Abstract
The question addressed by the study
Bronchiolitis is not only the leading cause of hospitalization in U.S. infants but also a major risk factor for asthma development. Growing evidence supports clinical heterogeneity within bronchiolitis. To identify metatranscriptome profiles of infant bronchiolitis, and examine their relationship with host transcriptome and subsequent asthma development.
Materials/patients and methods
As part of multicentre prospective cohort study of infants (age <12 months) hospitalized for bronchiolitis, we integrated virus and nasopharyngeal metatranscriptome (species-level taxonomy and function) data measured at hospitalization. We applied network-based clustering approaches to identify metatranscriptome profiles. We then examined their association with host transcriptome at hospitalization and risk for developing asthma.
Results
We identified five metatranscriptome profiles of bronchiolitis (n=244): A) virusRSVmicrobiomecommensals, B) virusRSV/RV-AmicrobiomeH.influenzae, C) virusRSVmicrobiomeS.pneumoniae, D) virusRSVmicrobiomeM.nonliquefaciens, and E) virusRSV/RV-CmicrobiomeM.catarrhalis. Compared with profile A, profile B infants were characterized by high proportion of eczema, H. influenzae abundance, and enriched virulence related to antibiotic resistance. These profile B infants also had upregulated TH17 and downregulated type I interferon pathways (FDR<0.005) and significantly higher risk for developing asthma (17.9% vs. 38.9%; adjOR, 2.81; 95%CI, 1.11–7.26). Likewise, profile C infants were characterized by high proportion of parental asthma, S. pneumoniae dominance, and enriched glycerolipid and glycerophospholipid metabolism of microbiome. These profile C infants had upregulated receptor for advanced glycation end products signalling pathway (FDR<0.005) and higher risk of asthma (17.9% vs. 35.6%; adjOR, 2.49; 95%CI, 1.10–5.87).
Answer to the question
Metatranscriptome and clustering analysis identified biologically-distinct metatranscriptome profiles that have differential risks of asthma.
INTRODUCTION
Bronchiolitis is the leading cause of hospitalization in U.S. infants, accounting for ~110,000 hospitalizations annually [1]. In addition to the substantial acute morbidity, ~30% of infants hospitalized for bronchiolitis (“severe bronchiolitis”) subsequently develop asthma [2]. However, the underlying mechanisms linking these two common conditions remain unclear. This major knowledge gap has hindered efforts to prevent asthma in this high-risk population.
Although bronchiolitis has traditionally been thought of as a single disease entity [3], growing evidence supports heterogeneity in the clinical manifestations [4] and pathobiology (e.g., as reflected by between-virus differences in the upper airway microbiome [via 16S rRNA gene sequencing]) [5–7]. Additionally, studies also suggest a complex interplay between viruses, microbiome, and host response in the airway, and its interrelationship with respiratory health [2, 8–14]. Despite the clinical and research significance, no study has integrated virus and airway microbiome (both taxonomy and function) data to investigate the metatranscriptome profiles of bronchiolitis, their relationship with the host response during the critical period of airway development (i.e., early infancy), and their contribution to incident asthma in later childhood.
To address this knowledge gap, we analysed data from a multicentre prospective cohort to 1) identify nasopharyngeal metatranscriptome profiles (or clusters) of bronchiolitis, and 2) investigate their association with host airway response and subsequent development of asthma.
METHODS
Study Design, Setting, and Participants
We analysed data from a multicentre prospective cohort study of infants hospitalized for bronchiolitis—the 35th Multicenter Airway Research Collaboration (MARC-35) study. Details of the study design, setting, participants, data collection, testing, and statistical analysis may be found in the Supplementary Methods of the Supplementary Materials.
Briefly, at 17 sites across 14 U.S. states (Supplementary Table S1), MARC-35 enrolled infants (age <1 year) who were hospitalized with an attending physician diagnosis of bronchiolitis during three bronchiolitis seasons in 2011–2014. The diagnosis of bronchiolitis was made according to the American Academy of Pediatrics bronchiolitis guidelines, defined as an acute respiratory illness with a combination of rhinitis, cough, tachypnoea, wheezing, crackles, or chest retractions. We excluded infants with a pre-existing heart and lung disease, immunodeficiency, immunosuppression, or gestational age of <32 weeks, those with a previous bronchiolitis hospitalization, or those who were transferred to a participating hospital >24 hours after initial hospitalization. All patients were treated at the discretion of the treating physicians. The institutional review board at each participating hospital approved the study with a written informed consent obtained from the parent or guardian.
Of 921 infants enrolled into the longitudinal cohort, the current analysis investigated 244 infants hospitalized for bronchiolitis who were randomly selected for nasopharyngeal metatranscriptome (microbiome) and transcriptome (host) testing (Figure 1 and Supplementary Figure S1).
Figure 1. Analytic workflow of metatranscriptome profiling.
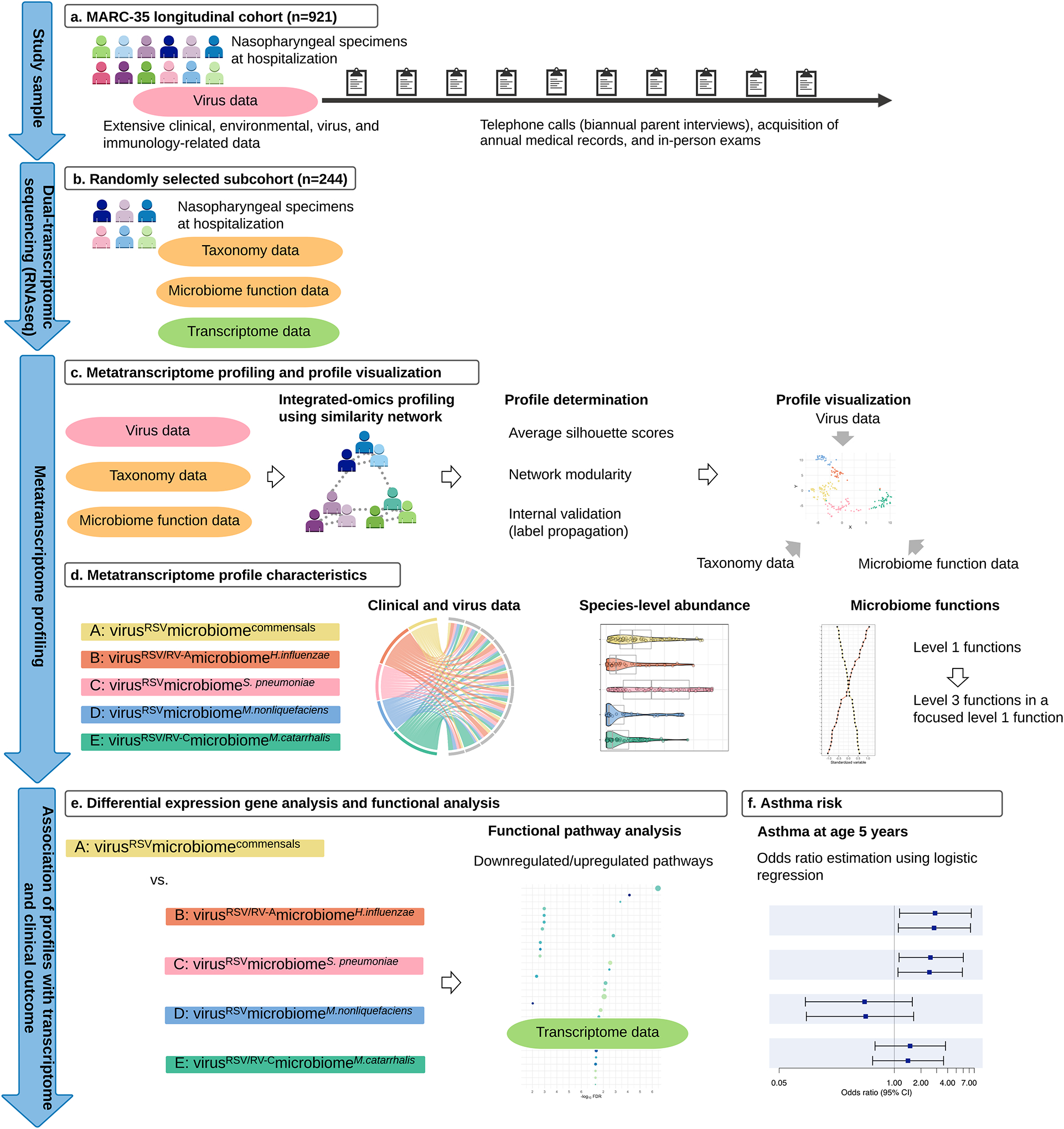
A. A total of 921 infants (age <1 year) hospitalized with bronchiolitis comprised the MARC-35 longitudinal cohort. At enrolment, the nasopharyngeal specimens were collected. These nasopharyngeal specimens were tested for respiratory viruses (e.g., RSV, RV species) and dual-transcriptome sequencing. These infants are followed with biannual parent interviews, acquisition of annual medical records, and in-person exams.
B. Randomly-selected 244 infants underwent dual-transcriptome sequencing (via RNAseq) of nasopharyngeal specimens to characterize the microbiome taxonomy and function as well as the host transcriptome.
