SUMMARY
The high incidence of norovirus (NoV) infections seems to be related to the emergence of new variants that evolved by genetic drift of the capsid gene. In this work, that represents a first effort to describe the molecular epidemiology of NoV in the northwest of Spain, a total of eight different NoV genotypes (GII.1, GII.3, GII.4, GII.6, GII.7, GII.12, GII.13, GII.14) were detected. The major genotypes observed were GII.4 (45·42%) and GII.14 (34·9%), being detected in all age groups. In addition, and although most of GII.4 sequences belonged to 2006b (7·2%) and 2010 (50·35%) variants, the presence of new NoV variants was observed. Phylogenetic analysis revealed that a high number of GII.4 sequences (35·24%) could be assigned to the newly emerging Sydney 2012 variant, even during late 2010. The high prevalence of NoV GII.14 observed in this study may indicate the emergence of this genotype in Spain.
Key words: Epidemiology, evolution, genotyping, norovirus
INTRODUCTION
Noroviruses (NoVs) are the major cause of outbreaks and sporadic cases of acute non-bacterial gastroenteritis in patients of all ages [1, 2]. It has been estimated that they are responsible for 218000 deaths each year in children aged <5 years and 1·1 million of hospitalizations worldwide [1, 2]. NoV transmission occurs by the faecal–oral route, either by direct contact or via contaminated surfaces, food or water. NoVs are extremely contagious, owing to a very low infectious dose (estimated median infectious dose of 18 viral particles) [3] and to high levels of shedding in stools (>1 × 1010 RNA copies/g). Additionally, although patients recover from symptoms in 2–3 days, shedding may last several weeks [4]. No or limited long-term immunity results from infection, so one person may be repeatedly infected [5].
NoVs, classified in the family Caliciviridae, are a group of non-enveloped, icosahedral viruses, with a 7·5–7·7 kb positive-sense single-stranded RNA genome. NoVs are genetically diverse, classified within five genogroups (GI–GV) [6]. A novel genogroup, affecting dogs has been recently proposed [7], although it has not been agreed internationally yet. These genogroups are further divided into at least 33 different genotypes based on <15% amino acid variation in the VP1 region [6, 8]. Genogroups II, I and rarely IV are, in order of greatest to lowest, responsible for human infections [6]. Within genogroup II, strains belonging to genotype II.4 are the most common cause of gastroenteritis outbreaks, including pandemics, and sporadic cases since the mid-1990s [9].
The high incidence of NoV infections seems to be related to the emergence of new GII.4 variants that evolved by genetic drift of the capsid gene [10, 11]. NoVs, and especially GII.4 variants, have been increasingly studied over the past 10 years. It has been shown for the first time that NoV GII.4 variant US95/96 was identified and was predominant worldwide during the winter of 1995–1996 [12]. This variant was detected from outbreaks for the next 7 years. In 2002, a new type of GII.4, the 2002 variant named Farmington Hills, was detected and became predominant [13]. The main feature of this strain was the insertion of 1 amino acid in the hypervariable region of the capsid [13]. Since 2002, a new GII.4 variant has become predominant every 2 or 3 years. Indeed, the 2002 strain was replaced in 2004 by the 2004 variant [14], which was then replaced by the 2006a and 2006b variants. Both 2006 variants co-circulated during 2006 and 2007 [15]. During recent years, a 2010 variant (New Orleans_2009 strain) was identified. This variant replaced the 2006 variants, accounting for 60% of outbreaks [16]. Recent studies have reported the detection of a new 2012 variant (Sydney variant), that could be emerging in different regions throughout the world, being responsible for most outbreaks during late 2012 [17, 18]. The classification of these emerging GII.4 variants is based on >5% amino acid variation in the VP1 gene from other GII.4 strains [8, 14].
In addition, occasional increases in prevalence of other genotypes aside from GII.4 have been reported [19–21]. Recent studies revealed that other NoV genotypes evolved similarly to GII.4, suggesting the possibility of future emergence and epidemic spread of other genotype variants [22].
In this work, NoV detected from patients with acute gastroenteritis in northwest (NW) Spain, during a 1-year period, from July 2010 to June 2011, were characterized with the aim of determining the molecular epidemiology of NoV in this region, as well as the possibility of newly emerging variant strains.
