Abstract
Pseudomonas fluorescens Pf-5 is a soil bacterium that suppresses plant pathogens due in part to its production of the antibiotic pyoluteorin. Previous characterization of Pf-5 revealed three global regulators, including the stationary-phase sigma factor ςS and the two-component regulators GacA and GacS, that influence both antibiotic production and stress response. In this report, we describe the serine protease Lon as a fourth global regulator influencing these phenotypes in Pf-5. lon mutants overproduced pyoluteorin, transcribed pyoluteorin biosynthesis genes at enhanced levels, and were more sensitive to UV exposure than Pf-5. The lon gene was preceded by sequences that resembled promoters recognized by the heat shock sigma factor ς32 (ςH) of Escherichia coli, and Lon accumulation by Pf-5 increased after heat shock. Therefore, ςH represents the third sigma factor (with ςS and ς70) implicated in the regulation of antibiotic production by P. fluorescens. Lon protein levels were similar in stationary-phase and exponentially growing cultures of Pf-5 and were not positively affected by the global regulator ςS or GacS. The association of antibiotic production and stress response has practical implications for the success of disease suppression in the soil environment, where biological control organisms such as Pf-5 are likely to encounter environmental stresses.
Pseudomonas fluorescens is a ubiquitous soil microorganism that inhabits the surfaces of seeds and roots. Some strains of P. fluorescens, when growing in association with plants, can protect them from infection by plant pathogens (50). One such strain, P. fluorescens Pf-5, produces a number of antibiotics, including pyoluteorin (Plt) (21), pyrrolnitrin (Prn) (20), and 2,4-diacetylphloroglucinol (Phl) (39). Of the three antibiotics, Plt is most toxic to the oomycete Pythium ultimum (34), which can infect seeds and roots of many plant hosts and cause seedling death and root rot (33).
Antibiotic production by P. fluorescens is controlled by several global regulatory genes that influence multiple phenotypes, including stress tolerance, and also by regulatory genes linked to antibiotic biosynthesis gene clusters. For example, Plt production by Pf-5 is controlled by pltR, a member of the LysR family of transcriptional activators that is linked to the Plt biosynthesis genes pltLABCDEFG and pltM (38), and the global regulatory genes gacA, gacS, and rpoS. GacA and GacS constitute a two-component regulatory system that is required for the production of antibiotics, exoenzymes, and virulence factors by many Pseudomonas spp. (8, 25). Derivatives of Pf-5 harboring mutations in gacA and gacS do not produce Plt, Prn, or Phl (8, 53) and are impaired in their tolerance to oxidative stress (53). The stationary-phase sigma factor ςS, encoded by rpoS, has a differential effect on antibiotic production by Pf-5; an rpoS::Tn5 mutant does not produce Prn but overproduces Plt and Phl and is superior to Pf-5 in suppressing Pythium damping-off of cucumber (46). The rpoS::Tn5 mutant is also impaired in its tolerance to oxidative and osmotic stress (46). GacA and GacS are necessary for the timely expression and accumulation of ςS during the transition between exponential growth and stationary phase, indicating that GacA and GacS influence a regulatory circuit in which ςS is a participant (53).
Characterization of gacA, gacS, and rpoS in Pf-5 has provided a glimpse into the intricate regulatory networks controlling antibiotic production in P. fluorescens. In this study, we cloned and sequenced a fourth global regulatory gene influencing antibiotic production by Pf-5. We identified the gene as a homolog of lon, which encodes an ATP-dependent serine protease (17, 18) found in diverse organisms, including bacteria, plants, and animals. In Escherichia coli, Lon is a heat shock protein that nonspecifically degrades denatured or nonfunctional intracellular proteins (17, 18). Lon also functions in gene regulation by specifically degrading unstable regulatory proteins (17, 18). In Bacillus subtilis, lon expression is induced by salt and oxidative stress (43) as well as starvation (19). Because resistance to salt, oxidation, and starvation is regulated by ςS in Pf-5 (46; V. O. Stockwell, unpublished data) and lon::Tn5 and rpoS::Tn5 mutants are similar in their overproduction of Plt (27, 46), we evaluated the influence of ςS on Lon accumulation. Lon protein levels did not increase during stationary phase and were not reduced in rpoS mutants of Pf-5, as would be expected for proteins positively regulated by rpoS. We also evaluated two stress responses that are influenced by Lon in other bacterial genera. Lon of Pf-5 was required for optimal tolerance of Pf-5 to UV irradiation, and Lon protein was induced by heat shock.
MATERIALS AND METHODS
Bacterial strains, plasmids, and culture conditions.
The bacterial strains and plasmids used are listed in Table 1. P. fluorescens was grown at 27°C with shaking at 200 rpm in King's medium B (KB) broth (24) for routine culturing; in nutrient broth (NB) (Difco Laboratories, Detroit, Mich.) supplemented with 2% (wt/vol) glucose or 1% (wt/vol) glycerol for antibiotic extractions; in NB supplemented with 1% (wt/vol) glycerol for ice nucleation assays; in M9 minimal medium (M9) supplemented with 0.4% glucose (45) for Western analysis; or in Luria-Bertani medium (LB) (45) for UV stress tests and Western analysis. Cultures of E. coli were grown in LB at 37°C. For cultures of E. coli, antibiotic concentrations were 100 μg of ampicillin (Ap) per ml, 12 μg of gentamicin (Gm) per ml, 50 μg of kanamycin (Km) per ml, and 20 μg of tetracycline (Tc) per ml.
TABLE 1.
