SUMMARY
Two-hundred MRSA strains from inpatients with healthcare-associated (HA) and 100 MRSA strains from outpatients with community-associated (CA) skin and soft tissue infections (SSTIs) were tested for antimicrobial susceptibility, staphylococcal cassette chromosome mec (SCCmec) typing, Panton–Valentine leucocidin (PVL) toxin, seh and arcA genes. Based on SCCmec typing, HA-MRSA isolates were further divided into HA-SCCmec I/II/III MRSA and HA-SCCmec IV/V MRSA, and CA-MRSA isolates into CA-SCCmec I/II/III MRSA and CA-SCCmec IV/V MRSA. SCCmec types were further characterized by pulsed-field gel electrophoresis, spa typing and multi-locus sequence typing. Seventy-five (37·5%) HA-MRSA isolates and 83/100 CA-MRSA isolates were SCCmec IV/V genotype. HA-SCCmec IV/V MRSA was associated with malignancy (P = 0·03) and bone fractures (P = 0·02) compared to CA-SCCmec IV/V MRSA. HA-SCCmec IV/V MRSA was associated with PVL gene carriage compared to HA-SCCmec I/II/III MRSA (P < 0·001). ST22-MRSA-IV (EMRSA-15), ST772-MRSA-V, and ST36-MRSA-IV and ST239:EMRSA-I:III were the major clones identified. Our study documents the emergence of SCCmec IV and SCCmec V MRSA clones in an Indian hospital.
Key words: Methicillin-resistant Staphylococcus aureus, MLST, Panton–Valentine leukocidin genes, PFGE, spa typing, SCCmec
INTRODUCTION
Methicillin-resistant Staphylococcus aureus (MRSA) is widely recognized as a major cause of nosocomial infection worldwide and the risk factors for infection with these pathogens in hospital populations are well established [1]. Although, traditionally considered to be a nosocomial pathogen, it is evident that the epidemiology of MRSA infections is rapidly changing.
During the 1990s, various reports of community-associated MRSA (CA-MRSA) infections in healthy individuals appeared in the literature, caused by novel strains which were genetically distinct from traditional healthcare-associated MRSA (HA-MRSA) [2]. CA-MRSA were traditionally regarded as MRSA strains causing infection in previously healthy young patients without prior healthcare contact, generally susceptible to non-β-lactam antibiotics, often carrying Panton–Valentine leukocidin (PVL) encoding genes, and of staphylococcal cassette chromosome mec (SCCmec) types IV or V [3]. By contrast HA-MRSA predominantly caused infections in patients exposed to healthcare settings, exhibited resistance to most non-β-lactam antibiotics and harboured SCCmec types I, II and III [4].
Consequently, as the microbiology and epidemiology of CA-MRSA have evolved, these traditional definitions have broken down, arguing in favour of genotypic classification of strains. As a result there is a growing consensus to define MRSA strains by combinations of genotyping methods such as multi-locus sequence typing (MLST), spa gene type and/or pulsed-field gel electrophoresis (PFGE) with SCCmec analysis to infer their likely epidemiological origin [5].
Numerous lineages of CA-MRSA have since emerged on every continent, several of which have spread internationally [6, 7]. Currently, more than 20 distinct genetic lineages have been identified worldwide [8], five of which are globally distributed, including ST1-IV (WA-1, USA400), ST8-IV (USA300), ST30-IV (South West Pacific clone), sequence type (ST)59-V (Taiwan clone), and ST80-IV (European clone) [6, 7, 9]. Among the latter, ST8-IV and ST30-IV may be considered pandemic, as they have been isolated repeatedly from every continent [7, 10].
CA-MRSA strains are increasingly implicated in nosocomial infections. Outbreaks of HA infections caused by CA-MRSA strains have been reported from Australia and the USA suggesting that such strains are spreading in healthcare settings and are replacing traditional HA-MRSA strains in some places [11, 12]. Various lineages typically associated with HA-MRSA, such as ST22-IV (EMRSA-15), are also increasingly identified in CA-MRSA infections [9, 10]. In recent years, ST772-V (Bengal Bay clone) has emerged as a virulent and unusually resistant CA-MRSA strain in Bangladesh and India [13–15], and its spread to the UK and Europe has been documented [10, 16].
MRSA is a widespread problem in Indian hospitals, where strains have been well characterized [17]. Previous molecular typing of nosocomial MRSA strains recovered from our hospital revealed a similar scenario to that documented for several Indian hospitals where the multi-resistant Brazilian and Hungarian epidemic clone (ST239-MRSA-III) was dominant [17–20]. Recently, MRSA strains carrying SCCmec types IV and V were identified in Indian hospitals in and around Bengaluru and Mumbai where ST772-MRSA-V along with ST22-MRSA-IV are increasingly prevalent and appear to have progressively displaced the previously predominant nosocomial ST239-MRSA-III clone [13–15]. However, these studies did not determine the differences in clinical, demographic and microbiological characteristics of SCCmec type IV/V MRSA strains isolated from patients with and without healthcare exposure. The aim of the present study was to determine whether CA-MRSA SCCmec type IV and type V strains have emerged in our tertiary-care hospital, located in New Delhi, the capital city of India and to analyse the clinical characteristics and clonal diversity of SCCmec IV and SCCmec V MRSA strains from patients with HA skin and soft tissue infections (SSTIs). In addition, we investigated the effects of hospital exposure on SCCmec IV/V MRSA isolates by comparing them with SCCmec IV/V MRSA genotypes isolated from outpatients not exposed to the healthcare setting.
METHODS
This descriptive study was conducted from July 2009 to December 2011 at the All India Institute of Medical Sciences, a 2500-bed teaching hospital providing tertiary care, located in the city of New Delhi, India. The study was approved by the Ethics Committee of the All India Institute of Medical Sciences, New Delhi, India (No. A-25).
Epidemiological definitions
A HA-MRSA SSTI case was defined as one that met one or more of the following criteria: the organism was isolated >48 h after admission to the hospital, the patient had been hospitalized or undergone surgery in the year prior to the MRSA-positive culture results; or an in-dwelling device or percutaneous catheter was present at the time the SSTI specimen for culture was obtained [21]. A case of SSTI infection was classified as CA-MRSA if MRSA was identified in the outpatient setting or <48 h after hospital admission in an individual with no medical history of MRSA infection or colonization, admission to a healthcare facility, dialysis, surgery or insertion of in-dwelling devices in the past year [5].
