutral sites (like 4-fold degenerate sites) to infer the demographic history and then estimates the DFE at selected sites conditional on the estimated demography and assuming that the selection coefficients are gamma distributed.
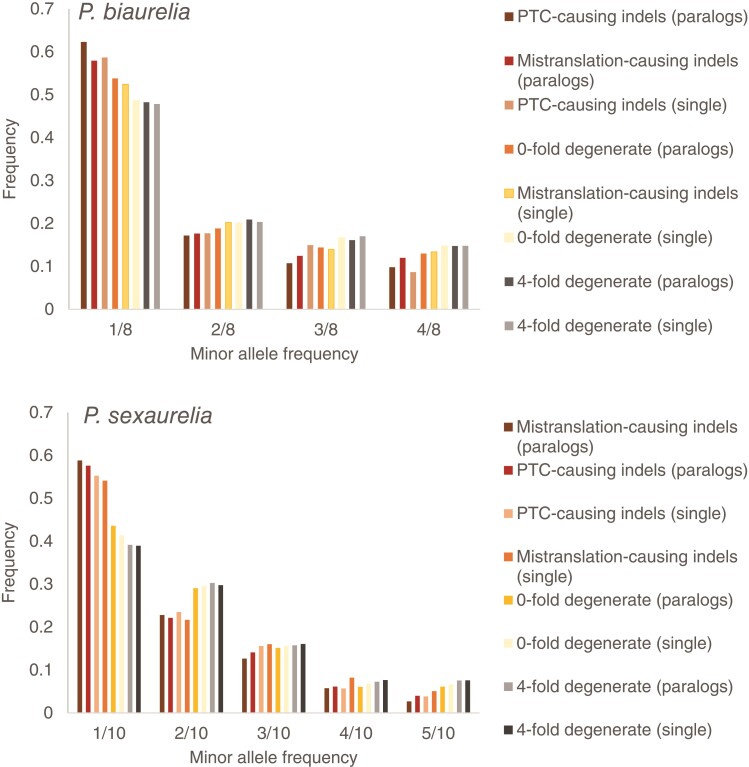
Fig. 4.
The SFS of 4-fold degenerate sites, 0-fold degenerate sites, frameshift-causing indels that result in PTCs, and frameshift-causing indels that do not result in PTCs in P. biaurelia and P. sexaurelia. The SFS is shown for both WGD paralogs and single-copy genes separately.
This analysis was restricted to P. biaurelia and P. sexaurelia, the two species with sufficient numbers of individuals to infer the DFE (see Materials and Methods). Using the 4-fold degenerate sites from both WGD paralogs and single-copy genes, we inferred an extremely recent 9.3-fold expansion in population size in P. biaurelia and a less recent 2-fold expansion in P. sexaurelia (details provided in supplementary table S2, Supplementary Material online). Although it is possible that both of these species have experienced population growth, it should be noted that unknown subpopulation structure (Chikhi et al. 2010; Mazet et al. 2015, 2016), effects of background selection (Ewing and Jensen 2016; Johri et al. 2021), and selection on synonymous sites (Johri et al. 2020) could all lead to a false inference of recent population growth. However, inference of the DFE has been shown to be relatively robust to the presence of above-mentioned confounding factors (Keightley and Eyre-Walker 2007; Huber et al. 2017; Kim et al. 2017; Huang et al. 2021).
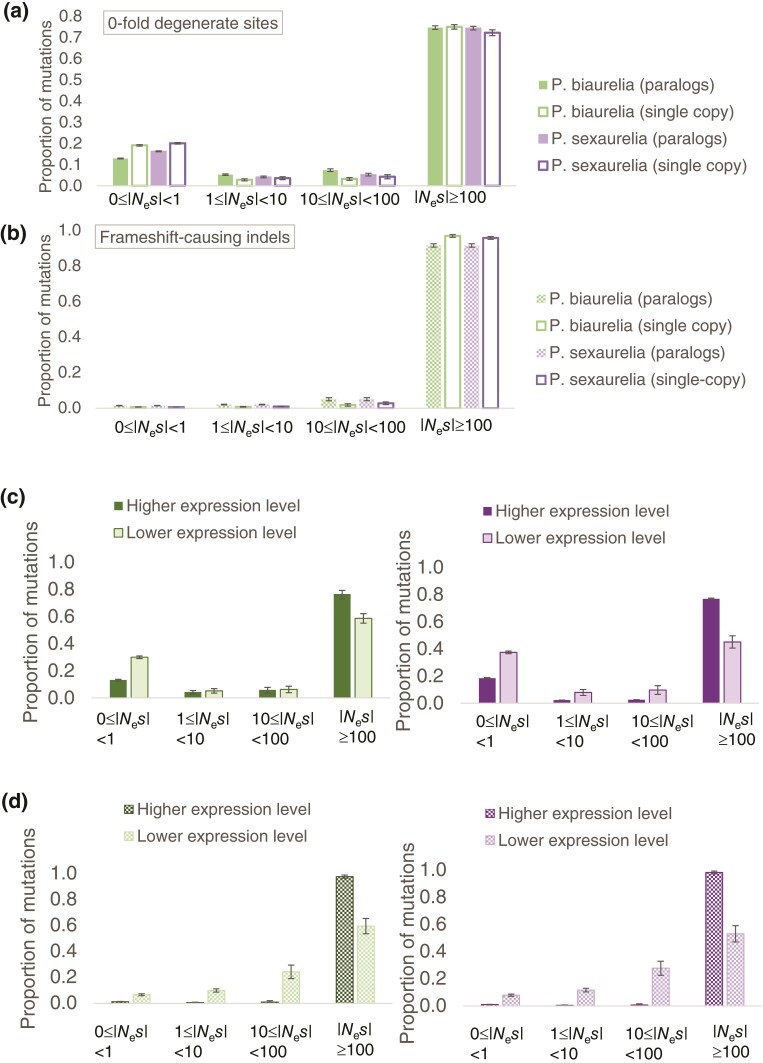
To test our hypothesis, we contrasted the DFE of WGD paralogs with a >5-fold difference in their level of expression. Paralogs with 5-fold lower expression than their counterparts have a much larger proportion (∼40% vs. 20%) of amino-acid changing mutations with no or effectively neutral fitness effects (i.e., |Nes| < 1; fig. 5c). Moreover, whereas ∼80% of frameshift-causing indels are strongly deleterious in highly expressed paralogs, only ∼60% are strongly deleterious in the lower expressed paralogs, with most of them being moderately and mildly deleterious instead (fig. 5d). We observe this pattern consistently in both P. biaurelia and P. sexaurelia (fig. 5). In addition, the DFE observed in the highly expressed paralogs is similar to the DFE of well-conserved genes (i.e., genes that have identifiable orthologs in all three P. aurelia species)—with both those that retain paralogs and those present as single copies having very similar DFE shapes (fig. 5a and b).
