SUMMARY
Genetic association studies of the cytokine interleukin-10 (IL-10) and sepsis have provided inconsistent results. This work attempts to further quantitatively assess the association of three widely evaluated polymorphisms of IL-10 (−592C/A, −819C/T, −1082A/G) with sepsis susceptibility through a meta-analysis. A search of Pubmed, Web of Science and EMBASE databases was performed. Overall, the three polymorphisms have no strong association with sepsis risk. Subgroup analysis by ethnicity showed there was association between sepsis susceptibility with −592C/A in Caucasians (A vs. C: OR 0·78, 95% CI 0·62–1·00, P = 0·05; AA + CA vs. CC: OR 0·75, 95% CI 0·56–1·00, P = 0·05), and with −1082A/G in Asians (G vs. A: OR 1·41, 95% CI 1·04–1·91, P = 0·03; GG + AG vs. AA: OR 2·11, 95% CI 1·07–4·16, P = 0·03). This meta-analysis suggests that −592C/A and −1082A/G polymorphisms are associated with sepsis susceptibility in Caucasian, and Asian populations, respectively.
Key words: Genetic polymorphism, interleukin-10, sepsis
INTRODUCTION
Sepsis is defined as the systemic inflammatory response caused by infection [1]. Despite modern resuscitation strategies and new anti-infective options, sepsis and its sequelae remain a common cause of acute illness and death in patients with community-acquired and nosocomial infections [2]. Therefore, predictive markers to identify high-risk patients are urgently needed for early detection and preventive care. Increasing evidence suggests that genetic variation especially single nucleotide polymorphisms (SNPs) in the innate immune system may influence the risk of patients for serious infection [3]. Delineating the variation in genes and associated differences in response to sepsis may contribute to the development of new genetically tailored diagnostic and therapeutic interventions to improve outcome in patients with sepsis susceptibility.
Interleukin-10 (IL-10) has been identified as one of the key anti-inflammatory cytokines in the inflammatory cascade as it decreases the production of inflammatory molecules, such as TNF-α, interferon (IFN)-γ, IL-12, reactive nitric oxide metabolites, major histocompatibility complex molecules [4] and inhibits antigen-specific cytotoxic T cells [5]. IL-10 has been shown to be elevated after trauma [6] and is well correlated with the development of sepsis and outcome in patients with major trauma [7, 8]. Previous studies have demonstrated that variation in IL-10 production is largely genetically determined through variations in the promoter region [9, 10]. The IL-10 5′-flanking region, which controls transcription, is polymorphic, with two microsatellites between −4000 and −1100, and three SNPs (−1082, −819, −592) [11]. Moreover, the IL-10 −1082AA genotype is related to lower production of IL-10, and carriers with heterozygous AG genotype and homozygous GG genotype have significantly increased inducibility of IL-10 production after stimulation [12–14].
To date, several studies have investigated the association of IL-10 polymorphisms with sepsis susceptibility but their findings lack consistency and remain inconclusive. Only one meta-analysis has investigated the association between the IL-10 −1082A/G polymorphism and sepsis risk [15]. This paper from 2013, included 11 studies published before September 2012, and concluded that the IL-10 −1082A/G polymorphism has an association with susceptibility to sepsis but the inclusion of overlapping studies [16, 17] might have biased their findings. We performed an initial systematic literature search which revealed several other studies on the −1082A/G polymorphism not cited in [15]. This therefore prompted a meta-analysis to investigate further associations between three widely evaluated variants of IL-10: −592C/A (rs1 800 872), −819C/T (rs1 800 871), and −1082A/G (rs1 800 896), and sepsis susceptibility.
METHODS
Literature search
A systematic literature search for published papers was performed in Pubmed, EMBASE, and Web of Science for the period up to 8 October 2013. The following key terms were used: (interleukin-10 OR IL10 OR IL-10) AND (polymorphism OR mutation OR variant) AND (sepsis OR septic). The references of original research reports and review articles were also searched. Studies fulfilling all of the following selection criteria were included: (1) they evaluated the association between polymorphisms in the IL-10 gene and risk of sepsis; (2) they were case-control or cohort studies; (3) they provided data on the genotype or allele counts of at least one polymorphism. Studies were excluded if any of the following features were identified: (1) they were not relevant to IL-10 polymorphisms and sepsis; (2) they were non-clinical; (3) they were reviews or comments. Only English-language publications were included. For overlapping studies, the one with the largest sample size was included.
Qualitative assessment and data extraction
Two investigators (W.P. and A.Q.Z.) independently extracted data, and made a quality assessment of the retrieved studies. Discrepancies were resolved in a consensus meeting. The Newcastle–Ottawa Scale (NOS) [18] was applied to evaluate the qualities of the included studies. A scoring system of 0–9 was used to judge and score data study quality based on three broad perspectives: selection, comparability, and outcome or finding of interest. Studies with scores of ⩾7 were considered to be of high quality and scores were summed to quantitatively compare the study quality.
The following information was extracted from each study: authors, year of publication, country, ethnicity of participants, source of controls, sample size, genotype distribution, sepsis type and genotyping method.