Patients
To find patients with HA-MRSA SSTIs, we identified all skin and soft tissue cultures obtained >48 h after hospitalization that grew as MRSA. A total of 237 patients including both adults and children were eligible for enrolment, but 28 were unavailable to study personnel because of discharge before contacting them; another nine patients refused to give their consent. Thus, a total of 200 patients with HA-MRSA SSTIs were enrolled in the study. To find patients with CA-MRSA SSTIs, we identified all skin and soft tissue cultures obtained from outpatients that grew as MRSA and had no HA risk factor in the past year prior to soft tissue culture. A total of 112 patients were eligible for enrolment; however, seven were unavailable to study personnel because of delay in contacting them and another five patients refused to give their consent. Thus, a total of 100 patients with CA-MRSA SSTIs were enrolled. For analysis, we examined only data from the first positive culture. The medical records of each patient were reviewed for patient demographics and clinical information.
Bacterial strains
A total of 300 MRSA strains (200 HA-MRSA, 100 CA-MRSA) were available for the study. All staphylococci were identified by standard biochemical tests. Susceptibility to oxacillin was determined by the cefoxitin (30 μg) disc diffusion test as recommended by Clinical and Laboratory Standards Institute (CLSI) [22]. Strains were confirmed as MRSA by detection of the femB gene and mecA gene using multiplex PCR [23].
Susceptibility testing
Susceptibility testing was performed on all MRSA isolates using the disc diffusion method to 15 antimicrobial agents: amikacin (30 μg), gentamicin (10 μg), netilimicin (30 μg), chloramphenicol (30 μg), ciprofloxacin (5 μg), levofloxacin (5 μg), clindamycin (2 μg), co-trimoxazole (1·25/23·75 μg), erythromycin (15 μg), fusidic acid (10 μg), linezolid (30 μg), mupirocin (200 μg), rifampicin (5 μg), teicoplanin (30 μg) and tetracycline (30 μg). Antimicrobial susceptibility was interpreted according to CLSI criteria. A vancomycin (6 μg/ml) agar screen was used to detect intermediate resistance to vancomycin [22]. Multi-drug resistance (MDR) was defined as resistance to ⩾3 non-β-lactam antibiotic classes [24].
Molecular typing
All MRSA isolates were subjected to SCCmec typing and detection of PVL genes. SCCmec elements (I–V) and PVL genes luk F–lukS were identified as described previously [25, 26]. Based on SCCmec types, the HA-MRSA isolates were further divided into HA-SCCmec I/II/III MRSA and HA-SCCmec IV/V MRSA. Similarly, CA-MRSA isolates were grouped into CA-SCCmec I/II/III MRSA and CA-SCCmec IV/V MRSA. Multiplex PCR detection of the seh gene (as a marker for CA-MRSA of clonal lineage ST1/USA400 and arcA gene as part of the ACME (arginine catabolic mobile element) cluster for ST8/t008/USA300 were performed on all SCCmec IV isolates as described previously [27]. Controls for the assay used were previously characterized reference isolates 05-012 90t127/ST1/she lukPV and 06-011 72t008/ST8/arcA lukPV (kindly provided by B. Strommenger, Robert Koch Institute, Germany).
PFGE was performed using SmaI-digested DNA as described previously [28] with selected MRSA strains of HA-SCCmec III, HA-SCCmec IV/V and CA-SCCmec IV/V having distinct resistance profiles. Comparison and grouping of the PFGE patterns were performed with InfoQuest™ FP Software v. 5·4 (Bio-Rad Laboratories, USA). PFGE patterns were compared on an unweighted pair-group method with averages (UPGMA) dendrogram based on Dice coefficients, where optimization and band position tolerance were set at 1·0% and 2·3%, respectively [29]. A similarity coefficient of 80% was selected to define the patterns [30].
MLST and spa typing were performed on selected representative isolates of major PFGE patterns in different SCCmec types as described previously [31, 32]. Isolates were assigned a ST according to the MLST website (http://www.mlst.net) and sequencing scape (Seqscape software, Applied Biosystems, USA).
Spa typing, based on DNA sequencing of the protein A gene variable region was performed, and isolates assigned as described on the Ridom website (http://spa.ridom.de/).
Statistical analysis
Each study variable was compared between SCCmec types. Quantitative variables were summarized as mean ± s.d. and qualitative variables as proportions (%). Odds ratios (ORs) were calculated from logistic regression analysis for SCCmec type IV and type V, along with the 95% confidence intervals (CIs). The ORs for drug resistance were calculated separately for HA-SCCmec I/II/II vs. HA-SCCmec IV/V; and CA-SCCmec IV/V vs. HA-SCCmec IV/V. The proportion of strains resistant to different drugs in different major clones of CA-MRSA was compared using Fisher's exact test. Multivariable logistic regression analysis was performed to identify the variables independently associated with HA-SCCmec IV/V vs. HA-SCCmec I/II/II, and CA-SCCmec IV/V. All tests were two-tailed and a P value ⩽0·05 was considered to be statistically significant. Stata v. 12·1 (StataCorp., USA) was used for all analyses.
RESULTS
SCCmec type, PVL genes and characteristics of patients with HA-MRSA SSTIs
The majority of the 200 HA-MRSA patient isolates investigated carried SCCmec III (n = 116, 58%), followed by SCCmec V (n = 46, 23%) and SCCmec IV (n = 29, 14·5%). Very few isolates carried SCCmec II (n = 5, 2·5%) and SCCmec I (n = 4, 2%). Thus, of this patient cohort classified epidemiologically as having HA-MRSA SSTIs, 125 (62·5%) were infected with HA-SCCmec I/II/III MRSA and 75 (37·5%) with MRSA of HA-SCCmec IV/V lineages. Forty (20%) isolates carried the genes for PVL, which was strongly associated with SCCmec IV and V MRSA strains (86·2% and 26·1%, respectively, carried PVL genes; P < 0·001).