Bacterial strains and plasmids used in this study
| Strain or plasmid | Description | Reference(s) |
|---|---|---|
| P. fluorescens: | ||
| Pf-5 | Rhizosphere isolate | 20 |
| JL3985 | rpoS::Tn5 derivative of Pf-5, Kmr | 42, 46 |
| JL4292 | lon::Tn5 derivative of Pf-5, Kmr | 27 |
| JL4389 | pltB::Tn3-nice derivative of Pf-5, Plt− Ina+ Kmr | 28 |
| JL4479 | lon::Tn5 derivative of Pf-5 obtained by marker exchange mutagenesis, Kmr | This study |
| JL4490 | rpoS::lacZ derivative of Pf-5 | 53 |
| JL4590 | ΔhupB::aphI derivatives of Pf-5 obtained by marker exchange mutagenesis, Kmr | This study |
| JL4591 | ΔhupB::aphI derivatives of Pf-5 obtained by marker exchange mutagenesis, Kmr | This study |
| JL4594 | lon::aacC1 derivative of JL4389 obtained by marker exchange mutagenesis, Ina+ Gmr Kmr | This study |
| JL4600 | rpoS::lacZ derivative of JL4389 obtained by marker exchange mutagenesis, Ina+ Lac+ Kmr | This study |
| JL4601 | JL4389 harboring pJEL5649, Ina+ Tcr | This study |
| JL4619 | lon::aacC1 derivative of Pf-5 obtained by marker exchange mutagenesis, Gmr Kmr | This study |
| JL4620 | lon::aacC1 rpoS::lacZ derivative of Pf-5 obtained by marker exchange mutagenesis, Gmr Kmr | This study |
| JL4621 | lon::aacC1 rpoS::lacZ derivative of JL4389 obtained by marker exchange mutagenesis, Ina+ Gmr Kmr | This study |
| E. coli | ||
| DH5α | F−endA1 hsdR17 (rk− mk+) supE44 thi-1 recA1 gyrA96 relA1 φ80dlacZΔM15 λ− | 45 |
| SG20781 | lon+cps::lac | 6 |
| Plasmids | ||
| pBR322 | ColE1 replicon, Apr Mob+ Tcr | 45 |
| pRK415 | IncP1 replicon, polylinker of pUC19, Mob+ Tcr | 23 |
| pRK2013 | Mobilizing plasmid, Tra+ Kmr | 14 |
| pLAFR3 | IncP1 replicon; cos Mob+ Tcr | 29 |
| pMGm | ColE1 replicon, source of aacC1 (Gmr) cassette on a 2.0-kb SmaI fragment, Gmr Tcr | 37 |
| pMKm | ColE1 replicon, source of aphI (Kmr) cassette on a 1.7-kb blunted XhoI fragment, Kmr Tcr | 37 |
| pUC19 | ColE1 replicon, Apr | 45 |
| pUC18 | ColE1 replicon, Apr | 45 |
| pJEL01 | Stably maintained in E. coli or Pseudomonas spp., replicons from pVSP1 and pACYC184, Mob+ Tcr | 46 |
| pJEL5500 | 2.9-kb EcoRI fragment containing rpoS from Pf-5 cloned in pUC19, Apr | 46 |
| pJEL5649 | 2.9-kb EcoRI fragment containing rpoS from Pf-5 cloned in pJEL01, Mob+ Tcr | 46 |
| pJEL5913 | 9.7-kb EcoRI genomic fragment from JL4292 containing Tn5 and adjacent DNA cloned into pBR322, Apr Tcr | This study |
| pJEL5922 | 3.6-kb HindIII-EcoRI genomic fragment from pJEL5913 containing 1.1 kb of Tn5 and flanking DNA from JL4292 cloned in pBR322, Apr Tcr | This study |
| pJEL5926 | rpoS::lacZ transcriptional fusion cloned in pRK415, Mob+ Tcr | 53 |
| pJEL6023 | 4.3-kb HindIII genomic fragment from Pf-5 containing lon and hupB genes in pUC19, Apr | This study |
| pJEL6161 | 5.7-kb HindIII fragment containing ΔhupB::aphI cloned in pUC19, Apr | This study |
| pJEL6195 | 2.3-kb EcoRI genomic fragment from Pf-5 containing the upstream regulatory region of lon cloned into pUC18, Apr | This study |
| pJEL6197 | 6.3-kb HindIII genomic fragment containing aacC1 cassette from pMGm inserted into blunted SunI site internal to lon in pRK415, Mob+ Gmr Tcr | This study |
| pJEL6206 | Cosmid library clone of Pf-5 containing lon and hupB cloned in pLAFR3, Tcr | This study |
Recombinant DNA techniques.
Genomic DNA was isolated by cetyltrimethylamonium bromide with isopropanol precipitation (3). Plasmids were purified by an alkaline lysis procedure (45). Methods for transformations, digestions with restriction enzymes, and gel electrophoresis were standard (45). Enzymes were from Gibco-BRL Life Technologies (Gaithersburg, Md.). Ends of restriction fragments were blunted with the large subunit of DNA polymerase (45), and thermal cycling was used for blunt-end ligations (32).
Cloning of lon and the linked gene hupB from Pf-5.
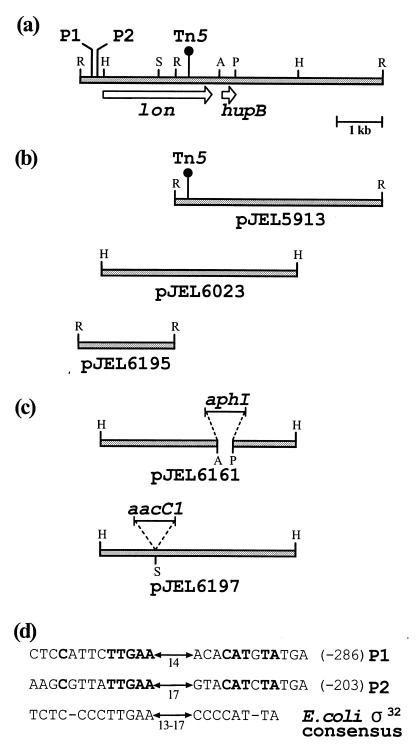
A pLAFR3 genomic library of Pf-5 (42) was screened by colony hybridization (16) to identify cosmids containing wild-type DNA corresponding to the mutagenized locus in the Plt-overproducing mutant JL4292 (Fig. 1a). The probe was a 9.7-kb EcoRI fragment containing Tn5 and flanking DNA from the genome of JL4292, which was cloned in pJEL5913 (Fig. 1b). The probe was labeled with digoxigenin-11-dUTP using a nick translation kit (Gibco-BRL Life Technologies). Qiabrane filters (Qiagen, Chatsworth, Calif.) were prepared and hybridized following the methods for the Genius system of Boehringer-Mannheim (Indianapolis, Ind.). Cosmids were isolated from colonies that hybridized to the probe, digested with HindIII or EcoRI, and analyzed in Southern blots. A 4.3-kb HindIII fragment from a cosmid that hybridized to the probe was identified by Southern analysis (data not shown) and cloned into pUC19 to construct pJEL6023 (Fig. 1b). From the same cosmid, an overlapping 2.3-kb EcoRI fragment that hybridized to the digoxigenin-11-dUTP-labeled 4.3-kb HindIII fragment from pJEL6023 was cloned into pUC18 to create pJEL6195.
FIG. 1.
Schematic representation of the genomic region of Pf-5 containing lon and hupB. (a) Restriction map of the locus and location of Tn5 in the lon mutants JL4292 and JL4479. Restriction enzymes: A, AflII; H, HindIII; P, HpaI; R, EcoRI; S, SunI. (b) Cloned genomic fragments used in sequence analysis and as probes for Southern blot analysis or for colony hybridization. (c) Schematic representation of cloned genomic fragments with insertions in lon and hupB. (d) Two potential promoters, P1 at −286 and P2 at −203 relative to the translational start site of lon in Pf-5. Nucleotides in bold are identical to the ς32 promoter consensus sequence of E. coli. Numbers beneath arrows represent nucleotides separating the consensus regions.
Sequence analysis.
DNA sequencing and oligonucleotide syntheses were performed at the Center for Gene Research and Biotechnology at Oregon State University in Corvallis. Sequencing of double-stranded templates was performed on an ABI model 373A automated DNA sequencer using a Taq DyeDeoxy Terminator Cycle sequencing kit (Applied Biosystems, Inc., Foster City, Calif.) according to the manufacturer's protocol. Oligonucleotide primers were synthesized on an ABI model 380B DNA synthesizer using phosphoramidite chemistry (1). Sequencing of lon and hupB cloned in pJEL6023 was initiated using oligonucleotide primers complementary to pUC19 DNA on either side of the polylinker and continued using primers complementary to sequenced DNA. Sequencing of the region upstream of lon was completed with primers complementary to previously sequenced DNA. The precise location of Tn5 in mutant JL4292 was determined from primers complementary to a terminal region of Tn5 using pJEL5922, a subclone of pJEL5913, as a template. Analyses of DNA and deduced protein sequences and comparisons with sequences in the GenBank database were accomplished with software from the Genetics Computer Group, Inc., Madison, Wis. (10) and the Staden software package (4).
Antibiotic quantification.