Fig. 5.
The DFE of new deleterious mutations in P. biaurelia (green) and P. sexaurelia (purple). (a) The DFE of single nucleotide mutations at 0-fold degenerate sites at WGD paralogs retained in all species (filled bars) and genes that are present in single copy in all species (empty bars). (b) The DFE of frameshift-causing indels of 1 and 2 bp at WGD paralogs retained in all species (patterned bars) and genes that are present in single copy in all species (empty bars). (c) The DFE of mutations at 0-fold degenerate sites of WGD paralogs where there is a 5-fold difference in their expression levels. (d) The DFE of frameshift-causing indels at WGD paralogs where there is a 5-fold difference in their expression levels. In both (c) and (d), darker colors represent paralogs with higher expression, whereas lighter colors represent paralogs with lower expression levels. The DFE of mutations at 0-fold degenerate sites is shown as solid bars and that of frameshift-causing indels is shown as patterned bars.
The DFE of nonsynonymous mutations of well-conserved genes is skewed toward highly deleterious mutations in both species, with ∼70% of all mutations being strongly deleterious with Nes < −100, and with a small proportion (∼10–20%) of mutations that are effectively neutral (fig. 5a). The DFE of frameshift-causing indels is skewed even more excessively toward strongly deleterious mutations in all WGD paralogs as well as in single-copy genes, with ∼90–95% of all mutations predicted to have Nes < −100 (fig. 5b), which is expected because frame-shifting indels are likely to have a larger effect on the protein-coding sequence than base mutations. The large proportion of the class of strongly deleterious nonsynonymous mutations is consistent with the expectation that populations with large effective sizes have a smaller proportion of effectively neutral mutations, because the efficacy of selection is higher in populations with large long-term Ne. The high levels of observed silent-site diversity in Paramecium species (Johri et al. 2017), along with observations of very low mutation rates (Sung et al. 2012) suggest large population sizes in these species. Although it is possible that our sample sizes are too small to accurately obtain the proportions of weakly and moderately deleterious mutations, the observation of reduced fitness cost of amino-acid changing and loss-of-function mutations in lowly expressed paralogs implies that paralogs with lower expression levels experience less stringent purifying selection, most likely because of a lower contribution to the total function of the encoded protein.
Discussion
Duplicate-Gene Loss is Irreversible and Occurs via Selection on the Total Activity of a Protein Encoded by Paralogs
Paralogs resulting from WGDs have been studied phylogenetically for decades and much has been learned about patterns of retention and loss from various eukaryotic species (Lynch 2007). It is still unclear why and how certain gene duplicates are retained, and a complete understanding of this issue will require multiple approaches, including genetic and molecular techniques to differentiate functions of paralogs. Taking a population-genetic approach to better understand the processes of loss of paralogs arising from WGD, we find that often one of the two WGD paralogs is under relaxed purifying selection, revealed by an increased probability of harboring loss-of-function polymorphisms. Moreover, the same locus was found to have accumulated nonsynonymous substitutions and experienced a concomitant decrease in expression level over a long period (∼236Ne generations in P. tetraurelia, corresponding to the expected time to the common ancestor of P. tetraurelia and P. biaurelia; see Materials and Methods), thereby sealing its eventual fate down a long-term path toward complete silencing.
These observations collectively imply that the slow accumulation of slightly deleterious mutations in one duplicate locus is a major path to nonfunctionalization. Earlier studies of duplicates created by WGDs in Paramecium (Gout and Lynch 2015) and in plants (Schnable et al. 2012) have invoked the quantitative subfunctionalization model of duplicate-gene loss (Force et al. 1999; Thompson et al. 2016), wherein selection acts on the sum of expression levels of the two loci, such that over time the two loci can evolve differences in expression level, with the paralog with lower expression being eventually lost. However, the current study suggests that nonfunctionalization likely proceeds via mutations in both regulatory and coding regions, as selection must act on the total performance of the encoded protein. In other words, the total performance of a protein must be the product of the expression level as well as the functional efficiency of the protein itself, governed separately by sequences at the intergenic and coding regions respectively. Thus, the two paralogs are allowed to wander both in expression and in protein-sequence space as long as the total activity performed by the sum of the gene products remains equivalent to that of the ancestor. The implication is that eventually one copy degenerates enough in sequence and/or expression that its contribution to the total activity is not significant, allowing loss-of-function mutations to segregate and eventually fix in the population.
Consistently, we find that the DFE of new nonsynonymous mutations and frameshift-causing indels in the paralog with lower levels of expression is skewed toward neutral or mildly deleterious mutations relative to their highly expressed counterparts. Although the quantitative subfunctionalization model assumes that changes in levels of expression precede those in coding regions, we here propose that deterioration of functional efficiency encoded by the coding sequence could be equally likely to initiate the process of duplicate-gene loss (as higher quantities of an inefficient protein product is less likely to be favored by selection). The question of whether duplicate-gene loss proceeds via fixations of deleterious mutations in protein-coding regions or regulatory regions first remains open and is probably dependent on their mutational target size as well the DFE of both coding and noncoding regions. However, it is clear that the process of duplicate-gene loss eventually becomes nearly irreversible, that is, once one of the two paralogs has begun to fix an excess of slightly deleterious mutations impairing its function or expression, that paralog is far more likely to be lost in the future, although eventual silencing still requires a very long time.
The proposed model here also suggests that the degeneration of one paralog could accompany the improvement of the corresponding paralog, resulting in either increase in levels of expression or an increased functional efficiency. Although detecting improvement in functional efficiency would be challenging, increase in levels of expression of the “better” paralog might be possible. However, only a slight increase in expression in the intact paralog was observed (see fig. 3). A rigorous phylogenetic study that takes into account the noise in the measurement of expression levels across species would be required in the future to explicitly test this hypothesis. It is also possible that the increase in expression of the highly expressed paralog is either significantly less likely or simply not observable. For instance, the fixation of weakly beneficial mutations that increase levels of expression or functional efficiency in the highly expressed paralog might be much less probable because beneficial mutations are extremely rare (Eyre-Walker and Keightley 2007). Moreover, new mutations that increase levels of expression of highly expressed genes are likely to be even rarer (e.g., Metzger et al. 2015) and most WGD paralogs currently retained belong to highly expressed genes. Thus, an increase in expression might not occur in all retained paralogs. Additionally, fixations of weakly beneficial mutations do not leave detectable signatures in patterns of polymorphism (Kim and Stephan 2002; Crisci et al. 2013) and would thus be difficult to identify using population-genomic data. However, divergence-based tests such as the McDonald–Kreitman (McDonald and Kreitman 1991) and Hudson-Kreitman-Aguadé (HKA) test (Hudson et al. 1987) performed in the coding and intergenic region, respectively, could be used to test this hypothesis more explicitly in the future. Adding to the above difficulties, the highly expressed paralog might instead fix mutations in the coding region that improve the functional efficiency of the protein product, which might not result in detectable increase in expression level.