The clinical and epidemiological data of the two groups of inpatients carrying HA-SCCmec IV/V (75) or HA-SCCmec I/II/III (125) were compared to assess associations between carriage of these types and particular risk factors (Table 1). Multivariable analysis confirmed that isolation of HA-SCCmec IV/V MRSA was associated with PVL gene carriage (OR 40·81, 95% CI 11·38–146·32, P < 0·001) and HA-SCCmec I/II/III MRSA was associated with burn wounds (OR 0·09, 95% CI 0·008–1·00, P = 0·050).
Table 1.
Characteristics of patients with HA-MRSA, skin and soft tissue infection as per SCCmec type isolated (n = 200)
| Characteristics | HA-SCCmec I/II/III MRSA (n = 125) | HA-SCCmec IV/V MRSA (n = 75) | OR | 95% CI | P |
|---|---|---|---|---|---|
| Age (years), mean ± s.d. | 34·58 ± 18·84 | 31·42 ± 22·87 | 0·99 | 0·98–1·01 | 0·29 |
| Sex | |||||
| Male | 83 (66·4) | 52 (69·3) | 1·0 | ||
| Female | 42 (33·6) | 23 (30·7) | 0·87 | 0·47–1·62 | 0·67 |
| Patient location | |||||
| Medical ward | 56 (44·8) | 31 (41·3) | 1·0 | ||
| Surgical ward | 56 (44·8) | 37 (49·3) | 1·02 | 0·37–2·85 | 0·96 |
| ICU | 13 (10·4) | 7 (9·3) | 1·22 | 0·45–3·36 | 0·69 |
| Duration of hospital stay (days) before culture | 18·5 ± 18·89 | 15·65 ± 11·96 | 0·99 | 0·97–1·01 | 0·26 |
| 0–9 | 41 (32·8) | 27 (36·0) | 1·0 | ||
| 10–19 | 34 (27·2) | 22 (29·3) | 0·98 | 0·48–2·03 | 0·96 |
| ⩾20 | 38 (30·4) | 23 (30·7) | 0·92 | 0·45–1·87 | 0·82 |
| Unknown | 12 (9·6) | 3 (4·0) | 0·38 | 0·10–1·47 | 0·16 |
| Panton–Valentine leukocidin | 3 (2·4) | 37 (49·3) | 39·6 | 11·56–135·7 | <0·001 |
| Risk factors associated | |||||
| Intravenous catheter in use at time of culture | 89 (70·2) | 51 (68·0) | 0·86 | 0·46–1·60 | 0·63 |
| Urinary catheter in use at time of culture | 52 (41·6) | 28 (37·3) | 0·84 | 0·46–1·51 | 0·55 |
| Inter-hospital transfer | 19 (15·2) | 9 (12·0) | 0·76 | 0·32–1·78 | 0·53 |
| Intra-hospital transfer | 38 (30·4) | 31 (41·3) | 1·6 | 0·89–2·93 | 0·12 |
| Previous hospitalization (<6 months) | 55 (44·0) | 37 (49·3) | 1·24 | 0·70–2·20 | 0·46 |
| ICU admission before culture | 35 (28·0) | 25 (33·3) | 1·29 | 0·69–2·39 | 0·43 |
| Endotracheal intubations | 26 (20·8) | 9 (12·0) | 0·52 | 0·23–1·18 | 0·12 |
| Prosthetic devices | 3 (2·4) | 8 (10·7) | 4·86 | 1·25–18·92 | 0·023 |
| Underlying diseases/conditions | |||||
| Diabetes mellitus | 12 (9·6) | 9 (12·0) | 1·28 | 0·51–3·21 | 0·59 |
| Bone fracture | 7 (5·6) | 8 (10·7) | 2·01 | 0·70–5·80 | 0·16 |
| Osteomyelitis | 9 (7·2) | 1 (1·3) | 0·17 | 0·02–1·40 | 0·1 |
| Malignancies/leukaemia | 17 (13·6) | 10 (13·3) | 0·98 | 0·42–2·26 | 0·96 |
| Chronic renal disease | 2 (1·6) | 5 (6·7) | 4·39 | 0·83–23·34 | 0·08 |
| HIV infection | 0 (0·0) | 1 (1·3) | 0·0 | 0·0 | 0·2 |
| Sepsis | 19 (15·2) | 10 (13·3) | 0·86 | 0·38–1·96 | 0·72 |
| Multi-organ failure | 6 (4·8) | 2 (2·7) | 0·54 | 0·11–2·76 | 0·46 |
| Respiratory tract infections | 7 (5·6) | 8 (10·7) | 2·01 | 0·67–5·80 | 0·2 |
| Non-infectious dermatosis* | 19 (15·2) | 14 (18·7) | 1·28 | 0·60–2·74 | 0·52 |
| Infected burns | 15 (12·0) | 1 (1·3) | 0·1 | 0·01–0·77 | 0·027 |
| Recent antimicrobial therapy† | |||||
| β-lactams | 99 (79·2) | 56 (74·7) | 0·77 | 0·39–1·52 | 0·46 |
| Aminoglycosides | 56 (44·8) | 31 (41·3) | 0·87 | 0·49–1·55 | 0·63 |
| Metronidazole | 36 (28·8) | 13 (17·3) | 0·52 | 0·25–1·06 | 0·07 |
| Quinolones | 32 (25·6) | 17 (23·7) | 0·85 | 0·43–1·67 | 0·64 |
| Linezolid | 23 (18·4) | 14 (18·7) | 1·02 | 0·49–2·13 | 0·96 |
| Glycopeptides | 17 (13·6) | 13 (17·3) | 1·33 | 0·61–2·93 | 0·48 |
| Clindamycin | 8 (6·4) | 2 (2·7) | 0·4 | 0·08–1·94 | 0·26 |
| Co-trimoxazole | 3 (2·4) | 2 (2·7) | 1·11 | 0·18–6·83 | 0·91 |
| Mupirocin | 2 (1·6) | 1 (1·3) | 0·83 | 0·07–9·32 | 0·88 |
| Total number of antibiotics received | |||||
| Monotherapy | 17 (13·6) | 17 (22·7) | 1·0 | ||
| Dual combination therapy | 37 (29·6) | 20 (26·7) | 1·61 | 0·73–3·52 | 0·24 |
| Triple or multiple combination therapy | 61 (48·8) | 38 (50·7) | 0·87 | 0·44–1·71 | 0·68 |
| No antibiotic therapy | 10 (8·0) | 0 (0·0) | — | — | — |
| Mortality | 6 (4·8) | 2 (2·7) | 0·54 | 0·11–2·76 | 0·46 |
OR, Odds ratio; CI, confidence interval; ICU intensive care unit.