Antibiotics were extracted from cells and spent medium of cultures grown in triplicate by described methods (46). Plt and Prn concentrations were quantified from cultures grown for 2 days at 20°C in 5 ml of NB containing 1% glycerol, a medium that favors their production. The concentration of Phl was quantified from cultures grown for 4 days in 5 ml of NB containing 2% glucose, a medium that favors its production. After centrifugation of the cultures at 5,000 × g for 5 min, the bacterial pellet was suspended in 4 ml of acetone, and the suspension was sonicated in an ultrasonic cleaner for 30 s. Cell suspensions were centrifuged at 5,000 × g for 10 min, and the acetone supernatant was removed and dried under reduced pressure. Culture supernatants were adjusted to pH 2.0 with 1 M HCl and extracted twice with 2 ml of ethyl acetate. The organic phases were combined and dried under reduced pressure. Extracts dissolved in methanol were analyzed by thin-layer chromatography (TLC) or high-performance liquid chromatography (HPLC). Organic extracts were separated on TLC plates (KC18F; Whatman International Ltd., Maidstone, England) in chloroform-acetone (9:1, vol/vol) and sprayed with diazotized sulfanilic acid for visualization of compounds (44). Antibiotics were detected by their characteristic colors and Rf values, which conformed to those of authentic standards on TLC plates: Plt, Rf = 0.32, brown; Prn, Rf = 0.81, maroon; and Phl, Rf = 0.64,, yellow. Antibiotics were separated by HPLC with a Waters Nova-pak radial compression cartridge (0.8 by 10 cm) with an 18-min linear gradient from 10 to 100% acetonitrile with 0.1% acetic acid in water at a flow rate of 1 ml/min (Plt and Prn) or with water-acetonitrile-methanol (45:30:25, vol/vol) at a flow rate of 1.5 ml/min (Phl). Antibiotics were detected with a UV photodiode array detector at 310 nm (Plt), 225 nm (Prn), and 278 nm (Phl) and quantified against authentic standards. Antibiotics, reported as micrograms per milliliter ± standard deviation, were quantified by HPLC from two replicated experiments with similar results, and data from one experiment are presented.
Derivation of lon, rpoS, and hupB mutants by marker exchange mutagenesis. (i) lon::Tn5 mutant.
A 9.7-kb Tn5-containing EcoRI fragment from the genome of JL4292 was cloned into pBR322, which does not replicate in Pseudomonas spp., to create pJEL5913 (Fig. 1b). pJEL5913 was mobilized from E. coli DH5α donors into Pf-5 in a triparental mating with E. coli DH5α containing the helper plasmid pRK2013. Transconjugants from this mating were selected on KB containing streptomycin (100 μg/ml) (to counterselect against E. coli donors) and kanamycin (50 μg/ml). The resultant marker-exchanged mutant JL4479 had a Tn5 insertion in the same region as the original mutant JL4292, as determined by Southern analysis (data not shown) with the 9.7-kb EcoRI fragment from pJEL5913 as a probe.
(ii) lon::aacC1 mutants.
We evaluated the effect of a mutation in lon on pltB transcription using existing fusions of the promoterless ice nucleation reporter gene in Tn3-nice to the promoter of pltB (pltB::Tn3-nice) (28, 29). Because Tn5 and Tn3-nice both confer resistance to kanamycin, the 2.0-kb SmaI fragment containing aacC1 from pMGm, which confers gentamicin resistance, was cloned into the blunted SunI site internal to lon in pJEL6023 (Fig. 1). The resulting 6.3-kb HindIII fragment, containing lon::aacC1, was cloned into pRK415, which confers resistance to tetracycline and is not stably maintained in Pf-5, to create pJEL6197. pJEL6197 was mobilized into Pf-5 as described for the lon::Tn5 mutant, and transconjugants were selected on KB supplemented with tetracycline (200 μg/ml), which selects for pRK415 in Pf-5 and counterselects against E. coli donors. To allow loss of the plasmid, Tcr Gmr transconjugants were grown in KB broth without antibiotics at 27°C with shaking for 1 to 3 days with daily subculturing. The resultant culture was spread on KB containing gentamicin (40 μg/ml), and individual Gmr colonies were screened for loss of pJEL6197 by lack of growth on KB containing tetracycline (200 μg/ml). lon::aacC1 was introduced into Pf-5 and JL4389, which contains pltB::Tn3-nice. In the resultant marker-exchanged mutants JL4619 and JL4594, respectively, lon::aacC1 replaced lon, as determined from Southern analysis (data not shown) with the 4.3-kb HindIII fragment from JL6203 as a probe (Fig. 1b).
(iii) rpoS::lacZ mutant.
Plasmid pJEL5926, which contains an rpoS::lacZ fusion (53), was used for marker exchange mutagenesis of JL4389, which contains pltB::Tn3-nice, as described for lon::aacC1. The resultant marker-exchanged mutant JL4600 was confirmed by Southern analysis (data not shown) with a 2.9-kb EcoRI fragment from JL5500 as a probe.
(iv) rpoS::lacZ lon::aacC1 double mutants.
The lon::aacC1 mutation was exchanged with lon in the genome of two rpoS::lacZ derivatives of Pf-5, strain JL4490 (53) and strain JL4600, which contains pltB::Tn3-nice, as described above. The resultant marker-exchanged mutants, JL4620 and JL4621, respectively, were confirmed by Southern analysis (data not shown) with the 4.3-kb HindIII fragment from JL6203 as a probe (Fig. 1b).
(v) ΔhupB::aphI mutant.
The hupB gene cloned in pJEL6023 was deleted by digesting plasmid DNA with AflII and HpaI, blunting the ends of the digested DNA, and inserting the 1.7-kb aphI cassette from pMKm to derive pJEL6161 (Fig. 1c). The resulting 5.7-kb HindIII fragment was cloned into pRK415 and used to mutagenize Pf-5 as described for lon::aacC1. Replacement of hupB with ΔhupB::aphI in two independently derived marker-exchanged mutants, JL4590 and JL4591, was confirmed by Southern analysis (data not shown) with the 4.3-kb HindIII fragment of pJEL6023 as a probe (Fig. 1b).
Transcription of Plt biosynthetic genes assessed with an ice nucleation reporter gene in Tn3-nice.
The effect of rpoS and lon on transcription of the Plt biosynthetic genes was determined by comparing ice nucleation activity expressed by Pf-5 containing genomic insertions of Tn3-nice in pltB (JL4389) (28) to derivative strains with rpoS::lacZ (JL4600) (53), with lon::aacC1 (JL4594), with both rpoS::lacZ and lon::aacC1 mutations (JL4621), or with multiple plasmid-borne copies of rpoS (JL4601). Ice nucleation activity was quantified by a droplet-freezing assay at −5°C as described previously (31) from cultures grown for 2 days at 20°C with shaking at 200 rpm in NB amended with 1% glycerol. Cultures were grown in triplicate, the experiment was done twice, and the results of a representative experiment are presented.
Western analysis of Lon and ςS.