Statistical analysis
Data were entered from studies into a computerized spreadsheet (Microsoft Excel, USA). Hardy–Weinberg equilibrium (HWE) – to measure whether the observed genotype frequencies in a population differ from the frequencies predicted by the equation – was tested by means of χ2 test (significant at the 0·05 level). The odds ratios (ORs) and 95% confidence intervals (CIs) as the effect measure were estimated for each study. The significance of pooled ORs was tested by Z test (P < 0·05) and pooled ORs were evaluated for the allele comparison model (B vs. A), the co-dominant model (BB vs. AA), the dominant model (BB + AB vs. AA), and the recessive model (BB vs. AA + AB); A and B represent the major and minor alleles, respectively. As heterogeneity was anticipated between observational studies, pooled ORs were calculated using a random-effects model for all analyses. Consistent with common practice, we performed a statistical test for heterogeneity between studies using Cochran's Q; a significant Q statistic (P < 0·10) indicated heterogeneity across studies. The I2 statistic was used for estimation of the degree of heterogeneity in meta-analyses and a study with an I2 of >50% was interpreted as having a high degree of heterogeneity [19]. In addition, several factors, such as ethnicity, age, and sepsis severity, were closely related to susceptibility of sepsis, thus subgroup analyses were performed by ethnicity, sepsis type and HWE. Sensitivity analysis was performed through sequentially excluded individual studies to assess the stability of the results. Potential publication bias was examined visually in a funnel plot of log (OR) against its standard error (s.e.), and the degree of asymmetry was tested using Egger's test [20]. All statistical tests were performed using Revman 5.2 software (Nordic Cochrane Center, Denmark) and Stata v. 11.0 software (Stata Corporation, USA).
RESULTS
Study characteristics
The search of databases retrieved 482 articles and three additional articles were identified from the reference lists of relevant studies giving a total of 485 relevant titles and abstracts for review. After removing 177 duplications, a further unrelated 245 articles were excluded based on title and abstract. After a full text review we excluded a further 43, leaving 37 studies for inclusion in the meta-analysis (Fig. 1).
Fig. 1.
Flow diagram of study identification and selection.
The main characteristics of the 37 eligible studies and genotype distributions of the three polymorphisms (−592C/A, −819C/T, −1082A/G) under study are listed in Table 1. Twenty-four studies were performed in Caucasian populations and 13 in Asian populations. Genotypes were determined by various methodologies: polymerase chain reaction–restriction fragment length polymorphism (PCR–RFLP) (n = 16), amplification refractory mutation system (ARMS) (n = 5), TaqMan probes (n = 7), SNaPshot (n = 2), fluorescence-labelled probes (n = 6), and single-stranded peptide–PCR (SSP–PCR) (n = 1). The quality scores of most studies ranged from 5 to 7, suggesting moderate quality.
Table 1.
Characteristics of the studies included in the meta-analysis
| Study | Country | Ethnicity | Sepsis type | Source of controls | Genotyping method | Sample size (cases/controls) | Cases (11/12/22)* | Controls (11/12/22)* | NOS score | HWE |
|---|---|---|---|---|---|---|---|---|---|---|
| rs1800872 −592C/A | ||||||||||
| Shimada, 2011 [21] | Japan | Asian | Sepsis | Critically ill | Fluorescence probe | 123/101 | 62/47/14 | 38/45/18 | 6 | Yes |
| Davis, 2010 [22] | America | Caucasian | Sepsis | Healthy | TaqMan probe | 26/53 | 18/8/0 | 30/17/6 | 7 | Yes |
| Gu, 2010 [16] | China | Asian | Sepsis | Trauma | Fluorescence probe | 153/154 | 17/66/70 | 26/57/71 | 7 | No |
| Huebinger, 2010 [23] | America | Caucasian | Sepsis | Burn | TaqMan probe | 50/215 | 22/24/4 | 95/98/22 | 6 | Yes |
| Nakada, 2005 [24] | Japan | Asian | Sepsis | Critically ill | PCR–RFLP | 86/111 | 8/38/40 | 19/47/45 | 6 | Yes |
| Balding, 2003 [25] | Ireland | Caucasian | Sepsis | Healthy | PCR–RFLP | 183/389 | 125/52/6 | 235/139/15 | 5 | Yes |
| Lowe, 2003 [26] | UK | Caucasian | Sepsis | Critically ill | PCR–RFLP | 41/36 | 30/10/1 | 23/13/0 | 6 | Yes |
| Shu, 2003 [27] | China | Asian | Severe sepsis | Healthy | PCR–RFLP | 116/141 | 71/38/7 | 91/39/11 | 6 | No |
| rs1800871 −819C/T | ||||||||||
| Palumbo, 2012 [28] | Italy | Caucasian | Sepsis | Burn | PCR–RFLP | 16/26 | 10†/6‡ | 12†/14‡ | 7 | n.a. |
| Shimada, 2011 [21] | Japan | Asian | Sepsis | Critically ill | Fluorescence probe | 123/101 | 62/47/14 | 38/46/17 | 6 | Yes |