Data are no. (%) of patients, unless otherwise specified.
For example, psoriasis, eczema, pemphigus vulgaris, etc.
Preceding antibiotics used for the condition for which the patient was hospitalized.
P values <0·05 are highlighted in bold.
SCCmec type, PVL genes and characteristics of patients with CA-MRSA SSTIs
The most common SCCmec types among 100 isolates from this patient cohort were types V (n = 45), IV (n = 38) and III (n = 17). Interestingly, 65·8% of SCCmec IV isolates carried the PVL gene compared to only 26·7% of type V (P < 0·001). The 83 isolates of types IV and V were designated as CA-SCCmec IV/V MRSA and subjected to further analysis. The demographic and clinical characteristics of these patients were compared with the 75 patients with HA-SCCmec IV/V MRSA SSTIs (Table 2). The mean age of all patients in both cohort groups was 27·95 ± 20·28 years and 104 (65·8%) were male. Multivariable regression analysis identified malignancy (OR 10·57, 95% CI 1·23–90·49, P = 0·03) and bone fractures (OR 12·33, 95% CI 1·47–103·34, P = 0·02) as independent risk factors significantly associated with HA-SCCmec IV/V MRSA SSTIs.
Table 2.
Association of study characteristics in patients with CA-SCCmec type IV/V and HA-SCCmec type IV/V infections
| Characteristics | CA-SCCmec IV/V MRSA (n = 83) | HA-SCCmec IV/V MRSA (n = 75) | OR | 95% CI | P |
|---|---|---|---|---|---|
| Age (years), mean ± s.d. | 24·7 ± 17·1 | 31·4 ± 22·9 | — | — | 0·04 |
| Sex | |||||
| Male | 52 (62·7) | 52 (69·3) | 1·0 | ||
| Female | 31 (37·4) | 23 (30·7) | 0·74 | 0·38–1·44 | 0·38 |
| Panton–Valentine leukocidin | 40 (48·2) | 37 (49·3) | 1·05 | 0·56–1·95 | 0·89 |
| Underlying diseases/conditions | |||||
| Non-infectious dermatosis* | 1 (1·2) | 14 (18·7) | 18·82 | 2·41–147·01 | 0·005 |
| Bone fracture | 1 (1·2) | 8 (10·7) | 9·79 | 1·19–80·26 | 0·034 |
| Osteomyelitis | 5 (6·0) | 1 (1·3) | 0·21 | 0·024–1·85 | 0·16 |
| Malignancies/leukaemia | 1 (1·2) | 10 (13·3) | 12·62 | 1·57–101·10 | 0·017 |
| Respiratory tract infections | 0 (0·0) | 8 (10·7) | — | — | 0·002 |
| Chronic renal disease | 0 (0·0) | 5 (6·67) | — | — | 0·02 |
| Wound infection | 4 (4·8) | 3 (4·0) | 0·82 | 0·18–3·80 | 0·80 |
| Infected ulcer | 2 (2·4) | 2 (2·7) | 1·11 | 0·15–8·08 | 0·92 |
| Abscess | 11 (13·3) | 3 (4·0) | 0·27 | 0·07–1·02 | 0·05 |
| Cellulites | 2 (2·3) | 2 (2·7) | 1·12 | 0·15–8·08 | 0·92 |
| Infected burns | 0 (0·0) | 1 (1·3) | — | — | 0·48 |
OR, Odds ratio; CI, confidence interval.
Data are no. (%) of patients, unless otherwise specified.
For example, psoriasis, eczema, pemphigus vulgaris, etc.
P values <0·05 are highlighted in bold.
Antimicrobial resistance
The distribution of resistance to antimicrobials of the MRSA isolates grouped by healthcare exposure status and SCCmec type are presented in Table 3. Compared with HA-SCCmec I/II/III MRSA isolates, a greater proportion of HA-SCCmec IV/V isolates were susceptible to all the antibiotics tested. However, compared to CA-SCCmec IV/V MRSA isolates, a higher proportion of HA-SCCmec IV/V isolates were antibiotic resistant. Ninety-five (76·0%) HA-SCCmec I/II/III isolates and 32 (42·7%) HA-SCCmec IV/V isolates were MDR (P < 0·001) whereas only five (6·0%) of CA-SCCmec IV/V MRSA isolates were MDR (P < 0·001).
Table 3.
Antimicrobial resistance patterns of methicillin-resistant Staphylococcus aureus (MRSA) isolates from patients with skin and soft tissue infections grouped by SCCmec type and healthcare exposure status*
| Antimicrobial agents (μg) | HA-SCCmec I/II/III MRSA (n = 125) | HA-SCCmec IV/V MRSA (n = 75) | OR | 95% CI | P† | CA-SCCmec IV/V MRSA (n = 83) | OR‡ | 95% CI‡ | P†‡ |
|---|---|---|---|---|---|---|---|---|---|
| Amikacin | 69 (55·2) | 28 (37·3) | 0·48 | 0·27–0·87 | 0·015 | 4 (4·8) | 11·77 | 3·88–35·63 | <0·001 |
| Gentamicin | 62 (49·6) | 25 (33·3) | 0·51 | 0·28–0·92 | 0·026 | 17 (20·5) | 1·94 | 0·95–3·98 | 0·07 |
| Netilimicin | 23 (18·4) | 10 (13·3) | 0·68 | 0·31–1·53 | 0·35 | 1 (1·2) | 12·62 | 1·57–101·10 | 0·017 |
| Chloramphenicol | 14 (11·2) | 6 (8·0) | 0·69 | 0·25–1·88 | 0·47 | 3 (3·6) | 2·32 | 0·56–9·62 | 0·25 |
| Ciprofloxacin | 91 (72·8) | 42 (56·0) | 0·48 | 0·26–0·87 | 0·016 | 20 (24·1) | 4·01 | 2·03–7·90 | <0·001 |
| Levofloxacin | 87 (69·6) | 40 (53·3) | 0·50 | 0·28–0·90 | 0·022 | 20 (24·1) | 3·60 | 1·83–7·09 | <0·001 |
| Clindamycin | 82 (65·6) | 43 (57·3) | 0·70 | 0·39–1·27 | 0·24 | 17 (20·5) | 5·22 | 2·58–10·53 | <0·001 |
| Co-trimoxazole | 88 (70·4) | 32 (42·7) | 0·31 | 0·17–0·57 | <0·001 | 42 (50·6) | 0·73 | 0·39–1·36 | 0·32 |
| Erythromycin | 106 (84·8) | 53 (70·7) | 0·43 | 0·22–0·87 | 0·018 | 33 (39·8) | 3·65 | 1·88–7·09 | <0·001 |
| Fusidic acid | 25 (20·0) | 14 (18·7) | 0·92 | 0·44–1·90 | 0·82 | 3 (3·6) | 6·12 | 1·68–22·25 | 0·006 |
| Mupirocin | 7 (5·6) | 3 (4·1) | 0·71 | 0·18–2·84 | 0·63 | 0 (0·0) | — | — | 0·064 |
| Rifampicin | 44 (35·2) | 16 (21·3) | 0·50 | 0·26–0·97 | 0·040 | 3 (3·6) | 7·23 | 2·01–25·96 | 0·002 |
| Tetracycline | 87 (69·6) | 34 (45·3) | 0·36 | 0·20–0·66 | 0·001 | 5 (6·0) | 12·94 | 4·70–35·59 | <0·001 |
OR, Odds ratio; CI, confidence interval.