Sodium dodecyl sulfate-polyacrylamide gel electrophoresis and transblotting of total protein extracts for Western analysis were done using the manufacturer's protocols (Bio-Rad Laboratories, Hercules, Calif.) and reported methods (11, 51). Bacterial cultures were grown in LB or M9 supplemented with 0.4% glucose at 27°C with shaking at 200 rpm. For each P. fluorescens culture, 30 μg of protein (Bio-Rad DC protein assay) from whole-cell extracts was boiled in sample buffer containing 2-mercaptoethanol and separated on a sodium dodecyl sulfate–8% polyacrylamide gel. A 10-μg amount of protein (Bio-Rad DC protein assay) from whole-cell extracts of E. coli was loaded on the gel as a positive control. Protein samples were transferred from the gel onto a nitrocellulose membrane, and the membrane was incubated with antibodies to the Lon protein from E. coli (11), which were detected by enhanced chemiluminesence as specified by the manufacturer (Amersham Pharmacia Biotech Inc., Piscataway, N.J.). The amount of Lon in strains of P. fluorescens was quantified by using a Molecular Dynamics Personal Densitometer model SI and ImageQuant software V4.1 (Sunnyvale, Calif.) and confirmed to be within the linear range of detection, as described previously (53). The amount of Lon is reported relative to the amount detected in Pf-5 growing exponentially at 27°C. ςS was quantified from these and similar blots with 10 μg of protein per sample after blots were stripped according to the manufacturer's protocols (Amersham) and reprobed with antibodies to ςS from E. coli (49). Each experiment was replicated with similar results.
Sensitivity to UV irradiation.
Cultures were grown with shaking at 27°C in LB, and stationary-phase cells were obtained 4 h after the optical density (λ = 600 nm) of cultures stopped increasing. Cells were pelleted, washed, serially diluted in 10 mM phosphate buffer (pH 7), and spread onto duplicate or triplicate LB agar plates. Agar plates were exposed to UV irradiation (λ = 254 nm) at a level of 10 erg/mm2 for various durations. Colonies arising from surviving cells were counted following 48 h of incubation in the dark. The experiment was done twice with similar results.
Nucleotide sequence accession number.
The GenBank accession number for the DNA sequence of the lon and hupB genes of P. fluorescens Pf-5 is AF250140.
RESULTS
Identification of a lon::Tn5 derivative of Pf-5 that overproduces Plt.
JL4292, a derivative of Pf-5 obtained following random Tn5 mutagenesis, overproduces the antibiotic Plt (27). To confirm that the Tn5 insertion caused overproduction of Plt by JL4292, the transposon was reintroduced into the same site in the genome of Pf-5 by marker exchange mutagenesis to create JL4479. JL4292 and JL4479 each contained a single Tn5 insertion in a 9.7-kb EcoRI fragment of genomic DNA, as determined from Southern analysis (data not shown) using the Tn5-containing EcoRI fragment from the genome of JL4292 as a probe (Fig. 1b). Both JL4292 and JL4479 produced a higher concentration of Plt than did Pf-5 (Table 2), confirming that Plt overproduction by JL4292 was associated with the Tn5 insertion and not due to secondary mutations at other loci. Pf-5, JL4292, and JL4479 did not differ significantly in their production of Phl in two replicate experiments (data not shown).
TABLE 2.
Antibiotic production by P. fluorescens Pf-5 and derivativesa
| Strain | Relevant characteristics | Mean production (μg/ml) ± SD (n = 3)
|
|
|---|---|---|---|
| Plt | Prn | ||
| Pf-5 | Wild type | 5.3 ± 1.2 | 1.04 ± 0.26 |
| JL4292 | lon::Tn5 | 117 ± 11 | 1.42 ± 0.10 |
| JL4479 | lon::Tn5 | 101 ± 14 | 1.44 ± 0.12 |
| JL4619 | lon::aacC1 | 96 ± 4 | 1.31 ± 0.13 |
| JL4620 | lon::aacC1 rpoS::lacZ | 164 ± 13 | 0.32 ± 0.07 |
| JL4490 | rpoS::lacZ | 145 ± 13 | 0.11 ± 0.02 |
| JL3985 | rpoS::Tn5 | 149 ± 9 | 0.38 ± 0.04 |
| JL4590 | ΔhupB::aphI | 4.2 ± 0.6 | 0.83 ± 0.18 |
Plt and Prn were extracted from cells and spent medium of cultures grown for 2 days at 20°C in 5 ml of NB containing 1% glycerol.
We began our analysis of the locus disrupted in JL4292 (Fig. 1a) by identifying cosmids containing the corresponding wild-type DNA from an extant genomic library of Pf-5 (42). Cosmids that hybridized to the Tn5-containing EcoRI fragment from the genome of JL4292 were identified. From one such cosmid, a 4.3-kb HindIII fragment that hybridized to the probe was cloned (pJEL6023) (Fig. 1b) and used as a template for sequencing. Within the 4.3-kb HindIII fragment, an open reading frame (ORF) of 2,397 bases was identified, with the putative ribosome-binding site GAGG located 8 bases upstream of an ATG start codon. In JL4292, Tn5 disrupted the ORF at base 1798 of the coding sequence. Once translated, the ORF was predicted to encode a peptide of 798 amino acids, with 70% identity to Lon of E. coli (7). Within the deduced amino acid (a.a.) sequence of Lon from Pf-5, we located all of the characteristic Lon protein domains. From a.a. residues 206 to 266, we located a charged region (a.a. 206 to 221, 53% basic; a.a. 227 to 249, 52% acidic; and a.a. 251 to 266, 31% basic). This region is predicted to form a coiled-coil structure in E. coli (13), a common motif involved in protein-protein interactions. Within the acidic portion of the charged region, we located a sequence identical to the discriminator activity consensus (KAIQKELGD from a.a. 232 to 240) from Lon of E. coli (13). A mutation within this sequence abolishes activity of Lon towards two specific substrates in E. coli (13). ATP-binding motifs (15, 52) that matched the corresponding motifs in Lon of E. coli (7) were located from a.a. 352 to 359 (GPPGVGKT) and from a.a. 405 to 420 (KVGVRNPLFLLD). A putative catalytic site serine (2) is at a.a. residue 674. In JL4292, Tn5 spatially separates the conserved serine residue from the ATP-binding motif and the discriminator activity consensus.
The upstream regulatory region of lon, including its promoter, was not located within the 4.3-kb HindIII fragment cloned in pJEL6023. Therefore, an overlapping EcoRI fragment was cloned into pUC18 to create pJEL6195 (Fig. 1b), which was a template for sequence analysis. In the sequence upstream of lon, we identified two potential promoters (Fig. 1d), one 203 bases upstream and the other 286 bases upstream of the start codon. The sequences of both promoter regions resembled the consensus sequence recognized by the ς32 (ςH) form of RNA polymerase, which is located upstream of various heat shock-inducible genes in E. coli, including lon (55).
ΔhupB::aphI derivative of Pf-5 overproduced Phl but did not overproduce Plt.
A second ORF with a predicted GTG start codon was identified 148 bases downstream of the lon ORF. The deduced protein encoded by the ORF is 90 amino acids in length and is 80% identical to the histone-like protein HU from Pseudomonas aeruginosa (9) encoded by hupB. The hupB gene in E. coli encodes one of two subunits of the heterodimeric protein that is involved in regulating transcription by constraining DNA supercoils and DNA accessibility to regulatory proteins (12). In the genome of E. coli, hupB is located downstream of lon, which further supports our designations for the ORFs as lon and hupB homologs. Within the intergenic region between lon and hupB, a putative rho factor-independent terminator-like sequence was identified (5) as a run of consecutive thymine residues. However, a discernible region of dyad symmetry that characterizes terminators did not immediately precede the sequence. Without more convincing evidence of a terminator between lon and hupB, we chose to test the possibility that a polar effect of lon::Tn5 on hupB was responsible for Plt overproduction in JL4292 by deleting hupB from Pf-5 and testing the resulting strains for Plt production.