Retention Due to Dosage or Dosage Compensation can Result in a Long Time to Duplicate-Gene Loss
The time to loss of a duplicate gene can shed light on mechanisms responsible for retention. Paramecium species have retained ∼50% of WGD paralogs for ∼320 Ma, such that the synonymous site divergence between paralogs is saturated, and it remains unclear why paralogs have been retained for such a long period of time. To answer this question, we require an expectation for the time to loss of gene duplicates once a polyploid has fixed in a population. This mean time to fixation of null alleles in gene duplicates, which has attracted much attention in population genetics (Bailey et al. 1978; Kimura and King 1979; Takahata and Maruyama 1979; Li 1980; Watterson 1983; Walsh 1995), is expected to be long in populations of large effective size, when the null mutation rate per locus is low, and when the selective disadvantage of null alleles is large. Consistently, previous work has suggested that base-pair mutation rates in Paramecium species are among the lowest in eukaryotes (Sung et al. 2012; Lynch et al. 2016; Long et al. 2018) and that their effective population sizes (∼107–108) are very large (Catania et al. 2009; Johri et al. 2017). In addition, the current study has concluded that the fitness effects of loss of a single locus are strongly deleterious relative to the power of drift (i.e., 2Nes < −100). Moreover, because ciliates use only one stop codon, the rate of input of null alleles is further reduced relative to other species with similar population sizes, suggesting potentially long times to fixation of null alleles in gene duplicates.
On accounting for large population sizes and mutation rates, and assuming that the selective disadvantage of the double homozygote null follows the DFEs of frameshift-causing indels inferred in this study (whereas all other genotypes have equal fitness; with the caveat that the DFE was inferred assuming additive effects), it is expected to take on average ∼16Ne, ∼20Ne, and ∼40Ne generations (with ∼300Ne generations for the shortest genes) for one of the two gene duplicate loci to be lost (obtained using eqs. 1 and 2 in Materials and Methods) when null mutation rates are 1.0×, 0.1×, and 0.01× the average base-pair mutation rate in Paramecium populations, respectively (table 1). Considering different possible values of mean selection against the double homozygote null (table 1), it is clear that genes under stronger purifying selection and those with a smaller null mutation rate (i.e., smaller total length) take much longer to be lost in a population than genes that are less important and/or physically longer (table 1). However, only ∼13% and 15% of WGD paralogs have been lost in the P. tetraurelia and P. biaurelia lineages, respectively, after their split about 236Ne generations ago. On the one hand, this raises the possibility that most of the gene duplicates currently retained (∼50%) have undergone neofunctionalization or subfunctionalization (which seems less likely from supplementary fig. S8, Supplementary Material online). However, the more likely possibility is that the relatively simplistic model assumptions about only the homozygous nulls having a selective disadvantage (i.e., there is a fitness cost only when both diploid loci of the two paralogs have a loss-of-function mutation) do not hold well and all functional copies of the WGD paralogs are often required to maintain wild-type (i.e., ancestral) fitness, in which case the mean expected time to loss would greatly increase (Takahata and Maruyama 1979) and be more consistent with observations. Further theoretical models need to be developed to obtain appropriate null expectations of time to duplicate-gene loss accounting for more complex fitness scenarios. For instance, in this study, WGD paralogs were observed to develop differences in expression levels gradually over a long evolutionary time, and were found to have different distributions of selection coefficients of new mutations, depending on their specific levels of expression. Thus, incorporation of (1) a DFE which is a function of the level of expression, (2) population-size changes, and (3) more accurate estimates of rates of new mutations that result in null alleles, will bring us closer to more realistic null expectations of the time to fix a null allele in a gene duplicate.
Table 1.
Expected Time to Fixation of Null Alleles in Gene Duplicates in P. tetraurelia and P. biaurelia Assuming Different Rates of Mutation to a Null Allele per Gene and Assuming That the Fitness Disadvantage of the Double Homozygote is s Compared With All the Other Genotypes (which have equal fitness).
| 2Nes (of Homozygote Null) | Mean Time to Fixation of a Loss-of-function Allele in Ne Generations (Minimum, Maximum) | ||
|---|---|---|---|
| Null Mutation Rate = Base-Pair Mutation Rate | Null Mutation Rate = 0.1 × Base-Pair Mutation Rate | Null Mutation Rate = 0.01 × Base-Pair Mutation Rate | |
| P. tetraurelia | |||
| −10 | 1.0 (0, 5.7) | 4.8 (0.5, 35.5) | 27.3 (3.6, 330.0) |
| −100 | 3.3 (0.4, 8.0) | 7.1 (2.8, 37.8) | 29.6 (5.9, 332.0) |
| −1,000 | 5.6 (2.7, 10.3) | 9.4 (5.1, 40.1) | 31.9 (8.2, 334.3) |
| −2Ne | 17.6 (14.6, 22.3) | 21.4 (17.0, 52.1) | 43.9 (20.1, 346.2) |
| DFE of indels | 16.6 (13.6, 21.3) | 20.4 (16.0, 51.1) | 42.9 (19.1, 345.2) |
| P. biaurelia | |||
| −10 | 0.8 (0.0, 4.5) | 3.8 (0, 24.6) | 19.1 (2.8, 220.7) |
| −100 | 2.9 (0.0, 6.8) | 6.1 (2.3, 26.9) | 21.4 (5.1, 223.0) |
| −1,000 | 5.2 (2.3, 9.1) | 8.4 (4.6, 29.2) | 23.7 (7.4, 225.3) |
| −2Ne | 17.3 (14.4, 21.2) | 20.5 (16.7, 41.3) | 35.8 (19.6, 237.5) |
| DFE of indels | 16.3 (13.4, 20.2) | 19.5 (15.7, 40.3) | 34.9 (18.6, 236.5) |
note.—The population-scaled fitness of the double homozygote is presented above as 2Nes where Ne is the effective population size of the Wright-Fisher diploid population and 2Nes ≫ 0. The “DFE of indels” corresponds to expected time to loss when the distribution of 2Nes is assumed to follow the inferred DFEs of frameshift-causing indels from this study. Mean, minimum, and maximum values of expected time to loss correspond to those obtained using the mean, maximum, and minimum lengths of all protein-coding sequences respectively in P. tetraurelia, P. biaurelia, P. sexaurelia, and P. caudatum genomes.