| Carregaro, 2010 [29] | Brazil | Caucasian | Sepsis | Blood donor | TaqMan probe | 97/207 | 47/38/12 | 82/102/23 | 6 | Yes |
| Davis, 2010 [22] | America | Caucasian | Sepsis | Healthy | TaqMan probe | 28/53 | 18/10/0 | 30/17/6 | 7 | Yes |
| Emonts, 2010 [30] | Netherlands | Caucasian | Sepsis | Healthy | SNaPshot | 79/460 | 49/24/6 | 283/145/32 | 6 | No |
| Gu, 2010 [16] | China | Asian | Sepsis | Trauma | Fluorescence probe | 148/160 | 12/49/87 | 17/73/70 | 7 | Yes |
| Huebinger, 2010 [23] | America | Caucasian | Sepsis | Burn | TaqMan probe | 50/215 | 22/24/4 | 95/98/22 | 6 | Yes |
| Nakada, 2005 [24] | Japan | Asian | Sepsis | Critically ill | PCR–RFLP | 86/111 | 8/38/40 | 19/47/45 | 6 | Yes |
| Shu, 2003 [27] | China | Asian | Severe sepsis | Healthy | PCR–RFLP | 116/141 | 71/38/7 | 91/39/11 | 6 | No |
| rs1800896 −1082A/G | ||||||||||
| Palumbo, 2012 [28] | Italy | Caucasian | Sepsis | Burn | PCR–RFLP | 16/26 | 12§/4¶ | 5§/21¶ | 7 | n.a. |
| Shimada, 2011 [21] | Japan | Asian | Sepsis | Critically ill | Fluorescence probe | 123/101 | 111/12/0 | 93/8/0 | 6 | Yes |
| Carregaro, 2010 [29] | Brazil | Caucasian | Sepsis | Blood donor | TaqMan probe | 97/207 | 44/44/9 | 82/103/22 | 6 | Yes |
| Davis, 2010 [22] | America | Caucasian | Sepsis | Healthy | TaqMan probe | 28/53 | 7/16/5 | 16/25/12 | 7 | Yes |
| Emonts, 2010 [30] | Netherlands | Caucasian | Sepsis | Healthy | SNaPshot | 82/459 | 17/44/21 | 117/219/123 | 6 | Yes |
| Gu, 2010 [16] | China | Asian | Sepsis | Trauma | Fluorescence probe | 147/161 | 106/37/4 | 118/36/7 | 7 | Yes |
| Abdel, 2009 [31] | Egypt | Caucasian | Sepsis | Neonates | PCR–RFLP | 54/70 | 11/37/6 | 14/47/9 | 5 | No |
| McDaniel, 2007 [32] | America | Caucasian | Sepsis | Trauma | ARMS | 31/37 | 7/13/11 | 18/11/8/ | 5 | No |
| Baier study1, 2006 [12] | America | Caucasian | Sepsis | VLBW | ARMS | 114/119 | 48/57/9 | 40/61/18 | 6 | Yes |
| Baier study2, 2006 [12] | America | Caucasian | Sepsis | VLBW | ARMS | 31/26 | 13/13/5 | 6/12/8/ | 6 | Yes |
| Garnacho, 2006 [33] | Spain | Caucasian | Sepsis | Healthy | PCR–RFLP | 224/101 | 92/99/33 | 36/50/15 | 7 | Yes |
| Stanilova, 2006 [34] | Bulgaria | Caucasian | Severe sepsis | Healthy | ARMS | 33/53 | 17/15/1 | 12/32/9 | 5 | Yes |
| Nakada, 2005 [24] | Japan | Asian | Sepsis | Critical ill | PCR–RFLP | 86/111 | 75/11/0 | 103/8/0 | 6 | Yes |
| Zhang, 2005 [35] | China | Asian | Septic shock | Critical ill | PCR–RFLP | 33/76 | 19/14/0 | 62/14/0 | 6 | Yes |
| Jaber, 2004 [36] | America | Caucasian | Sepsis | ARF | SSP–PCR | 38/21 | 13a/25b | 4a/17b | 5 | NA |
| Balding, 2003 [25] | Ireland | Caucasian | Sepsis | Healthy | PCR–RFLP | 183/389 | 36/95/52 | 86/180/123 | 5 | Yes |
| Lowe, 2003 [26] | UK | Caucasian | Sepsis | Critical ill | PCR–RFLP | 31/36 | 8/18/5 | 11/17/8 | 6 | Yes |
| Schaaf, 2003 [13] | Germany | Caucasian | Sepsis | Infection | ARMS | 51/18 | 17/20/14 | 8/8/2/ | 6 | Yes |
| Shu, 2003 [27] | China | Asian | Severe sepsis | Healthy | PCR–RFLP | 116/141 | 8/77/31 | 42/56/43 | 6 | No |
| Treszl, 2003 [37] | Hungary | Caucasian | Sepsis | VLBW | PCR–RFLP | 33/70 | 10/17/6 | 11/40/19 | 6 | Yes |
ARF, Acute renal failure; ARMS: amplification refractory mutation system; HWE, Hardy–Weinberg equilibrium; n.a., not available; NOS, Newcastle–Ottawa Scale; PCR, polymerase chain reaction; PCR–RFLP, PCR–restriction fragment length polymorphism; SSP–PCR, single-stranded peptide–PCR; VLBW, very-low-birth-weight neonates.
For −592C/A polymorphism (11 = CC, 12 = CA, 22 = AA); for −819C/T polymorphism (11 = CC, 12 = CT, 22 = TT); for −1082A/G polymorphism (11 = AA, 12 = AG, 22 = GG).
Genotype represented by 11.
Genotype represented by 12 + 22.
Genotype represented by 11 + 12.
Genotype represented by 22.
Quantitative data synthesis
For the IL-10 −592C/A polymorphism, eight studies encompassing 778 cases and 1200 controls were identified [16, 21–27]. Overall, there was no significant association between this polymorphism and sepsis risk (A vs. C: OR 0·91, 95% CI 0·74–1·11, P = 0·36; AA vs. CC: OR 0·94, 95% CI 0·60–1·48, P = 0·79; AA vs. CC + CA: OR 0·92, 95% CI 0·70–1·22, P = 0·56; AA + CA vs. CC: OR 0·92, 95% CI 0·69–1·25, P = 0·61) (Table 2). In subgroup analysis, the results showed significant association in Caucasians under two comparison models (A vs. C: OR 0·78, 95% CI 0·62–1·00, P = 0·05; AA + CA vs. CC: OR 0·75, 95% CI 0·56–1·00, P = 0·05) (Table 2). However, no significant association was found in subgroup analyses based on sepsis type and HWE under all genetic models.