According to the CDC definition [7].
All P values determined using odds ratios and not derived from t test or χ2 test. P values <0·05 highlighted in bold.
Analyses of antimicrobial resistance patterns of HA-SCCmec-IV/V and CA-SCCmec-IV/V MRSA isolates. All strains were susceptible to vancomycin (screen agar method), linezolid and teicoplanin.
P values <0·05 are highlighted in bold.
Clonality of CA-SCCmec IV/V, HA-SCCmec IV/V MRSA and HA-SCCmec III isolates
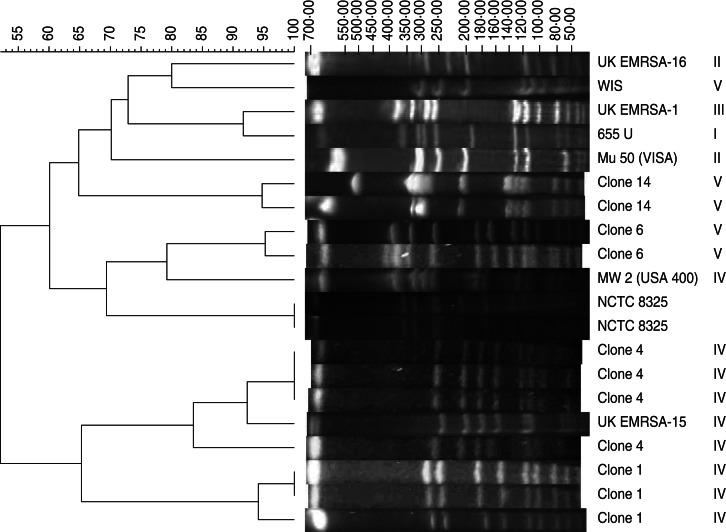
The SmaI macrorestriction fragment profiles of 48 HA-SCCmec IV/V, 41 CA-SCCmec IV/V MRSA and 20 HA-SCCmec III MRSA isolates revealed 19 clones with four major clusters predominating (clone 4, n = 32, 29·4%; clone 14, n = 22, 20·2%; clone 6, n = 15, 13·8%; clone 1, n = 11, 10·1%) and together representing 80% of all isolates tested (Fig. 1). Each major clone comprised 2–10 subtypes. Except for clone 6, which was exclusive to the HA-SCCmec III MRSA group, the remaining three major clones were found in each patient cohort with SCCmec IV/V MRSA isolates. Clone 4 isolates carried only SCCmec IV and were detected in both CA- (40·6%) and HA- (59·4%) SCCmec IV isolates, often exhibiting PVL carriage (75%). Similarly, clone 14 was highly associated with SCCmec type V (86·4%) and less so with SCCmec type IV (13·2%) isolates; 50% of this clone carried PVL. Clone 1 comprised only SCCmec IV isolates and these often exhibited PVL (81·8%).
Fig. 1.
Dendrogram based on similarities derived from the unweighted pair group method with arithmetic mean (UPGMA) and Dice coefficients, using InfoQuest™ FP software (Bio-Rad). The scale at the top represents similarity. Pulsed-field gel electrophoretic patterns of SmaI macrorestriction fragments of EMRSA-1, -15, -16, MW2, WIS, 655 U, Mu-50 along with representative strains of the SCCmec types IV/V major clones 1, 4, 6 and 14.
Comparison of local with international clones
PFGE was performed with epidemic MRSA strains (EMRSA)-1, -15, -16 from the UK, MW2, WIS, 655 U and Mu-50 (vanomycin-intermediate S. aureus) along with representative strains of major clones found here (Fig. 1). The predominant clone 4 showed close similarity (>90%) in DNA pattern to the epidemic UK EMRSA-15 clone. Of note, none of the isolates showed PFGE patterns of the USA400 CA-MRSA lineage. In addition, all HA-SCCmec IV and CA-SCCmec IV MRSA isolates were negative for arcA and seh genes.
Antimicrobial resistance patterns in different clones
The resistance to various antimicrobials varied from clone to clone and there was no correlation with resistance profiles and specific clones. Nevertheless, two broad groups were evident: one with isolates resistant only to β-lactams or 1–3 other antibiotics (clones 1, 2, 4, 12–17), and those resistant to almost all antibiotics tested (clones 3, 5–11, 18, 19). Isolates of clone 6 showed more resistance than other clones, with ~80% resistance to fluroquinolones, tetracycline, erythromycin and clindamycin; clone 4 was uniformly susceptibility to gentamicin, netlimicin, amikacin and co-trimoxazole.
Spa typing and MLST
MLST and spa typing were performed on 38 representative strains of the major clones 1, 4, 6 and 14 (n = 30) and sporadic clones (n = 8). Table 4 shows that the strains were assigned to 14 STs and 17 spa types. The 12 SCCmec V strains belonged to four STs and spa types with eight of them identified as ST772-SCCmec-V-t657 and the remainder were each unique. The SCCmec IV strains (13) clustered in four STs and the dominant ST22 consisted of diverse spa types. The 13 SCCmec III strains were grouped into six STs and spa types with 10 strains belonging to ST239 or the closely related ST241, ST1097 and ST1310.