Moreover, whereas fixation of null alleles has been more thoroughly investigated, broader theoretical models of duplicate-gene loss are needed that allow for more gradual accumulation of single-base mutations with mildly deleterious effects. A recent study by Thompson et al. (2016) used a diffusion-based approach to account for mutations of weaker effects (although they assumed a DFE constant in time) and modeled the change in levels of expression of the two paralogs such that selection acts only on the sum of their expression levels (i.e., dosage compensation). They showed that the time to gene-duplicate loss can increase substantially under such scenarios, with about half of the duplicates expected to be lost in ∼6,000N generations. We believe that we have caught this gradual loss of duplicates in progress and observe that it can take a long time for complete nonfunctionalization of one locus. Interestingly, when the strength of dosage compensation is stronger, the time to loss of duplicate loci is longer and the probability of neofunctionalization is higher (Thompson et al. 2016), possibly in the same locus that contributes less to the total activity of the protein, and is therefore allowed to explore previously inaccessible genotypes (Ohno 1970). Although this remains to be tested empirically, it seems clear that gene-duplicates created by WGDs can take an extremely long time to pseudogenize due to selective constraints such as dosage compensation. Although that could increase the probability of neofunctionalization in the long term, retention of gene duplicates for a long evolutionary time does not necessarily imply preservation by gain or change of function.
Materials and Methods
Obtaining Paramecium Reference Genomes and Expression Level Data
Complete genomes of P. tetraurelia, P. biaurelia, P. sexaurelia, and P. caudatum were downloaded from ParameciumDB (Arnaiz et al. 2007; https://paramecium.i2bc.paris-saclay.fr/download/Paramecium/; Arnaiz and Sperling 2011) along with the annotations (ptetraurelia_mac_51_annotation_v1.0.gff3; biaurelia_V1-4_annotation_v1.gff3; sexaurelia_AZ8-4_annotation_v1.gff3; caudatum_43c3d_annotation_v1.gff3). Ortho-paralog relationships as well as functional annotations were retrieved from the supplementary information, Supplementary Material online provided in McGrath, Gout, Doak, et al. (2014) (supplementary files S2 and S10, Supplementary Material online). RNAseq data for all four species were downloaded and processed as in Gout and Lynch (2015).
Reference genomes, annotations, and RNAseq data of P. octaurelia, P. decaurelia, and P. dodecaurelia were downloaded from ParameciumDB. Reads were mapped to the reference genome using tophat2/bowtie2 (Kim et al. 2013) and FPKM (Fragments Per Kilobase of transcript per Million mapped reads) values were obtained with cufflinks 2.2 (Trapnell et al. 2010). Expression levels are defined as log(FPKM + 0.01). The small 0.01 value is added to avoid performing a log-transform of zero values. For between-species comparisons, log-transformed expression levels were normalized using the quantile methods implemented in Bioconductor limma (Ritchie et al. 2015) so that the distributions of expression levels perfectly overlap for all the species considered.
Variant Calling
Whole-genome sequencing raw reads from five to ten isolates of P. tetraurelia, P. biaurelia, P. sexaurelia, P. caudatum, and P. multimicronucleatum were downloaded from SRA (SRA accession: SRR8698631–SRR8698604; Bioproject: PRJNA525710; Biosample: SAMN11059622–SAMN11086832), and SNPs were called as described by Johri et al. (2017). Briefly, reads were trimmed using Trimmomatic (version 0.36; Bolger et al. 2014) and mapped to reference genomes using bwamem (0.7.12; Li and Durbin 2010) under default parameters. Duplicate reads were marked using picard (2.8.0; https://broadinstitute.github.io/picard/). Sites were only considered for further analysis if the mapping quality was above 30, base quality was above 20, per-base alignment quality was above 15, and the sum of the depth of coverage for all individuals was about five times the number of individuals and less than twice the average population coverage. Variants were called using bcftools (Li et al. 2008) and filtered using vcftools (Danecek et al. 2011). An additional script vcfutils.pl provided by vcftools was used to filter variants (perl vcfutils.pl varFilter -d 50 -D 800 -1 -2 -3 -4 species_snps.vcf > species_snpsfiltered1.vcf), which was the only filter applied only to variants and not all sites in the genome. Only those sites were considered whose quality value (–minQ) was above 20. Genotypes whose genotype quality score (–minGQ) was lower than 30 or those that were supported by <4 reads (–minDP) were excluded or considered missing. In addition, protein-coding genes that had more than one heterozygous indel present in the same strain were excluded from further analyses in order to avoid false positives due to mis-mapping.
Detecting Genes with Inactivating Polymorphisms
The final filtered list of SNPs, indels, and all sites that passed the above filters were used to obtain nucleotide sequences for all protein-coding genes in each strain in every species separately, with missing or ambiguous genotypes encoded as “N.” Sequences where indels were detected to be larger than 30 bp and not a multiple of 3, were considered to have “large deletions” leading to loss-of-function mutations. If indels were smaller than 30 bp, all nucleotide sequences were translated to identify the presence of stop codons that appeared before the stop codon present in the reference genome, which was assumed to represent the longest open-reading frame. Such sequences were identified to have PTCs. Cases where multiple indels did not result in a PTC (i.e., the sum of numbers of nucleotides inserted/deleted in a gene were a multiple of 3) were not included in this analysis. Gene sequences with indel lengths that did not add up to multiples of 3 but also did not have PTCs were categorized as sequences with frameshift mutations only. Sequences with missing start and stop codons were also considered to have loss-of-function mutations. In order to avoid false positives generated by potential misalignments of the intergenic sequence upstream, gene sequences that did not have any in-frame start codons within the first 15 nucleotides at the 5′ end were identified to have “missing start codons.”
Loss-of-function mutations and larger deletions that were fixed in the population (i.e., allele frequency = 1.0) were excluded from any analyses in order to reduce detection of false positives due to reference genome assembly errors. We also restricted our analysis to genes where the average depth of coverage per individual for a gene was always >10× and where the average depth of coverage per individual for that gene was not higher than 2-fold of the mean coverage. These filters resulted in a total of 40,184; 37,764; 34,828; and 18,409 genes in P. tetraurelia, P. biaurelia, P. sexaurelia, and P. caudatum, respectively, with the mean coverage per individual per gene as ∼31, ∼50, ∼45, and ∼80 for each species, respectively.