METHODS
NoV detection
In a previous study [23] conducted in NW Spain during a 1-year period (July 2010 to June 2011) a total of 2643 specimens from both hospital outpatients (~90%) and inpatients (~10%) of all ages affected with sporadic cases of acute gastroenteritis were examined for NoV presence. Stool samples included in the study were obtained from Complejo Hospitalario Universitario de A Coruña, Galicia (NW Spain). This hospital complex serves more than 550 000 people in an area of 2750 km2. NoV GI and GII detection was performed by real-time reverse transcription–polymerase chain reaction (qRT–PCR) with TaqMan probes in all the samples. A total of 747 NoV strains were detected from analysed stool samples [23].
NoV typing
Genotyping of the detected NoV strains was performed on the basis of the partial capsid gene sequence. For this purpose, a semi-nested RT–PCR protocol with specific primers for NoV GI and NoV GII was performed. RT was performed with the Expand Reverse transcriptase (Roche, Germany) and 5 μl purified RNA and 100 μm random hexamer stock as reverse primer according to the manufacturer's specifications. First-round PCR was conducted using primers GOG1F and G1SKR for NoV GI and GOG2F and G2SKR for NoV GII [24, 25] (Supplementary Table S1). Second-round PCR for NoV GI was conducted with primers G1SKF and G1SKR. For NoV GII, the second round of PCR was performed with primers G2SKF and G2SKR (Supplementary Table S1). All the PCRs were conducted using the Expand High Fidelity PCR System (Roche, Germany) following the manufacturer's specifications. RT–PCR amplicons for NoV GI and GII were 330 bp and 344 bp, respectively. Amplification products were analysed by electrophoresis in 1·5% agarose gels and visualized with ethidium bromide. Amplicons were purified from gel with QIAQuick Gel Extraction kit (Qiagen, USA) and sequenced.
Phylogenetic analysis NoV strains
From the NoV partial capsid gene sequences obtained, 306 were appropriated for phylogenetic analysis. Sequence analysis was performed with the DNASTAR Lasergene SeqMan program (DNASTAR, USA). Phylogenetic reconstructions were performed based on the sequences of NoV partial capsid gene. Sequences of NoV reference strains were retrieved from Genbank. Neighbour-Joining (NJ) phylogenetic trees were constructed with MEGA5 software using the Kimura two-parameter model, and a bootstrap of 1000 replicates [26].
To compare the obtained phylogenetic results, sequences obtained were also analysed with the Norovirus Genotyping Tool version 1.0 [27].
RESULTS