Table 2.
Overall and subgroup analyses of the IL-10 −592C/A polymorphism with sepsis susceptibility
| Groups | Studies | Sample size | Test of association | Model | Heterogeneity | |||
|---|---|---|---|---|---|---|---|---|
| Cases | Controls | P value | OR (95% CI) | P value | I2 (%) | |||
| Total | ||||||||
| A vs. C | 8 | 778 | 1200 | 0·36 | 0·91 (0·74–1·11) | R | 0·12 | 39 |
| AA vs. CC | 8 | 778 | 1200 | 0·79 | 0·94 (0·60–1·48) | R | 0·20 | 29 |
| AA vs. CC + CA | 8 | 778 | 1200 | 0·56 | 0·92 (0·70–1·22) | R | 0·66 | 0 |
| AA + CA vs. CC | 8 | 778 | 1200 | 0·61 | 0·92 (0·69–1·25) | R | 0·09 | 44 |
| Asian | ||||||||
| A vs. C | 4 | 478 | 507 | 0·95 | 1·01 (0·75–1·36) | R | 0·07 | 57 |
| AA vs. CC | 4 | 478 | 507 | 0·87 | 1·06 (0·55–2·03) | R | 0·07 | 58 |
| AA vs. CC + CA | 4 | 478 | 507 | 0·76 | 0·95 (0·70–1·29) | R | 0·43 | 0 |
| AA + CA vs. CC | 4 | 478 | 507 | 0·59 | 1·15 (0·69–1·94) | R | 0·04 | 64 |
| Caucasian | ||||||||
| A vs. C | 4 | 300 | 693 | 0·05 | 0·78 (0·62–1·00) | R | 0·59 | 0 |
| AA vs. CC | 4 | 300 | 693 | 0·38 | 0·73 (0·36–1·47) | R | 0·59 | 0 |
| AA vs. CC + CA | 4 | 300 | 693 | 0·47 | 0·77 (0·39–1·54) | R | 0·57 | 0 |
| AA + CA vs. CC | 4 | 300 | 693 | 0·05 | 0·75 (0·56–1·00) | R | 0·73 | 0 |
| HWE | ||||||||
| A vs. C | 6 | 509 | 905 | 0·18 | 0·83 (0·64–1·08) | R | 0·12 | 43 |
| AA vs. CC | 6 | 509 | 905 | 0·55 | 0·83 (0·44–1·54) | R | 0·17 | 35 |
| AA vs. CC + CA | 6 | 509 | 905 | 0·60 | 0·90 (0·62–1·32) | R | 0·44 | 0 |
| AA + CA vs. CC | 6 | 509 | 905 | 0·13 | 0·79 (0·58–1·07) | R | 0·23 | 27 |
| Sepsis | ||||||||
| A vs. C | 7 | 662 | 1059 | 0·31 | 0·89 (0·70–1·12) | R | 0·08 | 46 |
| AA vs. CC | 7 | 662 | 1059 | 0·87 | 0·96 (0·56–1·63) | R | 0·14 | 38 |
| AA vs. CC + CA | 7 | 662 | 1059 | 0·56 | 0·94 (0·70–1·25) | R | 0·56 | 0 |
| AA + CA vs. CC | 7 | 662 | 1059 | 0·50 | 0·89 (0·63–1·25) | R | 0·08 | 47 |
OR, Odds ratio; CI, confidence interval; HWE, Hardy–Weinberg equilibrium.
For the IL-10 −819C/T polymorphism, nine studies comprised 743 cases and 1474 controls [16, 21–24, 27–30]. Overall, no evidence of a statistically significant association of this polymorphism with sepsis susceptibility was found (T vs. C: OR 0·99, 95% CI 0·80–1·23, P = 0·95; TT vs. CC: OR 0·99, 95% CI 0·67–1·48, P = 0·97; TT vs. CC + CT: OR 1·09, 95% CI 0·78–1·53, P = 0·59; TT + CT vs. CC: OR 0·92, 95% CI 0·73–1·16, P = 0·48) (Table 3). Similarly, subgroup analyses by ethnicity, sepsis type and HWE revealed no significant association under all genetic models.
Table 3.