Detecting Deletions and Duplications with CNVnator
We used CNVnator (0.3.2) (Abyzov et al. 2011) to identify isolate-specific duplications and deletions that are larger than 150 bp (length of our paired-end reads). Bam files were created using Samtools. Bin sizes were chosen such that the ratio of average and standard deviation of the read depth was around 4 (as instructed by the CNVnator manual). We used bin sizes of 100 for P. tetraurelia, 50 for P. biaurelia, 100 for P. sexaurelia, and 400 for P. caudatum. Because the average length of a protein-coding gene in Paramecium species is about 1,350 bp, we restricted all analyses of deletions to <2,000 bp. Most deletions within a gene were found in a single individual, with the frequency spectrum following a typical exponential distribution (supplementary fig. S10, Supplementary Material online).
Tajima’s Relative Rate Test
Protein-coding sequences of both the WGD paralogs from the reference genomes of each species were aligned to the P. caudatum ortholog using MUSCLE (Edgar 2004). Tajima’s relative test was conducted using MEGA (megacc-7.0.26; Kumar et al. 2016) with default parameters for obtaining asymmetry in rates of amino-acid change between the two paralogs with respect to the outgroup (P. caudatum). The total number of unique amino-acid differences in each WGD paralog, as calculated by the relative rate test, was normalized by the total length of the protein to calculate the number of amino-acid changes per codon for figure 3.
Calculation of Generations Since the Split Between P. tetraurelia and P. biaurelia
The time (Tg) in number of generations since the split between P. tetraurelia and P. biaurelia was calculated using dS = 2Tgμ, where dS is the average value of divergence at synonymous sites between P. tetraurelia and P. biaurelia estimated to be 0.8 (McGrath, Gout, Johri, et al. 2014), whereas μ is the mutation rate per site/generation. Mutation rate was assumed to be the average (=2.19 × 10−11 per site per generation) of rates in P. tetraurelia and P. biaurelia measured by mutation accumulations studies—2.44 × 10−11 and 1.94 × 10−11 per site per generation respectively (Sung et al. 2012; Long et al. 2018). Tg can also be expressed in terms of the number of Ne generations, where Ne is the effective population size of P. tetraurelia and was estimated assuming equilibrium conditions using πneu ∼ 4Neμ, where πneu is nucleotide site diversity at 4-fold degenerate sites measured previously to be 0.006 (Johri et al. 2017). Under these assumptions the Ne of P. tetraurelia was estimated to be 7.7 × 107 and Tg is obtained to be ∼236Ne generations.
Identifying Putative Candidates for Expression Neofunctionalization
Raw RNAseq data generated under vegetative condition, starvation, and different time points during autogamy in P. tetraurelia were downloaded from the SRA BioProject PRJEB19343 (https://www.ncbi.nlm.nih.gov//bioproject/PRJEB19343). The reads were mapped to protein-coding sequences using kallisto (Bray et al. 2016) to obtain expression levels in TPM (transcript per million). A value of 1.1 was added to the TPM values and they were log transformed, in order to have only positive and nonzero numbers. The relative difference in expression levels for each paralog pair for every condition was obtained as Δrel, condition = (xppara1,condition − xppara2, condition)/(xppara1, condition + xppara2, condition). For every condition (other than vegetative), the distribution of ΔΔ = Δrel,vegetative − Δrel, condition was obtained and all paralogs that lay in the tails of this distribution (defined as all values >2 × SD[ΔΔ]), were identified as putative candidates for neofunctionalization. It should be noted that this is a very generous list of candidates and will most likely contain a large number of false positives.
Calculating the SFS and the DFE
The SFS was calculated from distinct individuals, that is, individuals that seemed identical were excluded from this analysis as seen in PCA plots in Johri et al. (2017). SFS was calculated using the following strains from P. biaurelia—Sample_256-UB2, Sample_31, Sample_379, Sample_44, Sample_45, Sample_562alpha, Sample_76, Sample_7K, Sample_USBL-36I1. For P. sexaurelia, the following individuals were used—Sample_126, Sample_127, Sample_128, Sample_130, Sample_131, Sample_134, Sample_137, Sample_265, Sample_Indo1-7I, Sample_Moz13BIII. Folded SFS was calculated for 4-fold degenerate sites, 0-fold degenerate sites, and for indels of 1 or 2 bp which cause frameshift mutations (as determined above). In order to calculate the number of sites in the bin where allele frequency is zero (i.e., fixed sites), all sites that passed the depth of coverage cut-offs were used in the case of 4-fold and 0-fold degenerate sites. For indels, the total number of fixed sites (i.e., the “0” class) was multiplied by 0.1 in order to account for the difference between the mutation rate of base-pair mutations and indels. It was assumed that the mutation rate of indels is 0.1-fold that of base mutations, a number close to the average observed across eukaryotic species from previous mutation accumulation studies (Sung et al. 2016) including the one in P. tetraurelia (Sung et al. 2012). The “0” class for frameshift mutations for each protein-coding gene was obtained by calculating the total number of insertions/deletions of one or two base-pair that would result in a PTC downstream. The DFE was inferred by est_dfe in DFE-α (Keightley and Eyre-Walker 2007) assuming 1-epoch changes in population history, where time of change was variable. For every scenario, the DFE was estimated, five bootstrapped replicates were run with starting value of mean s of −0.01 and β of 0.5; another five set of bootstrapped replicates with starting values of −0.01 (mean s) and 2.0 (β); another five bootstrapped replicates with starting values of −0.001 (mean s) and 0.5 (β); another set of five bootstrapped replicates with starting values of −0.001 (mean s) and two (β). Thus, for every specific SFS, 20 replicates were used to infer the DFE and to obtain standard deviation for each class of mutations.
In order to compare across species, the DFE was estimated for WGD paralogs that have been retained in all P. aurelia species, as well as genes that are present in single copies in all three P. aurelia species. When identifying paralogs with differential expression, paralogs that had expression levels in FPKM, 5-fold higher or lower were chosen, conditional on both paralogs being expressed (i.e., paralogs where one copy was not expressed were excluded from this analysis).
Calculating Time to Fixation of Null Mutations in Gene Duplicates
The expected time to fixation (τ) of null alleles in gene duplicates provided by Watterson (1983) was calculated using
| (1) |
where Ne is the effective population size (calculated in a previous section), s is the selective disadvantage of a double homozygote null, μnull is the mutation rate of loss-of-function mutations, and ψ is the digamma function (Abramowitzm and Stegun 1970). This mathematical expression assumes that populations are finite, that there is no back mutation from a null allele to a functional gene sequence, and that only one complete gene copy is required to have wild-type function. The time to fixation accounting for a DFE (eq. 2) is derived in the Appendix.