Visual inspection of the tree built with the 306 sequences of NoV revealed that all sequences belonged to genogroup II, being distributed within eight different genotypes. Each of the different genotypes constituted a robust monophyletic group with bootstrap values between 95% and 99% (Supplementary Fig. S1). The major genotypes observed in the studied NoV population were GII.4 and GII.14, with 139 (45·4%) and 107 (34·9%) of cases, respectively. The other genogroups detected were: GII.1 (0·6%), GII.3 (1·9%), GII.6 (1·3%), GII.7 (6·9%), GII.12 (1·9%) and GII.13 (7·2%). NoV GII.4 was the major genotype detected in all age groups, with prevalences higher than 33%. GII.14 was the second highest genotype detected in patients from all ages. It was observed in more than 18% of the samples from each age group. Only in children aged <2 years and in adults aged 19–59 years were all the eight genotypes detected. Results of the characterization of the NoV strains detected from each age group are show in Table 1.
Table 1.
Prevalence of the different norovirus genotypes in each of the established age groups
| Age group | ||||||||
|---|---|---|---|---|---|---|---|---|
| 0–2 yr | 3–5 yr | 6–12 yr | 13–18 yr | 19–59 yr | >60 yr | Unknown age | Total | |
| No. of positive samples | 281 | 41 | 76 | 20 | 180 | 146 | 3 | 747 |
| Sequences analysed | 117 (41·6%) | 17 (41·5%) | 39 (51·3%) | 11 (55%) | 70 (38·9%) | 52 (35·6%) | — | 306 |
| GII.1 | 1 (0·8%) | — | — | — | 1 (1·4%) | — | 2 (0·6%) | |
| GII.3 | 2 (1·7%) | — | 2 (5·1%) | — | 1 (1·4%) | — | 5 (1·3%) | |
| GII.4 | 53 (45·3%) | 7 (41·1%) | 13 (33·4%) | 8 (72·7%) | 30 (42·8%) | 28 (53·8%) | 139 (45·4%) | |
| GII.6 | 1 (0·8%) | — | 1 (2·6%) | — | 2 (2·8%) | — | 4 (1·3%) | |
| GII.7 | 11 (9·4%) | 1 (5·9%) | 5 (12·8%) | 1 (9·1%) | 2 (2·8%) | 1 (1·9%) | 21 (6·9%) | |
| GII.12 | 4 (3·4%) | — | 1 (2·6%) | — | 1 (1·4%) | — | 6 (1·9%) | |
| GII.13 | 13 (11·2%) | — | 3 (7·7%) | — | 4 (5·7%) | 1 (1·9%) | 22 (7·2%) | |
| GII.14 | 32 (27·3%) | 8 (47%) | 14 (35·9%) | 2 (18·2%) | 29 (41·4%) | 22 (43·3%) | 107 (34·9%) | |
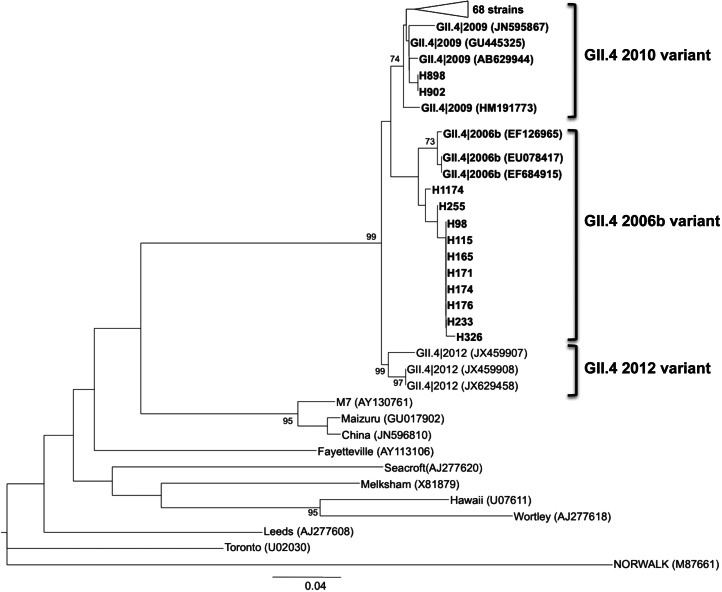
To assign the GII.4 sequences to a specific GII.4 variant, the Norovirus Genotyping Tool version 1.0 was employed. Of the 139 GII.4 sequences, 10 (7·19%) were characterized as 2006b variant, detected between August and December 2010. No NoV GII.4 sequences were detected in July 2010. In the period from July 2010 to June 2011, 70 (50·35%) of the strains were characterized as 2010 variant whereas the other 59 (42·44%) strains could not be assigned to any specific variant. A phylogenetic tree for NoV GII.4 sequences assigned to 2006b and 2010 variants was obtained based on the NJ method (Fig. 1).
Fig. 1.
Phylogenetic reconstruction based on the partial capsid gene sequences of norovirus GII.4 2006b and 2010 variant strains (using the Norovirus Genotyping Tool version 1.0) and reference strains by the neighbour-joining method. Bar, expected nucleotide substitutions per site. Only bootstrap values >70% are shown (1000 re-samplings) at each branch point. GenBank accession numbers of reference sequences are given in parentheses.