C. After individually computing an affinity matrix for each of three datasets (i.e., virus, microbiome taxonomy and function data), we generated a fused affinity matrix by similarity network fusion. Then, we used the fused affinity matrix to identify metatranscriptome profiles by spectral clustering. To choose an optimal number of profiles, we used a combination of silhouette scores, network modularity, profile size, and clinical and biological plausibility. We visualized the five profiles by using the t-distributed stochastic neighbour embedding (t-sne) method.
D. To visualize the between-metatranscriptome profile differences in the major clinical and virus variables, microbiome taxonomy, and functions, we used chord diagrams, pirate plots, and ranked plots.
E. To examine the relationship between the metatranscriptome profiles and host function (transcriptome data), we performed differential gene expression analyses and functional pathway analyses. As infants with a profile A clinically resembled “classic” bronchiolitis and had the largest profile size, this group served as the reference group.
F. To examine the relationship of the metatranscriptome profiles with the risk for developing asthma (binary outcome), we constructed unadjusted and adjusted logistic regression models.
Abbreviations: RSV, respiratory syncytial virus; RV-A, rhinovirus A; RV-C, rhinovirus C
Data Collection and Measurement of Virus, Metatranscriptome, and Transcriptome
Clinical data (patients’ demographic characteristics, and family, environmental, and medical history, and details of the acute illness) were collected via structured interview and chart reviews using a standardized protocol. Additionally, nasopharyngeal airway specimens were collected within 24 hours of hospitalization using a standardized protocol [12]. The details of the data collection and measurement methods are described in the Supplementary Methods. The nasopharyngeal specimens were tested for a) respiratory viruses (e.g., respiratory syncytial virus [RSV] and rhinoviruses [RVs]), b) metatranscriptome (microbiome taxonomy at the species-level and function), and c) transcriptome (host function).
RNAseq for Nasopharyngeal Dual-transcriptome (Metatranscriptome and Transcriptome)
The details of RNA extraction, RNAseq, and quality control are described in the Supplementary Methods. Briefly, after total RNA extraction, DNase treatment, and rRNA reduction, we performed RNAseq with Illumina NovaSeq6000 using an S4 100PE Flowcell (Illumina, San Diego, CA). All RNAseq samples had high sequence coverage (a mean of 8,067,019 pair-end reads/sample) after quality control. We filtered and trimmed raw reads for adapters and contaminants using the k-mers strategy in bbduck. We characterized the active (via RNA transcripts) bacterial component of the microbiome (species-level) coupling PathoScope 2.0 with the expanded Human Oral Microbiome Database. To characterize the microbiome function, we used SUPER-FOCUS and Diamond. To annotate proteins that implement a specific biological process or structural complex into subsystems, we used the SEED database. This database comprises three-level hierarchical microbiome functions: the level 1 subsystem with 35 functions, followed by the level 2 subsystem with 194 functions and the level 3 subsystem with 1,290 functions. Lastly, we estimated host transcript abundances in Salmon using the human genome (hg38) and the mapping-based mode.
Functional and Clinical Outcomes
The outcomes of interest are a) host function (the nasopharyngeal transcriptome) at index hospitalization for bronchiolitis and b) asthma development by age 5 years. The definition of asthma was based on a commonly-used epidemiologic definition of asthma—physician-diagnosis of asthma by age 5 years, plus either asthma medication use (e.g., albuterol inhaler, inhaled corticosteroids, montelukast) or asthma-related symptoms in the preceding year.
Statistical Analysis
The objectives of the current study are a) to identify biologically distinct metatranscriptome profiles among infants with bronchiolitis (description [clustering]), and b) to relate them to the host transcriptome and the risk of asthma development (association). The analytic workflow is summarized in Figure 1. The details of the statistical analysis can be found in the Supplementary Methods.
Briefly, we first computed a distance matrix for each of three datasets—virus (including the genomic load of RSV, RV-A, and RV-C), microbiome taxonomy (species-level), and microbiome function data: Euclidean distance for the virus data, Bray-Curtis distance for the taxonomy data, and Pearson distance for the function data. Then, we computed an affinity matrix of each dataset separately, and generated a fused affinity matrix by similarity network fusion using the SNFtool package. To identify mutually exclusive metatranscriptome profiles, we applied spectral clustering to the fused affinity matrix. To choose an optimal number of profiles, we used a combination of the silhouette scores (Supplementary Figure S2, Panel A), network modularity (Supplementary Figure S2, Panel B), profile size (n=33–67), and clinical and biological plausibility. The network modularity measures how well-separated subnetworks are given a particular partitioning (i.e., profiles) of the network. To test the stability of profiles (i.e., internal validation), we computed the accuracy of profiles using semi-supervised label propagation methods (Supplementary Figure S3). To complement these approaches, we also used a priori knowledge by confirming that the derived profiles are consistent with earlier studies [2].
After deriving the metatranscriptome profiles, we examined their relationships with both functional (host transcriptome) and clinical (asthma) outcomes. First, we conducted differential expression gene and functional pathway analyses by comparing the reference profile with each of the other profiles. To investigate whether genes for specific biological pathways are enriched among the large positive or negative fold changes, we conducted a functional class scoring analysis using clusterProfiler package. Second, to determine the longitudinal association of the profiles with asthma at age 5 years (binary outcome), we constructed unadjusted and adjusted logistic regression models accounting for patient clustering within sites. In the sensitivity analysis, we examined the robustness of profile-outcome associations by repeating the analysis using a different number of profiles. We analysed the data using R version 3.6.1 (R Foundation for Statistical Computing, Vienna, Austria). All P-values were two-tailed, with P<0.05 considered statistically significant. We corrected for multiple testing using the Benjamini-Hochberg false discovery rate (FDR) method.
RESULTS
Of the infants enrolled into this longitudinal cohort, the current study focused on 244 randomly selected infants with bronchiolitis who underwent testing for nasopharyngeal airway microbial metatranscriptome and host transcriptome (Figures 1 and Supplementary Figure S1). The analytic cohort and non-analytic cohorts did not differ in patient characteristics (P≥0.05; Supplementary Table S2), except for daycare use and solo-RSV infection. Among the analytic cohort, the median age was 3 (IQR, 2–6) months, 40.2% were female, and 41.8% were non-Hispanic white. Overall, 91.0% were RSV infection with solo-RSV infection in 65.2% and RSV/RV coinfection in 11.9% (Table 1).