The null mutation rate (μnull) in P. tetraurelia is difficult to estimate precisely as null mutations could include multiple types of mutations (for instance larger deletions) and was assumed to be 1×, 0.1×, and 0.01× the base-pair mutation rate of 1.94 × 10−11 per site per generation (Sung et al. 2012). Because the distribution of length of protein-coding sequences was found to be extremely similar between all Paramecium species included in this study, the mean (1,360 bp), minimum (102 bp), and maximum (22,932 bp) lengths of the genes across all four species were used to obtain the upper and lower bounds of time to fixation of null alleles in a protein-coding gene.
Supplementary Material
Supplementary data are available at Molecular Biology and Evolution online.
Supplementary Material
Acknowledgments
The authors would like to acknowledge Jeffrey Jensen, Kerry Geiler-Samerotte, and Daniel Schrider for helpful conversations related to this project, two anonymous reviewers for improving the manuscript, and Indiana University Research Computing for all computational resources. This research was supported by National Science Foundation grants MCB-1518060 and DEB-1927159 to M.L.
Appendix
Expected Time to Fixation of Null Alleles When Selection Against the Null Allele Follows a Uniform Distribution
A randomly mating Wright-Fisher diploid population is assumed. Two unlinked loci are of interest. At the first locus two alleles are segregating—A (functional) and a (null allele), whereas at the second locus the two alleles are B (functional) and b (null allele) such that functional alleles can mutate to null alleles at rate μnull irreversibly. It is assumed that the mutant double homozygote (aabb) is at a selective disadvantage s with respect to all other genotypes, which have equal fitness. The expected time to fixation of a null allele (τ) in one of the two duplicates, when the selection coefficient (s) of the double homozygous null was a constant was calculated using the approximation provided by Watterson (1983) as
where Ψ is the digamma function (Abramowitzm and Stegun 1970). Now we assume that selection against the null allele is given by a distribution ϕ(s) and that this distribution comprises of four nonoverlapping uniform distributions in the following bins: 0 ≤ 2Nes < 1, 1 ≤ 2Nes < 10, 10 ≤ 2Nes < 100, and 100 ≤ 2Nes < 2Ne such that f0, f1, f2, and f3 represent the proportion of all mutations that have fitness effects uniformly drawn from those bins respectively. In this case, on integrating τ with respect to s, we obtain
| (2) |
Contributor Information
Parul Johri, School of Life Sciences, Arizona State University, Tempe, AZ 85287, USA.
Jean-Francois Gout, Department of Biological Sciences, Mississippi State University, Mississippi State, MS 39762, USA.
Thomas G Doak, Department of Biology, Indiana University, Bloomington, IN 47405, USA; National Center for Genome Analysis Support, Indiana University, Bloomington, IN 47405, USA.
Michael Lynch, Center for Mechanisms of Evolution, The Biodesign Institute, Arizona State University, Tempe, AZ 85287, USA.
Data availability
All scripts and files used to perform analyses (including a list of genes with loss-of-function variants and putative candidates for neofunctionalization) are publicly available at https://github.com/paruljohri/WGD_Popgen_Paramecium. In addition, the following files have been made publicly available at https://figshare.com/projects/WGD_Popgen_Paramecium/136079—(1) reference genomes of all species (P. tetraurelia, P. biaurelia, P. sexaurelia, and P. caudatum) used to map resequenced data and call variants, (2) annotation of all reference genomes, (3) the final list of SNPs, (4) the final list of indels, (5) the list of all utilizable sites when calling variants, (6) nucleotide diversity per site across the genome, (7) nucleotide sequences of all protein-coding genes for each sequenced strain, and (8) the list of all genes with loss-of-function variants (including which type).
References
- Abramowitzm M, Stegun IA. 1970. Handbook of mathematical functions with formulas, graphs, and mathematical tables. Washington, DC.: National Bureau of Standards. [Google Scholar]
- Abyzov A, Urban AE, Snyder M, Gerstein M. 2011. CNVnator: an approach to discover, genotype, and characterize typical and atypical CNVs from family and population genome sequencing. Genome Res. 21:974–984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arnaiz O, Cain S, Cohen J, Sperling L. 2007. ParameciumDB: a community resource that integrates the Paramecium tetraurelia genome sequence with genetic data. Nucleic Acids Res. 35:D439–D444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arnaiz O, Sperling L. 2011. ParameciumDB in 2011: new tools and new data for functional and comparative genomics of the model ciliate Paramecium tetraurelia. Nucleic Acids Res. 39:D632–D636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arnaiz O, Van Dijk E, Betermier M, Lhuillier-Akakpo M, de Vanssay A, Duharcourt S, Sallet E, Gouzy J, Sperling L. 2017. Improved methods and resources for Paramecium genomics: transcription units, gene annotation and gene expression. BMC Genomics 18:483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aury JM, Jaillon O, Duret L, Noel B, Jubin C, Porcel BM, Segurens B, Daubin V, Anthouard V, Aiach N, et al. 2006. Global trends of whole-genome duplications revealed by the ciliate Paramecium tetraurelia. Nature 444:171–178. [DOI] [PubMed] [Google Scholar]
- Bailey GS, Poulter RT, Stockwell PA. 1978. Gene duplication in tetraploid fish: model for gene silencing at unlinked duplicated loci. Proc Natl Acad Sci U S A. 75:5575–5579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blanc G, Wolfe KH. 2004. Functional divergence of duplicated genes formed by polyploidy during Arabidopsis evolution. Plant Cell 16:1679–1691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolger AM, Lohse M, Usadel B. 2014. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 30:2114–2120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braasch I, Gehrke AR, Smith JJ, Kawasaki K, Manousaki T, Pasquier J, Amores A, Desvignes T, Batzel P, Catchen J, et al. 2016. The spotted gar genome illuminates vertebrate evolution and facilitates human-teleost comparisons. Nat Genet. 48:427–437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bray NL, Pimentel H, Melsted P, Pachter L. 2016. Near-optimal probabilistic RNA-seq quantification. Nat Biotechnol. 34:525–527. [DOI] [PubMed] [Google Scholar]