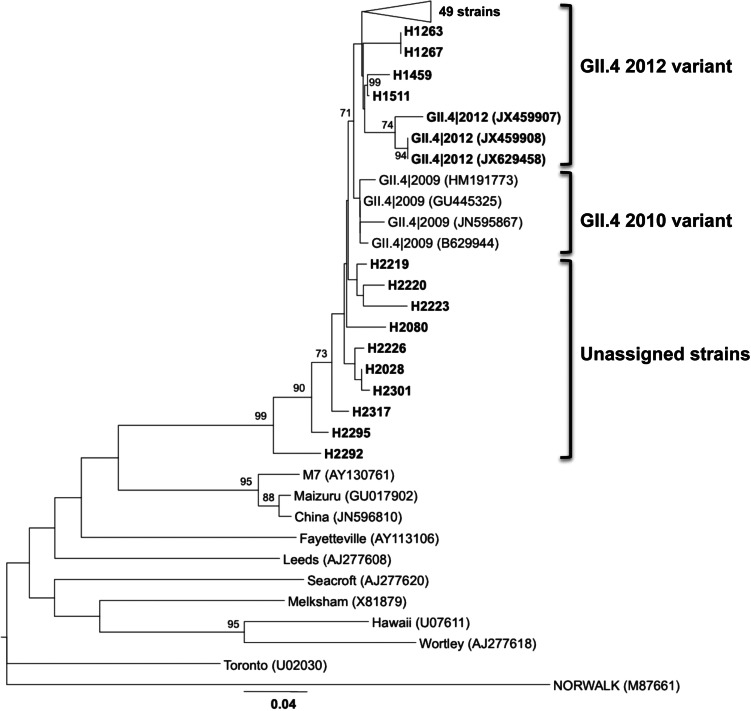
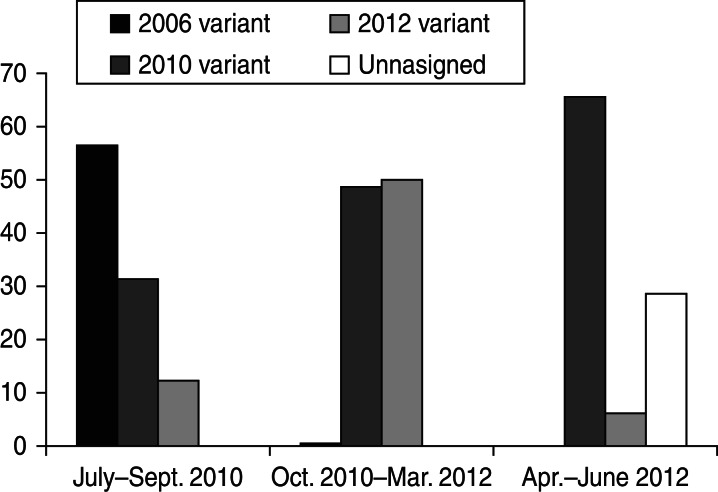
A phylogenetic tree was also constructed with the 59 unassigned sequences together with sequences of the newly emerging 2012 variant (Sydney strain). Visual inspection of the tree revealed that 49 of the analysed sequences and the 2012 variant sequences clustered together in a well-differentiated branch. The other 10 sequences included in the analysis were not located in any specific cluster (Fig. 2). The detection of the strains assigned to the new 2012 variant was higher during the winter season 2010/2011. Between October 2010 and March 2011 a total of 88 NoV GII.4 strains were detected, 44 (50%) of them characterized as the new 2012 variant and 43 (48·8%) characterized as 2010 variant. In the warm months of 2010 (August–September), five (31·25%) of the 16 NoV GII.4 detected strains corresponded to the 2010 variant and two (12·5%) to the 2012 variant. In the warm months of 2011 (April–June) of the 35 NoV GII.4 strains detected, 23 (65·7%) were 2010 variant, two (5·75%) were 2012 variant, and 10 (34·28%) were unassigned variants (Fig. 3).
Fig. 2.
Phylogenetic reconstruction based on the partial capsid gene sequences of norovirus GII.4 2012 variant strains, unassigned variant and reference strains by the neighbour-joining method. Bar, expected nucleotide substitutions per site. Only bootstrap values >70% are shown (1000 re-samplings) at each branch point. GenBank accession numbers of reference sequences are given in parentheses.
Fig. 3.
Percentage of the different GII.4 variants detected during the warm and cold months.
Similarities in sequences belonging to 2010 and 2012 variants ranged from 94·6% to 98·4%, and in sequences belonging to 2010 and 2006b variants from 94·8% to 98·9%. Slightly lower similarities were observed in 2012 and 2006b variants, with values ranging from 94·4% to 96·3%. Within the 2010 variant, similarity in sequences ranged from 96·8% to 100%, from 96·4% to 100% within the 2012 variant and from 94·9% to 100% within the 2006 variant.
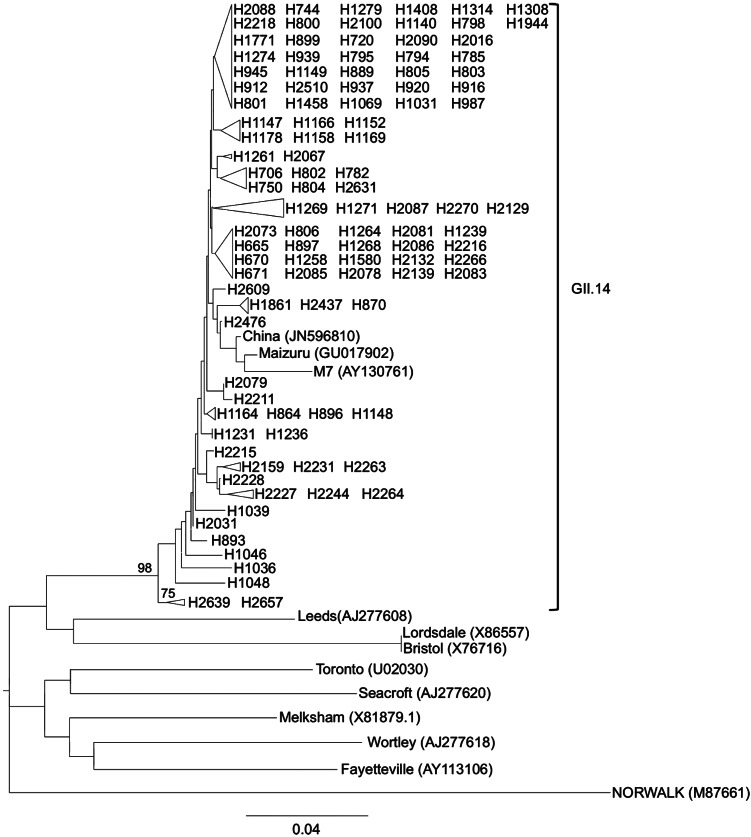
The NoV GII.14 phylogenetic tree revealed that, although in general terms bootstrap values obtained were not high, the 107 analysed sequences belonging to GII.14 constituted a robust monophyletic group with a bootstrap value of 98% (Fig. 4). The similarity within the GII.14 sequences ranged from 86·6% to 100%. With regard to NoV GII.4, NoV GII.14 strains were detected in higher numbers during the cold months. Of the 107 GII.14 strains, 70 (65·42%) were detected between October 2010 and March 2011. The other 37 (34·57%) strains were detected in April–June 2011. No NoV GII.14 strains were detected during July–September 2010.