Two ΔhupB::aphI mutants, JL4590 and JL4591, were generated by marker exchange mutagenesis and confirmed not to overproduce Plt, as assessed by TLC (data not shown). Both JL4590 and JL4591 were more mucoid on KB plates and more viscous in NB culture than Pf-5. Strain JL4590 was further evaluated by HPLC analysis, which revealed that the ΔhupB::aphI mutant did not differ from Pf-5 in its production of Plt (Table 2). Therefore, the possibility that the Tn5 insertion into lon enhanced Plt production by blocking readthrough transcription of hupB was discounted. JL4590 produced more Phl than was produced by Pf-5 in parallel cultures (38.6 ± 8.3 and 13.1 ± 0.7 μg/ml, respectively) grown in NB containing 2% glucose.
Lon protease and ςS influenced pltB biosynthetic gene transcription.
The influence of Lon and ςS on the transcription of pltB was assessed with transcriptional fusions of the ice nucleation reporter gene in Tn3-nice to pltB. Ice nucleation activity expressed by JL4389, which has an insertion of Tn3-nice in the genomic pltB gene, was compared to the activity of derivative strains with a lon::aacC1 or rpoS::lacZ mutation, with both the lon::aacC1 and rpoS::lacZ mutations, or with multiple plasmid-borne copies of rpoS (Table 3). Ice nucleation activity is expressed as log10 (ice nuclei per cell); therefore, increasingly positive values represent higher pltB transcription, whereas increasingly negative values represent lower pltB transcription. In derivatives of JL4389, mutations in rpoS and lon significantly increased pltB transcription compared to strains with functional rpoS and lon, and transcription in a strain harboring both mutations was further enhanced. Multiple plasmid-borne copies of rpoS significantly reduced pltB transcription compared to that in strains with a single genomic copy of rpoS.
TABLE 3.
Influence of lon and rpoS on transcription of the Plt biosynthesis gene pltB, assessed with an ice nucleation reporter gene in Tn3-nice
| Strain | Relevant characteristicsa | Ice nucleation activity [log10 (ice nuclei per cell)] ± SD |
|---|---|---|
| JL4389 | lon+rpoS+ | −1.7 ± 0.6 |
| JL4594 | lon::aacC1 rpoS+ | −0.39 ± 0.1 |
| JL4600 | lon+rpoS::lacZ | −0.67 ± 0.1 |
| JL4621 | lon::aacC1 rpoS::lacZ | 0.17 ± 0.3 |
| JL4601 | lon+rpoS++ | −3.1 ± 0.4 |
rpoS++ designates the presence of both a genomic copy of rpoS and pJEL5649, a plasmid containing the cloned rpoS from Pf-5.
Lon accumulation increased after heat shock.
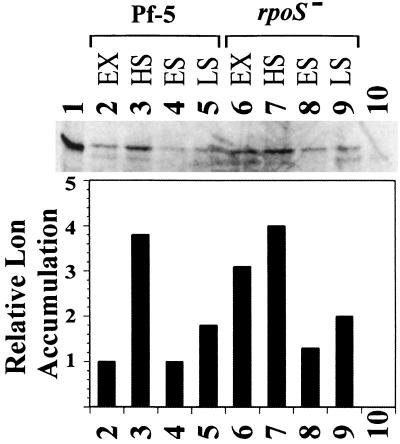
Western analysis identified the Lon protein in Pf-5 (Fig. 2, lanes 2 to 5) but identified no detectable Lon in JL4292 (lane 10). When exponentially growing cultures of Pf-5 at an optical density at 600 nm (OD600) of 0.2 (lane 2) were shifted from 27 to 42°C for 25 min to simulate heat shock (lane 3), Lon accumulation increased. In stationary-phase cultures at an OD600 of 2.0 (lane 4) or grown overnight to an OD600 of 2.0 to 4.0 (lane 5), Lon accumulation was not considerably greater than in exponentially growing cultures of Pf-5. An rpoS::Tn5 mutant of Pf-5 still showed heat shock induction of Lon (lanes 6 and 7) and had higher levels of Lon than did Pf-5 in both the exponential (lane 6) and stationary (lanes 8 and 9) phases. Lon was detected and induced by heat shock in cells lacking GacS (data not shown). The amounts of ςS in Pf-5 and the lon::Tn5 mutant JL4292 were indistinguishable on these and other blots (data not shown). Results were similar when strains were grown in LB (data not shown).
FIG. 2.
Detection of Lon from Pf-5 grown in M9 minimal medium (45) with antiserum to Lon of E. coli. Sample numbers correspond to extracts from cells growing exponentially (EX) at 27°C at an OD600 of 0.2 (lanes 1, 2, and 6), cells heat shocked (HS) at 42°C for 25 min (lanes 3, 7, and 10), early-stationary-phase cultures (ES) at an OD600 of 2 (lanes 4 and 8), and cultures grown overnight to a final OD600 of 2 to 4 (late stationary phase [LS]) (lanes 5 and 9). Lanes: 1, E. coli SG20781; 2 to 5, Pf-5; 6 to 9, JL3985 (rpoS::Tn5); 10, JL4292 (lon::Tn5). Experiments were repeated with similar results. Scanned images were prepared for publication using Adobe Photo Shop version 4.0 (Adobe Systems Incorporated, San Jose, Calif.).
lon::Tn5 derivative was more sensitive than Pf-5 to UV irradiation.
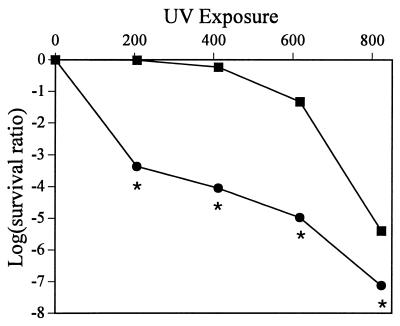
We tested Pf-5 and its lon::Tn5 derivative for their abilities to survive exposure to UV, because lon mutants of E. coli are more sensitive than lon+ strains to UV irradiation (17, 18). The survival ratio of JL4292 was 1,000 times lower than that of Pf-5 after UV exposure at 206 erg/mm2 and maintained that difference up to an exposure of 618 erg/mm2 (Fig. 3). Cells of JL4292 were also noticeably elongated both before and after UV exposure, consistent with the lethal filamentation phenotype associated with lon in E. coli.
FIG. 3.
Sensitivity of cells to UV irradiation. Stationary-phase cells of P. fluorescens Pf-5 (■) and JL4292 (●) were exposed to UV radiation (λ = 254 nm), expressed as ergs per square millimeter, for various durations. Colonies arising from surviving cells were counted following 48 h of incubation in the dark. Data were transformed as log survival ratio. Statistical analysis of variance was completed with SAS statistical software (Statistical Analysis Systems Institute, Cary, N.C.). Asterisks indicate data points where Pf-5 and JL4292 differ significantly (P = 0.05) as determined by Fisher's least-squares difference. The experiment was repeated with similar results.
DISCUSSION
We cloned, sequenced, and partially characterized the lon homolog in P. fluorescens and demonstrated its role in the regulation of the antibiotic Plt. Like the stationary-phase sigma factor ςS (46), Lon is a global regulator that negatively influences Plt production, so we evaluated the interactions of these regulators in Pf-5. Accumulation of Lon in cells of Pf-5 was not positively influenced by ςS or GacS, a member of a two-component regulatory system that controls ςS levels in this bacterium (53). Therefore, Lon does not appear to be an intermediate in the regulatory circuit involving GacA, GacS, and ςS. Furthermore, levels of ςS were similar in the lon::Tn5 mutant and Pf-5, indicating that Lon and ςS influence plt biosynthetic gene transcription and Plt production through separate regulatory circuits. It is possible that these circuits could converge through a plt pathway-specific regulator, which could integrate signals from diverse sensory transduction pathways.