Table 4.
SCCmec, PVL status, MLST and spa typing for selected strains of the major and sporadic PFGE clones
| SCCmec (n) | PVL status | MLST profile (no. of isolates) | ST | spa type (no. of isolates) | PFGE clones (no. of isolates) | Epidemic clone |
|---|---|---|---|---|---|---|
| V (12) | 1 | 6, 2 , 6 , 2 , 7, 13, 5 (1) | ST689 | t019 (1) | 2 (1) | |
| 8 | 1, 1, 1, 1, 22, 1, 1 (9) | ST772 | t657 (9) | 14 (9) | Asia (SLV of ST1) | |
| 1 | 22, 1, 14, 109, 12, 4, 3 (1) | ST1289 | t2526 (1) | 16 (1) | ||
| 1 | 1, 1, 1, 159, 1, 1, 1 (1) | ST2039 | t386 (1) | 8 (1) | ||
| IV (13) | 3 | 7, 6, 1, 5, 8, 8, 6 (6) | ST22 | t005 (1), t891 (2), t1152 (2), t3107 (1) | 4 (6) | United Kingdom (EMRSA-15) |
| 3 | 2, 2, 2, 2, 3, 3, 2 (3) | ST36 | t4410 (3) | 1 (3) | ||
| 1 | 7, 6, 1, 5, 8, 5, 6 (2) | ST217 | t1328 (2) | 4 (2) | ||
| 0 | 2, 2, 2, 2, 6, 3, 2 (2) | ST30 | t021 (2) | Southwest Pacific (USA1100) | ||
| III (13) | 1 | 2, 3, 1, 1, 4, 4, 3 (6) | ST239 | t030 (6) | 6 (6) | Brazilian (SLV of ST8) |
| 0 | 2, 3, 1, 1, 4, 4, 30 (2) | ST241 | t037 (2) | 6 (1), 9 (1) | ||
| 0 | 1, 4, 1, 8, 4, 4, 3 (1) | ST72 | t347 (1) | 8 (1) | ||
| 0 | 2, 3, 6, 1, 4, 4, 3 (2) | ST1097 | t2952 (2) | 3 (2) | ||
| 0 | 2, 3, 1, 1, 4, 4, 30 (1) | ST1310 | t4410 (1) | 8 (1) | ||
| 0 | 1, 3, 1, 1, 4, 4, 11. 1) | ST673 | t1309 (1) | 16 (1) |
PVL, Panton–Valentine leukocidin; MLST, multi-locus sequence typing; ST, sequence type; PFGE, pulsed-field gel electrophoresis; SLV, single locus variant.
DISCUSSION
Our study documents the emergence of MRSA isolates typical of CA genotypes in patients with HA-MRSA SSTIs in an Indian hospital. Remarkably, at our institute a large proportion (37·5%) of isolates classified epidemiologically as HA-MRSA had a CA genotype. These data confirm the reported spread of CA-MRSA SCCmec IV/V strains in hospital settings in Europe, the USA [33, 34] and India [13, 15]. Given the vulnerable population within the hospital setting, it is unclear how infections with isolates that contain SCCmec IV/V may differ in symptoms and severity from those caused by the traditional HA-SCCmec I/II/III isolates. The introduction of SCCmec IV/V strains into our hospital population did not result in a change in the spectrum or severity of illness in this group. Compared to patients with CA-SCCmec IV/V SSTIs, it was observed that HA-SCCmec IV/V SSTIs were significantly associated with malignancy and bone fractures. Patients with these conditions are predisposed to use healthcare facilities, are generally exposed to antibiotics, tend to have interventions performed, and hence present opportunities to contract MRSA in the healthcare facility. Exactly why the SCCmec IV/V strains are successful in hospital settings such as ours remains unknown but mathematical models predict the replacement of traditional HA-MRSA strains by CA-MRSA strains, due to their higher growth rate and greater genetic fitness [35, 36].
In this study, as expected, HA-SCCmec IV/V isolates were more susceptible to multiple antibiotics compared to HA-SCCmec I/II/III. However, similar to previous reports [11], HA-SCCmec IV/V strains demonstrated higher antimicrobial resistance rates compared to CA-SCCmec IV/V strains. Differences in antimicrobial susceptibility indicate that, in patients with healthcare contact, SCCmec IV/V MRSA strains exhibit characteristics of traditional HA-MRSA strains. Our findings support the hypothesis of Gonzalez et al. [37] that as typical CA-MRSA strains proliferate in healthcare settings, where antimicrobial selection pressures are high, they will continue to acquire additional antimicrobial resistance genetic elements, causing them to appear similar to more traditional MRSA isolates, with respect to resistance profiles. Our finding of PVL genes in 49% of HA-SCCmec IV/V isolates is not surprising since SCCmec IV/V strains have been associated largely with SSTIs and PVL production [12]. However, the sporadic (2·6%) PVL-positive SCCmec III HA-MRSA isolates is an issue of potential concern and could result in the emergence of MDR HA-MRSA isolates with increased virulence, despite the fact that the role of PVL as a virulence factor is a matter of much debate.
Molecular typing revealed that 73% of the isolates belonged to four major genetic lineages, with the remaining sporadic isolates showing a high degree of genetic diversity, suggesting the possibility of new strains being imported into the hospital from the expanding community reservoir. Interestingly, the highly infectious CA-MRSA strains USA300 and USA400 were not detected in our hospital or community. Previous characterization of nosocomial MRSA strains recovered in our hospital from patients with SSTIs identified a major MRSA clone closely related to the ST239:EMRSA-I:III which was MDR [17]. In the present study, selected strains of the major PFGE clone 6 also showed genetic relatedness to the lineage ST239:EMRSA-I:III with spa-type t030. ST239 is the major endemic HA-MRSA clone in many Asian countries, although recent studies show that it is being gradually replaced by emerging CA-MRSA clones [13–15] as was also observed in our study.