Fig. 4.
Phylogenetic reconstruction based on the partial capsid gene sequences of norovirus GII.14 strains and reference strains by the neighbour-joining method. Bar, expected nucleotide substitutions per site. Only bootstrap values >70% are shown (1000 re-samplings) at each branch point. GenBank accession numbers of reference sequences are given in parentheses.
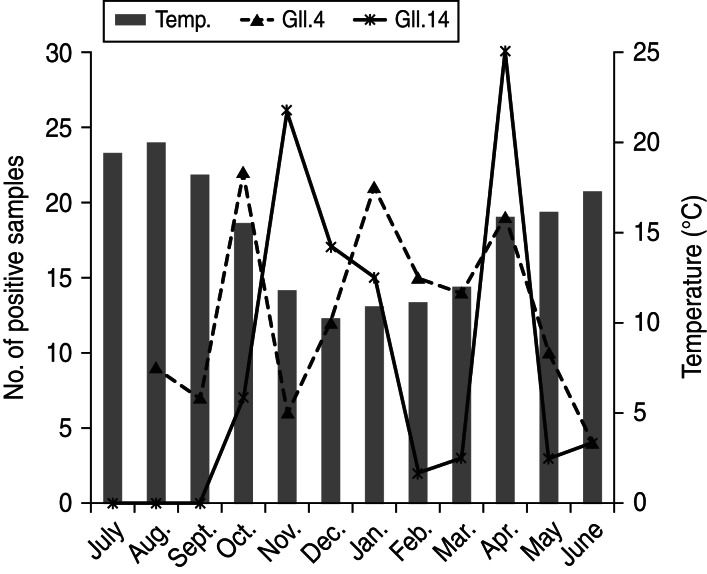
During the cold months between October 2010 and March 2011 both NoV GII.4 and GII.14 presented a peak in prevalence. However, it appears that both genotypes showed alternate detection during these months as shown in Figure 5.
Fig. 5.
Monthly prevalence of norovirus GII.4 and GII.14 strains.
A total of 21 sequences were assigned to NoV GII.7 and 22 sequences to GII.13. Regarding the other genotypes, phylogenetic trees were obtained based on the NJ method. Both GII.7 and GII.13 strains constituted robust monophyletic groups with bootstrap values of 99% and 100%, respectively (Supplementary Fig. S2). The similarity percentage within the GII.7 sequences ranged from 96·1% to 100%, whereas for GII.13 it ranged from 98·8% to 100%. A potential link with temperature or season was observed for GII.7 strains, as the strains belonging to this genotype were more prevalent during the warm months (contrary to GII.4 and GII.14) (Supplementary Fig. S3). Differences in prevalence linked with temperature were not observed for NoV GII.13 during the time period analysed (Supplementary Fig. S3).
DISCUSSION
NoV are the leading cause of outbreaks and sporadic cases of acute non-bacterial gastroenteritis in patients of all ages [1, 2]. A study analysing the role of NoV in gastroenteritis development during a 1-year period in NW Spain [23] revealed that an aetiological agent, viral and/or bacterial, could be detected in 39·5% of the patients studied, and NoV was responsible for 28·3% of those with an identified aetiology. This finding is in agreement with previous studies. In patients in Germany in 2004, the aetiology of gastroenteritis could be determined in 35% of the examined samples [28], with viral pathogens detected far more often than bacterial pathogens. These authors proposed that their results might be representative for Germany and other European countries. On the other hand, a study in one US state in 2004/2005 [29] also found NoV as the leading cause of acute gastroenteritis, although with a much lower prevalence (12%). An exclusive distinction between infectious and non-infectious intestinal disease cannot be made on the basis of clinical signs, therefore the cases in which no pathogen could be detected may be due to a non-infectious disease. Moreover, these cases may be caused by intestinal microorganisms not included in the study or by non-intestinal infections, such as influenza [30].