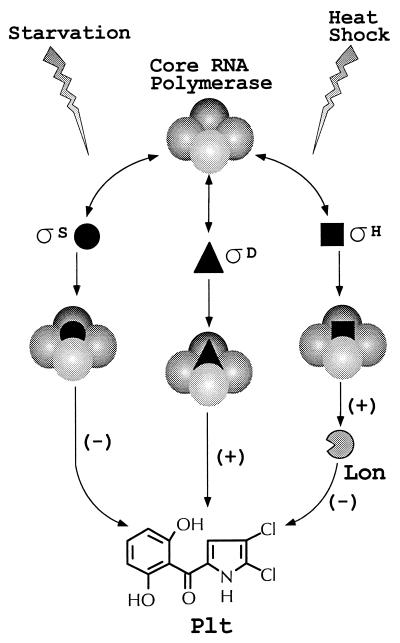
Competition between sigma factors for limited core RNA polymerase is implicated in regulation of Lon accumulation (40) and Plt production (46, 47). In Pf-5, lon is preceded by ς32-like promoter sequences, suggesting that transcription of lon is initiated by the ς32 homolog ςH. Furthermore, Lon accumulation increased after heat shock of Pf-5, as is typical of heat shock proteins transcribed from ςH promoters. In an rpoS::Tn5 derivative of Pf-5, Lon accumulation exceeded wild-type levels, indicating that the stationary-phase sigma factor ςS negatively influences Lon. One possible explanation for this result is that expression of lon increases with the concentration of the ςH RNA polymerase holoenzyme, which is likely to be enhanced in the absence of competing sigma factors such as ςS. A precedent for this explanation exists in E. coli, in which the induction of heat shock proteins is observed in a strain with decreased levels of the housekeeping sigma factor ς70 (40). In light of recent evidence that Lon degrades ςH in Bacillus subtilis under conditions in which the sigma factor has low activity (30), we considered the possibility that Lon repressed Plt production by degrading a sigma factor required for plt biosynthetic gene transcription. Close examination of the plt biosynthetic operon revealed no ς32-like promoter sequences; therefore, it is unlikely that ςH initiates plt transcription or that Lon represses plt transcription by degrading ςH and reducing the amount of ςH RNA polymerase holoenzyme. Previously, two other sigma factors were implicated in the regulation of Plt. Multiple copies of rpoD, encoding the housekeeping sigma factor ςD, enhanced production of Plt in P. fluorescens strain CHA0 (47), a phenotype reminiscent of rpoS mutations in Pf-5 (46, 53). The identification of Lon as a regulator of antibiotic biosynthesis fits into an evolving model that proposes the involvement of multiple sigma factors in the regulation of Plt production (Fig. 4). Direct examination of the role of sigma factor competition in Plt regulation, not included in this study, is warranted by these and previous data.
FIG. 4.
Proposed model depicting regulation of Plt by several identified global regulators. On one level, competition between ςS, ςD, and ςH for limited RNA polymerase core enzyme determines the suite of genes expressed and ultimately the level of Plt produced. In the absence of any one sigma factor, gene expression controlled by remaining sigma factors may increase due to decreased competition. In the presence of GacA and GacS, starvation rapidly induces ςS expression (53), which negatively influences Plt production. In response to heat shock, ςH mediates the induction of Lon, which negatively influences Plt production.
An alternative mechanism for regulation of Plt by Lon is through degradation of a positive regulator of Plt production, as has been described for many phenotypes regulated by Lon (17, 18). For example, Lon degrades the transcriptional regulator RcsA, which, in association with the activator RcsB, positively regulates colonic acid capsular polysaccharide (cps) gene expression in E. coli (6, 17, 18). In the absence of Lon, RcsA has an enhanced half-life, resulting in overexpression of cps genes (17, 18). RcsA and other previously described targets of Lon degradation are not known to participate in regulatory circuits controlling Plt production in P. fluorescens. Nevertheless, Lon could repress Plt production by degrading an activator of plt biosynthetic gene transcription. We propose that one possible candidate for Lon degradation is PltR, a transcriptional activator that is linked to the plt biosynthetic operon (38). Comparisons between Lon substrates have failed to identify any likely consensus sequence, but motifs that are recognized by Lon may be defined by structure (18). Gottesman (17) has proposed that susceptibility of proteins to degradation is controlled by sequestration of target motifs within an active protein or protein complex. If a substrate of Lon that functions in activation of Plt production was identified and target motifs were characterized, the Plt activator could possibly be altered to be less susceptible to degradation by Lon. This could provide an opportunity to enhance Plt production by Pf-5 when lon is induced by certain stresses, consequently improving the activity of Pf-5 as a biological control agent.
In addition to the novel phenotype of antibiotic regulation, two phenotypes of lon mutants in E. coli, filamentation and enhanced UV sensitivity, were found in lon mutants of Pf-5. In E. coli, Lon specifically degrades SulA, a repressor of cell division that is induced during the cell's SOS response to severe DNA damage (35). When exposed to UV irradiation, lon mutants of E. coli accumulate SulA and consequently fail to divide, become filamentous, and die. Conservation of Lon function in regulating UV tolerance was observed among the two bacterial species, although direct involvement of SulA in P. fluorescens was not investigated.
The influence of hupB, located immediately downstream of lon, on antibiotic production by Pf-5 was also investigated in this study. Deleting the entire hupB gene reduced Plt production, increased Phl production, and resulted in a mucoid morphology. In P. aeruginosa, other histone-like proteins, including AlgP (22, 26) and integration host factor (36, 54), influence alginate production; similarly, the hupB product HU is implicated in the mucoidy phenotype of E. coli (41). We are uncertain why the ΔhupB::aphI derivative produced more Phl than Pf-5 or if Phl overproduction and mucoidy are related, but it is possible that increased culture viscosity influenced antibiotic production. Indeed, in P. fluorescens F113, researchers noted that increasing the viscosity of broth cultures by adding 1.5% agar increases Phl production by 10-fold (48). Alternatively, through its influence on DNA conformation, HU may influence transcription of genes required for antibiotic production.
Lon is the fourth global regulator in Pf-5, along with GacA, GacS, and ςS, that influences both stress response and antibiotic production by Pf-5. Both lon::aacC1 and rpoS::lacZ mutants exhibit enhanced pltB transcription, implying that transcription of plt biosynthetic genes may be repressed under the stress conditions in which ςH and ςS are the dominant sigma factors directing transcription. Understanding the regulation of Plt production, both positive and negative, will allow manipulation of the strain to improve its consistency and performance as a biological control organism in the soil-root interface, where many stresses may be encountered.
ACKNOWLEDGMENTS
We thank H. Floss, C. Keel, and B. Nowak-Thompson for authentic standards of Prn, Phl, and Plt, respectively; R. Bonsall, N. Chaney, and M. Brodhagen for optimization of HPLC methods for antibiotic quantification; J. Trempy for the antibody to Lon; and K. Tanaka for antibody to ςS. We also thank J. Trempy for helpful discussions and protocols and L. S. Pierson III, J. Trempy, and C. Yeager for critical reviews of the manuscript. We are grateful to M. D. Henkels for assistance in preparing the figures.