Table 1.
Baseline characteristics and clinical course of infants with bronchiolitis, according to metatranscriptome profiles
| Characteristics | Overall (n=244; 100%) |
Profile A (n=67; 27.5%) |
Profile B (n=36; 14.8%) |
Profile C (n=59; 24.2%) |
Profile D (n=33; 13.5%) |
Profile E (n=49; 20.1%) |
P-value |
|---|---|---|---|---|---|---|---|
| Demographics | |||||||
| Age (month), median (IQR) | 3 (2–6) | 3 (1–6) | 4 (2–7) | 3 (2–6) | 4 (2–7) | 3 (2–6) | 0.36 |
| Female sex | 98 (40.2) | 29 (43.3) | 13 (36.1) | 22 (37.3) | 17 (51.5) | 17 (34.7) | 0.54 |
| Race/ethnicity | 0.33 | ||||||
| Non-Hispanic white | 102 (41.8) | 29 (43.3) | 14 (38.9) | 21 (35.6) | 16 (48.5) | 22 (44.9) | |
| Non-Hispanic black | 57 (23.4) | 12 (17.9) | 6 (16.7) | 21 (35.6) | 8 (24.2) | 10 (20.4) | |
| Hispanic | 76 (31.1) | 24 (35.8) | 15 (41.7) | 15 (25.4) | 9 (27.3) | 13 (26.5) | |
| Other or unknown | 9 (3.7) | 2 (3.0) | 1 (2.8) | 2 (3.4) | 0 (0.0) | 4 (8.2) | |
| Prematurity (32–36.9 weeks) | 47 (19.3) | 15 (22.4) | 6 (16.7) | 11 (18.6) | 9 (27.3) | 6 (12.2) | 0.48 |
| Birth weight (kg), median (IQR) | 3.20 (2.89–3.57) | 3.20 (2.90–3.52) | 3.17 (2.70–3.42) | 3.23 (2.86–3.69) | 3.09 (2.79–3.43) | 3.31 (3.00–3.64) | 0.43 |
| Mode of birth (caesarean delivery) | 84 (35.0) | 19 (28.8) | 15 (42.9) | 22 (37.3) | 11 (33.3) | 17 (36.2) | 0.69 |
| Previous breathing problems (count) | 0.92 | ||||||
| 0 | 204 (83.6) | 55 (82.1) | 31 (86.1) | 50 (84.7) | 27 (81.8) | 41 (83.7) | |
| 1 | 30 (12.3) | 8 (11.9) | 3 (8.3) | 8 (13.6) | 4 (12.1) | 7 (14.3) | |
| 2 | 10 (4.1) | 4 (6.0) | 2 (5.6) | 1 (1.7) | 2 (6.1) | 1 (2.0) | |
| Previous ICU admission | 4 (1.6) | 3 (4.5) | 0 (0.0) | 0 (0.0) | 1 (3.0) | 0 (0.0) | 0.19 |
| History of eczema | 31 (12.7) | 3 (4.5) | 8 (22.2) | 10 (16.9) | 2 (6.1) | 8 (16.3) | 0.03 |
| Palivizumab use | 8 (3.8) | 1 (1.7) | 2 (6.2) | 3 (6.1) | 0 (0.0) | 2 (4.9) | 0.48 |
| Lifetime antibiotic use* | 79 (32.4) | 25 (37.3) | 17 (47.2) | 16 (27.1) | 4 (12.1) | 17 (34.7) | 0.02 |
| Ever attended daycare | 71 (29.1) | 15 (22.4) | 11 (30.6) | 16 (27.1) | 13 (39.4) | 16 (32.7) | 0.46 |
| Cigarette smoke exposure at home | 34 (13.9) | 9 (13.4) | 4 (11.1) | 11 (18.6) | 6 (18.2) | 4 (8.2) | 0.52 |
| Maternal smoking during pregnancy | 34 (14.2) | 9 (13.6) | 7 (20.0) | 7 (11.9) | 3 (9.1) | 8 (17.0) | 0.69 |
| Parental history of asthma | 76 (31.1) | 14 (20.9) | 12 (33.3) | 24 (40.7) | 11 (33.3) | 15 (30.6) | 0.19 |
| Parental history of eczema | 46 (18.9) | 9 (13.4) | 12 (33.3) | 12 (20.3) | 8 (24.2) | 5 (10.2) | 0.06 |
| Clinical presentation | |||||||
| Weight (kg), median (IQR) | 6.07 (4.60–7.99) | 5.50 (4.36–7.10) | 6.80 (5.16–8.12) | 6.28 (4.52–7.53) | 6.40 (4.75–8.20) | 6.20 (4.80–8.45) | 0.22 |
| Respiratory rate (per minute), median (IQR) | 48 (40–60) | 48 (40–56) | 48 (40–56) | 48 (39– 60) | 52 (44–64) | 50 (41–60) | 0.45 |
| Oxygen saturation | 0.19 | ||||||
| <90% | 18 (7.6) | 5 (7.9) | 4 (11.8) | 2 (3.4) | 1 (3.1) | 6 (12.2) | |
| 90–93% | 29 (12.2) | 12 (19.0) | 4 (11.8) | 4 (6.8) | 2 (6.2) | 7 (14.3) | |
| ≥94% | 190 (80.2) | 46 (73.0) | 26 (76.5) | 53 (89.8) | 29 (90.6) | 36 (73.5) | |
| Blood eosinophilia (≥4%) | 21 (10.1) | 4 (7.0) | 4 (13.3) | 5 (10.0) | 2 (6.9) | 6 (14.3) | 0.73 |
| IgE sensitization | 51 (20.9) | 11 (16.4) | 9 (25.0) | 13 (22.0) | 6 (18.2) | 12 (24.5) | 0.77 |
| Clinical course | |||||||
| Positive pressure ventilation use † | 18 (7.4) | 6 (9.0) | 3 (8.3) | 5 (8.5) | 3 (9.1) | 1 (2.0) | 0.55 |
| Intensive treatment use‡ | 42 (17.2) | 13 (19.4) | 6 (16.7) | 12 (20.3) | 3 (9.1) | 8 (16.3) | 0.72 |
| Length of stay (day), median (IQR) | 2 (1–3) | 2 (1–4) | 2 (1–4) | 2 (1–4) | 2 (1–3) | 2 (1–3) | 0.89 |
| Antibiotic use during hospitalization | 80 (32.8) | 26 (38.8) | 15 (41.7) | 19 (32.2) | 8 (24.2) | 12 (24.5) | 0.29 |
| Corticosteroid use during hospitalization | 29 (11.9) | 8 (11.9) | 7 (19.4) | 10 (16.9) | 1 (3.0) | 3 (6.1) | 0.11 |
| Respiratory virus | |||||||
| RSV infection | 222 (91.0) | 61 (91.0) | 31 (86.1) | 59 (100.0) | 33 (100.0) | 38 (77.6) | <0.001 |
| RSV-A§ | 153 (62.7) | 46 (68.7) | 22 (61.1) | 41 (69.5) | 15 (45.5) | 29 (59.2) | 0.15 |
| RSV-B§ | 71 (29.1) | 16 (23.9) | 9 (25.0) | 19 (32.2) | 18 (54.5) | 9 (18.4) | 0.01 |
| Solo-RSV infection | 159 (65.2) | 42 (62.7) | 21 (58.3) | 48 (81.4) | 24 (72.7) | 24 (49.0) | 0.007 |
| RV infection | |||||||
| RV-A | 26 (10.7) | 8 (11.9) | 7 (19.4) | 5 (8.5) | 2 (6.1) | 4 (8.2) | 0.41 |
| RV-B | 4 (1.6) | 1 (1.5) | 1 (2.8) | 0 (0.0) | 1 (3.0) | 1 (2.0) | 0.67 |
| RV-C | 21 (8.6) | 2 (3.0) | 0 (0.0) | 1 (1.7) | 2 (6.1) | 16 (32.7) | <0.001 |
| Solo-RV infection | 13 (5.3) | 4 (6.0) | 1 (2.8) | 0 (0.0) | 0 (0.0) | 8 (16.3) | 0.002 |
| RSV/RV coinfection | 29 (11.9) | 5 (7.5) | 3 (8.3) | 6 (10.2) | 5 (15.2) | 10 (20.4) | 0.25 |
| Other coinfection pathogens with RSV¶ | 34 (13.9) | 14 (20.9) | 7 (19.4) | 5 (8.5) | 4 (12.1) | 4 (8.2) | 0.17 |
| Chronic outcome | |||||||
| Asthma at age 5 years |
62 (25.4) | 12 (17.9) | 14 (38.9) | 21 (35.6) | 3 (9.1) | 12 (24.5) | 0.009 |
Abbreviations: IQR, interquartile range; ICU, intensive care unit; RSV, respiratory syncytial virus; RV, rhinovirus; IgE, immunoglobulin E
Data are no. (%) of infants unless otherwise indicated. Percentages may not equal 100, because of rounding and missingness.
Any systemic antibiotic use from birth up to the index hospitalization for bronchiolitis.
Infants with bronchiolitis who underwent continuous positive airway ventilation and/or mechanical ventilation.
Infants with bronchiolitis who were admitted to ICU and/or who underwent positive pressure ventilation.
Two infants had coinfection of RSV-A and -B.
Infants with coinfection of RSV and non-RV virus(es) include adenovirus infection (n=7), bocavirus (n=8), endemic coronavirus (n=15), enterovirus (n=1), influenza virus (n=1), human metapneumovirus (n=4), Mycoplasma pneumonia (n=1), and parainfluenza virus (n=3). Since 6 infants have co-infection with ≥3 infecting agents, the total number is not equal to 34.
Integrated Omics Approach Identified Distinct Metatranscriptome Profiles
To derive biologically distinct metatranscriptome profiles of infant bronchiolitis, we applied integrative network and clustering approaches to the virus, microbiome taxonomy (species-level), and microbiome function data (Figure 1). Both of the average silhouette scores and network modularity found that a 5-class model was the optimal fit (Supplementary Figure S2), with the 5 profiles called A, B, C, D, and E. The semi-supervised label propagation methods also indicated that the stability was also highest with the 5-class model (Supplementary Figure S3).
The 5 distinct metatranscriptome profiles (Figure 2A and Supplementary Figure S4) were chiefly characterized by the identified virus(es) and the major bacteria species of the nasopharyngeal airway microbiome: A) virusRSVmicrobiomecommensals (27.5%), B) virusRSV/RV-AmicrobiomeH.influenzae (14.8%), C) virusRSVmicrobiomeS.pneumoniae (24.2%), D) virusRSVmicrobiomeM.nonliquefaciens (13.5%), and E) virusRSV/RV-CmicrobiomeM.catarrhalis (20.1%) (Table 1; Figures 2B and 3).
Figure 2. Metatranscriptome profiles among infants with bronchiolitis, and their relationship with major clinical and virus variables.