- Catania F, Wurmser F, Potekhin AA, Przybos E, Lynch M. 2009. Genetic diversity in the Paramecium aurelia species complex. Mol Biol Evol. 26:421–431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chikhi L, Sousa VC, Luisi P, Goossens B, Beaumont MA. 2010. The confounding effects of population structure, genetic diversity and the sampling scheme on the detection and quantification of population size changes. Genetics 186:983–995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crisci JL, Poh YP, Mahajan S, Jensen JD. 2013. The impact of equilibrium assumptions on tests of selection. Front Genet. 4:235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Danecek P, Auton A, Abecasis G, Albers CA, Banks E, DePristo MA, Handsaker RE, Lunter G, Marth GT, Sherry ST, et al. 2011. The variant call format and VCFtools. Bioinformatics 27:2156–2158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis JC, Petrov DA. 2004. Preferential duplication of conserved proteins in eukaryotic genomes. PLoS Biol. 2:E55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dehal P, Boore JL. 2005. Two rounds of whole genome duplication in the ancestral vertebrate. PLoS Biol. 3:e314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drummond DA, Bloom JD, Adami C, Wilke CO, Arnold FH. 2005. Why highly expressed proteins evolve slowly. Proc Natl Acad Sci U S A. 102:14338–14343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edgar RC. 2004. MUSCLE: a multiple sequence alignment method with reduced time and space complexity. BMC Bioinform. 5:113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ewing GB, Jensen JD. 2016. The consequences of not accounting for background selection in demographic inference. Mol Ecol. 25:135–141. [DOI] [PubMed] [Google Scholar]
- Eyre-Walker A, Keightley PD. 2007. The distribution of fitness effects of new mutations. Nat Rev Genet. 8:610–618. [DOI] [PubMed] [Google Scholar]
- Force A, Lynch M, Pickett FB, Amores A, Yan YL, Postlethwait J. 1999. Preservation of duplicate genes by complementary, degenerative mutations. Genetics 151:1531–1545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freeling M, Thomas BC. 2006. Gene-balanced duplications, like tetraploidy, provide predictable drive to increase morphological complexity. Genome Res. 16:805–814. [DOI] [PubMed] [Google Scholar]
- Gout J-F, Johri P, Arnaiz O, Doak TG, Bhullar S, Couloux A, Guérin F, Malinsky S, Sperling L, Labadie K, et al. 2019. Universal trends of post-duplication evolution revealed by the genomes of 13 Paramecium species sharing an ancestral whole-genome duplication. bioRxiv:573576; doi: 10.1101/573576. [DOI]
- Gout J-F, Kahn D, Duret L, Paramecium Post-Genomics Consortium . 2010. The relationship among gene expression, the evolution of gene dosage, and the rate of protein evolution. PLoS Genet. 6:e1000944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gout J-F, Lynch M. 2015. Maintenance and loss of duplicated genes by dosage subfunctionalization. Mol Biol Evol. 32:2141–2148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hahn MW. 2009. Distinguishing among evolutionary models for the maintenance of gene duplicates. J Hered 100:605–617. [DOI] [PubMed] [Google Scholar]
- Hakes L, Pinney JW, Lovell SC, Oliver SG, Robertson DL. 2007. All duplicates are not equal: the difference between small-scale and genome duplication. Genome Biol. 8:R209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang X, Fortier AL, Coffman AJ, Struck TJ, Irby MN, James JE, León-Burguete JE, Ragsdale AP, Gutenkunst RN. 2021. Inferring genome-wide correlations of mutation fitness effects between populations. Mol Biol Evol. 38:4588–4602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huber CD, Kim BY, Marsden CD, Lohmueller KE. 2017. Determining the factors driving selective effects of new nonsynonymous mutations. Proc Natl Acad Sci U S A. 114:4465–4470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hudson RR, Kreitman M, Aguadé M. 1987. A test of neutral molecular evolution based on nucleotide data. Genetics 116:153–159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Innan H, Kondrashov F. 2010. The evolution of gene duplications: classifying and distinguishing between models. Nat Rev Genet. 11:97–108. [DOI] [PubMed] [Google Scholar]
- Inoue J, Sato Y, Sinclair R, Tsukamoto K, Nishida M. 2015. Rapid genome reshaping by multiple-gene loss after whole-genome duplication in teleost fish suggested by mathematical modeling. Proc Natl Acad Sci U S A. 112:14918–14923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaillon O, Aury JM, Brunet F, Petit J-L, Stange-Thomann N, Mauceli E, Bouneau L, Fischer C, Ozouf-Costaz C, Bernot A, et al. 2004. Genome duplication in the teleost fish Tetraodon nigroviridis reveals the early vertebrate proto-karyotype. Nature 431:946–957. [DOI] [PubMed] [Google Scholar]
- Jiao Y, Li J, Tang H, Paterson AH. 2014. Integrated syntenic and phylogenomic analyses reveal an ancient genome duplication in monocots. Plant Cell 26:2792–2802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiao YN, Wickett NJ, Ayyampalayam S, Chanderbali AS, Landherr L, Ralph PE, Tomsho LP, Hu Y, Liang HY, Soltis PS, et al. 2011. Ancestral polyploidy in seed plants and angiosperms. Nature 473:97–113. [DOI] [PubMed] [Google Scholar]
- Johri P, Charlesworth B, Jensen JD. 2020. Toward an evolutionarily appropriate null model: jointly inferring demography and purifying selection. Genetics 215:173–192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johri P, Krenek S, Marinov GK, Doak TG, Berendonk TU, Lynch M. 2017. Population genomics of Paramecium species. Mol Biol Evol. 34:1194–1216. [DOI] [PubMed] [Google Scholar]
- Johri P, Riall K, Becher H, Excoffier L, Charlesworth B, Jensen JD. 2021. The impact of purifying and background selection on the inference of population history: problems and prospects. Mol Biol Evol. 38:2986–3003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keightley PD, Eyre-Walker A. 2007. Joint inference of the distribution of fitness effects of deleterious mutations and population demography based on nucleotide polymorphism frequencies. Genetics 177:2251–2261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim BY, Huber CD, Lohmueller KE. 2017. Inference of the distribution of selection coefficients for new nonsynonymous mutations using large samples. Genetics 206:345–361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim D, Pertea G, Trapnell C, Pimentel H, Kelley R, Salzberg SL. 2013. TopHat2: accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions. Genome Biol. 14:R36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim Y, Stephan W. 2002. Detecting a local signature of genetic hitchhiking along a recombining chromosome. Genetics 160:765–777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimura M, King JL. 1979. Fixation of a deleterious allele at one of two “duplicate” loci by mutation pressure and random drift. Proc Natl Acad Sci U S A. 76:2858–2861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar S, Stecher G, Tamura K. 2016. MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol Biol Evol. 33:1870–1874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee YC, Reinhardt JA. 2012. Widespread polymorphism in the positions of stop codons in Drosophila melanogaster. Genome Biol Evol. 4:533–549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li W-H. 1980. Rate of gene silencing at duplicate loci: a theoretical study and interpretation of data from tetraploid fishes. Genetics 95:237–258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Durbin R. 2010. Fast and accurate long-read alignment with Burrows-Wheeler transform. Bioinformatics 26:589–595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Ruan J, Durbin R. 2008. Mapping short DNA sequencing reads and calling variants using mapping quality scores. Genome Res. 18:1851–1858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Long H, Doak TG, Lynch M. 2018. Limited mutation-rate variation within the Paramecium aurelia species complex. G3 (Bethesda) 8:2523–2526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lynch M. 2007. The origins of genome architecture. Sunderland (MA): Sinauer Associates. [Google Scholar]