The genotyping of NoV strains detected in clinical samples from sporadic gastroenteritis cases in NW Spain showed the appearance, spread and monthly prevalence of different NoV genogroups and the changes in NoV molecular epidemiology in a limited geographical region. Although all the detected strains were submitted for typing analysis, in only 41% was a proper sequence obtained. No differences in the proportions of sequenced strains detected from inpatients or outpatients were observed. In addition, RNA quantification levels obtained from qRT–PCR detection showed no differences in genotyped and ungenotyped strains. One possible reason for these results is that NoV strain diversity is extremely high. The 5′ end of the NoV capsid region is not the most heterogeneous region in the NoV genome and it is used to amplify a wide range of NoV GI and GII genotypes [24] using a conserved set of primers. Even in this relatively conserved region, a variety of single nucleotide polymorphisms (SNPs) were found, which makes a suitable primer and probe design difficult, as well as assay optimization and verification extremely important [24, 31, 32].
Molecular characterization of NoV strains demonstrated that GII.4 was the dominant genotype throughout the 1-year period of the study and in patients of all ages. This finding is consistent with other recent molecular epidemiological and surveillance studies that have found GII.4 to be responsible for the majority of NoV infections and outbreaks worldwide [9, 11, 33, 34]. It was also observed in the present work that strains belonging to the GII.4 2006b variant were replaced by the 2010 variant. Similar findings were reported in studies developed in Asia, Europe and America [16, 34–36].
Lindesmith et al. [37] studied the molecular mechanism of GII.4 NoV evolution resulting in the persistence and emergence of new strains. The emerging variants appear to have a transmissibility advantage and increased virulence. It has been hypothesized that NoV GII.4 persist by altering their ABH histo-blood group antigens' carbohydrate-binding target over time, allowing them to escape host susceptibility. An alternative explanation is that evolving strains drift under immune selective pressure until mutations have accumulated to a point where a novel genetic variant phenotype becomes established and evades pre-established host immunity [10]. It is important to highlight the high number of sequences that could not be assigned to any GII.4 variant by using the Norovirus Genotyping tool version 1.0 [27]. Phylogenetic analysis by NJ performed with these unassigned strains and the recently described 2012 variant strains [17, 18] allowed most of these sequences to be aligned with the newly emerging 2012 variant. The Sydney 2012 variant has been described as being responsible for a high peak of NoV outbreaks during late 2012 [17, 18]. However, results obtained from the genotyping of Galician NoV strains revealed that this 2012 variant could have been already circulating since the last months of 2010.
On the other hand, the ten sequences that could not be assigned to any specific variant by phylogenetic analysis or by Norovirus Genotyping Tool version 1.0, might constitute newly emerging variants or recombinant strains. The approach used in this work utilizing the N-terminus of the VP1 protein (region C) as a target region for NoV genotyping has been described elsewhere [25] and the N/S domain was shown to be able to separate the strains into internal clusters or genotypes [38]. However, the use of the full-length capsid gene would increase the phylogenetic signal and would allow more accurate variant assignment [27]. Therefore, further studies sequencing the complete VP1 capsid coding region and the ORF1–ORF2 overlapping region (a known hotspot for NoV recombination) are needed.
The emergence of non-GII.4 strains has also been reported in the recent literature [19–21]. The winter of 2009/2010 saw the emergence of a GII.12 strain that was ultimately responsible for 16% of outbreaks in the USA that year [36]. In the present study, from July 2010 to June 2011, besides GII.4 seven other genotypes were detected, in order of importance GII.14, GII.7 and GII.13, respectively. These non-GII.4 strains suddenly appeared, spread and decreased in the short term. This behaviour might be attributed to population immunity, as described in an earlier report [22]. To our knowledge, this is the first time in which a high prevalence of NoV GII.14 has been observed in Spain. Previous studies in Spain reported NoV GII.4 as a major cause of gastroenteritis, and GII.1, GII.2, GII.6 to a lesser extent [37–39]. Further studies would confirm the emergence of this genotype and its importance as an aetiological agent of diarrhoeal disease.