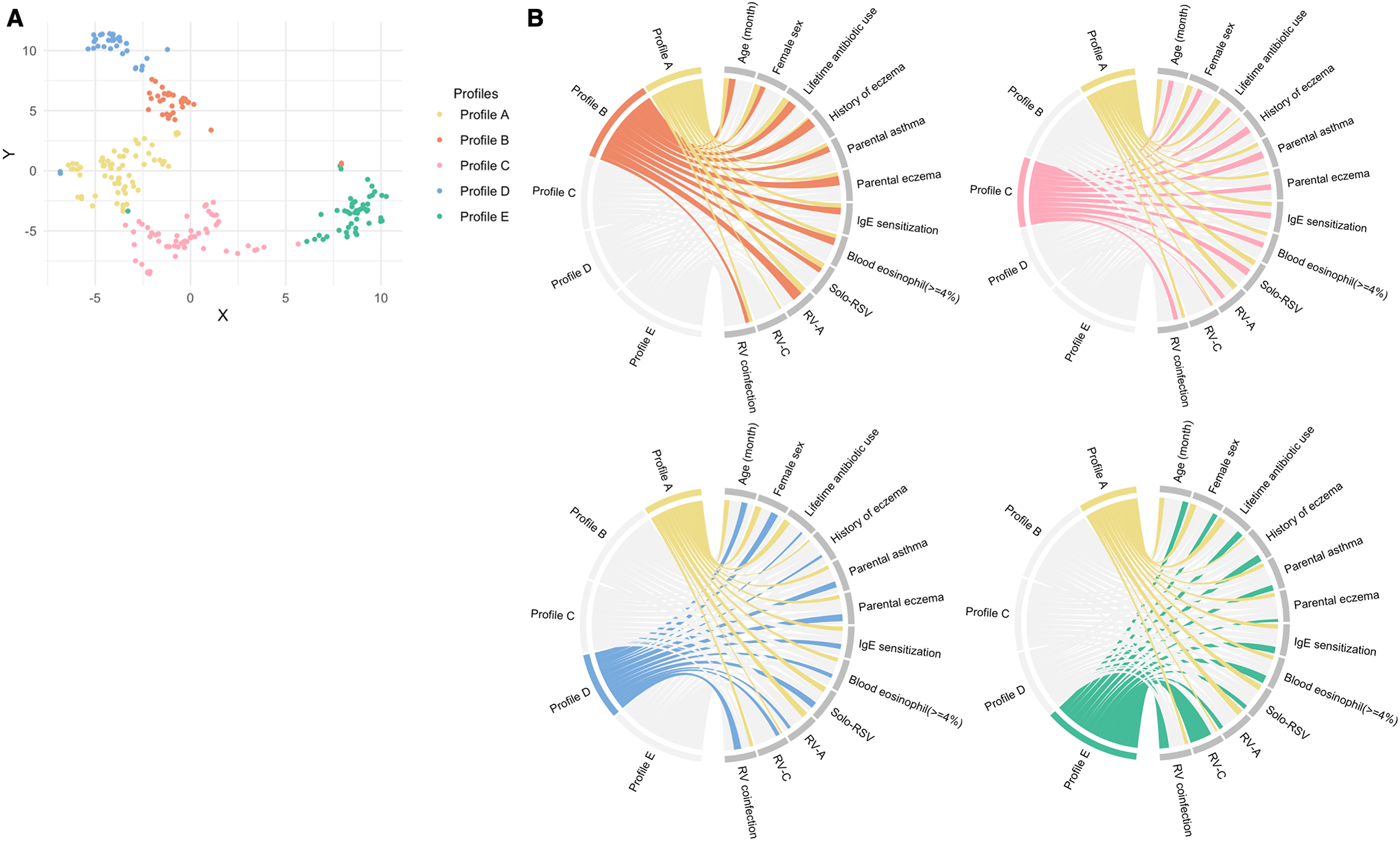
A. T-distributed stochastic neighbor embedding of nasopharyngeal metatranscriptome profiles To visualize the metatranscriptome profiles, the t-distributed stochastic neighbour embedding method was applied to the five eigenvectors in the spectral clustering. Each dot represents the metatranscriptome of a single infant in a low-dimensional space. The infants cluster together according to their metatranscriptome profiles.
B. Major clinical and virus characteristics according to metatranscriptome profiles The ribbons connect from the individual metatranscriptome profiles to the major clinical and virus characteristics. The width of the ribbon represents the proportion of infants within the profile who have the corresponding clinical or virus characteristic, which was scaled to a total of 100%. For example, the profile B infants (light orange) had a high proportion of lifetime antibiotics use, history of eczema, parental eczema, IgE sensitization, blood eosinophilia, and coinfection with RV-A. Profile C (pink) infants had a high proportion of parental asthma and solo-RSV infection.
Abbreviations: IgE, immunoglobulin E; RSV, respiratory syncytial virus; RV, rhinovirus; RV-A, rhinovirus A; RV-C, rhinovirus C
Figure 3. Between-profile differences in relative abundance of ten most abundant nasopharyngeal microbial species among infants with bronchiolitis.
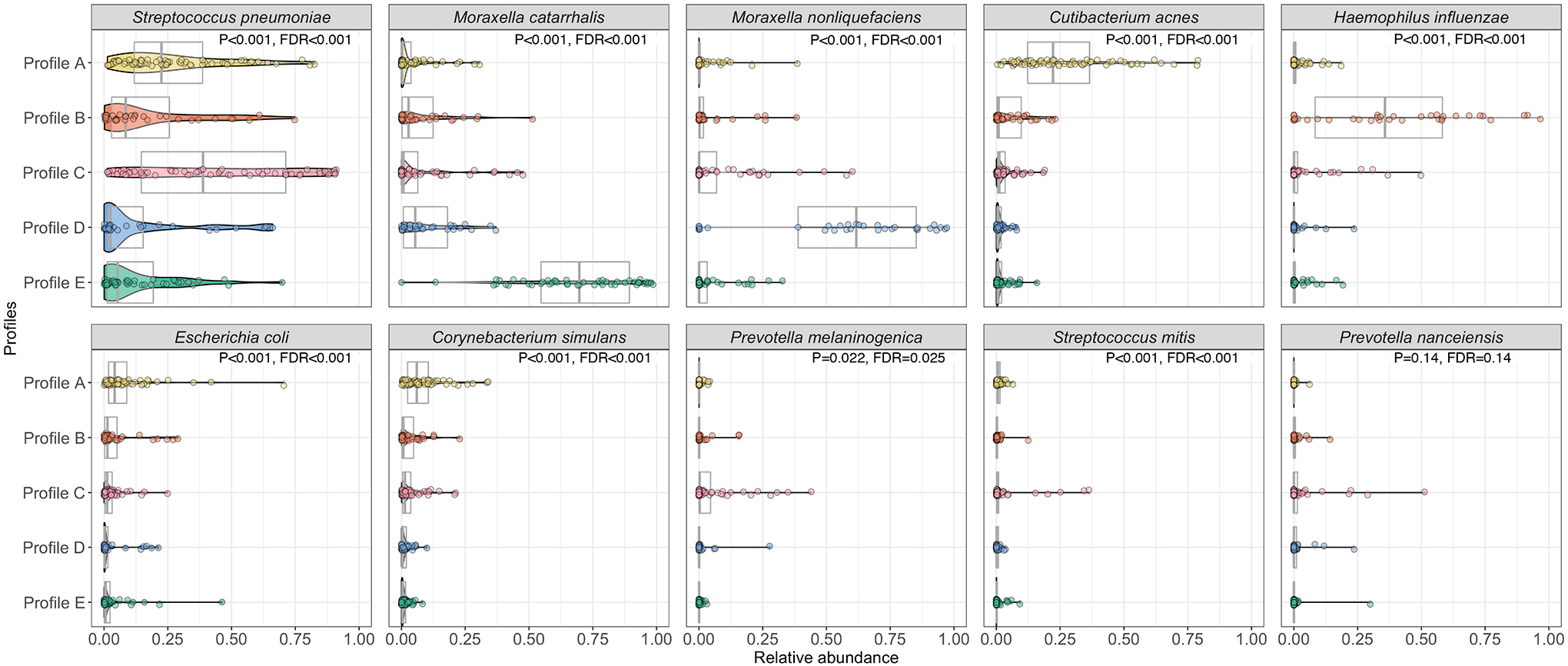
The pirate plots (a combination of boxplots and violin plots) show the distribution of the ten most abundant species in the nasopharyngeal microbiome, according to the five metatranscriptome profiles. In the overlying violin plots, the width represents the probability that infants in a profile take on a specific relative abundance. The between-profile differences in the relative abundance were tested by Kruskal-Wallis test.
Abbreviation: FDR, false discovery rate
Descriptively, infants with a profile A were characterized by a young age, a low proportion of parental asthma and eczema and personal history of eczema, and a high proportion of RSV infection (Figures 2, and Supplementary Figures S5, S6, and S7). In many respects they resembled “classic” bronchiolitis. These infants also had a higher abundance of commensals (e.g., Corynebacterium, Cutibacterium; both FDR<0.001; Figure 3 and Supplementary Figure S7). Infants with a profile B were characterized by a high proportion of lifetime antibiotics use, history of eczema, parental eczema, IgE sensitization, and coinfection with RV-A, and a higher abundance of H. influenzae (FDR<0.001when compared to those with a profile A). Infants with a profile C were characterized by a high proportion of parental asthma and solo-RSV infection as well as a higher abundance of S. pneumoniae (FDR<0.001). Infants with a profile D were characterized by a low proportion of hypoxemia, a high proportion of RSV infection, and a higher abundance of M. nonliquefaciens (FDR<0.001). Infants with a profile E were characterized by a high proportion of RV-C coinfection and a higher abundance of M. catarrhalis (FDR<0.001). These virus and microbiome variables that characterized the profiles had high-ranked normalized mutual information scores, indicating large contributions to the similarity network (Supplementary Figure S8).
Metatranscriptome Profiles Had Distinct Microbiome Function Pathways
These metatranscriptome profiles of infant bronchiolitis also had distinct microbiome functions. For example, compared to infants with profile A (virusRSVmicrobiomecommensals) who clinically resembled “classic” bronchiolitis and were the largest group, those with a profile B (virusRSV/RV-AmicrobiomeH.influenzae) had enriched virulence and iron acquisition and metabolism (FDR<0.05; Figure 4). More specifically, the profile B infants had an upregulated virulence function related to antibiotic resistance (e.g., multidrug resistance efflux pumps) (FDR<0.05; Supplementary Figure S9A) and iron metabolism function related to hemin transport (FDR<0.05; Supplementary Figure S9B). The profile C (virusRSVmicrobiomeS.pneumoniae) infants had enriched fatty acid, lipid, and isoprenoid metabolism (FDR<0.05; Figure 4)—e.g., upregulated glycerolipid and glycerophospholipid metabolism (FDR<0.05; Supplementary Figure S10). In contrast, the profile D (virusRSVmicrobiomeM.nonliquefaciens) infants had downregulated glycerolipid and glycerophospholipid function in fatty acid, lipid, and isoprenoid metabolism (FDR<0.05; Figure 4 and Supplementary Figure S11). Lastly, the profile E (virusRSV/RV-CmicrobiomeM.catarrhalis) infants had upregulated stress response metabolism (FDR<0.05; Figure 4)—e.g., cold shock CspA protein family (FDR<0.05; Supplementary Figure S12).