Overall and subgroup analyses of the IL-10 −819C/T polymorphism with sepsis susceptibility
| Groups | Studies | Sample size | Test of association | Model | Heterogeneity | |||
|---|---|---|---|---|---|---|---|---|
| Cases | Controls | P value | OR (95% CI) | P value | I2 (%) | |||
| Total | ||||||||
| T vs. C | 8 | 727 | 1448 | 0·95 | 0·99 (0·80–1·23) | R | 0·04 | 52 |
| TT vs. CC | 8 | 727 | 1448 | 0·97 | 0·99 (0·67–1·48) | R | 0·22 | 26 |
| TT vs. CC + CT | 8 | 727 | 1448 | 0·59 | 1·09 (0·78–1·53) | R | 0·19 | 29 |
| TT + CT vs. CC | 9 | 743 | 1474 | 0·48 | 0·92 (0·73–1·16) | R | 0·33 | 12 |
| Asian | ||||||||
| T vs. C | 4 | 473 | 513 | 0·61 | 1·10 (0·76–1·60) | R | 0·01 | 72 |
| TT vs. CC | 4 | 473 | 513 | 0·77 | 1·11 (0·56–2·18) | R | 0·07 | 58 |
| TT vs. CC + CT | 4 | 473 | 513 | 0·61 | 1·14 (0·69–1·85) | R | 0·08 | 56 |
| TT + CT vs. CC | 4 | 473 | 513 | 0·74 | 1·09 (0·66–1·78) | R | 0·07 | 57 |
| Caucasian | ||||||||
| T vs. C | 4 | 254 | 935 | 0·31 | 0·89 (0·71–1·11) | R | 0·65 | 0 |
| TT vs. CC | 4 | 254 | 935 | 0·62 | 0·88 (0·52–1·48) | R | 0·58 | 0 |
| TT vs. CC + CT | 4 | 254 | 935 | 0·90 | 0·97 (0·59–1·60) | R | 0·51 | 0 |
| TT + CT vs. CC | 5 | 270 | 961 | 0·23 | 0·84 (0·64–1·12) | R | 0·84 | 0 |
| HWE | ||||||||
| T vs. C | 6 | 532 | 847 | 0·86 | 0·97 (0·72–1·32) | R | 0·01 | 65 |
| TT vs. CC | 6 | 532 | 847 | 0·98 | 0·99 (0·57–1·73) | R | 0·10 | 46 |
| TT vs. CC + CT | 6 | 532 | 847 | 0·65 | 1·10 (0·72–1·69) | R | 0·11 | 44 |
| TT + CT vs. CC | 6 | 532 | 847 | 0·52 | 0·89 (0·64–1·26) | R | 0·17 | 36 |
| Sepsis | ||||||||
| T vs. C | 7 | 611 | 1307 | 0·89 | 0·98 (0·76–1·26) | R | 0·03 | 58 |
| TT vs. CC | 7 | 611 | 1307 | 0·95 | 1·02 (0·64–1·61) | R | 0·16 | 35 |
| TT vs. CC + CT | 7 | 611 | 1307 | 0·51 | 1·13 (0·79–1·62) | R | 0·17 | 34 |
| TT + CT vs. CC | 8 | 627 | 1333 | 0·34 | 0·88 (0·69–1·14) | R | 0·32 | 14 |
OR, Odds ratio; CI, confidence interval; HWE, Hardy–Weinberg equilibrium.
For the IL-10 −1082A/G polymorphism, 20 studies of 1551 cases and 2275 controls were identified [12, 13, 16, 21, 22, 24–37]. The overall result suggested no statistically significant association of this polymorphism with sepsis risk (G vs. A: OR 1·13, 95% CI 0·89–1·44, P = 0·31; GG vs. AA: OR 0·91, 95% CI 0·61–1·35, P = 0·64; GG vs. AG + AA: OR 0·83, 95% CI 0·68–1·02, P = 0·08; GG + AG vs. AA: OR 1·10, 95% CI 0·82–1·47, P = 0·52) (Table 4). However, by subgroup analysis based on ethnicity, there was a statistically significant association in Asians under two comparison models (G vs. A: OR 1·41, 95% CI 1·04–1·91, P = 0·03; GG + AG vs. AA: OR 2·11, 95% CI 1·07–4·16, P = 0·03) (Table 4). No similar association was evident under all comparison models in other subgroup analyses based on sepsis type and HWE.
Table 4.
Overall and subgroup analyses of the IL-10 −1082A/G polymorphism with sepsis susceptibility
| Groups | Studies | Sample size | Test of association | Model | Heterogeneity | |||
|---|---|---|---|---|---|---|---|---|
| Cases | Controls | P value | OR (95% CI) | P value | I2 (%) | |||
| Total | ||||||||
| G vs. A | 18 | 1497 | 2228 | 0·31 | 1·13 (0·89–1·44) | R | <0·00 001 | 76 |
| GG vs. AA | 18 | 1497 | 2228 | 0·64 | 0·91 (0·61–1·35) | R | 0·008 | 53 |
| GG vs. AA + AG | 19 | 1513 | 2254 | 0·08 | 0·83 (0·68–1·02) | R | 0·65 | 0 |
| GG + AG vs. AA | 19 | 1535 | 2249 | 0·52 | 1·10 (0·82–1·47) | R | <0·0001 | 65 |
| Asian | ||||||||
| G vs. A | 5 | 354 | 468 | 0·03 | 1·41 (1·04–1·91) | R | 0·25 | 25 |
| GG vs. AA | 5 | 354 | 468 | 0·58 | 1·64 (0·29–9·41) | R | 0·02 | 81 |
| GG vs. AA + AG | 5 | 354 | 468 | 0·36 | 0·79 (0·48–1·31) | R | 0·67 | 0 |
| GG + AG vs. AA | 5 | 354 | 468 | 0·03 | 2·11 (1·07–4·16) | R | 0·006 | 73 |
| Caucasian | ||||||||
| G vs. A | 13 | 1143 | 1760 | 0·82 | 1·03 (0·77–1·39) | R | <0·00 001 | 81 |
| GG vs. AA | 13 | 1143 | 1760 | 0·31 | 0·82 (0·57–1·19) | R | 0·07 | 39 |
| GG vs. AA + AG | 14 | 1159 | 1786 | 0·15 | 0·85 (0·69–1·06) | R | 0·56 | 0 |
| GG + AG vs. AA | 14 | 1181 | 1781 | 0·34 | 0·88 (0·67–1·15) | R | 0·04 | 44 |
| HWE | ||||||||
| G vs. A | 15 | 1296 | 1980 | 0·6 | 1·08 (0·82–1·42) | R | <0·00 001 | 78 |
| GG vs. AA | 15 | 1296 | 1980 | 0·10 | 0·75 (0·54–1·06) | R | 0·19 | 26 |
| GG vs. AA + AG | 15 | 1296 | 1980 | 0·06 | 0·80 (0·64–1·01) | R | 0·64 | 0 |
| GG + AG vs. AA | 15 | 1296 | 1980 | 0·84 | 0·97 (0·75–1·26) | R | 0·02 | 48 |
| Sepsis | ||||||||
| G vs. A | 15 | 1315 | 1958 | 0·35 | 1·13 (0·88–1·46) | R | <0·00 001 | 75 |
| GG vs. AA | 15 | 1315 | 1958 | 0·36 | 0·87 (0·63–1·18) | R | 0·23 | 21 |
| GG vs. AA + AG | 16 | 1331 | 1984 | 0·17 | 0·86 (0·69–1·07) | R | 0·75 | 0 |
| GG + AG vs. AA | 16 | 1353 | 1979 | 0·92 | 0·99 (0·81–1·21) | R | 0·21 | 22 |
OR, Odds ratio; CI, confidence interval; HWE, Hardy–Weinberg equilibrium.