This article represents a first effort to describe the molecular epidemiology of NoV in NW Spain. Gastroenteritis caused by NoV continues to have a great impact on public health worldwide. Since NoV vaccines, if they are to be developed [40], will need to protect against the different emerging GII.4 variants or other emerging genotypes, regional, national and global analysis of NoV infections and a better knowledge of NoV evolution will help in the development of effective measures for their control and prevention.
ACKNOWLEDGEMENTS
We gratefully acknowledge Germán Bou from the Complejo Hospital Universitario de A Coruña (Galicia, Spain), for all the samples included in the analysis. This work was supported in part by grant 10MMA200010PR from the Consellería de Economía e Industria, Xunta de Galicia (Spain). C.F.M. acknowledges the Xunta the Galicia for a research fellowship.
Supplementary material
For supplementary material accompanying this paper visit http://dx.doi.org/10.1017/S0950268814000740.
click here to view supplementary material
DECLARATION OF INTEREST
None.
REFERENCES
- 1.Patel MM, et al. Systematic literature review of role of noroviruses in sporadic gastroenteritis. Emerging Infectious Diseases 2008; 14: 1224–1231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Patel MM, et al. Noroviruses: a comprehensive review. Journal of Clinical Virology 2009; 44: 1–8. [DOI] [PubMed] [Google Scholar]
- 3.Teunis PF, et al. Norwalk virus: how infectious is it? Journal of Medical Virology 2008; 80: 1468–1476. [DOI] [PubMed] [Google Scholar]
- 4.Tu ET, et al. Norovirus excretion in an aged-care setting. Journal of Clinical Microbiology 2008; 46: 2119–2121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Johnson PC, et al. Multiple-challenge study of host susceptibility to Norwalk gastroenteritis in US adults. Journal of Infectious Diseases 1990; 161: 18–21. [DOI] [PubMed] [Google Scholar]
- 6.Zheng DP, et al. Norovirus classification and proposed strain nomenclature. Virology 2006; 346: 312–323. [DOI] [PubMed] [Google Scholar]
- 7.Mesquita JR, et al. Novel norovirus in dogs with diarrhea. Emerging Infectious Diseases 2010; 16: 980–982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zheng DP, et al. Molecular epidemiology of genogroup II-genotype 4 noroviruses in the United States between 1994 and 2006. Journal of Clinical Microbiology 2010; 48: 168–177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Siebenga JJ, et al. Norovirus illness is a global problem: emergence and spread of norovirus GII.4 variants, 2001–2007. Journal of Infectious Diseases 2009; 200: 802–812. [DOI] [PubMed] [Google Scholar]
- 10.Siebenga JJ, et al. Epochal evolution of GGII.4 norovirus capsid proteins from 1995 to 2006. Virology 2007; 81: 9932–9941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bull RA, et al. Rapid evolution of pandemic noroviruses of the GII.4 lineage. PLoS Pathogens 2010; 6: e1000831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Vinjé J, Altena SA, Koopmans MP. The incidence and genetic variability of small round-structured viruses in outbreaks of gastroenteritis in The Netherlands. Journal of Infectious Diseases 1997; 176: 1374–1378. [DOI] [PubMed] [Google Scholar]
- 13.Widdowson MA, et al. Outbreaks of acute gastroenteritis on cruise ships and on land: identification of a predominant circulating strain of norovirus – United States, 2002. Journal of Infectious Diseases 2004; 190: 27–36. [DOI] [PubMed] [Google Scholar]
- 14.Bull RA, et al. Emergence of a new norovirus genotype II.4 variant associated with global outbreaks of gastroenteritis. Journal of Clinical Microbiology 2006; 44: 327–333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tu ET, et al. Epidemics of gastroenteritis during 2006 were associated with the spread of norovirus GII.4 variants 2006a and 2006b. Clinical Infectious Disease 2008; 46: 413–420. [DOI] [PubMed] [Google Scholar]
- 16.Vega E, et al. Novel surveillance network for norovirus gastroenteritis outbreaks, United States. Emerging Infectious Diseases 2011; 17: 1389–1395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.van Beek J, et al. Indications for worldwide increased norovirus activity associated with emergence of a new variant of genotype II.4, late 2012. Eurosurveillance 2013; 18(1). [PubMed] [Google Scholar]
- 18.Bennett S, et al. Increased norovirus activity in Scotland in 2012 is associated with the emergence of a new norovirus GII.4 variant. Eurosurveillance 2013; 18(2). [PubMed] [Google Scholar]