Figure 4. Between-profile differences in nasopharyngeal microbiome function among infants with bronchiolitis.
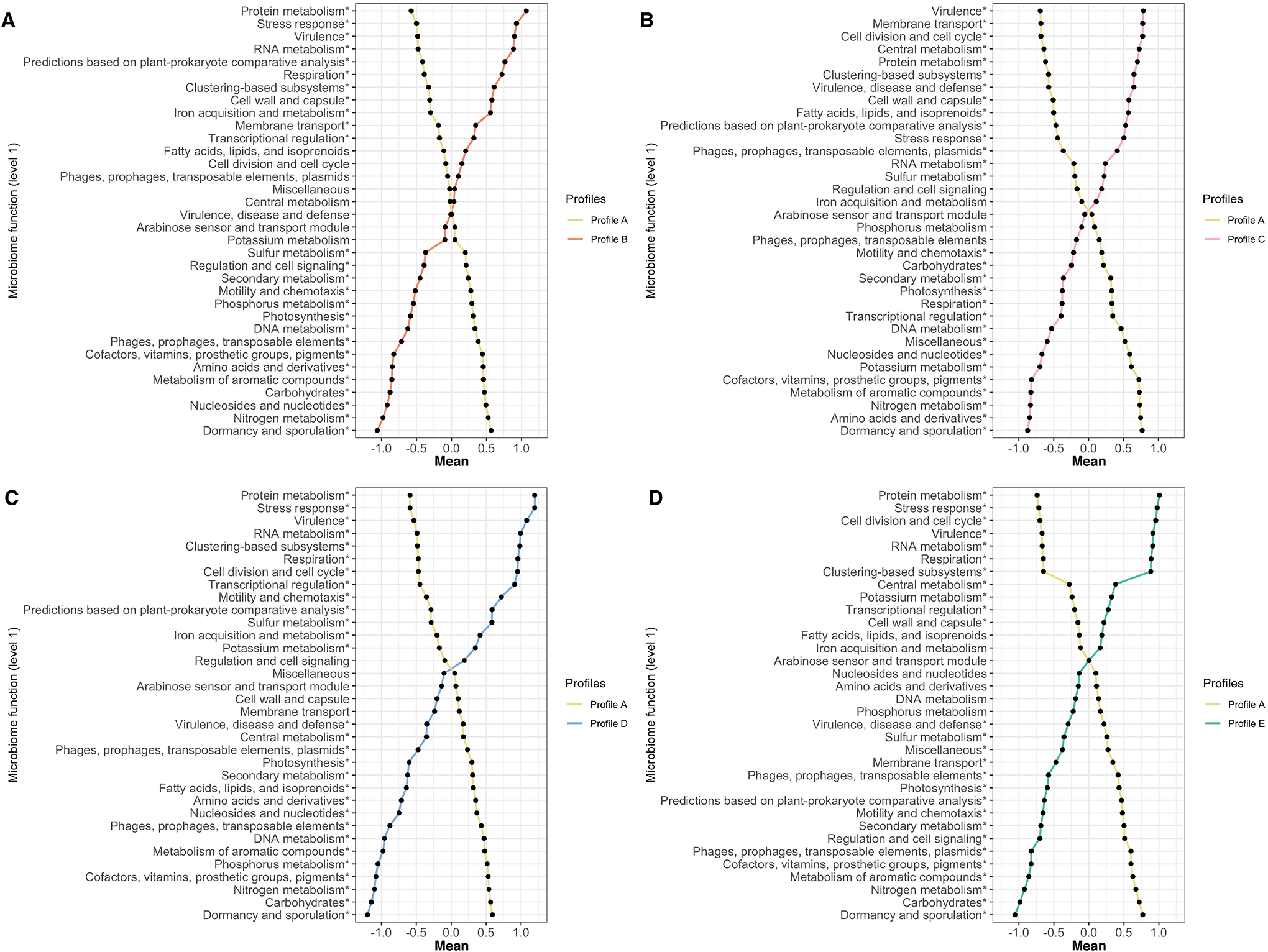
In all comparisons (metatranscriptome profile A vs. each of the other profiles), the mean values of microbiome function variables (35 level-1 functions) in the corresponding profiles are plotted. The microbiome function variables are standardized by using auto-scaling after variance stabilizing transformation. The differences in more detailed microbiome functions (level-3 functions) of specific level-1 functions are presented in Figures E8–E11.
* False discovery rate<0.05
Abbreviations; DNA, deoxyribonucleic acid; RNA, ribonucleic acid
A. Profiles A vs. B comparison
B. Profiles A vs. C comparison
C. Profiles A vs. D comparison
D. Profiles A vs. E comparison
Metatranscriptome Profiles Had Distinct Host Transcriptome Characteristics During Infancy and Differential Risk for Developing Asthma
To better understand the relationship between the metatranscriptome profiles and the host response (represented by the transcriptome) during infancy, we conducted differential expression gene and functional pathway analyses. Compared with the profile A, the profile B had 63 differentially enriched pathways (FDR<0.05; Supplementary Figure S13)—e.g., upregulated TH17 and downregulated type I interferon pathways. Similarly, the profile C had 45 differentially enriched pathways (FDR<0.05)—e.g., an upregulated receptor for advanced glycation end products (RAGE) signalling pathway (Supplementary Figure S14). For the profiles A vs. D and A vs. E comparisons, the detailed differences are summarized in Supplementary Figures S15 and S16.
The metatranscriptome profiles also had differential risks for developing asthma by age 5 years (Figure 5). For example, compared with profile A infants, profile B infants had a significantly higher risk (17.9% vs. 38.9%; adjOR, 2.81; 95%CI, 1.11–7.26; P=0.030). Likewise, profile C infants also had a significantly higher risk of asthma (35.6%; adjOR, 2.49; 95%CI, 1.10–5.87; P=0.031) while profile D infants had a non-significantly lower risk of asthma (9.1%; adjOR, 0.47; 95%CI, 0.10–1.65; P=0.28). In the stratification by the development of recurrent wheeze by age 3 years, the results were similar (Supplementary Figures S17).
Figure 5. Association between nasopharyngeal metatranscriptome profiles of infant bronchiolitis and risk for developing asthma.
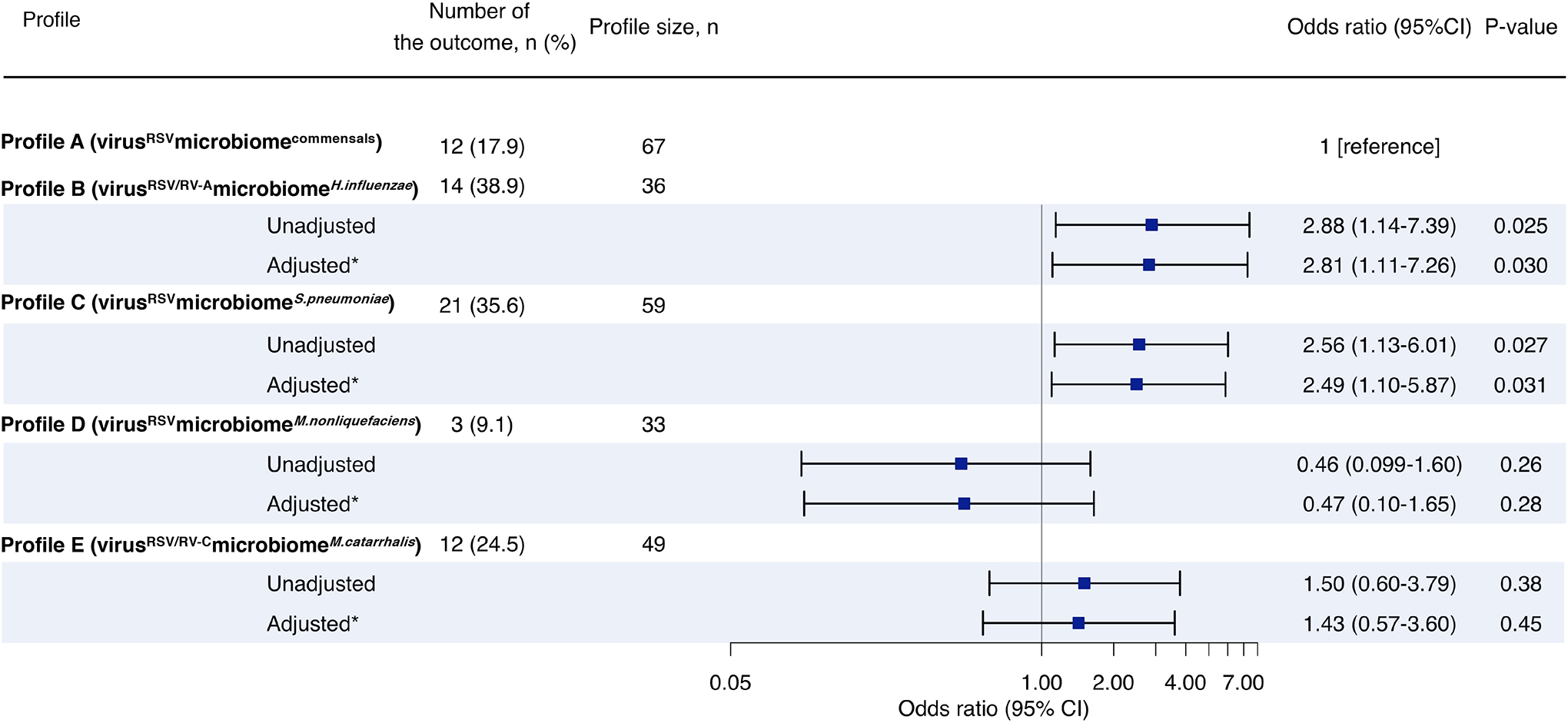
Asthma (binary outcome) was defined as physician-diagnosis of asthma at age 5 years, plus either asthma medication use (e.g., albuterol inhaler, inhaled corticosteroids, montelukast) or asthma-related symptoms in the preceding year. To examine the association between bronchiolitis profiles (profile A as the reference) and the risk of developing childhood asthma, unadjusted and adjusted logistic regression models were fit.