This research was supported in part by fellowships to C.A.W. from the U.S. Environmental Protection Agency, Science to Achieve Results Fellowship Program (U-91523-01), and the U.S. Department of Agriculture, National Needs Fellowship Program (93-28420-8573), and by grant 95-37312-1655 from the U.S. Department of Agriculture, National Research Initiatives, Competitive Grants Program.
REFERENCES
- 1.Alvarado-Urbina G, Chiarello R, Roberts E, Vilain G, Jurik F, Christensen L, Carmona C, Fang L, Watterson M, Crea R. Rapid automated synthesis via diisopropylphosporamidite in situ activation: chemical synthesis and cloning of a calmodulin gene. Biochem Cell Biol. 1986;64:548–555. doi: 10.1139/o86-077. [DOI] [PubMed] [Google Scholar]
- 2.Amerik A Y, Antonov V K, Gorbalenya A E, Kotova S A, Rotanova T V, Shimbarevich E V. Site-directed mutagenesis of La protease. FEBS Lett. 1991;287:211–214. doi: 10.1016/0014-5793(91)80053-6. [DOI] [PubMed] [Google Scholar]
- 3.Ausubel F, Brent R, Kingston R E, Moore D D, Seidman J G, Smith J A, Struhl K, editors. Current protocols in molecular biology. New York, N.Y: Wiley and Sons, Inc.; 1990. [Google Scholar]
- 4.Bonfield J K, Smith K F, Staden R. A new DNA sequence assembly program. Nucleic Acids Res. 1995;23:4992–4999. doi: 10.1093/nar/23.24.4992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Brendel V, Trifonov E N. A computer algorithm for testing potential prokaryotic terminators. Nucleic Acids Res. 1984;12:4411–4427. doi: 10.1093/nar/12.10.4411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Brill J A, Quinlan-Walshe C, Gottesman S. Fine-structure mapping and identification of two regulators of capsule synthesis in Escherichia coli K-12. J Bacteriol. 1988;170:2599–2611. doi: 10.1128/jb.170.6.2599-2611.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chin D T, Goff S A, Webster T, Smith T, Goldberg A L. A heat-shock gene which encodes the ATP-dependent protease LA. J Biol Chem. 1988;263:11718–11728. [PubMed] [Google Scholar]
- 8.Corbell N, Loper J E. A global regulator of secondary metabolite production in Pseudomonas fluorescens Pf-5. J Bacteriol. 1995;177:6230–6236. doi: 10.1128/jb.177.21.6230-6236.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Delic-Attree I, Toussaint B, Vignais P M. Cloning and sequence analyses of the genes coding for the integration host factor (IHF) and HU proteins of Pseudomonas aeruginosa. Gene. 1995;154:61–64. doi: 10.1016/0378-1119(94)00875-s. [DOI] [PubMed] [Google Scholar]
- 10.Devereux J, Haeberli P, Smithies O. A comprehensive set of sequence analysis programs for the VAX. Nucleic Acids Res. 1984;12:387–395. doi: 10.1093/nar/12.1part1.387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dierksen K P, Marks J, Chen D D, Trempy J E. Evidence for structural conservation of Lon and RcsA. J Bacteriol. 1994;176:5126–5130. doi: 10.1128/jb.176.16.5126-5130.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Drlica K, Rouvière-Yaniv J. Histonelike proteins of bacteria. Microbiol Rev. 1987;51:301–319. doi: 10.1128/mr.51.3.301-319.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ebel W, Skinner M M, Dierksen K P, Scott J M, Trempy J E. A conserved domain in Escherichia coli Lon protease is involved in substrate discriminator activity. J Bacteriol. 1999;181:2236–2243. doi: 10.1128/jb.181.7.2236-2243.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Figurski K H, Helinski D R. Replication of an origin-containing derivative of plasmid RK2 dependent on a plasmid function provided in trans. Proc Natl Acad Sci USA. 1979;76:1648–1652. doi: 10.1073/pnas.76.4.1648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gay N J, Walker J E. Homology between human bladder carcinoma oncogene product and mitochondrial ATP-synthase. Nature (London) 1983;301:262–264. doi: 10.1038/301262a0. [DOI] [PubMed] [Google Scholar]
- 16.Gergen J P, Stern R H, Wensink P C. Filter replicas and permanent collections of recombinant DNA plasmids. Nucleic Acids Res. 1979;7:2115–2136. doi: 10.1093/nar/7.8.2115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gottesman S. Proteases and their targets in Escherichia coli. Annu Rev Genet. 1996;30:465–506. doi: 10.1146/annurev.genet.30.1.465. [DOI] [PubMed] [Google Scholar]
- 18.Gottesman S, Maurizi M R. Regulation by proteolysis: energy-dependent proteases and their targets. Microbiol Rev. 1992;56:592–621. doi: 10.1128/mr.56.4.592-621.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hecker M, Schumann W, Völker U. Heat-shock and general stress response in Bacillus subtilis. Mol Microbiol. 1996;19:417–428. doi: 10.1046/j.1365-2958.1996.396932.x. [DOI] [PubMed] [Google Scholar]
- 20.Howell C R, Stipanovic R D. Control of Rhizoctonia solani in cotton seedlings with Pseudomonas fluorescens and with an antibiotic produced by the bacterium. Phytopathology. 1979;69:480–482. [Google Scholar]
- 21.Howell C R, Stipanovic R D. Suppression of Pythium ultimum induced damping-off of cotton seedlings by Pseudomonas fluorescens and its antibiotic pyoluteorin. Phytopathology. 1980;70:712–715. [Google Scholar]
- 22.Kato J, Misra T K, Chakrabarty A M. AlgR3, a protein resembling eukaryotic histone H1, regulates alginate synthesis in Pseudomonas aeruginosa. Proc Natl Acad Sci USA. 1990;87:2887–2891. doi: 10.1073/pnas.87.8.2887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Keen N T, Tamaki S, Kobayashi D, Trollinger D. Improved broad host range plasmids for DNA cloning in Gram-negative bacteria. Gene. 1988;70:191–197. doi: 10.1016/0378-1119(88)90117-5. [DOI] [PubMed] [Google Scholar]
- 24.King E O, Ward M K, Raney D E. Two simple media for the demonstration of pyocyanin and fluorescin. J Lab Clin Med. 1954;44:301–307. [PubMed] [Google Scholar]
- 25.Kitten T, Kinscherf T G, McEvoy J L, Willis D K. A newly identified regulator is required for virulence and toxin production in Pseudomonas syringae. Mol Microbiol. 1998;28:917–929. doi: 10.1046/j.1365-2958.1998.00842.x. [DOI] [PubMed] [Google Scholar]
- 26.Konyecsni W M, Deretic V. DNA sequence and expression analysis of algP and algQ, components of the multigene system transcriptionally regulating mucoidy in Pseudomonas aeruginosa: algP contains multiple direct repeats. J Bacteriol. 1990;172:2511–2520. doi: 10.1128/jb.172.5.2511-2520.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kraus J, Loper J E. Lack of evidence for a role of antifungal metabolite production by Pseudomonas fluorescens Pf-5 in biological control of Pythium damping-off of cucumber. Phytopathology. 1992;82:264–271. [Google Scholar]