Heterogeneity analysis
For the −592C/A polymorphism, heterogeneity was significant in the subgroup analyses of Asians and sepsis type. Since the genotype data for two studies were not in HWE [16, 27], a subgroup analysis was performed based on this equation. The exclusion of these latter studies rendered the data more homogeneous (Table 2).
For the −819C/T polymorphism, statistically significant heterogeneity between studies was found in the pooling analyses of total available studies and in the subgroup analyses of Asians and sepsis type. However, results were similar after excluding studies which were not in HWE [27, 28, 30], (Table 3).
For the −1082A/G polymorphism, a significant level of heterogeneity between studies was observed in the overall analyses. Some of the heterogeneity was resolved by ethnicity-specific analyses. Subgroup analyses by HWE and sepsis type were used to pool the data and the results became more homogeneous (Table 4).
Sensitivity analysis
Sensitivity analysis was performed by excluding one study in the meta-analysis at a time to reflect the influence of the individual dataset to the pooled ORs for each of the studied polymorphisms. The corresponding pooled ORs were not significantly altered, which suggested the robustness of the results (data not shown).
Publication bias
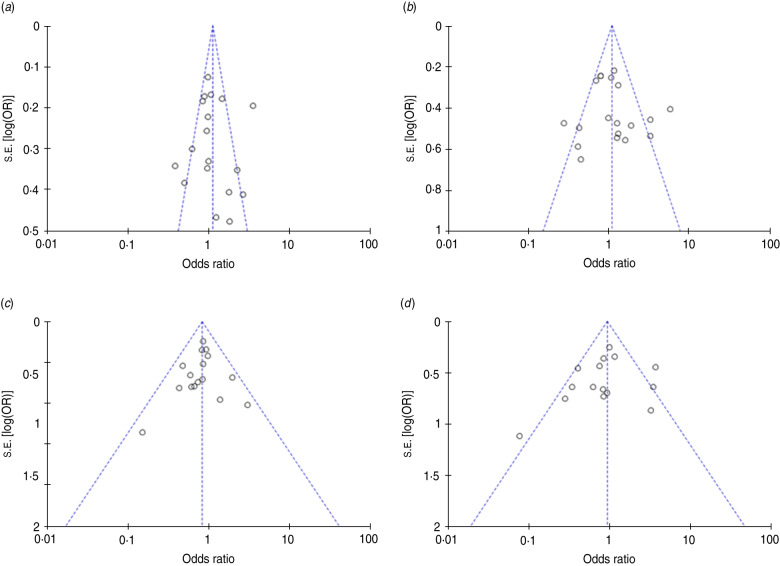
Funnel plots and Egger's test were performed to estimate publication bias of the literature. The shapes of the funnel plots appeared slightly asymmetrical (Fig. 2) but Egger's test revealed no publication bias for studies published on the −1082A/G polymorphism (G vs. A: P = 0·939; GG vs. AA: P = 0·096; GG vs. AG + AA: P = 0·456; GG + AG vs. AA: P = 0·754). Due to the limitation of smaller study sizes, funnel plots and Egger's test were not performed for −592C/A and −819C/T polymorphisms.
Fig. 2.
Funnel plot for IL-10 −1082A/G polymorphism and sepsis risk under the (a) G vs. A model, (b) GG + AG vs. AA model, (c) GG vs. AG + AA model, (d) GG vs. AA model.
DISCUSSION
In this meta-analysis, we found that the pooled ORs and 95% CIs under different comparison models did not show any associations of the A allele of the −592C/A polymorphism, T allele of −819C/T polymorphism and G allele of −1082A/G polymorphism with susceptibility to sepsis in the overall study population. However, subgroup analysis by ethnicity indicated a statistical association between sepsis susceptibility and the −592C/A polymorphism in Caucasian, and the −1082A/G polymorphism in Asian populations. It therefore appears that there is a race-specific effect in the association between the two polymorphisms and sepsis susceptibility in different ethnicities. However, it should be noted that there were only four eligible studies for −592C/A and five studies for −1082A/G available in the current meta-analysis and such small samples with a limited number of subjects may be open to selection bias, and do not categorically support or deny an association. It is therefore critical that larger and well-designed studies are performed to re-evaluate the apparent associations with ethnicity.