* Multivariable random-effect logistic model adjusted for age, sex, and clustering within hospitals
Abbreviation: CI, confidence interval; RSV, respiratory syncytial virus; RV, rhinovirus
Sensitivity Analysis
To address the robustness of these findings, we examined different numbers of profiles. Alluvial plot (Supplementary Figure S18) demonstrates the consistency of the original profiles (profiles A-E) across the different numbers chosen (4 and 6 profiles). For example, with the use of 4-class models (that had the second-highest accuracy in label propagation methods), the first and fourth profiles had >90% concordance with the original profiles A and E (Supplementary Table S3). In contrast, the second profile had a mixture of the original profiles B and C. Similar to the primary analysis, these four profiles were also characterized by virus and microbiome taxonomy (e.g., S. pneumoniae, M. catarrhalis) (Supplementary Table S3; Supplementary Figure S19). Lastly, compared to profile 1 (which is concordant with profile A), profile 2 (concordant with profiles B and C) infants with distinct microbiome functions (e.g., enriched by virulence, iron acquisition and metabolism, and fatty acid, lipid and isoprenoid metabolism; Supplementary Figure S20) had upregulated TH17 (FDR=0.002) and RAGE signalling (FDR=0.001) pathways. These infants also had a significantly higher risk for developing asthma (18.8% vs. 34.6%; adjOR, 2.25; 95%CI, 1.05–5.00; P=0.041; Supplementary Figure S21).
DISCUSSION
By integrating the virus and nasopharyngeal metatranscriptome (both microbiome taxonomy and function) data from a multicentre prospective cohort study of 244 infants with severe bronchiolitis, we identified five biologically distinct metatranscriptome profiles. In particular, infants with profile B (virusRSV/RV-AmicrobiomeH.influenzae) not only had distinct microbiome function (e.g., an upregulated virulence function) but also were associated with unique host response in the nasopharyngeal airway (e.g., upregulated TH17 pathways) at the time of bronchiolitis. Additionally, infants with profile C (virusRSVmicrobiomeS.pneumoniae) also had distinctive microbiome function (e.g., an enriched lipid metabolism) and host response (e.g., an upregulated RAGE signalling pathways). Furthermore, these two metatranscriptome profiles had a significantly higher risk for developing childhood asthma. To the best of our knowledge, this is the first study that has identified metatranscriptome profiles in bronchiolitis and demonstrated their relationship with host airway response and subsequent development of asthma.
Recent research has suggested the relationship between the virus, airway microbiome, host response, and respiratory disease. For example, studies have reported the association between respiratory viruses and unique upper airway microbiome (measured either via 16S rRNA gene sequencing or quantitative PCR) in infants with bronchiolitis [5, 6, 15] and school-age children [16]. In these studies, RV-A infection was associated with Haemophilus-dominant profile and RV-C with Moraxella-dominant profile, which is consistent with our metatranscriptomics findings. Recent studies have also shown the potential role of Haemophilus and Streptococcus genera in the upper airway—both among infants with or without bronchiolitis—in the host immune response [17, 18] and the development of wheeze illness and asthma [18–20]. Furthermore, research has suggested that the interaction of microbiome-host functions—via downstream metabolic regulation—contributes to the pathobiology of bronchiolitis and asthma [21]. Indeed, studies of upper airway metabolome among infants with bronchiolitis have reported the associations of altered lipid metabolism with disease severity [12] and asthma risk [21]. The current study corroborates these earlier reports, and extends them not only by identifying metatranscriptome profiles through the integrated omics approach but also by demonstrating their relationship with unique host immune response and asthma development.
There are several potential mechanisms linking the metatranscriptome profiles to the host airway response and subsequent asthma risk. First, we observed the relationship of profile B (virusRSV/RV-AmicrobiomeH.influenzae)—characterized by a high likelihood of previous antibiotics exposure, atopy/allergic sensitization, H. influenzae abundance, and enriched virulence related with antibiotic resistance and iron metabolism function—with upregulated TH17 and downregulated type I interferon pathways. Consistently, study has reported that antibiotic exposures during early infancy lead to Haemophilus-dominant maturation of nasal microbiome during the first two years of life [22]. Likewise, RV-A infection is also associated with a high abundance of Haemophilus in young children [7, 16]. Additionally, H. influenzae requires hemin transport function and X factor for its aerobic growth [23]. Furthermore, animal models have reported that H. influenzae induces early IL-17 responses from lung macrophages and neutrophils, followed by later responses from Th17 cells in lungs and mediastinal lymph nodes, leading to neutrophil influx into the airways [24]. Studies have also shown the roles of TH17 pathway in neutrophilic inflammation, steroid insensitivity, and airway remodelling in both allergic and non-allergic asthma [25, 26]. In addition to the upregulated TH17 pathway, the profile B also had downregulated type I interferon pathways. Recent research has reported immature type I interferon response to RV-A infection [27]. Reduced anti-viral response (e.g., interferons) to RV infection impairs phagocytosis of Haemophilus influenzae among patients with chronic lung disease [28]. Studies have also demonstrated that type I interferon response to RV infection is impaired among infants with allergic sensitization [29] and that the use of anti-IgE monoclonal antibody improves interferon-α response and reduces asthma exacerbation risks [30]. These prior data are in line with our findings in the profile B.
Second, the mechanisms linking profile C (virusRSVmicrobiomeS.pneumoniae)—characterized by a high likelihood of solo-RSV infection, S. pneumoniae dominance, and enriched glycerolipid and glycerophospholipid metabolism—to the unique host response (e.g., RAGE signalling) and asthma risk warrants further clarification. Research has shown that RSV infection increases the virulence of S. pneumoniae [31]. S. pneumoniae produces phosphatidic acid, a precursor to all membrane glycerophospholipids [32]. Studies have also suggested the pro-inflammatory role of glycerophospholipid (e.g., activation of natural killer T cells) in the pathobiology of asthma [33, 34]. Additionally, S. pneumoniae is associated with an upregulated RAGE expression in the lung [35]. Mendelian randomization study has also demonstrated the causal role of RAGE in asthma pathobiology [36]. Furthermore, compared with RAGE knockout mice, wild mice have developed more pronounced airway inflammation and mucus metaplasia when intranasally administered recombinant type 2 cytokines [37]. While it is intriguing to observe the abundance of S pneumoniae and its potential pathobiological effect in the post-pneumococcal conjugate vaccine (PCV) era, research has shown that the introduction of PCV-13 has led to the changes in pneumococcal serotypes, genotypes, and antimicrobial resistance [38].
In contrast, we observed that the profile D (virusRSVmicrobiomeM.nonliquefaciens) was characterized by downregulated glycerolipid and glycerophospholipid function and had the lowest risk for developing asthma. Studies have reported that M. nonliquefaciens is less-pathogenic [39] and associated with a lower risk of incident asthma [40]. Besides, the low bronchiolitis severity (suggested by the low proportion of hypoxemia) in the profile D may also have contributed to the decreased asthma risk. Lastly, the profile E (virusRSV/RV-CmicrobiomeM.catarrhalis) had upregulated Casp A family proteins, which induces uspA1 gene expression and prolongs survival of M. catarrhalis [41]. M. catarrhalis’s lipopolysaccharides activate both MyD88-dependent and TRIF-dependent signaling pathways [42]. These pathways activate proinflammatory downstream signalling (e.g., NFκB, mitogen-activated protein kinases, interferon regulatory factors) that play roles in asthma [43]. Notwithstanding the complexity of these mechanisms, the observed interrelations between the metatranscriptome profiles, host immune response, and asthma development are important findings. Our data should not only advance research that will disentangle the complex web but they also inform the development of microbiome- (or endotype-) specific strategies for the primary prevention of asthma.