The ST22-SCCmec IV MRSA (EMRSA-15) strain is a global pandemic HA-MRSA clone and interestingly it was recovered from both inpatients and outpatients in our hospital. This suggests that outpatients may represent an important reservoir for MRSA dissemination within the hospital, when admitted as inpatients and reinforces the observation of Alcoceba et al. [38] regarding the movement of EMRSA-15 from the community to the hospital setting.
The SCCmec V MRSA isolates that were genotyped were of spa type t657 and ST772, similar to the types reported recently in Mumbai and elsewhere in India [13–15]. The decreasing prevalence of the HA-MRSA strain ST239-MRSA-III in hospital in India since 2006, coupled with an increase in prevalence of ST772-MRSA-V and ST22-MRSA-IV, has led to the suggestion that these strains may be replacing the ST239-MRSA strain in Indian hospitals [13–15]. The emergence of the MDR clone ST772-MRSA-V in our hospital is a worrying development, and is indicative of the need for enhanced surveillance to ensure that these strains do not spread.
Another relevant finding of the present study concerns the presence of the ST36:EMRSA-16:IV (major PFGE clone 1) clones of spa-type t4410. Similar to the finding of Söderquist et al. [39], it was found to have the same ST as the EMRSA-16 (ST36-MRSA-II) strain, which is one of the predominant EMRSA strains in Europe. Furthermore, the described isolate was PVL positive, as were the majority of CA-MRSA strains, but this differs from EMRSA-16 ST36, which is PVL negative. The PFGE pattern of the isolate differed by >6 bands compared to EMRSA-16. To our knowledge, this is the first report of the ST36:EMRSA-16:IV clone in an Asian hospital.
This study had certain limitations. All of the observations were derived from a single hospital and therefore, may only reflect local trends and, thus, might not be representative of other settings in India or other countries. Further, only representative SCCmec IV/V strains of the major PFGE clones were genotyped by MLST and spa typing. Nevertheless, our data clearly demonstrate that the SCCmec type IV and type V epidemic clones of ST22 (EMRSA-15) and ST772 have entered into our hospital and now cause a substantial proportion of serious HA-MRSA infections.
In summary, the high proportions of HA-MRSA strains carrying SCCmec types IV and V, together with the considerable occurrence of PVL-positive MRSA strains, confirm the extensive infiltration of CA-MRSA genotypes in our hospital. We found that, following healthcare exposure, SCCmec IV/V strains have characteristics similar to those of other HA-MRSA strains isolated from these patients. Moreover, HA-SCCmec IV/V strains exhibited an antimicrobial susceptibility pattern in-between those of CA-SCCmec IV/V and HA-SCCmec I/II/III. This finding raises concerns that CA-SCCmec IV/V and HA-SCCmec IV/V strains may exchange genetic material resulting in an organism uniquely adapted to produce aggressive SSTIs similar to CA-MRSA strains which carry PVL genes as well expressing resistance to multiple antimicrobial agents generally associated with the current SCCmec I/II/III strains. The dissemination of these epidemic CA-MRSA clones in both inpatients and outpatients represents a significant challenge to infection control.
ACKNOWLEDGEMENTS
We thank K. Hiramatsu, H. de Lencastre and B. Strommenger for kindly donating some of the prototype and reference strains used in this study. Financial support was received from the Indian Council of Medical Research (ICMR), New Delhi, India.
DECLARATION OF INTEREST
None.
REFERENCES
- 1.Lowy FD. Staphylococcus aureus infections. New England Journal of Medicine 1998; 20: 520–532. [DOI] [PubMed] [Google Scholar]
- 2.Chambers HF, Deleo FR. Waves of resistance: Staphylococcus aureus in the antibiotic era. Nature Reviews Microbiology 2009, 7: 629–641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Naimi TS, et al. Comparison of community- and healthcare-associated methicillin-resistant Staphylococcus aureus infection. Journal of the American Medical Association 2003; 290: 2976–2984. [DOI] [PubMed] [Google Scholar]
- 4.Zetola N, et al. Community-acquired methicillin-resistant Staphylococcus aureus: an emerging threat. Lancet Infectious Diseases 2005; 5: 275–286. [DOI] [PubMed] [Google Scholar]
- 5.Otter JA, French GL. Community-associated meticillin-resistant Staphylococcus aureus: the case for a genotypic definition. Journal of Hospital Infection 2012; 81: 143–148. [DOI] [PubMed] [Google Scholar]
- 6.David MZ, Daum RS. Community-associated methicillin-resistant Staphylococcus aureus: epidemiology and clinical consequences of an emerging epidemic. Clinical Microbiology Reviews 2010; 23: 616–687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Deleo FR, et al. Community-associated meticillin-resistant Staphylococcus aureus. Lancet 2010; 375: 1557–1568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mediavilla JR, et al. Global epidemiology of community-associated methicillin resistant Staphylococcus aureus (CA-MRSA). Current Opinion in Microbiology 2012; 15: 588–595. [DOI] [PubMed] [Google Scholar]
- 9.Witte W. Community-acquired methicillin-resistant Staphylococcus aureus: what do we need to know? Clinical Microbiology and Infection 2009; 15: 17–25. [DOI] [PubMed] [Google Scholar]
- 10.Monecke S, et al. A field guide to pandemic, epidemic and sporadic clones of methicillin-resistant Staphylococcus aureus. PLoS ONE 2011; 6: e17936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Maree CL, et al. Community–associated methicillin-resistant Staphylococcus aureus isolates causing healthcare-associated infections. Emerging Infectious Diseases 2007; 13: 236–242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Valsesia G, et al. Emergence of SCCmec type IV and SCCmec type V methicillin-resistant Staphylococcus aureus containing the Panton-Valentine leukocidin genes in a large academic teaching hospital in central Switzerland: external invaders or persisting circulators? Journal of Clinical Microbiology 2010; 48: 720–727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.D'Souza M, Rodrigues C, Mehta A. Molecular characterization of methicillin-resistant Staphylococcus aureus with emergence of epidemic clones of sequence types (ST) 22 and ST772 in Mumbai, India. Journal of Clinical Microbiology 2010; 48: 1806–1811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Nadig S, et al. Staphylococcus aureus eye infections in two Indian hospitals: emergence of ST772 as a major clone. Clinical Ophthalmology 2012; 6: 165–173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Shambat S, et al. Clonal complexes and virulence factors of Staphylococcus aureus from several cities in India. BMC Microbiology 2012; 12: 64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Brennan GI, et al. Emergence of hospital- and community-associated Panton-Valentine leukocidin-positive methicillin-resistant Staphylococcus aureus genotype ST772-MRSA-V in Ireland and detailed investigation of an ST772-MRSA-V cluster in a neonatal intensive care unit. Journal of Clinical Microbiology 2012; 50: 841–847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gadepalli R, et al. Clinical and molecular characteristics of nosocomial methicillin-resistant Staphylococcus aureus skin and soft tissue isolates from three Indian hospitals. Journal of Hospital Infection 2009; 73: 253–263. [DOI] [PubMed] [Google Scholar]
- 18.Arakere G et al. Genotyping of methicillin-resistant Staphylococcus aureus strains from two hospitals in Bangalore, South India. Journal of Clinical Microbiology 2005; 43: 3198–3202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Shabir S et al. Epidemiological typing of meticillin-resistant Staphylococcus aureus isolates from Pakistan and India. Journal of Medical Microbiology 2010; 59: 330–337. [DOI] [PubMed] [Google Scholar]
- 20.Abimanyu N, Murugesan S, Krishnan P. Emergence of methicillin-resistant Staphylococcus aureus ST239 with high-level mupirocin and inducible clindamycin resistance in a tertiary care center in Chennai, South India. Journal of Clinical Microbiology 2012; 50: 3412–3413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Centers for Disease Control and Prevention. Community associated MRSA information for clinicians. Infection control topics. Centers for Disease Control and Prevention, Atlanta, GA, 2005. (www.cdc.gov/ncidod/dhqp/ar_mrsa_ca_clinicians.html).