- 19.Lewis DC, et al. Epidemiology of Mexico virus, a small round-structured virus in Yorkshire, United Kingdom, between January 1992 and March 1995. Journal of Infectious Diseases 1997; 175: 951–954. [DOI] [PubMed] [Google Scholar]
- 20.Koopmans M, et al. Molecular epidemiology of human enteric caliciviruses in The Netherlands. Journal of Infectious Diseases 2000; 181: 262–269. [DOI] [PubMed] [Google Scholar]
- 21.Iritani N, et al. Molecular epidemiology of noroviruses detected in seasonal outbreaks of acute nonbacterial gastroenteritis in Osaka City, Japan, from 1996–1997 to 2008–2009. Journal of Medical Virology 2010; 82: 2097–2105. [DOI] [PubMed] [Google Scholar]
- 22.Iritani N, et al. Genetic analysis of the capsid gene of genotype GII.2 noroviruses. Journal of Virology 2008; 82: 7336–7345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Manso CF, Romalde JL. Role of Norovirus in acute gastroenteritis in the Northwest of Spain. Journal of Medical Virology 2013; 85: 2009–2015. [DOI] [PubMed] [Google Scholar]
- 24.Kageyama T, et al. Broadly reactive and highly sensitive assay for norwalk-like viruses based on real-time quantitative reverse transcription-PCR. Journal of Clinical Microbiology 2003; 41: 548–557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kojima S, et al. Genogroup-specific PCR primers for detection of Norwalk-like viruses. Journal of Virological Methods 2002; 100: 107–114. [DOI] [PubMed] [Google Scholar]
- 26.Balboa S, et al. Proteomics and multilocus sequence analysis confirm intraspecific variability of Vibrio tapetis. FEMS Microbiology Letters 2011; 324: 80–87. [DOI] [PubMed] [Google Scholar]
- 27.Kroneman A, et al. An automated genotyping tool for enteroviruses and noroviruses. Journal of Clinical Virology 2011; 51: 121–125. [DOI] [PubMed] [Google Scholar]
- 28.Karsten C, et al. Incidence and risk factors for community-acquired acute gastroenteritis in north-west Germany in 2004. European Journal of Clinical Microbiology and Infectious Diseases 2009; 28: 935–943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hall AJ, et al. Incidence of acute gastroenteritis and role of norovirus, Georgia, USA, 2004–2005. Emerging Infectious Diseases 2011; 17: 1381–1388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.de Wit MA, et al. Gastroenteritis in sentinel general practices, The Netherlands. Emerging Infectious Diseases 2001; 7: 82–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Loisy, et al. Real-time RT-PCR for norovirus screening in shellfish. Journal of Virological Methods 2005; 123: 1–7. [DOI] [PubMed] [Google Scholar]
- 32.Trujillo, et al. Use of TaqMan real-time reverse transcription-PCR for rapid detection, quantification, and typing of Norovirus. Journal of Clinical Microbiology 2006; 44: 1405–1412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.McAllister G, et al. Molecular epidemiology of norovirus in Edinburgh healthcare facilities, Scotland 2007–2011. Epidemiology and Infection 2012; 140: 2273–2281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lopman BA, et al. Clinical manifestation of norovirus gastroenteritis in health care settings. Clinical Infectious Disease 2004; 39: 318–324. [DOI] [PubMed] [Google Scholar]
- 35.Kroneman A, et al. Analysis of integrated virological and epidemiological reports of norovirus outbreaks collected within the foodborne viruses in Europe network from 1 July 2001 to 30 June 2006. Journal of Clinical Microbiology 2008; 46: 2956–2965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kaplan NM, et al. Detection and molecular characterisation of rotavirus and norovirus infections in Jordanian children with acute gastroenteritis. Archives of Virology 2011; 156: 1477–1480. [DOI] [PubMed] [Google Scholar]
- 37.Lindesmith LC, et al. Mechanisms of GII.4 norovirus persistence in human populations. PLoS Medicine 2008; 5: 31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Katayama K, et al. Phylogenetic analysis of the complete genome of 18 Norwalk-like viruses. Virology 2002; 299: 225–239. [DOI] [PubMed] [Google Scholar]
- 39.Vega E, Vinjé J. Novel GII.12 norovirus strain, United States, 2009–2010. Emerging Infectious Diseases 2011; 17: 1516–1518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Domínguez A, et al. Aetiology and epidemiology of viral gastroenteritis outbreaks in Catalonia (Spain) in 2004–2005. Journal of Clinical Virology 2008; 43: 126–131. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
For supplementary material accompanying this paper visit http://dx.doi.org/10.1017/S0950268814000740.
click here to view supplementary material