Previously, only one meta-analysis has investigated the association between the IL-10 −1082A/G polymorphism and risk of sepsis [15]. This included 11 studies and concluded that the IL-10 −1082A/G polymorphism has an association with susceptibility of sepsis (for G vs. A: OR 0·83; for GG vs. AA: OR 0·67). However, their findings could be heavily biased as a consequence of the methods used. The inclusion of overlapping studies [16, 17] which used the same sample could lead to an underestimation of an association. Moreover, this meta-analysis [15] appeared to lack a robust quality assessment and quantitative data analysis as quality of studies was assessed by HWE in controls and a fixed-effects model (Mantel–Haenszel method) was used in quantitative data analysis. Last, several studies of IL-10 −1082A/G were not included in the analysis [21–23, 25, 30–32, 36, 37], and since their publication a new study of IL-10 gene polymorphisms in burn sepsis has appeared [28].
Statistical heterogeneity between studies is common in meta-analysis of genetic association studies (GAS) and it can be taken into account by performing a random-effects model [38]. In addition, deviations from HWE in control subjects, which can be due to genotyping errors, population stratification, selection bias in the choice of controls and confounding factors unaccounted for, may bias the estimates of genetic effects in GAS and meta-analysis [39], thus we performed subgroup analysis by HWE. No consensus is achieved currently for whether or not to include the studies departing from HWE. But if the results are different before and after removing studies not in HWE, it is suggested that the analysis without studies not conforming to HWE would be more valid [40].
Our meta-analysis has several limitations. The number of included studies was relatively small for the stratified analysis and low sample size could also introduce bias into the analysis of ethnic populations, sepsis types and HWE. Further, we did not address the effect of gene–gene and gene–environment interactions due to a lack of the related information in the analysis and finally, our results were based on unadjusted genotype data. A more precise analysis should be made by adjusting other potentially confounding factors such as age, sex, and environmental factors. Nevertheless, to our knowledge, this is the first comprehensive study that has quantitatively synthesized the association between the three common polymorphisms of IL-10: −592C/A, −819C/T, −1082A/G and sepsis. We suggest that such an approach of combining the results of association studies may help us to better understand the effect of polymorphisms on disease risk.
ACKNOWLEDGEMENTS
This work was supported by the National Key Technology R&D Programme (2012BAI11B01), the Major State Basic Research Development Programme of China (2012CB518104), the National Natural Science Fund for Distinguished Young Scholars (81201462), and the Open Fund of the State Key Laboratory of Trauma, Burns and Combined Injury, Third Military Medical University (SKLZZ201104). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
DECLARATION OF INTEREST
None.
REFERENCES
- 1.Bone RC, et al. Definitions for sepsis and organ failure and guidelines for the use of innovative therapies in sepsis. The ACCP/SCCM Consensus Conference Committee. American College of Chest Physicians/Society of Critical Care Medicine. Chest 1992; 101: 1644–1655. [DOI] [PubMed] [Google Scholar]
- 2.Martin GS, et al. The epidemiology of sepsis in the United States from 1979 through 2000. New England Journal of Medicine 2003; 348: 1546–1554. [DOI] [PubMed] [Google Scholar]
- 3.Bochud PY, et al. Innate immunogenetics: a tool for exploring new frontiers of host defence. Lancet Infectious Diseases 2007; 7: 531–542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Mosmann TR. Properties and functions of interleukin-10. Advances in Immunology 1994; 56: 1–26. [PubMed] [Google Scholar]
- 5.Mosser DM, Zhang X. Interleukin-10: new perspectives on an old cytokine. Immunological Reviews 2008; 226: 205–218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Neidhardt R, et al. Relationship of interleukin-10 plasma levels to severity of injury and clinical outcome in injured patients. Journal of Trauma 1997; 42: 863–870. [DOI] [PubMed] [Google Scholar]
- 7.Maier B, et al. Early versus late onset of multiple organ failure is associated with differing patterns of plasma cytokine biomarker expression and outcome after severe trauma. Shock 2007; 28: 668–674. [PubMed] [Google Scholar]
- 8.McDaniel DO, et al. Molecular analysis of inflammatory markers in trauma patients at risk of postinjury complications. Journal of Trauma 2007; 63: 147–157. [DOI] [PubMed] [Google Scholar]
- 9.Westendorp RG, et al. Genetic influence on cytokine production and fatal meningococcal disease. Lancet 1997; 349: 170–173. [DOI] [PubMed] [Google Scholar]
- 10.Stewart D, et al. Genetic contribution to the septic response in a mouse model. Shock 2002; 18: 342–347. [DOI] [PubMed] [Google Scholar]
- 11.Turner DM, et al. An investigation of polymorphism in the interleukin-10 gene promoter. European Journal of Immunogenetics 1997; 24: 1–8. [DOI] [PubMed] [Google Scholar]
- 12.Baier RJ, Loggins J, Yanamandra K. IL-10, IL-6 and CD14 polymorphisms and sepsis outcome in ventilated very low birth weight infants. BMC Medicine 2006; 4: 10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Schaaf BM, et al. Pneumococcal septic shock is associated with the interleukin-10-1082 gene promoter polymorphism. American Journal of Respiratory and Critical Care Medicine 2003; 168: 476–480. [DOI] [PubMed] [Google Scholar]
- 14.Jin X, et al. Association of interleukin-10-1082 G/G genotype with lower mortality of acute respiratory distress syndrome in a Chinese population. Genetic Testing and Molecular Biomarkers 2011; 15: 203–206. [DOI] [PubMed] [Google Scholar]
- 15.Ouyang L, et al. Quantitative analysis of the association between interleukin-10 1082A/G polymorphism and susceptibility to sepsis. Molecular Biology Reports 2013; 40: 4327–4332. [DOI] [PubMed] [Google Scholar]
- 16.Gu W, et al. Clinical relevance of 13 cytokine gene polymorphisms in Chinese major trauma patients. Intensive Care Medicine 2010; 36: 1261–1265. [DOI] [PubMed] [Google Scholar]
- 17.Zeng L, et al. Clinical relevance of the interleukin 10 promoter polymorphisms in Chinese Han patients with major trauma: genetic association studies. Critical Care 2009; 13: R188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wells OCB, et al. The Newcastle–Ottawa Scale (NOS) for assessing the quality of nonrandomised studies in meta-analyses, 2006. (www.ohri.ca/programs/clinical_epidemiology/oxford.htm).