Our study has several potential limitations. First, bronchiolitis involves inflammation of the lower airways in addition to the upper airways. While the present study is based on nasopharyngeal samples, studies have shown that upper airway sampling provides a reliable representation of the lung microbiome [43] and transcriptome [44]. Furthermore, the use of upper airway specimens is preferable because lower airway sampling (e.g., bronchoscopy) would be quite invasive in young infants. Second, the nasopharyngeal samples were obtained at a single time-point. While longitudinal molecular data are also informative, the study objective was to identify metatranscriptome profiles of bronchiolitis. However, even with single-time point data, we successfully identified biologically distinct profiles that are longitudinally associated with asthma risk. Third, it is possible that asthma diagnosis is misclassified and that some children are going to develop asthma at a later age. To address these points, the study sample is currently being followed up to age 9 years. Fourth, the present study did not have healthy “controls”. However, our study objective was not to evaluate metatranscriptome profiles related to bronchiolitis development (i.e., bronchiolitis yes vs. no) but to examine the relationship between the metatranscriptome profile of infants with bronchiolitis and their asthma risk. Fifth, while this hypothesis-generating study derives novel and well-calibrated hypotheses that facilitate future experiments, our findings warrant further validation. Lastly, the study sample consisted of racially/ethnically and geographically diverse infants hospitalized for bronchiolitis. Our findings may not be generalizable to infants with mild-to-moderate bronchiolitis or a sample with different respiratory virus proportions. Regardless, our data remain relevant for the 110,000 infants hospitalized yearly in the U.S. [1], a vulnerable population with substantial morbidity burden.
CONCLUSION
In summary, by applying an integrated omics approach to data from a multicentre prospective cohort of 244 infants with severe bronchiolitis, we identified five biologically distinct and clinically meaningful metatranscriptome profiles. These profiles were associated not only with distinct host airway response during bronchiolitis but also with differential risks for developing asthma. Our data suggest a complex interplay between the respiratory virus, airway microbiome, and host immune response, and their integrated contributions to the subsequent development of asthma. For clinicians, our findings may provide an evidence base for the early identification of high-risk children during an important period of airway development—early infancy. For researchers, our data should facilitate further investigations into the development of microbiome profile (or endotype)-specific strategies for asthma prevention.
Supplementary Material
Take Home Message:
Our data suggest a complex interplay between the respiratory virus, airway microbiome, and host immune response in infants with severe bronchiolitis, and their integrated contributions to the subsequent development of childhood asthma.
Acknowledgments:
This study was supported by grants from the National Institutes of Health (Bethesda, MD): U01 AI-087881, R01 AI-114552, R01 AI-108588, R01 AI-134940, and UG3/UH3 OD-023253. M.P.-L. was partially supported by the Margaret Q. Landenberger Research Foundation, the NIH National Center for Advancing Translational Sciences (Award Number UL1TR001876), and the Fundação para a Ciência e a Tegnologia (T495756868-00032862). The content of this manuscript is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health. The funding organizations were not involved in the collection, management, or analysis of the data; preparation or approval of the manuscript; or decision to submit the manuscript for publication.
We thank the MARC-35 study hospitals and research personnel for their ongoing dedication to bronchiolitis and asthma research (Supplementary Table S1 in the Supplementary Materials), and Ashley F. Sullivan, MS, MPH and Janice A. Espinola, MPH (Massachusetts General Hospital, Boston, MA) for their many contributions to the MARC-35 study. We also thank Alkis Togias, MD, at the National Institutes of Health (Bethesda, MD) for helpful comments about the study results.
Footnotes
Conflicts of Interest Statement:
Dr. Bochkov has patents on production methods of rhinoviruses. Dr. Gern is a paid consultant to AstraZeneca and Meissa Vaccines Inc., has stock options in Meissa Vaccines Inc., and has patents on production methods of rhinoviruses. The other authors have no financial relationships relevant to this article to disclose.
Twitter feed comments:
Metatranscriptome profiles identified by clustering analysis suggest a complex interplay between the respiratory virus, airway microbiome, and host immune response in infants with severe bronchiolitis, and their integrated contributions to the subsequent risk of childhood asthma.
REFERENCES
- 1.Fujiogi M, Goto T, Yasunaga H, Fujishiro J, Mansbach JM, Camargo CA, Hasegawa K. Trends in bronchiolitis hospitalizations in the United States: 2000–2016. Pediatrics 2019; 144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hasegawa K, Dumas O, Hartert TV, Camargo CA. Advancing our understanding of infant bronchiolitis through phenotyping and endotyping: clinical and molecular approaches. Expert Rev Respir Med 2016; 10: 891–899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ralston SL, Lieberthal AS, Meissner HC, Alverson BK, Baley JE, Gadomski AM, Johnson DW, Light MJ, Maraqa NF, Mendonca EA, Phelan KJ, Zorc JJ, Stanko-Lopp D, Brown MA, Nathanson I, Rosenblum E, Sayles S, Hernandez-Cancio S, American Academy of Pediatrics. Clinical practice guideline: the diagnosis, management, and prevention of bronchiolitis. Pediatrics 2014; 134: e1474–1502. [DOI] [PubMed] [Google Scholar]
- 4.Dumas O, Hasegawa K, Mansbach JM, Sullivan AF, Piedra PA, Camargo CA. Severe bronchiolitis profiles and risk of recurrent wheeze by age 3 years. J. Allergy Clin. Immunol 2019; 143: 1371–1379.e7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Rosas-Salazar C, Shilts MH, Tovchigrechko A, Schobel S, Chappell JD, Larkin EK, Shankar J, Yooseph S, Nelson KE, Halpin RA, Moore ML, Anderson LJ, Peebles RS, Das SR, Hartert TV. Differences in the nasopharyngeal microbiome during acute respiratory tract infection with human rhinovirus and respiratory syncytial virus in infancy. J. Infect. Dis 2016; 214: 1924–1928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Mansbach JM, Hasegawa K, Henke DM, Ajami NJ, Petrosino JF, Shaw CA, Piedra PA, Sullivan AF, Espinola JA, Camargo CA. Respiratory syncytial virus and rhinovirus severe bronchiolitis are associated with distinct nasopharyngeal microbiota. J Allergy Clin Immunol 2016; 137: 1909–1913.e4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Toivonen L, Camargo CA, Gern JE, Bochkov YA, Mansbach JM, Piedra PA, Hasegawa K. Association between rhinovirus species and nasopharyngeal microbiota in infants with severe bronchiolitis. J. Allergy Clin. Immunol 2019; 143: 1925–1928.e7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Rosas-Salazar C, Shilts MH, Tovchigrechko A, Schobel S, Chappell JD, Larkin EK, Gebretsadik T, Halpin RA, Nelson KE, Moore ML, Anderson LJ, Peebles RS, Das SR, Hartert TV. Nasopharyngeal Lactobacillus is associated with a reduced risk of childhood wheezing illnesses following acute respiratory syncytial virus infection in infancy. J Allergy Clin Immunol 2018; 142: 1447–1456.e9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Turi KN, Shankar J, Anderson LJ, Rajan D, Gaston K, Gebretsadik T, Das SR, Stone C, Larkin EK, Rosas-Salazar C, Brunwasser SM, Moore ML, Peebles RS, Hartert TV. Infant viral respiratory infection nasal immune-response patterns and their association with subsequent childhood recurrent wheeze. Am. J. Respir. Crit. Care Med 2018; 198: 1064–1073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Raita Y, Camargo CA, Bochkov YA, Celedón JC, Gern JE, Mansbach JM, Rhee EP, Freishtat RJ, Hasegawa K. Integrated-omics endotyping of infants with rhinovirus bronchiolitis and risk of childhood asthma. J Allergy Clin Immunol 2021; 147: 2108–2117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hasegawa K, Mansbach JM, Ajami NJ, Petrosino JF, Freishtat RJ, Teach SJ, Piedra PA, Camargo CA. The relationship between nasopharyngeal CCL5 and microbiota on disease severity among infants with bronchiolitis. Allergy 2017; 72: 1796–1800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Stewart CJ, Mansbach JM, Wong MC, Ajami NJ, Petrosino JF, Camargo CA, Hasegawa K. Associations of nasopharyngeal metabolome and microbiome with severity among infants with bronchiolitis. A Multiomic Analysis. Am. J. Respir. Crit. Care Med 2017; 196: 882–891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fujiogi M, Camargo CA, Raita Y, Bochkov YA, Gern JE, Mansbach JM, Piedra PA, Hasegawa K. Association of rhinovirus species with nasopharyngeal metabolome in bronchiolitis infants: A multicenter study. Allergy 2020; 75: 2379–2383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tang HHF, Lang A, Teo SM, Judd LM, Gangnon R, Evans MD, Lee KE, Vrtis R, Holt PG, Lemanske RF, Jackson DJ, Holt KE, Inouye M, Gern JE. Developmental patterns in the nasopharyngeal microbiome during infancy are associated with asthma risk. J Allergy Clin Immunol 2021; 147: 1683–1691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Stewart CJ, Mansbach JM, Piedra PA, Toivonen L, Camargo CA, Hasegawa K. Association of respiratory viruses with serum metabolome in infants with severe bronchiolitis. Pediatr Allergy Immunol 2019; 30: 848–851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bashir H, Grindle K, Vrtis R, Vang F, Kang T, Salazar L, Anderson E, Pappas T, Gangnon R, Evans MD, Jackson DJ, Lemanske RF, Bochkov YA, Gern JE. Association of rhinovirus species with common cold and asthma symptoms and bacterial pathogens. J. Allergy Clin. Immunol 2018; 141: 822–824.e9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Følsgaard NV, Schjørring S, Chawes BL, Rasmussen MA, Krogfelt KA, Brix S, Bisgaard H. Pathogenic bacteria colonizing the airways in asymptomatic neonates stimulates topical inflammatory mediator release. Am. J. Respir. Crit. Care Med 2013; 187: 589–595. [DOI] [PubMed] [Google Scholar]