- 22.Clinical and Laboratory Standards Institute. Performance standards for antimicrobial susceptibility testing; twenty-first informational supplement, M100-S21. CLSI, 2011. Wayne, PA: USA. [Google Scholar]
- 23.Perez-Roth E, Claverie-Martin F, Batista N. Mupirocin resistance in methicillin-resistant Staphylococcus aureus clinical isolates in a Spanish hospital. Co-application of multiplex PCR assay and conventional microbiology methods. Diagnostic Microbiology and Infectious Disease 2002; 43: 123–128. [DOI] [PubMed] [Google Scholar]
- 24.Fey PD, et al. Comparative molecular analysis of community- or hospital-acquired methicillin-resistant Staphylococcus aureus. Antimicrobial Agents and Chemotherapy 2003; 47: 196–203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Milheirico C, Oliveira DC, de Lencastre H. Update to the multiplex PCR strategy for assignment of mec element types in Staphylococcus aureus. Antimicrobial Agents and Chemotherapy 2007; 51: 3374–3377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lina G, et al. Involvement of Panton-Valentine leukocidin-producing Staphylococcus aureus in primary skin infections and pneumonia. Clinical Infectious Diseases 1999; 29: 1128–1132. [DOI] [PubMed] [Google Scholar]
- 27.Strommenger B, et al. Multiplex PCR for rapid detection of Staphylococcus aureus isolates suspected to represent community-acquired strains. Journal of Clinical Microbiology 2008; 46: 582–587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Udo EE, Al-Mufti S, Albert MJ. The prevalence of antimicrobial resistance and carriage of virulence genes in Staphylococcus aureus isolated from food handlers in Kuwait City restaurants. BMC Research Notes 2009; 2: 108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Faria NA, et al. Epidemiology of emerging methicillin-resistant Staphylococcus aureus (MRSA) in Denmark: a nationwide study in a country with low prevalence of MRSA infection. Journal of Clinical Microbiology 2005; 43:1836–1842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.McDougal LK, et al. Pulsed-field gel electrophoresis typing of oxacillin-resistant Staphylococcus aureus isolates from the United States: establishing a national database. Journal of Clinical Microbiology 2003; 41: 5113–5120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Enright MC, et al. Multi locus sequence typing for characterization of methicillin-resistant and methicillin-susceptible clones of Staphylococcus aureus. Journal of Clinical Microbiology 2000; 38: 1008–1015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Harmsen D, et al. Typing of methicillin-resistant Staphylococcus aureus in a University hospital setting using a novel software for Spa repeat determination and database management. Journal of Clinical Microbiology 2003; 41: 5442–5448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Klevens RM, et al. National nosocomial infections surveillance system. Changes in the epidemiology of methicillin-resistant Staphylococcus aureus in intensive care units in US hospitals, 1992–2003. Clinical Infectious Diseases 2006; 42: 389–391. [DOI] [PubMed] [Google Scholar]
- 34.Otter JA, French GL. Molecular epidemiology of community-associated methicillin-resistant Staphylococcus aureus in Europe. Lancet Infectious Diseases 2010; 10: 227–239. [DOI] [PubMed] [Google Scholar]
- 35.D'Agata EMC, et al. Modeling the invasion of community-acquired methicillin-resistant Staphylococcus aureus into hospitals. Clinical Infectious Diseases 2009; 48: 274–284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Popovich KJ, Weinstein RA, Hota B. Are community-associated methicillin-resistant Staphylococcus aureus (MRSA) strains replacing traditional nosocomial MRSA strains? Clinical Infectious Diseases 2008; 46: 787–794. [DOI] [PubMed] [Google Scholar]
- 37.Gonzalez BE, et al. Community-associated strains of methicillin-resistant Staphylococcus aureus as the cause of healthcare-associated infection. Infection Control and Hospital Epidemiology 2006; 27: 1051–1056. [DOI] [PubMed] [Google Scholar]
- 38.Alcoceba E, et al. Molecular epidemiology of methicillin-resistant Staphylococcus aureus in Majorcan hospitals: high prevalence of the epidemic clone EMRSA-15. Clinical Microbiology Infection 2007; 13: 599–605. [DOI] [PubMed] [Google Scholar]
- 39.Söderquist B, Berglund C, Strålin K. Community-acquired pneumonia and bacteremia caused by an unusual methicillin-resistant Staphylococcus aureus (MRSA) strain with sequence type 36, staphylococcal cassette chromosome mec type IV and Panton-Valentine leukocidin genes. European Journal of Clinical Microbiology and Infectious Disease 2006; 25: 604–606. [DOI] [PubMed] [Google Scholar]