- 19.Higgins JP, et al. Measuring inconsistency in meta-analyses. British Medical Journal 2003; 327: 557–560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Egger M, et al. Bias in meta-analysis detected by a simple, graphical test. British Medical Journal 1997; 315: 629–634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Shimada T, et al. Outcome prediction in sepsis combined use of genetic polymorphisms – a study in Japanese population. Cytokine 2011; 54: 79–84. [DOI] [PubMed] [Google Scholar]
- 22.Davis SM, et al. The association of innate immune response gene polymorphisms and puerperal group A streptococcal sepsis. American Journal of Obstetrics and Gynecology 2010; 202: e301–308. [DOI] [PubMed] [Google Scholar]
- 23.Huebinger RM, et al. IL-10 polymorphism associated with decreased risk for mortality after burn injury. Journal of Surgical Research 2010; 164: E141–E145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Nakada TA, et al. Influence of toll-like receptor 4, CD14, tumor necrosis factor, and interleukine-10 gene polymorphisms on clinical outcome in Japanese critically ill patients. Journal of Surgical Research 2005; 129: 322–328. [DOI] [PubMed] [Google Scholar]
- 25.Balding J, et al. Genomic polymorphic profiles in an Irish population with meningococcaemia: is it possible to predict severity and outcome of disease? Genes and Immunity 2003; 4: 533–540. [DOI] [PubMed] [Google Scholar]
- 26.Lowe PR, et al. Influence of interleukin-10 polymorphisms on interleukin-10 expression and survival in critically ill patients. Critical Care Medicine 2003; 31: 34–38. [DOI] [PubMed] [Google Scholar]
- 27.Shu Q, et al. IL-10 polymorphism is associated with increased incidence of severe sepsis. Chinese Medical Journal 2003; 116: 1756–1759. [PubMed] [Google Scholar]
- 28.Palumbo AA, et al. Analysis of IL-6, IL-10 and IL-17 genetic polymorphisms as risk factors for sepsis development in burned patients. Burns 2012; 38: 208–213. [DOI] [PubMed] [Google Scholar]
- 29.Carregaro F, et al. Polymorphisms IL10-819 and TLR-2 are potentially associated with sepsis in Brazilian patients. Memorias do Instituto Oswaldo Cruz 2010; 105: 649–656. [DOI] [PubMed] [Google Scholar]
- 30.Emonts M, et al. Polymorphisms in PARP, IL1B, IL4, IL10, C1INH, DEFB1, and DEFA4 in meningococcal disease in three populations. Shock 2010; 34: 17–22. [DOI] [PubMed] [Google Scholar]
- 31.Abdel H, et al. Genetic polymorphisms of IL-6-174 and IL-10-1082 in full term neonates with late onset blood stream infections. Journal of Pediatric Infectious Diseases 2009; 4: 357–365. [Google Scholar]
- 32.McDaniel DO, et al. Molecular analysis of inflammatory markers in trauma patients at risk of postinjury complications. Journal of Trauma – Injury, Infection and Critical Care 2007; 63: 147–157. [DOI] [PubMed] [Google Scholar]
- 33.Garnacho-Montero J, et al. Timing of adequate antibiotic therapy is a greater determinant of outcome than are TNF and IL-10 polymorphisms in patients with sepsis. Critical Care 2006; 10: R111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Stanilova SA, et al. Interleukin-10-1082 promoter polymorphism in association with cytokine production and sepsis susceptibility. Intensive Care Medicine 2006; 32: 260–266. [DOI] [PubMed] [Google Scholar]
- 35.Zhang DL, et al. Association of polymorphisms of IL and CD14 genes with acute severe pancreatitis and septic shock. World Journal of Gastroenterology 2005; 11: 4409–4413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Jaber BL, et al. Cytokine gene promoter polymorphisms and mortality in acute renal failure. Cytokine 2004; 25: 212–219. [DOI] [PubMed] [Google Scholar]
- 37.Treszl A, et al. Genetic variants of TNF-(alpha), IL-1(beta), IL-4 receptor (alpha)-chain, IL-6 and IL-10 genes are not risk factors for sepsis in low-birth-weight infants. Biology of the Neonate 2003; 83: 241–245. [DOI] [PubMed] [Google Scholar]
- 38.Munafo MR, et al. Meta-analysis of genetic association studies. Trends in Genetics 2004; 20: 439–444. [DOI] [PubMed] [Google Scholar]
- 39.Zintzaras E. Impact of Hardy-Weinberg equilibrium deviation on allele-based risk effect of genetic association studies and meta-analysis. European Journal of Epidemiology 2010; 25: 553–560. [DOI] [PubMed] [Google Scholar]
- 40.Thakkinstian A, et al. A method for meta-analysis of molecular association studies. Statistics in Medicine 2005; 24: 1291–1306. [DOI] [PubMed] [Google Scholar]