- 28.Kraus J, Loper J E. Characterization of a genomic region required for production of the antibiotic pyoluteorin by the biological control agent Pseudomonas fluorescens Pf-5. Appl Environ Microbiol. 1995;61:849–854. doi: 10.1128/aem.61.3.849-854.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lindgren P B, Frederick R, Govindarajan A G, Panopoulos N J, Staskawicz B J, Lindow S E. An ice nucleation reporter gene system: identification of inducible pathogenicity genes in Pseudomonas syringae pv. phaseolicola. EMBO J. 1989;8:1291–1301. doi: 10.1002/j.1460-2075.1989.tb03508.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Liu J, Cosby W M, Zuber P. Role of Lon and ClpX in the posttranslational regulation of a sigma subunit of RNA polymerase required for cellular differentiation in Bacillus subtilis. Mol Microbiol. 1999;33:415–428. doi: 10.1046/j.1365-2958.1999.01489.x. [DOI] [PubMed] [Google Scholar]
- 31.Loper J E, Lindow S E. Reporter gene systems useful in evaluating in situ gene expression by soil and plant-associated bacteria. In: Hurst C J, Knudsen G R, McInerney M J, Stetzenbach L D, Walter M V, editors. Manual of environmental microbiology. Washington, D.C.: ASM Press; 1997. pp. 482–492. [Google Scholar]
- 32.Lund A H, Duch M, Pedersen F S. Increased cloning efficiency by temperature-cycle ligation. Nucleic Acids Res. 1996;24:800–801. doi: 10.1093/nar/24.4.800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Martin F N, Loper J E. Soilborne plant diseases caused by Pythium spp.: ecology, epidemiology, and prospects for biological control. Crit Rev Plant Sci. 1999;18:111–181. [Google Scholar]
- 34.Maurhofer M, Keel C, Haas D, Défago G. Pyoluteorin production by Pseudomonas fluorescens strain CHA0 is involved in the suppression of Pythium damping-off of cress but not of cucumber. Eur J Plant Pathol. 1994;100:221–232. [Google Scholar]
- 35.Mizusawa S, Gottesman S. Protein degradation in Escherichia coli: the lon gene controls the stability of SulA protein. Proc Natl Acad Sci USA. 1983;80:358–362. doi: 10.1073/pnas.80.2.358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mohr C D, Deretic V. In vitro interactions of the histone-like protein IHF with the algD promoter, a critical site for control of mucoidy in Pseudomonas aeruginosa. Biochem Biophys Res Commun. 1992;189:837–844. doi: 10.1016/0006-291x(92)92279-7. [DOI] [PubMed] [Google Scholar]
- 37.Murillo J H, Shen D, Gerhold D, Sharma A, Cooksey D A, Keen N T. Characterization of pPT23b, the plasmid involved in syringolide production by Pseudomonas syringae pv. tomato PT23. Plasmid. 1994;31:275–287. doi: 10.1006/plas.1994.1029. [DOI] [PubMed] [Google Scholar]
- 38.Nowak-Thompson B, Chaney N, Wing J S, Gould S J, Loper J E. Characterization of the pyoluteorin biosynthetic gene cluster of Pseudomonas fluorescens Pf-5. J Bacteriol. 1999;181:2166–2174. doi: 10.1128/jb.181.7.2166-2174.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Nowak-Thompson B, Gould S J, Kraus J, Loper J E. Production of 2,4-diacetylphloroglucinol by the biocontrol agent Pseudomonas fluorescens Pf-5. Can J Microbiol. 1994;40:1064–1066. [Google Scholar]
- 40.Osawa T, Yura T. Effects of reduced amount of RNA polymerase sigma factor on gene expression and growth of Escherichia coli: studies of the rpoD40 (amber) mutation. Mol Gen Genet. 1981;184:166–173. doi: 10.1007/BF00272900. [DOI] [PubMed] [Google Scholar]
- 41.Painbéni E, Mouray E, Gottesman S, Rouvière-Yaniv J. An imbalance of HU synthesis induces mucoidy in Escherichia coli. J Mol Biol. 1993;234:1021–1037. doi: 10.1006/jmbi.1993.1656. [DOI] [PubMed] [Google Scholar]
- 42.Pfender W F, Kraus J, Loper J E. A genomic region from Pseudomonas fluorescens Pf-5 required for pyrrolnitrin production and inhibition of Pyrenophora tritici-repentis in wheat straw. Phytopathology. 1993;83:1223–1228. [Google Scholar]
- 43.Riethdorf S, Völker U, Gerth U, Winkler A, Engelmann S, Hecker M. Cloning, nucleotide sequence, and expression of the Bacillis subtilis lon gene. J Bacteriol. 1994;176:6518–6527. doi: 10.1128/jb.176.21.6518-6527.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Roitman J N, Mahoney N E, Janisiewicz W J. Production and composition of phenylpyrrole metabolites produced by Pseudomonas cepacia. Appl Microbiol Biotechnol. 1990;34:381–386. [Google Scholar]
- 45.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1989. [Google Scholar]
- 46.Sarniguet A, Kraus J, Henkels M D, Muehlchen A M, Loper J E. The sigma factor RpoS affects antibiotic production and biological control activity of Pseudomonas fluorescens Pf-5. Proc Natl Acad Sci USA. 1995;92:12255–12259. doi: 10.1073/pnas.92.26.12255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Schnider U, Keel C, Blumer C, Troxler J, Défago G, Haas D. Amplification of the housekeeping sigma factor in Pseudomonas fluorescens CHA0 enhances antibiotic production and improves biocontrol abilities. J Bacteriol. 1995;177:5387–5392. doi: 10.1128/jb.177.18.5387-5392.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Shanahan P, O'Sullivan J, Simpson P, Glennon J D, O'Gara F. Isolation of 2,4-diacetylphloroglucinol from a fluorescent pseudomonad and investigation of physiological parameters influencing its production. Appl Environ Microbiol. 1992;58:353–358. doi: 10.1128/aem.58.1.353-358.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Tanaka K, Takahashi H. Cloning, analysis, and expression of an rpoS homologue gene from Pseudomonas aeruginosa PAO1. Gene. 1994;150:81–85. doi: 10.1016/0378-1119(94)90862-1. [DOI] [PubMed] [Google Scholar]
- 50.Thomashow L, Weller D. Current concepts in the use of introduced bacteria for biological control: mechanisms and antifungal metabolites. In: Stacey G, Keen N T, editors. Plant-microbe interactions. New York, N.Y: Chapman and Hall; 1995. pp. 187–235. [Google Scholar]
- 51.Towbin H, Staehelin T, Gordon J. Electrophoretic transfer of proteins from polyacrylamide gels to nitrocellulose sheets: procedure and some applications. Proc Natl Acad Sci USA. 1979;76:4350–4354. doi: 10.1073/pnas.76.9.4350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Walker J E, Saraste M, Runswick M J, Gay N J. Distantly related sequences in the α- and β-subunits of ATP synthase, myosin, kinases and other ATP-requiring enzymes and a common nucleotide binding fold. EMBO J. 1982;1:945–951. doi: 10.1002/j.1460-2075.1982.tb01276.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Whistler C A, Corbell N A, Sarniguet A, Ream W, Loper J E. The two-component regulators GacS and GacA influence accumulation of the stationary-phase sigma factor RpoS and the stress response in Pseudomonas fluorescens Pf-5. J Bacteriol. 1998;180:6635–6641. doi: 10.1128/jb.180.24.6635-6641.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Wozniak D J. Integration host factor and sequences downstream of the Pseudomonas aeruginosa algD transcription start site are required for expression. J Bacteriol. 1994;176:5068–5076. doi: 10.1128/jb.176.16.5068-5076.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Yura T, Nagai H, Mori H. Regulation of the heat-shock response in bacteria. Annu Rev Microbiol. 1993;47:321–350. doi: 10.1146/annurev.mi.47.100193.001541. [DOI] [PubMed] [Google Scholar]