- Lynch M, Ackerman MS, Gout J-F, Long H, Sung W, Thomas WK, Foster PL. 2016. Genetic drift, selection and the evolution of the mutation rate. Nat Rev Genet. 17:704–714. [DOI] [PubMed] [Google Scholar]
- Lynch M, Conery JS. 2000. The evolutionary fate and consequences of duplicate genes. Science 290:1151–1155. [DOI] [PubMed] [Google Scholar]
- MacArthur DG. 2012. A systematic survey of loss-of-function variants in human protein-coding genes. Science 336:296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maere S, De Bodt S, Raes J, Casneuf T, Van Montagu M, Kuiper M, Van de Peer Y. 2005. Modeling gene and genome duplications in eukaryotes. Proc Natl Acad Sci U S A. 102:5454–5459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mazet O, Rodriguez W, Chikhi L. 2015. Demographic inference using genetic data from a single individual: separating population size variation from population structure. Theor Popul Biol. 104:46–58. [DOI] [PubMed] [Google Scholar]
- Mazet O, Rodriguez W, Grusea S, Boitard S, Chikhi L. 2016. On the importance of being structured: instantaneous coalescence rates and human evolution—lessons for ancestral population size inference? Heredity 116:362–371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McDonald JH, Kreitman M. 1991. Adaptive protein evolution at the Adh locus in Drosophila. Nature 351:652–654. [DOI] [PubMed] [Google Scholar]
- McGrath CL, Gout JF, Doak TG, Yanagi A, Lynch M. 2014. Insights into three whole-genome duplications gleaned from the Paramecium caudatum genome sequence. Genetics 197:1417–1428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGrath CL, Gout JF, Johri P, Doak TG, Lynch M. 2014. Differential retention and divergent resolution of duplicate genes following whole-genome duplication. Genome Res. 24:1665–1675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Metzger BP, Yuan DC, Gruber JD, Duveau F, Wittkopp PJ. 2015. Selection on noise constrains variation in a eukaryotic promoter. Nature 521:344–347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer A, Van de Peer Y. 2005. From 2R to 3R: evidence for a fish-specific genome duplication (FSGD). Bioessays 27:937–945. [DOI] [PubMed] [Google Scholar]
- Morin RD, Chang E, Petrescu A, Liao N, Griffith M, Chow W, Kirkpatrick R, Butterfield YS, Young AC, Stott J, et al. 2006. Sequencing and analysis of 10,967 full-length cDNA clones from Xenopus laevis and Xenopus tropicalis reveals post-tetraploidization transcriptome remodeling. Genome Res. 16:796–803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohno S. 1970. Evolution by gene duplication. Germany: Springer-Verlag, Heidelberg. [Google Scholar]
- Postlethwait JH, Woods IG, Ngo-Hazelett P, Yan Y-L, Kelly PD, Chu F, Huang H, Hill-Force A, Talbot WS. 2000. Zebrafish comparative genomics and the origins of vertebrate chromosomes. Genome Res. 10:1890–1902. [DOI] [PubMed] [Google Scholar]
- Ritchie ME, Phipson B, Wu D, Hu Y, Law CW, Shi W, Smyth GK. 2015. Limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 43:e47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scannell DR, Wolfe KH. 2008. A burst of protein sequence evolution and a prolonged period of asymmetric evolution follow gene duplication in yeast. Genome Res. 18:137–147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schnable JC, Wang X, Pires JC, Freeling M. 2012. Escape from preferential retention following repeated whole genome duplications in plants. Front Plant Sci. 3:94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shields DC, Wolfe KH. 1997. Accelerated evolution of sites undergoing mRNA editing in plant mitochondria and chloroplasts. Mol Biol Evol. 14:344–349. [DOI] [PubMed] [Google Scholar]
- Simillion C, Vandepoele K, Van Montagu MC, Zabeau M, Van de Peer Y. 2002. The hidden duplication past of Arabidopsis thaliana. Proc Natl Acad Sci U S A. 99:13627–13632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sonneborn TM. 1975. Paramecium aurelia complex of 14 sibling species. Trans Am Microsc Soc. 94:155–178. [Google Scholar]
- Sung W, Ackerman MS, Dillon MM, Platt TG, Fuqua C, Cooper VS, Lynch M. 2016. Evolution of the insertion–deletion mutation rate across the tree of life. G3 (Bethesda) 6:2583–2591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sung W, Tucker AE, Doak TG, Choi E, Thomas WK, Lynch M. 2012. Extraordinary genome stability in the ciliate Paramecium tetraurelia. Proc Natl Acad Sci U S A. 109:19339–19344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tajima F. 1993. Simple methods for testing the molecular evolutionary clock hypothesis. Genetics 135:599–607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahata N, Maruyama T. 1979. Polymorphism and loss of duplicate gene expression: a theoretical study with application of tetraploid fish. Proc Natl Acad Sci U S A. 76:4521–4525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson A, Zakon HH, Kirkpatrick M. 2016. Compensatory drift and the evolutionary dynamics of dosage-sensitive duplicate genes. Genetics 202:765–774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trapnell C, Williams BA, Pertea G, Mortazavi A, Kwan G, van Baren MJ, Salzberg SL, Wold BJ, Pachter L. 2010. Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat Biotechnol. 28:511–515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van de Peer Y, Maere S, Meyer A. 2010. 2R or not 2R is not the question anymore. Nat Rev Genet. 11:166. [DOI] [PubMed] [Google Scholar]
- Walsh JB. 1995. How often do duplicated genes evolve new functions? Genetics 139:421–428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watterson GA. 1983. On the time for gene silencing at duplicate loci. Genetics 105:745–766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang JZ. 2003. Evolution by gene duplication: an update. Trends Ecol Evol. 18:292–298. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All scripts and files used to perform analyses (including a list of genes with loss-of-function variants and putative candidates for neofunctionalization) are publicly available at https://github.com/paruljohri/WGD_Popgen_Paramecium. In addition, the following files have been made publicly available at https://figshare.com/projects/WGD_Popgen_Paramecium/136079—(1) reference genomes of all species (P. tetraurelia, P. biaurelia, P. sexaurelia, and P. caudatum) used to map resequenced data and call variants, (2) annotation of all reference genomes, (3) the final list of SNPs, (4) the final list of indels, (5) the list of all utilizable sites when calling variants, (6) nucleotide diversity per site across the genome, (7) nucleotide sequences of all protein-coding genes for each sequenced strain, and (8) the list of all genes with loss-of-function variants (including which type).