- 18.Larsen JM, Brix S, Thysen AH, Birch S, Rasmussen MA, Bisgaard H. Children with asthma by school age display aberrant immune responses to pathogenic airway bacteria as infants. J. Allergy Clin. Immunol 2014; 133: 1008–1013. [DOI] [PubMed] [Google Scholar]
- 19.Mansbach JM, Luna PN, Shaw CA, Hasegawa K, Petrosino JF, Piedra PA, Sullivan AF, Espinola JA, Stewart CJ, Camargo CA. Increased Moraxella and Streptococcus species abundance after severe bronchiolitis is associated with recurrent wheezing. J. Allergy Clin. Immunol 2020; 145: 518–527.e8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bisgaard H, Hermansen MN, Buchvald F, Loland L, Halkjaer LB, Bønnelykke K, Brasholt M, Heltberg A, Vissing NH, Thorsen SV, Stage M, Pipper CB. Childhood asthma after bacterial colonization of the airway in neonates. N. Engl. J. Med 2007; 357: 1487–1495. [DOI] [PubMed] [Google Scholar]
- 21.Fujiogi M, Camargo CA, Raita Y, Zhu Z, Celedón JC, Mansbach JM, Spergel JM, Hasegawa K. Integrated associations of nasopharyngeal and serum metabolome with bronchiolitis severity and asthma: A multicenter prospective cohort study. Pediatr Allergy Immunol 2021; 32: 905–916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Raita Y, Toivonen L, Schuez-Havupalo L, Karppinen S, Waris M, Hoffman KL, Camargo CA, Peltola V, Hasegawa K. Maturation of nasal microbiota and antibiotic exposures during early childhood: a population-based cohort study. Clin Microbiol Infect 2021; 27: 283.e1–283.e7. [DOI] [PubMed] [Google Scholar]
- 23.Whitby PW, Seale TW, VanWagoner TM, Morton DJ, Stull TL. The iron/heme regulated genes of Haemophilus influenzae: comparative transcriptional profiling as a tool to define the species core modulon. BMC Genomics 2009; 10: 6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Essilfie A-T, Simpson JL, Horvat JC, Preston JA, Dunkley ML, Foster PS, Gibson PG, Hansbro PM. Haemophilus influenzae infection drives IL-17-mediated neutrophilic allergic airways disease. PLoS Pathog 2011; 7: e1002244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chesné J, Braza F, Mahay G, Brouard S, Aronica M, Magnan A. IL-17 in severe asthma. Where do we stand? Am J Respir Crit Care Med 2014; 190: 1094–1101. [DOI] [PubMed] [Google Scholar]
- 26.Ricciardolo FLM, Sorbello V, Folino A, Gallo F, Massaglia GM, Favatà G, Conticello S, Vallese D, Gani F, Malerba M, Folkerts G, Rolla G, Profita M, Mauad T, Di Stefano A, Ciprandi G. Identification of IL-17F/frequent exacerbator endotype in asthma. J Allergy Clin Immunol 2017; 140: 395–406. [DOI] [PubMed] [Google Scholar]
- 27.Wark PAB, Grissell T, Davies B, See H, Gibson PG. Diversity in the bronchial epithelial cell response to infection with different rhinovirus strains. Respirology 2009; 14: 180–186. [DOI] [PubMed] [Google Scholar]
- 28.Finney LJ, Belchamber KBR, Fenwick PS, Kemp SV, Edwards MR, Mallia P, Donaldson G, Johnston SL, Donnelly LE, Wedzicha JA. Human rhinovirus impairs the innate immune response to bacteria in alveolar macrophages in chronic obstructive pulmonary disease. Am J Respir Crit Care Med 2019; 199: 1496–1507. [DOI] [PubMed] [Google Scholar]
- 29.Durrani SR, Montville DJ, Pratt AS, Sahu S, DeVries MK, Rajamanickam V, Gangnon RE, Gill MA, Gern JE, Lemanske RF, Jackson DJ. Innate immune responses to rhinovirus are reduced by the high-affinity IgE receptor in allergic asthmatic children. J Allergy Clin Immunol 2012; 130: 489–495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Teach SJ, Gill MA, Togias A, Sorkness CA, Arbes SJ, Calatroni A, Wildfire JJ, Gergen PJ, Cohen RT, Pongracic JA, Kercsmar CM, Khurana Hershey GK, Gruchalla RS, Liu AH, Zoratti EM, Kattan M, Grindle KA, Gern JE, Busse WW, Szefler SJ. Preseasonal treatment with either omalizumab or an inhaled corticosteroid boost to prevent fall asthma exacerbations. J Allergy Clin Immunol 2015; 136: 1476–1485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Smith CM, Sandrini S, Datta S, Freestone P, Shafeeq S, Radhakrishnan P, Williams G, Glenn SM, Kuipers OP, Hirst RA, Easton AJ, Andrew PW, O’Callaghan C. Respiratory syncytial virus increases the virulence of Streptococcus pneumoniae by binding to penicillin binding protein 1a. A new paradigm in respiratory infection. Am. J. Respir. Crit. Care Med 2014; 190: 196–207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gullett JM, Cuypers MG, Frank MW, White SW, Rock CO. A fatty acid-binding protein of Streptococcus pneumoniae facilitates the acquisition of host polyunsaturated fatty acids. J Biol Chem 2019; 294: 16416–16428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Fazlollahi M, Lee TD, Andrade J, Oguntuyo K, Chun Y, Grishina G, Grishin A, Bunyavanich S. The nasal microbiome in asthma. J Allergy Clin Immunol 2018; 142: 834–843.e2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kinjo Y, Wu D, Kim G, Xing G-W, Poles MA, Ho DD, Tsuji M, Kawahara K, Wong C-H, Kronenberg M. Recognition of bacterial glycosphingolipids by natural killer T cells. Nature 2005; 434: 520–525. [DOI] [PubMed] [Google Scholar]
- 35.van Zoelen MAD, Schouten M, de Vos AF, Florquin S, Meijers JCM, Nawroth PP, Bierhaus A, van der Poll T. The receptor for advanced glycation end products impairs host defense in pneumococcal pneumonia. J Immunol 2009; 182: 4349–4356. [DOI] [PubMed] [Google Scholar]
- 36.Raita Y, Zhu Z, Freishtat RJ, Fujiogi M, Liang L, Patregnani JT, Camargo CA, Hasegawa K. Soluble receptor for advanced glycation end products (sRAGE) and asthma: Mendelian randomisation study. Pediatr Allergy Immunol 2021; 32: 1100–1103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Perkins TN, Oczypok EA, Dutz RE, Donnell ML, Myerburg MM, Oury TD. The receptor for advanced glycation end products is a critical mediator of type 2 cytokine signaling in the lungs. J Allergy Clin Immunol 2019; 144: 796–808.e12. [DOI] [PubMed] [Google Scholar]
- 38.Lo SW, Gladstone RA, van Tonder AJ, Lees JA, du Plessis M, Benisty R, Givon-Lavi N, Hawkins PA, Cornick JE, Kwambana-Adams B, Law PY, Ho PL, Antonio M, Everett DB, Dagan R, von Gottberg A, Klugman KP, McGee L, Breiman RF, Bentley SD, Global Pneumococcal Sequencing Consortium. Pneumococcal lineages associated with serotype replacement and antibiotic resistance in childhood invasive pneumococcal disease in the post-PCV13 era: an international whole-genome sequencing study. Lancet Infect Dis 2019; 19: 759–769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.LaCroce SJ, Wilson MN, Romanowski JE, Newman JD, Jhanji V, Shanks RMQ, Kowalski RP. Moraxella nonliquefaciens and M. osloensis are important Moraxella species that cause ocular infections. Microorganisms 2019; 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Raita Y, Pérez-Losada M, Freishtat RJ, Harmon B, Mansbach JM, Piedra PA, Zhu Z, Camargo CA, Hasegawa K. Integrated omics endotyping of infants with respiratory syncytial virus bronchiolitis and risk of childhood asthma. Nat Commun 2021; 12: 3601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Heiniger N, Spaniol V, Troller R, Vischer M, Aebi C. A reservoir of Moraxella catarrhalis in human pharyngeal lymphoid tissue. J Infect Dis 2007; 196: 1080–1087. [DOI] [PubMed] [Google Scholar]
- 42.Hassan F Molecular mechanisms of moraxella catarrhalis-induced otitis media. Curr Allergy Asthma Rep 2013; 13: 512–517. [DOI] [PubMed] [Google Scholar]
- 43.Marsh RL, Kaestli M, Chang AB, Binks MJ, Pope CE, Hoffman LR, Smith-Vaughan HC. The microbiota in bronchoalveolar lavage from young children with chronic lung disease includes taxa present in both the oropharynx and nasopharynx. Microbiome 2016; 4: 37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Poole A, Urbanek C, Eng C, Schageman J, Jacobson S, O’Connor BP, Galanter JM, Gignoux CR, Roth LA, Kumar R, Lutz S, Liu AH, Fingerlin TE, Setterquist RA, Burchard EG, Rodriguez-Santana J, Seibold MA. Dissecting childhood asthma with nasal transcriptomics distinguishes subphenotypes of disease. J. Allergy Clin. Immunol 2014; 133: 670–678.e12. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


